Abstract
Classically, blood arises from stem cells through a series of oligopotent progenitors that become increasingly restricted to unipotent progenitors, each slotted into a hierarchical layer based on their differentiation potential. The presence of oligopotent cells is critical to the standard model of blood differentiation as they define the path from stem cells to unipotent progenitors. We developed a new cell-sorting scheme to resolve myeloid (My), erythroid (Er) and megakaryocytic (Mk) fates from single CD34+ cells and then mapped the progenitor hierarchy across human development. Fetal liver contained large numbers of distinct oligopotent progenitors with entangled My, Er and Mk fates. Unexpectedly in adult bone marrow, few oligopotent progenitor intermediates were present with multipotent and unipotent progenitors predominating, and now Er-Mk lineages emerged from multipotent cells. The developmental shift to an adult ‘two-tier’ hierarchy challenges current dogma and provides a new framework to understand normal and disease states of human hematopoiesis.
For decades, hematopoiesis has been described as a cellular hierarchy maintained by self-renewing hematopoietic stem cells (HSCs) that reside at the apex of its pyramidal structure (1, 2). This differentiation scheme highlights key features of the blood system and has been critical to our understanding of how stem cells manage life-long blood production. In general, self-renewing cell types with extended lifespan like long term HSC (LT-HSC), as well as short-term HSC (ST-HSC) and multipotent progenitors (MPPs) are rare and remain closer to the conceptual peak of the hierarchy; oligopotent and unipotent progenitors below have shorter lifespans, increase numerically, and become gradually restricted into more than ten functional blood cell types. In the standard model of hematopoiesis, hierarchical differentiation commences from HSCs with the production of stem cell intermediates with less durable self-renewal potential that culminate with the generation of MPPs, the penultimate step before lineage specification. From MPPs, the common lineages for myelopoiesis (common myeloid progenitor – CMP) and lymphopoiesis (common lymphoid progenitor – CLP) are segregated. In My differentiation, oligopotent CMPs undergo further restriction into bivalent granulocyte-monocyte progenitor (GMPs) that go on to make granulocytes and monocytes, and megakaryocyte-erythroid progenitors (MEPs) that go on to make platelets and red blood cells (RBCs). Thus, CMPs represent the critical oligopotent progenitor from which all My (defined herein as granulocyte/monocyte), Er and Mk cells arise. Although the standard model is still used extensively as an operational paradigm, further cell purification and functional clonal assays have led to key revisions to the model. In mouse, the identification of lymphoid-primed multipotent progenitors (LMPP) argued that megakaryocyte-erythroid (Mk-Er) potential must be the first lineage branch lost in lympho-myeloid specification of HSCs (3, 4). Recently, paired-daughter analysis monitoring HSC cell divisions have demonstrated that Mk-Er progenitors can be derived from HSC directly without progressing through conventional MPPs and CMPs (5). Although these data challenge the standard model, clear consensus on a revised model of hematopoiesis is still lacking. Human hematopoiesis is widely regarded as following the mouse hematopoiesis (reviewed in (6)). Early work involving cell purification and methylcellulose (MC) colony-forming cell (CFC) assays yielded an identical scheme as the mouse including CMP and CLP (7-10). However, purification schemes to resolve My, Er, Mk and Ly fates remained poor. Through the development of more efficient assays to monitor Ly fates in single-cell stromal assays and an improved sorting scheme, we identified human multilymphoid progenitors (MLP) as the earliest lymphoid differentiation precursor with concomitant lymphoid (T, B, NK) and myelomonocytic potential, rather than CLP (11, 12). Considerable uncertainty remains concerning the myelo-erythro-megakaryocytic branch of human hematopoiesis since clonogenic CFC assays do not read out My, Er and Mk fates efficiently, nor contemporaneously making it difficult to account for all cells within phenotypically pure populations of CMPs and MEPs. A comprehensive analysis of human myelo-erythro-megakaryocytic development has not been undertaken and so it is really only by default that the standard model applies.
Much of our understanding of the molecular basis of cellular differentiation and lineage commitment is derived from the assumptions implicit in the standard model. For example, simultaneous expression of molecular factors associated with My-Er-Mk lineages at low levels is considered to maintain CMPs as the origin of the common lineage for myelopoiesis (7). During restriction to GMPs and MEPs, progressive upregulation of particular lineage factors initiate feedforward and feedback molecular controls that lock-in a granulocyte/monocyte or a Mk-Er differentiation program. An important axiom that arises from this molecular view of the standard model is that cellular differentiation is gradual. However, transcriptional studies of highly purified or single cell murine HSPC has established that molecular programs corresponding to My-Er-Mk fates can directly emerge in multipotent cells, arguing that cellular differentiation is not gradual and that myeloid differentiation can occur without progressing through an intermediate CMP stage (4, 5, 13-17). Naik et al. have demonstrated that nearly half of the LMPP compartment is biased towards dendritic cell commitment, a lineage previously thought to come from the CMP to GMP route (15). Molecular factors associated with Mk-Er differentiation have been shown to be active in LT-HSCs (13, 14), and prospective isolation of platelet-biased LT-HSCs strongly supports that this lineage is not derived from the CMP to MEP route (16). Whether molecular programs that regulate My-Er-Mk fates arise at the level of HSCs in humans is not known. Where the Er and Mk lineage branching occurs in the human hematopoietic hierarchy is important to understand as these lineages comprise 99% of the cellular component of blood, and represent the bulk of the 300 billion blood cells that turnover daily in humans. Mapping the cellular origins of Er and Mk lineages in the human blood hierarchy represents a critical step to define the molecular basis of their fate commitment.
The cellular roadmap describing the blood hierarchy has been built on two core experimental pillars: cell purification and clonal assays. Human studies reporting on sorting schemes for the myelo-erythroid progenitor hierarchy (CMPs, GMPs and MEPs) have often assumed that ‘marker-pure’ subsets are synonymous with ‘functionally-pure’ subsets (9). In other words, each cell within the purified subset possesses the same differentiation potential (Fig. 1A, ①). This interpretation is primarily derived from clonogenic assessment of human CMPs, GMPs and MEPs using standard CFC assays. In CFC assays purified subsets of CMP typically generate My, Er or Mk colonies, GMPs give rise to My colonies only, and MEPs give rise to Er and/or Mks (9). Since CMPs displayed all lineage readouts whereas GMPs and MEPs did not, CMPs were interpreted as both marker-pure and functionally pure on the basis that the CFC assay was inefficient in being able to readout mixed differentiation potential. This reasoning underpins the basic bifurcating scheme of human CMPs to GMPs and MEPs. However, an alternate interpretation exists if we assume that the CFC assay is actually efficient. In this case, human CMPs are marker-pure but are functionally heterogeneous, consisting of diverse unipotent progenitors (Fig. 1A, ②). This contention could only be proven if a new sorting strategy is able to isolate, in functionally pure form, each type of the unilineage progenitor from the starting CMP population. To distinguish between the two alternatives, we need 1) a new sorting strategy for human myelo-erythroid progenitors, and 2) a more sensitive assay to assess mixed cell potential from them. Until both scenarios can be experimentally resolved, there is considerable uncertainty that clouds the classical view of the human hematopoietic hierarchy. We attempted to address both of these issues by developing a novel cell-sorting scheme and an optimized single cell assay to efficiently readout My, Er and Mk fates from putative multilineage cell types.
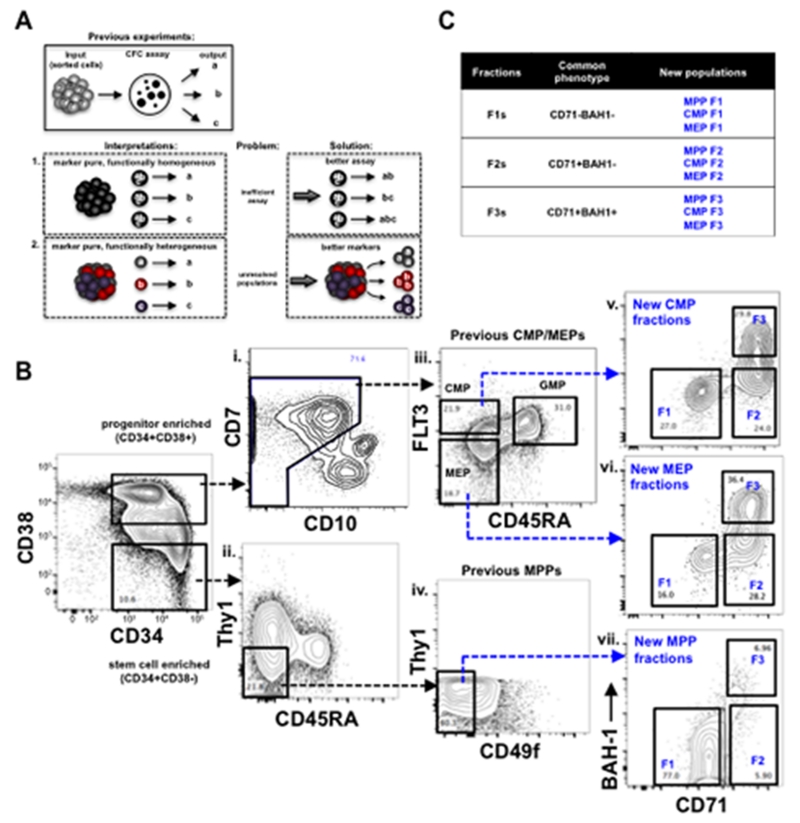
Figure 1. Use of novel phenotypic markers revealed heterogeneity in human hematopoietic hierarchy.
(A) A review of classic interpretations from previous work using CFC assays to measure the clonal outputs from a sorted, marker-pure population of cells. In scenario 1, each cell within a marker-pure population has the potential to give rise to three functional outputs (a, b, and c), but only gives rise to one of them in the assay. In this scenario, diverse functional outputs from a marker-pure population are interpreted to be derived from a pure population of a multilineage cells. In the alternate scenario 2, a marker pure population is comprised of three distinct unipotent cell types that give rise to their respective functional progeny. Solutions to each one of these scenarios are presented on the right. (B) The gating scheme of defining MPPs (CD34+CD38−Thy1−CD45RA−CD49f−), CMPs (CD34+CD38+CD10−FLT3+CD45RA−) and MEPs (CD34+CD38+CD10−FLT3CD45RA−) from FL is shown with black-dashed arrows. Each one of these compartments was further divided into F1 (CD71−BAH1−), F2 (CD71+BAH1−), and F3 (CD71+BAH1+) shown with blue-dashed arrows. Full gating scheme for FL, CB, and BM CD34+ cells is presented in fig. S1. (C) Summary of the new subsets used in this study.
RESULTS
Novel populations lie within currently defined human MPPs, CMPs and MEPs
We developed a novel cell-sorting scheme to examine the cellular heterogeneity within the CD34+ compartment of human blood. To a previous 7-parameter design (CD34, CD38, CD7, CD10, FLT3, CD45RA, Thy1) (11), we distinguished HSCs from MPPs by adding CD49f (18), identified Er-Mk progenitors by adding cMPL (CD110) (19, 20) (designated here as BAH1 to be consistent with the antibody clone used to detect this antigen) (21)(22) and CD71 (Transferrin receptor), and distinguished B-lymphoid committed progenitors from My progenitors such as GMPs in the CD45RA+ fraction by adding CD19. Upon evaluation in human fetal liver (FL), neonatal cord blood (CB) and adult bone marrow (BM), this 11 parameter cell-sorting layout provided a high resolution view of the phenotypic heterogeneity that exists within CD34+ cells across all developmental stages (Fig. 1B and fig. S1). To our knowledge, this sorting design is the most advanced scheme available for the human blood system.
Many distinct cell types, such as HSCs, MPPs and MLPs, reside within the CD34+CD38−/lo (simplified as CD34+CD38− herein) stem cell enriched compartment of human blood. We first explored if CD71 or BAH1 expression corresponded to a known cell type within this compartment in the FL. Approximately 10% of the FL CD34+CD38− expressed CD71, and half of these CD71+ cells also expressed BAH1 (fig. S1A, panel 2). Neither CD71 nor BAH1 was expressed on Thy1+ HSCs (fig. S2A) or on CD45RA+ MLPs (fig S2B) suggesting that these markers identify a novel cell type within the MPP compartment. We redefined the current MPP compartment into three fractions (F1, F2 and F3) based on CD71 and BAH1 expression: MPP F1 cells were CD71−BAH1−, MPP F2 cells were CD71+BAH1− and MPP F3 cells were CD71+BAH1+ (Fig. 1B.vii). The expression of these molecules in the CD34+CD38− compartment was unexpected as the onset of Mk-Er lineage commitment according to the standard model occurs at the level of CMPs and MEPs that are found in the progenitor enriched CD34+CD38+ compartment. These data suggest that the detection of functional Er and Mk differentiation molecules on a subset of CD34+CD38− cells represents a unique branch point of this lineage within the multilineage compartment.
We next analyzed CD71 and BAH1 expression in the CD34+CD38+ progenitor compartment. Co-expression of FLT3 and the lymphoid antigen CD7 in FL CD34+CD38+ cells (fig. S2C) indicated that CD7 expression is not exclusive to lymphoid progenitors as reported previously in CB (23). Thus, CD7 expressing cells were not excluded in our sorting layout. In lieu of CD7, we used CD10 expression to exclude Ly progenitors (Fig. 1B.i; fig. S1, panels 6). In the FL CD34+CD38+CD10− cell compartment, FLT3 and CD45RA expression was used to identify commonly defined CMPs (FLT3+CD45RA−), GMPs (FLT3+CD45RA+), and MEPs (FLT3−CD45RA−) (Fig. 1B.iii; fig. S1, panels 7). Addition of CD71 and BAH1 to CMP and MEP populations uncovered striking phenotypic heterogeneity within these populations previously considered to be homogeneous. In line with the nomenclature we employed for the redefined MPP compartment described above, CMP and MEP compartments were also subdivided into three fractions with CD71 and BAH1 (F1: CD71−BAH1−, F2: CD71+BAH1−, F3: CD71+BAH1+; Fig. 1B.v-vii and fig. S1, panels 8,10). We repeated the same analysis in CB and adult BM to determine if CB and adult BM samples were similarly heterogeneous. Remarkably, all the major cell populations identified in the FL were also observed in CB and BM albeit at different degrees (table S1) indicating that the cellular heterogeneity uncovered by CD71 and BAH1 existed across all developmental stages (fig. S1B,C). Thus, previously defined human CMPs and MEPs that were considered homogeneous are in fact phenotypically heterogeneous when Er and Mk markers are applied.
In summary, previously defined MPPs, CMPs and MEPs contain three distinct cellular fractions: F1s: lacking CD71 and BAH1; F2s: expressing CD71 but lacking BAH1; and F3s: expressing both molecules. A total of 33 distinct cellular classes from FL, CB and BM (11 per developmental stage) were functionally interrogated to evaluate their lineage fate potential. To facilitate the review of the results below, a legend and complete phenotype is provided in Fig. 1C and table S1.
An optimized single cell assay for human My-Er-Mk progenitors
To evaluate the functional potential of the novel cellular subsets identified above, we developed a single-cell in vitro assay that overcame the shortcomings of prior approaches to characterize human myeloid progenitors. An ideal assay would support the ability of single cells to simultaneously commit along My, Er and/or Mk fates as well as provide the conditions for their differentiated progeny to survive, propagate and expand to permit detection. Standard MC assays do not strictly fulfill these criteria. For example, CD49f+ HSCs exclusively generated CFU-My in MC (fig. S3A and Supplementary text), yet single HSC sustain multilineage hematopoiesis in vivo (18). Also, the limited self-renewal potential of downstream progenitors makes them difficult to readout in vivo at clonal resolution, further highlighting the need to develop more sensitive in vitro methodology to assess the lineage potential of progenitors. We found that serum-free conditions supplemented with growth factors (SCF, FLT3, TPO, EPO, IL-6, IL-3, IL-11, GM-CSF, LDL) and stroma was highly efficient at assaying My, Er and Mk lineage potential from single CD34+ cells. Single cell derived clones were analyzed by flow cytometry after a 2 to 3 weeks culture period for Mk (CD41, CD42b), Er (GlyA) and My (CD14, CD15, CD33) cells. One example of the efficiency of this new assay comes from the analysis of CD49f+ HSC subset, which previously could not be read out in vitro as single cells (11). Under these new conditions, 77% of FL, 72% of CB and 48% of BM single CD49f+ HSCs were able to produce a clone (fig. S5A, left). Whereas only CFU-My were produced from HSC in MC, mixed clonogenic potential was now readily detectable with this assay. We used this assay to functionally map the lineage potential of all the newly defined CD34+ subsets from all three developmental time points.
Unipotent progenitors dominate the blood hierarchy by adulthood
To gain a global perspective of the functional differences in the blood hierarchy across ontogeny, we first combined all 11 CD34+ subsets from each developmental timepoint into one analysis of nearly 3000 single cells. Cloning efficiency was highest for FL and decreased gradually in CB and BM (FL – 74%, CB – 69%, BM – 55%; Fig. 2A). A simple stratification based on whether a single cell gave rise to one (unilineage) or more (multilineage) cell lineages revealed that 40% of FL CD34+ cells were multilineage compared to 27% of CB (p < 10−3, Fisher’s exact test) and 18% of BM (p < 10−3, Fisher’s exact test) CD34+ cells (Fig. 2B). Thus, the ratio of multilineage to unilineage progenitors changes en bloc in development within the CD34+ population, a result that is independent of the complex marker scheme we used.
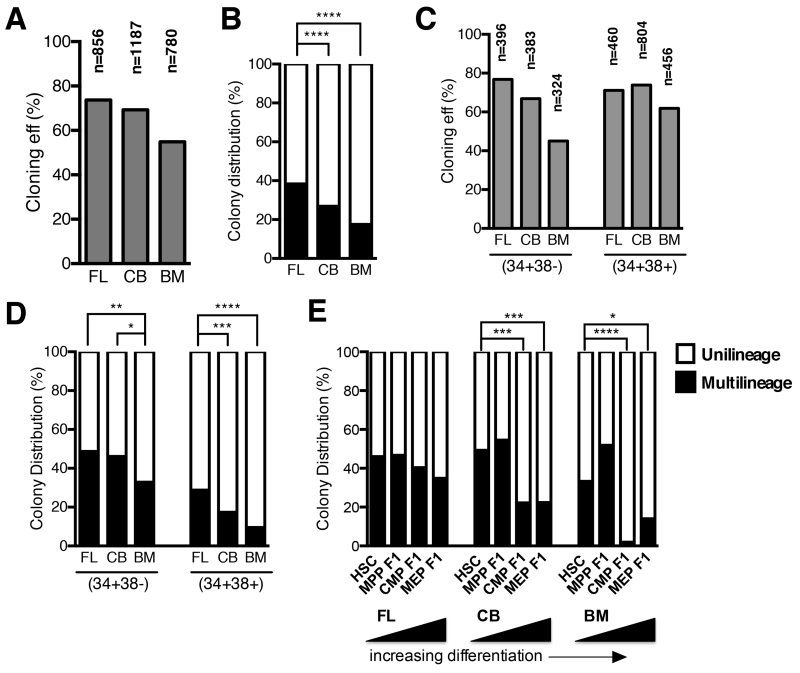
Figure 2. Assessment of multilineage and unilineage cell potential of single CD34+ cells from FL, CB and BM.
(A-D) Single cells from subsets shown in Fig. 1B and fig. S1 were deposited by FACS and cultured for several weeks. Emergent clones were analyzed by flow cytometry for My, Er and Mk lineages (Fig. 3A). To gain a global perspective of the functional differences, subsets were combined into one analysis of the CD34+ compartment (A&B), or stem (CD34+CD38−) and progenitor (CD34+CD38+) (C&D) compartments. Multilineage (black) potential was defined as any single cell that gave rise to more than one lineage (any two of My, E, Mk), and unipotent (white) potential as a single cell that gave rise to one lineage only (My or E or Mk) (B&D). Overall cloning efficiency is shown in grey (A&C). (E) Distribution of multilineage and unilineage cell potential from populations that lack expression of CD71 and BAH-1 (F1s) in FL, CB and BM. Shown by increasing levels of differentiation (HSC > MPP F1 > CMP F1 > MEP F1). Asterisks indicate significance based on Fisher’s exact test (* p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001).
To continue exploring the organizational relationships of progenitors across developmental timepoints, we investigated the proportion of multilineage to unilineage cells in the stem cell enriched (CD34+CD38−) and the progenitor enriched (CD34+CD38+) subsets. Within CD34+CD38− cells, FL and CB had a significantly higher proportion of multilineage cells compared to BM (FL vs. BM: 48.6% vs. 32.9%, p=0.0016; CB vs. BM: 46.1% vs. 32.9%, p=0.011; Fisher’s exact test). These proportional differences were more pronounced in the CD34+CD38+ progenitor compartment. In CD34+CD38+ cells, BM displayed three fold fewer multilineage cells compared to FL (FL: 28.8%; CB: 17.3%; 9.6%; p<0.0001, Fisher’s exact test) (Fig. 2C,D). Thus, both the stem and progenitor compartments from each developmental stage exhibited a proportional change in the percentage of multilineage cell types.
We made further attempts to localize the differentiation stages most affected by the loss of multilineage progenitors. We reasoned that HSCs and subsets that lack differentiation markers CD71 and BAH1 (MPP F1, CMP F1 and MEP F1) would be enriched for cells with multilineage cell potential. Remarkably in FL, the ratio of multilineage to unilineage progenitors remained nearly constant across these subsets (Fig. 2E). By contrast, only HSC and MPP F1 subsets from CB and BM were highly enriched for multilineage cells, whereas their corresponding CMP/MEP F1s were comprised mostly of unilineage cell types (Fig. 2E; p < 0.05, Fisher’s exact test). In BM, virtually all multilineage cells were restricted to the CD34+CD38− stem cell compartment (Fig. 2E; p < 0.05, Fisher’s exact test). Hence, multilineage cell potential extends into the progenitor compartment in FL, but in BM, this potential is restricted to the CD34+CD38− stem cell compartment. In parallel, the progenitor compartment in BM is dominated by unilineage cell types.
Mk-Er lineage branching in the blood hierarchy is developmentally defined
To gain a detailed understanding of the differentiation potential of each progenitor subset identified by our sorting scheme applied to FL, CB and BM, we classified the functional potential of each single cell in our data set. Five distinct clonal outputs were classified: Mk only, Er only, My only, Mk/Er, and mixed (bipotent: Er/My or Mk/My; tripotent: Er/Mk/My)(Fig. 3A). The cloning efficiency of all subsets was high in MPP, CMP and MEP fractions (50-80%; Fig. 3B; fig. S5A).
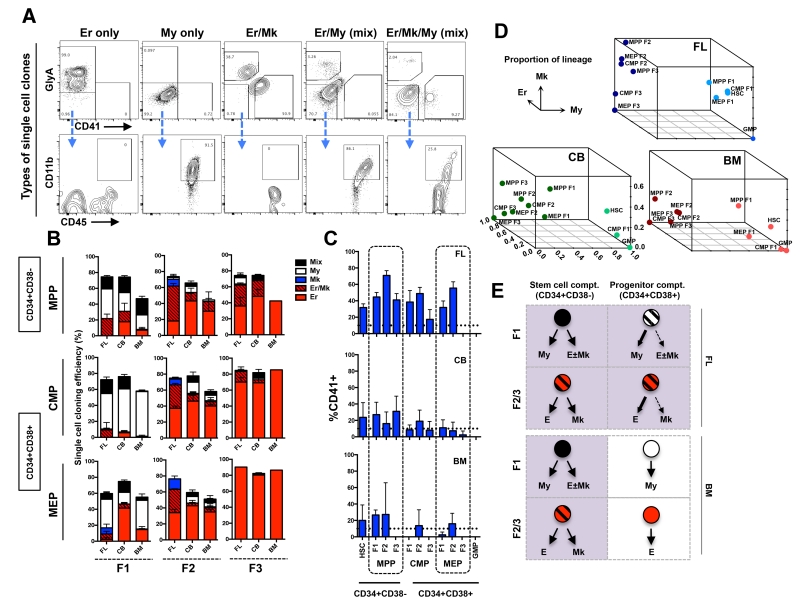
Figure 3. Lineage analysis of single cell derived clones from newly defined subsets of human MPPs, CMPs and MEPs.
(A) Single cell clones were analyzed by flow cytometry and binned into 5 distinct lineage outcomes. Erythroid clones were defined as GlyA+ only (Er only). Myeloid clones were identified as GlyA−CD41−, but CD45+CD11b+ (My only). Erythroid-megakaryocyte clones were defined as GlyA+ and CD41+ but negative for CD11b (Er/Mk). Mix clones were defined as the My cells (CD45+CD11b+) and Er cells (GlyA+) or Mk (CD41+) (column 4), or both (column 5). (B) Cloning efficiency and lineage outcomes of single cells from newly defined MPP, CMP, and MEP fractions (F1s, F2s and F3s) from FL, CB and BM. (C) Total Mk output (CD41+) from all newly defined subsets from FL, CB and BM. Bars indicate mean ± standard error. Total number of independent experiments: n=3, 6 and 4 for FL, CB and BM respectively. (D) 3-D summary of lineage outputs (My, Er and Mk) from all cellular subsets in FL, CB and BM presented in Fig. 3B. (E) Pictorial summary of the predominant lineage outcomes from stem (CD34+CD38−) and progenitor (CD34+CD38+) cell compartments in FL and BM data.
The highest percentage of mixed clones was found amongst the HSC subsets (FL: 46.1%; CB: 49.3; BM: 33.3%; fig. S5A), except in BM where MPP F1 harbored higher mixed clone potential than HSC (51.9%), although this was not significant (BM HSC vs. MPP F1: p=0.1, Fisher’s exact test) (Fig. 2E). FL HSC and MPP F1 subsets had a statistically higher distribution of tripotent versus bipotent mixed clones compared to CB and BM HSC and MPP F1 (fig. S5B). In FL and CB, Mk-Er only clones appeared at the MPP F1 stage (FL: 29% of total; CB: 18% of total; Fig 3B, column1, row1). In BM, Mk-Er clones from MPP F1 were rare (~2%), rather 15% of all clones from this subset were Er-only. Importantly, Mk activity from BM MPP F1 was detected as a component of mixed clones that also contained My cells (further discussed below).
We next interrogated the CD34+CD38+ compartment. Based on the classical view of the blood hierarchy, we would expect that true CMPs reside in a subset that lacks expression of differentiation markers such as CD71 and BAH1 (CMP F1). Only 15% of FL and CB CMP F1 clones were mixed, and no mixed clones from this subset were detected in BM (Fig. 3B, column 1, row2). Thus, we conclude that previously defined BM CMPs are not homogeneous for cells with multilineage My-Er-Mk potential, rather they heterogeneous composed of subpopulations of unilineage My, Er and Mk progenitors (Fig 3B, row 2). To determine if the small percentage of mixed clones from FL and CB CMPs F1 population were derived from bona fide CMPs, we evaluated their My-Er-Mk potential. More than 80% of the mixed clones from FL and CB were bipotent (either Er/My or Mk/My) without concurrent Mk-Er-My potential (fig S5C). This data demonstrates that “bona-fide” CMPs are a rare component of the human hematopoietic tree, irrespective of developmental stage.
MC and megacult colony assays indicated that subsets defined by CD71 and BAH1 expression (F2 and F3) were highly enriched for Mk and Er activity (fig. S3A-D and Supplementary text). However, these assays cannot formally rule out that Er and Mk potential was derived from independent unilineage progenitors. We first tracked Mk-Er activity from single cells within the MEP subsets (MEP F1 through F3; Fig. 3B, row 3). MEP F1 was highly heterogeneous across developmental timepoints and comprised mostly of My progenitors in FL and BM (60% or more; Fig. 3B, column 1, row 3) that functionally resemble other F1 subsets from MPP and CMP. In CB and BM, ~70% of clones from MEP F2 were Er-only and 10% or less were Mk-Er clones (Fig. 3B, column 2, row3). MEP F2 is likely the subset within classical MEPs that gave rise to low-level Mk colonies in previous studies. Strikingly, MEP F3, the numerically dominant cell population within the classical MEPs, uniformly produced Er-only clones in FL, CB and BM (Fig. 3B, column 3, row3). Thus, classically defined MEPs are principally comprised of Er-only progenitors when analyzed at single cell resolution and are not Mk-Er progenitors as previously thought.
As only rare cells within the MEP fractions give rise to Mks, Mk potential must lie elsewhere in the blood hierarchy. We found that most Mk-Er activity came from the CD34+CD38− stem cell compartment (fig. S3C) and was particularly enriched within one of our newly defined MPP subsets (Fig. 3B, column2, row1). In FL, a striking 60% of clones from MPP F2 were of Mk-Er type and the remainder of this subset was composed of Mk-only or Er-only clones (Fig. 3B, column2, row1). Notably Mk activity was enriched but not restricted to the stem cell compartment in the FL. Since we did not find strong evidence for FL CMPs, we expect that FL Mk-Er progenitors arise from the stem cell compartment, specifically from MPP F2. In CB and BM, Mk-Er clones represented one-quarter of the total clonal output from this subset (Fig. 3B, column2, row1) and the rest were Er-only clones. In CB and more evident in BM, Mks predominantly emerged as part of mixed clones from HSCs and MPP F1, supporting the hypothesis that Mk branching occurs directly from a multipotent cell as predicted by the murine studies (5). These unexpected data suggest that both Mk-Er and multilineage potential are restricted to the stem cell compartment by adulthood, whereas unilineage fates predominate the progenitor compartment forming a simple ‘two-tier’ hierarchy, with few intervening oligopotent intermediates (Fig. 3D,E).
In vivo analysis establishes hierarchical relationships between novel progenitor subsets
In the blood hierarchy cell types near the peak of the hierarchy, such as HSCs and MPPs, are rarer but possess higher proliferative potential. Lineage differentiation typically correlates with loss of proliferative potential;. Although proportionally HSC and MPP subsets (F1-F3) are minor populations, they yielded 5-10× more cells compared to more abundant populations from the CMP and MEP subsets in vitro (fig. S6A).
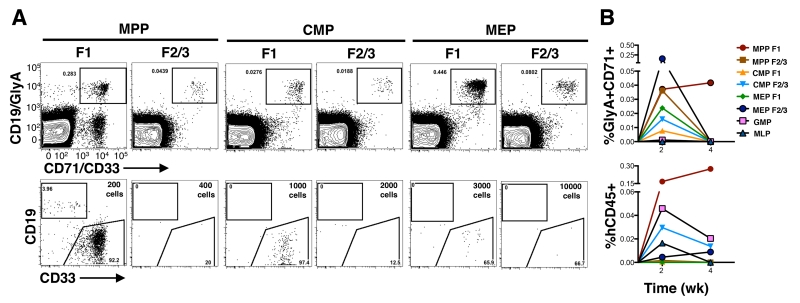
We then transplanted our cellular subsets in vivo and measured graft durability and size, as well as its lineage composition to establish the hierarchical relationships of our newly defined progenitor subsets. Due to tissue availability, only CB was used. As there is limited data on the engraftment capacity of human progenitors in the NOD-Scid-Il2rgnull (NSG) model, we first scrutinized the repopulations kinetics of human blood cells in this model. Using HSCs, we observed low-level lympho-myeloid as well as erythro-megakaryocytic engraftment as early as two weeks after transplant (fig. S6B-C) consistent with prior studies using NOD-Scid mice (24). We used two weeks as the standpoint from which to assess progenitor cell engraftment in vivo. One thousand CMP F1 cells and 3000 MEP F1 cells generated a myelo-erythroid restricted that did not persist beyond two weeks (Fig. 4A, column 3 and 5; fig. S6E). By contrast, 200 MPP F1 cells were able to sustain a robust and systemic multilineage graft (My-Er-Ly) beyond two weeks, consistent with the functional potential of true MPPs (Fig. 4A, column 1; Fig. 4B; fig. S6E) (18).
Figure 4. In vivo potential of progenitor subsets.
(A) Freshly sorted populations from CB were intrafemorally transplanted into sublethally irradiated NOD-scid-IL2Rgnull (NSG) mice. BM cells from injected femur and non-injected bones were analyzed by flow cytometry 2 weeks post transplant. Average transplanted cell dose is shown as an inset in the flow plot. Top row indicates Er engraftment (GlyA+CD71+). Bottom row depicts the CD45+ cells, B-lymphoid cells and My cells were detected using CD19 and CD33, respectively. (B) Kinetic analysis of engraftment from progenitor subsets. Mean levels of erythroid (GlyA+CD71+) and human cell engraftment (CD45+) are shown.
To gather enough cell numbers for in vivo detection of Er-enriched subsets, we combined the F2 and F3 subsets from MPP, CMP and MEP for transplantation since they shared similar functional potential in vitro. The combined F2/F3 subsets from MPP, CMP and MEP all gave rise to prominent Er grafts in vivo, concordant with their in vitro potential (Fig. 4A columns 2,4,6; fig S6D top panels). MPP F2/F3 cells were highly proliferative and generated a robust Er graft with only 400 transplanted cells (Fig. 4A, column 2), whereas CMP F2/F3 and MEP F2/F3 required 5 to 25 fold higher cell doses to generate an in vivo graft (Fig. 4A, columns 4 and 6). Although platelets were difficult to detect in vivo from progenitor subsets, we did observe them in rare mice engrafted with either MPP F1 and MPP F2/3 cells (fig. S6C). Only MPP F2/F3 but not CMP F2/3 and MEP F2/3 were able to migrate systemically to non-transplanted bones and resemble the proliferative potential of MPP F1 (fig. S6E). Collectively, when combine with the in vitro analyses of these subsets, these in vivo experiments support that Mk-Er enriched MPP F2/F3 are derived from HSCs or MPPs directly without invoking a lineage route via a CMP intermediate.
A transcriptionally defined erythroid progenitor subnetwork in the CD34 hierarchy
Lineage commitment coincides with the expression of key molecules that aid to ‘lock-in’ a differentiation program (25). Using low cell input RNA sequencing methodology (26), we first analyzed the expression profile of canonical lineage factors in bulk CB subsets. Genes associated with My specification such as MPO and CSF2RA (GM-CSFR) were highly expressed in GMPs, whereas genes associated with the Er lineage such as GATA-1 and EPOR, were highly expressed in F2/F3 subsets from MPP, CMP and MEP populations (fig. S7A). Mk differentiation markers, CD41 (ITGA2B) and CD42b (GP1BA), were highly expressed in MPP F2, in line with functional potential of this subset. This data provides an independent line of evidence that supports committed Mk progenitors reside within the stem cell compartment. Low-level expression of CD41 and CD42b was also detected in MPP F3 and MEP F2 consistent with the residual Mk activity from these CB subsets (fig. S7A).
Unsupervised hierarchical clustering and principal component analysis of the entire data set revealed two major molecular subgroups: those with Er-enriched potential (MPP, CMP and MEP F2/F3s), and another with multilineage (HSC, MPP F1) or My-enriched potential (CMP F1 or MEP F1; fig. S7B). F2 and F3 subsets within MPP, CMP and MEP populations clustered together, suggesting these pairs are more closely related within each broader compartment (fig. S7Bi). Due to their close transcriptional and functional relationship, we merged F2 and F3 subsets from MPP, CMP and MEP subsets to compare global transcriptional differences amongst these Er-enriched subsets. We found that 230 genes were differentially expressed between MPP (F2/F3) versus CMP (F2/F3), and 52 genes between CMP (F2/F3) versus MEP (F2/F3). These differentially expressed genes were highly enriched in cell cycle, and DNA replication and metabolic processes, and coincide with the extensive proliferation that Er progenitors undergo during specification (fig. S7C). These data reinforce the idea that the close functional relationship between Mk-Er and Er enriched subsets is likely due to shared molecular programs.
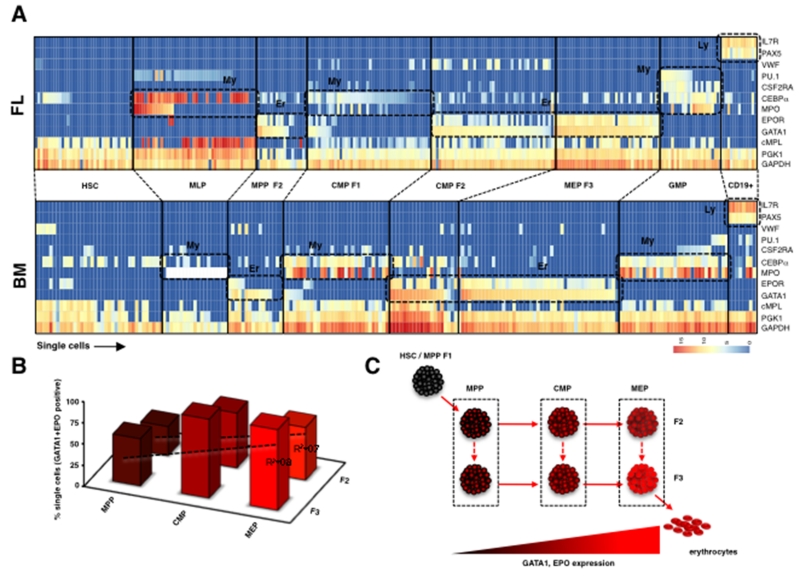
Molecular heterogeneity amongst single cells within a purified subset is commonly lost in a population-level analysis. We tracked the expression of key lineage factors amongst single cells in our progenitor subsets. Only FL and BM were used in this experiment since they represent the two ends of the development timepoints used in this study. In both FL and BM, single cells from F2 and F3 from MPP, CMP and MEP displayed a dominant Er gene expression program often co-expressing both GATA1 and EPOR genes in the same cell (Fig. 5A, fig. S7D). This data confirms that our new sorting can essentially resolve the Er-committed progenitors within the CD34+ hierarchy. We observed that GATA1 expression was present amongst most single cells in MPP F2 and MPP F3, but EPOR expression was only present in a subset of GATA1 positive cells (Fig. 5A,B). Since GATA1 precedes EPOR expression in Er differentiation, this data strongly supports the conclusion that MPP F2/F3 cells represent the earliest erythroid differentiation precursor in the human blood hierarchy. The percentage of single cells that co-expressed GATA1 and EPOR increased in proportion amongst F2 and F3 subsets from MPP, CMP and MEP in both FL and BM (Fig. 5B). These molecular factors are considered a surrogate of the degree of Er differentiation (Fig. 5B) and when considered jointly with our in vitro and in vivo analyses, these data indicate that these subsets are already Er specified but vary mostly in their proliferative potential. We hypothesize that these subsets compose a hierarchical subnetwork of Er progenitors within the CD34+ compartment (Fig. 5C). A candidate network map is shown in Fig. 5C. Considering that erythrocytes comprise nearly 99% of all blood cells, this network may offer a high degree of flexibility to synthesize erythrocytes under homeostatic and emergency erythropoiesis without HSC input.
Figure 5. Single cell gene expression profiling from FL and BM subsets.
(A) Single cells from FL (top) and BM (bottom) subsets were sorted and analyzed for expression of genes associated with My, Er, and lymphoid (Ly) lineages (shown on right) on the Fluidigm platform. My, Er or Ly gene clusters are shown as dashed boxes. (B) Percent of single cells that co-express GATA1 and EPOR in F2 and F3 from MPP, CMP, and MEP subsets. BM and FL F2 and F3 subsets were combined in this analysis. (C) A theoretical subnetwork of Er progenitors within the CD34+ compartment.
Dramatic loss of Er progenitors compared to My progenitors in a hematologic condition of HSC deficiency
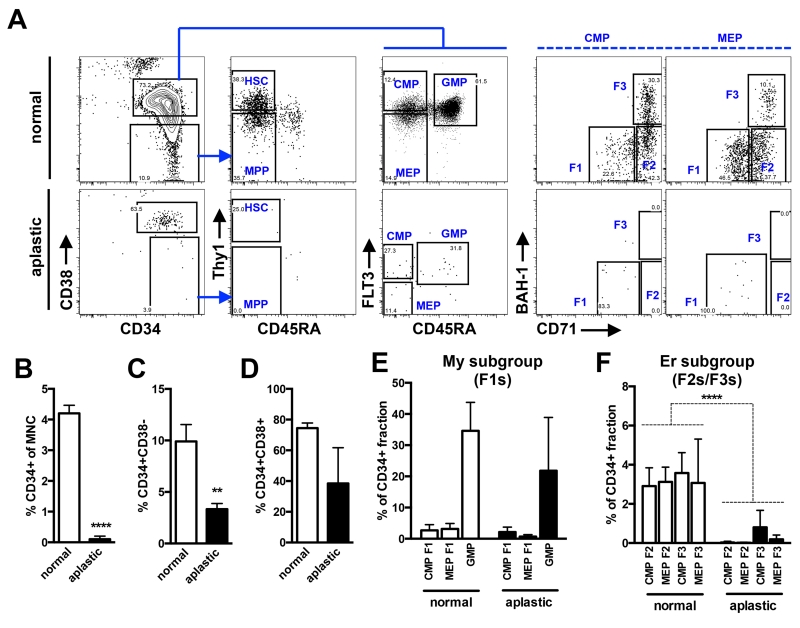
Recent HSC fate-mapping analyses in mouse have provocatively shown that progenitors and not HSCs, are fundamental for on-going hematopoiesis under homeostatic conditions (27, 28). Physiological studies to experimentally test this question are not possible in normal human subjects. However, certain disease states permit a glimpse into the consequences of HSC loss on progenitor cell architecture and may shed light on the role of HSCs in human blood synthesis under non-transplant conditions. In aplastic anemia (AA), HSCs are damaged likely due to an autoimmune response and unable to contribute to on-going hematopoiesis (29, 30). This effect seems to be specific to HSCs as all mature blood lineages are depressed in AA (31-33). We examined the progenitor architecture of the CD34+ cells in three cases of AA by applying our sorting scheme (Fig. 6). Consistent with previous reports, the proportion of CD34+ cells within the overall mononuclear cell (MNC) pool was significantly lower in AA compared to normal BM (0.1% vs. 4.2%, p<0.0001, t-test) (Fig. 6B) (31, 32). The CD34+CD38− stem cell compartment in AA patients was more significantly depleted compared to the CD34+CD38+ progenitor compartment (Fig. 6A, left column; Fig. 6C,D). Moreover, HSCs and MPPs were virtually undetectable in the residual CD34+CD38− compartment, confirming that HSCs are specifically lost in this clinical condition (Fig. 6A, column 2), at least at the phenotypic level. Despite the loss of phenotypic HSCs, the CD34+CD38+ compartment was detectable in all cases. We quantified the subsets within the CD34+CD38+ compartment to determine if all cell types are indiscriminately affected. Based on our single cell functional readouts, we grouped progenitors enriched for myeloid (CMP F1, MEP F1, GMP) and erythroid (CMP F2/F3, MEP F2/F3) differentiation potential to increase the power of the analysis due to relative loss of CD34+ cells in AA. Despite significant depletion of HSCs, the percentage of myeloid progenitors was stable compared to normal BM (Fig. 6E). In contrast, erythroid progenitors were significantly lost, like HSCs, in all three patients analyzed (Fig. 6F, p<0.0001, t-test). These results suggest that on-going erythropoiesis is more reliant on HSC input compared to myelopoiesis. Although we cannot rule out a specific Er lineage defect in AA, the pan-lineage deficiencies observed in AA likely rule out this possibility and support the idea that the Er progenitor loss is most likely a repercussion of HSC depletion. Since all HSC types (LT/IT/ST) seem to be broadly lost in AA, our results cannot distinguish which type of HSC is crucial to maintain erythropoiesis. Recognizing that hematopoiesis in AA is not normal, the revised hierarchy model predicted from our experimental data does appear to have physiological relevance in the human setting.
Figure 6. Analysis of My and Er progenitors in patients with aplastic anemia.
(A) To examine the consequences of HSC loss on progenitor subsets, BM cells from three AA patients and normal controls were subjected to the new sorting design shown in Fig. 1B. Representative flow plots from a single AA case and a control are shown. (B) Quantification of total CD34+ cells as a fraction of mononuclear cell (MNC) pool from controls (empty bars) versus AA (filled bars). (C-D) The CD34+ subset from controls and AA was further sub-divided into the stem (CD34+CD38−) and progenitor (CD34+CD38+) cell compartments. (E) Analysis of My enriched subsets (CMP F1, MEP F1 and GMP) in control and patients with AA. (F) Analysis of Er enriched subsets (CMP F2/F3, MEP F2/F3) in controls and AA. Bars indicate mean ± standard error from 3 controls and 3 cases of AA. Asterisks indicate significance based on t-test (** p<0.01, **** p<0.0001).
DISCUSSION
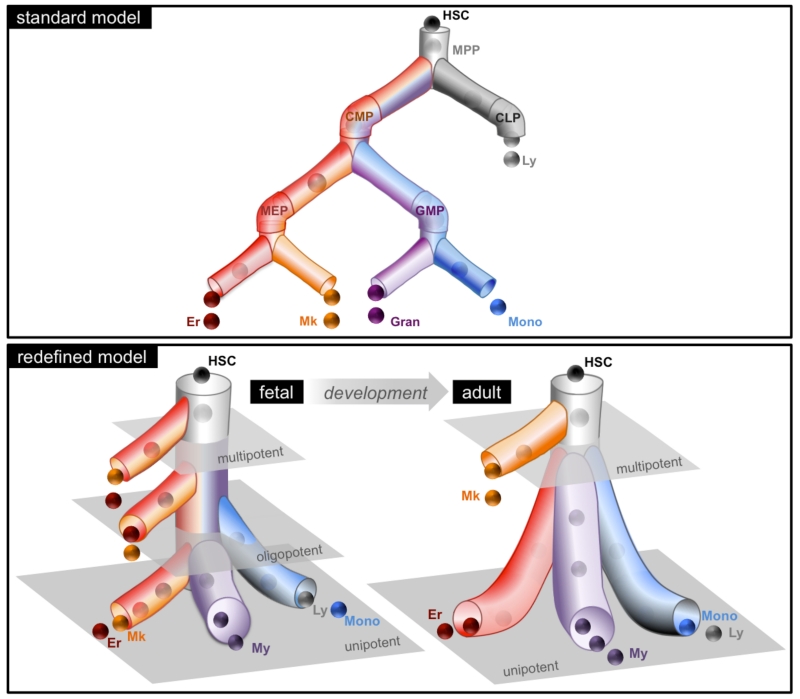
Our study challenges the current view that human blood development occurs progressively through a series of multipotent, oligopotent and then unilineage progenitor stages. By subjecting the classically defined progenitor subsets to a new sorting scheme that efficiently resolved My, Er and Mk lineage fates, combined with single-cell functional analysis, we made two unexpected findings. First, we found that the blood progenitor architecture is not identical across development. In FL, oligopotent progenitors with My-Er-Mk and Er-Mk activity were a prominent component of the hierarchy. By contrast, the BM was dominated by unilineage progenitors with primarily My or Er potential. This shift in progenitor classes demarcates a fundamental readjustment in the blood hierarchy during in utero to adulthood timepoints. The absence of oligopotent intermediates that become gradually restricted to unilineage progenitors in BM cannot be reconciled under the standard model of HSC differentiation. Instead, our data support a hierarchy composed mainly of two-tiers in adults: a top-tier which contains multipotent cells such as HSCs and MPPs, and a bottom-tier composed of committed unipotent progenitors (Fig. 7). We cannot formally rule out the presence of a highly transient adult CMP-like progenitor stage that exists when multipotent cells differentiate into unipotent progenitors, however our in vitro and in vivo assays surveyed large cell numbers (~3000) and such a progenitor was not detected. If lineage restricted cell types able to generate a subset or full spectrum of myeloid cells do exist in the stem cell compartment of adult marrow, they most probably represent the murine counterparts of myeloid-bias or myeloid-restricted HSC subtypes (5, 34). Second, the origins of the Mk lineage branch change from FL to BM. In FL, Mk progenitors were enriched but not restricted to the stem cell compartment whereas in BM, the Mk lineage was closely tied to the fate of multipotent cells. These data are not consistent with the principal tenet of the standard model that My, Er and Mk lineages originate from a common lineage progenitor such as CMP.
Figure 7. Redefined model of human blood development.
A graphical representation of the redefined model that encompasses the predominant lineage potential of the newly defined progenitor subsets; the standard model is shown for comparison. The redefined model envisions a developmental shift in the progenitor cell architecture resulting in a two-tier hierarchy by adulthood.
Historically, the first major lineage bifurcation step in the blood hierarchy was considered to be the segregation of myeloid (My-Er-Mk) and lymphoid fates, with CMPs occupying the lineage fork that gives rise to the entire myeloid arm. The co-emergence of My, Er and Mk lineages was central to the description of CMPs. Our results reveal that the originally defined CMPs are highly heterogeneous, primarily composed of unipotent My or Er progenitors with little Mk activity. In the absence of CMPs, how can the origins of lineage-restricted progenitors such as GMPs and MEPs be reconciled? Our previous clonal analysis suggested that myelomonocytic lineages originate from MLPs, which we suspect is the most probable precursor of GMPs due to their shared functional and transcriptional profiles (11, 12, 35). In this study, we found that GATA-1 positive Mk-Er committed progenitors exist in the stem cell compartment suggesting that MEPs are derived from multipotent cells. In the murine bone marrow niche, up to a quarter of LT-HSCs lie directly adjacent to Mks (36-38). Mks play a dual role in HSC regulation. Under normal conditions, Mk-HSC contact is essential to preserve the quiescent nature of adult LT-HSCs. Interestingly, after myeloablation, this effect is temporarily abrogated and Mks secrete growth factors that permit HSCs to expand (37). Thus, direct differentiation of Mks from HSCs may represent a physical mechanism to regulate blood stem cell functionality in the niche. Overall, the first major bifurcation step in blood differentiation is far more complex than a simple segregation of myeloid and lymphoid lineages. In humans, we suspect this first step splits the Mk-Er lineage from the myelomonocytic lineage that co-segregates with the lymphoid fate (11). Why the myelomonocytic lineage but not the granulocytic lineage is tied to the lymphoid fate will be a critical area of future investigation (39, 40). The ability to isolate developmental populations reported here provides a critical experimental framework to facilitate such studies in humans.
Our data may also have implications for our understanding of lineage specification at the molecular level. vWF, a key molecular marker strongly associated with Mk differentiation, was expressed in a subset of BM HSCs and may identify an HSC subtype primed for platelet production as shown in mouse (16). Interestingly, molecular factors involved in Mk (vWF), Er (EPOR) and My (CSF2RA) differentiation were expressed in small pockets of single cells from undifferentiated cells, like BM HSCs, in a near mutually exclusive manner. This data is consistent with the notion that TFs associated with myeloid lineage specification are individually but not simultaneously primed at the level of stem cells. It is difficult to preclude that this type of molecular heterogeneity reflects impurities in human HSC isolation. However, analysis of purer murine HSC compartments supports that molecular factors associated with lineage commitment is stochastically activated at the level of HSCs (16, 17). Maintenance of a multipotent state is thought to occur via low-level lineage priming where TFs of different lineages are co-expressed in the same cell. In this model, commitment towards a particular lineage occurs by the mutual antagonism of these TFs where one TF eventually wins, locking-in a differentiation program. However, the near mutually exclusive expression of vWF, CSFR2A and EPOR in BM HSCs does not agree with this logic. The hypothesis that lineage commitment occurs in the absence of a coordinated differentiation program (17, 25, 41) is more consistent with our data.
Ultimately, a better understanding of normal blood differentiation programs will be critical to decipher how such programs go awry in disease. Indeed in AA, we observed that Mk-Er progenitors are lost alongside HSC, but the My progenitor pool can continue to persist. Recent evidence from murine in situ tracking experiments showed that My progenitors can be sustained long term without contribution from HSCs (27, 28). If human My progenitors are similarly long-lived, as our data suggests, they would have a higher probability of acquiring mutations that could lead to clonal expansion and eventual My lineage leukemias. The short-lived nature of Mk-Er progenitors and their dependency on HSC input reduces their probability of accumulating enough mutations leading to leukemia. Clinical evidence that acute leukemia of the Mk and Er lineages are extremely rare compared to My leukemia is consistent with this idea. Our work on AA highlights one example of the clinical utility of a high-resolution developmental roadmap of normal hematopoiesis. The adaptable nature of our new sorting scheme should similarly inform on other hematological conditions.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all members of the Dick Laboratory for the critical review of the manuscript especially Jean CY Wang, Eric Lechman, and Melissa Cooper; N. Simard and Sherry Zhao and members of the Sickkids-UHN Flow facility for technical support. We thank K. Moore and the obstetrics unit of Trillium Hospital (Mississauga, Ontario) for providing CB. We thank Jurg Roher from BD Biosciences for supplying BAH-1 clone. This work was supported by Postdoctoral Fellowship Awards from Canadian Institute of Health Research (CIHR) to FN and SZ. Work in the Dick laboratory is supported by grants from the CIHR, Canadian Cancer Society, Terry Fox Foundation, Genome Canada through the Ontario Genomics Institute, Ontario Institute for Cancer Research with funds from the province of Ontario, a Canada Research Chair and the Ontario Ministry of Health and Long Term Care (OMOHLTC). The views expressed do not necessarily reflect those of the OMOHLTC.
Footnotes
References and Notes
- 1.TILL JE, McCULLOCH EA. Hemopoietic stem cell differentiation. Biochim. Biophys. Acta. 1980;605:431–459. doi: 10.1016/0304-419x(80)90009-8. [DOI] [PubMed] [Google Scholar]
- 2.Spangrude GJ, et al. Mouse hematopoietic stem cells. Blood. 1991;78:1395–1402. [PubMed] [Google Scholar]
- 3.Adolfsson J, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Månsson R, et al. Molecular Evidence for Hierarchical Transcriptional Lineage Priming in Fetal and Adult Stem Cells and Multipotent Progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Yamamoto R, et al. Clonal Analysis Unveils Self-Renewing Lineage-Restricted Progenitors Generated Directly from Hematopoietic Stem Cells. 2013;154:1112–1126. doi: 10.1016/j.cell.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 6.Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10:120–136. doi: 10.1016/j.stem.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 7.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 8.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 9.Manz MG, Miyamoto T, Akashi K, Weissman IL. Prospective isolation of human clonogenic common myeloid progenitors. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11872–11877. doi: 10.1073/pnas.172384399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galy A, Travis M, Cen D, Chen B. Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity. 1995;3:459–473. doi: 10.1016/1074-7613(95)90175-2. [DOI] [PubMed] [Google Scholar]
- 11.Doulatov S, et al. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. Nat. Immunol. 2010;11:585–593. doi: 10.1038/ni.1889. [DOI] [PubMed] [Google Scholar]
- 12.Laurenti E, et al. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat. Immunol. 2013:1–11. doi: 10.1038/ni.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pronk CJH, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 14.Guo G, et al. Mapping cellular hierarchy by single-cell analysis of the cell surface repertoire. Cell Stem Cell. 2013;13:492–505. doi: 10.1016/j.stem.2013.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Naik SH, et al. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496:229–232. doi: 10.1038/nature12013. [DOI] [PubMed] [Google Scholar]
- 16.Sanjuan-Pla A, et al. Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature. 2013:1–17. doi: 10.1038/nature12495. [DOI] [PubMed] [Google Scholar]
- 17.Pina C, et al. Inferring rules of lineage commitment in haematopoiesis. Nature cell biology. 2012;14:287–294. doi: 10.1038/ncb2442. [DOI] [PubMed] [Google Scholar]
- 18.Notta F, et al. Isolation of single human hematopoietic stem cells capable of long-term multilineage engraftment. Science. 2011;333:218–221. doi: 10.1126/science.1201219. [DOI] [PubMed] [Google Scholar]
- 19.Qian H, et al. Critical Role of Thrombopoietin in Maintaining Adult Quiescent Hematopoietic Stem Cells. Cell Stem Cell. 2007;1:671–684. doi: 10.1016/j.stem.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Solar GP, et al. Role of c-mpl in early hematopoiesis. Blood. 1998;92:4–10. [PubMed] [Google Scholar]
- 21.Deng B, et al. An agonist murine monoclonal antibody to the human c-Mpl receptor stimulates megakaryocytopoiesis. Blood. 1998;92:1981–1988. [PubMed] [Google Scholar]
- 22.Edvardsson L, Dykes J, Olofsson T. Isolation and characterization of human myeloid progenitor populations—TpoR as discriminator between common myeloid and megakaryocyte/erythroid progenitors. Experimental hematology. 2006;34:599–609. doi: 10.1016/j.exphem.2006.01.017. [DOI] [PubMed] [Google Scholar]
- 23.Kohn LA, et al. Lymphoid priming in human bone marrow begins before expression of CD10 with upregulation of L-selectin. Nat. Immunol. 2012;13:963–971. doi: 10.1038/ni.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazurier F, Doedens M, Gan OI, Dick JE. Rapid myeloerythroid repopulation after intrafemoral transplantation of NOD-SCID mice reveals a new class of human stem cells. Nature medicine. 2003;9:959–963. doi: 10.1038/nm886. [DOI] [PubMed] [Google Scholar]
- 25.Enver T, Pera M, Peterson C, Andrews PW. Stem cell states, fates, and the rules of attraction. Cell Stem Cell. 2009;4:387–397. doi: 10.1016/j.stem.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 26.Ramsköld D, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nature biotechnology. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun J, et al. Clonal dynamics of native haematopoiesis. Nature. 2014;514:322–327. doi: 10.1038/nature13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Busch K, et al. Fundamental properties of unperturbed haematopoiesis from stem cells in vivo. Nature. 2015;518:542–546. doi: 10.1038/nature14242. [DOI] [PubMed] [Google Scholar]
- 29.Sankaran VG, Weiss MJ. Anemia: progress in molecular mechanisms and therapies. Nature medicine. 2015;21:221–230. doi: 10.1038/nm.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maciejewski JP, Risitano A. Hematopoietic stem cells in aplastic anemia. Arch. Med. Res. 2003;34:520–527. doi: 10.1016/j.arcmed.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Marsh JC, Chang J, Testa NG, Hows JM, Dexter TM. The hematopoietic defect in aplastic anemia assessed by long-term marrow culture. Blood. 1990;76:1748–1757. [PubMed] [Google Scholar]
- 32.Rizzo S, et al. Stem cell defect in aplastic anemia: reduced long term culture-initiating cells (LTC-IC) in CD34+ cells isolated from aplastic anemia patient bone marrow. Hematol. J. 2002;3:230–236. doi: 10.1038/sj.thj.6200187. [DOI] [PubMed] [Google Scholar]
- 33.Matsui WH, Brodsky RA, Smith BD, Borowitz MJ, Jones RJ. Quantitative analysis of bone marrow CD34 cells in aplastic anemia and hypoplastic myelodysplastic syndromes. Leukemia. 2006;20:458–462. doi: 10.1038/sj.leu.2404119. [DOI] [PubMed] [Google Scholar]
- 34.Benz C, et al. Hematopoietic stem cell subtypes expand differentially during development and display distinct lymphopoietic programs. Cell Stem Cell. 2012;10:273–283. doi: 10.1016/j.stem.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida T, Ng SY-M, Zuñiga-Pflücker J-C, Georgopoulos K. Early hematopoietic lineage restrictions directed by Ikaros. Nat. Immunol. 2006;7:382–391. doi: 10.1038/ni1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heazlewood SY, et al. Megakaryocytes co-localise with hemopoietic stem cells and release cytokines that up-regulate stem cell proliferation. Stem cell research. 2013;11:782–792. doi: 10.1016/j.scr.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 37.Zhao M, et al. Megakaryocytes maintain homeostatic quiescence and promote post-injury regeneration of hematopoietic stem cells. Nature medicine. 2014;20:1321–1326. doi: 10.1038/nm.3706. [DOI] [PubMed] [Google Scholar]
- 38.Bruns I, et al. Megakaryocytes regulate hematopoietic stem cell quiescence through CXCL4 secretion. Nature medicine. 2014;20:1315–1320. doi: 10.1038/nm.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Görgens A, et al. Revision of the Human Hematopoietic Tree: Granulocyte Subtypes Derive from Distinct Hematopoietic Lineages. CellReports. 2013;3:1539–1552. doi: 10.1016/j.celrep.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 40.Mori Y, et al. Identification of the human eosinophil lineage-committed progenitor: revision of phenotypic definition of the human common myeloid progenitor. Journal of Experimental Medicine. 2009;206:183–193. doi: 10.1084/jem.20081756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rieger MA, Hoppe PS, Smejkal BM, Eitelhuber AC, Schroeder T. Hematopoietic cytokines can instruct lineage choice. Science. 2009;325:217–218. doi: 10.1126/science.1171461. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.