Abstract
Objectives:
Colorectal cancer initially lies dormant as dysplasia, a premalignant state that provides an opportunity for early cancer detection. Dysplasia can be flat in morphology, focal in size, and patchy in distribution, and thus it appears “invisible” on conventional wide-field endoscopy.
Aims:
We aim to develop and validate a peptide that is specific for epidermal growth factor receptor (EGFR), a cell surface target that is overexpressed in colonic adenomas and is readily accessible for imaging.
Methods:
We expressed and purified the extracellular domain of EGFR for use with phage display to identify a peptide QRHKPRE that binds to domain 2 of this target. A near-infrared fluorescence endoscope was used to perform in vivo imaging to validate specific peptide binding to spontaneous colonic adenomas in a mouse model with topical administration. We also validated specific peptide binding to human colonic adenomas on immunohistochemistry and immunofluorescence.
Results:
After labeling with Cy5.5, we validated specific peptide binding to EGFR on knockdown and competition studies. Peptide binding to cells occurred within 2.46 min and had an affinity of 50 nm. No downstream signaling was observed. We measured a target-to-background ratio of 4.0±1.7 and 2.7±0.7, for polyps and flat lesions, respectively. On immunofluorescence of human colonic specimens, greater intensity from peptide binding to dysplasia than normal was found with a 19.4-fold difference.
Conclusions:
We have selected and validated a peptide that can be used as a specific contrast agent to identify colonic adenomas that overexpress EGFR in vivo on fluorescence endoscopy.
INTRODUCTION
Colorectal cancer (CRC) is one of the most common causes of cancer-related mortality worldwide. Approximately 1,361,000 new cases were diagnosed globally in 2012, resulting in ~694,000 annual deaths.1 These numbers are expected to nearly double over the next 20 years as obesity grows at epidemic levels and more developing countries are adopting a Western diet.2 Greater emphasis on early detection of premalignant lesions is needed.3 Imaging with colonoscopy is widely accepted by patients and referring physicians, and it is the preferred method for screening of CRC.4 Currently, white-light illumination is used to detect adenomas based on structural changes. Unfortunately, a significant miss rate of >25% has been found on back-to-back exams for grossly visible adenomas.5, 6, 7 Moreover, premalignant lesions (dysplasia) that are flat can also give rise to cancer,8 and they have been found with a prevalence as high as 36%.9 Flat lesions may be more biologically aggressive than polyps,10 and five times more likely to harbor either in situ or submucosal adenocarcinoma in some patient populations.11 Sessile serrated adenomas (SSAs) can also be flat or slightly raised in appearance, and they can also progress to cancer.12 Thus, imaging methods with improved contrast and sensitivity to molecular rather than morphological properties may improve early detection and prevention of CRC.
Epidermal growth factor receptor (EGFR) is a transmembrane tyrosine kinase that stimulates normal epithelial cell growth and differentiation.13 Ligand binding to the extracellular domains (ECD) 1 and 3 of EGFR results in receptor dimerization and autophosphorylation.14 This cell surface receptor has an important role in the development of a number of epithelial-derived cancers,15 and it is an important target for CRC therapy.16, 17 Overexpression of EGFR has been reported in as high as 97% of colonic adenocarcionomas, and it is a validated biomarker for CRC.18, 19 Adenomas with high-grade dysplasia and villous features on histology have been shown to exhibit increased expression of EGFR on immunohistochemistry.20 Furthermore, EGFR gene copy number has been found to increase with histological progression of disease.21, 22 In animals, EGFR signaling was required to form adenomas in azoxymethane-induced mouse models of CRC,23 and it was shown to promote flat lesions in aberrant crypt foci in rat colon.24 These findings support further development of EGFR as a promising imaging target for early detection of flat colonic lesions.
Fluorescence provides images with high contrast to visualize molecular expression of neoplastic lesions in real time. Antibodies,25 enzyme-activated probes,26 and lectins27 are being developed as specific imaging agents to target premalignant lesions on endoscopy. We have recently shown that peptides are promising for use as clinical diagnostic imaging agents.28, 29 Peptides have low molecular weight and can achieve high specificity with binding affinities on the nanomolar scale. This probe platform has flexibility to be labeled with a broad range of fluorophores,30 and it is inexpensive to produce in large quantities. These features are well-suited to provide effective surveillance of large patient populations in procedure units that perform endoscopic procedures in high volume. We hypothesize that a peptide specific for EGFR can be developed with high specificity and rapid binding for detection of premalignant colonic lesions with topical administration.
METHODS
Cells, chemicals, and materials
Human CRC cells (HT29, SW480, and SW620) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). We used McCoy's Medium for HT29 cells and Dulbecco's Modified Eagle Medium for SW480 and SW620 cells. All cells were cultured at 37 °C in 5% CO2, and supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin. Penicillin–streptomycin was omitted for the small interfering RNA (siRNA) knockdown studies. The cells were passaged using 0.25% EDTA containing trypsin (Mediatech, Manassas, VA). The number of cells were counted on a hemocytometer. For Chinese Hamster Ovary cells, we used minimum essential media α (Life Technologies, Carlsbad, CA, #12561) with 10 μg/ml glycine, 2 mm/l glutamine, 15 μg/ml hypoxanthine, and 5 μg/ml thymidine for culture and serum-free cell culture media (Thermo Scientific, Waltham, MA, HyClone SFM4CHO) to produce the EGFR–ECD protein. Peptide synthesis reagents were obtained from Anaspec (Anaspec, Fremont, CA) or AAPPTEC (AAPPTEC, Louisville, KY), were of the highest grade available (>99% purity), and were used without further purification. Solvents and other chemical reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise mentioned.
Expression of EGFR ECD
The ECD of EGFR (amino acids 1–645 in domains 1–4) was cloned into the pDual GC mammalian expression vector (Stratagene, La Jolla, CA, #214503).31 The gene was inserted between two Eam1104 I restriction sites, resulting in one directional ligation. A cytomegalovirus promoter drives protein expression in mammalian cells. Myc and His tags were expressed in-frame on the C terminus of the recombinant EGFR–ECD protein for use in characterization and purification, respectively. A thrombin recognition site between EGFR–ECD and myc-His allows for the tags to be cleaved after use. Correct construction was verified on DNA sequencing. The construct was first transiently transfected into HEK293T cells and verified on western blot. The construct was then introduced into Chinese Hamster Ovary cells. Stable clones were established by Geneticin selection, and those with the highest expression levels were expanded and cultured in serum-free cell medium to produce EGFR–ECD in microgram quantities for biopanning. The recombinant proteins were purified with cobalt affinity chromatography using a TALON metal affinity resin (Clontech, Mountain View, CA, #635503). The elution was concentrated with an Amicon Ultra-15 centrifugal filter unit with Ultracel-30 membrane (Millipore, Billerica, MA, #UFC03024) and dialyzed in thrombin cleavage buffer (Sigma-Aldrich, T9685). The myc and His tags were removed using a thrombin CleanCleave kit (Sigma-Aldrich, #RECOMT). ECD–EGFR was further purified with a gel filtration column (GE Healthcare, Little Chalfont, UK, HiLoad 16/600 Superdex, 200 pg). The final protein was concentrated, dialyzed in 0.1 m NaHCO3 buffer, and quantified by SDS–PAGE using bovine serum albumin as control.
Peptide specific for EGFR
We used phage display to select candidates that bind specifically to EGFR–ECD.32 A library of M13 bacteriophage with >109 unique sequences was incubated with the EGFR–ECD recombinant protein to identify high-affinity binding interactions. We synthesized Cy5.5-labeled peptides using standard Fmoc-mediated solid-phase synthesis.33 We used Fmoc- and Boc-protected l-amino acids, and synthesis was assembled on rink amide MBHA resin. The peptide was synthesized on a PS3 automatic synthesizer (Protein Technologies, Tucson, AZ). The C-terminal lysine was incorporated as Fmoc-Lys (ivDde)-OH, and the N-terminal amino acid was incorporated with Boc protection to avoid unwanted Fmoc removal during deprotection of the ivDde moiety before fluorophore labeling. Upon complete assembly of the peptide, the resin was transferred to a reaction vessel for manual labeling with the dye. The ivDde side-chain-protecting group was removed with 5% hydrazine in dimethylformamide (3 × 10 min) with continuous shaking at room temperature (RT). The resin was washed with dimethylformamide and dichloromethane three times each for 1 min. The protected resin-bound peptide was incubated overnight with Cy5.5-N-hydroxysuccinimide ester (Lumiprobe, Hallandale Beach, FL) with N,N-diisopropylethylamine, and the completion of the reaction was monitored by a qualitative Ninhydrin test. Upon completion of labeling, the peptide was cleaved from the resin using trifluoroacetic acid:triisopropylsilane:H2O (95:2.5:2.5v/v/v; Sigma-Aldrich) for 4 h with shaking in the dark at RT. After separation of the peptide from the resin, the filtrate was evaporated with N2 gas followed by precipitation with chilled diethyl ether and stored overnight at −20 °C. The precipitate was centrifuged at 3000 r.p.m. for 5 min and washed with diethyl ether three times and centrifuged in between each washing step. The crude peptides were dissolved in 1:1 Acetonitrile/H2O (v/v) and purified by preparative high-performance liquid chromatography with a C18 column (Waters, Milford, MA) using a water (0.1% trifluoroacetic acid)-acetonitrile (0.1% trifluoroacetic acid) gradient. The final purity of the peptides was confirmed by analytical C18-column. Further characterization was performed with either electrospray ionization (Waters) or quadrupole time-of-flight (Agilent Technologies, Santa Clara, CA) mass spectrometry.
siRNA knockdown of EGFR
We used siRNA to knockdown the expression of EGFR in HT29 cells to validate specific peptide binding. We used ON-TARGETplus human EGFR siRNA and ON-TARGETplus nontargeting pool (Thermo Scientific) as per the manufacturer's protocol. siRNA (5 μl) at a 5 μm/l concentration was transfected into HT29 cells using DharmaFECT (Thermo Scientific). QRH*-Cy5.5 and PEH*-Cy5.5 were incubated with HT29 cells transfected with small interfering RNA EGFR (siEGFR) and small interfering RNA control (siCL). The cells were fixed with either 4% PFA or methanol. A 1:1000 dilution of primary monoclonal mouse anti-EGFR antibody (Thermo Scientific, #MS-396, clone 199.12, immunoglobulin G2a isotype) was incubated overnight at 4 °C. Afterward, the cells were washed three times with phosphate-buffered saline (PBS) and further incubated with 1:500 dilution of AF488-labeled secondary goat anti-mouse immunoglobulin G antibody (Life Technologies, #A-11029) for 1 h at RT, washed thrice, and then mounted on glass slides with ProLong Gold reagent containing DAPI (Invitrogen, Waltham, MA). Confocal fluorescence images were collected using a 63 × oil-immersion objective.
Competition for peptide binding
Specific binding of QRH*-Cy5.5 to HT29 cells was validated on competitive inhibition with unlabeled QRH* peptide. A total of ~103 HT29 cells were grown to ~70% confluence on coverslips in triplicate. Unlabeled QRH* and PEH* at 0, 50, 100, 150, 250, and 500 μm were added and incubated with the cells for 30 min at 4 °C. The cells were washed and incubated with 5 μm QRH*-Cy5.5 for another 30 min at 4 °C. The cells were washed and fixed with 4% PFA for 5 min. The cells were washed with PBS and mounted with ProLong Gold reagent containing DAPI (Invitrogen). Fluorescence images were collected at each concentration using a 63 × objective (Zeiss Axioskop 2 plus microscope), and intensities from five cells in two independent images were quantified using the custom Matlab (Mathworks, Natick, MA) software.
Confocal fluorescence microscopy
HT29, SW480, and SW620 cells (~103) were grown on coverslips to ~80% confluence. The cells were washed once with PBS and incubated with 5 μm QRH*-Cy5.5 and PEH*-Cy5.5 for 3 min at RT. The cells were then washed three times in PBS, fixed with 4% PFA for 5 min, washed with 1 × PBS, and then mounted on glass slides with ProLong Gold reagent containing DAPI (Invitrogen). Confocal microscopy (Leica Inverted SP5X) was performed using a 63 × oil-immersion objective. Fluorescence intensities from five cells in two independent images were quantified using custom Matlab (Mathworks) software.
Effect of peptide on cell signaling
HT29 cells were seeded in 12-well flat-bottom plates with 500 μl of serum-free medium for 16 h. EGF (#E9644, Sigma) was reconstituted to a concentration of 1 mg/ml using 10 mm acetic acid, diluted with 0.1% bovine serum albumin, and added to the HT29 cells at concentrations of 100 ng/ml for 10, 20, and 120 min in separate wells. In addition, QRH*-Cy5.5 at concentrations of 5 and 100 μm for 10, 20, and 120 min was added to separate wells. The cells were washed with PBS and lysed in RIPA buffer containing protease inhibitors (#11836170001, Roche, Basel, Switzerland). Lysates were separated by gel electrophoresis, transferred to polyvinylidene difluoride membranes (#ISEQ00010, Millipore), and detected by immunoblotting using an enhanced chemiluminescence system (#RPN2106, GE Healthcare, Little Chalfont, UK). Anti-EGFR antibody (#2232S, Cell Signaling Technology), anti-phospho-EGFR sampler kit (#9922s, Cell Signaling Technology), anti-AKT (#4691P, Cell Signaling Technology, Danvers, MA), anti-ERK1/2 (#4695P, Cell Signaling Technology), anti-phospho-AKT (pS473; #4060P, Cell Signaling Technology), anti-phospho-ERK1/2 (#4370P, Cell Signaling Technology), and anti-tubulin (#32–2600, Invitrogen) were used as per the manufacturer's instructions.
Characterization of peptide binding
We measured the apparent dissociation constant for peptide binding to HT29 cells as an assessment of affinity. QRH*-Cy5.5 was serially diluted in PBS at concentrations of 0, 10, 25, 50, 75, 100, 125, 150, and 200 nm. HT29 cells (~105) were incubated with QRH*-Cy5.5 at 4 °C for 1 h, and washed with cold PBS. The mean fluorescence intensities were measured by flow cytometry. The equilibrium dissociation constant kd=1/ka was calculated by performing a least squares fit of the data to the nonlinear equation I=(I0+Imaxka[X])/(I0+ka[X]). I0 and Imax are the initial and maximum fluorescence intensities, corresponding to no peptide and at saturation, respectively, and [X] represents the concentration of the bound peptide.34 Origin 6.1 data analysis software (OriginLab, Northampton, MA) was used to calculate kd.
We measured the apparent association time constant for peptide binding to HT29 cells as an assessment of time scale. HT29 cells were grown to ~80% confluence in 10- cm dishes, and detached with PBS-based cell dissociation buffer (Invitrogen). Cells (~105) were incubated with 5 μm QRH*-Cy5.5 at 4 °C for various intervals ranging from 0 to 20 min. The cells were centrifuged, washed with cold PBS, and fixed with 4% PFA. Flow cytometric analysis was performed as described above, and the median fluorescence intensity (y) was taken as a ratio with that of HT29 cells without the addition of peptide at different time points (t) using the Flowjo software (LLC, Ashland, OR). The rate constant k was calculated by fitting the data to a first-order kinetics model, y(t)=Imax[1-exp(−kt)], where Imax=maximum value,35 using the Prism 5.0 software (GraphPad, La Jolla, CA).
In vivo imaging of colon in CPC;Apc mouse
We used CPC;Apc mice that are genetically engineered with a Cre recombinase under the control of the Cdx2 promoter (CDX2P-9.5NLS-Cre) and a floxed allele of the APC gene.36 This Cre-regulated somatic mutation in one of the Apc alleles causes adenomas to develop spontaneously in the distal colon of the mouse. Mouse imaging studies were performed with approval of the University of Michigan Committee on the Use and Care of Animals. The mice were housed in pathogen-free conditions and supplied water ad libitum under controlled conditions of humidity (50±10%), light (12/12 h light/dark cycle) and temperature (25 °C). Anesthesia was induced and maintained via a nose cone with inhaled isoflurane mixed with oxygen at a concentration of 2–4% at a flow rate of ∼0.5 l/min. White-light illumination was used first to remove mucous and debris by rinsing with water. The Cy5.5-labeled peptides were delivered at a concentration of 100 μm in a volume of 1.5 ml through the three Fr instrument channel. After 5 min of incubation, the colon was rinsed with water to remove the unbound peptides. The distance from the anus and clockwise location were used to record the presence of any polyps or flat lesions seen on fluorescence. After imaging was completed, the mice were killed. The colon was resected and divided longitudinally. Flat lesions and polyps were identified on fluorescence, excised perpendicular to the mucosal surface, and processed for histology (hematoxylin and eosin (H&E) staining). We collected images from mice (n=5) that ranged in age from 7 to 10 months.
Imaging was performed using a small-animal endoscope (Karl Storz Veterinary Endoscopy, Goleta, CA).37 A xenon light source provides white-light illumination via a fluid light cable. A diode-pumped solid-state laser (TechnicaLaser, Orlando, FL) provides excitation at λex=671 nm. The laser beam is expanded to a diameter of 3 mm to fill the aperture of the light cable. The laser power at the tip of the endoscope is <2 mW. The white-light images reflect off a dichroic mirror (Semrock, Rochester, NY, #FF685-Di02–25 × 36, λc=685 nm), and they are focused by an achromatic doublet (Edmund Optics, Barrington, NJ, #32–323). They pass through a neutral density filter (optical density 1), and they are detected by a color camera (Point Gray Research, Richmond, British Columbia, Canada, #GX-FW-28S5C-C). The fluorescence images pass through a dichroic and band-pass filter (Semrock, #FF01–716/40–25, λC=716 nm, Δλ=40 nm), and are focused on a monochrome camera (Point Gray Research, #GX-FW-28S5M-C). All videos (1932 × 1452 resolution) are recorded at 15 frames per sec via a firewire connection.
White-light and fluorescence videos were exported in avi format with 24 (RGB) and 8 (grayscale) bit digital resolution for white-light and fluorescence images, respectively. Streams that showed minimum motion artifact and absence of debris (stool, mucus) were selected for quantification. Individual frames were exported using the custom Matlab software.
EGFR expression in flat and polypoid dysplasia on immunohistochemistry
Formalin-fixed sections of mouse colonic mucosa were deparaffinized, and antigen retrieval was performed using standard methods. Briefly, the sections were incubated in xylene for 3 min three times, washed with 100% ethanol for 2 min two times, and washed with 95% ethanol for 2 min two times. Rehydration was performed by washing the sections twice in dH2O for 5 min. Antigen unmasking was performed by heating the slides in 10 mM sodium citrate buffer with 0.05% Tween at pH 6.0, and then maintaining at a sub-boiling temperature for 15 min. The slides were cooled for 30 min. The sections were washed three times in dH2O for 3 min, and then incubated in 3% H2O2 in H2O for 10 min. The sections were washed three times in dH2O for 2 min and in PBST for 5 min. We used two primary anti-EGFR antibodies that cross-react with both mouse and human tissues, including 1:1000 dilution of monoclonal goat anti-mouse antibody (GαM) and 1:500 dilution of polyclonal goat anti-rabbit antibody (GαR; Cell Signaling Technology, #2232). Blocking was performed with either tris-buffered saline and Tween 20/5% normal goat serum (GαR) or DAKO protein blocking agent (X0909, DAKO, Carpinteria, CA) for 45 min at RT. The sections were incubated overnight at 4 °C and then washed in PBS for 5 min three times. A 1:200 dilution of secondary antibody (goat anti-mouse or anti-rabbit immunoglobulin G) was added to each section and incubated for 30 min at RT. The secondary antibody solution was removed by washing three times with PBS for 5 min. Premixed Elite Vectastain ABC reagent (Vector Labs, Burlingame, CA) was added to each section and incubated for 30 min at RT. The sections were washed three times in PBST for 5 min, and developed with 3,3'-diaminobenzidine substrate. The reaction was monitored for 3 min, and then quenched by immersing the slides in dH2O. Hematoxylin was added as a counterstain for ~20 s, and the sections were dehydrated in increasing concentrations of ethyl alcohol (70%, 80%, 95% 2 × , 100% 2 × ). Coverslips were attached using permount mounting medium (Fisher, Pittsburgh, PA, #SP15–100) in xylene. Serial sections were processed for histology (H&E). Controls were prepared using secondary antibody, Elite Vectastain ABC reagent, Vector Labs and 3,3'-diaminobenzidine (without primary anti-EGFR antibody).
EGFR expression in flat and polypoid dysplasia on immunofluorescence
Specimens of flat dysplasia in mouse colon were formalin-fixed and processed as described previously. Specimens of polypoid dysplasia in mouse colon were frozen in optimal cutting temperature (Sakura Finetek, Torrance, CA), cut in 10 -μm sections, and incubated with 1:1000 dilution of primary goat anti-mouse anti-EGFR antibody and AF488-labeled secondary antibody, as described previously. The sections were washed three times with PBS, fixed with 4% PFA for 10 min, washed with PBS once, and mounted with Prolong Gold reagent containing DAPI (Invitrogen). Confocal microscopy was performed. Adjacent sections were processed for histology (H&E).
Binding of EGFR peptide and antibody to human colonic dysplasia
Specimens of human colonic mucosa were obtained from biopsy during routine colonoscopy, frozen in optimal cutting temperature, cut into 10 -μm sections, and incubated with QRH*-Cy5.5 (5 μm) in 1 × PBS for 10 min at RT. The sections were washed three times with PBS and incubated overnight at 4 °C with 1:1000 dilution of primary monoclonal rabbit anti-EGFR antibody (Cell Signaling Technology, #4267, isotype immunoglobulin G). The sections were washed three times with PBS and incubated with 1:500 dilution of Alexa Fluor 488-labeled secondary goat anti-rabbit antibody (Invitrogen) for 1 h at RT. The sections were washed three times with PBS, and fixed with 4% PFA for 10 min. The sections were then washed once with PBS, and mounted with ProLong Gold reagent containing DAPI (Invitrogen). Confocal microscopy (Leica TCS SP5 Microsystems) was performed with a 20 × objective. The mean fluorescence intensities from three boxes (dimensions of 30 × 30 μm2) located completely within the surface epithelium of each specimen were measured. Regions that showed intensity saturation were avoided. Adjacent sections were processed for routine histology (H&E) that was reviewed by a gastrointestinal pathologist (SRO).
Statistical analysis
For the in vivo fluorescence images of mouse colon, the target-to-background ratios for flat dysplasia and polyps were log-transformed to improve normality and to stabilize variance. The fold-change between classification pairs was estimated using the anti-log of the difference in the log-transformed data. Colocalization of peptide and antibody binding was evaluated with Pearson's correlation coefficient.
RESULTS
Expression of EGFR ECD
We used the pDual GC mammalian system to express the ECD of EGFR, Supplementary Figure S1A online. EGFR–ECD represents the exposed region of the target that is accessible to imaging, and it was linked in frame via a thrombin cleavage site to a myc-His tag. We used myc to confirm (transient) the expression of the recombinant protein in HEK293T cells on western blotting, Supplementary Figure S1B, before a stable transfection was performed in Chinese Hamster Ovary cells. Clones with the highest levels of protein expression (solid boxes) on the western blot were expanded; see Supplementary Figure S1C. Affinity purification was performed using His-tag-based cobalt chromatography. The elution was first verified on SDS–PAGE and then cleaved with thrombin to remove the myc-His tag. Further purification was performed on gel filtration, and the EGFR–ECD peak (dashed box) was identified, Supplementary Figure S1D. The corresponding bands (solid box) on SDS–PAGE were selected and concentrated, Supplementary Figure S1E. We produced 600 μg of EGFR–ECD with >90% purity, Supplementary Figure S1F.
Peptide specific for EGFR
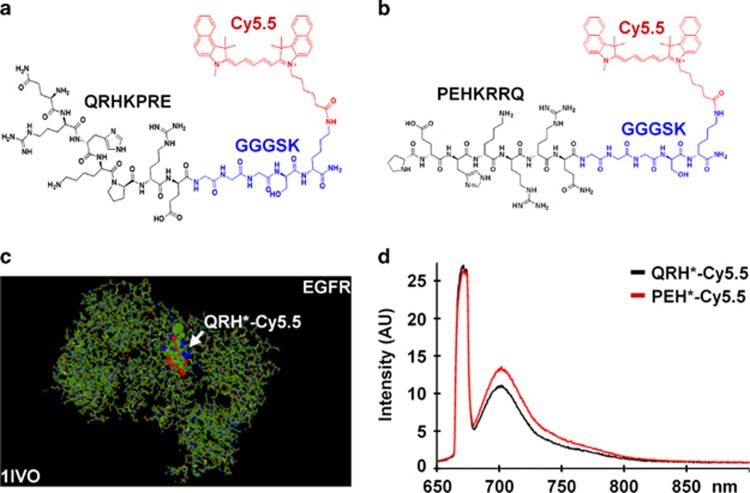
After four rounds of biopanning with phage display, 10 sequences showed enrichment. We evaluated the binding of each candidate on a structural model,38, 39 and found a minimum energy of Et=−504.1 for docking of QRHKPRE labeled with Cy5.5 to EGFR (1IVO) (http://www.rcsb.org/pdb). We synthesized this sequence (black) and attached a Cy5.5 (red) fluorophore via a GGGSK linker (blue) on the C terminus, hereafter QRH*-Cy5.5, to prevent steric hindrance, Figure 1a. Cy5.5 was chosen for its high-quantum yield and photostability.40 We used this model to develop a scrambled sequence PEHKRRQ for control, hereafter PEH*-Cy5.5, Figure 1b, and found Et=−493.1. In the model, QRH*-Cy5.5 binds to amino acids 230–310 of EGFR (domain 2), Figure 1c. The fluorescence spectra of QRH*-Cy5.5 and PEH*-Cy5.5 at 10 μm concentration in PBS with λex=671 nm excitation revealed a peak emission at 710 nm, Figure 1d. We purified the Cy5.5-labeled peptides to >97% on high-performance liquid chromatography, and measured an experimental mass-to-charge (m/z) ratio on mass spectrometry of 1900.05 for both QRH*-Cy5.5 and PEH*-Cy5.5 that agreed with expected values, Supplementary Figure S2A, B.
Figure 1.
Peptide specific for EGFR. (a) Chemical structure of QRHKPRE peptide (black) with GGGSK linker (blue) and Cy5.5 fluorophore (red). (b) Scrambled peptide PEHKRRQ (control). (c) QRH*-Cy5.5 was found on the structural model to bind domain 2 of EGFR (1IVO). (d) Fluorescence spectra of Cy5.5-labeled peptides with λex=671 nm shows peak emission near 710 nm. AU, arbitrary unit; EGFR, epidermal growth factor receptor.
siRNA knockdown of EGFR
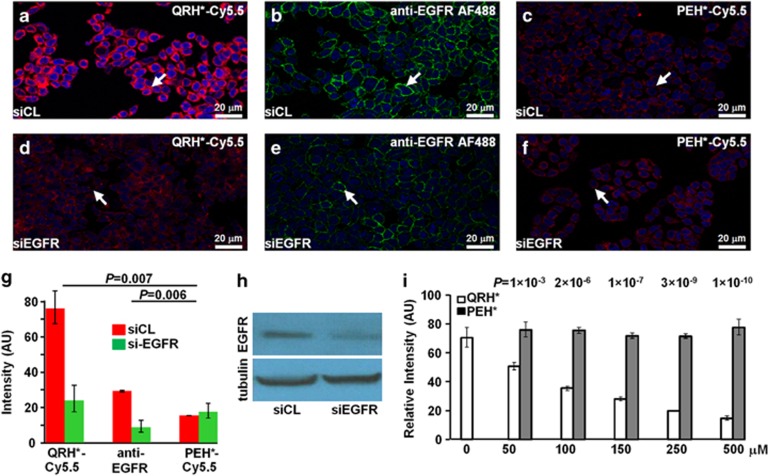
We validated specific binding of QRH*-Cy5.5 to EGFR on siRNA knockdown in HT29 cells. On confocal microscopy, QRH*-Cy5.5- (red), Figure 2a, and AF488-labeled anti-EGFR (green), Figure 2b, bind strongly to the surface (arrows) of control HT29 cells, transfected with siCL nontargeting siRNA. PEH*-Cy5.5 shows minimal binding, Figure 2c. Reduced fluorescence intensity was observed for HT29 knockdown cells, transfected with siEGFR-targeting siRNA, Figure 2d, e. PEH*-Cy5.5 shows minimal binding, Figure 2f. QRH*-Cy5.5 and anti-EGFR antibody showed significantly higher intensities for siCL-transfected HT29 cells than for those treated with siEGFR, whereas PEH*-Cy5.5 showed a small nonsignificant increase, Figure 2g. Differences for siCL vs. siEGFR for QRH*-Cy5.5 and anti-EGFR antibody were significantly greater than those for PEH*-Cy5.5. EGFR expression is shown on the western blot, Figure 2h.
Figure 2.
Validation of specific peptide binding to EGFR. On confocal microscopy, we found strong binding of (a) QRH*-Cy5.5 peptide (red) and (b) AF488-labeled anti-EGFR (green) to the surface (arrow) of control HT29 cells (siCL). (c) PEH*-Cy5.5 (red) binding is minimal. (d–f) The fluorescence intensities are significantly reduced in knockdown of HT29 cells (siEGFR). (g) Quantified results for QRH*-Cy5.5 and anti-EGFR show significantly higher intensities for siCL- vs. siEGFR-transfected cells (3.2- and 3.4-fold change, P=0.0021 and 0.0017, respectively), whereas PEH*-Cy5.5 showed a nonsignificant decrease (0.87 fold-change, P=0.57). Differences for siCL vs. siEGFR for QRH*-Cy5.5 and anti-EGFR were significantly greater than those for PEH*-Cy5.5 (P=0.007 and 0.006, respectively). We fit two-way ANOVA models with the terms for six conditions and two replicate slides on log-transformed data. Measurements were on an average of five randomly chosen cells on two slides for each condition. (h) Western blot shows EGFR expression levels. (i) On competition, we found a significant difference in binding of QRH*-Cy5.5 to HT29 cells with the addition of unlabeled QRH* and PEH* at the concentrations of 50 μm and higher. Nonsignficant difference was found at 0 μm. We fit two-way ANOVA models with the terms for the labeled peptide, concentrations of the unlabeled peptides, and their interactions on log-transformed data. P-values shown here compare the difference in the intensity between unlabeled QRH* and PEH* at each dose with that at 0 μm. Measurements are on an average of five randomly chosen cells on two slides for each condition. ANOVA, analysis of variance; EGFR, epidermal growth factor receptor.
Competition for peptide binding
We evaluated the binding of QRH*-Cy5.5 to HT29 cells on competition with the addition of unlabeled QRH* and PEH*. No significant difference was found at 0 μm, P=0.18. Significant differences were observed at concentrations of 50 μm and higher. These differences were significantly larger than those at 0 μm (P-values shown in the above data), Figure 2i. Addition of unlabeled QRH* reduced intensities in a concentration-dependent manner. These results support peptide rather than fluorophore binding to the surface of HT29 cells.
Confocal fluorescence microscopy
On confocal microscopy, we assessed the binding of QRH*-Cy5.5 and PEH*-Cy5.5 to a panel of three CRC cells that vary in the level of EGFR expression. We observed different strengths of QRH*-Cy5.5 binding to the surface (arrows) of HT29, SW480, and SW620 cells (high, medium, and low EGFR expression, respectively), Supplementary Figure S3A–C. Minimal binding was observed for PEH*-Cy5.5 (control), Supplementary Figure S3D–F. We observed significant differences in binding between QRH*-Cy5.5 and PEH*-Cy5.5 for HT29, P=4.7 × 10−4, and SW480, P=4.3 × 10−4, but not for SW620, P=0.08, Supplementary Figure S3G. The QRH*-Cy5.5 vs. PEH*-Cy5.5 differences were significantly larger for HT29 and SW480 than for SW620 (P=0.003 in both cases). EGFR expression is shown on the western blot, Supplementary Figure S3H.
Effect of peptide on cell signaling
HT29 cells incubated with QRH-Cy5.5 showed no evidence of either EGFR phosphorylation or downstream phosphorylation of Akt and Erk1/2 at the concentrations used for imaging, Supplementary Figure S4.
Characterization of peptide binding
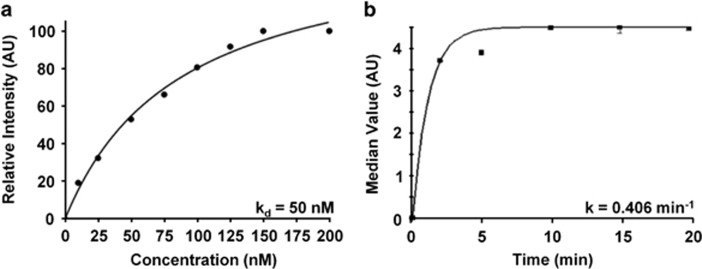
We measured an apparent dissociation constant of kd=50 nm, R2=0.95 for QRH*-Cy5.5 to HT29 cells on flow cytometry, Figure 3a. This result provides a measure of binding affinity. In addition, we measured an apparent association time constant of k=0.406/min for QRH*-Cy5.5 to HT29 cells on flow cytometry, Figure 3b. This result provides a time scale of 2.46 min for binding. Both results are representative of six independent experiments.
Figure 3.
Characterization of EGFR peptide-binding parameters. (a) Apparent dissociation constant kd=50 nm, R2=0.95 was measured for binding of QRH*-Cy5.5 to HT29 cells. (b) Apparent association time constant k=0.406/min (2.46 min) was measured for binding of QRH*-Cy5.5 to HT29 cells. Both results are representative of six independent experiments.
In vivo imaging of colon in CPC;Apc mouse
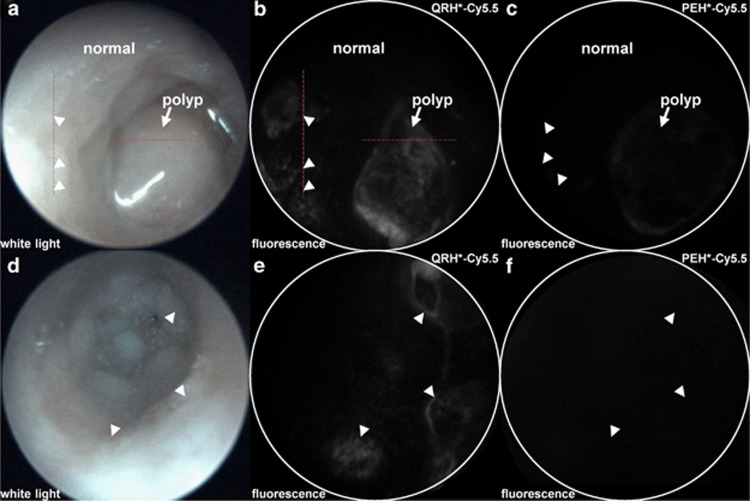
We evaluated specific binding of QRH*-Cy5.5 in vivo to spontaneous adenomas in CPC;Apc mice (n=5) using a wide-field small-animal endoscope that is sensitive to near-infrared fluorescence. This mouse was genetically engineered to somatically delete an Apc allele under Cre regulation, and it develops both flat and polypoid adenomas in the distal colon.36 This model is representative of human disease because Apc mutations are found in >80% of sporadic CRCs.41 A white-light image of a polyp (arrow) is shown (Supplementary Video S1) in Figure 4a. QRH*-Cy5.5 was topically administered in the distal colon, and allowed to incubate for 5 min. Pathology was later evaluated along the red lines. The unbound peptides were rinsed away, and the fluorescence image shows increased intensity from the polyp in a heterogeneous pattern (Supplementary Video S2), Figure 4b. Flat lesions (arrowheads) can also be seen that are not apparent on white light and were found later on pathology to be low-grade dysplasia. Normal colonic mucosa shows minimal background. Using distance from the anus and clockwise location, images were collected using PEH*-Cy5.5 (control) from the same region of colon in this mouse. Minimal signal was found for either type of lesion (Supplementary Video S3), Figure 4c. A white-light image from the colon of a different animal shows no grossly visible polyps (Supplementary Video S4), Figure 4d. Several flat lesions are seen on fluorescence after the administration of QRH*-Cy5.5 (Supplementary Video S5), Figure 4e. Minimal signal was observed with the PEH*-Cy5.5 control (Supplementary Video S6), Figure 4f. EGFR overexpression can be seen within the individual dysplastic crypts for polyp, Supplementary Figure S5A–C, and flat lesion, Supplementary Figure S5D–F.
Figure 4.
In vivo imaging of colon in CPC;Apc mouse. (a) White-light image of colon in CPC;Apc mouse shows the presence of polyp (arrow). Pathology was evaluated along (dashed) red lines. (b) NIR fluorescence image after topical administration of QRH*-Cy5.5 shows increased intensity from polyp (arrow) and several flat lesions (arrowheads) with heterogeneous pattern. (c) Image with PEH*-Cy5.5 shows minimal signal. (d) White-light image shows no grossly visible lesions (polyps). (e) NIR fluorescence image with QRH*-Cy5.5 shows the presence of flat lesions (arrowheads). (f) Image with PEH*-Cy5.5 shows minimal signal. NIR, near-infrared.
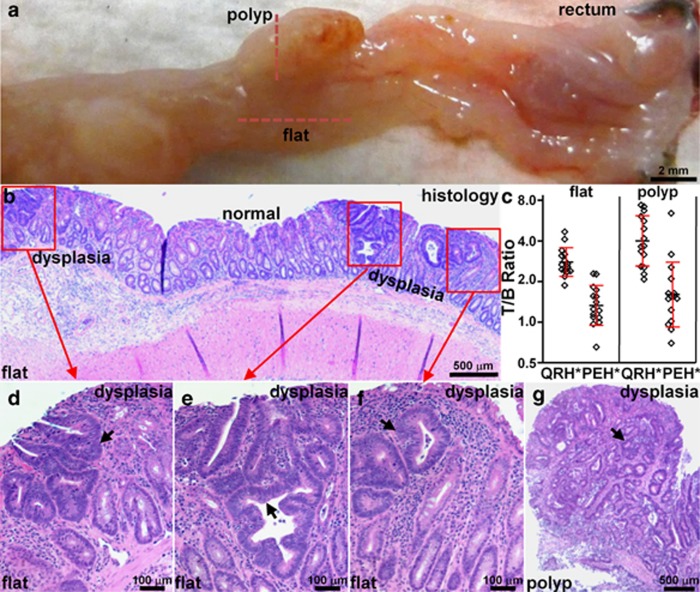
After completion of imaging, the animals were killed. The colon was excised and divided longitudinally. The gross specimen with the polyp and flat lesions from Figure 4 are shown, Figure 5a. Fluorescence imaging was used to guide a perpendicular section of the mucosal surface along the horizontal and vertical red lines. The flat lesions show mucosa with nonpolypoid morphology on histology (H&E), Figure 5b. Foci of dysplasia (red boxes) can be seen separated by regions of normal. The average fluorescence intensity from three regions of interest with dimensions of 25 × 25 pixels were picked at random from areas of “high” intensity and adjacent areas of “low” intensity. The target-to-background ratios were determined by taking ratios of the means of these results. We measured significantly greater fluorescence intensity from both polyps (n=15) and flat lesions (n=15) compared with that from the adjacent normal mucosa, target-to-background ratio 4.0±1.7 and 2.7±0.7, respectively. The difference in results between QRH*-Cy5.5 and PEH*-Cy5.5 was significant. A high-magnification view of histology from the flat regions (red boxes) show features of low-grade dysplasia (arrows), including collections of irregular crypts lined by epithelium with crowded, elongated, and hyperchromatic nuclei, Figure 5d–f. Histology of the polyp also shows identical features of dysplasia, Figure 5g.
Figure 5.
Validation of colonic dysplasia on pathology. (a) Excised colon from Figure 4 shows locations of flat lesions and polyp (dashed red lines), Bar = 2 mm. (b) Histology (H&E) of flat lesion shows nonpolypoid mucosal morphology and foci of low-grade adenomatous dysplasia (red boxes) separated by intervening regions of normal mucosa, Bar=500 μm. (c) Greater mean fluorescence intensities from polyps (n=15) and flat lesions (n=15) were found compared with those from adjacent normal mucosa, T/B ratio 4.0±1.7 and 2.7±0.7, respectively. For polyps, mean±s.d. of the T/B ratio (log2) for QRH*-Cy5.5 and PEH*-Cy5.5 was 1.90±0.60 and 0.62±0.77, P=4.1 × 10−4 by paired, two-sided t-test, respectively, and mean fold difference was 2.43. For flat lesions, the results were 1.39±0.34 and 0.36±0.47, P=7.4 × 10−6 by paired, two-sided t-test, and mean fold difference was 2.05. (d–f) Magnified view of red boxes in b shows histological features of low-grade dysplasia (arrows). (g) Histology (H&E) of polyp along vertical red line in a shows identical histological features of dysplasia. H&E, hematoxylin and eosin; T/B, target-to-background.
EGFR expression in flat and polypoid dysplasia on immunohistochemistry
We found increased expression of EGFR in dysplasia compared with normal in flat colonic lesions on immunohistochemistry using primary goat anti-rabbit anti-EGFR antibody (GαR), Supplementary Figure S6A. A magnified view of dashed box is shown in Supplementary Figure S6B. EGFR staining appears membranous with strongest positivity on the apical and basolateral aspects of dysplastic cells. An adjacent section was stained with secondary antibody but not primary antibody (control), Supplementary Figure S6C. Corresponding histology (H&E), Supplementary Figure S6D. We also found high expression of EGFR in dysplastic polyps, Supplementary Figure S6E. Magnified view of dashed box is shown in Supplementary Figure S6F. An adjacent section (control) is shown in Supplementary Figure S6G. Corresponding histology (H&E) is shown in Supplementary Figure S6H. We also used primary goat anti-mouse anti-EGFR antibody (GαM) to confirm increased expression of EGFR in dysplastic polyp, Supplementary Figure S6I. Magnified view of dashed box is shown in Supplementary Figure S6J. An adjacent section (control) is shown in Supplementary Figure S6K. Corresponding histology (H&E) is shown in Supplementary Figure S6L. Minimal reactivity was found for normal colonic mucosa on low and high magnification, as shown in Supplementary Figure S6M,N, respectively. An adjacent section (control) is shown in Supplementary Figure S6O. Corresponding histology (H&E) is shown in Supplementary Figure S6P.
EGFR expression in flat and polypoid dysplasia on immunofluorescence
We found increased expression of EGFR in colonic dysplasia on immunofluorescence using formalin-fixed specimens. Contrast between flat dysplasia and normal mucosa can be seen at lesion border, Supplementary Figure S7A. A magnified view of the solid boxes in Supplementary Figure S7A shows greater fluorescence intensity for dysplasia compared with that of normal, Supplementary Figure S7B, C. Corresponding histology (H&E) is shown in Supplementary Figure S7D–F. We also observed increased fluorescence intensity on binding of QRH*-Cy5.5 to dysplastic crypts (arrow) in polyps using optimal cutting temperature-embedded specimens, Supplementary Figure S7G. A magnified view of the dashed box in Supplementary Figure S7G shows cell surface staining, Supplementary Figure S7H. Minimal EGFR expression was seen in normal colonic mucosa, Supplementary Figure S7I. A magnified view of the dashed box in Supplementary Figure S7I is shown, Supplementary Figure S7J.
Binding of EGFR peptide and antibody to human colonic dysplasia
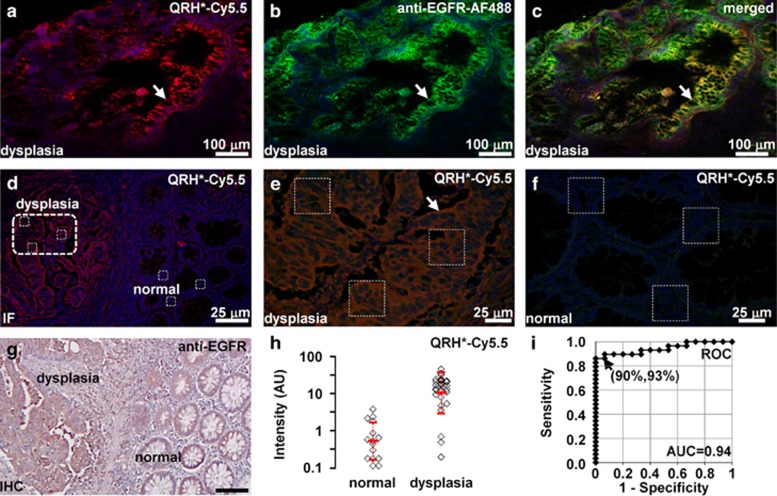
On confocal microscopy, we observed strong binding of QRH*-Cy5.5 (red) and AF488-labeled anti-EGFR (green) to the surface (arrows) of dysplastic colonocytes in human colonic specimens, Figure 6a, b, respectively. Colocalization of peptide and antibody binding can be seen on merged image, P=0.71, Figure 6c. Contrast between dysplasia and normal for peptide binding can be appreciated at the lesion border, Figure 6d. A high-magnification view of white boxes in Figure 6d is shown for dysplasia, Figure 6e, and normal, Figure 6f. Fluorescence intensities were measured from sets of three (dashed white) boxes with dimensions of 30 × 30 μm2 to calculate the target-to-background ratio. Corresponding immunohistochemistry from Figure 6d shows increased reactivity for EGFR in dysplasia, Figure 6g. Dysplasia (n=29) showed a significantly higher mean fluorescence intensity compared with that of normal (n=15) by 19.4-fold, Figure 6h. The receiver operating characteristic curve shows 90% sensitivity and 93% specificity for distinguishing dysplasia from normal with an area under curve of 0.94.
Figure 6.
Binding of EGFR peptide and antibody to human colonic neoplasia. On confocal microscopy, binding of (a) QRH*-Cy5.5 peptide (red) co-localizes with that of (b) AF488-labeled anti-EGFR antibody (green) on surface of dysplastic colonocytes (arrow), shown in (c) merged image, P=0.71. (d) Image contrast can be appreciated at lesion border. Magnified view of boxes in d is shown for (e) dysplasia and (f) normal. (g) Corresponding immunohistochemistry from a shows increased reactivity for EGFR in dysplasia. (h) Dysplasia (n=29) showed significantly higher fluorescence intensities than normal (n=15) by an average of 19.4-fold, P=1.7 × 10−9 by two-sample t-test on log-transformed data. (i) Receiver operating characteristic curve shows 90% sensitivity and 93% specificity with area under curve (AUC) of 0.94 for distinguishing dysplasia from normal using peptide.
We then evaluated cell surface expression of EGFR in human specimens of flat adenomas excised from the proximal colon to assess future clinical relevance. These lesions are known to have unique and more aggressive biology.10 On immunohistochemistry with anti-EGFR, Supplementary Figure S8A,B shows no staining from representative sections of normal and hyperplastic polyps. Supplementary Figure S8C,D shows intense cell surface staining (arrows) for SSA and adenoma (dysplasia). Supplementary Figure S8E shows that EGFR is overexpressed in proximal lesions by greater than four-fold for dysplasia (n=26) and greater than three-fold for SSA (n=6) compared with normal (n=13) and hyperplastic polyps (n=12). Supplementary Figure S8F shows that EGFR can be used to distinguish premalignant (dysplasia and SSA) from benign (hyperplastic polyps and normal) colon in the proximal colon with 94% sensitivity and 92% specificity.
DISCUSSION
We have identified the QRHKPRE peptide that binds to domain 2 of EGFR. This location has a β-hairpin that forms dimers rather than stimulate mitogenic activity, which occurs when EGF docks between domains 1 and 3.14 We confirmed lack of cytoplasmic signaling with peptide binding in HT29 cell lines, Supplementary Figure S4. The ECD of EGFR for human and mouse has 97.5% homology, and it is 100% conserved in domain 2.42 We used a structural model to optimize the sequence to achieve high binding affinity of kd=50 nm. In addition, binding occurs rapidly <2.5 min (k=0.406/min) with topical administration. This time scale is compatible with clinical use during colonoscopy. We demonstrated in vivo use of this peptide to detect flat and polypoid colonic adenomas that were diagnosed as low-grade dysplasia on pathology in a spontaneous mouse model of CRC. The flat lesions were not seen on white-light endoscopy. Furthermore, this peptide was found to bind human colonic dysplasia with 90% sensitivity and 93% specificity.
These results were confirmed on immunohistochemistry using a known antibody, and they support the development of EGFR as an imaging target for early detection of premalignant colonic lesions that may otherwise go undetected on conventional white-light colonoscopy. With topical administration, peptides can be delivered in high concentrations to the mucosa at a risk of harboring disease to maximize binding interactions and to achieve high image contrast with little risk for toxicity. This approach results in a rapid binding with minimal background, and it avoids undesired biodistribution of the exogenous agent to other tissues, such as what occurs with the intravenous administration. Because of their small size, peptides have reduced immunogenicity and lower large-scale production costs. Peptides specific for EGFR have been previously developed for use as therapeutic agents with systemic administration for metastatic disease.43, 44, 45
For in vivo imaging, we used a mouse model that spontaneously develops colonic adenomas that may have either flat or polypoid architecture. We performed repetitive imaging using a near-infrared fluorescence endoscope to localize the premalignant lesions. Cy5.5 was used because this fluorophore emits in a spectral regime that is less sensitive to hemoglobin absorption and tissue scattering, minimizes background from tissue autofluorescence, and provides the maximum light penetration depth.40 We confirmed expression of EGFR in dysplastic mouse crypts on immunohistochemistry using two validated antibodies. Imaging of EGFR has been performed previously in a mouse orthotopic xenograft model of CRC. Human recombinant EGF was labeled with IRDye 800 CW (N-hydroxysuccinimide ester), and it was shown to bind to a mouse xenograft tumor that overexpressesed EGFR on whole-body fluorescence imaging. Peak signal was reached 2 days after injection.46 In vivo imaging has also been performed with a handheld confocal endomicroscope in the cecum of a xenograft mouse model using an FITC-labeled anti-EGFR antibody by exposing the tumor with an abdominal incision.47 For future clinical imaging, a wide-field endoscope that is sensitive to fluorescence can be used, and it may be able to distinguish sporadic and SSAs from hyperplastic polyps based on the EGFR expression level. We expect to find a much higher EGFR expression level in human adenomas, as shown in Figure 6.
We have previously demonstrated a peptide VRPMPLQ that was identified using human biopsy specimens for selection with phage display. This peptide was labeled with FITC and used to detect human dysplastic colorectal polyps in vivo with confocal endomicroscopy.28 Because this peptide was selected empirically, the target is unknown and its clinical use may not be widely generalizable. By comparison, EGFR is a known target that is overexpressed by many cancers of epithelial origin, including lung,48 breast,49 pancreas,50 head and neck,51 and esophagus.52 This peptide is promising for in vivo use as an imaging agent to target premalignant lesions in the proximal colon that are flat in appearance and go undetected on conventional white-light colonoscopy that may otherwise lead to preventable cancers, and may also have broad use for early cancer detection in other imaging applications.
Study Highlights

Acknowledgments
This study was funded in part by National Institutes of Health U54 CA163059, R01 CA142750, U54 CA13642, and P50 CA93990 (TDW).
Footnotes
Supplementary Information accompanies this paper on the Clinical and Translational Gastroenterology website (http://www.nature.com/ctg)
Guarantor of the article: Thomas D. Wang, MD, PhD.
Specific author contributions: JZ, BPJ, ERF, and TDW designed the research; JZ, BPJ, AP, XD, and TDW performed the research; JZ, BPJ, XD, and TDW contributed new reagents or analytic tools; JZ, BPJ, LZ, RK, SRO, and TDW analyzed the data; JZ, BPJ, and TDW wrote the manuscript. JZ, BPJ, and TDW are co-inventors on a patent disclosure submitted to the University of Michigan on the peptide presented in this manuscript.
Financial support: None.
Supplementary Material
References
- Ferlay J, Soerjomataram I, Ervik M et al. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No 11 [Internet]Lyon, France: International Agency for Research on Cancer, 2013. [Google Scholar]
- Ferlay J, Shin HR, Bray F et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127: 2893–2917. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE et al. Cancer genome landscapes. Science 2013; 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeff LC, Richards TB, Shapiro JA et al. How many endoscopies are performed for colorectal cancer screening? Results from CDC's survey of endoscopic capacity. Gastroenterology 2004; 127: 1670–1677. [DOI] [PubMed] [Google Scholar]
- Heresbach D, Barrioz T, Lapalus MG et al. Miss rate for colorectal neoplastic polyps: a prospective multicenter study of back-to-back video colonoscopies. Endoscopy 2008; 40: 284–290. [DOI] [PubMed] [Google Scholar]
- Leufkens AM, van Oijen MG, Vleggaar FP et al. Factors influencing the miss rate of polyps in a back-to-back colonoscopy study. Endoscopy 2012; 44: 470–475. [DOI] [PubMed] [Google Scholar]
- Rex DK, Cutler CS, Lemmel GT et al. Colonoscopic miss rates of adenomas determined by back-to-back colonoscopies. Gastroenterology 1997; 112: 24–28. [DOI] [PubMed] [Google Scholar]
- Muto T, Kamiya J, Sawada T et al. Small "flat adenoma" of the large bowel with special reference to its clinicopathologic features. Dis Colon Rectum 1985; 28: 847–851. [DOI] [PubMed] [Google Scholar]
- Rembacken BJ, Fujii T, Cairns A et al. Flat and depressed colonic neoplasms: a prospective study of 1000 colonoscopies in the UK. Lancet 2000; 8 355 1211–1214. [DOI] [PubMed] [Google Scholar]
- Voorham QJ, Carvalho B, Spiertz AJ et al. Chromosome 5q loss in colorectal flat adenomas. Clin Cancer Res 2012; 18: 4560–4569. [DOI] [PubMed] [Google Scholar]
- Soetikno RM, Kaltenbach T, Rouse RV et al. Prevalence of nonpolypoid (flat and depressed) colorectal neoplasms in asymptomatic and symptomatic adults. JAMA 2008; 299: 1027–1035. [DOI] [PubMed] [Google Scholar]
- Mäkinen MJ. Colorectal serrated adenocarcinoma. Histopathology 2007; 50: 131–150. [DOI] [PubMed] [Google Scholar]
- Citri A, Yarden Y. EGF-ERBB signalling: towards the systems level. Nat Rev Mol Cell Biol 2006; 7: 505–516. [DOI] [PubMed] [Google Scholar]
- Ogiso H, Ishitani R, Nureki O et al. Crystal structure of the complex of human epidermal growth factor and receptor extracellular domains. Cell 2002; 110: 775–787. [DOI] [PubMed] [Google Scholar]
- Bianco R, Gelardi T, Damiano V et al. Rational bases for the development of EGFR inhibitors for cancer treatment. Int J Biochem Cell Biol 2007; 39: 1416–1431. [DOI] [PubMed] [Google Scholar]
- Van Cutsem E, Köhne CH, Hitre E et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 2009; 360: 1408–1417. [DOI] [PubMed] [Google Scholar]
- Seymour MT, Brown SR, Middleton G et al. Panitumumab and irinotecan versus irinotecan alone for patients with KRAS wild-type, fluorouracil-resistant advanced colorectal cancer (PICCOLO): a prospectively stratified randomised trial. Lancet Oncol 2013; 14: 749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano JP, Lagorce C, Atlan D et al. Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann Oncol 2005; 16: 102–108. [DOI] [PubMed] [Google Scholar]
- Porebska I, Harlozińska A, Bojarowsi T. Expression of the tyrosine kinase activity growth factor receptors (EGFR, ERB B2, ERB B3) in colorectal adenocarcinomas and adenomas. Tumour Biol 2000; 21: 105–115. [DOI] [PubMed] [Google Scholar]
- Bansal A, Liu X, McGregor DH et al. Correlation of epidermal growth factor receptor with morphological features of colorectal advanced adenomas: a pilot correlative case series. Am J Med Sci 2010; 340: 296–300. [DOI] [PubMed] [Google Scholar]
- Flora M, Piana S, Bassano C et al. Epidermal growth factor receptor (EGFR) gene copy number in colorectal adenoma-carcinoma progression. Cancer Genet 2012; 205: 630–635. [DOI] [PubMed] [Google Scholar]
- Rego RL, Foster NR, Smyrk TC et al. Prognostic effect of activated EGFR expression in human colon carcinomas: comparison with EGFR status. Br J Cancer 2010; 102: 165–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichera A, Little N, Jagadeeswaran S et al. Epidermal growth factor receptor signaling is required for microadenoma formation in the mouse azoxymethane model of colonic carcinogenesis. Cancer Res 2007; 67: 827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty U, Sehdev A, Cerda S et al. Epidermal growth factor receptor controls flat dysplastic aberrant crypt foci development and colon cancer progression in the rat azoxymethane model. Clin Cancer Res 2008; 14: 2253–2262. [DOI] [PubMed] [Google Scholar]
- Liu J, Zuo X, Li C et al. In vivo molecular imaging of epidermal growth factor receptor in patients with colorectal neoplasia using confocal laser endomicroscopy. Cancer Lett 2013; 330: 200–207. [DOI] [PubMed] [Google Scholar]
- Mitsunaga M, Kosaka N, Choyke PL et al. Fluorescence endoscopic detection of murine colitis-associated colon cancer by topically applied enzymatically rapid-activatable probe. Gut 2013; 62: 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird-Lieberman EL, Neves AA, Lao-Sirieix P et al. Molecular imaging using fluorescent lectins permits rapid endoscopic identification of dysplasia in Barrett's esophagus. Nat Med 2012; 18: 315–321. [DOI] [PubMed] [Google Scholar]
- Hsiung PL, Hardy J, Friedland S et al. Detection of colonic dysplasia in vivo using a targeted heptapeptide and confocal microendoscopy. Nat Med 2008; 14: 454–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm MB, Joshi BP, Lu S et al. Targeted imaging of esophageal neoplasia with a fluorescently labeled peptide: first-in-human results. Sci Transl Med 2013; 5: 184ra61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SJ, Lee CM, Joshi BP et al. Targeted detection of murine colonic dysplasia in vivo with flexible multispectral scanning fiber endoscopy. J Biomed Opt 2012; 17: 021103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padgett KA, Sorge JA. Creating seamless junctions independent of restriction sites in PCR cloning. Gene 1996; 168: 31–35. [DOI] [PubMed] [Google Scholar]
- Li M, Anastassiades CP, Joshi BP et al. Affinity peptide for targeted detection of dysplasia in Barrett's esophagus. Gastroenterology 2010; 139: 1472–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields GB, Noble RL. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int J Pept Protein Res 1990; 35: 161–214. [DOI] [PubMed] [Google Scholar]
- Thomas R, Chen J, Roudier MM et al. In vitro binding evaluation of 177Lu-AMBA, a novel 177Lu-labeled GRP-R agonist for systemic radiotherapy in human tissues. Clin Exp Metastasis 2009; 26: 105–119. [DOI] [PubMed] [Google Scholar]
- Joshi BP, Liu Z, Elahi SF et al. Near-infrared-labeled peptide multimer functions as phage-mimic for high affinity, specific targeting of colonic adenomas in vivo. Gastrointest Endosc 2012; 76: 1197–1206 e1-e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoi T, Akyol A, Theisen BK et al. Mouse model of colonic adenoma-carcinoma progression based on somatic Apc inactivation. Cancer Res 2007; 67: 9721–9730. [DOI] [PubMed] [Google Scholar]
- Liu Z, Miller SJ, Joshi BP et al. Targeted wide-field fluorescence imaging of colonic dysplasia with near-infrared octapeptide. Gut 2013; 62: 395–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsalaki E, Stark A, García-Urdiales E et al. Accurate prediction of peptide binding sites on protein surfaces. PLoS Comput Biol 2009; 5: e1000335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macindoe G, Mavridis L, Venkatraman V et al. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res 2010; 38: W445–W449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, Zhang E, Su Y et al. A review of NIR dyes in cancer targeting and imaging. Biomaterials 2011; 32: 7127–7138. [DOI] [PubMed] [Google Scholar]
- Rowan AJ, Lamlum H, Ilyas M et al. APC mutations in sporadic colorectal tumors: A mutational "hotspot" and interdependence of the "two hits". Proc Natl Acad Sci USA 2000; 97: 3352–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Yoon S, Kim KL et al. Computational Design of Binding Proteins to EGFR Domain II. PLoS One 2014; 9: e92513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai S, Duan J, Liu X et al. Biological evaluation of a novel doxorubicin-peptide conjugate for targeted delivery to EGF receptor-overexpressing tumor cells. Mol Pharm 2011; 8: 375–386. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhao R, Wu X et al. Identification and characterization of a novel peptide ligand of epidermal growth factor receptor for targeted delivery of therapeutics. FASEB J 2005; 19: 1978–1985. [DOI] [PubMed] [Google Scholar]
- Song S, Liu D, Peng J et al. Novel peptide ligand directs liposomes toward EGF-R high-expressing cancer cells in vitro and in vivo. FASEB J 2009; 23: 1396–1404. [DOI] [PubMed] [Google Scholar]
- Cohen G, Lecht S, Arien-Zakay H et al. Bio-imaging of colorectal cancer models using near infrared labeled epidermal growth factor. PLoS One 2012; 7: e48803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Ziebart A, Foersch S et al. In vivo molecular imaging of colorectal cancer with confocal endomicroscopy by targeting epidermal growth factor receptor. Gastroenterology 2010; 138: 435–446. [DOI] [PubMed] [Google Scholar]
- Hirsch FR, Varella-Garcia M, Bunn PA Jr et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol 2003; 21: 3798–3807. [DOI] [PubMed] [Google Scholar]
- Bhargava R, Gerald WL, Li AR et al. EGFR gene amplification in breast cancer: correlation with epidermal growth factor receptor mRNA and protein expression and HER-2 status and absence of EGFR-activating mutations. Mod Pathol 2005; 18: 1027–1033. [DOI] [PubMed] [Google Scholar]
- Jimeno A, Tan AC, Coffa J et al. Coordinated epidermal growth factor receptor pathway gene overexpression predicts epidermal growth factor receptor inhibitor sensitivity in pancreatic cancer. Cancer Res 2008; 68: 2841–2849. [DOI] [PubMed] [Google Scholar]
- Reuter CW, Morgan MA, Eckardt A. Targeting EGF-receptor-signalling in squamous cell carcinomas of the head and neck. Br J Cancer 2007; 96: 408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawa M, Suzuki S, Dobashi Y et al. EGFR protein overexpression and gene amplification in squamous cell carcinomas of the esophagus. Int J Cancer 2006; 118: 1173–1180. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.