Abstract
Estrogen-receptor-positive (ER+) tumors employ complex signaling that engages in crosstalk with multiple pathways through genomic and non-genomic regulation. A greater understanding of these pathways is important for developing improved biomarkers that can better determine treatment choices, risk of recurrence and cancer progression. Deficiencies in DNA repair capacity (DRC) is a hallmark of breast cancer (BC); therefore, in this work we tested whether ER signaling influences DRC. We analyzed the association between ER positivity (% receptor activation) and DRC in 270 BC patients, then further stratified our analysis by HER2 receptor status. Our results show that among HER2 negative, the likelihood of having low DRC values among ER- women is 1.92 (95% CI: 1.03, 3.57) times the likelihood of having low DRC values among ER+ women, even adjusting for different potential confounders (p<0.05); however, a contrary pattern was observed among HER2 positives women. In conclusion, there is an association between DRC levels and ER status, and this association is modified by HER2 receptor status. Adding a DNA repair capacity test to hormone receptor testing may provide new information on defective DNA repair phenotypes, which could better stratify BC patients who have ER+ tumors. ER+/HER2- tumors are heterogeneous, incompletely defined, and clinically challenging to treat; the addition of a DRC test could better characterize and classify these patients as well as help clinicians select optimal therapies, which could improve outcomes and reduce recurrences.
Introduction
Worldwide, breast cancer (BC) is a growing health problem, both in its increasing incidence [1] and resistance to treatment [2]. Between 2008 and 2013, the worldwide incidence of BC rose by 20% [3]. Additionally, 30% of all estrogen-receptor-positive (ER+) breast tumors exhibit de novo resistance, and 40% of patients who initially respond to treatment will acquire resistance despite remaining ER+ [4,5].
ER status is an established therapeutic and prognostic biomarker for BC [6–10]. Although hormone receptor status is a mainstay for molecular and clinicopathological classification of BC [11–15], it provides an incomplete molecular model for classifying BC tumors. Although significant progress has been made in the clinical management of ER+ tumors, prevailing challenges are associated with their molecular heterogeneity, resistance to standard endocrine therapy, and the risk of late recurrence. These challenges are driving intense research efforts to find predictors of chemosensitivity in ER+ BC.
The majority of BC deaths ultimately occur in ER+ women. ER signaling is complex and highly regulated; it affects both proliferation and survival [16]. Understanding the scope and influence of ER signaling is challenging—including how it affects therapeutic response. Approximately 75% of all breast tumors are ER+ [17–21], but only about half of all ER+ tumors respond to anti-estrogen therapy [19,22–24]. Additionally, only 20% of those tumors stop expressing ER when they become endocrine resistant [25,26]. There is therefore an urgent need to find effective interventions [27,28].
ER signaling works through both genomic and non-genomic mechanisms that engage in crosstalk [25,29]. ER signaling has a propensity to increase proliferation, which can lead to mutations when DNA repair is dysregulated. Such dysregulation is a hallmark of breast carcinogenesis [30–34] and confers a phenotype of increased cellular division or decreased apoptosis [35]. Indeed, recent studies have documented interactions between ER and DNA damage response/repair [36,37]. Moreover, our previous work showed that DNA repair capacity (DRC) in BC patients is approximately half that of women without the disease (p<0.001) [38], and deficiency in the nucleotide excision repair (NER) pathway figures prominently in the pathobiology of sporadic BC [39].
The relationship between ER activation/signaling and DNA damage/repair is only starting to be investigated. This manuscript: (1) explains our recent findings regarding the association between ER status and the defective DNA repair phenotype, controlling for different variables including HER2 receptor status [38,39], (2) outlines potential mechanisms behind what we found for future mechanistic studies, and (3) proposes how our findings could form the basis for a new combination of biomarker testing that may better characterize BC tumors, which could improve diagnostic, prognostic, and therapeutic efforts.
Results
Study population
The average age of women with BC was 56.4 years (±12.2) at the time of initial diagnosis. All of the women were untreated BC cases. Approximately, 18.5% were younger than 45 years and 24.4% were older than 65 years. Nearly 25% of the women with BC studied reported having consumed multivitamins in the last 5 years, 17.6% reported having consumed calcium in the last 5 years. Approximately 65% were menopausal women, and 24.8% of patients had a family history of BC. About 52% (n = 127) of the women studied with BC had grade II tumors, followed by 36% of the women with grade III tumors (n = 88), only 13% (n = 31) had grade I tumors (Table 1).
Table 1. Characteristics of study group consisting of women with breast cancer.
Women with BC history, menopausal and their tumor grade and receptor status (estrogen, HER2, progesterone) classification expressed as numerical and % values (n = 270).
| Variables | Summary measures(N = 270) |
|---|---|
| N (%) | |
| DRC (%)* | 3.05 ± 2.8 |
| Median DRC (P25, P75) | 2.1 (1.24%, 4.07%) |
| DRC (tertiles) | |
| < 1.6% | 90 (33.3) |
| 1.6–3.2% | 90 (33.3) |
| > 3.2% | 90 (33.3) |
| Age | |
| < 45 | 50 (18.5) |
| 46–55 | 79 (29.3) |
| 56–65 | 75 (27.8) |
| > 65 | 66 (24.4) |
| Median Age (SD) | 56.4 ± 12.2 |
| History of breast cancer | |
| Yes | 67 (24.8) |
| No | 203 (75.2) |
| Menopausal status | |
| Yes | 174 (64.4) |
| No | 96 (35.6) |
| Multivitamin consumption in the last 5 years | |
| Yes | 66 (25.2) |
| No | 204 (74.8) |
| Calcium consumption in the last 5 years | |
| Yes | 46 (17.6) |
| No | 224 (82.4) |
| Tumor grade | |
| I | 31 (12.6) |
| II | 127 (51.6) |
| III | 88 (35.8) |
| Missing | 24 |
| Receptor status | |
| Estrogen Receptor | |
| Negative | 73 (27.0) |
| Positive | 197 (73.0) |
| HER2 Receptor | |
| Negative | 206 (76.3) |
| Positive | 64 (23.7) |
| Progesterone Receptor | |
| Negative | 100 (37.0) |
| Positive | 170 (63.0) |
* Value of % DNA repair capacity (DRC) measured in lymphocytes expressed as mean (), ± 1 standard deviation (SD) and median values including percentile 25 (P25) and percentile 75 (P75).
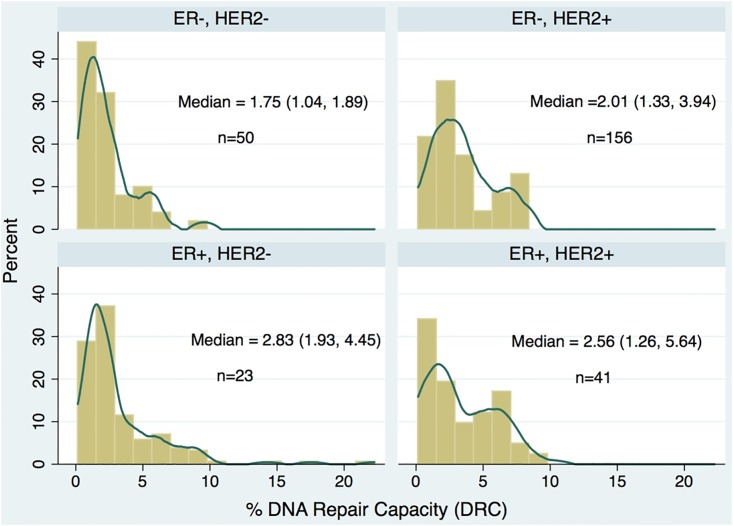
The overall mean value DRC was 3.0% (±2.8% S.D.) and the median DRC value was 2.1%, with an interquartile range of 2.8% (75th percentile minus 25th percentile) (Table 1). Nearly 73% of the BC patients had tumors classified as ER positive, 23.7% had HER2 positive tumors and 63% were progesterone positive (Table 1). When the DRC distribution was described by HER2 (+,-) and ER (+,-) status among women with BC, different patterns were observed. In all four combinations of the categories of these two receptors, the DRC values showed a positive skew (were highly concentrated at low values) (Fig 1). The highest median DRC value (2.83%) was in the ER-HER2+ group, the ER-HER2- group had a DRC value (1.75%), representing a reduction of approximately 38%. However, among HER2 negatives, a higher number of women had low DRC values, independently of the ER status (Fig 1).
Fig 1. Distribution of levels of % DNA repair capacity (DRC) in 270 women with breast cancer separated in four groups according to estrogen receptor (ER) and HER2 status.
The height of the rectangle denotes the percentage of women with a specific %DRC level. The line denotes the Kernel density estimation (KDE) to show the smoothing distribution of the %DRC. Between parentheses indicates percentile 25 and percentile 75.
Ordinal Logistic regression model
The ordinal logistic model was utilized to analyze the data with different cutoff points in the DRC according to the HER2 status. This was done in order to more precisely assess the relationship between DRC expressed as a categorical variable in tertiles and ER status. Different patterns were observed, as presented in Table 2. These were divided into two groups:
Table 2. Magnitude of the association between tertiles of levels % DNA repair capacity (DRC) according to estrogen receptor (ER) and HER2 receptor status (n = 270).
| HER2 | ER | %DRC | Adjusted# | Adjusted# | Adjusted# | ||
|---|---|---|---|---|---|---|---|
| <1.6% | 1.6–3.2% | >3.2% | |||||
| Negative | Negative | 24 | 14 | 12 | 2.23* (1.13, 4.38) | 1.42 (.67, 2.99) | 1.92* (1.03, 3.57) |
| (n = 206) | Positive | 46 | 60 | 50 | 1 | 1 | 1 |
| Positive | Negative | 5 | 8 | 10 | 0.38 (0.11, 1.32) | 0.92 (0.31,2.73) | 0.67 (0.25,1.79) |
| (n = 64) | Positive | 15 | 8 | 18 | 1 | 1 | 1 |
| Total | Negative | 29 | 22 | 22 | 1.48 (0.84, 2.61) | 1.26 (0.70, 2.27) | 1.38 (0.83, 2.29) |
| (n = 270) | Positive | 61 | 68 | 68 | 1 | 1 | 1 |
#Adjusted for menopausal status, breast cancer history age, multivitamin and/or calcium consumption in the last 5 years
*P-value < 0.05
(1) Only 1 OR is reported because the proportional odds assumption was met (p>0.05).
Women with HER2 negative breast cancer tumors
The results indicate that the likelihood of having a DRC below 1.6% among women with ER- tumors is 2.23 (95% CI: 1.13, 4.68) times the likelihood of having a DRC below 1.6% among ER+ women. After adjusting for different potential confounders, this excess likelihood was statistically significant (p<0.05). When the cutoff point of the DRC of 3.2% is used (Table 2), the likelihood of having a DRC below 3.2% among ER- women is 1.42 (95% CI: 0.67, 2.99) times the likelihood of having a DRC below 3.2% among ER+ women, even adjusting for different potential confounders; however, this excess was not statistically significant (p>0.05). When we assessed the proportional odds assumption of the ordinal logistic model, the results were not significant (p> 0.05); hence, only one OR can be estimated without depending on the cutoff point. Therefore, the results indicate that the likelihood of having low DRC values among ER- women is 1.92 (95% CI: 1.03, 3.57) times the likelihood of having low DRC values among ER+ women, even adjusting for different potential confounders; this excess likelihood was statistically significant (p<0.05).
Women with HER2 positive breast cancer tumors
Women with HER2 BC tumors showed the opposite pattern, the likelihood of having low DRC values among ER- women is 33% (OR: 0.67, 95% CI: 0.25, 1.79) lower than the likelihood of having low DRC values among ER+ women, even adjusting for different potential confounders; however, this reduction was not statistically significant (p>0.05).
Discussion
We found that DRC and ER levels are associated and that this association is modified by HER2 receptor status in women whose BC tumors are HER2-. This association suggests that DRC and ER levels are linked: as DRC levels increase, ER levels also increase, and vice versa. Although previous studies have established that a defective DNA repair phenotype is commonly found in women with BC [38–40], to our knowledge, this is the first study to show that ER status is associated with a defective DNA repair phenotype. Our previous study (Matta et al. 2012) using a case control design with 824 women showed the usefulness of DRC level as a measure of BC risk. For every percent unit of decrease in DRC, there is 64% more likelihood of having BC. Thus, the subtle differences in DRC in women with BC divided into the four groups presented in Fig 1, provide additional evidence on how DRC is associated with BC risk. For example, the two groups with the lowest DRC (ER-/HER2-, ER+/HER2-) represented 76% of the study group comprised of 270 women. DNA repair genes, particularly those involved in base excision repair may have a significant effect on estrogen-driven breast cancer specific survival (BCSS) as recently shown by Abdel-Fatah et al. (2014). They studied 1,406 women with ER positive early stage BCs with 20 years long term clinical follow-up data. Multivariate Cox proportional hazards model was used to calculate a DNA repair prognostic index and correlated to clinicopathological variables and survival outcomes. Key base excision repair proteins including XRCC1, APE1, SMUG1 and FEN1 were independently associated with poor BCSS [36].
Background related to findings
Most ER+ breast tumors are classified as luminal [13–15,41]. Luminal A is characterized by ER+/PR+/HER2-/low Ki-67 [42] and typically responds well to endocrine therapy [28]. In contrast, Luminal B is either HER2+ or HER2- but is ER+/PR+/high Ki-67 [42]; such tumors are complex diagnostically and therapeutically [28]. Luminal A tumors are considered as more differentiated, indolent, and sensitive to endocrine therapy, whereas Luminal B tumors are more aggressive and resistant to endocrine therapy [43]. Indeed, Lips’ 2012 analysis of biopsies from 211 treatment-naïve ER+/HER2- primary breast tumors could not define a subgroup that would be most likely to benefit from neoadjuvant therapy, much less predict chemosensitivity or treatment outcome. Biopsies of the same patients taken post-treatment but prior to surgery were equally unrevealing [41].
Despite today’s availability of comprehensive gene signature assays (see review by Issa et al. 2014) and our increasing knowledge of which genes are dysregulated in BC and how, the information has not always resulted in increased clinical utility or better prognostic tools [44]. Santarpia’s 2013 research on a 145-gene expression profile to try to characterize ER+/HER2- subgroups yielded interesting but inconclusive information [37]. But problems exist even with genes that have been studied in depth for years. For example, EGFR is amplified in many BCs, and its crosstalk with ER signaling is well established. Yet large randomized trials of treatment-naïve ER+/HER- patients did not respond any better to treatment when an EGFR signaling inhibitor was coupled with an aromatase inhibitor [41]. Similar results occurred in studies using a fibroblast growth factor receptor inhibitor [41]. ASCO’s 2014 guidelines acknowledge this knowledge gap and note that no optimal first- or second-line treatment exists for advanced ER+/HER2- BC [8]. Collectively this underscores the need for better molecular classification [19,23,41,45], and ER+/HER2- breast tumors may be the “canary in the coal mine” pointing us to a more effective predictive model: a combination of receptor assays and DRC. Even though research has made great strides in the molecular characterization of DNA repair pathways—and efforts to map NER and two other pathways were awarded 2015 Nobel Prizes in Chemistry—our knowledge still remains incomplete. Because of this we propose that the combination of molecular signatures data, including the DNA repair prognostic index recently developed by Abdel-Fatah et al. (2014), with DNA repair phenotypic data can enable more accurate diagnostic and therapeutic decisions [36].
Biological plausibility of our findings
In healthy people, ER signaling generally downregulates DNA damage response (DDR) [46] while simultaneously promoting proliferation [46,47]. Seventy percent of ER activity is governed by highly regulated genetics that keep ER’s influence in check [23]. However, dysregulated ER signaling confers a phenotype of increased proliferation or decreased apoptosis [35]. Sustained ER signaling permits accumulation of low-level DNA damage, an event documented early in tumorigenesis [48].
ER activation leads to transcriptional responses both in genes with and without estrogen response elements. Additionally, ER activation can elicit non-transcriptional cellular responses, all of which favor cell proliferation and survival [21,49]. This diversity points to traditional (canonical) genomic signaling in the nucleus, as well as plasma-membrane-bound (non-canonical) signaling. The former involves ER binding directly to a cognate ligand on the promoter region of a gene. In the latter, ER “tethers” to “pioneer factors” that enable ER to influence pathways that eventually act on downstream target transcription factors (such as ER’s interactions with VEGF and AP-1) [21]. Both types of activities result in altered gene expression, and both can lead to point mutations and uncontrolled cellular proliferation—despite the former being genomic while the latter is not [45]. More importantly, sustained ER signaling can lead to greater expression of proliferation-related genes that may rely less on expression of traditional ER genes but more on alternate pathways for activation and tumor growth [28].
Our finding that women with BC and ER negative tumors have lower DRC values among HER2- seems counterintuitive at first. However, this molecular response may be an early event in tumorigenesis—perhaps ER’s attempt to induce DNA repair to offset increasing replication errors of early mutagenesis.
This raises the question of what factor(s) could modulate that association, and how might that affect treatment options? If such factors could be determined, more effective biomarker panels and therapeutic regimens could be devised. Although the DRC assay is currently used primarily as a method of estimating BC risk [38], its combination with hormone receptor status as described here may expand its applications.
Despite the variety of gene signature assays available today, a 2014 analysis [50] shows that even the most comprehensive assay is not cost-effective in pinpointing optimal adjuvant treatment or outcomes for ER+ BCs [50]. New combinations of molecular markers such as DRC plus hormone receptor status may allow better tumor differentiation within the same molecular subtype—and lead to more effective treatment.
Why only ER+/HER2- tumors?
Breast cells with estrogen-induced DNA damage can either delay damage response/repair or can promote cell survival at the expense of repair [51]. So why this DRC/ER relationship would be detectable only in the ER+/HER2- subpopulation is a tantalizing question.
ER appears to influence virtually every DNA repair pathway [46,48,52–54]. While direct evidence for some of those pathways is sparse, dysfunction and downregulation of the NER pathway is prominent and well-documented in BCs [38–40]. Abnormal expression of two NER proteins (RPA, a damage sensor; and PCNA, a progressivity factor) is associated with ER+/HER2- breast tumors [37].
Two linchpins for ER’s influence on DNA repair pathways may be p53 and PI3K. ER appears to have a bidirectional, yin-yang relationship with p53, which influences many repair pathways. But more evidence for how ER can shift repair signaling appears to be associated with the PI3K pathway—a superhighway for survival signaling [26,55,56]. Hyperactivation of the PI3K-Akt-mTOR pathway is common in BC [57]. Dysregulated expression of growth factor receptors including ER can activate the Ras/Raf/MEK/ERK pathway, Ras/PI3K/PTEN/Akt/mTOR and other signaling pathways, creating a self-sustaining feed-forward loop of persistent signaling [58], which could lead to resistance to hormonal therapy [59]. S1 Fig depicts our vision of how estrogen affects DNA repair.
Strengths and limitations of this study
To our knowledge, we are the first to explore the relationship between DRC and ER in this manner. Additionally, we believe we are the first to explore how HER2 status affects the DRC/ER relationship. This novel approach has great potential utility for personalized medicine. For example, with ER emerging as a contributor to dysregulated DNA repair; it makes sense to include testing for a DNA repair phenotype along with hormone receptor status testing. Based on our results, our proposal using these two types of testing in combination can provide further molecular characterization of breast tumors—with possible insights into better treatment choices—particularly in the more clinically challenging BCs including, but not limited to ER+/HER2- cases. Our study has robust sample size and power [38]; results were consistent after adjustment of different sources of variability (menopausal status, history of BC, and HER2 status) in the ordinal logistic regression model.
Conclusions
The heterogeneity of ER+ tumors, their unequal response to anti-estrogen therapy, and ER’s capacity to influence gene expression collectively point to the need for more comprehensive molecular characterization of BC tumors. In BC, adding a test for DRC to a genome signature allows identification of the defective DNA repair phenotype and may enhance our capacity to predict which Luminal B tumors are likely to be more aggressive. This allows selection of better treatment regimens, especially for diagnostically and therapeutically complex molecular BC subtypes such as ER+/HER2-. Our findings indicate a previously undetected, subtle difference in ER+ tumors, which could also help explain the lack of correlation between end points among some clinical trials for these tumors. The availability of single-cell and single-molecule assays puts the exciting possibility of better molecular profiling within our grasp.
Methods
Patient Recruitment
The study was approved by the Ponce Health Sciences University Institutional Review Board (IRB #120207-JM). Patients for this study were selected from our larger BC study (1,181 patients and controls; recruited 2006–2012), which has been previously described [38,60,61]. The cases selected for this study were women with primary diagnosis of BC and were recruited from gynecological and oncology clinics throughout Puerto Rico.
Inclusion criteria were patients who: (1) were recently diagnosed histopathologically with primary BC, (2) were treatment-naïve (had not received chemotherapy, blood transfusions, or radiotherapy), and (3) had pathology reports that included hormone receptor information. We obtained consent forms that allowed us to interview the participants, obtain blood samples, and review their pathology reports. Among the cases, 344 women were eligible to participate in the study; however, only 270 had complete hormone receptor status information. Therefore, our final study group comprised 270 BC cases.
DNA Repair Capacity (DRC) Measurements
The host cell reactivation (HCR) assay with a luciferase reporter gene that we utilized to measure DRC levels in lymphocytes has been described in previously published molecular epidemiological studies of cancer [40,62–68]. We have previously published details about the variability, stability of plasmid transfection and differences between cryopreserved versus fresh blood samples for the HCR [38]. This assay measures the total DRC of transfected lymphocytes. Results reflect the host cells’ overall repair capacity, although HCR primarily detects activity of the nucleotide excision repair (NER) pathway [38].
Blood collection and isolation of lymphocytes from women
We used peripheral blood samples to obtain lymphocytes, which were separated, purified and grown from each patient sample [38]. Those cells were used as surrogate markers of the patients’ overall DRC [69,70]. Approximately 30 mL of peripheral blood was obtained from each participant and stored in heparinized tubes. The lymphocytes were then isolated by the Ficoll gradient technique and suspended in 2 mL of freezing media containing 10% dimethyl sulfoxide, 40% RPMI 1640 medium, 50% fetal bovine serum, and 1% antibiotic/antimycotic. Aliquots were stored in a −80°C freezer for 1–3 weeks. The lymphocytes were later thawed in batches of 5–7 samples for the HCR assay (details follow). Collection periods were approximately the same for patients and women without BC because recruitment was conducted concurrently.
The HCR was performed on the peripheral blood lymphocytes to measure in vivo DRC, as described in previous studies [38,40,60,61,71]. The late Dr. Lawrence Grossman (Johns Hopkins School of Public Health, Baltimore, MD) provided the luciferase plasmid for the HCR assay and the protocol for its use. A nonreplicating plasmid expression vector (pCMVluc) of 4,863 base pairs was genetically engineered to contain a bacterial luciferase reporter gene that is not present in a mammalian cell. The gene was damaged by ultraviolet C radiation (254 nm) exposure in a controlled, quantitative manner (dose—response curve) so that the level of its expression was a direct measure of the repair capacity of the host mammalian cell. The plasmid construct containing the luciferase gene (LUC) was irradiated at 0, 350, and 700 J/m2 using a 254-nm UVC lamp (38). This plasmid construct and its validation have been described previously [72]. The controlled, quantitative UV exposure produced a dose—response curve so that the level of its expression was a direct measure of the repair capacity of the host mammalian cell. After transfection into lymphocytes, repair-transcription-blocking damage was introduced exogenously on foreign DNA; then overall DRC was measured via HCR [71]. This approach measured the unaffected phenotype, which reflects the cells’ inherent DRC, measured primarily in terms of their NER activity [71]. Keeping the time constant for the lymphocytes to complete the repair mirrored the true cellular process [70].
To calculate the DRC, undamaged plasmid DNA was compared to repair of in-vitro-damaged plasmid DNA; the results were expressed as the percentage of residual luciferase reporter gene expression (% luciferase activity in luminescence units). The amount of gene expression reflected DRC, expressed as a percentage. A detailed description of the assay, including separation of DRC by tertiles, is found in Matta et al. 2012 [38]. This study builds upon our laboratory’s 17 years of experience in performing the HCR assay to measure DRC and its validation of the sensitivity, specificity, and usefulness as a measure of BC risk [38].
Hormone Receptor Status
We reviewed the patients’ medical records to collect receptor status data on estrogen (ER), progesterone (PR) and human epidermal growth factor receptor 2 (HER2). Ten private laboratories in Puerto Rico performed the receptor status assays on patients’ formalin fixed tumor biopsies, using immunohistochemistry (IHC) methods per ASCO (American Society of Clinical Oncology) and CAP (College of American Pathologists) guidelines [10,73]. ER and PR analyses included the percentage of positive-staining cells, the intensity of staining (weak, moderate, or strong), and an interpretation (“receptor positive” meant ≥1% of invasive tumor cells stained positive for ER/PR; “receptor negative” meant <1% of invasive tumor cells stained positive for ER/PR) [10].
HER2 testing also utilized IHC-based assays. All laboratories used FDA-approved IHC assays. When an assay yielded equivocal results (2+ or 1+), Fluorescence In Situ Hybridization (FISH) was used to determine whether those samples were HER2+ or HER2-, based on the quantity of HER2 gene copies per nucleus [10]. For our analysis, we categorized HER2 status as a dichotomous variable of ‘‘positive” (all 3+ results) or ‘‘negative” (2+ to 0 results).
Statistical Analysis
We used descriptive statistics to summarize quantitative variables; while for categorical variables we used percentages in each category of these variables. To assess the possibility of selection bias, a significance test was performed using the normal approach to compare the mean DRC among women with complete (n = 270) versus incomplete (n = 74) receptor status information. The results from this comparison did not show any significant difference (p>0.05).
An ordinal logistic regression model (OLRM) was used to assess the relationship between DRC and ER status [74]. The OLRM is an extension of the logistic regression model that applies to dichotomous dependent variables, allowing for more than two (ordered) response categories. In order to apply this model to our analysis, the DRC was categorized in different groups, using as a cut-off points the observed DRC tertiles (<1.6%, 1.6–3.2%, >3.2%). The expression of this model is as follows:
where P≤k indicates the prevalence of women with DRC equal or below tertile k, P>k indicates the prevalence of women with DRC above tertile k, βEk indicates the coefficient associated to ER when the kth-DRC tertile is used, ER is a dummy variables to indicate the ER status (+, -), and βi indicates the coefficient associated to each potential confounding variables, Ci. This model provides an estimate of the magnitude of the association () between DRC and ER at different cut-off point of the DRC, as follows:
where indicates the standard error of . However, if the assumption of proportional odds ( = ) is met, then only one OR is estimated without depending of the cutoff point of the DRC. Our data showed that this assumption was met (p>0.1); therefore, only one estimation of the OR was reported.
Supporting Information
Estrogen can have a variety of effect on the tumor cells some of which may involve DNA repair signaling specially among HER2(-) tumors. (A) In the canonical pathway, ER binds to the promoter regions of genes involved in cell cycle progression. (B) Non-canonical pathways involve many other signaling cascades such as p53, mTOR and ERK which can also lead to increased proliferation. Both of these pathways can lead to low levels of DNA damage that may “switch on” DNA repair mechanisms (evaluated in in this study in the patients’ circulating lymphocytes).
(DOCX)
Acknowledgments
Special thanks go to Mrs. Lana Christian (CreateWrite Inc.) for her outstanding assistance editing the manuscript and her critical analysis of the manuscript as well. Bob Ritchie (PHSU) provided helped in manuscript formatting. The authors wish to acknowledge Dr. James Robb for the critical review of this manuscript and for valuable feedback from Dr. Timothy Rebbeck, University of Pennsylvania. We also acknowledge the support from: Dr. Diego E. Zavala Zegarra, Dr. Guillermo Tortolero Luna, Naydi Pérez-Ríos, MS, and Carlos R. Torres-Cintrón, MPH from the Puerto Rico Central Cancer Registry for their assistance and access to the pathology reports of participants.
Abbreviations
- BC
breast cancer
- ER
estrogen receptor
- HER2
HER2neu receptor
- DRC
DNA repair capacity
- NER
nucleotide excision repair
- OR
odds ratio
- OLRM
Ordinal logistic regression model
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by grants from the NCI Center to Reduce Health Disparities and NIH-MBRS Program grant # S06 GM008239-20 and 9SC1CA182846-04 to Ponce School of Medicine through Dr. J. Matta and PSM-MCC Partnership grant # 5U54CA163071-04. Carmen Ortiz was supported by MBRS-RISE GM082406 [NIH-NIGMS]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, et al. (2013) GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide:IARC CancerBase No. 11. In: Cancer IAfRo, editor. Lyon, France. [Google Scholar]
- 2.Ross DD, Nakanishi T (2010) Impact of breast cancer resistance protein on cancer treatment outcomes. Methods Mol Biol 596: 251–290. 10.1007/978-1-60761-416-6_12 [DOI] [PubMed] [Google Scholar]
- 3.American Cancer Society (2013) Breast Cancer Facts and Figures 2013-2014. Atlanta: American Cancer Society. [Google Scholar]
- 4.Rahal OM, Simmen RC (2011) Paracrine-acting adiponectin promotes mammary epithelial differentiation and synergizes with genistein to enhance transcriptional response to estrogen receptor beta signaling. Endocrinology 152: 3409–3421. 10.1210/en.2011-1085 [DOI] [PubMed] [Google Scholar]
- 5.Gnant M (2012) Overcoming endocrine resistance in breast cancer: importance of mTOR inhibition. Expert Rev Anticancer Ther 12: 1579–1589. 10.1586/era.12.138 [DOI] [PubMed] [Google Scholar]
- 6.Chen JJ, Wang Y, Xue JY, Chen Y, Chen YL, et al. (2014) A clinicopathological study of early-stage synchronous bilateral breast cancer: a retrospective evaluation and prospective validation of potential risk factors. PLoS One 9: e95185 10.1371/journal.pone.0095185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosa FE, Caldeira JR, Felipes J, Bertonha FB, Quevedo FC, et al. (2008) Evaluation of estrogen receptor alpha and beta and progesterone receptor expression and correlation with clinicopathologic factors and proliferative marker Ki-67 in breast cancers. Hum Pathol 39: 720–730. 10.1016/j.humpath.2007.09.019 [DOI] [PubMed] [Google Scholar]
- 8.Partridge AH, Rumble RB, Carey LA, Come SE, Davidson NE, et al. (2014) Chemotherapy and targeted therapy for women with human epidermal growth factor receptor 2-negative (or unknown) advanced breast cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol 32: 3307–3329. 10.1200/JCO.2014.56.7479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan I, Salazar J, Majem M, Pallares C, Del Rio E, et al. (2014) Pharmacogenetics of the DNA repair pathways in advanced non-small cell lung cancer patients treated with platinum-based chemotherapy. Cancer Lett 353: 160–166. 10.1016/j.canlet.2014.07.023 [DOI] [PubMed] [Google Scholar]
- 10.Hammond ME, Hayes DF, Wolff AC, Mangu PB, Temin S (2010) American society of clinical oncology/college of american pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Oncol Pract 6: 195–197. 10.1200/JOP.777003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang DY, Done SJ, McCready DR, Boerner S, Kulkarni S, et al. (2011) A new gene expression signature, the ClinicoMolecular Triad Classification, may improve prediction and prognostication of breast cancer at the time of diagnosis. Breast Cancer Research 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Belle V, Van Calster B, Brouckaert O, Vanden Bempt I, Pintens S, et al. (2010) Qualitative assessment of the progesterone receptor and HER2 improves the Nottingham Prognostic Index up to 5 years after breast cancer diagnosis. J Clin Oncol 28: 4129–4134. 10.1200/JCO.2009.26.4200 [DOI] [PubMed] [Google Scholar]
- 13.Ellis MJ, Perou CM (2013) The genomic landscape of breast cancer as a therapeutic roadmap. Cancer Discov 3: 27–34. 10.1158/2159-8290.CD-12-0462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prat A, Perou CM (2011) Deconstructing the molecular portraits of breast cancer. Mol Oncol 5: 5–23. 10.1016/j.molonc.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.TCGA (2012) Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70. 10.1038/nature11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osborne CK, Schiff R (2011) Mechanisms of endocrine resistance in breast cancer. Annu Rev Med 62: 233–247. 10.1146/annurev-med-070909-182917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clark GM, Osborne CK, McGuire WL (1984) Correlations between estrogen receptor, progesterone receptor, and patient characteristics in human breast cancer. J Clin Oncol 2: 1102–1109. [DOI] [PubMed] [Google Scholar]
- 18.Nadji M, Gomez-Fernandez C, Ganjei-Azar P, Morales AR (2005) Immunohistochemistry of estrogen and progesterone receptors reconsidered: experience with 5,993 breast cancers. Am J Clin Pathol 123: 21–27. [DOI] [PubMed] [Google Scholar]
- 19.Huynh KT, Chong KK, Greenberg ES, Hoon DS (2012) Epigenetics of estrogen receptor-negative primary breast cancer. Expert Rev Mol Diagn 12: 371–382. 10.1586/erm.12.26 [DOI] [PubMed] [Google Scholar]
- 20.Jemal A, Fedewa SA (2012) Is the prevalence of ER-negative breast cancer in the US higher among Africa-born than US-born black women? Breast Cancer Res Treat 135: 867–873. 10.1007/s10549-012-2214-2 [DOI] [PubMed] [Google Scholar]
- 21.Martin HL, Smith L, Tomlinson DC (2014) Multidrug-resistant breast cancer: current perspectives. Breast Cancer (Dove Med Press) 6: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liang J, Shang Y (2013) Estrogen and cancer. Annu Rev Physiol 75: 225–240. 10.1146/annurev-physiol-030212-183708 [DOI] [PubMed] [Google Scholar]
- 23.Raha P, Thomas S, Munster PN (2011) Epigenetic modulation: a novel therapeutic target for overcoming hormonal therapy resistance. Epigenomics 3: 451–470. 10.2217/epi.11.72 [DOI] [PubMed] [Google Scholar]
- 24.Stone A, Cowley MJ, Valdes-Mora F, McCloy RA, Sergio CM, et al. (2013) BCL-2 hypermethylation is a potential biomarker of sensitivity to antimitotic chemotherapy in endocrine-resistant breast cancer. Mol Cancer Ther 12: 1874–1885. 10.1158/1535-7163.MCT-13-0012 [DOI] [PubMed] [Google Scholar]
- 25.Magnani L, Brunelle M, Gevry N, Lupien M (2012) Chromatin landscape and endocrine response in breast cancer. Epigenomics 4: 675–683. 10.2217/epi.12.64 [DOI] [PubMed] [Google Scholar]
- 26.Musgrove EA, Sutherland RL (2009) Biological determinants of endocrine resistance in breast cancer. Nat Rev Cancer 9: 631–643. 10.1038/nrc2713 [DOI] [PubMed] [Google Scholar]
- 27.Sutherland RL (2011) Endocrine resistance in breast cancer: new roles for ErbB3 and ErbB4. Breast Cancer Res 13: 106 10.1186/bcr2878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang MH, Man HT, Zhao XD, Dong N, Ma SL (2014) Estrogen receptor-positive breast cancer molecular signatures and therapeutic potentials (Review). Biomed Rep 2: 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Becerra R, Santos N, Diaz L, Camacho J (2012) Mechanisms of Resistance to Endocrine Therapy in Breast Cancer: Focus on Signaling Pathways, miRNAs and Genetically Based Resistance. Int J Mol Sci 14: 108–145. 10.3390/ijms14010108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelley M (2012) Introduction and Overview of DNA Repair Targets: from Bench to Clinic In: Kelley MR, editor. DNA Repair in Cancer Therapy: Molecular targets and Clinical Aplications. First ed. United States of America: Elsevier Inc; pp. 1–16. [Google Scholar]
- 31.Annunziata CM, O'Shaughnessy J (2010) Poly (ADP-ribose) polymerase as a novel therapeutic target in cancer. Clin Cancer Res 16: 4517–4526. 10.1158/1078-0432.CCR-10-0526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tobin LA, Robert C, Nagaria P, Chumsri S, Twaddell W, et al. (2012) Targeting abnormal DNA repair in therapy-resistant breast cancers. Mol Cancer Res 10: 96–107. 10.1158/1541-7786.MCR-11-0255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alli E, Ford JM (2012) Breast cancers with compromised DNA repair exhibit selective sensitivity to elesclomol. DNA Repair (Amst) 11: 522–524. [DOI] [PubMed] [Google Scholar]
- 34.Abbotts R, Thompson N, Madhusudan S (2014) DNA repair in cancer: emerging targets for personalized therapy. Cancer Manag Res 6: 77–92. 10.2147/CMAR.S50497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robertson NM, Yigit MV (2014) The role of microRNA in resistance to breast cancer therapy. Wiley Interdiscip Rev RNA 5: 823–833. 10.1002/wrna.1248 [DOI] [PubMed] [Google Scholar]
- 36.Abdel-Fatah TM, Perry C, Arora A, Thompson N, Doherty R, et al. (2014) Is there a role for base excision repair in estrogen/estrogen receptor-driven breast cancers? Antioxid Redox Signal 21: 2262–2268. 10.1089/ars.2014.6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santarpia L, Iwamoto T, Di Leo A, Hayashi N, Bottai G, et al. (2013) DNA repair gene patterns as prognostic and predictive factors in molecular breast cancer subtypes. Oncologist 18: 1063–1073. 10.1634/theoncologist.2013-0163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matta J, Echenique M, Negron E, Morales L, Vargas W, et al. (2012) The association of DNA Repair with breast cancer risk in women. A comparative observational study. BMC Cancer 12: 490 10.1186/1471-2407-12-490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Latimer JJ, Johnson JM, Kelly CM, Miles TD, Beaudry-Rodgers KA, et al. (2010) Nucleotide excision repair deficiency is intrinsic in sporadic stage I breast cancer. Proc Natl Acad Sci U S A 107: 21725–21730. 10.1073/pnas.0914772107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramos JM, Ruiz A, Colen R, Lopez ID, Grossman L, et al. (2004) DNA repair and breast carcinoma susceptibility in women. Cancer 100: 1352–1357. [DOI] [PubMed] [Google Scholar]
- 41.Lips EH, Mulder L, de Ronde JJ, Mandjes IA, Vincent A, et al. (2012) Neoadjuvant chemotherapy in ER+ HER2- breast cancer: response prediction based on immunohistochemical and molecular characteristics. Breast Cancer Res Treat 131: 827–836. 10.1007/s10549-011-1488-0 [DOI] [PubMed] [Google Scholar]
- 42.Good P (2006) Resampling Methods: A Practical Guide to Data Analysis: Birkhauser; Boston. [Google Scholar]
- 43.Jeselsohn R, Buchwalter G, De Angelis C, Brown M, Schiff R (2015) ESR1 mutations-a mechanism for acquired endocrine resistance in breast cancer. Nat Rev Clin Oncol 12: 573–583. 10.1038/nrclinonc.2015.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Issa-Nummer Y, Loibl S, von Minckwitz G, Denkert C (2014) Tumor-infiltrating lymphocytes in breast cancer: A new predictor for responses to therapy. Oncoimmunology 3: e27926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wajed SA, Laird PW, DeMeester TR (2001) DNA methylation: an alternative pathway to cancer. Ann Surg 234: 10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Caldon CE (2014) Estrogen signaling and the DNA damage response in hormone dependent breast cancers. Front Oncol 4: 106 10.3389/fonc.2014.00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trimarchi MP, Mouangsavanh M, Huang TH (2011) Cancer epigenetics: a perspective on the role of DNA methylation in acquired endocrine resistance. Chin J Cancer 30: 749–756. 10.5732/cjc.011.10128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Medunjanin S, Weinert S, Schmeisser A, Mayer D, Braun-Dullaeus RC (2010) Interaction of the double-strand break repair kinase DNA-PK and estrogen receptor-alpha. Mol Biol Cell 21: 1620–1628. 10.1091/mbc.E09-08-0724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bjornstrom L, Sjoberg M (2005) Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol Endocrinol 19: 833–842. [DOI] [PubMed] [Google Scholar]
- 50.Bonastre J, Marguet S, Lueza B, Michiels S, Delaloge S, et al. (2014) Cost effectiveness of molecular profiling for adjuvant decision making in patients with node-negative breast cancer. J Clin Oncol 32: 3513–3519. 10.1200/JCO.2013.54.9931 [DOI] [PubMed] [Google Scholar]
- 51.Pedram A, Razandi M, Evinger AJ, Lee E, Levin ER (2009) Estrogen inhibits ATR signaling to cell cycle checkpoints and DNA repair. Mol Biol Cell 20: 3374–3389. 10.1091/mbc.E09-01-0085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hilakivi-Clarke L (2000) Estrogens, BRCA1, and breast cancer. Cancer Res 60: 4993–5001. [PubMed] [Google Scholar]
- 53.Likhite VS, Cass EI, Anderson SD, Yates JR, Nardulli AM (2004) Interaction of estrogen receptor alpha with 3-methyladenine DNA glycosylase modulates transcription and DNA repair. J Biol Chem 279: 16875–16882. [DOI] [PubMed] [Google Scholar]
- 54.Curtis CD, Thorngren DL, Ziegler YS, Sarkeshik A, Yates JR, et al. (2009) Apurinic/apyrimidinic endonuclease 1 alters estrogen receptor activity and estrogen-responsive gene expression. Mol Endocrinol 23: 1346–1359. 10.1210/me.2009-0093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di Cosimo S, Baselga J (2010) Management of breast cancer with targeted agents: importance of heterogeneity. [corrected]. Nat Rev Clin Oncol 7: 139–147. 10.1038/nrclinonc.2009.234 [DOI] [PubMed] [Google Scholar]
- 56.Serra V, Scaltriti M, Prudkin L, Eichhorn PJ, Ibrahim YH, et al. (2011) PI3K inhibition results in enhanced HER signaling and acquired ERK dependency in HER2-overexpressing breast cancer. Oncogene 30: 2547–2557. 10.1038/onc.2010.626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jerusalem G, Rorive A, Collignon J (2014) Use of mTOR inhibitors in the treatment of breast cancer: an evaluation of factors that influence patient outcomes. Breast Cancer (Dove Med Press) 6: 43–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davis NM, Sokolosky M, Stadelman K, Abrams SL, Libra M, et al. (2014) Deregulation of the EGFR/PI3K/PTEN/Akt/mTORC1 pathway in breast cancer: possibilities for therapeutic intervention. Oncotarget 5: 4603–4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller TW, Hennessy BT, Gonzalez-Angulo AM, Fox EM, Mills GB, et al. (2010) Hyperactivation of phosphatidylinositol-3 kinase promotes escape from hormone dependence in estrogen receptor-positive human breast cancer. J Clin Invest 120: 2406–2413. 10.1172/JCI41680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morales L, Alvarez-Garriga C, Matta J, Ortiz C, Vergne Y, et al. (2013) Factors associated with breast cancer in Puerto Rican women. J Epidemiol Glob Health 3: 205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vergne Y, Matta J, Morales L, Vargas W, Alvarez-Garriga C, et al. (2013) Breast Cancer and DNA Repair Capacity: Association With Use of Multivitamin and Calcium Supplements. Integr Med (Encinitas) 12: 38–46. [PMC free article] [PubMed] [Google Scholar]
- 62.Matta JL, Villa JL, Ramos JM, Sanchez J, Chompre G, et al. (2003) DNA repair and nonmelanoma skin cancer in Puerto Rican populations. J Am Acad Dermatol 49: 433–439. [DOI] [PubMed] [Google Scholar]
- 63.Cheng L, Eicher SA, Guo Z, Hong WK, Spitz MR, et al. (1998) Reduced DNA repair capacity in head and neck cancer patients. Cancer Epidemiol Biomarkers Prev 7: 465–468. [PubMed] [Google Scholar]
- 64.D'Errico M, Calcagnile A, Iavarone I, Sera F, Baliva G, et al. (1999) Factors that influence the DNA repair capacity of normal and skin cancer-affected individuals. Cancer Epidemiol Biomarkers Prev 8: 553–559. [PubMed] [Google Scholar]
- 65.Hu JJ, Hall MC, Grossman L, Hedayati M, McCullough DL, et al. (2004) Deficient nucleotide excision repair capacity enhances human prostate cancer risk. Cancer Res 64: 1197–1201. [DOI] [PubMed] [Google Scholar]
- 66.Wei Q, Cheng L, Hong WK, Spitz MR (1996) Reduced DNA repair capacity in lung cancer patients. Cancer Res 56: 4103–4107. [PubMed] [Google Scholar]
- 67.Wei Q, Matanoski GM, Farmer ER, Hedayati MA, Grossman L (1993) DNA repair and aging in basal cell carcinoma: a molecular epidemiology study. Proc Natl Acad Sci U S A 90: 1614–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Murray D, Begg AC (2004) Relationship Among DNA Repair Genes, Cellular Radiosensitivity, and the Response of Tumors and Normal Tissues to Radiotherapy In: Panasci LC, Alaoui-Jamali MA, editors. DNA Repair in Cancer Therapy. Totowa, NJ: Humana Press; pp. 211–256. [Google Scholar]
- 69.Mendez P, Taron M, Moran T, Fernandez MA, Requena G, et al. (2011) A modified host-cell reactivation assay to quantify DNA repair capacity in cryopreserved peripheral lymphocytes. DNA Repair (Amst) 10: 603–610. [DOI] [PubMed] [Google Scholar]
- 70.Athas WF, Hedayati MA, Matanoski GM, Farmer ER, Grossman L (1991) Development and field-test validation of an assay for DNA repair in circulating human lymphocytes. Cancer Res 51: 5786–5793. [PubMed] [Google Scholar]
- 71.Wang L, Wei Q, Shi Q, Guo Z, Qiao Y, et al. (2007) A modified host-cell reactivation assay to measure repair of alkylating DNA damage for assessing risk of lung adenocarcinoma. Carcinogenesis 28: 1430–1436. [DOI] [PubMed] [Google Scholar]
- 72.Qiao Y, Spitz MR, Guo Z, Hadeyati M, Grossman L, et al. (2002) Rapid assessment of repair of ultraviolet DNA damage with a modified host-cell reactivation assay using a luciferase reporter gene and correlation with polymorphisms of DNA repair genes in normal human lymphocytes. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis 509: 165–174. [DOI] [PubMed] [Google Scholar]
- 73.Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, et al. (2014) Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med 138: 241–256. 10.5858/arpa.2013-0953-SA [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hosmer D, Lemeshow S, Sturdivant R (2013) Applied Logistic Regression.: John Wiley and Sons. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Estrogen can have a variety of effect on the tumor cells some of which may involve DNA repair signaling specially among HER2(-) tumors. (A) In the canonical pathway, ER binds to the promoter regions of genes involved in cell cycle progression. (B) Non-canonical pathways involve many other signaling cascades such as p53, mTOR and ERK which can also lead to increased proliferation. Both of these pathways can lead to low levels of DNA damage that may “switch on” DNA repair mechanisms (evaluated in in this study in the patients’ circulating lymphocytes).
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.