Abstract
Recent lineage tracing studies support the existence of prostate luminal progenitors that possess extensive regenerative capacity, but their identity remains unknown. We show that Sca-1 (Stem Cell Antigen-1) identifies a small population of murine prostate luminal cells that reside in the proximal prostatic ducts adjacent to the urethra. Sca-1+ luminal cells do not express Nkx3.1. They do not carry the secretory function, although they express the androgen receptor. These cells are enriched in the prostates of castrated mice. In the in vitro prostate organoid assay, a small fraction of the Sca-1+ luminal cells are capable of generating budding organoids that are morphologically distinct from those derived from other cell lineages. Histologically, this type of organoid is composed of multiple inner layers of luminal cells surrounded by multiple outer layers of basal cells. When passaged, these organoids retain their morphological and histological features. Finally, the Sca-1+ luminal cells are capable of forming small prostate glands containing both basal and luminal cells in an in vivo prostate regeneration assay. Collectively, our study establishes the androgen-independent and bipotent organoid-forming Sca-1+ luminal cells as a functionally distinct cellular entity. These cells may represent a putative luminal progenitor population and serve as a cellular origin for castration resistant prostate cancer.
Keywords: Progenitor cells, Sca-1, prostate, organoid
Introduction
Prostate epithelia are composed of three types of cells. Luminal epithelial cells carry the secretory functions and form a single layer surrounding the lumen filled with eosinophilic secretions, whereas basal epithelial cells encapsulate luminal cells, acting as a layer to protect epithelial integrity [1, 2]. Neuroendocrine cells are very rare and are less understood compared to the other two cell types [3]. Several recent studies using a lineage tracing approach demonstrated that prostate basal cells and luminal cells are independently sustained in adult mice [4–7]. However, basal cells are functionally plastic and are able to generate luminal cells when stimulated by embryonically-derived urogenital sinus mesenchymal (UGSM) cells [8–13] or under certain pathological conditions such as prostatitis [14].
The observation that the basal and luminal cell lineages in adult mice are independently sustained implies the existence of respective stem cells or progenitors in these two lineages. A lineage hierarchy within basal cells has been revealed. For example, the surface antigen Trop2 was shown to be able to enrich for stem cell activity in basal cells using various in vitro and in vivo prostate stem cell assays [12]. In contrast, the lineage hierarchy in the luminal cell lineage remains unclear. There has been some evidence suggesting the existence of phenotypic and functional heterogeneities within the luminal cell lineage. For example, Tsujimura et al. showed by BrdU label retention that luminal cells in the murine prostatic ducts that are adjacent to the urethra, i.e. proximal prostatic ducts, divide less frequently than those at the tips of prostatic ducts [15]. In addition, we showed previously that despite the activation of Notch signaling in adult murine luminal cells, only a small fraction of luminal cells are capable of acquiring the capacity for anoikis resistance and prostate sphere formation in vitro [16]. Finally, in in vitro organoid assays developed very recently, only a small fraction (less than 1%) of prostate luminal cells are able to generate organoids containing both basal cells and luminal cells [17, 18]. Although these studies further support the existence of a functional hierarchy within the prostate luminal cell lineage, the identity of the putative luminal progenitors remains undefined. In this study, we identify a small population of Sca-1-expressing luminal epithelial cells that reside in the proximal prostatic ducts in mice. We further demonstrate that they represent a bona fide cellular entity that possesses a distinct functional capacity as compared to the rest of the luminal epithelial cells.
Results
Stem cell antigen-1 identifies a distinct fraction of murine prostate luminal cells
Several lineage tracing studies including ours have demonstrated that prostate luminal cells in adult mice are self-sustained when prostate epithelia are induced to undergo several cycles of involution and regeneration by alternate androgen-depletion and replacement [4–7]. These studies suggest the existence of androgen-independent luminal progenitors, but their identity remains undefined. We reasoned that the luminal progenitors should be enriched in the prostate tissues of castrated mice and sought to identify this cell population based on their surface antigen expression profiles. Previously, major prostate cell lineages have been successfully fractionated based on the expression of Sca-1, CD49f, and several lineage markers (CD45;CD31;Ter119) (Fig. 1A). Basal cells are Lin−Sca-1+CD49fhigh, luminal cells are Lin−Sca-1−CD49flow, and stromal cells are Lin−Sca-1+CD49f− [9, 10]. After analyzing the luminal cells in intact versus castrated mice, we discovered that luminal cells in castrated mice express relatively higher levels of Sca-1 (Fig. 1B). More interestingly, the contour plots indicate the existence of a distinct cell population in castrated mice that is Sca-1+CD49flow (approximately 9.22% of total cells). When androgen was replaced in castrated mice, the androgen-dependent Sca-1−CD49flow luminal cells repopulated, whereas the percentage of Sca-1+CD49flow cells dropped back to 1.83% (Supplementary Fig. 1A).
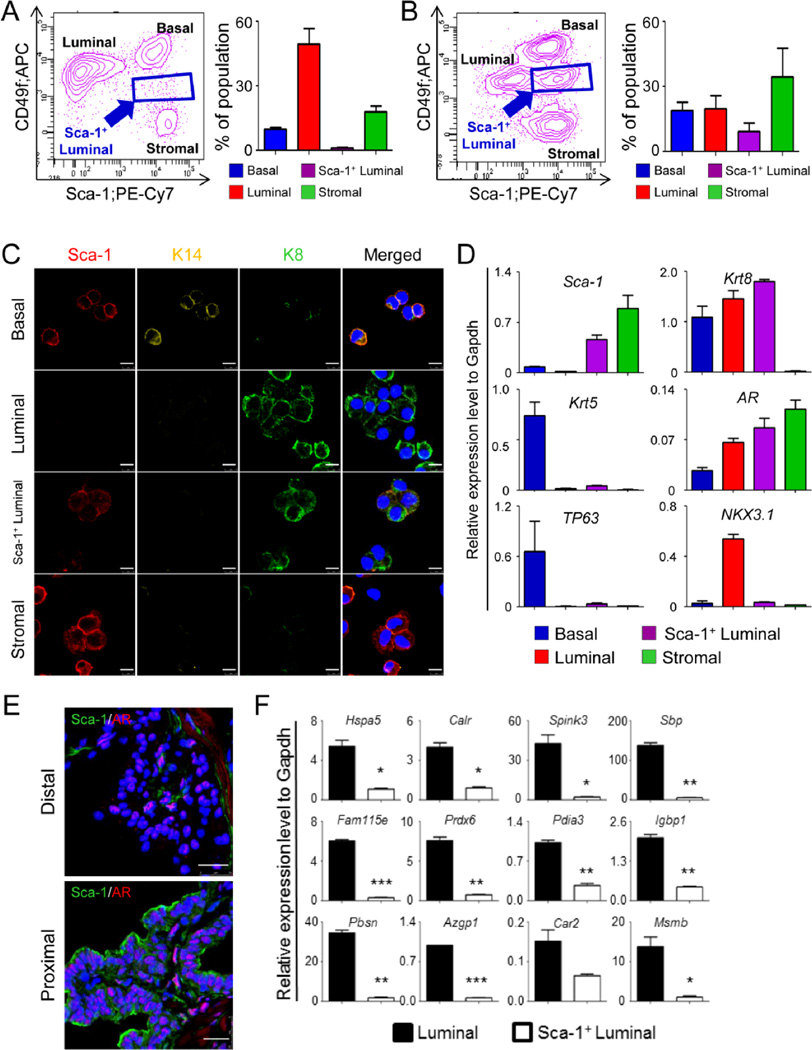
Figure. 1. Sca-1 defines a distinct population of prostate luminal cells.
A–B: FACS plots of prostate cell lineages in intact (A) and castrated (B) adult mice. Bar graphs show means ± s.d. of percentages of individual cell lineages from 3 independent experiments. C: Co-immunostaining of Sca-1, cytokeratin 14 (K14), and cytokeratin 8 (K8) on cytospins of individual FACS-sorted prostate lineages. Bars=10µm. D: qRT-PCR analysis of lineage marker expressions in individual FACS-sorted prostate cell lineages. Results show means ± s.d. from 3 independent experiments. E: Co-immunostaining of Sca-1 and androgen receptor (AR) in proximal and distal prostatic ducts. F: qRT-PCR analysis of expression of prostate secretory proteins in FACS-sorted Sca-1+ and Sca-1− luminal cells. Results show means ± s.d. from 3 independent experiments. *:p<0.05, **:p<0.01, ***:p<0.001.
To characterize the identity of this unique cell population, we prepared cytospun fractions from FACS-isolated cells and examined the expression of lineage markers by immunostaining. More than 70% of these cells display a luminal cell phenotype as they only express the luminal cell marker cytokeratin 8 (CK8), but not the basal cell marker cytokeratin 5 (CK5) or the stromal cell marker α smooth muscle actin (αSMA)(Supplementary Fig. 1B). We also confirmed the existence of the Sca-1+CK5− and Sca-1+CK8+ cells in the prostate tissues in vivo using co-immunostaining (Supplementary Fig. 1C–D).
We reasoned that the Sca-1+CD49flow luminal cells may represent luminal progenitors and sought to verify whether this cell population represents a real entity in intact mice. Fig. 1A shows that about 1.4% of the cells in 8–12 week old intact mice are Sca-1+CD49flow. This cell population is detectable in all three murine prostate lobes at similar frequencies, ranging from 0.9% in the dorsolateral lobes to 1.7% in the ventral lobes (Supplementary Fig. 1E). Immunostaining analysis of FACS-sorted cells confirmed that 66.8% of these cells express Sca-1 and CK8 but not the basal cell marker cytokeratin 14 (CK14) (Fig. 1C), whereas the rest of the cells represent contaminating stromal cells (23.6%), basal cells (5.0%) and Sca-1− luminal (4.6%) cells (Supplementary Fig. 1F). QRT-PCR analysis further corroborated that the Sca-1+CD49flow cells express Sca-1 and Krt8, but very low levels of basal cell markers, Krt5 and TP63 (Fig. 1D). More interestingly, unlike the Sca-1− luminal cells, Sca-1+CD49flow cells do not express Nkx3.1, a homeobox gene abundantly present in the prostate luminal cells [19]. Finally, although the Sca-1+CD49flow cells express the androgen receptor at a similar level as compared to the Sca-1− luminal cells (Fig. 1E), they do not express major androgen-regulated prostatic secretory proteins or express them at substantially lower levels (Fig. 1F). This suggests that these cells do not carry the secretory function. Collectively, these results imply that the Sca-1+CD49flow luminal cells represent a distinctive cell population in adult murine prostates.
Sca-1+ luminal cells are enriched in the proximal prostatic ducts
BrdU label-retention represents a property of replication-quiescent stem cells in some organs [20]. Tsujimura et al. showed previously that a small fraction of BrdU-label retaining cells are enriched in the proximal regions of prostatic ducts that are adjacent to the urethra [15]. These BrdU label-retaining cells include both basal cells and luminal cells and are suspected to be the putative stem cells in the prostate. We sought to investigate whether the Sca-1+CD49flow luminal cells are enriched in the proximal prostatic ducts. As shown in Fig. 2A, anterior prostates were dissected longitudinally into three equal portions, representing the distal, middle, and proximal prostatic ducts, and dissociated into single cells for FACS analysis. Consistent with a previous report [10], Lin−Sca-1+CD49high basal cells were enriched by 2-fold in the proximal prostatic ducts as compared to the distal ducts (Figs. 2B and 2C). Interestingly, the Sca-1+CD49flow cells constitute 7.6% of the cells in proximal ducts. In contrast, only 0.6 and 1.2% of the cells in the distal and middle ducts of the anterior prostate lobes are Sca-1+CD49flow cells, respectively. IHC analysis of Sca-1, CK5, and CK8 further confirmed that the Sca-1+CK5−CK8+ luminal cells are mostly localized in the ducts immediately adjacent to the urethra, which represent approximately 1/4–1/5 of total prostatic ducts (Figs. 2D and 2E). The Sca-1+ luminal cells were also detected in the proximal ducts of other prostatic lobes (Supplementary Fig. 2A). In addition, IHC analysis shows that Sca-1-expressing luminal cells do not express Nkx3.1 (Fig. 2F), corroborating the RNA analysis (Fig. 1D). Finally, neuroendocrine cells are not enriched in the Sca-1+CD49flow cell fraction as determined the expression of the neuroendocrine cell marker Synaptophysin using qRT-PCR (Supplementary Fig. 2B).
Figure. 2. Sca-1+ luminal cells reside in proximal prostatic ducts.
A: Schematic illustration of distal, middle, and proximal anterior prostatic ducts. B: FACS plots of prostate cell lineages in proximal, middle, and distal anterior prostatic ducts. C: Bar graphs show means ± s.d. of percentages of individual cell lineages from 3 independent FACS experiments. D–F: Co-immunostaining of Sca-1 and K5 (D), Sca-1 and K8 (E), Sca-1 and Nkx3.1 (F) in anterior prostate lobes. Yellow bars=100µm. White bars=25µm.
Sca-1 sub-fractionates the CD24highCD49flow luminal cells
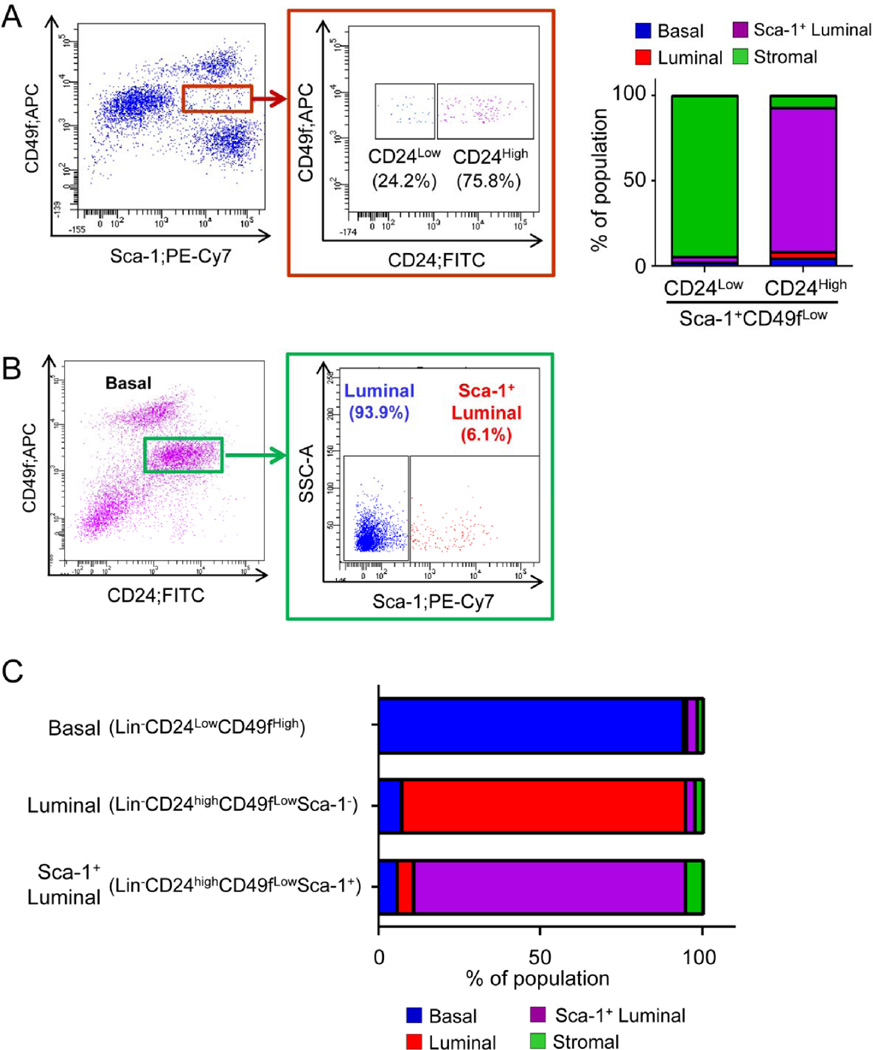
Only about 66.8% of FACS-sorted Lin−Sca-1+CD49flow cells are phenotypically CK5−CK8+ luminal cells, whereas the remaining cells are mainly contaminating Sca-1+ stromal cells (Supplementary Fig. 1F). We sought to take advantage of additional surface antigens to further purify the Sca-1+ luminal cells. Previously, prostate luminal cells have been shown to express high levels of CD24 (Lin−CD24highCD49flow)[10]. We reasoned that inclusion of CD24 would reduce the contamination of stromal cells in FACS-sorted Sca-1+CD49flow luminal cells. Fig. 3A shows that 75.8% of Sca-1+ luminal cells are CD24+. Immunostaining of cytospun FACS-sorted cells reveals that more than 94.5% of the Lin−CD24− CD49flowSca-1+ cells are contaminating stromal cells that express αSMA, whereas 84.5% of the Lin−CD24highCD49flowSca-1+ cells are phenotypically luminal cells (CK8+CK5−). This validates CD24 as an efficient surface marker to further enrich the Sca-1+ luminal cells. Fig. 3B shows that about 6.1% of Lin−CD24highCD49flow luminal cells are Sca-1+. Interestingly, Sca-1+ luminal cells appear to express CD24 at an even higher level than the Sca-1− luminal cells (Supplementary Fig. 3). Cytospin analysis further corroborated efficient separation of different epithelial cell lineages by this combination of surface markers (94.0%, 87.5% and 83.9% for Lin−CD24lowCD49fhigh basal cells, Lin−CD24highCD49flowSca-1− luminal cells, and Lin−CD24highCD49flowSca-1+ luminal cells, respectively) (Fig. 3C). Therefore, we used FACS-separated individual prostate epithelial cell lineages based on this combination of surface antigens throughout the following functional studies.
Figure. 3. Sca-1 fractionates CD24lowCD49fhigh luminal cells.
A: FACS plot shows that CD24 separates stromal cell contamination from Sca-1-expressing luminal cells. Bar graph shows the identity of FACS-sorted cells based on immunostaining of cytospins. B: FACS plot shows that Sca-1 fractionates CD24lowCD49fhigh luminal cells. C: Bar graph representing the purity of individual FACS-sorted prostate lineages quantified based on immunostaining of cytospins.
Different prostate epithelial lineages generate distinct forms of organoids in vitro
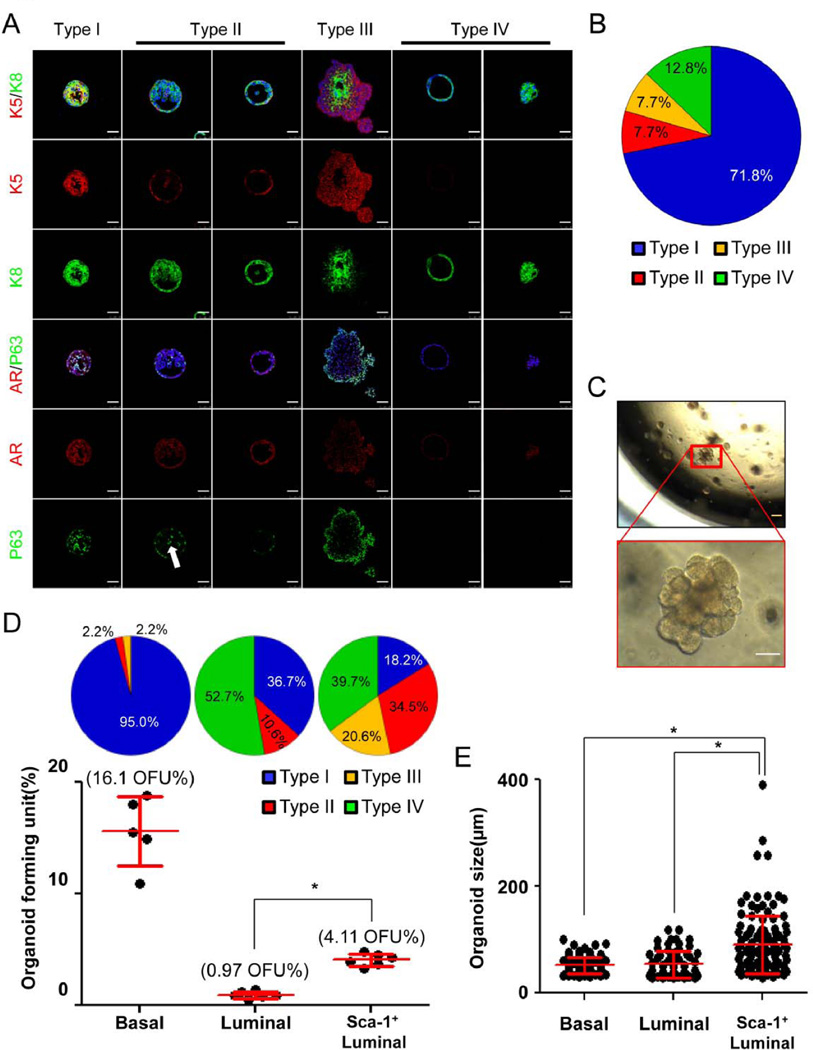
We cultured dissociated prostate cells from 8–12 week-old mice using the organoid assay established by Karthaus et al [18]. Approximately 1.8% of dissociated prostate cells are capable of generating organoids with diameters ranging from 30–200 µm after a 10-day culture. We performed organoid assays using a mixture of unlabeled prostate epithelial cells from C57Bl/6 mice and an mTmG transgenic mouse line that expresses red fluorescent protein in the whole body. There were no chimeric organoids formed in the assay, suggesting that the organoids are clonally derived (Supplementary Fig. 4A–B). Immunostaining of lineage markers revealed the existence of four major types of organoids (Figs. 4A–B and Supplementary Fig. 4C). Type I organoids constitute approximately 71.8% of total organoids. Cells in type I organoids not only express CK5 and P63 but also CK8 and AR. Type II organoids (7.7% of total organoids) consist of lumens surrounded by one or multiple layers of CK8-expressing luminal cells that are further encapsulated by a layer of CK5-expressing basal cells. In type II organoids with multiple layers of luminal cells, it is common to observe a few basal cells distributed sporadically within the luminal cells (white arrow, Fig. 4A). Type III organoids (7.7% of total organoids) are similar to type II organoids. However, there are multiple outer layers of CK5+CK8− cells in type III organoids. Cells in type III organoids express AR at a lower level as compared to cells in other types of organoids (Fig. 4A). In addition, a unique feature of type III organoids is that they usually start to display a budding phenotype at approximately 7 days in culture (Fig. 4C). Most budding structures do not canalize and appear to be solid. This budding capability is cell-intrinsic, as other types of organoids do not bud even upon prolonged culture. As a result of this budding morphology, type III organoids appear the largest in size in the culture (Fig. 4C). The remaining 12.8% of organoids are type IV organoids that consist of cells that only express CK8 and AR, but not the basal cell markers CK5 and P63. These organoids are either solid or contain an empty lumen surrounded by a single layer of cells. Exclusion of either R-spondin1 or Noggin significantly reduces organoid-forming activity (Supplementary Fig. 4D). In contrast, organoid formation is not dependent on androgen signaling. Although exclusion of di-hydro-testosterone from the organoid culture eliminates nuclear AR staining, it does not alter organoid size, morphology, or the number of organoid-forming unit (OFU) (Supplementary Fig. 5A–D). Enzalutamide treatment does not affect organoid-forming activity, either (Supplementary Fig. 5E)
Figure. 4. Different lineages of prostate epithelial cells generate distinct organoids in vitro.
A: IHC analysis of lineage marker expression in the 4 types of organoids. White arrow points to P63-expressing basal cells inside an organoid. Bars=50µm. B: Pie chart quantifies composition of organoids derived from wild type prostate epithelial cells. C: Transilluminating images of prostate organoids. Enlarged inset shows a type III budding organoid. Yellow bar=100µm; white bar=50µm. D: Individual prostate epithelial lineages form different types of organoids at distinct efficiencies. Dot plot shows means ± s.d. of organoid-forming units of different lineages from 3 independent experiments. Pie charts quantify the types of organoids in different cultures. *:p<0.001. E: Dot plot shows means ± s.d. of organoid size in different cultures. *:p<0.001.
We reasoned that these different types of organoids have distinct cellular origins. To test this hypothesis, we cultured FACS-isolated Lin−CD24lowCD49fhigh basal cells, Lin−CD24highCD49flowSca-1− (Sca-1−) luminal cells, and Lin−CD24highCD49flowSca-1+ (Sca-1+) luminal cells separately. Basal cells form organoids at a much higher frequency (OFU = 16.1%) than both types of luminal cells (dot plot, Fig. 4D). The OFU of the Sca-1+ luminal cells (4.11%) is 4.23-fold more than that of the Sca-1− luminal cells (0.97%). Basal cells form mostly type I organoids. Both Sca-1− and Sca-1+ luminal cells are able to generate type II and type IV organoids. However, type III organoids are mainly observed in the Sca-1+ luminal cell culture (Pie charts, Fig. 4D). Because the bigger type III organoids were mainly produced by the Sca-1+ luminal cells, the size of organoids in the Sca-1+ organoid culture was more heterogeneous than the other two groups (Fig. 4E).
The observation that type I organoids also formed in the cultures of the Sca-1+ and Sca-1− luminal cells probably reflects an imperfect FACS separation of individual cell lineages. For example, the organoid forming unit in the Sca-1+ luminal cell culture was about 4.11%, among which 18.2% were type I organoids (Fig. 4D). Fig. 3C shows that there were about 5% contaminating basal cells in the Sca-1+ luminal cells. As the organoid forming activity of basal cells is about 16.1% (Fig. 4D), the amount of the type I organoids generated by the contaminating basal cells (5% × 16.1%) is roughly the same as that of the type I organoids in the Sca-1+ luminal cell culture (4.11% × 18.2%). This indirectly supports that the type I organoids in the Sca-1+ luminal cell culture were derived from contaminating basal cells, while the other types of organoids were generated by the Sca-1+ luminal cells. Of course, we cannot exclude the possibility that the Sca-1+ luminal cells are also capable of generating type I organoids in this assay. But overall, we can conclude that the Sca-1+ luminal cells represent a functionally distinct cellular entity.
Distinct types of organoids retain their morphological and histological features upon passaging
To determine whether prostate organoid-forming cells possess the capacity for self-renewal, we dissociated prostate organoids derived from total prostate cells into single cells and replated them in the organoid assay. Fig. 5A shows that 3.3%–4.3% of dissociated organoid cells were capable of forming organoids upon serial passage. Interestingly, the type II organoids expanded during serial passaging (Pie charts, Fig. 5A). This result suggests that the type II organoids replate more efficiently than the other types of organoids in this culture condition. Or alternatively, cells in other types of organoids can generate type II organoids upon passaging. To distinguish these possibilities, we attempted to passage individual types of organoids. Relatively pure type I organoids (>95%) were obtained from organoid cultures of FACS-isolated basal cells. Type III organoids were picked by pipette under a microscope based on their distinctive budding morphology. As we were unable to isolate pure type II and IV organoids, we did not passage them individually. Fig. 5B shows that both the type I and the type III organoids were able to be replated, but type I organoids replated more efficiently than type III organoids. Interestingly, cells from these two types of organoids only generated the same types of organoids as their parental organoids. From these analyses, we infer that type II and IV organoids are also capable of replating, although we are unable to conclude whether these two types of organoids can generate each other upon passaging. In summary, these studies demonstrate that prostate organoids retain their morphological and histological features upon passaging, which implies that the capabilities to form distinct prostate organoids reflect the differences in intrinsic biological properties among different prostate epithelial lineages. In addition, this study also shows that the type II organoids replate more efficiently under the condition of the prostate organoid assay.
Figure. 5. Distinct types of organoids retain their histological and morphological features upon passaging.
A: Prostate organoid cells can be serially passaged as dissociated cells. Dot plot shows means ± s.d. of organoid-forming units during serial passaging. Pie charts show composition of organoid types. B: Type I and type III organoids can be passaged and only generate organoids of parental types. Dot plot shows means ± s.d. of OFUs from 3 independent experiments.
Sca-1+ luminal epithelial cells are capable of regenerating prostate glands in vivo
We further investigated whether the Sca-1+ luminal cells possess the bipotent potential in vivo using a prostate regeneration assay. In this assay, dissociated adult murine or human prostate epithelial cells were mixed with embryonically-derived urogenital sinus mesenchymal cells and incubated in immunodeficient male host mice [21]. Epithelial cells with stem or progenitor activities are capable of forming glandular structures that contain both basal and luminal epithelial cells. It has been shown previously that both the basal epithelial stem cells in intact mice and the castration-resistant Nkx3.1-expressing cells (CARN) identified in castrated mice are capable of regenerating prostate tissues (8–10, 12, 13, 22, 23). Briefly, 1.5×104 FACS-isolated Sca-1+ luminal cells, Sca-1− luminal cells, and basal cells were mixed with 1×105 UGSM cells separately and transplanted in immunodeficient mice subcutaneously. UGSM cells alone were also transplanted as a control. Consistent with previous reports, almost no discernable glandular structures formed in the UGSM cell alone group or the Sca-1− luminal cell group. In contrast, 0.057% of basal cells and 0.043% of Sca-1+ luminal cells were capable of generating glandular structures (Fig. 6A). The regenerated glands in these two groups are histologically similar. They all contain lumens surrounded by a layer of CK8+ luminal cells that is encapsulated by a discontinuous layer of CK5+ basal cells (Fig. 6B). Interestingly, the average diameter of the regenerated glands from the Sca-1+ luminal cells is 38% smaller than that of the basal cell group (Fig. 6C). Although there were approximately 5% contaminating basal cells in the FACS-sorted Sca-1+ luminal cells, it is unlikely that all the regenerated glands in the Sca-1+ luminal cell group were derived from the contaminating basal cells, because only 0.057% basal cells are able to regenerate prostate glands. In addition, there were even more contaminating basal cells in the Sca-1− luminal cells (Fig. 3C), but very few glands were regenerated in this group (Fig. 6A). Therefore, our result suggests that the Sca-1+ luminal cells also possess the bipotent differentiation potential in vivo. However, it should be noted that this capacity for bipotent differentiation may represent a facultative function because previous lineage tracing studies showed that the basal and luminal cell lineages are independently sustained in vivo in adult mice [4–7]. But this facultative bipotent potential implies that this cell population lies at a different stage of differentiation from the rest of the luminal epithelial cells.
Figure. 6. The Sca-1+ luminal cells possess in vivo bipotent differentiation capacity in the prostate regeneration assay.
A: Bar graph shows means ± s.e.m. of gland-forming capacity of FACS-sorted basal cells, Sca-1− luminal cells, and Sca-1+ luminal cells. UGSM cells serve as negative control. B: H&E staining and IHC analysis of K5 and K8 in regenerated glands. C: Dot plot shows means ± s.e.m. of diameters of regenerated glands. Bars=50µm. ***:p<0.001.
Discussion
Are Sca-1+ luminal cells the progenitor cells in the luminal cell lineage?
In this study, we establish the Sca-1+ luminal cells as a cellular entity that is functionally distinct from the rest of the luminal epithelial cells. Multiple lines of evidence support that some of these cells may represent a progenitor population in the luminal cell lineage. They are enriched in the proximal prostatic ducts where the BrdU label-retaining cells are located [15]; they survive androgen ablation and are enriched in the prostates of castrated mice; they do not carry the secretory function, a feature of terminally differentiated luminal epithelial cells; a fraction of the Sca-1+ luminal cells display bipotent potential in both the in vitro organoid assay and the in vivo prostate regeneration assay. It should be noted that the Sca-1+ luminal cells do not express Nkx3.1, hence are distinct from the previously reported castration resistant Nkx3.1-expressing (CARN) cells [23].
However, it still remains a question whether the Sca-1+ luminal cells represent a bona fide population of stem cells or progenitors. By definition, stem cells should possess the capacity to generate all of the different cell lineages in the organs from which they were derived [24]. To this end, there is no compelling evidence that phenotypically luminal epithelial cells in intact adult rodents are able to generate all of the different cell lineages in the prostate in vivo under physiological conditions. Therefore, although in the organoid and the prostate regeneration assays Sca-1+ cells are able to generate basal and luminal cells, this capacity most likely reflects a facultative function of these cells under those experimental conditions. These cells are more likely committed luminal progenitors. It is interesting that Sca-1 is also expressed in a fraction of luminal epithelial cells in the mammary gland, and Sca-1+ mammary luminal epithelial cells have been considered as ductal progenitors in the mammary gland [25–27]. Given the many similarities between the prostate and the mammary glands in terms of their lineage hierarchy [28], it is tempting to hypothesize that Sca-1+ luminal cells may serve as proximal prostatic ductal progenitors. To substantiate this hypothesis, future in vivo lineage tracing studies should be performed to investigate how Sca-1+ luminal cells contribute to the maintenance of the luminal cell lineage in adult rodents. In addition, since there is no human paralogue of murine Sca-1, a detailed gene expression profile analysis of the murine Sca-1+ luminal cells should help identify useful markers to isolate the functionally equivalent cell population in human prostate tissues.
Developmentally, how do the Sca-1+ luminal cells originate? They are unlikely derived directly from basal epithelial cells in adult rodents, as previous lineage tracing studies showed that adult murine prostate basal cells rarely generate luminal cells. An interesting observation is that all epithelial cells in the embryonic urogenital sinus express Sca-1 (unpublished observation). Therefore, it is more likely that these cells and basal cells originate from the same embryonic prostate epithelial stem cells. The commitment, survival, and expansion of these cells may be affected by altered prostatic microenvironmental signaling. For example, it was shown that activation of prolactin signaling in the prostate results in an expansion of the Sca-1+ luminal cells [29]. Understanding the molecular mechanisms that regulate the homeostasis of this cell population may help dissect how prostate cancer is initiated.
Do Sca-1+ cells serve as a cell-of-origin for prostate cancer?
Several studies, including ours, showed previously that when the tumor suppressor Pten was deleted in the Keratin-8 expressing luminal epithelial cells, mice developed prostate adenocarcinoma (5–7, 30). These studies demonstrate that luminal epithelial cells can serve as a cell-of-origin for prostate cancer. Our current study raises the question of whether the Sca-1+ luminal cells serve as a preferred cell-of-origin for prostate cancer as compared to the Sca-1− luminal cells. Several lines of evidence support that the Sca-1+ luminal cells may act as favored cells of origin for prostate cancer. First, Zhou et al showed that in a mouse model for prostate cancer with P53 and Rb deletion, tumors developed preferentially from the proximal regions in the prostate [31]. Second, prostate cancer often recurs and becomes castration resistant after treatments. As the Sca-1+ luminal cells survive androgen ablation, they may act as a cell-of-origin for prostate cancer and serve as the root of castration-resistant tumor cells. Finally, prostate tumor cells in mouse models for prostate cancer often express Sca-1 unanimously [9] and human prostate cancer cells also display reduced secretory function as compared to normal epithelial cells. These phenotypical changes suggest an expansion of murine Sca-1+ cells or their human counterparts during tumor initiation and progression, but of course, could also be a consequence of altered Sca-1 expression or dedifferentiation of mature luminal cells during carcinogenesis.
Microenvironmental cues at the proximal prostatic ducts
The Sca-1+ luminal cells are enriched in the proximal prostatic ducts, implying that the micro-environmental cues at the proximal prostatic ducts may shape some of their molecular features. Although the Sca-1+ luminal cells express the androgen receptor, they do not carry the secretory function and do not express major AR-regulated proteins in the prostate. This feature is similar to that of the luminal epithelial cells in a mouse model with prostate-specific activation of Notch signaling [16], suggesting that the Notch pathway may regulate the biology of these cells. We showed previously that the Notch ligand Jagged1 is mainly expressed by prostate basal cells [32]. Jagged1 expressed by the basal cells can activate Notch signaling in the luminal cells via the Notch receptors expressed by the luminal cells. Salm et al showed that stromal cells in the proximal region express higher levels of TGFβ [33], and we showed that TGFβ can upregulate Jagged1 in prostate basal cells [34]. Therefore, basal cells in the proximal prostatic ducts are not only high in density but also may express Jagged1 at a higher level than those at distal regions; hence, the Notch activity in the luminal cells at the proximal region may be higher than those in other prostatic ductal regions. Notch downstream target genes, such as Hey1 and HeyL, have been shown to suppress AR transcriptional activity [35, 36]. However, our previous study showed that Notch loss of function does not affect the regenerative capacity of the prostate tissues [32]; therefore, it is unlikely that Notch signaling is essential for the progenitor activity of Sca-1+ luminal cells, or alternatively, the Sca-1+ luminal cells only serve as the proximal prostatic ductal progenitors and are not the only essential cell population for the maintenance of the luminal cell lineage. Gene expression profiling of the Sca-1+ luminal cells and in vivo lineage tracing studies in the future will address these issues definitively.
Materials and Methods
Mice
C57BL/6 mice were purchased from Charles River (Wilmington, MA). The mTmG fluorescence reporter mouse line was generated by the group of Dr. Liqun Luo at Stanford University [37]. Experimental mice were castrated at the age of 8 weeks using standard techniques as described previously [7]. Testosterone pellets (10mg/pellet) were inserted into castrated mice subcutaneously to restore serum testosterone levels. All animal work were approved by and performed under the regulation of the Institutional Animal Care Use Committee for the Baylor College of Medicine.
Dissociation of primary prostates and flow cytometry
Prostate tissues were digested in DMEM/F12/Collagenase/Hyaluronidase/FBS (Invitrogen, Carlsbad, CA) for 3 hours at 37°C, followed by an additional 1 hour of digestion in 0.25% Trypsin-EDTA (Invitrogen, Carlsbad, CA) on ice. Subsequently, digested cells were suspended in Dispase (Invitrogen, Carlsbad, CA, 5 mg/mL) and DNase I (Roche Applied Science, Indianapolis, IN, 1 mg/mL), and pipetted vigorously to dissociate cell clumps. Dissociated cells were then passed through 70 µm cell strainers (BD Biosciences, San Jose, CA) to get single cells. For FACS analysis and sorting, singled murine prostate cells were stained with Pacific blue-conjugated anti CD31, CD45, and Ter119 antibodies (eBioscience, San Diego, CA), PE-Cy7-conjugated anti Sca-1 antibody (eBioscience, San Diego, CA), FITC-conjugated CD24 antibody (BD Biosciences, San Jose, CA), and Alexa 647-conjugated anti CD49f antibody (Biolegend, San Diego, CA). Cell sorting and analyses were performed using Aria II and LSR II, respectively (BD Biosciences, San Jose, CA).
Immunocytochemistry and immunostaining
To prepare cytospin from FACS-sorted cells, cells were loaded into cytospin slide chambers of Shannon Cytospin 4 (Thermal Scientific, Odessa, TX) and centrifuged at a speed of 800rpm at room temperature for 5 minutes. H&E and immunofluorescence staining were performed using standard protocols on 5-µm paraffin sections or cytospin slides. Primary antibodies and the dilutions used are listed below. Slides were incubated with 5% normal goat serum (Vector Labs, Burlingame, CA) and with primary antibodies diluted in 2.5% normal goat serum overnight at 4°C. Primary antibodies used in this study were mouse anti-K14 (LL002, Biogenex, San Ramon, CA), rabbit anti-K5 (#PRB-160P, Covance, Berkeley, CA), mouse anti-K8 (#MMS-162P, Covance), rat anti-K8 (Troma-1, DSHB, Iowa City, IA), mouse anti-P63 (4A4, Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-AR (SC-816, Santa Cruz Biotechnology), mouse anti-smooth muscle actin(Sigma-Aldrich, St. Louis, MO), rabbit anti-Synaptophysin (Invitrogen, Carlsbad, CA), rabbit anti-vimentin (#5741, Cell Signaling Technology, Beverly, MA), rabbit anti-Ki67 (NCL-Ki67-P, Novocastra, Newcastle, UK), Rat anti-Sca-1 (#557403, BD PharMingen, San diego, CA), Rabbit anti-NKX3.1 (#0315, Athena Enzyme Systems, Baltimore, MD), and mouse anti-BrdU (DSHB, Iowa City, IA, USA). Slides then were incubated with secondary antibodies (diluted 1:500 in PBST) labeled with Alexa Fluor 488, 594, or 633 (Invitrogen/Molecular Probes, Eugene, OR). Sections were counterstained with 4,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, St. Louis, MO). Immunofluorescence staining was imaged using an Olympus BX60 fluorescence microscope (Olympus Optical Co Ltd, Tokyo, Japan) or a Leica EL6000 confocal microscope (Leica Microsystems, Wetzlar, Germany).
RNA isolation and qRT–PCR
Total RNA was isolated using the mRNA Kit (Qiagen, Gaithersburg, MD). Reverse transcription was performed using the superscript Kit (BioRad, Hercules, CA). qRT-PCR was performed using the SYBR GreenER qPCR mix (BioRad, Hercules, CA) on a StepOne plus Real-Time PCR system (Applied Biosystems, Grand Island, NY). The relative amount of specific mRNA was normalized to Gapdh. Primers for qRT–PCR are listed in Supplementary Table1.
Prostate organoid culture and prostate regeneration assay
The organoid culture was performed following the previous study [18]. Briefly, dissociated prostate cells from 8–12 week-old C57Bl/6 mice were cultured in DMEM/F12 supplemented with B27 (Life technologies, Grand Island, NY), 10 mM HEPES, Glutamax (Life technologies, Grand Island, NY), Penicillin/Streptomycin, and the following growth factors: EGF 50 ng/ml (Peprotech, Rocky Hill, NJ), 500 ng/ml recombinant R-spondin1 (Peprotech, Rocky Hill, NJ), 100 ng/ml recombinant Noggin (Peprotech, Rocky Hill, NJ), 200 µM TGF-β/Alk inhibitor A83-01 (Tocris, Ellisville, MO), and 10 µM Y-27632 (Tocris, Ellisville, MO). Dihydrotestosterone (Sigma, St. Louis, MO) was added at 1 nM final concentration. Cells were mixed with growth factor reduced matrigel (Corning, Corning, NY) by 1:1 ratio and plated in 96-well plates. In some experiments, enzalutamide (Selleckchem, Houston, TX) was added to a final concentration of 10 µM.
To collect prostate organoids, culture media was removed and 100 µl of 1 mg/ml Dispase solution (Invitrogen, Carlsbad, CA) was added and incubated for 1 hour at 37°C. Samples were then transferred to 1.6 ml Eppendorf tubes and centrifuged at 850×g for 2 minutes. Organoids were fixed with 10% formalin for 10 minutes and resuspended in 100 µl of HistoGel (Richard-Allan Scientific, Kalamazoo, MI) for preparation of paraffin embedded blocks.
To dissociate prostate organoids, organoids released from matrigel were resuspended in 300 µl of chilled Trypsin-Versene (Lonza, Walkersville, MD) and were gently passed through 28-gauge insulin syringes approximately 10 times for 5 minutes. It usually took an extra two minutes to dissociate Type III organoids due to their bigger sizes. Subsequently, 200 µl of organoid culture media was added and centrifuged at 850×g for 2 minutes. Dissociated cells were resuspended in culture media. Viable single cells were enumerated under a microscope and plated in 96-well plates with growth factor reduced matrigel (Corning, Corning, NY).
Prostate regeneration assays were performed as described previously [21]. Briefly, 1.5×104 FACS-sorted specific prostate epithelial cell fractions were mixed with 1×105 urogenital sinus mesenchymal cells, mixed with growth factor reduced matrigel (Corning, Corning, NY) (1:1 v/v), and transplanted subcutaneously in immunodeficient male NOD/SCID mice for 6 weeks. Regenerated tissues were collected, fixed in 10% buffered formalin, and embedded in paraffin for IHC analysis. Because the regenerated tissues were small and flat, we cut and collected the sections that have the biggest tissue area for quantification of regenerated glands. Glands from three sequential sections for each sample were enumerated and mean values were obtained. Therefore, the gland-forming unit is underestimated, especially for that of the Sca-1+ luminal cells because the regenerated glands from them were smaller. But this does not affect the conclusion of our study.
Statistics
All experiments were performed at least 3 times using independent experimental materials (cells or mice). Data are presented as mean ± SD. Student’s t test was used to determine significance between groups. For all statistical tests, the 0.05 level of confidence was accepted for statistical significance.
Supplementary Material
Significance Statement.
The prostate epithelial lineage hierarchy remains inadequately defined. We discovered a distinct population of luminal epithelial cells that express the Stem Cell Antigen-1 (Sca-1). These cells are enriched in specific anatomic niches in adult murine prostates. They can survive androgen deprivation and possess bipotent differentiation capacity. These cells may represent the luminal progenitors and serve as the cells-of-origin for castration resistant prostate cancer.
Acknowledgments
We thank Dr. Jeffrey Rosen for critical comments, the technical support by the Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from the NIH (P30 AI036211, P30 CA125123, and S10 RR024574) and the expert assistance of Joel M. Sederstrom. This work is supported by NIH R01 DK092202 (L.X.), NIH R01 CA190378 (L. X.), and NIH P30 CA125123 (the Cancer Center Shared Resources Grant).
Source of support: NIDDK, NCI
REFERENCES
- 1.Abate-Shen C, Shen MM. Molecular genetics of prostate cancer. Genes & development. 2000;14(19):2410–2434. doi: 10.1101/gad.819500. [DOI] [PubMed] [Google Scholar]
- 2.El-Alfy M, Pelletier G, Hermo LS, Labrie F. Unique features of the basal cells of human prostate epithelium. Microscopy research and technique. 2000;51(5):436–446. doi: 10.1002/1097-0029(20001201)51:5<436::AID-JEMT6>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 3.Vashchenko N, Abrahamsson PA. Neuroendocrine differentiation in prostate cancer: implications for new treatment modalities. European urology. 2005;47(2):147–155. doi: 10.1016/j.eururo.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Qin J, et al. The PSA(−/lo) prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell stem cell. 2012;10(5):556–569. doi: 10.1016/j.stem.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang ZA, et al. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nature cell biology. 2013;15(3):274–283. doi: 10.1038/ncb2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu TL, et al. Conditionally ablated Pten in prostate basal cells promotes basal-to-luminal differentiation and causes invasive prostate cancer in mice. Am J Pathol. 2013;182(3):975–991. doi: 10.1016/j.ajpath.2012.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Choi N, Zhang B, Zhang L, Ittmann M, Xin L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell. 2012;21(2):253–265. doi: 10.1016/j.ccr.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burger PE, et al. Sca-1 expression identifies stem cells in the proximal region of prostatic ducts with high capacity to reconstitute prostatic tissue. Proc Natl Acad Sci U S A. 2005;102(20):7180–7185. doi: 10.1073/pnas.0502761102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xin L, Lawson DA, Witte ON. The Sca-1 cell surface marker enriches for a prostate-regenerating cell subpopulation that can initiate prostate tumorigenesis. Proc Natl Acad Sci U S A. 2005;102(19):6942–6947. doi: 10.1073/pnas.0502320102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawson DA, Xin L, Lukacs RU, Cheng D, Witte ON. Isolation and functional characterization of murine prostate stem cells. Proc Natl Acad Sci U S A. 2007;104(1):181–186. doi: 10.1073/pnas.0609684104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xin L, Lukacs RU, Lawson DA, Cheng D, Witte ON. Self-renewal and multilineage differentiation in vitro from murine prostate stem cells. Stem Cells. 2007;25(11):2760–2769. doi: 10.1634/stemcells.2007-0355. [DOI] [PubMed] [Google Scholar]
- 12.Goldstein AS, et al. Trop2 identifies a subpopulation of murine and human prostate basal cells with stem cell characteristics. Proc Natl Acad Sci U S A. 2008;105(52):20882–20887. doi: 10.1073/pnas.0811411106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leong KG, Wang BE, Johnson L, Gao WQ. Generation of a prostate from a single adult stem cell. Nature. 2008;456(7223):804– 808. doi: 10.1038/nature07427. [DOI] [PubMed] [Google Scholar]
- 14.Kwon OJ, Zhang L, Ittmann MM, Xin L. Prostatic inflammation enhances basal-to-luminal differentiation and accelerates initiation of prostate cancer with a basal cell origin. Proc Natl Acad Sci U S A. 2013 doi: 10.1073/pnas.1318157111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsujimura A, et al. Proximal location of mouse prostate epithelial stem cells: a model of prostatic homeostasis. J Cell Biol. 2002;157(7):1257–1265. doi: 10.1083/jcb.200202067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwon OJ, et al. Increased Notch signalling inhibits anoikis and stimulates proliferation of prostate luminal epithelial cells. Nature communications. 2014;5:4416. doi: 10.1038/ncomms5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chua CW, et al. Single luminal epithelial progenitors can generate prostate organoids in culture. Nature cell biology. 2014;16(10):951–961. 951–954. doi: 10.1038/ncb3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karthaus WR, et al. Identification of Multipotent Luminal Progenitor Cells in Human Prostate Organoid Cultures. Cell. 2014 doi: 10.1016/j.cell.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhatia-Gaur R, et al. Roles for Nkx3.1 in prostate development and cancer. Genes & development. 1999;13(8):966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cotsarelis G, Sun TT, Lavker RM. Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell. 1990;61(7):1329–1337. doi: 10.1016/0092-8674(90)90696-c. [DOI] [PubMed] [Google Scholar]
- 21.Xin L, Ide H, Kim Y, Dubey P, Witte ON. In vivo regeneration of murine prostate from dissociated cell populations of postnatal epithelia and urogenital sinus mesenchyme. Proc Natl Acad Sci U S A. 2003;100(Suppl 1):11896–11903. doi: 10.1073/pnas.1734139100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, et al. ROCK inhibitor Y-27632 suppresses dissociation-induced apoptosis of murine prostate stem/progenitor cells and increases their cloning efficiency. PLoS One. 2011;6(3):e18271. doi: 10.1371/journal.pone.0018271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461(7263):495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weissman IL, Anderson DJ, Gage F. Stem and progenitor cells: origins, phenotypes, lineage commitments, and transdifferentiations. Annual review of cell and developmental biology. 2001;17:387–403. doi: 10.1146/annurev.cellbio.17.1.387. [DOI] [PubMed] [Google Scholar]
- 25.Welm BE, et al. Sca-1(pos) cells in the mouse mammary gland represent an enriched progenitor cell population. Developmental biology. 2002;245(1):42–56. doi: 10.1006/dbio.2002.0625. [DOI] [PubMed] [Google Scholar]
- 26.Shehata M, et al. Phenotypic and functional characterisation of the luminal cell hierarchy of the mammary gland. Breast cancer research : BCR. 2012;14(5):R134. doi: 10.1186/bcr3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes & development. 2014;28(11):1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Risbridger GP, Davis ID, Birrell SN, Tilley WD. Breast and prostate cancer: more similar than different. Nature reviews. Cancer. 2010;10(3):205–212. doi: 10.1038/nrc2795. [DOI] [PubMed] [Google Scholar]
- 29.Sackmann-Sala L, et al. Prolactin-induced prostate tumorigenesis links sustained Stat5 signaling with the amplification of basal/stem cells and emergence of putative luminal progenitors. Am J Pathol. 2014;184(11):3105–3119. doi: 10.1016/j.ajpath.2014.07.020. [DOI] [PubMed] [Google Scholar]
- 30.Xin L. Cells of origin for cancer: an updated view from prostate cancer. Oncogene. 2013;32(32):3655–3663. doi: 10.1038/onc.2012.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Z, Flesken-Nikitin A, Nikitin AY. Prostate cancer associated with p53 and Rb deficiency arises from the stem/progenitor cell-enriched proximal region of prostatic ducts. Cancer Res. 2007;67(12):5683–5690. doi: 10.1158/0008-5472.CAN-07-0768. [DOI] [PubMed] [Google Scholar]
- 32.Valdez JM, et al. Notch and TGFbeta form a reciprocal positive regulatory loop that suppresses murine prostate basal stem/progenitor cell activity. Cell stem cell. 2012;11(5):676–688. doi: 10.1016/j.stem.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salm SN, et al. TGF-{beta} maintains dormancy of prostatic stem cells in the proximal region of ducts. J Cell Biol. 2005;170(1):81–90. doi: 10.1083/jcb.200412015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valdez JM, Xin L. The dual nature of Notch in tissue homeostasis and carcinogenesis. Cell Cycle. 2013;12(4):541. doi: 10.4161/cc.23671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lavery DN, et al. Repression of androgen receptor activity by HEYL, a third member of the Hairy/Enhancer-of-split-related family of Notch effectors. The Journal of biological chemistry. 2011;286(20):17796–17808. doi: 10.1074/jbc.M110.198655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Belandia B, et al. Hey1, a mediator of notch signaling, is an androgen receptor corepressor. Molecular and cellular biology. 2005;25(4):1425–1436. doi: 10.1128/MCB.25.4.1425-1436.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.