Abstract
CRISPR-Cas systems have rapidly transitioned from intriguing prokaryotic defense systems to powerful and versatile biomolecular tools. This article reviews how these systems have been translated into technologies to manipulate bacterial genetics, physiology, and communities. Recent applications in bacteria have centered on multiplexed genome editing, programmable gene regulation, and sequence-specific antimicrobials, while future applications can build on advances in eukaryotes, the rich natural diversity of CRISPR-Cas systems, and the untapped potential of CRISPR-based DNA acquisition. Overall, these systems have formed the basis of an ever-expanding genetic toolbox and hold tremendous potential for our future understanding and engineering of the bacterial world.
Keywords: Antimicrobials, Cas9, Genetic control, Genetic circuits, Genome engineering, Undomesticated microbes
Introduction
Almost three decades ago, Japanese researchers identified an unusual set of repetitive DNA sequences in the genome of the bacterium Escherichia coli (Ishino et al., 1987). These repeats were later found to be part of an expansive family of repetitive DNA sequences termed clustered regularly interspaced short palindromic repeats, or CRISPRs (Jansen et al., 2002). While the repeats attracted initial attention, the intervening “spacer” sequences turned out to be the critical elements in initially defining the function of CRISPR-Cas systems. These spacers were found to be homologous to foreign plasmid and bacteriophage sequences (Bolotin et al., 2005; Mojica et al., 2005), which hinted at a defensive function for CRISPR. The major breakthrough came in 2007, with the report that bacteriophage-resistant strains had acquired spacer sequences that matched the bacteriophage genome (Barrangou et al., 2007). Critically, the acquired spacer and the flanking CRISPR-associated (Cas) genes were essential to confer immunity to the bacteriophage. This seminal work quickly led to our current understanding of these diverse adaptive defense systems in bacteria and archaea now known as CRISPR-Cas systems.
CRISPR-Cas systems consist of two general components: CRISPR RNAs (crRNAs) and Cas proteins. The crRNAs base pair with complementary DNA or RNA sequences associated with an invader, and the Cas proteins clear the recognized genetic material. Because base pairing is straightforward to predict and to design, the biotechnology community was interested in the capacity of these systems to bind and cleave user-defined sequences. The catalyst for the CRISPR-Cas revolution, however, came with the demonstration that a single protein, Cas9, could be harnessed for site-specific DNA binding and cleavage (Gasiunas et al., 2012; Jinek et al., 2012). In the few short years since this demonstration, CRISPR-Cas systems have emerged as powerful and versatile tools in applications ranging from genome editing to molecular imaging. While most of these advances have been reported in eukaryotes, CRISPR-Cas systems also offer promising tools for understanding and engineering bacteria. This article discusses recent applications of CRISPR-Cas systems in bacteria in the realms of genome editing, gene regulation, and antimicrobials. The review then forecasts upcoming opportunities and challenges associated with further exploiting these versatile prokaryotic immune systems.
A PRIMER ON CRISPR-CAS SYSTEMS
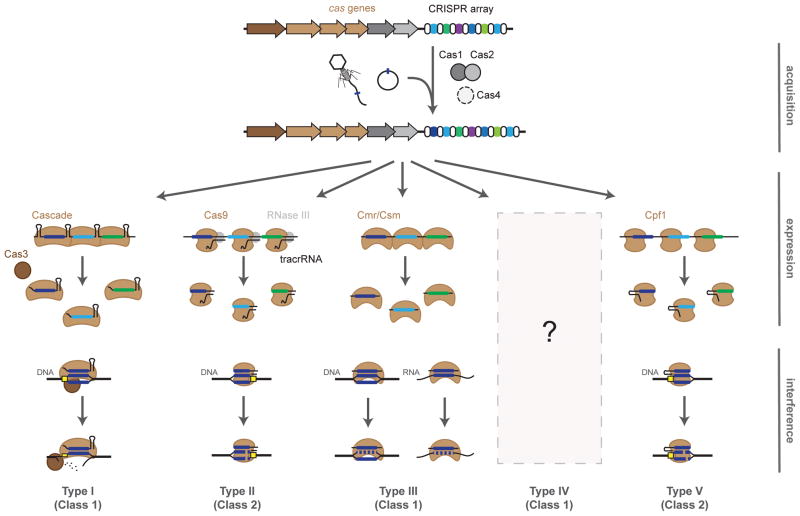
CRISPR-Cas systems naturally protect bacteria and archaea from foreign genetic elements such as plasmids or bacteriophages. Immunity proceeds in three stages: acquisition, expression, and interference (Figure 1). For acquisition, a spacer generated from a short sequence of invading DNA is incorporated at the leading edge of the CRISPR locus. Next, for expression, the array of alternating repeats and spacers is transcribed and subsequently processed by the Cas proteins and accessory factors into individual crRNAs. Finally, for interference, a ribonucleoprotein complex of the Cas protein(s) and an individual crRNA binds and cleaves nucleic acids that are complementary to the spacer portion of the crRNA. More details on the mechanisms of CRISPR-based immunity can be found in other recent reviews (Barrangou and Marraffini, 2014; Bondy-Denomy and Davidson, 2014; Van der Oost et al., 2014).
Figure 1.
Overview of adaptive immunity by CRISPR-Cas systems. Immunity is conferred through three steps: acquisition, expression, and interference. Acquisition: a small piece of the invader DNA is integrated as a new spacer within the CRISPR array. Expression: the CRISPR array is transcribed and undergoes processing by the Cas proteins and accessory factors to form the CRISPR RNA (crRNA). Interference: the spacer portion of the crRNA serves as a recognition element for the Cas proteins to target invading DNA (Type I, II, III, V) or RNA (Type III). Type I, II, and V systems require a protospacer-adjacent motif (PAM, yellow box) for target recognition. The current understanding of Type IV systems is limited to bioinformatics analyses.
CRISPR-Cas systems are remarkably widespread and diverse. To date, the CRISPRdb online database (Grissa et al., 2007) has identified 1302 bacterial and archaeal strains with putative CRISPR arrays out of 2762 genomes analyzed. Each of these arrays is associated with differing families of cas genes that necessitated a standard system for their classification and nomenclature. The latest classification divides CRISPR-Cas systems into two classes according to the configuration of their effector modules (Makarova et al., 2015). Class 1 systems are defined by multisubunit effector complexes while Class 2 systems utilize a single effector protein. Within these two classes, CRISPR-Cas systems can be further divided into five types with sixteen total subtypes, defined based on the distinct proteins that facilitate CRISPR-Cas activities (Chylinski et al., 2014; Jiang and Doudna, 2015; Makarova et al., 2011; Makarova et al., 2015). Class 1 CRISPR-Cas systems include Type I, Type III, and putative Type IV systems; Class 2 systems include Type II systems and putative new Type V systems. These types (and subtypes) vary in the number of Cas proteins, the mechanism of crRNA processing and targeting, and whether the target is DNA or RNA (Table 1). These attributes offer distinct capabilities, some of which have been co-opted for biotechnology.
Table 1.
The five standard types of CRISPR-Cas systems. The naming conventions are based on the most recent nomenclature for CRISPR-Cas systems (Makarova et al., 2015). The types are grouped into two classes based on the use of multiple proteins (Class 1) or a single protein (Class 2) for interference. Cascade: CRISPR associated complex for adaptive defense. PAM: protospacer-adjacent motif.
| Class | Type | Subtypes | Signature gene | # non-acquisition cas genes | Processing factors | Targeting | Self/non-self recognition |
|---|---|---|---|---|---|---|---|
| 1 | I | A,B,C,D,E,F,U | cas3 | 4 – 8 | Cascade | Nicks, degrades DNA | PAM 5′ of matching target |
| III | A,B,C,D | cas10 | 6 – 8 | Cas6 | Cleaves DNA/RNA | Base pairing with 5′ handle | |
| IV (putative) | n/a | csf1 | 3 – 4 | Unknown | Unknown | Unknown | |
| 2 | II | A,B,C | cas9 | 1 | tracrRNA, RNase III | Cleaves DNA | PAM 3′ of matching target |
| V (putative) | n/a | cpf1 | 1 | Cpf1 | Cleaves DNA | PAM 5′ of matching target |
Type I CRISPR-Cas systems, the most abundant type in both bacteria and archaea, are defined by the presence of the signature Cas3 protein (Jackson et al., 2014b; Sinkunas et al., 2011). Cas3 works together with a large multimeric protein complex termed the CRISPR-associated complex for antiviral defense (Cascade) whose composition is unique to each subtype (I-A through I-F, I-U) (Brendel et al., 2014; Jackson et al., 2014a; Jore et al., 2011; Makarova et al., 2015; Nam et al., 2012; Wiedenheft et al., 2011; Zhao et al., 2014). Cascade is responsible for both processing the crRNAs as well as locating and binding the target sequence (Brouns et al., 2008; Westra et al., 2012). Once Cascade binds to the recognized DNA site, Cas3 is prompted to unwind, cleave, and degrade one strand of the DNA (Sinkunas et al., 2011).
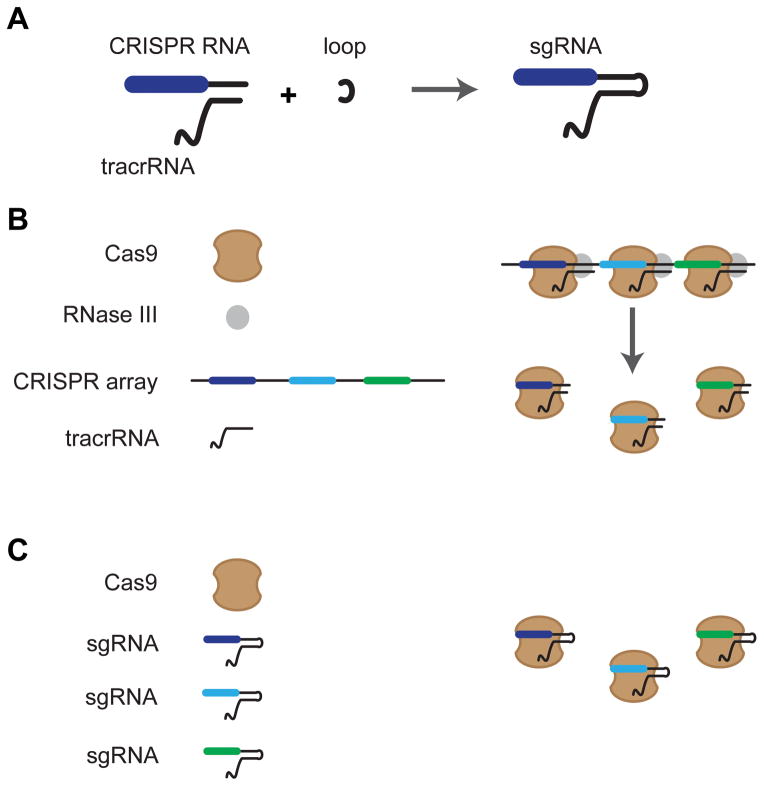
Type II systems represent the least abundant systems, including no known examples in archaea. These systems have three subtypes (II-A, II-B, II-C) that are united by the signature Cas9 protein and are differentiated by the Cas proteins involved in acquisition (Chylinski et al., 2014; Makarova et al., 2015). In general, Type II systems are very compact, as they require only Cas9 for interference. Cas9 generates double-stranded DNA breaks via its HNH-nuclease and RuvC-like nuclease domains, which cleave the target strand and non-target strand, respectively (Gasiunas et al., 2012; Jiang et al., 2015a; Jinek et al., 2012; Nishimasu et al., 2014). Along with Cas9, Type II systems require a trans-activating crRNA (tracrRNA) and RNase III as part of crRNA processing (Chylinski et al., 2013; Deltcheva et al., 2011). To simplify this system for practical use, Jinek and coworkers fused the processed version of the tracrRNA and the crRNA from Streptococcus pyogenes into a chimeric RNA, termed a single-guide RNA (sgRNA) (Jinek et al., 2012) (Figure 2). This sgRNA design eliminated the need for RNase III processing and allowed the expression of a single RNA molecule for DNA targeting. The downside was that the inherent capability of the CRISPR arrays for multiplexing—targeting multiple sequences with a single transcript—was lost. However, various groups have engineered platforms to encode multiple sgRNAs from a single transcript (Nissim et al., 2014; Tsai et al., 2014; Xie et al., 2015).
Figure 2.
Design of the sgRNA. (A) The natural crRNA and tracrRNA are connected by a loop to form a single-guide RNA (sgRNA). (B) Processing of the natural Type II CRISPR array requires two additional factors: RNase III and tracrRNA. (C) Use of sgRNAs bypasses the requirement for RNase III and tracrRNA.
Type III systems are found in both bacteria and archaea and are typified by the signature cas10 gene (Makarova et al., 2011; Makarova et al., 2015). These systems drew initial interest because of their natural ability to target RNA (Hale et al., 2009; Staals et al., 2014; Tamulaitis et al., 2014). However, other early accounts indicated that Type III systems instead target DNA (Hatoum-Aslan et al., 2014; Marraffini and Sontheimer, 2008). While these systems were originally thought to target either DNA or RNA, there is emerging evidence that some systems can simultaneously target both nucleic acids. For instance, the Type III-B system in the archaeon Sulfolobus islandicus was shown to be capable of targeting DNA and RNA (Peng et al., 2013; Peng et al., 2014) whereas the Type III-A system in the pathogenic bacterium Staphylococcus epidermidis cleaves RNA and transcriptionally active DNA via independent active sites within the protein effector complex (Samai et al. 2015). In addition to the III-A and III-B subtypes discussed here, there are two additional type III subtypes—III-C and III-D—that are less understood (Makarova et al., 2015). Interestingly, Type III systems are phylogenetically related to Type I systems and even share structural similarities between the Csm (III-A) or Cmr (III-B) complex and the Cascade complex from Type I systems (Osawa et al., 2015; Rouillon et al., 2013; Spilman et al., 2013; Staals et al., 2013; Taylor et al., 2015).
Lastly, two new types have been proposed: a Class 1 Type IV system and a Class 2 Type V system (Makarova et al., 2015). The putative Type IV system encodes a predicted minimal multi-subunit effector complex with a unique large subunit, Csf1, which serves as the signature protein for this system. The suggested Type V system utilizes Cpf1 as a single protein for interference. To date, only one Type V system has been characterized. Zetsche et al. (2015) demonstrated that the Type V system of Francisella novicida U112 does not require a tracrRNA for crRNA maturation and Cpf1-crRNA complexes are sufficient to cleave DNA target molecules. Furthermore, Cpf1 introduces a staggered double-stranded DNA break with overhangs unlike the blunt-ended break of Type II Cas9.
One of the original conundrums of CRISPR-Cas systems was how crRNAs could differentiate between their own genomic spacer sequence and the identical target sequence present in the invader. The answer was not found within the target sequence (termed the protospacer), but immediately flanking it. For Type I, Type II, and Type V systems, this flanking feature is called a protospacer adjacent motif (PAM) (Mojica et al., 2009; Szczelkun et al., 2014; Zetsche et al., 2015) and is typically 2 – 5 base pairs. For Type I systems, the PAM is located on the 5′ end of the protospacer (herein defined as the strand matching the spacer) and is recognized by Cascade (Sashital et al., 2012; Westra et al., 2013). For Type II systems, the PAM is located on the 3′ end of the protospacer and is recognized by Cas9 (Deveau et al., 2008; Gasiunas et al., 2012; Jinek et al., 2012). For example, the widely used S. pyogenes Cas9 recognizes an NGG PAM sequence (where N is any nucleotide) located on the 3′ end of the protospacer. The Type V systems contain a T-rich PAM on the 5′ end of the protospacer (Zetsche et al., 2015). In contrast, Type III systems do not rely on a PAM. Instead, these systems evaluate base pairing between the target and the 5′ handle of the crRNA (Marraffini and Sontheimer, 2010). Extensive base pairing between the handle and the protospacer results in no targeting while limited base pairing results in targeting. These requirements result in two simple rules when designing crRNAs: (1) identify a PAM (Types I, II, V) or a sequence with poor base-pairing potential to the 5′ portion of the crRNA repeat (Type III) and (2) use the flanking 20 – 30 nucleotides as the spacer portion of the crRNA. These remarkably straightforward rules have helped drive the implementation of CRISPR-Cas systems in a wide range of applications.
CURRENT APPLICATIONS
A number of new applications have been developed in bacteria that take advantage of the programmability and sequence specificity of CRISPR-Cas systems. Cas9 has dominated these applications based on its compactness, although the use of the Type I systems and Type III systems is beginning to gain traction.
Bacterial genome editing
Genome editing has been one of the most visible and celebrated applications of CRISPR to date, with an overwhelming focus on eukaryotes. In 2013, genome editing was demonstrated in human cells (Cho et al., 2013; Cong et al., 2013; Jinek et al., 2013; Mali et al., 2013b) and was quickly expanded to eukaryotic organisms ranging from fungi to monkeys (Bassett et al., 2013; DiCarlo et al., 2013; Dickinson et al., 2013; Friedland et al., 2013; Hwang et al., 2013; Li et al., 2013; Nekrasov et al., 2013; Niu et al., 2014). In these eukaryotic cells and organisms, Cas9 was used to introduce a double-stranded break into a defined location in the genome, followed by repair through the endogenous non-homologous end joining (NHEJ) or homology-directed repair pathways.
Although Cas9-based genome editing in bacteria was also first reported in 2013 (Jiang et al., 2013a), only a handful of publications have ensued. The disparity between these few publications and the volumes published for eukaryotes can be explained by the relatively poor capacity of bacteria to repair double-stranded breaks by CRISPR-Cas systems. Unlike eukaryotes, bacteria generally cannot repair breaks caused by CRISPR. The lethality of genome targeting is supported by the natural co-occurrence of self-targeting CRISPRs and degenerate cas genes (Stern et al., 2010); a putatively large evolutionary change following the natural acquisition of a self-targeting spacer (Aklujkar and Lovley, 2010); the cytotoxicity of expressing a genome-targeting crRNA (Edgar and Qimron, 2010; Vercoe et al., 2013); and the subsequent appearance of disruptive mutations in the cas genes, self-targeting crRNA, or the target location to escape self-targeting cell death (Gomaa et al., 2014; Jiang et al., 2013b).
A simple explanation for the lethality of bacterial genome targeting is that, given the defensive function of CRISPR-Cas systems, repairing targeting events would be wholly counterproductive. If bacteriophage or plasmid DNA was attacked by CRISPR and the break was repaired, then the invader would persist. It is worth noting that bacteria often possess multiple pathways for repairing DNA damage (Selle and Barrangou, 2015). We speculate that CRISPR-Cas systems evolved to block many of these repair mechanisms in bacteria, whether by Cas9 staying tightly bound to the DNA end following cleavage (Gasiunas et al., 2012) or by Cas3 degrading a strand of the target DNA through Cas3’s exonuclease activity (Westra et al., 2012). See Selle and Barrangou (2015) for a focused review describing mechanisms of genome repair in bacteria and how it relates to genome editing with CRISPR.
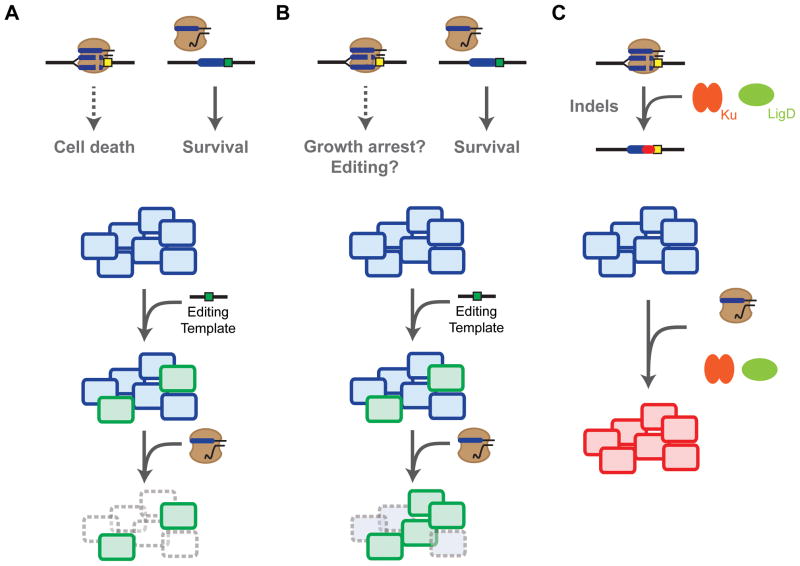
These obstacles were originally addressed in the first demonstration of genome editing in bacteria (Jiang et al., 2013a). In this work, Cas9 was used in combination with a CRISPR array and a tracrRNA to generate defined point mutations in Streptococcus pneumoniae and in Escherichia coli. In both cases, a template oligonucleotide designed for homologous recombination was introduced simultaneously with the CRISPR array. The oligonucleotide mutated the PAM of the target site, preventing Cas9 from recognizing the sequence. As a result, CRISPR-Cas9 served a cleanup role by eliminating cells that did not undergo recombination (Figure 3A). By either relying on high recombination rates in S. pneumoniae or introducing the λ-red recombination system E. coli, the authors reported remarkably high rates of recombination ranging between 65% and 100% of all screened colonies. The arrays could also be multiplexed, which allowed editing at multiple locations at one time.
Figure 3.
Genome editing with CRISPR-Cas9 in bacteria. (A) DNA cleavage by Cas9 is generally lethal, leading to clearance of cells that did not undergo recombination. (B) Employing a nicking Cas9 appears to either temper the lethality of genome targeting or drive genome editing. (C) Utilizing the bacterial non-homologous end-joining pathway composed of the Ku and LigD can rescue the lethality of Cas9-based genome targeting and drive indel formation.
Following the initial demonstration of genome editing in S. pneumoniae and in E. coli, Cas9-based genome editing has been reported in ranging bacteria of industrial relevance. These include Lactobacillus reuteri, a lactic acid bacterium with probiotic properties (Oh and van Pijkeren, 2014); Clostridium beijerinckii, a species widely used as a metabolic host for acetone, butanol, and ethanol production (Wang et al., 2015); multiple Streptomyces species, which synthesize natural products with antimicrobial activity (Cobb et al., 2015; Huang et al., 2015; Tong et al., 2015); and additional demonstrations in E. coli (Jiang et al., 2015b; Li et al., 2015; Pyne et al., 2015). These examples principally relied on the chimeric sgRNA design (Cobb et al., 2015; Jiang et al., 2015b; Li et al., 2015; Wang et al., 2015), although the dual-expression of tracrRNAs and crRNAs has also been implemented for genome editing (Jiang et al., 2013a; Oh and van Pijkeren, 2014; Pyne et al., 2015). The DNA templates were supplied as single-stranded oligonucleotides or as double-stranded DNA within the sgRNA plasmid. Between these two options, encoding the repair template within the sgRNA plasmid generally yielded greater editing efficiencies.
Many of these examples confirmed the need for high rates of transformation and recombination: transformation determines the number of cells subject to the DNA template and CRISPR-Cas9, whereas recombination determines the number of cells that will survive attack by Cas9. For instance, follow-up demonstrations in E. coli were limited to only three simultaneous editing events, even following overexpression of the λ-red recombination machinery (Jiang et al., 2015b; Li et al., 2015). Without high rates of both transformation and recombination, the background mutational rate of the crRNA or Cas9 (typically 10−5 – 10−4) (Gomaa et al., 2014; Jiang et al., 2013b) will overwhelmingly account for surviving transformants. The transformation barrier may be alleviated through inducible control of the crRNA or the Cas9, although tightly controlled inducible systems are not widely available. Furthermore, more universal recombination systems may be needed to drive high rates of homologous recombination in otherwise disparate organisms.
One potential strategy to improve the editing efficiency is using a Cas9 that harbors a point mutation in either the RuvC or HNH nuclease domain (D10A or H840A for the S. pyogenes Cas9) (Figure 3B). Because each nuclease domain cleaves either strand of the target DNA, the mutated Cas9 nicks the target without introducing a double-stranded break (Gasiunas et al., 2012; Jinek et al., 2012). Xu et al. (2015) hypothesized that a nicking Cas9 could improve editing in bacteria by reducing the lethality of DNA targeting. Supplying Clostridium cellulolyticum with a nicking Cas9, an sgRNA, and a template oligonucleotide resulted in small insertions or deletions, with editing efficiencies up to 95% for 200-nucleotide homology arms. In contrast, the regular Cas9 did not yield any colonies. While DNA nicking did promote greater editing frequencies than double-stranded breaks, the precise mechanism is not fully understood. On one hand, Cas9 and its nicking counterpart could similarly clear unedited members of the population, but the tempered lethality of nicks could allow more time to achieve recombination. On the other hand, the nicking Cas9 could be driving homologous recombination by accelerating strand invasion, similar to nickase activity in eukaryotes (Cong et al., 2013; Mali et al., 2013b). Once the mechanism of editing is fully elucidated, this nickase strategy could be extended to other bacterial organisms that are poorly equipped to repair double-stranded breaks introduced by Cas9.
Another intriguing strategy is recapitulating the NHEJ pathway in bacteria to repair double-stranded breaks by CRISPR-Cas9 (Figure 3C). Rather than attempt to import eukaryotic NHEJ pathways and their numerous components, NHEJ pathways found in some bacteria offered simpler opportunities. These bacterial pathways only require two proteins, Ku and LigD, and are found in a small but diverse fraction of bacteria (Aravind and Koonin, 2001; Bowater and Doherty, 2006). To date, only one recent report has coupled NHEJ and Cas9-based genome editing in bacteria (Tong et al., 2015). Here, genome editing was performed in the actinomycetes species Streptomyces coelicolor. The ligD gene was expressed from a related Streptomyces species to reconstitute the incomplete NHEJ pathway in this species. Co-expressing LigD along with Cas9 and a designed sgRNA led to high efficiencies of NHEJ-generated indels—insertions or deletions—within two genes from the biosynthetic pathway of the blue actinorhodin antibiotic. It will be interesting to see if a reconstituted NHEJ pathway along with a homologous template will increase recombination rates or if indel formation will dominate over homology directed repair. If the NHEJ pathway can be imported into other bacteria to alleviate the lethality of genome targeting and boost the overall efficiency of genome editing, Ku and LigD could become synonymous with CRISPR-Cas9 for genome editing in bacteria.
Bacterial gene regulation
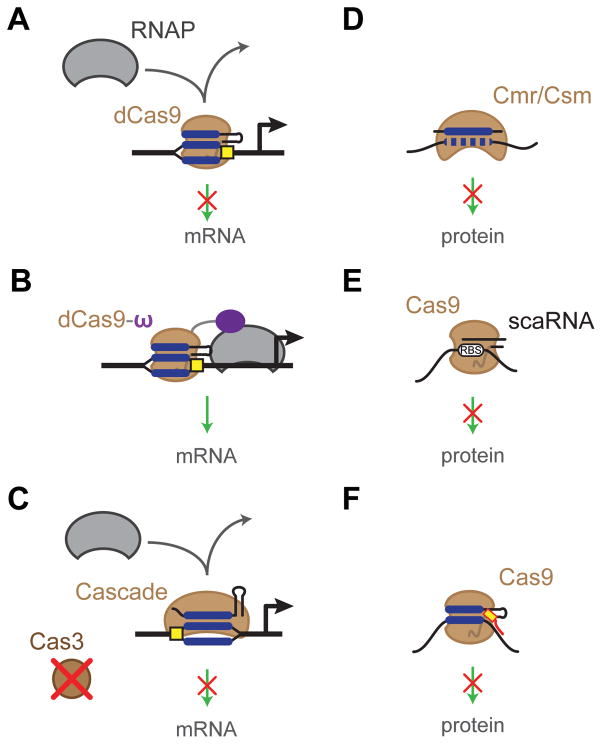
Despite the predominant focus on genome editing, CRISPR-Cas systems are also revolutionizing the programmable regulation of gene expression. This feat was first demonstrated with CRISPR-Cas9 in two landmark publications (Bikard et al., 2013; Qi et al., 2013). In both publications, the S. pyogenes Cas9 nuclease was converted into a DNA-binding protein by point mutations to the RuvC and HNH domains (D10A and H840A). This catalytically dead Cas9 (dCas9) lost the ability to cleave DNA but retained its DNA binding ability (Jinek et al., 2012). By directing dCas9 to bind to the promoter or open reading frame of a target gene, expression could be repressed by preventing transcription initiation or elongation (Bikard et al., 2013; Qi et al., 2013) (Figure 4A). The strongest levels of repression occurred when targeting the promoter, with up to 1,000-fold repression. Targeting the coding region also repressed gene expression, where targeting was much more effective for the non-template strand than the template strand (Bikard et al., 2013; Qi et al., 2013). Expressing multiple sgRNAs was also shown to silence multiple genes at one time or, when targeting the same gene, to further boost silencing as long as the targeted sites did not overlap (Qi et al., 2013). Either an sgRNA (Qi et al., 2013) or the combination of a CRISPR array and a tracrRNA (Bikard et al., 2013) were capable of gene repression; the sgRNA offered a more compact option whereas the CRISPR array:tracrRNA was better suited for multiplexing. Advances in gene regulation with dCas9 were reviewed recently (Bikard and Marraffini, 2013; Fineran and Dy, 2014; Sampson and Weiss, 2014) and a protocol is available based on sgRNAs (Larson et al., 2013).
Figure 4.
CRISPR-based gene regulation. (A) A catalytically dead Cas9 (dCas9) can be targeted to the promoter or coding region of a gene, blocking transcription. (B) A fusion between dCas9 and the ω subunit of RNA polymerase (dCas9-ω) can recruit RNA polymerase to activate transcription. (C) Eliminating the cas3 gene from Type I systems can allow targeted DNA binding and transcriptional repression with Cascade. (D) Type III systems can be readily co- opted to bind and cleave target mRNAs. (E) The Francisella novicida Cas9 utilizes a scaRNA to silence an endogenous gene through putative base-pairing interactions. (F) Introducing a DNA oligonucleotide PAMmer allows Cas9 to bind and cleave RNAs complementary to the guide portion of an sgRNA.
Beyond E. coli, dCas9 has been implemented in a growing collection of other bacterial strains. Bikard et al. (2013) implemented dCas9 in Streptococcus pneumoniae to repress β-galactosidase expression and observed up to a 14-fold reduction in activity. Choudhary et al. (2015) utilized dCas9 in mycobacteria to identify essential genes. Tong et al. (2015) applied dCas9 in the actinomycetes species Streptomyces coelicolor to reversibly control expression of a gene involved in production of the actinorhodin antibiotic. These examples underscore the broad applicability of dCas9 to gene repression in bacteria. For basic genetic studies and strain development, dCas9 may even be preferable over Cas9-based editing because multiple genes can be silenced at one time without the need to modify each genetic locus.
The dCas9 protein is also capable of bacterial gene activation (Figure 4B). In E. coli, this was demonstrated by fusing the ω subunit of RNA polymerase to dCas9 and then expressing this construct in cells lacking the ω subunit gene rpoZ (Bikard et al., 2013). This subunit co-purifies with RNA polymerase (RNAP) yet the subunit is dispensable, with no discernible deletion phenotypes (Dove and Hochschild, 1998). To optimize gene activation with dCas9, Bikard et al. (2013) assessed multiple permutations, including N- and C-terminal fusions, targeting the template or the non-template strand, varying the distance between the target site and the transcriptional start site, and testing promoters with varying transcriptional activities. The most successful combination was fusing the ω subunit to the C-terminus of Cas9 (Cas9-ω) and targeting 96 nucleotides upstream of the transcriptional start site of a weak promoter. While this configuration yielded ~23-fold activation (Bikard et al., 2013), most other configurations yielded less than 8-fold activation. Compared to >100-fold repression reported for dCas9 (Qi et al., 2013), gene activation with CRISPR-Cas9 has significant room for improvement.
Despite, the intense focus on Type II systems and dCas9, Type I systems are also capable of transcriptional regulation (Figure 4C). This capability was recently demonstrated for the Type I-E system in E. coli through the expression of Cascade and the elimination of the Cas3 nuclease (Luo et al., 2015; Rath et al., 2015). In the absence of Cas3, the Cascade complex still processes the CRISPR array and binds target DNA sequences. Cascade can be expressed off the endogenous locus in the genome (Luo et al., 2015) or introduced exogenously via a plasmid (Rath et al., 2015). Similar to Type II systems, the Type I-E system was also capable of >100-fold repression when targeting the promoter. Strand bias within the transcribed region was also observed for Cascade-driven repression, although the particular bias did not match between the two reports (Luo et al., 2015; Rath et al., 2015). Because Type I systems comprise a diverse set of subtypes, it will be interesting to see if other subtypes are capable of gene silencing and whether endogenous Type I systems in other bacteria can be repurposed for programmable gene regulation.
One draw of gene regulation using Type I systems is that multiple mature crRNA molecules can be generated from a single CRISPR array without the need for accessory processing factors. Using a single array, Luo et al. (2015) simultaneously silenced four independent endogenous sugar utilization pathways in E. coli, thereby generating a complex growth phenotype. When evaluating the potency of spacers within an array, it was observed that every additional spacer added to the array reduced the efficiency of each individual spacer (Luo et al., 2015). This points to Cascade being a limiting factor in Type I silencing and should be considered when targeting large numbers of genes. Similar analyses remain to be reported for gene regulation with dCas9.
Another emerging opportunity for CRISPR-based gene regulation is mRNA targeting. Hale et al., (2012) first demonstrated this capability by programming the Pyrococcus furiosus Type III-B complex to cleave RNAs in vitro. This demonstration was followed by Zebec et al. (2014) using the Type III-B system of Sulfolobus solfataricus to target mRNA degradation of specific chromosomal genes in vivo (Figure 4D). The Type III-A complex was also recently discovered to target RNA as demonstrated in vivo (Staals et al., 2014; Tamulaitis et al., 2014). Thus, the Type III-A Csm complex and the Type III-B Cmr complex hold promise as genetic tools to for the post-transcriptional regulation of chromosomal genes. Type III systems are also not subject to PAMs, potentially creating greater flexibility when selecting target sequences. A major downside was that gene silencing in vivo was relatively modest (~2-fold) in comparison to that achieved by dCas9 and Cascade (Zebec et al., 2014).
There is compounding evidence that some Cas9’s naturally target RNA (Figure 4E). Sampson et al. (2013) demonstrated that the Type II-B Cas9 from Francisella novicida naturally represses the expression of the FTN_1103 bacterial lipoprotein as part of immune avoidance during host infection. Silencing required the tracrRNA as well as a small, CRISPR-Cas-associated RNA (scaRNA) encoded adjacent to the CRISPR array. The scaRNA was predicted to hybridize to the ribosome-binding site and start codon of the FTN_1103 mRNA. Although the mechanism of silencing remains unclear, this general strategy allowed for the targeted silencing of the Hepatitis C virus in mammalian cells (Price et al., 2015). There is thus tremendous potential for targeted gene silencing with this Cas9 and a need to elucidate design principles.
Aside from the natural RNA-targeting ability of some Cas9 proteins, more traditional Cas9’s can be coaxed into targeting RNA. This capability was demonstrated in vitro by pairing the S. pyogenes Cas9 with PAM-presenting oligonucleotides (PAMmers) (O’Connell et al., 2014) (Figure 4F). When the PAMmer base pairs with the target RNA, Cas9 recognizes the PAM but proceeds to cleave the RNA base-paired to the sgRNA. While Cas9 also would be expected to recognize and cleave the associated DNA, selecting targets that lacked a PAM allowed Cas9 to only recognize and cleave the PAMmer-bound RNA. Interestingly, RNA cleavage was only observed using a deoxyribonucleotide-based PAMmer but not a ribonucleotide-based PAMmer, suggesting that Cas9 can differentiate the slight structural variations between deoxyribose and ribose moieties within the PAM. Whether the PAMmer can yield efficient gene silencing in vivo remains to be investigated.
Beyond the co-regulation of multiple genes, CRISPR-Cas systems are now being implemented for the design of genetic circuits. While the first examples were published in mammalian cells (Kiani et al., 2014; Nissim et al., 2014), there has been one report of circuit design in bacteria (Nielsen and Voigt, 2014). In this work, sgRNAs were designed to target promoters driving expression of other sgRNAs, allowing the construction of complex circuit topologies. The sgRNAs were designed to recognize distinct sequences, preventing any measurable crosstalk. The resulting circuits could interface with endogenous processes to control cellular phenotypes. Specifically, the authors connected an OR logic gate to the expression of the malT transcription factor, which regulates maltose utilization and also production of a lambda phage receptor. When at least one input (inducer) was present, the cells exhibited near-normal lambda phage infectivity. However, when both inputs were absent, the cells exhibited a 240-fold reduction in plaque formation. While these circuits all relied on a single Cas9, the availability of orthogonal Cas9’s that recognize different sgRNAs (Esvelt et al, 2013) opens the opportunity to create sophisticated gene circuits and functionalities.
Separately from gene regulation, CRISPR-Cas systems can be harnessed for RNA processing. Type I and Type III systems naturally process the transcribed CRISPR array into individual crRNAs through the activity of the Cas6 protein (Carte et al., 2008; Haurwitz et al., 2010; Li, 2015; Niewoehner et al., 2014). This protein specifically binds the hairpin within the CRISPR repeat, thereby cleaving at the 3′ base. Qi et al. (2012) exploited this activity in E. coli by encoding the hairpin sequence from the Pseudomonas aeruginosa Type I-F CRISPR locus between the 5′ untranslated region of a gene and its ribosome-binding site. Co-expressing the system’s Cas6 protein Csy4 resulted in efficient cleavage of the hairpin, separating the untranslated region from the coding region of the gene. The result was that the sequence of the untranslated region had little effect on translation—effectively insulating gene expression from the upstream sequence. This same approach yielded predictable gene expression for multi-gene operons when the hairpin was placed in the intervening untranslated regions. The extension of this strategy in the bacterium Bacillus subtilis, the eukaryote Saccharomyces cerevisiae (Qi et al., 2012), and mammalian cells (Nissim et al., 2014; Tsai et al., 2014) demonstrates the universality of Csy4 for directed RNA processing.
Next-generation antimicrobials
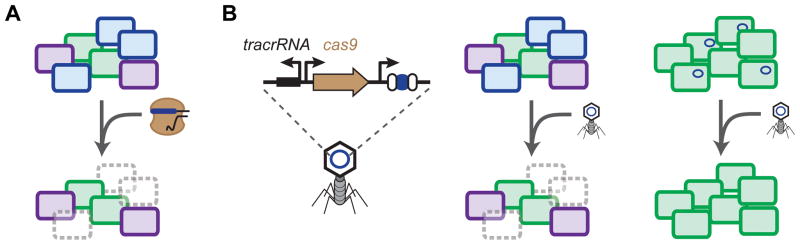
The emergence of multidrug resistance and the trickling pipeline for new small-molecule antibiotics has led to intense interest in the development of novel antimicrobials. While broad-spectrum antimicrobials have been the standard, the increasingly recognized importance of the human microbiota (Lemon et al., 2012; Neish, 2009; Walter and Ley, 2011) underscores the need to selectively eliminate pathogens while maintaining the natural microflora. The specificity, ease of design, and lethality of CRISPR created the possibility of exploiting these defense systems as the basis of programmable antimicrobials (Figure 5).
Figure 5.
CRISPR-based antimicrobials. (A) Targeting the bacterial genome leads to potent and sequence-specific cell killing. (B) Bacteriophages can be employed to deliver plasmids encoding CRISPR-Cas9, leading to targeted killing or plasmid clearance.
There was circumstantial evidence that CRISPR could serve as an antimicrobial based on the lethality and specificity of genome targeting (see section on Bacterial genome editing). However, the concept of CRISPR-based antimicrobials was first extended and experimentally demonstrated only recently (Gomaa et al., 2014). It was hypothesized that the nuclease of a CRISPR-Cas system could be directed to the bacterial genome, leading to genome cleavage and cell death; specificity would come from selecting target sequences present in some bacteria but not others. To demonstrate this concept, Gomaa et al. (2014) encoded a genome-targeting CRISPR array from the E. coli Type I-E system and transformed this plasmid into an E. coli strain expressing the associated Cas genes. The transformation efficiency of this plasmid was ~105-fold lower than that of a non-targeting control, indicating potent killing. Remarkably, every location tested—whether in coding or non-coding regions, essential or non-essential genes, top and bottom strands of the genome—resulted in a similar drop in the transformation efficiency. Furthermore, killing could be achieved using exogenous or endogenous CRISPR-Cas systems. The only requirements were the presence of the target sequence and the presence of a flanking PAM. This targeting flexibility was used to differentiate two highly similar K-12 and B lineages of E. coli as well as between E. coli K-12 and Salmonella. By using mixtures of targeting and non-targeting plasmids, the authors were able to quantitatively reduce the levels of one strain, opening the possibility of finely tuning the composition of a mixed microbial population rather than only removing individual members.
With the framework established for CRISPR-based antimicrobials, the next obstacle was delivery. A promising delivery vehicle involves bacteriophages. Bacteriophages naturally prey on bacteria by injecting their genetic material into cells and hijacking the host machinery to undergo replication. One way to co-opt bacteriophages as delivery vehicles is to incorporate the bacteriophage’s packing and replication elements into a plasmid —called a phagemid (Westwater et al., 2002). As part of the lytic cycle, the phagemid undergoes packaging in competition with the bacteriophage genome. The resulting bacteriophage particles then deliver the packaged phagemid to susceptible strains.
Two recent publications exploited this strategy to deliver a phagemid encoding the S. pyogenes cas9, tracrRNA, and designed CRISPR array (Bikard et al., 2014; Citorik et al., 2014). This setup delivers the complete CRISPR-Cas9 system, obviating the need for an endogenous, active system. Bikard et al. (2014) employed this strategy to selectively kill an antibiotic- resistant strain of Staphylococcus aureus. S. aureus was a notable target because methicillin- resistant S. aureus (MRSA) is a principal cause of antibiotic-resistant infections. The phagemid was generated by cloning the packaging site and the rinA, terS, and terL genes from the staphylococcal ΦNM1 bacteriophage along with the required elements of CRISPR-Cas9. The CRISPR array was designed to target an antibiotic resistance gene present in the genome of S. aureus. Incubating the resulting bacteriophage particles with the S. aureus cells resulted in up to a 104-fold reduction in the number of viable colonies. This degree of killing was impressive considering that no selective pressure was applied to retain the phagemid. This phagemid system was also tested in a mouse skin colonization model in which the mice were exposed to a mixture of a kanamycin-resistant strain and a kanamycin-sensitive strain of S. aureus. As expected, the phagemid targeting the kanR gene reduced the proportion of kanamycin-resistant S. aureus.
Citorik et al. (2014) similarly used M13-derived phagemids to selectively target antibiotic-resistant strains of E. coli. The CRISPR array was designed to target a mutation in the gyrA gene of E. coli that confers resistance to quinolone antibiotics. Exposing the resulting bacteriophage particles to E. coli cells harboring this mutation resulted in up to a 104-fold reduction in the number of viable colonies, paralleling that observed for S. aureus (Bikard et al., 2014). This same approach was used to selectively remove the gyrA mutants from a mixture of this strain and two other antibiotic-resistant strains of E. coli. To simulate treatment of an in vivo infection, a strain of enterohaemorrhagic E. coli (EHEC) was fed to larvae of the moth Galleria mellonella. The phagemid targeting a chromosomally encoded virulence factor was able to moderately improve survival.
Another potential use of CRISPR antimicrobials is the clearance of antibiotic resistance plasmids, as first demonstrated by Garneau et al., (2010). Bikard et al. (2014) designed the phagemid to target plasmid-borne antibiotic resistance genes harbored by a clinical strain of methicillin-resistant S. aereus (MRSA). Delivery of the ΦNM1 phagemid eliminated the plasmids without impacting cell viability and prevented any import of the same plasmid via conjugation. Citorik et al. (2014) targeted the pNDM-1 and pSHV-18 plasmids that confer resistance to β-lactam antibiotics. In both cases, delivery of the M13 phagemid resulted in a 103-fold reduction in the number of viable colonies. This reduction was associated with addiction systems encoded on the plasmid, which triggered cell death upon plasmid removal. This strategy was quite convenient by simultaneously removing antibiotic resistance and killing the host cells.
In a similar attempt to sensitize bacteria to antibiotics, Yosef et al. (2015) engineered a λ prophage to carry the E. coli Type I-E system as well as a CRISPR array encoding spacers that target the β-lactamase genes ndm-1 and ctx-M-15. In this case, the phage would form a stable lysogen in the host, and the encoded CRISPR-Cas system would both clear existing resistance genes and prevent the cells from acquiring resistance in the future. The authors demonstrated that the presence of the lysogen reduced the transformation efficiency of plasmids encoding the target sequences by three orders-of-magnitude. Furthermore, lysogens with the ndm-1 and ctx-M-15 spacers resisted T7 phages that housed ndm-1 and ctx-M-15 protospacers by over four orders-of-magnitude when compared to lysogens lacking the spacer array, demonstrating that these antibiotic sensitized bacteria have a selective advantage in their resistance to lytic phages.
Aside from bacteriophages, bacterial conjugation could be used to deliver DNA. Conjugation relies on cell-to-cell contact to transfer plasmids that often have wide host ranges. Citorik et al. (2014) found that conjugative transfer of the plasmid encoding the S. pyogenes tracrRNA, cas9, and targeting CRISPR array resulted in up to a 60-fold reduction of viable recipients in comparison to non-targeted cells. The limited reduction in the number of viable cells was attributed to the inefficiency of conjugal transfer, a common issue for this delivery method.
Delivery remains the largest hurdle to the implementation of CRISPR antimicrobials. Even in both examples cited above, the simple in vivo models exhibited greatly reduced efficacy in comparison to the in vitro experiments. Future efforts will need to focus on engineering the bacteriophages for improved delivery and exploring alternative delivery vehicles. The long-term success will also depend on identifying applications uniquely suited to CRISPR antimicrobials—particularly over traditional antibiotics, antimicrobial peptides, and lytic bacteriophages. These applications may extend well beyond human therapeutics and into the realms of diagnostics, agriculture, biomanufacturing, and research tools.
FUTURE PERSPECTIVES
CRISPR-Cas systems have already proven to be powerful tools for understanding and engineering bacteria. However, engineered CRISPR-Cas systems represent a three-year-old technology and much of their potential remains to be realized. Below, we posit next steps for the development of CRISPR technologies in bacteria and the ensuing opportunities and challenges.
Following in the footsteps of CRISPR in eukaryotes
Even though CRISPR-Cas systems are native to prokaryotes, advances in CRISPR technologies have focused almost entirely on eukaryotes. Regardless of the underlying reasons, advances in eukaryotes can serve as a guide for similar developments in prokaryotes—whether to drive existing applications or to institute entirely new ones.
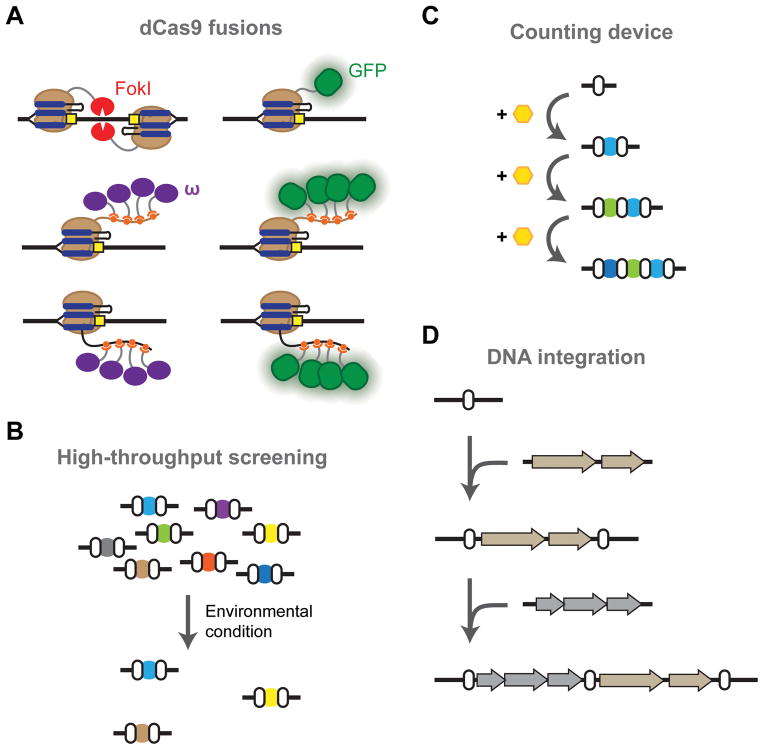
Genome editing remains the dominant and best-developed application of CRISPR-Cas systems in eukaryotes. Most of these developments have been driven by the need to express multiple sgRNAs, initiate homology-directed repair (HDR) over NHEJ, and limit unintended targeting (Hsu et al., 2014). Unintended targeting (or off-target effects) is less likely in bacteria because of their smaller genomes, and natural CRISPR arrays are well suited for multiplexing. However, many of the associated advances in eukaryotes could be useful in bacteria. For instance, fusions of dCas9 and the FokI endonuclease minimize unintended editing events by requiring a FokI dimerization event in order for DNA cleavage to occur (Guilinger et al., 2014; Tsai et al., 2014) (Figure 6A). In bacteria, this same fusion protein may generate double-stranded breaks in a manner that can more easily undergo repair by endogenous pathways. Separately, importing pathways for NHEJ or HDR could transition CRISPR from being a selective pressure to actively driving editing, as suggested by genome editing in actinomycetes (Tong et al., 2015). By improving the efficiency of genome editing in bacteria, CRISPR-based editing could be sufficiently effective to generate genome-wide modifications or to conduct high-throughput functional screens that identify genes associated with defined phenotypes (Koike-Yusa et al., 2013; Shalem et al., 2014; Wang et al., 2014; Zhou et al., 2014) (Figure 6B).
Figure 6.
Opportunities for CRISPR technologies in bacteria. (A) Building on advances in eukaryotes. The FokI nuclease can be fused to Cas9 as an alternative means of introducing double-stranded breaks as part of genome editing. Fusing binding domains to Cas9 or the 3′ end of the sgRNA can recruit other proteins to regulate gene expression (e.g. the ω subunit of RNA polymerase) or to dynamically image genomic loci (e.g. GFP). (B) Libraries of crRNAs or sgRNAs can be compiled and subjected to specific environmental conditions to rapidly screen for genes that either promote or inhibit growth. (C) Acquisition could be used to count transient events such as an environmental stimulus. (D) Acquisition could also be harnessed to integrate large pieces of synthetic DNA into the genome.
Gene regulation with CRISPR-Cas9 in eukaryotes has also witnessed major breakthroughs, with a focus on improving the system’s overall efficiency and utility. The most notable advance has been the development of protein fusions to drive gene activation and repression (Gilbert et al., 2013; Maeder et al., 2013; Mali et al., 2013a; Perez-Pinera et al., 2013) (Figure 6A). One of the key developments was transitioning from single-domain fusions, such as between dCas9 and the VP64 activating domain or the KRAB repressing domain, to extensions of dCas9 or the sgRNA that recruit the regulatory domains (Konermann et al., 2014; Shechner et al., 2015; Tanenbaum et al., 2014; Zalatan et al., 2014). For instance, the 3′ end of the sgRNA was extended to encode aptamers that recruited VP64 domains to efficiently activate transcription (Mali et al., 2013a). While gene repression with dCas9 and Cascade seems to be sufficiently potent in bacteria, there is ample room for improvement for gene activation.
Aside from improving existing applications in bacteria, a notable advancement in eukaryotes has not been reported in bacteria: DNA imaging. Here, dCas9 was fused to GFP (Anton et al., 2014; Chen et al., 2013; Ma et al., 2015) or recruited GFP (Tanenbaum et al., 2014) to dynamically image chromosomal structure and localization in mammalian cells (Figure 6A). Bacterial genomes are known to be highly structured and undergo major localization changes based on the environmental conditions (Libby et al., 2012). However, the associated imaging tools rely on previous technologies such as fusing GFP to transcription factors; incorporating dCas9 and GFP offers a more programmable tool that could drive our understanding of chromosomal dynamics in bacteria, shedding new light into the structure of bacterial cells and how this structure changes based on environmental conditions.
Extending beyond imported Cas9
CRISPR technologies have focused on importing Type II systems because of the ease in using Cas9 and an sgRNA. However, Type II systems represent merely one type of CRISPR-Cas systems, where the other systems individually offer distinct attributes and capabilities. For instance, Type I systems naturally process CRISPR arrays without the need for further engineering. Type I systems are also unique in that their signature Cas3 protein does not generate double-stranded breaks but rather is recruited to cleave and degrade DNA through the action of the protein’s 3′-to-5′ exonuclease activity (Sinkunas et al. 2013). Separately, Type III systems naturally target RNA and may offer targeting flexibility due to the absence of a PAM—whether for mRNA silencing or transcript processing. The challenge will be finding applications that are uniquely suited to a given system and counterbalance the added difficulty of expressing multiple proteins in defined ratios, rather than the single Cas9 protein for Type II systems. The newly categorized Type V systems and their Cpf1 proteins offer additional possibilities. Because Cpf1 processes crRNA arrays without any additional RNA species and the Cpf1-crRNA complex alone can achieve DNA cleavage, these systems may simplify the design of genome-editing tools (Zetsche et al., 2015). Alternatively, by implementing multiple, truly orthogonal types of CRISPR-Cas systems, multiple functions such as genome editing, plasmid curing, gene activation, gene repression, and invader defense can be performed simultaneously.
One potential opportunity beyond Cas9 is the utilization of endogenous systems already present in a microbe. CRISPR loci have been identified in the genomes of ~50% of bacteria and ~87% of archaea (Makarova et al., 2015), and the vast majority of these are associated with Type I systems and Type III systems. For bacteria harboring an active CRISPR-Cas system, the system may be readily co-opted by merely importing a synthetic CRISPR array. This approach was part of CRISPR antimicrobials (Gomaa et al., 2014), large deletions (Vercoe et al., 2013), and gene repression (Luo et al., 2015; Zebec et al., 2014). The benefit is that all of the protein components are already present—a major advantage for Class 1 systems that require multiple proteins (Table 1). In extreme thermophiles and hyperthermophiles, the traditional Cas9 proteins from mesophiles are unlikely to function at typical growth conditions. In contrast, Type I and Type III systems appear to be enriched in these microbes, arguing for their use in CRISPR-based applications. The downside to using endogenous systems is that the system must be confirmed to be active, which could be environment-dependent, and must undergo full characterization—namely, identifying the PAM for Type I, II, and V systems as well as determining the tracrRNA for Type II systems. Furthermore, co-opting the endogenous system may interfere with its natural functions such as in host defense. Accordingly, choosing to import a system (and which type) or to rely on an endogenous system will likely depend on the selected organism and the needs of the project. Finally, even if an imported Cas9 is selected, there is a choice between using CRISPR arrays or sgRNAs. The important takeaway is that importing Cas9 with an sgRNA is not the only option and, in many cases, may be less desirable than the alternatives.
Exploiting CRISPR-Cas acquisition
To date, CRISPR-derived genetic tools have centered on the final step in prokaryotic adaptive immunity—interference. However, an unexplored step is acquisition (Heler et al., 2014). Acquisition represents the first step of acquired immunity wherein the CRISPR-Cas system integrates a small piece of the foreign invader DNA (Figure 1). This step is associated with the near-universal proteins Cas1 and Cas2, with some involvement by Cas4 in some subtypes (Chylinski et al., 2014; Makarova et al., 2011; Makarova et al., 2015; Nuñez et al., 2014). Cas1 and Cas2 integrate new repeat-spacers in the CRISPR array, with a preference for stalled replication forks (Levy et al., 2015). However, the Cas proteins involved in interference appear to play a predominant role in spacer selection, whether by selecting spacers with a PAM (Heler et al., 2015; Paez-Espino et al., 2013) or by driving acquisition of sequences in the vicinity of a target sequence (Fineran et al., 2014; Richter et al., 2014). There is still plenty left to learn about the natural mechanisms that drive acquisition, although there may be sufficient information to begin engineering this untapped yet universal aspect of CRISPR-Cas systems.
We envision two avenues in which acquisition can be harnessed for tools in bacteria. In the first avenue, acquisition can be linked to an exogenous or intracellular signal such that transient induction leads to an acquisition event (Figure 6C). This could in turn be used as a permanent memory of prior transient events—much like natural CRISPR loci serve as memories of prior infections. This approach could offer a simpler strategy to track these events, particularly in comparison to existing synthetic counters and memory devices (Bonnet et al., 2012; Friedland et al., 2009; Siuti et al., 2013). Second, the system could be harnessed as site-specific recombinases in order to drive the efficient insertion of a synthetic DNA sequence (Figure 6D). While the natural systems prefer to integrate ~30-base sequences directly into the leading repeat, it may be possible to reengineer the system to accept larger or shorter sequences that can be inserted at other locations. This capability could greatly expand the existing repertoire of protein integrases and form the next generation of CRISPR technologies.
CONCLUSIONS
CRISPR-Cas systems have enjoyed tremendous popularity within recent years, owing to the versatility and power of these widespread defense systems. Bacterial applications thus far include streamlined genome engineering, programmable transcriptional regulation, typing and epidemiology of strains, vaccination of bacteria against mobile genetic elements, and smart antibiotics. Most of these applications are in their infancy and there still is ample room for further improvements. Nonetheless, the gains so far have begun reshaping how we pursue basic and applied research in bacteria and offer considerable potential for the treatment of bacterial infections.
Acknowledgments
We thank K. Selle, R. Barrangou, and G. Williams for critical reading of the manuscript. This work was supported by funding from the National Science Foundation (CBET-1403135 to C.L.B, MCB-1452902 to C.L.B.) and the National Institutes of Health (5T32GM008776-15 to R.T.L.). M.L.L. and C.L.B. are co-inventors on multiple patent applications related to CRISPR-Cas systems and their uses.
References
- Aklujkar M, Lovley DR. Interference with histidyl-tRNA synthetase by a CRISPR spacer sequence as a factor in the evolution of Pelobacter carbinolicus. BMC Evol Biol. 2010;10:230. doi: 10.1186/1471-2148-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton T, Bultmann S, Leonhardt H, Markaki Y. Visualization of specific DNA sequences in living mouse embryonic stem cells with a programmable fluorescent CRISPR/Cas system. Nucleus. 2014;5:163–172. doi: 10.4161/nucl.28488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11:1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Marraffini LA. CRISPR-Cas systems: Prokaryotes upgrade to adaptive immunity. Mol Cell. 2014;54:234–244. doi: 10.1016/j.molcel.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett AR, Tibbit C, Ponting CP, Liu JL. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Rep. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Euler CW, Jiang W, Nussenzweig PM, Goldberg GW, Duportet X, Fischetti VA, Marraffini LA. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat Biotechnol. 2014;32:1146–1150. doi: 10.1038/nbt.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, Marraffini LA. Control of gene expression by CRISPR-Cas systems. F1000Prime Rep. 2013;5:47. doi: 10.12703/P5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J, Davidson AR. To acquire or resist: The complex biological effects of CRISPR-Cas systems. Trends Microbiol. 2014;22:218–225. doi: 10.1016/j.tim.2014.01.007. [DOI] [PubMed] [Google Scholar]
- Bonnet J, Subsoontorn P, Endy D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc Natl Acad Sci U S A. 2012;109:8884–8889. doi: 10.1073/pnas.1202344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowater R, Doherty AJ. Making ends meet: Repairing breaks in bacterial DNA by non-homologous end-joining. PLoS Genet. 2006;2:e8. doi: 10.1371/journal.pgen.0020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel J, Stoll B, Lange SJ, Sharma K, Lenz C, Stachler AE, Maier LK, Richter H, Nickel L, Schmitz RA, Randau L, Allers T, Urlaub H, Backofen R, Marchfelder A. A complex of Cas proteins 5, 6, and 7 is required for the biogenesis and stability of clustered regularly interspaced short palindromic repeats (CRISPR)-derived RNAs (crRNAs) in Haloferax volcanii. J Biol Chem. 2014;289:7164–7177. doi: 10.1074/jbc.M113.508184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carte J, Wang R, Li H, Terns RM, Terns MP. Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev. 2008;22:3489–3496. doi: 10.1101/gad.1742908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Gilbert LA, Cimini BA, Schnitzbauer J, Zhang W, Li GW, Park J, Blackburn EH, Weissman JS, Qi LS, Huang B. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:230–232. doi: 10.1038/nbt.2507. [DOI] [PubMed] [Google Scholar]
- Choudhary E, Thakur P, Pareek M, Agarwal N. Gene silencing by CRISPR interference in mycobacteria. Nat Commun. 2015;6:6267. doi: 10.1038/ncomms7267. [DOI] [PubMed] [Google Scholar]
- Chylinski K, Makarova KS, Charpentier E, Koonin EV. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:6091–6105. doi: 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013;10:726–737. doi: 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat Biotechnol. 2014;32:1141–1145. doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobb RE, Wang Y, Zhao H. High-Efficiency Multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth Biol. 2015;4:723–728. doi: 10.1021/sb500351f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonté J, Fremaux C, Boyaval P, Romero DA, Horvath P, Moineau S. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. doi: 10.1128/JB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson DJ, Ward JD, Reiner DJ, Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dove SL, Hochschild A. Conversion of the omega subunit of Escherichia coli RNA polymerase into a transcriptional activator or an activation target. Genes Dev. 1998;12:745–754. doi: 10.1101/gad.12.5.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Qimron U. The Escherichia coli CRISPR system protects from λ lysogenization, lysogens, and prophage induction. J Bacteriol. 2010;192:6291–6294. doi: 10.1128/JB.00644-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fineran PC, Dy RL. Gene regulation by engineered CRISPR-Cas systems. Curr Opin Microbiol. 2014;18:83–89. doi: 10.1016/j.mib.2014.02.007. [DOI] [PubMed] [Google Scholar]
- Fineran PC, Gerritzen MJH, Suárez-Diez M, Künne T, Boekhorst J, van Hijum SAFT, Staals RHJ, Brouns SJJ. Degenerate target sites mediate rapid primed CRISPR adaptation. Proc Natl Acad Sci U S A. 2014;111:E1629–1638. doi: 10.1073/pnas.1400071111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, Lu TK, Wang X, Shi D, Church G, Collins JJ. Synthetic gene networks that count. Science. 2009;324:1199–1202. doi: 10.1126/science.1172005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedland AE, Tzur YB, Esvelt KM, Colaiácovo MP, Church GM, Calarco JA. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomaa AA, Klumpe HE, Luo ML, Selle K, Barrangou R, Beisel CL. Programmable removal of bacterial strains by use of genome-targeting CRISPR-Cas systems. MBio. 2014;5:e00928–13. doi: 10.1128/mBio.00928-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Majumdar S, Elmore J, Pfister N, Compton M, Olson S, Resch AM, Glover CVC, Graveley BR, Terns RM, Terns MP. Essential features and rational design of CRISPR RNAs that function with the Cas RAMP module complex to cleave RNAs. Mol Cell. 2012;45:292–302. doi: 10.1016/j.molcel.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale CR, Zhao P, Olson S, Duff MO, Graveley BR, Wells L, Terns RM, Terns MP. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatoum-Aslan A, Maniv I, Samai P, Marraffini La. Genetic characterization of antiplasmid immunity through a type III-A CRISPR-Cas system. J Bacteriol. 2014;196:310–7. doi: 10.1128/JB.01130-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haurwitz RE, Jinek M, Wiedenheft B, Zhou K, Doudna JA. Sequence- and structure-specific RNA processing by a CRISPR endonuclease. Science. 2010;329:1355–1358. doi: 10.1126/science.1192272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R, Marraffini LA, Bikard D. Adapting to new threats: the generation of memory by CRISPR-Cas immune systems. Mol Microbiol. 2014;93:1–9. doi: 10.1111/mmi.12640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heler R, Samai P, Modell JW, Weiner C, Goldberg GW, Bikard D, Marraffini LA. Cas9 specifies functional viral targets during CRISPR-Cas adaptation. Nature. 2015;519:199–202. doi: 10.1038/nature14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Zheng G, Jiang W, Hu H, Lu Y. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim Biophys Sin (Shanghai) 2015;47:231–243. doi: 10.1093/abbs/gmv007. [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, Peterson RT, Yeh JR, Joung JK. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Golden SM, van Erp PBG, Carter J, Westra ER, Brouns SJJ, van der Oost J, Terwilliger TC, Read RJ, Wiedenheft B. Crystal structure of the CRISPR RNA-guided surveillance complex from Escherichia coli. Science. 2014a;345:1473–1479. doi: 10.1126/science.1256328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RN, Lavin M, Carter J, Wiedenheft B. Fitting CRISPR-associated Cas3 into the Helicase family tree. Curr Opin Struct Biol. 2014b;24:106–114. doi: 10.1016/j.sbi.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- Jiang F, Doudna JA. The structural biology of CRISPR-Cas systems. Curr Opin Struct Biol. 2015;30:100–111. doi: 10.1016/j.sbi.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L, Gressel S, Doudna JA. A Cas9-guide RNA complex preorganized for target DNA recognition. Science. 2015a;348:1477–1481. doi: 10.1126/science.aab1452. [DOI] [PubMed] [Google Scholar]
- Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013a;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Maniv I, Arain F, Wang Y, Levin BR, Marraffini LA. Dealing with the evolutionary downside of CRISPR immunity: bacteria and beneficial plasmids. PLoS Genet. 2013b;9:e1003844. doi: 10.1371/journal.pgen.1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S. Multigene editing in the Escherichia coli genome using the CRISPR-Cas9 system. Appl Environ Microbiol. 2015b;81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:e00471. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jore MM, Lundgren M, van Duijn E, Bultema JB, Westra ER, Waghmare SP, Wiedenheft B, Pul U, Wurm R, Wagner R, Beijer MR, Barendregt A, Zhou K, Snijders APL, Dickman MJ, Doudna JA, Boekema EJ, Heck AJR, van der Oost J, Brouns SJJ. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat Struct Mol Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- Kiani S, Beal J, Ebrahimkhani MR, Huh J, Hall RN, Xie Z, Li Y, Weiss R. CRISPR transcriptional repression devices and layered circuits in mammalian cells. Nat Methods. 2014;11:723–726. doi: 10.1038/nmeth.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike-Yusa H, Li Y, Tan E-P, Velasco-Herrera MDC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2013;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, Nureki O, Zhang F. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2014;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon KP, Armitage GC, Relman DA, Fischbach MA. Microbiota-targeted therapies: an ecological perspective. Sci Transl Med. 2012;4:137rv5. doi: 10.1126/scitranslmed.3004183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A, Goren MG, Yosef I, Auster O, Manor M, Amitai G, Edgar R, Qimron U, Sorek R. CRISPR adaptation biases explain preference for acquisition of foreign DNA. Nature. 2015;520:505–510. doi: 10.1038/nature14302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Structural principles of CRISPR RNA processing. Structure. 2015;23:13–20. doi: 10.1016/j.str.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Lin Z, Huang C, Zhang Y, Wang Z, Tang Y, Chen T, Zhao X. Metabolic engineering of Escherichia coli using CRISPR-Cas9 meditated genome editing. Metab Eng. 2015;31:13–21. doi: 10.1016/j.ymben.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Libby EA, Roggiani M, Goulian M. Membrane protein expression triggers chromosomal locus repositioning in bacteria. Proc Natl Acad Sci U S A. 2012;109:7445–7450. doi: 10.1073/pnas.1109479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo ML, Mullis AS, Leenay RT, Beisel CL. Repurposing endogenous Type I CRISPR-Cas systems for programmable gene repression. Nucleic Acids Res. 2015;43:674–681. doi: 10.1093/nar/gku971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Naseri A, Reyes-Gutierrez P, Wolfe SA, Zhang S, Pederson T. Multicolor CRISPR labeling of chromosomal loci in human cells. Proc Natl Acad Sci USA. 2015;112:3002–3007. doi: 10.1073/pnas.1420024112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, Moineau S, Mojica FJM, Wolf YI, Yakunin AF, van der Oost J, Koonin EV. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJJ, Charpentier E, Haft DH, Horvath P, Moineau S, Mojica FJM, Terns RM, Terns MP, White MF, Yakunin AF, Garrett RA, van der Oost J, Backofen R, Koonin EV. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015 doi: 10.1038/nrmicro3569. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013a;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013b;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–1845. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. Self versus non-self discrimination during CRISPR RNA-directed immunity. Nature. 2010;463:568–571. doi: 10.1038/nature08703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- Mojica FJM, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- Nam KH, Haitjema C, Liu X, Ding F, Wang H, DeLisa MP, Ke A. Cas5d protein processes pre-crRNA and assembles into a Cascade-like interference complex in subtype IC/Dvulg CRISPR-Cas system. Structure. 2012;20:1574–1584. doi: 10.1016/j.str.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neish AS. Microbes in Gastrointestinal Health and Disease. Gastroenterology. 2009;136:65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov V, Staskawicz B, Weigel D, Jones JDG, Kamoun S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31:691–693. doi: 10.1038/nbt.2655. [DOI] [PubMed] [Google Scholar]
- Nielsen AA, Voigt CA. Multi-input CRISPR/Cas genetic circuits that interface host regulatory networks. Mol Syst Biol. 2014;10:763. doi: 10.15252/msb.20145735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niewoehner O, Jinek M, Doudna JA. Evolution of CRISPR RNA recognition and processing by Cas6 endonucleases. Nucleic Acids Res. 2014;42:1341–1353. doi: 10.1093/nar/gkt922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, Xiang AP, Zhou J, Guo X, Bi Y, Si C, Hu B, Dong G, Wang H, Zhou Z, Li T, Tan T, Pu X, Wang F, Ji S, Zhou Q, Huang X, Ji W, Sha J. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- Nuñez JK, Kranzusch PJ, Noeske J, Wright AV, Davies CW, Doudna Ja. Cas1–Cas2 complex formation mediates spacer acquisition during CRISPR–Cas adaptive immunity. Nat Struct Mol Biol. 2014;21:528–534. doi: 10.1038/nsmb.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, Doudna JA. Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature. 2014;516:263–266. doi: 10.1038/nature13769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J-H, van Pijkeren J-P. CRISPR-Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Res. 2014;42:e131. doi: 10.1093/nar/gku623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Oost J, Westra ER, Jackson RN, Wiedenheft B. Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014;12:479–492. doi: 10.1038/nrmicro3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa T, Inanaga H, Sato C, Numata T. Crystal Structure of the CRISPR-Cas RNA silencing Cmr complex bound to a target analog. Mol Cell. 2015;58:418–430. doi: 10.1016/j.molcel.2015.03.018. [DOI] [PubMed] [Google Scholar]
- Paez-Espino D, Morovic W, Sun CL, Thomas BC, Ueda K, Stahl B, Barrangou R, Banfield JF. Strong bias in the bacterial CRISPR elements that confer immunity to phage. Nat Commun. 2013;4:1430. doi: 10.1038/ncomms2440. [DOI] [PubMed] [Google Scholar]
- Peng W, Feng M, Feng X, Liang YX, She Q. An archaeal CRISPR type III-B system exhibiting distinctive RNA targeting features and mediating dual RNA and DNA interference. Nucleic Acids Res. 2014;43:406–417. doi: 10.1093/nar/gku1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W, Li H, Hallstrøm S, Peng N, Liang YX, She Q. Genetic determinants of PAM-dependent DNA targeting and pre-crRNA processing in Sulfolobus islandicus. RNA Biol. 2013;10:738–748. doi: 10.4161/rna.23798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, Guilak F, Crawford GE, Reddy TE, Gersbach CA. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AA, Sampson TR, Ratner HK, Grakoui A, Weiss DS. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc Natl Acad Sci U S A. 2015;112:6164–6169. doi: 10.1073/pnas.1422340112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyne ME, Moo-Young M, Chung DA, Chou CP. Coupling the CRISPR/Cas9 System with Lambda Red recombineering enables simplified chromosomal gene replacement in Escherichia coli. In: Kivisaar M, editor. Appl Environ Microbiol. Vol. 81. 2015. pp. 5103–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L, Haurwitz RE, Shao W, Doudna JA, Arkin AP. RNA processing enables predictable programming of gene expression. Nat Biotechnol. 2012;30:1002–1006. doi: 10.1038/nbt.2355. [DOI] [PubMed] [Google Scholar]
- Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath D, Amlinger L, Hoekzema M, Devulapally PR, Lundgren M. Efficient programmable gene silencing by Cascade. Nucleic Acids Res. 2015;43:237–246. doi: 10.1093/nar/gku1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter C, Dy RL, McKenzie RE, Watson BNJ, Taylor C, Chang JT, McNeil MB, Staals RHJ, Fineran PC. Priming in the Type I-F CRISPR-Cas system triggers strand-independent spacer acquisition, bi-directionally from the primed protospacer. Nucleic Acids Res. 2014;42:8516–8526. doi: 10.1093/nar/gku527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouillon C, Zhou M, Zhang J, Politis A, Beilsten-Edmands V, Cannone G, Graham S, Robinson CV, Spagnolo L, White MF. Structure of the CRISPR interference complex CSM reveals key similarities with Cascade. Mol Cell. 2013;52:124–134. doi: 10.1016/j.molcel.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samai P, Pyenson N, Jiang W, Goldberg GW, Hatoum-Aslan A, Marraffini LA. Co-transcriptional DNA and RNA cleavage during type III CRISPR-Cas immunity. Cell. 2015;161:1164–1174. doi: 10.1016/j.cell.2015.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Saroj SD, Llewellyn AC, Tzeng YL, Weiss DS. A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature. 2013;497:254–457. doi: 10.1038/nature12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson TR, Weiss DS. Exploiting CRISPR/Cas systems for biotechnology. Bioessays. 2014;36:34–38. doi: 10.1002/bies.201300135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Wiedenheft B, Doudna JA. Mechanism of foreign DNA selection in a bacterial adaptive immune system. Mol Cell. 2012;46:606–615. doi: 10.1016/j.molcel.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selle K, Barrangou R. Harnessing CRISPR-Cas systems for bacterial genome editing. Trends Microbiol. 2015;23:225–232. doi: 10.1016/j.tim.2015.01.008. [DOI] [PubMed] [Google Scholar]
- Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG, Zhang F. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shechner DM, Hacisuleyman E, Younger ST, Rinn JL. Multiplexable, locus-specific targeting of long RNAs with CRISPR-Display. Nat Methods. 2015;12:664–670. doi: 10.1038/nmeth.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkunas T, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. Cas3 is a single-stranded DNA nuclease and ATP-dependent helicase in the CRISPR/Cas immune system. EMBO J. 2011;30:1335–1342. doi: 10.1038/emboj.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siuti P, Yazbek J, Lu TK. Synthetic circuits integrating logic and memory in living cells. Nat Biotechnol. 2013;31:448–452. doi: 10.1038/nbt.2510. [DOI] [PubMed] [Google Scholar]
- Spilman M, Cocozaki A, Hale C, Shao Y, Ramia N, Terns R, Terns M, Li H, Stagg S. Structure of an RNA silencing complex of the CRISPR-Cas immune system. Mol Cell. 2013;52:146–152. doi: 10.1016/j.molcel.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RHJ, Agari Y, Maki-Yonekura S, Zhu Y, Taylor DW, van Duijn E, Barendregt A, Vlot M, Koehorst JJ, Sakamoto K, Masuda A, Dohmae N, Schaap PJ, Doudna JA, Heck AJR, Yonekura K, van der Oost J, Shinkai A. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol Cell. 2013;52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staals RHJ, Zhu Y, Taylor DW, Kornfeld JE, Sharma K, Barendregt A, Koehorst JJ, Vlot M, Neupane N, Varossieau K, Sakamoto K, Suzuki T, Dohmae N, Yokoyama S, Schaap PJ, Urlaub H, Heck AJR, Nogales E, Doudna JA, Shinkai A, van der Oost J. RNA Targeting by the Type III-A CRISPR-Cas Csm complex of Thermus thermophilus. Mol Cell. 2014;56:518–530. doi: 10.1016/j.molcel.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A, Keren L, Wurtzel O, Amitai G, Sorek R. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 2010;26:335–340. doi: 10.1016/j.tig.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczelkun MD, Tikhomirova MS, Sinkunas T, Gasiunas G, Karvelis T, Pschera P, Siksnys V, Seidel R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc Natl Acad Sci U S A. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamulaitis G, Kazlauskiene M, Manakova E, Venclovas C, Nwokeoji AO, Dickman MJ, Horvath P, Siksnys V. Programmable RNA Shredding by the Type III-A CRISPR-Cas System of Streptococcus thermophilus. Mol Cell. 2014;56:506–517. doi: 10.1016/j.molcel.2014.09.027. [DOI] [PubMed] [Google Scholar]
- Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD. A versatile protein tagging system for signal amplification in single molecule imaging and gene regulation. Cell. 2014;159:635–646. doi: 10.1016/j.cell.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor DW, Zhu Y, Staals RHJ, Kornfeld JE, Shinkai A, van der Oost J, Nogales E, Doudna JA. Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. Science. 2015;348:581–585. doi: 10.1126/science.aaa4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong Y, Charusanti P, Zhang L, Weber T, Lee SY. CRISPR-Cas9 based engineering of Actinomycetal genomes. ACS Synth Biol. 2015;4:1020–1029. doi: 10.1021/acssynbio.5b00038. [DOI] [PubMed] [Google Scholar]
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercoe RB, Chang JT, Dy RL, Taylor C, Gristwood T, Clulow JS, Richter C, Przybilski R, Pitman AR, Fineran PC. Cytotoxic Chromosomal Targeting by CRISPR/Cas Systems Can Reshape Bacterial Genomes and Expel or Remodel Pathogenicity Islands. PLoS Genet. 2013;9:e1003454. doi: 10.1371/journal.pgen.1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter J, Ley R. The Human Gut Microbiome: Ecology and Recent Evolutionary Changes. Annu Rev Microbiol. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang ZT, Seo SO, Choi K, Lu T, Jin YS, Blaschek HP. Markerless chromosomal gene deletion in Clostridium beijerinckii using CRISPR/Cas9 system. J Biotechnol. 2015;200:1–5. doi: 10.1016/j.jbiotec.2015.02.005. [DOI] [PubMed] [Google Scholar]
- Westra ER, van Erp PBG, Künne T, Wong SP, Staals RHJ, Seegers CLC, Bollen S, Jore MM, Semenova E, Severinov K, de Vos WM, Dame RT, de Vries R, Brouns SJJ, van der Oost J. CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell. 2012;46:595–605. doi: 10.1016/j.molcel.2012.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, Semenova E, Datsenko KA, Jackson RN, Wiedenheft B, Severinov K, Brouns SJJ. Type I-E CRISPR-cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS Genet. 2013;9:e1003742. doi: 10.1371/journal.pgen.1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westwater C, Schofield DA, Schmidt MG, Norris JS, Dolan JW. Development of a P1 phagemid system for the delivery of DNA into Gram-negative bacteria. Microbiology. 2002;148:943–950. doi: 10.1099/00221287-148-4-943. [DOI] [PubMed] [Google Scholar]
- Wiedenheft B, Lander GC, Zhou K, Jore MM, Brouns SJJ, van der Oost J, Doudna Ja, Nogales E. Structures of the RNA-guided surveillance complex from a bacterial immune system. Nature. 2011;477:486–489. doi: 10.1038/nature10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci U S A. 2015;112:3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Li Y, Shi Z, Hemme CL, Li Y, Zhu Y, Van Nostrand JD, He Z, Zhou J. Efficient genome editing in Clostridium cellulolyticum via CRISPR-Cas9 nickase. In: Spormann AM, editor. Appl Environ Microbiol. Vol. 81. 2015. pp. 4423–4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yosef I, Manor M, Kiro R, Qimron U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc Natl Acad Sci USA. 2015;112:7267–7272. doi: 10.1073/pnas.1500107112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalatan JG, Lee ME, Almeida R, Gilbert LA, Whitehead EH, La Russa M, Tsai JC, Weissman JS, Dueber JE, Qi LS, Lim WA. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2014;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebec Z, Manica A, Zhang J, White MF, Schleper C. CRISPR-mediated targeted mRNA degradation in the archaeon Sulfolobus solfataricus. Nucleic Acids Res. 2014;42:5280–5288. doi: 10.1093/nar/gku161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F. Cpf1 Is a Single RNA-guided endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015 doi: 10.1016/j.cell.2015.09.038. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Sheng G, Wang J, Wang M, Bunkoczi G, Gong W, Wei Z, Wang Y. Crystal structure of the RNA-guided immune surveillance Cascade complex in Escherichia coli. Nature. 2014;515:147–150. doi: 10.1038/nature13733. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, Wei W. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]