Abstract
There is a fine balance in the mutual relationship between the intestinal microbiota and its mammalian host. It is thought that disruptions in this fine balance contribute/account for the pathogenesis of many diseases. Recently, the significance of the relationship between gut microbiota and its mammalian host in the pathogenesis of obesity and the metabolic syndrome has been demonstrated. Emerging data has linked intestinal dysbiosis to several gastrointestinal diseases including inflammatory bowel disease, irritable bowel syndrome, nonalcoholic fatty liver disease, and gastrointestinal malignancy. This article is intended to review the role of gut microbiota maintenance/alterations of gut microbiota as a significant factor as a significant factor discriminating between health and common diseases. Based on current available data, the role of microbial manipulation in disease management remains to be further defined and a focus for further clinical investigation.
INTRODUCTION
The gastrointestinal tract is thought to house ~1014 microorganisms, which in its totality is comprised of ~1,000 distinct bacterial species.1 Recent advances in sequencing technologies have given researchers further insight into the symbiotic relationship between the intestinal microbiome and its mammalian host.2 The “normal” gut flora encompasses a wide variety of microbacteria that have a vital role in digestion, fermenting unused energy substrates, maintaining the immune system, and in the synthesis of vitamins and enzymes (such as vitamin K and biotin).3 The metabolic activity performed by the gut microflora equates to that of a virtual organ, which is why many refer to the gut microflora as the “forgotten organ”. Factors influencing this relationship such as the environment,4 diet,5, 6 and genetics7 alter its metabolic capabilities resulting in the pathogenesis of a variety of disease states. Here, we review the literature in microbiome studies, the emerging links between the microbiome and its effect on gastrointestinal health, and approaches to manipulate this symbiotic relationship to potentially improve overall gastrointestinal health.
MICROBIOTA AND ITS LINK TO OBESITY AND THE METABOLIC SYNDROME
There is significant overlap between obesity and the metabolic syndrome; however, the two should be viewed as two separate distinct entities. Obesity refers to accumulation of excess body fat whereas the metabolic syndrome is a disorder of energy storage and utilization resulting in central adiposity, hypertension, dyslipidemia, or insulin resistance. The two together carry significant morbidity and mortality and is thought to be one of the leading preventable causes of death.
The role of microbiota in the pathogenesis of obesity and the metabolic syndrome
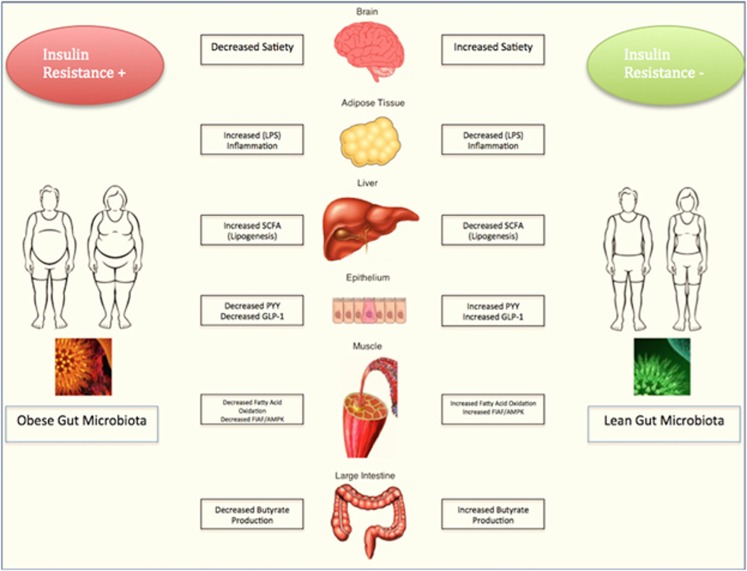
There are several underlying mechanisms thought to contribute to the pathogenesis of obesity and the metabolic syndrome. These underlying mechanisms are outlined in Figure 1.
Figure 1.
Gut microbiota and its influence on obesity.
Bacterial fermentation and its role in energy harvest
Metabolism of dietary polysaccharides is a complex process carried out by gut microbiota.5 Gut microbiota, namely methanogens, ferment dietary polysaccharides resulting in the production of metabolites, namely monosaccharides and short-chain fatty acids (SCFAs). These metabolites are then absorbed and act as an energy source by the host. In addition, SCFAs, via free fatty acid receptors 2 (FFAR2) and 3 (FFAR3), are thought to have a role in regulating gut hormones. A recent study sought to evaluate the effects of these SCFA in mice.8 Butyrate, propionate, and acetate were all shown to have protective effects against diet-induced obesity and insulin resistance. In addition, butyrate and propionate induced gut hormones and reduced overall intake. Similar effects of butyrate and propionate on body weight and food intake were seen in FFAR3 deficient mice, which indicated that there may be additional mediators necessary for these desired effects.
Initial studies demonstrated increased fermentation activity of gut microbiota in the obese population,9 with subsequent quantitative studies confirming these results.10 Fernandes et al.11 sought to compare fecal SCFA concentrations and gut microbial profiles in healthy and overweight/obese volunteers, defined as having a body mass index <25 and body mass index >25, respectively. The ratio of Bacteroides to Prevotella inversely correlated with total fecal SFCA (r=−0.32; P=0.002). The ratio of Firmicutes to Bacteroides/Prevotella positively correlated with total fecal SCFA (r=0.42; P<0.0001), which led authors to conclude that colonic fermentation patterns may be altered, resulting in different fecal SCFA concentrations in overweight/obese individuals as compared with their lean counterparts.
Impact on lipoprotein lipase and fasting-induced adipocyte factor
Lipoprotein lipase hydrolyzes triglycerides into two free fatty acids and one monoacylglycerol molecule.12 Once the fatty acids enter the adipocyte they are re-esterified into triglycerides and stored as fat. Angiopoietin-like-4 (fasting-induced adipocyte factor (Fiaf)) is a protein that is secreted predominately by the liver, which antagonizes the effects of lipoprotein lipase, ultimately preventing the storage of triglycerides as fat.5 Studies have demonstrated an increase in lipoprotein lipase activity with a simultaneous decrease in Fiaf expression to result in a net increase in body fat deposition.13 Bäckhead et al.14 sought to evaluate the effect of Fiaf in germ-free Fiaf deficient mice to germ-free wild-type mice when introduced to a western diet.14 The germ-free mice were not resistant to western-diet induced obesity in comparison with their germ-free wild-type cohort, which the investigators attributed to suppression of Fiaf expression secondary to over nutrition, thereby increasing lipoprotein lipase activity with resultant fat deposition in adipocytes.
Suppression of adenosine monophosphate-activated protein kinase
In times of metabolic stress, the sympathetic response upregulates adenosine monophosphate-activated protein kinase (AMPK) in order to offset any energy expenditure and prevent an energy deprived state. Primarily expressed by brain, liver, and skeletal muscle, AMPK acts to regulate energy homeostasis via stimulation of fatty acid oxidation, ketogenesis, glucose uptake, and insulin secretion while simultaneously inhibiting cholesterol and triglyceride synthesis along with lipogenesis.14, 15 Bäckhed et al.14 sought to evaluate whether AMPK is involved in mediating the resistance of germ-free mice to diet-induced obesity or if the gut microbiome alters the upregulation of AMPK. The levels of active AMPK, which is phosphorylated AMPK, harvested from gastrocnemius muscle in germ-free mice were compared with conventionalized animals on a western diet. Biochemical assays revealed significantly elevated levels of phosphorylated AMPK in gastrocnemius muscles of germ-free mice compared with their conventionalized counterpart with no difference in concentrations of adenosine triphosophate or adenosine monophosphate. Their findings suggested that the presence of altered gut microbiome suppressed skeletal muscle fatty acid oxidation through a metabolic pathway, which may involve activation of AMPK. In addition, the levels of AMPK in the livers of germ-free mice were also noted to be elevated. After being fed a western diet for 5 weeks, levels of hepatic glycogen and glycogen synthase levels were significantly reduced in germ-free mice compared with their conventionalized mice on a western diet, which was confirmed by glucose and insulin tolerance tests. Lastly, biochemical assays demonstrated that germ-free mice had 72% higher levels of nicotinamide adenine dinucleotide (NAD)+, which also upregulates AMPK. The authors concluded that collectively these findings suggest that increased AMPK activity in germ-free mice have a protective role against diet-induced obesity. The microbiome seems to have a suppressive effect on AMPK activity, thereby predisposing the host to obesity and insulin resistance.
The role of SCFAs and G-protein-coupled receptors
Glucagon-like peptide (GLP) and peptide YY (PYY) regulate satiety via the production and release of digestive enzymes.9 The two are co-secreted by intestinal L-cells, induced by gut microbiota16 after each meal and have been demonstrated to have anorectic effects, which can be additive in overweight and obese individuals.17 In mice models, mice lacking PYY had a tendency toward hyperphagia and obesity.18 Conversely, obese rodents that that were subjected to chronic PYY3-36 administration had reduced adiposity over time. Lastly, transgenic mice with increased circulating PYY were resistant to diet-induced obesity. Collectively, coupled with the retained responsiveness of obese subjects to the effects of PYY-36 demonstrate its role in hyperphagia and obesity.
G-protein-coupled receptors, namely FFAR2 and FFAR3, mediate the signaling cascades of SCFAs. As previously described propionate and butyrate have an affinity for FFAR3, whereas acetate appears to have more of an affinity for FFAR2. 8, 19 The primary role of FFAR2 is to promote energy storage. In the colon, both FFAR2 and FFAR3 work in concert to regulate satiety and intestinal motility via GLP-1.20 FFAR2 and FFAR3-deficient mice have both proven to have increase energy expenditure, with the net downstream effect being increased lean body mass and significantly lower fat body mass when compared with their wild-type counterpart.21, 22
Effect of impaired innate immunity
Toll-like receptors (TLRs) are a type of pattern recognition receptor, which has been demonstrated to increase blood glucose and nonesterified free fatty acids.5 Coupled with interleukin-1, it forms the “interleukin-1 receptor/TLR superfamily” which via the release of cytokines and reactive oxygen species produces a pro-inflammatory state, such as that seen in diabetes mellitus.23 TLR-5 has been extensively studied, and has been demonstrated to have a pivotal role in the activation of innate immunity via pattern recognition of microbe-associated molecular patterns seen on bacteria, viruses, and fungi.5, 24, 25 Thus the interaction between gut microbiota and TLR-5 results in the induction of inflammatory cascade and downstream transcription of various cytokines and inflammatory mediators, most notably NFκB, resulting in a low-grade inflammatory state associated with obesity. Additional studies have demonstrated that TLR-5-deficient mice as having a propensity to develop hallmark features of the metabolic syndrome.26
Therapeutic implications
Manipulation of gut intestinal microflora can be accomplished via antibiotics, prebiotics, probiotics, and lastly fecal microbiota transplant (FMT). There appears to be a correlation between early antibiotic exposure and childhood obesity27, 28, 29 however data evaluating this relationship are in its infancy. In the adult population, a recent study by Hwang et al. suggests that Firmicutes and Bacteroidetes may impact insulin resistance by mediating GLP-1 secretion in obesity.30 In mice models, antibiotic-induced depletion of Firmicutes and Bacteroidetes resulted in increased GLP-1 secretion, which ultimately in improved systemic glucose intolerance, hyperinsulinemia, and insulin resistance independent of obesity as compared with untreated controls when exposed to diet-induced obesity. In addition, depletion of Firmicutes and Bacteroidetes resulted in increases in metabolically beneficial gut-derived metabolites.
Prebiotics and probiotics differ in that prebiotics are non-digestible polysaccharides that stimulate growth of digestive bacteria, whereas probiotics are live microorganisms that when administered attempt to reconstitute the gut microflora. Probiotics and prebiotics typically impact populations of Bifidobacterium, Lactobacillus, Saccharomyces, Streptococcus, and Enterococcus. To date, prospective data evaluating the effect on obesity is lacking. There have been several studies that evaluated the effects of probiotics on obesity.31, 32, 33, 34 The most recent data comes from Wang et al.35 Three probiotic strains, Lactobacillus paracasei, Lactobacillius rhamnosus, and Bifidobacterium animalis were administered to mice subjected to a high fat diet for 12 weeks. Each of the 3 strains vitiated weight gain and markedly improved glucose-insulin homeostasis. Pyrosequencing demonstrated that use of all three probiotic strains shifted the gut microflora of mice in the high fat group to resemble that of lean fed mice and thus could potentially be used to attenuate the effects of high fat diet-induced obesity.
Although being utilized for over 50 years, FMT is recently gaining traction as a potential for therapy in obesity in the metabolic syndrome. Several recent studies have demonstrated that FMT from conventionally raised mice with a genetic predisposition to obesity into germ-free mice result in phenotypically obese mice.13, 36, 37 In addition, a recent case report described a patient who underwent successful FMT who developed new-onset obesity after receiving stool from an overweight donor.38 One double-blind controlled trial randomized 18 men with metabolic syndrome to FMT with their own stool (placebo; n=9) or stool from lean men (defined as having a body index <23 kg/m2; n=9).39 Those who received stool from lean donors developed significantly reduced fasting triglyceride levels post FMT. In addition, the nine men who received stool from lean donors had notable decreased hepatic and peripheral insulin sensitivity post FMT compared with those who were transplanted their own stool. Collectively, this suggests that obese microbiota is transmissible and manipulation of the intestinal microflora may be a therapeutic target in the battle against the obesity epidemic.
MICROBIOTA AND ITS LINK TO INFLAMMATORY BOWEL DISEASE
The role of microbiota in the pathogenesis of inflammatory bowel disease
Inflammatory bowel disease (IBD) is a multifaceted entity arising from both genetic and environmental factors, as evident by the discordance in several twin studies.40, 41 With that being said, the host genome may have a pivotal role in shaping the gut microbiota in IBD. The first identified susceptibility gene in Crohn's disease was NOD2,42 which has been demonstrated to perpetuate an immune response to bacterial cell wall.43 Patients with a diagnosis of Crohn's disease known to carry the NOD2 mutation have an increased population of mucosa-adherent bacteria, in particular Escherichia coli. Therefore the host microbiome has the theoretic potential to be reshaped based on genetic variants, although there are currently no available genome-wide studies validating this.
The colon has two distinct mucosal layers, a firmly attached inner mucus layer and an outer mucus layer of variable thickness compared with the small intestine, which is comprised of only one mucosal layer.44 The gut microbiome is intertwined within the pre-epithelial mucous layer, forming a system of “checks and balances”.45 In a normal host, mucins, trefoil peptides, immunoglobulin A make up the mucus layer, which acts to provide a barrier between the gut microflora and intraepithelial cells.45, 46, 47, 48 In the lesser dense small intestine, antimicrobial factors such as defensins and RegIII-γ prevent gut microbes from direct access to epithelial cells.47 Patients suffering from IBD are thought to have a compromised mucus layer, thus allowing luminal microflora to penetrate intraepithelial cells and drive inflammatory and proliferative processes.45 Fu et al.49 recently demonstrated mice deficient in mucins essential in maintaining intestinal mucosal integrity developed spontaneous colitis resembling that of human ulcerative colitis. Recent studies have demonstrated a higher abundance of Fusobacterium varium in patients with ulcerative colitis,50, 51which may in part be responsible for colonic mucosal erosion as seen in mice models52 due to compromised mucosal integrity.
Literature also suggests that intestinal dysbiosis may be responsible for the development of Crohn's disease.53, 54, 55, 56, 57 A recent large, multicenter study compared the microbiome of new-onset, treatment naïve pediatric Crohn's patients to a healthy cohort.58 The study comprised of 447 patients, aged 3–17, with newly diagnosed Crohn's disease. Biopsies obtained during colonoscopy of the rectum and terminal ileum prior to treatment were submitted for rRNA sequencing. In addition, a subset of 223 subjects also provided a fecal sample. Samples obtained from a control population comprised of 221 subjects with non-inflammatory gastrointestinal disease. There was a positive correlation between a diagnosis of Crohn's disease and the presence of Pasteurellacae (Haemophilus sp.), Veillonella parvula, Neisseriacaea corrodens, and Fusobacteriaceae nucleatum (which has been implicated in promoting a microenvironment beneficial toward progression of colorectal carcinoma in addition to being a suggested biomarker for IBD). Additionally, the prevalence of Pasteurellacea, Veillonellaceae parvula, and Rothia mucilaginosa correlated with deep ulcerations (ileal or colonic) seen during diagnostic colonoscopy. These findings were only seen on mucosal biopsies and not on fecal samples, which implies that dysbiosis within mucosa-associated bacterium may be responsible in new-onset Crohn's disease and not alterations within the intestinal lumen.
Therapeutic implications
Prospective data regarding the effect of prebiotics and probiotics in the course of IBD is still in its infancy. To date no prebiotic or probiotic regimen has been consistently beneficial in Crohn's disease, with the exception of Faealibacerium prausinitzii which has been demonstrated to have anti-inflammatory effects in vitro and in vivo in mice models.59, 60, 61 On the contrary, initial data in ulcerative colitis appears promising. A recent meta-analysis including 23 randomized controlled trials totaling 1,763 patients evaluated the effects of probiotics on in inducing remission and maintaining therapy in ulcerative colitis.62 The use of VSL#3 significantly increased remission rates in patients with active ulcerative colitis when compared with placebo (P=0.004, RR=1.74).
Antibiotics remain mainstay therapy in the treatment of septic complications of IBD, such as abscesses and wound infections. Their role in the treatment of primary disease remains controversial and to date its benefit has not been well established. Theoretically, the use of antibiotics in patients with IBD could decrease concentrations of maleficent luminal microorganisms, promote growth of beneficial microorganisms, and decrease bacterial translocation thus preventing its interaction with intraepithelial cells. Recognizably, antibiotics may via antimicrobial properties; they may also exert anti-inflammatory effects.63 The downfall is that the effects of antibiotics on colonic microflora are only transient and over time promotes the proliferation of antibiotic resistant strains. A meta-analysis by Khan et al.64 sought to evaluate the effects of antibiotics in inducing remission in patients with active IBD. For active Crohn's disease there were a total of 10 randomized controlled trials totaling 1,160 patients. The use of antibiotics was superior to use of placebo (P=0.03), however there was a moderate heterogeneity between results given the diverse number of antibiotics tested, either alone or in combination. Rifamycin derivatives, when used either alone or in combination, consistently had significant effect at inducing remission in active Crohn's disease. For active ulcerative colitis, there were nine randomized-controlled trials involving 662 patients. Similar to Crohn's disease with the use of antibiotics for inducing remission, there was moderate heterogeneity given the multitude of antibiotics used, whether it is alone or in combination. Clinically, most practitioners opt to use ciprofloxacin, metronidazole, a combination of ciprofloxacin and metronidazole, rifaxamin, or clarithromycin. Clinical results did not reach statistical significance or equated to placebo effect. 65, 66, 67, 68, 69, 70, 71, 72
There has long been a question of the role of mycobacteria in the pathogenesis of IBD. Thus, anti-tuberculosis treatment was tried in the IBD population. Initial data suggested an overall benefit with use of anti-tuberculosis treatment although it is unclear if response was derived from anti-tuberculosis treatment or anti-inflammatory properties of the antibiotics.73, 74, 75, 76, 77, 78 A subsequent prospective randomized controlled trial by Selby et al.69 randomized 213 patients with Crohn's disease to 2 years of clarithromycin, rifabutin, and clofazimine or placebo, in addition to a 16-week tapering course of prednisolone. At a 2-year follow-up, there was no evidence of sustained benefit with antibiotic use compared with placebo.
As evidence continues to link intestinal dysbiosis to IBD in genetically susceptible individuals, research efforts seek to find the role of FMT. To date, the data for FMT in ulcerative colitis seems to shows mixed results. A recent prospective, randomized controlled trial assigned 48 patients with active ulcerative colitis to FMT from a healthy donor (n=23) or their own feces (placebo; n=25).79 A total of 37 patients completed follow-up and 41% patients who received FMT from a healthy recipient vs. 25% of patients who received FMT placebo achieved clinical remission and endoscopic response (P=0.29), which led researchers to conclude that FMT was not superior to placebo for inducing clinical and endoscopic remission in moderately active ulcerative colitis patients. The use of FMT for refractory UC has been described in only three publications totaling nine patients, all of whom had severe, active, long-standing ulcerative colitis refractory to therapy with glucocorticoids, 5-aminosalicylates, and azathioprine.80, 81, 82 At 6 weeks, there was complete resolution of all symptoms without relapse. Clinical remission was maintained for up to 13 years, with endoscopic evaluation in eight out of the nine patients showing no evidence of active ulcerative colitis (n=6) or only mild chronic inflammation (n=2).81, 82, 83 There has also been heterogeneity in the use of FMT for Crohn's disease. Sunskind et al.84 was the first to demonstrate the potential therapeutic benefit of FMT for Crohn's disease. In this prospective, open-labeled trial, nine patients (12–19 years of age) with mild-to-moderate Crohn's disease (defined by a Pediatric Crohn's Disease Activity Index of 10–29) received FMT from a healthy donor. Follow-up evaluations were done at 2, 6, and 12 weeks with PCDAI, C-reactive protein, and fecal calprotectin measured at each visit. At 2 week follow-up, seven out of the nine patients were in clinical remission based on PCDAI scoring with five out of nine patients maintaining remission without any additional therapy at 6 and 12 week follow-up. At 2 week follow-up, eight out of the nine patients had improvement or normalization in their C-reactive protein levels, with the mean decreasing from 2.4±1.2 mg/dl at baseline to 1.5±0.6 mg/dl. At 6 and 12 weeks post FMT the mean C-reactive protein levels for those individuals who did not require additional medical therapy remained below baseline at 2.0±1.2 and 2.3±2.3 mg/dl, respectively. Lastly, stool calprotectin decreased or remain unchanged for eight out of nine patients at 2 week follow-up. Despite initial improvement in fecal calprotectin for most patients, at 12 week follow-up the levels rose for most patients. Collectively, the authors concluded this to be the first study to demonstrate FMT as a possible therapeutic option for Crohn's disease.
MICROBIOTA AND ITS LINK TO IRRITABLE BOWEL SYNDROME
The role of microbiota in the pathogenesis of irritable bowel syndrome
The hallmark of irritable bowel syndrome (IBS) lays in the heterogeneity of its clinical presentation, yet the underlying pathogenesis remains poorly understood.85, 86 The emergence of postinfectious IBS87, 88, 89, 90, 91 has prompted a paradigm shift toward investigating whether or not dysbiosis of the gut microbiota is a major contributor to the fluctuations in severity of clinical manifestations of IBS.
There are two schools of thought as to the role of the microbiota in IBS. One focus is on the relationship between small bacterial overgrowth (SIBO) and IBS, given its overlap in symptomatology. An open-label study by Pimental et al.92 sought to evaluate the link between SIBO and IBS and whether treatment of SIBO reduced IBS complaints. The study totaled 202 patients who were Rome I criteria positive for IBS underwent lactulose hydrogen breath test to assess for SIBO. Of the 202 patients, 157 had SIBO of which 47 patients had follow-up testing. Subjects were then treated with open label antibiotics after a positive breath test and were then asked to return for follow-up breath tests to confirm eradication. They were blinded to the results of their follow-up tests and asked to answer a questionnaire regarding their symptoms. The investigators found that eradication of SIBO eliminated IBS in 48% of subjects, which let them to conclude that there is an association between IBS and SIBO. Subsequently, Pyleris et al. went to quantitatively evaluate the relationship between IBS and SIBO.93 Consecutive patients presenting for upper endoscopic evaluation were eligible to participate, totaling 320 patients. Quantitative cultures, under aerobic conditions, of aspirates sampled from the third part of the duodenum during upper endoscopy were conducted. Rome II criteria were used to define IBS. Among those enrolled, SIBO was diagnosed in ~62 patients, 42 of whom were diagnosed with concomitant IBS (67.7%). In patients with diarrhea predominant IBS, SIBO was found in 60% compared with 27.3% without diarrhea (P=0.004), with E.coli, Enterococcus spp, and K.pneumoniae being the most common isolates, which led the investigators to conclude that SIBO by aerobic bacteria is independently linked with IBS.
A second focus is on the relationship between gut microbiota and host immune activation. The notion that dysbiosis drives a low-grade inflammatory state lies on the interaction between the gut microbiome and the innate immune system. The role of TLRs (discussed previously) appears to impact the pathogenesis of IBS. Several studies have demonstrated an upregulation of TLR in conjunction with altered TLR-mediated signaling in patients with IBS when compared with healthy controls.94, 95, 96
Therapeutic implications
To date, the available data for prebiotic use in IBS is limited and results are disappointing.60 There have been several controlled trials of probiotics in IBS,97, 98, 99, 100, 101, 102, 103, 104, 105, 106, 107 demonstrated favorable results however the majority are limited by suboptimal study design, small sample, and that they were short term studies. In addition, the magnitude of benefited in studies with positive results was only modest. A recent multicenter randomized, double blind, controlled trial sought to overcoming the shortcomings of previous trials and determine the dose related effects of a novel probiotic combination (I.31 which was mixture of equal parts of three probiotic bacteria:2 Lactobacillus plantarum and1 Pedicoccus acidilactici) on IBS-related quality of life.108 The formula was chosen because of its ability to survive gut motility and adhere to the intestinal mucus layer in vitro. In addition, this combination produced significant amounts of SCFAs, namely butyric, propionic acid, and acetic acid in a ration similar to that seen in a healthy gut. In this study, 84 patients with IBS-D (diarrhea predominant) according to Rome III criteria were randomly allocated to receive once daily I.31 at a high dose (n=28), I.31 at a low dose (n=27), and placebo (n=29). IBS-quality of life (IBS-QoL), Visceral Sensitivity Index (VSI), and global symptom relief questionnaires were filed out at baseline, and at weeks 3 and 6 of treatment. After 6 weeks of treatment the IBS-QoL was significantly increased in both the I.31 high dose and low dose groups compared placebo at 18±3 (P=0.041), 22±4 (0=0.023), and 9±3, respectively. In addition, there was significantly greater improvement in VSI after 6 weeks of treatments in in both the I.31 high dose and low dose groups compared placebo at 10±2 (P<0.05), 14±2 (P<0.05), and 7±1, respectively. There were no significant changes seen in the global symptom relief questionnaires amongst the three arms. This led to the conclusion that this new combination of probiotics was superior to placebo in improving IBS-related quality of life in patients with IBS-D.
Currently, diet modification (low diet low in fermentable oligo-, di-, and monosaccharides and polyols [FODMAPs]), antispasmodics, and neuromodulators (TCAs and SSRIs) remain the cornerstone of therapy in treating IBS. For those who fail, particularly in patients with bloating, a trial of antibiotic therapy ought to be considered. This recommendation stems from the largest randomized control trials to date by Pimental et al.109 totaling 1,260 patients with IBS (without constipation). Patients were randomized to receive rifaxamin 550 mg or placebo three times daily for a total of 2 weeks and were then followed up for an additional 10 weeks. At follow-up, significantly more patients in the rifaxamin group had adequate relief of global IBS symptoms (assessed by daily ratings of IBS symptoms, bloating, abdominal pain, and stool consistency) as compared with placebo with similar incidence of adverse events, which led the authors to conclude that among patients with IBS (without constipation), treatment for rifaximin for 2 weeks provides significant relief of global IBS symptoms. A subsequent meta-analysis by Menees et al. which included five randomized controlled trials evaluated the efficacy of rifaximin.110 Rifixamin when compared with placebo was more efficacious for global IBS symptom improvement (OR 1.57) and significantly more likely to be associated with decreased bloating (OR 1.55).
The microbiome of patients suffering with IBS differs from that of its healthy counterpart. Studies by Kerckhoffs et al.111 have demonstrated there to be decreased populations of Bifidobacteria catenulatum in both fecal and duodenal brush samples of IBS samples compared with healthy subjects, and significantly higher levels of Pseudomonas aeruginosa in duodenal brushings of IBS patients compared with healthy subjects.112 Studies have also demonstrated that IBS-C (constipation predominant) have increased sulfate-reducing bacteria when compared with healthy controls,113 particularly Methanobrevibacter smithii which has been isolated as the predominant methanogen in patients with IBS-C that have positive methane breath tests.114 Thus, FMT has been suggested as a potential therapeutic alternative in patients suffering with IBS. The data on its role, however, remains limited to predominantly anecdotal evidence.115, 116 The most recent data was recently presented by Pinn et al.,117 which included 13 patients with refractory IBS (IBS-D n=9, IBS-C n=3, IBS-A (alternating) n=1). Following FMT, 70% of patients had resolution or improvement in symptoms, which included abdominal pain (72%), bowel habit (69%), dyspepsia (67%), bloating (50%), flatus (42%), and overall quality of life (46%).
MICROBIOTA AND ITS LINK TO NONALCOHOLIC FATTY LIVER DISEASE/STEATOHEPATITIS
The role of microbiota in the pathogenesis of nonalcoholic fatty liver disease/steatohepatitis
Nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH) are hepatic manifestations of the metabolic syndrome with a propensity of NASH to progress to advanced fibrosis and cirrhosis in comparison with patients with NAFLD.118 The underlying pathogenesis of NAFLD/NASH remains unclear; however, alterations in gut microbiota are thought to be a major contributor in its development as depicted in Figure 2.
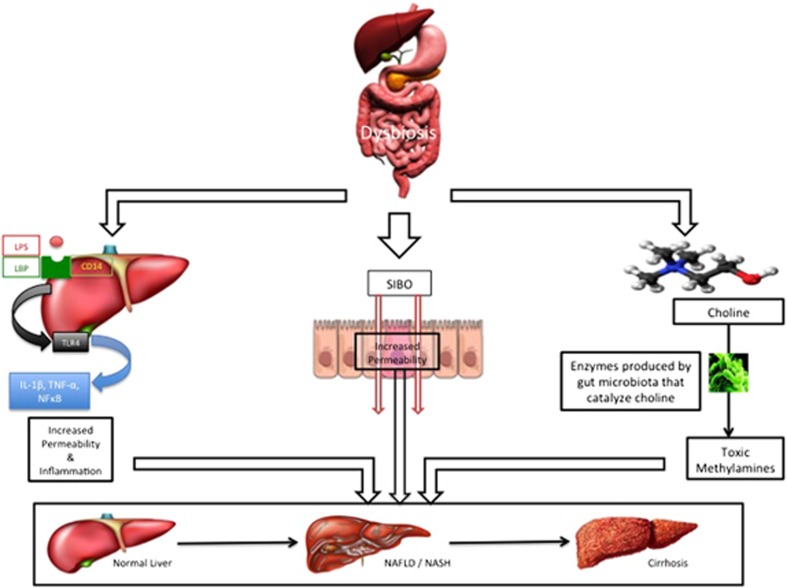
Figure 2.
Gut microbiota and its influence on nonalcoholic fatty liver disease and nonalcohlic steatohepatitis. Proposed mechanisms, working individually or in concert, by which intestinal dysbiosis results in nonalcoholic fatty liver disease and nonalcohlic steatohepatitis. Interaction between lipopolysaccharide and SIBO/CD14 upregulates pro-inflammatory cytokines resulting in increased permeability and inflammation. Bacterial translocation (from SIBO) occurs in lieu of increased permeability activating the innate immune response resulting in a pro-inflammatory state. Intestinal dysbiosis results in production of enzyme that catalyzes choline into toxic methylamines that are injurious to the host liver.
Metabolism by colonic bacteria produces volatile organic compounds that may have deleterious effects on the liver.119 Recent studies have utilized gas-chromatography-mass spectrometry to compliment pyrosequencing in order to differentiate microflora in patients with NAFLD/NASH compared with healthy controls.119, 120, 121, 122, 123, 124 The available data indicates that there is a significant increase in fecal volatile organic compounds with compositional shifts in the microbiome of obese patients with NAFLD/NASH compared with healthy controls. The taxonomic composition remains up for debate at this point with certain studies indicating there to be disproportionately low levels of bacteria from the Ruminococcacae family and high levels of Escherichia in patients with NAFLD/NASH122 where as other studies implicate lower levels of Bacteroides in patients with NASH.123
Metabolic endotoxemia
As previously described, TLRs are pattern recognition motifs that recognize highly conserved microbial molecules called “pathogen-associated molecular patterns” and endogenous products called “damage-associated molecular patterns”.124 Lipopolysaccharide (LPS), an endotoxin found on the cell membrane of Gram-negative bacteria,125 is the most frequently studied pathogen-associated molecular pattern. When bound to LPS-binding protein and CD14, LPS forms a complex that activates TLR-4 (found in Kupffer cells), which then in turn initiates a pro-inflammatory cascade by way of IL-1β, tumor necrosis factor-α (TNF-α), and NFκB.126
Bacterial translocation
The gut epithelium, comprised of epithelial cells linked via tight junctions, has a pivotal role in maintaining the integrity of the intestinal barrier and demarcating the gut microflora from the innate immune system.127 Alterations of gut permeability in the presence of SIBO have been linked to the development of NAFLD/NASH.128 This is thought to be a result of bacterial translocation from the intestinal lumen into portal circulation due to disruption in the intestinal barrier. Gäbele et al.129 sought to evaluate the role of altered intestinal permeability and bacterial translocation in the development of NASH. Mice separated into four groups were set to receive (I) standard chow, (II) a high fat diet, (III) standard chow with dextran sulfate (application of which results in damage to the intestinal barrier), or (IV) which constituted a high-fat diet in conjunction with dextran sulfate. They were then monitored for 12 weeks and pro-inflammatory cytokine levels measured. Endotoxin levels, pro-inflammatory markers (TLR4 and TLR9) were significantly elevated in mice fed a high-fat diet in conjunction with dextran sulfate. In line with this, histological examination demonstrated increased hepatic fibrosis in mice fed a high fat diet in conjunction with dextran sulfate, which led to the conclusion that induction of intestinal inflammation with resultant alteration of gut permeability promoted LPS translocation, hepatic inflammation, and fibrogenesis thought to be secondary to inhibition of intestinal antimicrobial peptides.
Alterations in choline metabolism
Choline, a phosopholipid component of the cell membrane, has a key role in the hepatic metabolism of fat.127 Recent studies have demonstrated gut microbiota to produce enzymes that catalyze the conversion of dietary choline into toxic methylamines.130, 131 Hepatic uptake of these toxic metabolites results in the induction of the inflammatory cascade with a net downstream effect of progression to NAFLD/NASH.
Therapeutic implications
The treatment of both NASH and NAFLD focus on dietary changes, which is a key determinant of the gut microbiota composition.132 Velayudham et al.135 were among the first long term studies, compared with its predecessors that offered mixed short-term data31, 133, 134, to evaluate the benefits of probiotics in patients with NASH. VSL#3 given to methionine-choline-deficient diet fed mice (propensity towards NASH) failed to ameliorate methionine-choline-deficient -induced liver steatosis or inflammation. Despite VSL#3 supplementation, methionine-choline-deficient diet feeding upregulated endotoxins and expression of pro-inflammatory cytokines, namely TLR-4, CD 14, and NF-κB, which led to the conclusion that VSL#3 does not protect the host from inflammation and steatosis in NASH. There was noted improvement in liver fibrosis with VSL#3 supplementation, which was consistent with results from previous studies likely secondary to its effect on collagen expression and modulation of apoptosis.31, 133, 134 Recent studies have evaluated several varying strains of probiotics including Bifidobacterium breve, Lactobacillus rhamnosus, and Lactobacillus johnsonii all of which showing promising results in mice models; however, long-term data and large randomized, controlled human trials are not currently available.136, 137, 138
The limited available data from human studies appears promising. A 2013 meta-analysis, which included four randomized controlled trials totaling 134 NASH patients, evaluated the effects of various probiotic therapies in NAFLD.139 The use of probiotic therapy significantly decreased aminotransferase levels, total cholesterol, high-density lipoprotein, TNF-α, and homeostasis model assessment of insulin resistance suggesting that modulation of gut microbiota with probiotic therapy may have a role in the treatment of NAFLD. The most recent data comes from Alisi et al. who randomized 44 obese children with biopsy-proven NAFLD to VSL#3 (n=22) or placebo (n=22).140 After 4 months of therapy, there were significant improvements in fatty liver severity assessed by ultrasound in the VSL#3 cohort compared with placebo controls. In addition, there was a noted decrease in body mass index and GLP-1 in the VSL#3-treated group (P<0.001 for all comparisons), which was thought to be due to correction of dysbiosis.
MICOBIOTA AND ITS LINK TO GASTROINTESTINAL MALIGNANCY
There has been a recent shift in focus onto microbiota and its potential implication in carcinogenesis, as dysbiosis has been proven to induce DNA damage, genetic alterations, or creating a pro-inflammatory state ultimately leading to tumor initiation, progression, and finally metastasis.
Gastric cancer
Helicobacter pylori are Gram-negative bacteria, thought to colonize the stomach in approximately half the global population.141 As such, H. pylori is accountable for ~66,000 new cases of gastric cancer annually,142 and rarely in mucosa-associated lymphoid tissue (MALT) lymphoma.143 The risk of developing neoplasia resides in the strain of H. pylori colonizing the host,144, 145, 146 duration of infection and interactions between the host and its environmental/microbial determinants. H.pylori are classified into cagA-positive and cagA-negative strains, which depend on the presence or absence of CagA, a terminal gene product that is secreted into host cells after bacterial attachment ultimately inducing cell morphological changes.147, 148 Studies have demonstrated that CagA is a marker of pathogenic disease as patients with elevated antibody titers against CagA have a higher incidence of gastric adenocarcinoma.149 Several subsequent animal models have demonstrated the oncogenic effects of CagA.150, 151 As H. pylori gains entry into the host cell, the CagA protein undergoes phosphorylation via Src152 and Abl153 (two well-known oncogenes). Phosphorylation occurs at specific sequence motifs, with certain sequence motifs having a higher propensity toward gastric adenocarcinoma than others.
H. pylori is not the only microbe linked to the development of gastric cancer. A few studies have attempted to explore the relationship between the upper digestive tract microbiota and its relationship to human health, however results were not conclusive. Yu et al.154 recently sought to evaluate this relationship by testing for the presence of 272 bacterial species in 333 upper digestive tract samples form a Chinese cancer-screening cohort. A serum enzyme-linked immunosorbent assay was used to determine serum pepsinogen I/pepsinogen II ratio with a lower ratio as predictor of gastric cancer risk. There was a demonstrable association with upper digestive tract bacteria and lower pepsinogen I/pepsinogen II ratios, which led to the conclusion that upper digestive microbiota may have a role in the development of gastric cancers. The depth and breadth of bacterial carcinogens responsible for the development of gastric cancers remains poorly understood. The data by Yu et al. suggests that there may be several species of microbes in play, either working individually or synergistically, which increase the risk of gastric cancer.
Esophageal cancer
In contrast to gastric cancer, H. pylori has been proven to have protective effect against the development of esophageal adenocarcinoma.155 There have been several mechanisms proposed to explain the paradox as to why H. pylori is carcinogenic in the stomach yet chemoprotective in the esophagus.156 One possible mechanism is that by inhibiting parietal cell function and/or inducing the development of atrophic gastritis, H. pylori blunts the acid secretion required to develop gastroesophageal reflux disease and its resultant sequelae, i.e., Barrett's esophagus and ultimately esophageal adenocarcinoma. A second possibility is that loss of H. plyori contributes to alterations in the gastric microflora, which ultimately results in reflux-mediated esophageal adenocarcinoma.
Yang et al.157 investigated the impact alterations in the esophageal microbiome had on the pathogenesis of esophageal disease. Biopsy samples were taken from the distal esophagus in 34 patients and defined histologically as being normal or evident of esophagitis, or intestinal metaplasia (Barrett's Esophagus). They were able to identify two distinct microbiome clusters. Type I, which was mainly concentrated in the phenotypically normal esophagus, consisted primarily of Gram-positive bacteria, namely Firmicutes. Type II, which correlated with esophagitis (odds ratio 15.4) and intestinal metaplasia (odds ratio 16.5), consisted primarily of Gram-negative bacteria, namely Bacteroides, Proteobacteria, Fusobacteria, and Spirochaetes. This led to the conclusion that there may be a role for dysbiosis in the pathogenesis of reflux-related disorders.
There are several mechanisms accounting for the role of dysbiosis in the pathogenesis of reflux-related disorders. Gram-negative organisms (type II microbiome) induce a pro-inflammatory signaling cascade by way of LPS, TLR4, and NFκB.158 The downstream result is an increase in levels of pro-inflammatory cytokines, namely IL1β, IL6, IL8, and TNF-α. Additionally, LPS found on Gram-negative bacteria upregulate nitric oxide synthase, which in turn decreases the lower esophageal sphincter.159 This ultimately increases the risk of reflux and resultant esophagitis and eventual metaplasia. Figure 3 is a schematic representation of the microbiome impact on the pathogenesis of esophageal adenocarcinoma.
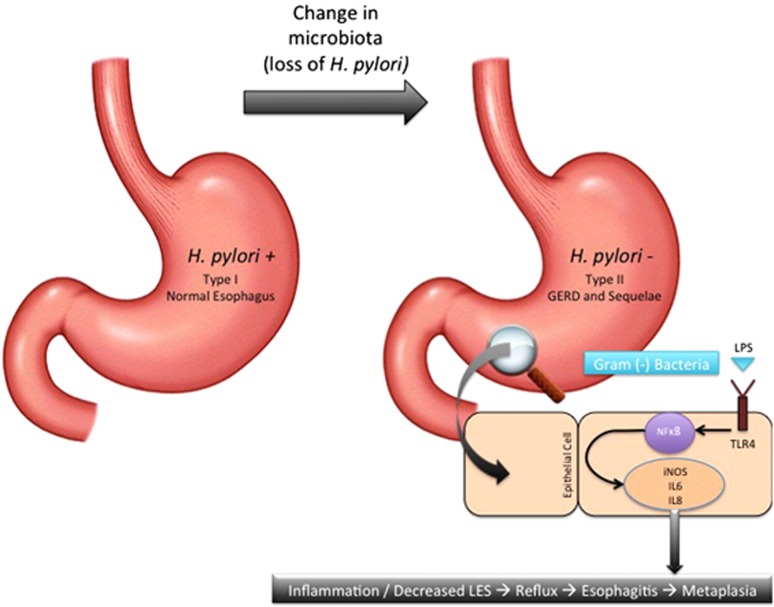
Figure 3.
Gut microbiota and its influence on esophageal adenocarcinoma. Type-microbiota (Gram-positive predominant) with H. pylori provides a neutral esophageal environment. Type-II microbiota (Gram-negative predominant) with loss of H. pylori invokes a pro-inflammatory state in two ways. First, loss of H. pylori allow for increased acid secretion resulting in gastroesophageal reflux disease and its sequelae. Second, predominance of Gram-positive bacteria upregulate the pro-inflammatory cascade due to the interaction between lipopolysaccharide and Toll-like receptor 4.
A recent study by Nasrollahzadeh et al.160 was the first to show the association between gastric microbiota with squamous-cell dysplasia and squamous-cell carcinoma. Gastric microbiota from patients with diagnosed stage I-II and squamous dysplasia were compared with a control group comprised of age and sex-matched subjects with esophagitis (disease control) and histologically normal esophagus (healthy control). Patients with squamous-cell carcinoma and squamous dysplasia had more abundant populations of Clostridiales and Erysipelotrichales not seen in the disease control or healthy control, which led to the conclusion that gastric microbiota differs in squamous-cell carcinoma and SD as compared with a healthy esophagus.
Colon cancer
Intestinal microbes, namely Bacteroidetes and Firmicutes, constitute the majority microbial population in the colon.161, 162 They have been proven to help maintain homeostatsis by contributing to immune development, preventing pathogen colonization, processing drug and toxic metabolites, and releasing nutrients and harvesting energy as previously described. There are several species of intestinal microbes that have recently been implicated as having pro-carcinogenic traits including Fusobacterium, Streptococcus gallolyticus (formerly known as S. bovis) and adherent-invasive E. coli.163, 164, 165, 166 Defective barrier function, such as that produced in a pro-inflammatory state, allow for colibactin-producing bacterial strains to damage host DNA by inducing double-strand breaks, activating damage signaling cascades, ultimately leading to chronic mitotic and chromosomal aberrations.167, 168 Additionally, the defective mucosal barrier allows for translocation of microbial process that leads to the activation of IL-23-producing myeloid cells ultimately promoting tumor growth.169
Sears and Garrett recently reviewed several theories outlining the relationship between the microbiome and its role in the initiation and/or progression of colonic carcinogenesis.170 Coined the “Casualty Theory”, microbiota are regarded as either primary (initiators) or secondary (fostering progression) contributors to colorectal cancer pathogenesis. Under this theory, the authors propose three distinct models, which may be the underlying mechanism promoting colonic carcinogenesis. Model 1 implicating specific microbes, model 2 implicates a microbial community, and model 3 refers to the two acting sequentially and/or synergistically. Carefully designed studies are required in order to link the role of the microbiome as a community and/or select microbes with oncogenic potential in order to discern which of these models promote colonic carcinogenesis.
CONCLUSION
There is clear evidence that the gut microbiome has a profound effect on the balance between health and disease. As we begin to understand how alterations in microbiota impact the pathogenesis of many disease states so to brings the possibility of potential targeted therapy. Unfortunately, studies to date have failed to provide long-term data. In addition, there have been only a select few studies taking into account genetic predisposition towards disease, as phenotypic disease states may be a culmination of genetic and environmental factors. Lastly, given the vast depth of the microbial environment future studies should not only focus on individual subspecies but also their interactions with surrounding microbes. Optimal disease management therefore remains to be defined and remains a significant focus for further clinical investigation.
Guarantor of the article: David A. Johnson, MD, MACG, FASGE, FACP.
Specific author contributions: Concept, initiation of study, and writing up of manuscript: Parth J. Parekh, and David A. Johnson; writing up of manuscript: Luis A. Balart.
Financial support: None.
Potential competing interests: None.
References
- Fujimura KE, Slusher NA, Cabana MD et al. Role of the gut microbiota in defining human health. Expert Rev Anti Infect Ther 2010; 8: 435–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-bello MG, Blaser MJ, Ley RE et al. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology 2011; 140: 1713–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner F, Malagelada JR. Gut flora in health and disease. Lancet 2003; 361: 512–19. [DOI] [PubMed] [Google Scholar]
- Ursell LK, Van treuren W, Metcalf JL et al. Replenishing our defensive microbes. Bioessays 2013; 35: 810–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh PJ, Arusi E, Vinik AI et al. The role and influence of gut microbiota in pathogenesis and management of obesity and metabolic syndrome. Front Endocrinol (Lausanne) 2014; 5: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014; 505: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson AK, Kelly SA, Legge R et al. Individuality in gut microbiota composition is a complex polygenic trait shaped by multiple environmental and host genetic factors. Proc Natl Acad Sci USA 2010; 107: 18933–18938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin HV, Frassetto A, Kowalik EJ et al. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS ONE 2012; 7: e35240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conterno L, Fava F, Viola R et al. Obesity and the gut microbiota: does up-regulating colonic fermentation protect against obesity and metabolic disease? Genes Nutr 2011; 6: 241–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwiertz A, Taras D, Schäfer K et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity (Silver Spring) 2010; 18: 190–195. [DOI] [PubMed] [Google Scholar]
- Fernandes J, Su W, Rahat-rozenbloom S et al. Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutr Diabetes 2014; 4: e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teusink B, Voshol PJ, Dahlmans VE et al. Contribution of fatty acids released from lipolysis of plasma triglycerides to total plasma fatty acid flux and tissue-specific fatty acid uptake. Diabetes 2003; 52: 614–620. [DOI] [PubMed] [Google Scholar]
- Bäckhed F, Ding H, Wang T et al. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA 2004; 101: 15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Manchester JK, Semenkovich CF et al. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA 2007; 104: 979–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winder WW, Hardie DG. AMP-activated protein kinase, a metabolic master switch: possible roles in type 2 diabetes. Am J Physiol. 1999; 277: E1–10. [DOI] [PubMed] [Google Scholar]
- Tremaroli V, Bäckhed F. Functional interactions between the gut microbiota and host metabolism. Nature 2012; 489: 242–249. [DOI] [PubMed] [Google Scholar]
- Field BC, Wren AM, Peters V et al. PYY3-36 and oxyntomodulin can be additive in their effect on food intake in overweight and obese humans. Diabetes 2010; 59: 1635–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karra E, Chandarana K, Batterham RL. The role of peptide YY in appetite regulation and obesity. J Physiol (Lond) 2009; 587: 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzi J, Frost GS, Robertson MD. Do SCFA have a role in appetite regulation? Proc Nutr Soc. 2011; 70: 119–128. [DOI] [PubMed] [Google Scholar]
- Cuche G, Cuber JC, Malbert CH. Ileal short-chain fatty acids inhibit gastric motility by a humoral pathway. Am J Physiol Gastrointest Liver Physiol. 2000; 279: G925–G930. [DOI] [PubMed] [Google Scholar]
- Bjursell M, Admyre T, Göransson M et al. Improved glucose control and reduced body fat mass in free fatty acid receptor 2-deficient mice fed a high-fat diet. Am J Physiol Endocrinol Metab. 2011; 300: E211–E220. [DOI] [PubMed] [Google Scholar]
- Samuel BS, Shaito A, Motoike T et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci USA 2008; 105: 16767–16772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasu MR, Ramirez S, Isseroff RR. Toll-like receptors and diabetes: a therapeutic perspective. Clin Sci. 2012; 122: 203–214. [DOI] [PubMed] [Google Scholar]
- Hayashi F, Smith KD, Ozinsky A et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 2001; 410: 1099–1103. [DOI] [PubMed] [Google Scholar]
- Rhee SH. Basic and translational understandings of microbial recognition by toll-like receptors in the intestine. J Neurogastroenterol Motil. 2011; 17: 28–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-kumar M, Aitken JD, Carvalho FA et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 2010; 328: 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajslev TA, Andersen CS, Gamborg M et al. Childhood overweight after establishment of the gut microbiota: the role of delivery mode, pre-pregnancy weight and early administration of antibiotics. Int J Obes (Lond) 2011; 35: 522–529. [DOI] [PubMed] [Google Scholar]
- Cox LM, Yamanishi S, Sohn J et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell 2014; 158: 705–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jess T. Microbiota, antibiotics, and obesity. N Engl J Med. 2014; 371: 2526–2528. [DOI] [PubMed] [Google Scholar]
- Hwang I, Park YJ, Kim YR et al. Alteration of gut microbiota by vancomycin and bacitracin improves insulin resistance via glucagon-like peptide 1 in diet-induced obesity. FASEB J. e-pub ahead of print. [DOI] [PubMed]
- Ma X, Hua J, Li Z. Probiotics improve high fat diet-induced hepatic steatosis and insulin resistance by increasing hepatic NKT cells. J Hepatol. 2008; 49: 821–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez M, Darimont C, Drapeau V et al. Effect of Lactobacillus rhamnosus CGMCC1.3724 supplementation on weight loss and maintenance in obese men and women. Br J Nutr. 2014; 111: 1507–1519. [DOI] [PubMed] [Google Scholar]
- Toral M, Gómez-guzmán M, Jiménez R et al. The probiotic Lactobacillus coryniformis CECT5711 reduces the vascular pro-oxidant and pro-inflammatory status in obese mice. Clin Sci. 2014; 127: 33–45. [DOI] [PubMed] [Google Scholar]
- Kadooka Y, Sato M, Imaizumi K et al. Regulation of abdominal adiposity by probiotics (Lactobacillus gasseri SBT2055) in adults with obese tendencies in a randomized controlled trial. Eur J Clin Nutr. 2010; 64: 636–643. [DOI] [PubMed] [Google Scholar]
- Wang J, Tang H, Zhang C et al. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J. 2015; 9: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013; 341: 1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006; 444: 1027–1031. [DOI] [PubMed] [Google Scholar]
- Alang N, Kelly CR. Acute weight gain after fecal microbiota transplantation. Open Forum Infect Dis 2015; 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrieze A, Van nood E, Holleman F et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2012; 143: 913–6.e7. [DOI] [PubMed] [Google Scholar]
- Brant SR. Update on the heritability of inflammatory bowel disease: the importance of twin studies. Inflamm Bowel Dis. 2011; 17: 1–5. [DOI] [PubMed] [Google Scholar]
- Halme L, Paavola-sakki P, Turunen U et al. Family and twin studies in inflammatory bowel disease. World J Gastroenterol. 2006; 12: 3668–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y, Bonen DK, Inohara N et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature 2001; 411: 603–606. [DOI] [PubMed] [Google Scholar]
- Philpott DJ, Girardin SE. Crohn's disease-associated Nod2 mutants reduce IL10 transcription. Nat Immunol. 2009; 10: 455–457. [DOI] [PubMed] [Google Scholar]
- Johansson ME, Larsson JM, Hansson GC. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci USA 2011; 108 (Suppl 1): 4659–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cario E. Microbiota and innate immunity in intestinal inflammation and neoplasia. Curr Opin Gastroenterol. 2013; 29: 85–91. [DOI] [PubMed] [Google Scholar]
- Shanahan F. The colonic microbiota in health and disease. Curr Opin Gastroenterol. 2013; 29: 49–54. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 2011; 334: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science 2012; 336: 1268–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Wei B, Wen T et al. Loss of intestinal core 1-derived O-glycans causes spontaneous colitis in mice. J Clin Invest. 2011; 121: 1657–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkusa T, Sato N, Ogihara T et al. Fusobacterium varium localized in the colonic mucosa of patients with ulcerative colitis stimulates species-specific antibody. J Gastroenterol Hepatol. 2002; 17: 849–853. [DOI] [PubMed] [Google Scholar]
- Ohkusa T, Yoshida T, Sato N et al. Commensal bacteria can enter colonic epithelial cells and induce proinflammatory cytokine secretion: a possible pathogenic mechanism of ulcerative colitis. J Med Microbiol. 2009; 58: 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss J, Kaplan GG, Beck PL et al. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011; 17: 1971–1978. [DOI] [PubMed] [Google Scholar]
- Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 2014; 146: 1489–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiodini RJ, Van kruiningen HJ, Thayer WR et al. Possible role of mycobacteria in inflammatory bowel disease. I. An unclassified Mycobacterium species isolated from patients with Crohn's disease. Dig Dis Sci. 1984; 29: 1073–1079. [DOI] [PubMed] [Google Scholar]
- Ott SJ, Musfeldt M, Wenderoth DF et al. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 2004; 53: 685–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MT, Lisby G, Moser C et al. Results of multiple diagnostic tests for Mycobacterium avium subsp. paratuberculosis in patients with inflammatory bowel disease and in controls. J Clin Microbiol. 2000; 38: 4373–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamboli CP, Neut C, Desreumaux P et al. Dysbiosis in inflammatory bowel disease. Gut 2004; 53: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall LJ, Walshaw J, Watson AJ. Gut microbiome in new-onset Crohn's disease. Gastroenterology 2014; 147: 932–934. [DOI] [PubMed] [Google Scholar]
- Müllner K, Miheller P, Herszényi L et al. Probiotics in the management of Crohn's disease and ulcerative colitis. Curr Pharm Des 2014; 20: 4556–4560. [DOI] [PubMed] [Google Scholar]
- Guandalini S, Cernat E, Moscoso D. Prebiotics and probiotics in irritable bowel syndrome and inflammatory bowel disease in children. Benef Microbes 2015; 6: 209–217. [DOI] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 2008; 105: 16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Zuo ZX, Mao AP. Effect of probiotics on inducing remission and maintaining therapy in ulcerative colitis, Crohn's disease, and pouchitis: meta-analysis of randomized controlled trials. Inflamm Bowel Dis. 2014; 20: 21–35. [DOI] [PubMed] [Google Scholar]
- Xu G, Fujita J, Negayama K et al. Effect of macrolide antibiotics on macrophage functions. Microbiol Immunol. 1996; 40: 473–479. [DOI] [PubMed] [Google Scholar]
- Khan KJ, Ullman TA, Ford AC et al. Antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol. 2011; 106: 661–673. [DOI] [PubMed] [Google Scholar]
- Steinhart AH, Feagan BG, Wong CJ et al. Combined budesonide and antibiotic therapy for active Crohn's disease: a randomized controlled trial. Gastroenterology 2002; 123: 33–40. [DOI] [PubMed] [Google Scholar]
- Prantera C, Zannoni F, Scribano ML et al. An antibiotic regimen for the treatment of active Crohn's disease: a randomized, controlled clinical trial of metronidazole plus ciprofloxacin. Am J Gastroenterol. 1996; 91: 328–332. [PubMed] [Google Scholar]
- Shafran I, Johnson LK. An open-label evaluation of rifaximin in the treatment of active Crohn's disease. Curr Med Res Opin. 2005; 21: 1165–1169. [DOI] [PubMed] [Google Scholar]
- Sutherland L, Singleton J, Sessions J et al. Double blind, placebo controlled trial of metronidazole in Crohn's disease. Gut 1991; 32: 1071–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selby W, Pavli P, Crotty B et al. Two-year combination antibiotic therapy with clarithromycin, rifabutin, and clofazimine for Crohn's disease. Gastroenterology 2007; 132: 2313–2319. [DOI] [PubMed] [Google Scholar]
- Mantzaris GJ, Archavlis E, Christoforidis P et al. A prospective randomized controlled trial of oral ciprofloxacin in acute ulcerative colitis. Am J Gastroenterol. 1997; 92: 454–456. [PubMed] [Google Scholar]
- Sato K, Chiba T, Ohkusa T. Serial changes of cytokines in active ulcerative colitis: effects of antibiotic combination therapy. Hepatogastroenterology 2009; 56: 1016–1021. [PubMed] [Google Scholar]
- Madden MV, Mcintyre AS, Nicholls RJ. Double-blind crossover trial of metronidazole versus placebo in chronic unremitting pouchitis. Dig Dis Sci. 1994; 39: 1193–1196. [DOI] [PubMed] [Google Scholar]
- Afdhal NH, Long A, Lennon J et al. Controlled trial of antimycobacterial therapy in Crohn's disease. Clofazimine versus placebo. Dig Dis Sci. 1991; 36: 449–453. [DOI] [PubMed] [Google Scholar]
- Prantera C, Kohn A, Mangiarotti R et al. Antimycobacterial therapy in Crohn's disease: results of a controlled, double-blind trial with a multiple antibiotic regimen. Am J Gastroenterol. 1994; 89: 513–518. [PubMed] [Google Scholar]
- Gui GP, Thomas PR, Tizard ML et al. Two-year-outcomes analysis of Crohn's disease treated with rifabutin and macrolide antibiotics. J Antimicrob Chemother. 1997; 39: 393–400. [DOI] [PubMed] [Google Scholar]
- Leiper K, Morris AI, Rhodes JM. Open label trial of oral clarithromycin in active Crohn's disease. Aliment Pharmacol Ther. 2000; 14: 801–806. [DOI] [PubMed] [Google Scholar]
- Shafran I, Kugler L, El-zaatari FA et al. Open clinical trial of rifabutin and clarithromycin therapy in Crohn's disease. Dig Liver Dis. 2002; 34: 22–28. [DOI] [PubMed] [Google Scholar]
- Borody TJ, Leis S, Warren EF et al. Treatment of severe Crohn's disease using antimycobacterial triple therapy–approaching a cure? Dig Liver Dis. 2002; 34: 29–38. [DOI] [PubMed] [Google Scholar]
- Rossen N, Fuentes S, van der Spek M et al. Abstract OP003. Presented at: 10th Congress of ECCO; 18-21 February 2015, Barcelona, Spain.
- Borody TJ, Warren EF, Leis SM et al. Bacteriotherapy using fecal flora: toying with human motions. J Clin Gastroenterol. 2004; 38: 475–483. [DOI] [PubMed] [Google Scholar]
- Borody TJ, Leis S, McGrath K et al. Treatment of chronic constipation and colitis using human probiotic infusions. Presented at: Probiotics, Prebiotics and New Foods Conference; Rome, Italy; September 2-4, 2001.
- Borody TJ, Warren EF, Leis S et al. Treatment of ulcerative colitis using fecal bacteriotherapy. J Clin Gastroenterol. 2003; 37: 42–47. [DOI] [PubMed] [Google Scholar]
- Borody TJ, George L, Andrews P et al. Bowel-flora alteration: a potential cure for inflammatory bowel disease and irritable bowel syndrome? Med J Aust. 1989; 150: 604. [DOI] [PubMed] [Google Scholar]
- Suskind DL, Brittnacher MJ, Wahbeh G et al. Fecal microbial transplant effect on clinical outcomes and fecal microbiome in active Crohn's disease. Inflamm Bowel Dis. 2015; 21: 556–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremonini F, Talley NJ. Irritable bowel syndrome: epidemiology, natural history, health care seeking and emerging risk factors. Gastroenterol Clin North Am. 2005; 34: 189–204. [DOI] [PubMed] [Google Scholar]
- Collins SM. A role for the gut microbiota in IBS. Nat Rev Gastroenterol Hepatol 2014; 11: 497–505. [DOI] [PubMed] [Google Scholar]
- King T. Psychometric scores and persistence of irritable bowel after infectious diarrhoea. Clin Nutr. 1996; 15: 143. [DOI] [PubMed] [Google Scholar]
- Marshall JK, Thabane M, Garg AX et al. Incidence and epidemiology of irritable bowel syndrome after a large waterborne outbreak of bacterial dysentery. Gastroenterology 2006; 131: 445–450. [DOI] [PubMed] [Google Scholar]
- Neal KR, Barker L, Spiller RC. Prognosis in post-infective irritable bowel syndrome: a six year follow up study. Gut 2002; 51: 410–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley EM. Bugs on the brain; brain in the gut–seeking explanations for common gastrointestinal symptoms. Ir J Med Sci. 2013; 182: 1–6. [DOI] [PubMed] [Google Scholar]
- Shanahan F, Quigley EM. Manipulation of the microbiota for treatment of IBS and IBD-challenges and controversies. Gastroenterology 2014; 146: 1554–1563. [DOI] [PubMed] [Google Scholar]
- Pimentel M, Chow EJ, Lin HC. Eradication of small intestinal bacterial overgrowth reduces symptoms of irritable bowel syndrome. Am J Gastroenterol. 2000; 95: 3503–3506. [DOI] [PubMed] [Google Scholar]
- Pyleris E, Giamarellos-bourboulis EJ, Tzivras D et al. The prevalence of overgrowth by aerobic bacteria in the small intestine by small bowel culture: relationship with irritable bowel syndrome. Dig Dis Sci. 2012; 57: 1321–1329. [DOI] [PubMed] [Google Scholar]
- Brint EK, Macsharry J, Fanning A et al. Differential expression of toll-like receptors in patients with irritable bowel syndrome. Am J Gastroenterol. 2011; 106: 329–336. [DOI] [PubMed] [Google Scholar]
- Mckernan DP, Gaszner G, Quigley EM et al. Altered peripheral toll-like receptor responses in the irritable bowel syndrome. Aliment Pharmacol Ther. 2011; 33: 1045–1052. [DOI] [PubMed] [Google Scholar]
- Macsharry J, O'mahony L, Fanning A et al. Mucosal cytokine imbalance in irritable bowel syndrome. Scand J Gastroenterol. 2008; 43: 1467–1476. [DOI] [PubMed] [Google Scholar]
- Tromm A, Niewerth U, Khoury M et al. The probiotic E. coli strain Nissle 1917 for the treatment of collagenous colitis: first results of an open-label trial. Z Gastroenterol. 2004; 42: 365–369. [DOI] [PubMed] [Google Scholar]
- O'sullivan MA, O'morain CA. Bacterial supplementation in the irritable bowel syndrome. A randomised double-blind placebo-controlled crossover study. Dig Liver Dis. 2000; 32: 294–301. [DOI] [PubMed] [Google Scholar]
- Nobaek S, Johansson ML, Molin G et al. Alteration of intestinal microflora is associated with reduction in abdominal bloating and pain in patients with irritable bowel syndrome. Am J Gastroenterol. 2000; 95: 1231–1238. [DOI] [PubMed] [Google Scholar]
- Whorwell PJ, Altringer L, Morel J et al. Efficacy of an encapsulated probiotic Bifidobacterium infantis 35624 in women with irritable bowel syndrome. Am J Gastroenterol. 2006; 101: 1581–1590. [DOI] [PubMed] [Google Scholar]
- Brenner DM, Moeller MJ, Chey WD et al. The utility of probiotics in the treatment of irritable bowel syndrome: a systematic review. Am J Gastroenterol. 2009; 104: 1033–1049. [DOI] [PubMed] [Google Scholar]
- O'mahony L, Mccarthy J, Kelly P et al. Lactobacillus and Bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology 2005; 128: 541–551. [DOI] [PubMed] [Google Scholar]
- Levri KM, Ketvertis K, Deramo M et al. Do probiotics reduce adult lactose intolerance? A systematic review. J Fam Pract. 2005; 54: 613–620. [PubMed] [Google Scholar]
- Francavilla R, Miniello V, Magistà AM et al. A randomized controlled trial of Lactobacillus GG in children with functional abdominal pain. Pediatrics 2010; 126: e1445–e1452. [DOI] [PubMed] [Google Scholar]
- Ki cha B, Mun jung S, Hwan choi C et al. The effect of a multispecies probiotic mixture on the symptoms and fecal microbiota in diarrhea-dominant irritable bowel syndrome: a randomized, double-blind, placebo-controlled trial. J Clin Gastroenterol. 2012; 46: 220–227. [DOI] [PubMed] [Google Scholar]
- Sisson G, Ayis S, Sherwood RA et al. Randomised clinical trial: a liquid multi-strain probiotic vs. placebo in the irritable bowel syndrome–a 12 week double-blind study. Aliment Pharmacol Ther. 2014; 40: 51–62. [DOI] [PubMed] [Google Scholar]
- Stevenson C, Blaauw R, Fredericks E et al. Randomized clinical trial: effect of Lactobacillus plantarum 299 v on symptoms of irritable bowel syndrome. Nutrition 2014; 30: 1151–1157. [DOI] [PubMed] [Google Scholar]
- Lorenzo-zúñiga V, Llop E, Suárez C et al. I.31, a new combination of probiotics, improves irritable bowel syndrome-related quality of life. World J Gastroenterol. 2014; 20: 8709–8716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel M, Lembo A, Chey WD et al. Rifaximin therapy for patients with irritable bowel syndrome without constipation. N Engl J Med. 2011; 364: 22–32. [DOI] [PubMed] [Google Scholar]
- Menees SB, Maneerattannaporn M, Kim HM et al. The efficacy and safety of rifaximin for the irritable bowel syndrome: a systematic review and meta-analysis. Am J Gastroenterol. 2012; 107: 28–35. [DOI] [PubMed] [Google Scholar]
- Kerckhoffs AP, Samsom M, Van der rest ME et al. Lower Bifidobacteria counts in both duodenal mucosa-associated and fecal microbiota in irritable bowel syndrome patients. World J Gastroenterol. 2009; 15: 2887–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerckhoffs AP, Ben-amor K, Samsom M et al. Molecular analysis of faecal and duodenal samples reveals significantly higher prevalence and numbers of Pseudomonas aeruginosa in irritable bowel syndrome. J Med Microbiol. 2011; 60: 236–245. [DOI] [PubMed] [Google Scholar]
- Chassard C, Dapoigny M, Scott KP et al. Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment Pharmacol Ther. 2012; 35: 828–838. [DOI] [PubMed] [Google Scholar]
- Kim G, Deepinder F, Morales W et al. Methanobrevibacter smithii is the predominant methanogen in patients with constipation-predominant IBS and methane on breath. Dig Dis Sci. 2012; 57: 3213–8. [DOI] [PubMed] [Google Scholar]
- Borody TJ, Paramsothy S, Agrawal G. Fecal microbiota transplantation: indications, methods, evidence, and future directions. Curr Gastroenterol Rep 2013; 15: 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borody TJ, Brandt LJ, Paramsothy S. Therapeutic faecal microbiota transplantation: current status and future developments. Curr Opin Gastroenterol. 2014; 30: 97–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinn DM, Aroniadis OC, Brandt LJ. Follow-up study of fecal microbiota transplantation (FMT) for the treatment of refractory irritable bowel syndrome (IBS) [abstract]. ACG 2013 Annual Scientific Meeting and Postgraduate Course; 11–16 October 2013, San Diego, CA: San Diego Convention Center.
- Vernon G, Baranova A, Younossi ZM. Systematic review: the epidemiology and natural history of non-alcoholic fatty liver disease and non-alcoholic steatohepatitis in adults. Aliment Pharmacol Ther. 2011; 34: 274–285. [DOI] [PubMed] [Google Scholar]
- Raman M, Ahmed I, Gillevet PM et al. Fecal microbiome and volatile organic compound metabolome in obese humans with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2013; 11: 868–75. e1-3. [DOI] [PubMed] [Google Scholar]
- Ahmed I, Greenwood R, Costello Bde L et al. An investigation of fecal volatile organic metabolites in irritable bowel syndrome. PLoS ONE 2013; 8: e58204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Heilig HG, Klaassens ES et al. Isolation of DNA from bacterial samples of the human gastrointestinal tract. Nat Protoc. 2006; 1: 870–873. [DOI] [PubMed] [Google Scholar]
- Garner CE, Smith S, De lacy costello B et al. Volatile organic compounds from feces and their potential for diagnosis of gastrointestinal disease. FASEB J. 2007; 21: 1675–1688. [DOI] [PubMed] [Google Scholar]
- Mouzaki M, Comelli EM, Arendt BM et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 2013; 58: 120–127. [DOI] [PubMed] [Google Scholar]
- Pisetsky D. Cell death in the pathogenesis of immune-mediated diseases: the role of HMGB1 and DAMP-PAMP complexes. Swiss Med Wkly. 2011; 141: w13256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beveridge TJ. Structures of gram-negative cell walls and their derived membrane vesicles. J Bacteriol. 1999; 181: 4725–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ohnishi H. Role of gut microbiota and Toll-like receptors in nonalcoholic fatty liver disease. World J Gastroenterol. 2014; 20: 7381–7391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aron-wisnewsky J, Gaborit B, Dutour A et al. Gut microbiota and non-alcoholic fatty liver disease: new insights. Clin Microbiol Infect. 2013; 19: 338–348. [DOI] [PubMed] [Google Scholar]
- Shanab AA, Scully P, Crosbie O et al. Small intestinal bacterial overgrowth in nonalcoholic steatohepatitis: association with toll-like receptor 4 expression and plasma levels of interleukin 8. Dig Dis Sci. 2011; 56: 1524–1534. [DOI] [PubMed] [Google Scholar]
- Gäbele E, Dostert K, Hofmann C et al. DSS induced colitis increases portal LPS levels and enhances hepatic inflammation and fibrogenesis in experimental NASH. J Hepatol. 2011; 55: 1391–1399. [DOI] [PubMed] [Google Scholar]
- Wang Z, Klipfell E, Bennett BJ et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011; 472: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer MD, Hamp TJ, Reid RW et al. Association between composition of the human gastrointestinal microbiome and development of fatty liver with choline deficiency. Gastroenterology 2011; 140: 976–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabl B, Brenner DA. Interactions between the intestinal microbiome and liver diseases. Gastroenterology 2014; 146: 1513–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Yang S, Lin H et al. Probiotics and antibodies to TNF inhibit inflammatory activity and improve nonalcoholic fatty liver disease. Hepatology 2003; 37: 343–350. [DOI] [PubMed] [Google Scholar]
- Esposito E, Iacono A, Bianco G et al. Probiotics reduce the inflammatory response induced by a high-fat diet in the liver of young rats. J Nutr. 2009; 139: 905–911. [DOI] [PubMed] [Google Scholar]
- Velayudham A, Dolganiuc A, Ellis M et al. VSL#3 probiotic treatment attenuates fibrosis without changes in steatohepatitis in a diet-induced nonalcoholic steatohepatitis model in mice. Hepatology 2009; 49: 989–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaza-diaz J, Gomez-llorente C, Abadia-molina F et al. Effects of Lactobacillus paracasei CNCM I-4034, Bifidobacterium breve CNCM I-4035 and Lactobacillus rhamnosus CNCM I-4036 on hepatic steatosis in Zucker rats. PLoS ONE 2014; 9: e98401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin J, Zeng D, Wang H et al. Preventing non-alcoholic fatty liver disease through Lactobacillus johnsonii BS15 by attenuating inflammation and mitochondrial injury and improving gut environment in obese mice. Appl Microbiol Biotechnol. 2014; 98: 6817–6829. [DOI] [PubMed] [Google Scholar]
- Ritze Y, Bárdos G, Claus A et al. Lactobacillus rhamnosus GG protects against non-alcoholic fatty liver disease in mice. PLoS ONE 2014; 9: e80169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma YY, Li L, Yu CH et al. Effects of probiotics on nonalcoholic fatty liver disease: a meta-analysis. World J Gastroenterol. 2013; 19: 6911–6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alisi A, Bedogni G, Baviera G et al. Randomised clinical trial: the beneficial effects of VSL#3 in obese children with non-alcoholic steatohepatitis. Aliment Pharmacol Ther. 2014; 39: 1276–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De bernard M, Josenhans C. Pathogenesis of Helicobacter pylori infection. Helicobacter 2014; 19 (Suppl 1): 11–18. [DOI] [PubMed] [Google Scholar]
- De martel C, Ferlay J, Franceschi S et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol 2012; 13: 607–615. [DOI] [PubMed] [Google Scholar]
- Bouzourene H, Haefliger T, Delacretaz F et al. The role of Helicobacter pylori in primary gastric MALT lymphoma. Histopathology 1999; 34: 118–123. [DOI] [PubMed] [Google Scholar]
- Peek RM, Crabtree JE. Helicobacter infection and gastric neoplasia. J Pathol. 2006; 208: 33–248. [DOI] [PubMed] [Google Scholar]
- Kwok T, Zabler D, Urman S et al. Helicobacter exploits integrin for type IV secretion and kinase activation. Nature 2007; 449: 862–866. [DOI] [PubMed] [Google Scholar]
- Shaffer CL, Gaddy JA, Loh JT et al. Helicobacter pylori exploits a unique repertoire of type IV secretion system components for pilus assembly at the bacteria-host cell interface. PLoS Pathog. 2011; 7: e1002237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wroblewski LE, Peek RM, Wilson KT. Helicobacter pylori and gastric cancer: factors that modulate disease risk. Clin Microbiol Rev. 2010; 23: 713–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Ding H, Li W. Role of Toll-like receptors in microbiota-associated gastrointestinal cancer metastasis. J Cancer Res Ther 2013; 9: S142–S149. [DOI] [PubMed] [Google Scholar]
- Wu AH, Crabtree JE, Bernstein L et al. Role of Helicobacter pylori CagA+ strains and risk of adenocarcinoma of the stomach and esophagus. Int J Cancer. 2003; 103: 815–821. [DOI] [PubMed] [Google Scholar]
- Ohnishi N, Yuasa H, Tanaka S et al. Transgenic expression of Helicobacter pylori CagA induces gastrointestinal and hematopoietic neoplasms in mouse. Proc Natl Acad Sci USA 2008; 105: 1003–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimuro H, Suzuki T, Nagai S et al. Helicobacter pylori dampens gut epithelial self-renewal by inhibiting apoptosis, a bacterial strategy to enhance colonization of the stomach. Cell Host Microbe. 2007; 2: 250–263. [DOI] [PubMed] [Google Scholar]
- Selbach M, Moese S, Hauck CR et al. Src is the kinase of the Helicobacter pylori CagA protein in vitro and in vivo. J Biol Chem. 2002; 277: 6775–6778. [DOI] [PubMed] [Google Scholar]
- Poppe M, Feller SM, Römer G et al. Phosphorylation of Helicobacter pylori CagA by c-Abl leads to cell motility. Oncogene 2007; 26: 3462–3472. [DOI] [PubMed] [Google Scholar]
- Yu G, Gail MH, Shi J et al. Association between upper digestive tract microbiota and cancer-predisposing states in the esophagus and stomach. Cancer Epidemiol Biomarkers Prev 2014; 23: 735–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ME, Bhatnagar S, George MD et al. The impact of Helicobacter pylori infection on the gastric microbiota of the rhesus macaque. PLoS ONE 2013; 8: e76375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abreu MT, Peek RM. Gastrointestinal malignancy and the microbiome. Gastroenterology 2014; 146: 1534–1546.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lu X, Nossa CW et al. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology 2009; 137: 588–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Francois F, Pei Z. Molecular pathways: pathogenesis and clinical implications of microbiome alteration in esophagitis and Barrett esophagus. Clin Cancer Res. 2012; 18: 2138–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan YP, Chakder S, Gao F et al. Inducible and neuronal nitric oxide synthase involvement in lipopolysaccharide-induced sphincteric dysfunction. Am J Physiol Gastrointest Liver Physiol. 2001; 280: G32–G42. [DOI] [PubMed] [Google Scholar]
- Nasrollahzadeh D, Malekzadeh R, Ploner A et al. Variations of gastric corpus microbiota are associated with early esophageal squamous cell carcinoma and squamous dysplasia. Sci Rep 2015; 5: 8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010; 464: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Ley R. The human gut microbiome: ecology and recent evolutionary changes. Annu Rev Microbiol. 2011; 65: 411–429. [DOI] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell 2010; 140: 883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JC, Perez-chanona E, Mühlbauer M et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012; 338: 120–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Rhee KJ, Albesiano E et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17T cell responses. Nat Med 2009; 15: 1016–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nougayrède JP, Homburg S, Taieb F et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 2006; 313: 848–851. [DOI] [PubMed] [Google Scholar]
- Cougnoux A, Dalmasso G, Martinez R et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 2014; 63: 1932–1942. [DOI] [PubMed] [Google Scholar]
- Secher T, Samba-louaka A, Oswald E et al. Escherichia coli producing colibactin triggers premature and transmissible senescence in mammalian cells. PLoS ONE 2013; 8: e77157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Wang K, Mucida D et al. Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 2012; 491: 254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sears CL, Garrett WS. Microbes, microbiota, and colon cancer. Cell Host Microbe. 2014; 15: 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]