Abstract
In subcellular light-sheet fluorescence microscopy (LSFM) of adherent cells, glass substrates are advantageously rotated relative to the excitation and emission light paths to avoid glass-induced optical aberrations. Because cells are spread across the sample volume, three-dimensional imaging requires a light-sheet with a long propagation length, or rapid sample scanning. However, the former degrades axial resolution and/or optical sectioning, while the latter mechanically perturbs sensitive biological specimens on pliant biomimetic substrates (e.g., collagen and basement membrane). Here, we use aberration-free remote focusing to diagonally sweep a narrow light-sheet along the sample surface, enabling multicolor imaging with high spatiotemporal resolution. Further, we implement a dithered Gaussian lattice to minimize sample-induced illumination heterogeneities, significantly improving signal uniformity. Compared with mechanical sample scanning, we drastically reduce sample oscillations, allowing us to achieve volumetric imaging at speeds of up to 3.5 Hz for thousands of Z-stacks. We demonstrate the optical performance with live-cell imaging of microtubule and actin cytoskeletal dynamics, phosphoinositide signaling, clathrin-mediated endocytosis, polarized blebbing, and endocytic vesicle sorting. We achieve three-dimensional particle tracking of clathrin-associated structures with velocities up to 4.5 μm/s in a dense intracellular environment, and show that such dynamics cannot be recovered reliably at lower volumetric image acquisition rates using experimental data, numerical simulations, and theoretical modeling.
Introduction
Fluorescence microscopy is the method of choice for analyzing cellular heterogeneity (1), the spatiotemporal dynamics of signal transduction (2), and the evolution of molecular architectures (3). However, the absorption of light by fluorophores and electronically conjugated intracellular constituents (e.g., NADH and flavins) produces chemically reactive species that compromise cellular health and degrade image contrast (4). Thus, to minimize photobleaching and phototoxicity, the excitation light should ideally be 1) kept at low intensities, 2) restricted to the focal plane that is being imaged, and 3) fluorescence should be efficiently captured. These three criteria are fulfilled by light-sheet fluorescence microscopy (LSFM), which illuminates only the part of the sample that is in focus and significantly reduces sample irradiance (5). Consequently, LSFM has enabled unprecedented long-term functional imaging of embryogenesis and neurotransmission in small and sufficiently transparent animals, including Drosophilia melanogaster, Danio rerio, and Caenorhabditis elegans (6).
Typically, one-dimensional (1D) and two-dimensional (2D) Gaussian beams are used to generate a light-sheet, as such beams most effectively confine the excitation power to the focal plane. An obvious drawback of Gaussian optics is that with an increasing field of view (FOV), restricted by ∼2 Rayleigh lengths of the beam, the beam waist grows and degrades the axial resolution (7). Various illumination strategies have been proposed to overcome this inherent trade-off between axial resolution and FOV, including Bessel beams and their coherent superposition (8, 9, 10, 11, 12), Airy beams (13), extended focusing (14, 15), fusion of multiple volumetric data sets (16, 17), and scanning a narrow light-sheet in the image plane (18). Although all of these techniques extend the FOV for a given axial resolution, they do so by drastically reducing illumination confinement. Consequently, important advantages of LSFM are lost, including the reduction of volumetric sample exposure and the minimization of photobleaching and phototoxicity. To simultaneously achieve high excitation confinement and axial resolution, it is preferable to illuminate the sample with the shortest light-sheet possible, regardless of the type of light-sheet (e.g., Gaussian, Bessel, or Airy) used.
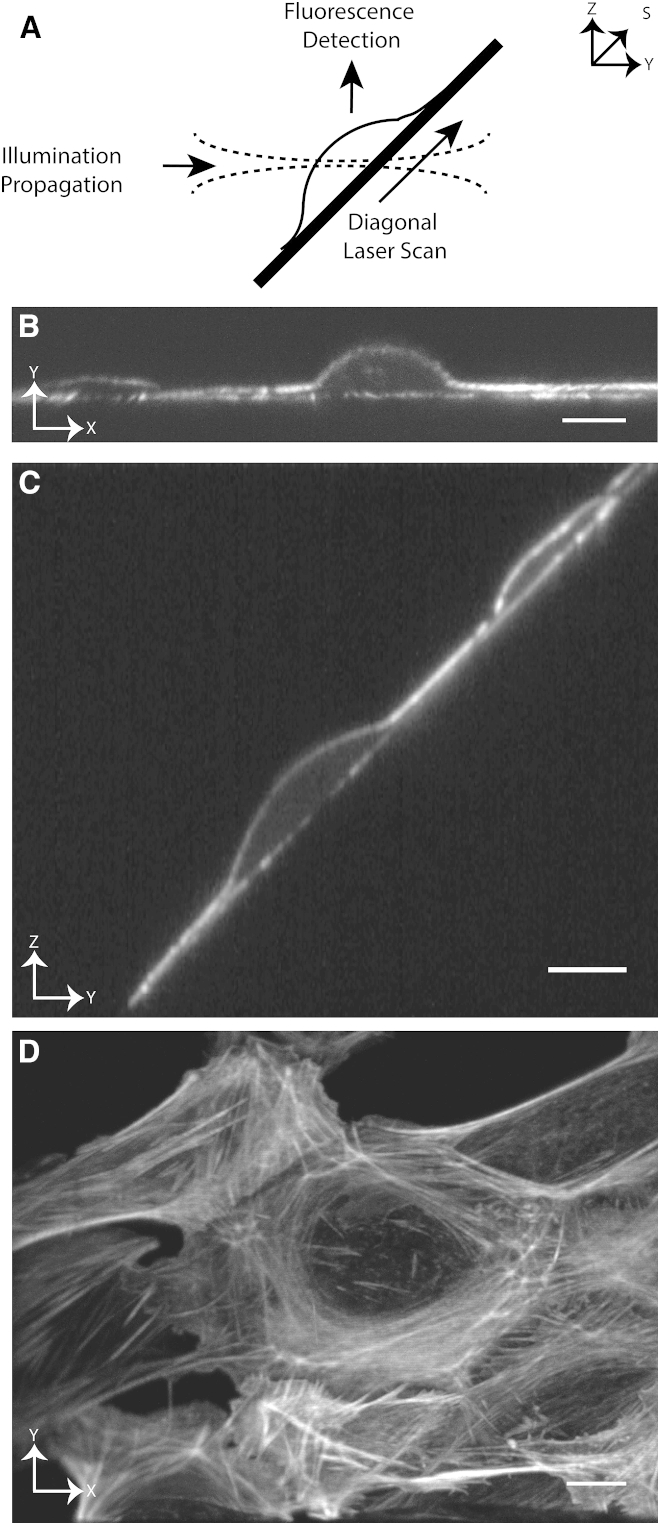
Many mammalian cell types adopt thin morphological profiles when adhered to rigid planar substrates (e.g., optical coverslips). If such a sample is mounted at 45° relative to the optical axes of the illumination and detection optics, a narrow light-sheet is sufficient to illuminate even the tallest part of the cell (Fig. 1 A). Furthermore, this arrangement is favorable for water-dipping objectives, as both the illumination and detection light paths are not aberrated by the glass coverslip. Nevertheless, for adherent cells oriented at 45°, a scan in the Z-direction is accompanied by a lateral shift of the sample relative to the beam waist in the Y direction (Fig. 1 A), necessitating a longer light-sheet at the cost of decreased axial resolution and/or illumination confinement. This problem can be overcome by diagonally scanning the sample itself through the beam waist (9). However, at rapid imaging speeds, sample scanning may perturb the biological sample, and each image slice has to be computationally shifted (deskewed) to its proper position in the three-dimensional (3D) image volume (11). As subpixel shifts are required, interpolation of the data cannot be avoided, resulting in local smoothing of the data and alteration of the signal statistics.
Figure 1.
(A) Schematic of DiaSLM. A dithered Gaussian lattice propagating in the Y-direction illuminates a cell oriented at 45°, and fluorescence is imaged in an orthogonal direction. The illumination beam is scanned diagonally in the S-direction along the coverslip, and an image is acquired at each Z plane. The beam length is configured to encompass the tallest region of the cell (e.g., the nucleus, ∼11 μm at FWHM). (B) Image of U2OS cells expressing CyOFP-Tractin, acquired at an intermediate Z plane. (C) A single plane in YZ, showing the 45° sample geometry and high degree of optical sectioning. (D) XY maximum intensity projection of the same U2OS cells expressing CyOFP-Tractin. Scale bar, 10 μm.
Previously, we reported a variant of LSFM, referred to as axially swept light-sheet microscopy (ASLM), that achieved isotropic sub-400 nm resolution throughout large imaging volumes (160 × 160 × 100 μm3) (18). In ASLM, a narrow light-sheet is swept along its propagation path (Y-direction) with a remote focusing scan mechanism and only the in-focus region is imaged. However, when equipped with galvanometric mirrors that scan the beam in both Z and X, the illumination train for ASLM is also capable of translating a light-sheet arbitrarily in 3D, including along a coverslip oriented at 45° relative to the optical axes. We exploited this capability to create a new imaging mode for ASLM, which we refer to as diagonally swept light-sheet microscopy (DiaSLM), that allows rapid multicolor and volumetric imaging (3.5 Hz) of cells on 2D surfaces. Since the light-sheet is placed at the optimal position in both the Z- and Y-directions for every Z-slice, sample scanning (and thus mechanical perturbation of the sample) is unnecessary. This scheme provides true 3D imaging in the sense that the raw sample information is in its proper Euclidean reference frame without the need for any signal-altering computational processing. Because our light-sheet is free of side lobes that propagate beyond the detection objective depth of focus, the raw data are devoid of out-of-focus blur. Thus, in contrast to most high-resolution LSFM techniques (9, 11, 14, 16, 19), computational deconvolution or reconstruction of the data is unnecessary, the statistical moments of the data are preserved, and the data can be viewed and analyzed in their raw form.
Here, we demonstrate potential applications of DiaSLM by volumetrically imaging rapid intracellular dynamics in live-cells for thousands of Z-stacks. These processes include actin and microtubule dynamics, clathrin-mediated endocytosis (CME), phosphoinositide signaling, endocytic trafficking of transferrin, membrane protrusions, ruffling, and polarized blebbing. We also perform 3D particle tracking of densely populated clathrin-coated structures with velocities up to 4.5 μm/s. Using simulations and a mathematical model that relates particle motion, particle density, and the volumetric image acquisition rate, we show that such dynamics cannot be reliably estimated below a 3D image acquisition rate of 3.1 Hz. Lastly, we perform mechanically nonperturbative imaging of cells on top of thick extracellular matrices (ECMs).
Materials and Methods
Microscope setup
The microscope configuration is presented in Fig. S1 in the Supporting Material. Two modifications of ASLM (18) were introduced to enable greater illumination intensity stability and uniformity. Illumination stability was improved by modulating the laser amplitude with a temperature-stabilized acousto-optical tunable filter (AOTFnC-400.650-TN; AA Opto-Electronic, Orsay, France). Illumination uniformity was improved by generating a lattice of Gaussian beams at the sample plane and dithering the lattice in the X-direction with a galvanometric mirror conjugate to the back-pupil plane of the excitation objective. The Gaussian lattice was generated with a custom-built binary amplitude grating (20% transmission, 60 μm pitch; Photo Sciences, Torrance, CA) conjugated to the sample plane. The grating was designed so that for both excitation wavelengths (488 and 560 nm), five diffraction orders (0, ±1, and ±2) fit into the back pupil of the excitation objective (40×, 0.8 NA; Nikon Instruments, Melville, NY). A low-spatial-duty cycle was selected to strengthen the higher diffraction orders relative to the zeroth diffraction order, thereby improving the illumination homogenization. The X-galvo (6210HSM40B; Cambridge Technologies, Bedford, MA) was conjugated to the Z-galvo (6210HSM40B; Cambridge Technologies) with a pair of 60 mm achromatic doublets (AC254-060A; ThorLabs, Newton, NJ). All downstream components were identical to those reported previously (18), with the exception of the scan lens (S4LFT4375; Sill Optics, Wendelstein, Germany), which was inadvertently reported to have a focal length of 60 mm (it is actually 80.2 mm).
At an NA of 0.8 (the maximum of our excitation objective), the dithered Gaussian lattice had a 20% and 30% larger beam waist and propagation length (defined as 2 Rayleigh lengths of the beam), respectively, than a canonical 1D Gaussian beam. This is because the higher diffraction orders (±1 and ±2) have a lower effective NA than the zeroth-order beam. In this work, we did not use the full NA of the illumination objective, a parameter that was controlled by a variable slit aperture (Fig. S1 A). By adjusting the slit width, we could arbitrarily control the NA of the illumination beam. Here, we routinely set the full width half maximum (FWHM) of the beam waist and propagation length to ∼800 nm and ∼11 μm, respectively, in the sample plane of the microscope. Using 100 nm fluorescent spheres (Fluoresbrite Plain YG; Polysciences, Warrington, PA), the axial and lateral resolution was found to be 725 ± 29 nm and 379 ± 18 nm, respectively. Because the detection path remained unchanged, the lateral resolution is the same as that reported for ASLM (18).
For optimal high-speed and high-resolution imaging, vibrations were identified with an accelerometer (41A19-1032; Meggitt Sensing Systems, Irvine, CA), an IEPE data acquisition system (cDAQ-9171 and NI 9234; National Instruments, Austin, TX), and custom software (LabView; The MathWorks, Natick, MA). Primary vibrations included the 488 nm laser fan (95 and 661 Hz, Sapphire; Coherent, Santa Clara, CA), camera fan (67 and 337 Hz, Flash 4.0; Hamamatsu, Bridgewater, NJ), piezo driver (86, 123, 617, and 863 Hz; E-509; Physik Instrumente, Auburn, MA), and building (2.5 Hz). With the exception of those induced by the building, vibrations were remedied by turning off the laser and camera fans, providing water cooling to the camera (ThermoCube 300; Solid State Cooling Systems, Wappingers Falls, NY), and removing the piezo driver from the optical table.
Sample scan experiments
To evaluate sample scanning, an objective scan unit (P-726.1CD; Physik Instrumente) was attached to a 3D stage (MP-285; Sutter Instruments) for sample positioning. Instead of the objective, a short lens tube (SM1L01; ThorLabs) was mounted to the piezo scanner with a thread adaptor. At the end of the lens tube, an SM1 threaded cage plate (CP08; ThorLabs) was attached. Our custom-fabricated 5 mm coverslip mounting unit (White Glove Machining, Upper Marlboro, MD) was mounted to the cage plate with a 0.75-inch-long 0.5-inch-diameter stainless steel optical post (TR075; ThorLabs). To evaluate whether the sample mount itself had measurable mechanical oscillations, we imaged 200 nm fluorescent microspheres that had adhered to glass coverslips in sample scanning and DiaSLM modes.
Sample preparation
All samples were positioned at 45° relative to the excitation and detection axes. Cellular imaging was performed on #1 thickness glass substrates (CS-5R; Warner Instruments) within a custom low-profile coverslip mounting system (White Glove Machining). A CAD drawing of the sample holder and its positioning relative to the objectives is shown in Fig. S2. ECM-based imaging was performed identically, with the exception that the coverslip was recessed 2 mm to provide a thick substrate. This allowed us to keep the cells far away from stiff surfaces, decreasing the likelihood of alterations to their mechanical microenvironment. ECMs were prepared by neutralizing pepsinized bovine collagen I at a final concentration of 2 mg/mL (Corning, Corning, NY). For the 50:50 collagen matrigel matrix, equal parts of matrigel and 2 mg/mL collagen I solution were mixed rapidly during polymerization of the matrix.
SK-MEL2 melanoma cells overexpressing human clathrin light chain A (CLCa) fused to TagRFP-T (20) have been previously described (21). We labeled F-actin in these cells by infecting them with lentivirus encoding Tractin-EGFP, a peptide that contains residues 9–52 of the F-actin-binding enzyme ITPKA (22). After infection, the cells were placed under selection with 0.1 mg/mL puromycin for 10 days, sorted for cells positive for both red and green expression, and amplified in the absence of puromycin. For imaging, cells were trypsinized and seeded at ∼75% confluency directly on uncoated flame-sterilized coverglass. Cells were imaged 18–24 h after seeding. Spontaneously arising retinal pigment epithelial (ARPE)-19 cells were infected with retrovirus generated from a pMIEG3 vector encoding EGFP-CLCa, as previously described (23). Then, 105 cells were seeded per well in a six-well plate containing 5 mm uncoated coverslips with Dulbecco’s modified Eagle medium/F12 containing 10% fetal bovine serum. MV3 cells were grown in Dulbecco’s modified Eagle medium supplemented with 10% fetal calf serum and 0.1% antibiotic, and stably transfected with the GFP-AktPH biosensor in a retroviral vector as described previously (24). Osteosarcoma cells (U2OS) were maintained in McCoy’s 5A medium with 10% fetal bovine serum and 0.2% antibiotic-antimycotic, and the filamentous actin was labeled with lentivirus encoding CyOFP-Tractin. CyOFP is a novel cyan-excitable orange fluorescent protein with peak excitation and emission at 505 and 588 nm, respectively, and will be described elsewhere.
Results
The concept of DiaSLM is schematically shown in Fig. 1 A and a technical drawing of the microscope is presented in Fig. S1 A. The sample, placed at 45° relative to the excitation and detection axes, is illuminated with a dithered Gaussian lattice (see below; beam propagation length ∼11 μm, beam waist ∼800 nm; Fig. S1, B and C) propagating in the Y-direction (25). When viewed on a camera, the cell thus appears as a narrow optical section, as shown in Fig. 1 B. To generate 3D data, the dithered light-sheet is scanned diagonally along the coverslip and an optical section of the cell is acquired for each plane in the Z-direction. Scanning of the illumination beam in the Y- and Z-directions is achieved with aberration-free remote focusing (18, 26, 27) and a galvanometric mirror, respectively. The detection objective is synchronously stepped with a piezo actuator.
Because the light-sheet is free of side lobes, a high degree of optical sectioning (i.e., the ability to reject light originating from outside of the detection objective’s depth of focus) is achieved, as shown in a YZ cross-section (Fig. 1 C). Alternatively, projecting the data along the Z axis provides a convenient view of the entire cell, albeit rotated 45° off-axis from the coverslip (Fig. 1 D). Affine transformation of the data can reorient the sample into the more familiar epifluorescence perspective (e.g., normal to the coverslip), but at the cost of interpolating voxel intensities.
The same image volume could be acquired with ASLM (18), but for this particular sample geometry, DiaSLM offers two important advantages. First, because the time-averaged Gaussian lattice in DiaSLM is approximately stationary during each camera exposure, the effective pixel dwell time is drastically longer (∼30× for a 512 × 512 image). Second, in DiaSLM, the remote focusing piezo actuator makes only one sweep during the acquisition of an entire Z-stack, whereas in ASLM it makes one sweep per acquired image plane (see schematic, Fig. S3). Therefore, the bandwidth requirements of the remote focusing piezo are relaxed by ∼2 orders of magnitude. Together, these advantages result in improved sensitivity, decreased sample irradiance (e.g., 0.3–30 μW in the back pupil of the excitation objective), diminished photobleaching and phototoxicity, and image acquisition rates approaching the camera-readout limited regime (400 planes per second for a 512 × 512 image). Consequently, DiaSLM enables routine multicolor observation of cells in 3D for thousands of Z-stacks at a volumetric image acquisition rate of up to 3.5 Hz, without stereotypic phototoxic signatures (Movie S1).
The effective pixel dwell time is a key parameter for imaging sensitivity in LSFM (11), but it must be balanced with illumination uniformity. For example, a 1D light-sheet formed by a cylindrical lens enables instantaneous illumination of the entire FOV (and thus the highest-illumination duty cycle). However, such a beam geometry is prone to shadow artifacts that arise from sample heterogeneity and imperfections in the optical train (25). The evenness of illumination can be greatly enhanced by rapidly sweeping a 2D Gaussian beam across the FOV to generate a time-averaged light-sheet. The improvement in illumination uniformity arises because a 2D Gaussian beam fills a much larger solid angle than a beam generated from a cylindrical lens. However, sweeping comes at the cost of a drastically decreased illumination duty cycle (9, 28). Alternatively, a 1D illumination beam can be rotated in the sample plane, but, for short beams, this results in an unacceptable amount of out of focus illumination and blur (25). In DiaSLM, we balance the spatial duty cycle and illumination heterogeneity by generating a discrete set of laterally superimposed 2D Gaussian beams, which we refer to as a Gaussian lattice. We produce this lattice by placing a custom amplitude grating conjugate to the sample plane in the illumination path (Fig. S1). The Gaussian lattice is dithered in the X-direction with a galvanometric mirror to produce a time-averaged light-sheet with greatly improved illumination homogeneity (Fig. S4).
As a result of the rapid image acquisition rate and sensitive detection, DiaSLM enabled us to image a wide variety of fast-timescale biological events in 3D, including GFP-AktPH, a biosensor that reports on local phosphatidylinositol-3-kinase (PI3K) activity. Typically, such translocation biosensors must be imaged by total internal reflection fluorescence (TIRF) microscopy (29), which is limited to the ventral surface of cells and precludes observation of large-scale membrane ruffles (24). We volumetrically imaged GFP-AktPH in MV3 melanoma cells and observed heightened PI3K activity in membrane ruffles (Fig. 2, A and B; Movies S2 and S3). Although PI3K activity is necessary for membrane ruffling and accordingly has been observed in ruffles close to adhesive surfaces via TIRF or at indeterminate locations via epifluorescence (30, 31, 32), this is, to our knowledge, the first direct observation of PI3K activity in dorsal membrane ruffles. We also observed the formation of transverse arcs, specialized actin filaments involved in cell migration (Fig. 2 C), and the dynamics of other actin cytoskeleton-related structures (e.g., lamellipodium) in U2OS osteosarcoma cells (33). Furthermore, by labeling microtubule +TIPs with EB3-mNeonGreen, DiaSLM clearly resolved microtubule polymerization and centrosome position in 3D (Fig. 2 D; Movie S4).
Figure 2.
Dynamic signal transduction and cytoskeletal reorganization imaged in 3D with DiaSLM. (A) Volume rendering of a GFP-AktPH biosensor in a MV3 melanoma cell, with enhanced phosphatidylinositol localization in active membrane ruffles. (B) Maximum intensity projection of another MV3 cell expressing the GFP-AktPH biosensor. (C) Transverse contractile actin stress fibers in an osteosarcoma U2OS cell labeled with tractin-CyOFP. (D) Microtubule +TIP dynamics in adjacent osteosarcoma U2OS cells labeled with EB3-mNeonGreen. Scale bar, 10 μm.
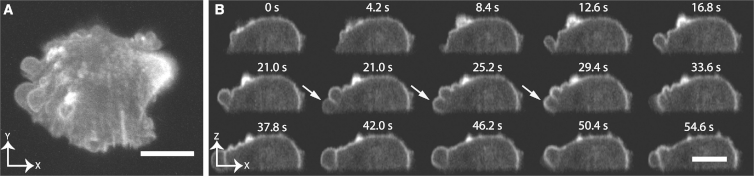
We also imaged MV3 melanoma cells expressing GFP-Tractin, which binds to filamentous actin, undergoing polarized blebbing (Fig. 3 A; Movie S5). Despite cellular heterogeneity (some cells spread and generated fibroblast-like morphological profiles (e.g., Fig. 2 B), many cells exhibited a stereotypical polarized amoeboid phenotype (34). Here, blebbing events were observed opposite to a stable uropod (35), with blebs and membrane appearing to flow backward toward the uropod. In some cases, the uropod was observed on the dorsal surface of the cell, and blebbing proceeded around the entire cell perimeter (data not shown). Because of the high temporal sampling (2.86 Hz), we were able to clearly resolve separation of the membrane from the actin cortex, rapid hemispherical expansion of the membrane, and polymerization of a nascent F-actin cortex within the newly protruded region (arrows in Fig. 3 B; Movie S6).
Figure 3.
Volumetric imaging at 2.86 Hz of an MV3 cell labeled with GFP-Tractin undergoing polarized blebbing. (A) XY maximum intensity projection. Blebs initiate adjacent to the coverslip on the left and flow toward a stable uropod, located in the upper right. (B) Montage of bleb formation and cortical flow. Every 12th time point of a single XZ cross section is shown. Arrows indicate membrane detachment from the actin cortex, polymerization of a nascent cortical actin meshwork, and flow toward the uropod (not visible in this cross section). Scale bar, 10 μm.
Because DiaSLM is based upon refractive optical elements, it is capable of operating in a simultaneous multicolor or pulse-interleaved illumination mode. In cases where fluorophore crosstalk is negligible, simultaneous multicolor illumination maximizes the image acquisition rate and enables the application of advanced computer vision frameworks, including econometric analysis of biological fluctuations and causality in signal transduction (2, 36, 37). In contrast, the pulsed-interleaved mode minimizes fluorescence crosstalk but introduces a small temporal delay between the acquisitions of each color channel.
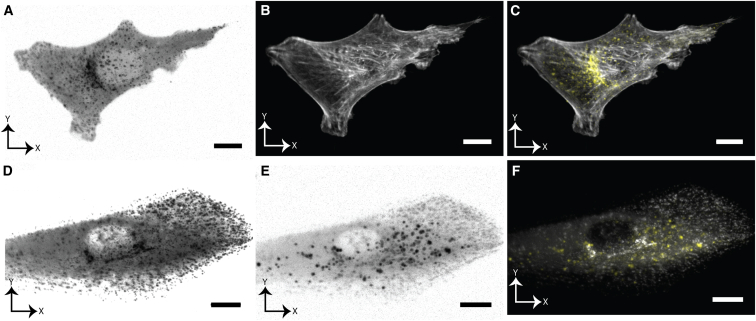
To demonstrate these imaging modes, we first imaged the dynamics of CME and the filamentous actin cytoskeleton using the simultaneous image acquisition mode in SK-MEL-2 melanoma cells (Fig. 4, A–C; Movie S7). The role of the actin cytoskeleton in CME remains controversial and is an active area of research, with investigators reporting contradictory observations depending on the cell type. Boulant et al. (38) suggested that actin’s main function during CME may be to counteract membrane tension, which hinders clathrin-coated pits from acquiring curvature. Since membrane tension depends on cellular adhesion and contractility, studying CME on both dorsal and ventral cell surfaces could provide significant insight into the molecular regulation and kinetics of CME.
Figure 4.
Volumetric multicolor imaging of the actin cytoskeleton, CME, transferrin uptake, and endosomal recycling. (A and B) XY maximum intensity projections of an SK-MEL-2 melanoma cell labeled with (A) CLCa-TagRFP-T and (B) EGFP-Tractin (image shown with a gamma correction of 0.7). (C) Merged color channels shown in (A) and (B). Images were acquired in a pulse-interleaved multicolor imaging mode at 0.19 Hz. (D and E) XY maximum intensity projections of human ARPE-19 cells labeled with (D) CLCa-EGFP and (E) Alexa Fluor 568-transferrin. (F) Merged color channels shown in (D) and (E). Images were acquired in a simultaneous multicolor imaging mode at 0.36 Hz. Scale bar, 10 μm.
We also imaged CLCa-EGFP-expressing cells in pulse-interleaved mode together with fluorescently labeled transferrin, which is transported into the cell by receptor-mediated endocytosis and is commonly used as a marker for CME. This allowed us to observe sites of transferrin uptake and their relation to CLCa-EGFP. In nonpolarized RPE cells, intracellular vesicles containing transferrin appeared to undergo periods of local diffusion followed by short bursts of rapid transport (Fig. 4, D–F; Movie S8). We also observed similar dynamics for clathrin-associated structures in polarized epithelial cells when we imaged Caco-2 human colon carcinoma cells, which adopt a columnar epithelial-tissue-like morphology. Biologically, these fast motions likely reflect large-scale motor-protein-driven transport along microtubules (39). Although the function of these clathrin-associated vesicles remains poorly understood, their rapid intracellular velocities are 1–2 orders of magnitude faster than the dynamics of clathrin-coated pits observed on the cell membrane (40).
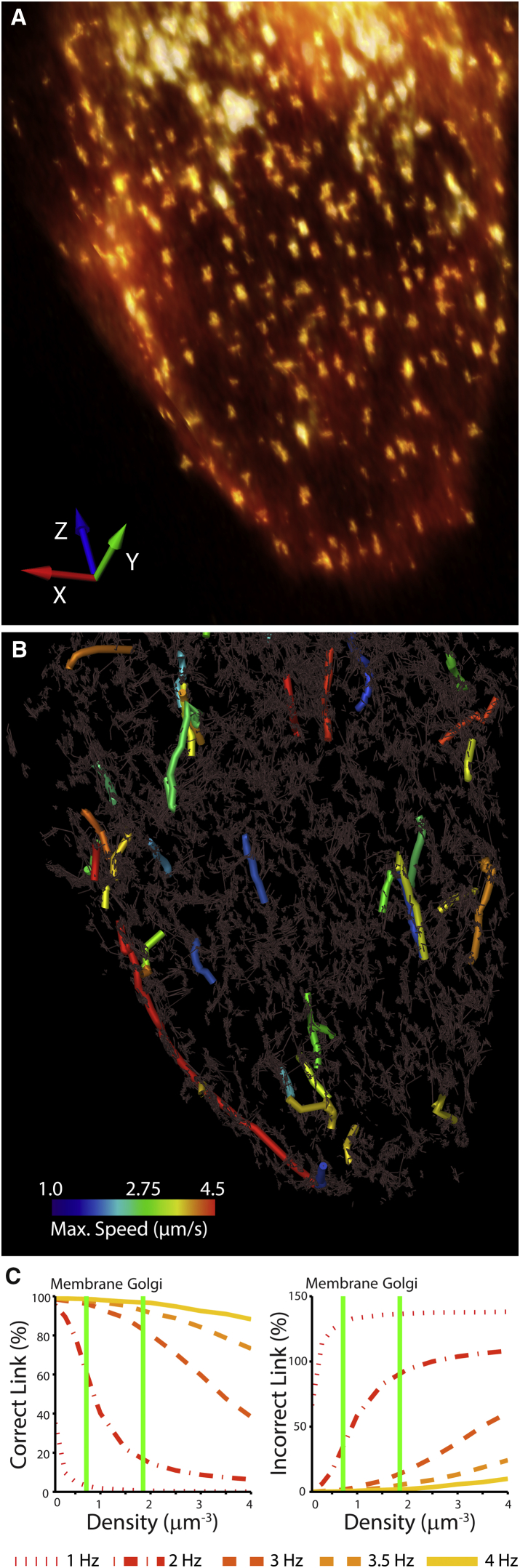
In an effort to quantify the fast intracellular transport of the clathrin-associated structures, we performed single-particle tracking with the automatic and locally adaptive u-track software (41, 42). At a volumetric image acquisition rate of 3.5 Hz, we were able to robustly track active transport of particles with velocities between 2.2 and 4.5 μm/s in a dense field of locally diffusing objects (0.7–1.9 objects per μm3, near the plasma membrane and Golgi apparatus, respectively; see Figs. 5, A and B and S5; Movie S9). However, despite optimization of the tracking parameters, at a volumetric image acquisition rate of 2.3 Hz, automated particle tracking failed (Movie S10; Note S1 in the Supporting Material).
Figure 5.
Volumetric imaging (3.5 Hz) of CLCa dynamics in Caco-2 epithelial cells. (A) Volume rendering of Caco2 epithelial cell expressing CLCa-EGFP. To aid in the visualization of dim structures, the raw data are merged with the adaptively thresholded mask used for particle detection. (B) Cumulative plot of clathrin tracks. Confined motions, representing CLCa at sites of endocytosis, are in grayscale. Intracellular clathrin-associated particles undergoing active transport, identified as particles with a diffusion coefficient greater than 100 μm3/s, are pseudocolored according to their maximum velocity. (C) Numerical simulations and tracking performance as a function of particle density and the volumetric image acquisition rate. Performance is only measured on tracks undergoing active transport (>2.2 μm/s), and percentages are relative to the simulation ground truth.
To investigate the capacity of our microscope to improve automated particle tracking robustness in dense intracellular environments, we simulated the observed active transport of diffraction-limited structures in a field of confined particles with local densities ranging from 0.1 to 4 particles per μm3 (Note S2 in the Supporting Material). To evaluate tracking robustness, we compared the particle trajectories with the simulation ground truth. In agreement with experimental observations, the simulations suggested that robust tracking was only feasible at volumetric image acquisition rates greater than 3 Hz (Fig. 5 C; Movie S11), even in the sparsest scenario (0.1 particles per μm3).
We also investigated the theoretical requirements for reliable 3D particle tracking by developing a mathematical model that deduces the relationship between the volumetric image acquisition rate, local object density, and object motion while considering the robustness and predictive capacity of conventional tracking algorithms (Note S3 in the Supporting Material). For a single actively transported particle, geometric considerations show that the volumetric image acquisition rate must increase linearly with the minimum particle velocity, with the square root of the particle’s instantaneous acceleration, and with the cube root of the particle density. Thus, given the densities observed for the intracellular clathrin-associated structures in Fig. 5, a volumetric image acquisition rate of at least 3.1 Hz is necessary to accurately track particle dynamics without introducing false positives. This prediction was supported by further numerical simulations, which evaluated tracking performance at volumetric image acquisition rates between 0.1 and 6.0 Hz (Fig. S6). These data underscore the importance of Nyquist sampling in both space and time to mitigate analytical artifacts (43). This model also provides us with a prediction of the required acquisition speed for a given biological process of interest and the density of labeled intracellular objects.
Lastly, in an attempt to more faithfully replicate the chemical and mechanical properties experienced by cells in vivo, we imaged human bronchial epithelial and U2OS cells on top of pliant ECMs. Indeed, the mechanochemical properties of the ECM have significant effects on cell morphology, cell fate, and cancer progression (44, 45, 46). However, the pliant nature of the ECM prohibits rapid sample scanning, as image quality is degraded by oscillations introduced into the sample (Fig. 6 A). These oscillations arise from the ECM and not from the sample holder, as we verified by imaging beads on a rigid glass coverslip (Fig. S7). Since DiaSLM keeps the sample stationary and diagonally scans the illumination beam without external mechanical perturbation, we were able to rapidly image the ECM without introducing sample oscillations (Fig. 6 B). Through this approach, we were able to observe cytoskeletal dynamics in a dense layer of nonpolarized RPE cells (Fig. 6 C; Movie S12) and the cell-spreading behavior of a U2OS cell (Fig. 6 D; Movie S13). Cell spreading is a mechanosensitive process and is one of the principal steps in polarity establishment and the adoption of a cell-type-specific morphological output. Thus, with DiaSLM, one may now study cellular behavior with high spatiotemporal resolution in more physiological microenvironments.
Figure 6.
High-speed imaging on pliant ECMs. (A) Rapid sample scanning induces vibrations in the collagen matrix that degrade image quality. Each plane was acquired in 5 ms and the final data were deskewed and affine transformed for visualization. A maximum intensity projection over a shallow volume is shown. (B) DiaSLM imaging of the same collagen area as in (A). Each plane was acquired in 5 ms and the final data were affine transformed for visualization. A maximum intensity projection over a shallow volume is shown. (C) XY maximum intensity projection of a confluent monolayer of nonpolarized human bronchial epithelial cells on an ECM consisting of equal parts of type II collagen (final concentration, 1 mg/mL) and matrigel. Cells were labeled with Tractin-GFP and volumetrically imaged at 0.17 Hz. (D) XY maximum intensity projection of a single U2OS cell undergoing cell spreading on a 2 mg/mL collagen matrix, volumetrically imaged at 1.38 Hz. Scale bar, 10 μm.
Discussion
We have introduced DiaSLM as a method for rapid and efficient 5D (X, Y, Z, time, and wavelength) imaging of adherent cells, and demonstrated its potential by volumetrically resolving cellular processes at image acquisition rates up to 3.5 Hz for thousands of Z-stacks. In contrast to sample scanning, DiaSLM avoids mechanical perturbation of the sample, which is particularly important for biological studies on soft substrates, including reconstituted ECM. However, faster acquisition rates will be necessary to follow some biological processes. For example, even at 3.5 Hz, some intracellular clathrin-coated vesicles proved particularly challenging for robust 3D tracking.
Several strategies can be used to accelerate the volumetric imaging rate beyond 3.5 Hz. As currently implemented in DiaSLM, Z-stepping of the detection objective is rate limiting. This is because the mass of the objective is approximately two orders of magnitude greater than the mass of the remote focusing mirror unit. Thus, to further improve temporal resolution, one must eliminate the objective’s physical movement. This can be achieved with the use of an electrotunable lens or a deformable mirror that refocuses the focal plane onto the camera (47), wavefront coding (48, 49), or aberration-free remote focusing in the detection arm of the microscope (26, 27). With these additions, and by adding a faster actuator for the remote focusing mirror (27), video-rate DiaSLM imaging should be feasible with optimal fluorescent probes, labeling strategies, and scientific cameras with accelerated readout speeds (50).
In summary, DiaSLM complements ASLM for rapid and efficient volumetric imaging of adherent cells on 2D surfaces. Owing to rapid progress in high-resolution LSFM, biological processes that hitherto have been only accessible to 2D imaging (e.g., TIRF microscopy) can now be performed in 3D, thereby greatly expanding the number of biological questions that can be addressed. Furthermore, DiaSLM enables biologists to move beyond the optical coverslip and into more physiologically meaningful environments. Thus, owing to its mechanical noninvasiveness, efficient use of short illumination beams, high optical sectioning strength, and simultaneous multicolor capability, as well as the fact that raw data can be visualized and analyzed directly in their raw form, DiaSLM is an enabling technique that will accelerate biological research.
Author Contributions
K.M.D. and R.F. conceived and built the microscope, analyzed the data, and wrote the manuscript. K.M.D., E.S.W., and R.F. performed the experiments. K.M.D., C.R.R., M.M., and R.F. designed the biological experiments. P.R. performed dynamic resolution modeling, simulations, and 3D particle detection and tracking.
Acknowledgments
We thank the BioHPC team at the University of Texas Southwestern Medical Center for providing the infrastructure and support for analysis of large data sets, Dr. Dick McIntosh (University of Colorado Boulder) for kindly providing us with U2OS cells, Dr. Kim Reed for administrative and scientific support, and Dr. Sandra Schmid for helpful discussion.
This work was supported by the Cancer Prevention Research Institute of Texas (R1225 to Dr. Gaudenz Danuser), the National Institutes of Health (GM73165 to Drs. Gaudenz Danuser and Sandra Schmid and F32GM117793 to K.M.D.), and the Human Frontier Science Program (fellowship LT000954/2015 to P.R.). The content of the manuscript is solely the responsibility of the authors and does not represent the official views of the National Institutes of Health, Cancer Prevention Research Institute of Texas, or Human Frontier Science Program. The LabView software used to control the microscope was custom developed by Coleman Technologies, building on a core set of functions licensed from the Howard Hughes Medical Institute’s Janelia Farm Research Campus.
Editor: Paul Wiseman.
Footnotes
Kevin M. Dean and Philippe Roudot contributed equally to this work.
Supporting Materials and Methods, seven figures, and thirteen movies are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(16)00139-9.
Supporting Material
Volumetric image acquisition rate of 0.37 Hz, 500 time points, 10 ms image acquisition per plane. The horizontal striping visible in the movie is likely due to laser intensity fluctuations. Scale bar, 10 μm.
Volumetric image acquisition rate of 0.37 Hz, 158 time points, 5 ms image acquisition per plane.
Volumetric image acquisition rate of 1 Hz, 500 time points, 5 ms image acquisition per plane. Scale bar, 10 μm.
Exponential photobleaching correction was applied. Volumetric image acquisition rate of 0.46 Hz, 200 time points, 20 ms image acquisition per plane. Scale bar, 10 μm.
XY maximum intensity projection of F-actin cytoskeleton imaged with Tractin-GFP. Volumetric image acquisition rate of 2.85 Hz, 1000 time points, 2.5 ms image acquisition per plane. Scale bar, 10 μm.
Same cell as in Movie S5. Volumetric image acquisition rate of 2.85 Hz, 1000 time points, 2.5 ms image acquisition per plane. Scale bar, 10 μm.
Yellow, clathrin-mediated endocytosis; gray, actin cytoskeleton. An XY maximum intensity projection of CLCa marks active sites of endocytosis at the plasma membrane and also labels intracellular structures associated with the Golgi apparatus, clearly visible in the perinuclear region of the cell. Color channels were acquired sequentially. Volumetric image acquisition rate of 0.20 Hz, 223 time points, 20 ms image acquisition per plane. Scale bar, 10 μm.
Gray, clathrin-mediated endocytosis; yellow, transferrin. Color channels were acquired simultaneously. Volumetric image acquisition rate of 0.36 Hz, 500 time points, 20 ms image acquisition per plane. Scale bar, 10 μm.
The vast majority of tracks show a confined displacement at the membrane and thus exhibit a low diffusion coefficient (red colored tracks). Rare events of active translocations can be detected with a maximum velocity 4.5 μm/s (shown in blue).
Upper left: 2.2 Hz sequence tracked with a maximum search radius of 1.8 μm/s. Upper right: 2.2 Hz sequence tracked with a maximum search radius of 2.8 μm/s. Lower left: 3.5 Hz sequence tracked with a maximum search radius of 2.8 μm/s.
Density is 0.72 object per μm3. Tracks are pseudocolored using their estimated diffusion coefficient (red tracks are below 0.5 μm3/s). The orange bounding box is 60 × 60 × 60 μm3.
XY maximum intensity projection of cells labeled with Tractin-GFP. Volumetric image acquisition rate of 0.17 Hz, 500 time points, 10 ms image acquisition per plane. Scale bar, 10 μm. The data were subjected to image registration to account for movement of the sample using the ImageJ function StackReg.
Volumetric image acquisition rate of 1.37 Hz, 3000 time points, 2 ms image acquisition per plane.
References
- 1.Schubert W., Bonnekoh B., Dress A.W.M. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat. Biotechnol. 2006;24:1270–1278. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- 2.Machacek M., Hodgson L., Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Engelenburg S.B., Shtengel G., Lippincott-Schwartz J. Distribution of ESCRT machinery at HIV assembly sites reveals virus scaffolding of ESCRT subunits. Science. 2014;343:653–656. doi: 10.1126/science.1247786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vegh R.B., Bravaya K.B., Solntsev K.M. Chromophore photoreduction in red fluorescent proteins is responsible for bleaching and phototoxicity. J. Phys. Chem. B. 2014;118:4527–4534. doi: 10.1021/jp500919a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scherf N., Huisken J. The smart and gentle microscope. Nat. Biotechnol. 2015;33:815–818. doi: 10.1038/nbt.3310. [DOI] [PubMed] [Google Scholar]
- 6.Huisken J., Swoger J., Stelzer E.H. Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science. 2004;305:1007–1009. doi: 10.1126/science.1100035. [DOI] [PubMed] [Google Scholar]
- 7.Born M., Wolf E., Hecht E. Principles of optics: electromagnetic theory of propagation, interference and diffraction of light. Phys. Today. 2000;53:77–78. [Google Scholar]
- 8.Fahrbach F.O., Rohrbach A. Propagation stability of self-reconstructing Bessel beams enables contrast-enhanced imaging in thick media. Nat. Commun. 2012;3:632. doi: 10.1038/ncomms1646. [DOI] [PubMed] [Google Scholar]
- 9.Planchon T.A., Gao L., Betzig E. Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods. 2011;8:417–423. doi: 10.1038/nmeth.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao M., Zhang H., Peng L. Cellular imaging of deep organ using two-photon Bessel light-sheet nonlinear structured illumination microscopy. Biomed. Opt. Express. 2014;5:1296–1308. doi: 10.1364/BOE.5.001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen B.C., Legant W.R., Betzig E. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fahrbach F.O., Rohrbach A. A line scanned light-sheet microscope with phase shaped self-reconstructing beams. Opt. Express. 2010;18:24229–24244. doi: 10.1364/OE.18.024229. [DOI] [PubMed] [Google Scholar]
- 13.Vettenburg T., Dalgarno H.I.C., Dholakia K. Light-sheet microscopy using an Airy beam. Nat. Methods. 2014;11:541–544. doi: 10.1038/nmeth.2922. [DOI] [PubMed] [Google Scholar]
- 14.Dean K.M., Fiolka R. Uniform and scalable light-sheets generated by extended focusing. Opt. Express. 2014;22:26141–26152. doi: 10.1364/OE.22.026141. [DOI] [PubMed] [Google Scholar]
- 15.Zong W., Zhao J., Chen L. Large-field high-resolution two-photon digital scanned light-sheet microscopy. Cell Res. 2014;25:254–257. doi: 10.1038/cr.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu Y., Wawrzusin P., Shroff H. Spatially isotropic four-dimensional imaging with dual-view plane illumination microscopy. Nat. Biotechnol. 2013;31:1032–1038. doi: 10.1038/nbt.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao L. Extend the field of view of selective plan illumination microscopy by tiling the excitation light sheet. Opt. Express. 2015;23:6102–6111. doi: 10.1364/OE.23.006102. [DOI] [PubMed] [Google Scholar]
- 18.Dean K.M., Roudot P., Fiolka R. Deconvolution-free subcellular imaging with axially swept light sheet microscopy. Biophys. J. 2015;108:2807–2815. doi: 10.1016/j.bpj.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao L., Shao L., Betzig E. Noninvasive imaging beyond the diffraction limit of 3D dynamics in thickly fluorescent specimens. Cell. 2012;151:1370–1385. doi: 10.1016/j.cell.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaner N.C., Lin M.Z., Tsien R.Y. Improving the photostability of bright monomeric orange and red fluorescent proteins. Nat. Methods. 2008;5:545–551. doi: 10.1038/nmeth.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doyon J.B., Zeitler B., Drubin D.G. Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat. Cell Biol. 2011;13:331–337. doi: 10.1038/ncb2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson H.W., Schell M.J. Neuronal IP3 3-kinase is an F-actin-bundling protein: role in dendritic targeting and regulation of spine morphology. Mol. Biol. Cell. 2009;20:5166–5180. doi: 10.1091/mbc.E09-01-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu A.P., Loerke D., Danuser G. Global and local regulation of clathrin-coated pit dynamics detected on patterned substrates. Biophys. J. 2009;97:1038–1047. doi: 10.1016/j.bpj.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Welf E.S., Ahmed S., Haugh J.M. Migrating fibroblasts reorient directionality by a metastable, PI3K-dependent mechanism. J. Cell Biol. 2012;197:105–114. doi: 10.1083/jcb.201108152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huisken J., Stainier D.Y.R. Even fluorescence excitation by multidirectional selective plane illumination microscopy (mSPIM) Opt. Lett. 2007;32:2608–2610. doi: 10.1364/ol.32.002608. [DOI] [PubMed] [Google Scholar]
- 26.Botcherby E.J., Juskaitis R., Wilson T. Aberration-free optical refocusing in high numerical aperture microscopy. Opt. Lett. 2007;32:2007–2009. doi: 10.1364/ol.32.002007. [DOI] [PubMed] [Google Scholar]
- 27.Botcherby E.J., Smith C.W., Wilson T. Aberration-free three-dimensional multiphoton imaging of neuronal activity at kHz rates. Proc. Natl. Acad. Sci. USA. 2012;109:2919–2924. doi: 10.1073/pnas.1111662109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keller P.J., Schmidt A.D., Stelzer E.H.K. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–1069. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 29.Haugh J.M., Codazzi F., Meyer T. Spatial sensing in fibroblasts mediated by 3′ phosphoinositides. J. Cell Biol. 2000;151:1269–1280. doi: 10.1083/jcb.151.6.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Innocenti M., Frittoli E., Scita G. Phosphoinositide 3-kinase activates Rac by entering in a complex with Eps8, Abi1, and Sos-1. J. Cell Biol. 2003;160:17–23. doi: 10.1083/jcb.200206079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deming P.B., Campbell S.L., Howe A.K. Protein kinase A regulates 3-phosphatidylinositide dynamics during platelet-derived growth factor-induced membrane ruffling and chemotaxis. J. Biol. Chem. 2008;283:35199–35211. doi: 10.1074/jbc.M804448200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao X., Xing D., Tang Y. H-Ras and PI3K are required for the formation of circular dorsal ruffles induced by low-power laser irradiation. J. Cell. Physiol. 2009;219:535–543. doi: 10.1002/jcp.21693. [DOI] [PubMed] [Google Scholar]
- 33.Burnette D.T., Manley S., Lippincott-Schwartz J. A role for actin arcs in the leading-edge advance of migrating cells. Nat. Cell Biol. 2011;13:371–381. doi: 10.1038/ncb2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu Y.-J., Le Berre M., Piel M. Confinement and low adhesion induce fast amoeboid migration of slow mesenchymal cells. Cell. 2015;160:659–672. doi: 10.1016/j.cell.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 35.Lorentzen A., Bamber J., Marshall C.J. An ezrin-rich, rigid uropod-like structure directs movement of amoeboid blebbing cells. J. Cell Sci. 2011;124:1256–1267. doi: 10.1242/jcs.074849. [DOI] [PubMed] [Google Scholar]
- 36.Welf E.S., Danuser G. Using fluctuation analysis to establish causal relations between cellular events without experimental perturbation. Biophys. J. 2014;107:2492–2498. doi: 10.1016/j.bpj.2014.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vilela M., Halidi N., Danuser G. Fluctuation analysis of activity biosensor images for the study of information flow in signaling pathways. Methods Enzymol. 2013;519:253–276. doi: 10.1016/B978-0-12-405539-1.00009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boulant S., Kural C., Kirchhausen T. Actin dynamics counteract membrane tension during clathrin-mediated endocytosis. Nat. Cell Biol. 2011;13:1124–1131. doi: 10.1038/ncb2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan T., Liu L., Chen L. Diacylglycerol guides the hopping of clathrin-coated pits along microtubules for exo-endocytosis coupling. Dev. Cell. 2015;35:120–130. doi: 10.1016/j.devcel.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 40.Kural C., Akatay A.A., Kirchhausen T. Asymmetric formation of coated pits on dorsal and ventral surfaces at the leading edges of motile cells and on protrusions of immobile cells. Mol. Biol. Cell. 2015;26:2044–2053. doi: 10.1091/mbc.E15-01-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaqaman K., Loerke D., Danuser G. Robust single-particle tracking in live-cell time-lapse sequences. Nat. Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aguet F., Antonescu C.N., Danuser G. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev. Cell. 2013;26:279–291. doi: 10.1016/j.devcel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Danuser G. Reply to “Acquisition frame rate affects microtubule plus-end tracking analysis”. Nat. Methods. 2014;11:220. doi: 10.1038/nmeth.2860. [DOI] [PubMed] [Google Scholar]
- 44.Discher D.E., Mooney D.J., Zandstra P.W. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324:1673–1677. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pickup M.W., Mouw J.K., Weaver V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014;15:1243–1253. doi: 10.15252/embr.201439246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Butcher D.T., Alliston T., Weaver V.M. A tense situation: forcing tumour progression. Nat. Rev. Cancer. 2009;9:108–122. doi: 10.1038/nrc2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fahrbach F.O., Voigt F.F., Huisken J. Rapid 3D light-sheet microscopy with a tunable lens. Opt. Express. 2013;21:21010–21026. doi: 10.1364/OE.21.021010. [DOI] [PubMed] [Google Scholar]
- 48.Olarte O.E., Andilla J., Loza-Alvarez P. Decoupled illumination detection in light sheet microscopy for fast volumetric imaging. Optica. 2015;2:702. [Google Scholar]
- 49.Abrahamsson S., Satoru U., Wilson T. A new approach to extended focus for high-speed, high-resolution biological microscopy. In: Conchello J.-A., Coqswell C.J., Wilson T., editors. Proceedings of SPIE. International Society for Optics and Photonics; 2006. [Google Scholar]
- 50.Dean K.M., Palmer A.E. Advances in fluorescence labeling strategies for dynamic cellular imaging. Nat. Chem. Biol. 2014;10:512–523. doi: 10.1038/nchembio.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Volumetric image acquisition rate of 0.37 Hz, 500 time points, 10 ms image acquisition per plane. The horizontal striping visible in the movie is likely due to laser intensity fluctuations. Scale bar, 10 μm.
Volumetric image acquisition rate of 0.37 Hz, 158 time points, 5 ms image acquisition per plane.
Volumetric image acquisition rate of 1 Hz, 500 time points, 5 ms image acquisition per plane. Scale bar, 10 μm.
Exponential photobleaching correction was applied. Volumetric image acquisition rate of 0.46 Hz, 200 time points, 20 ms image acquisition per plane. Scale bar, 10 μm.
XY maximum intensity projection of F-actin cytoskeleton imaged with Tractin-GFP. Volumetric image acquisition rate of 2.85 Hz, 1000 time points, 2.5 ms image acquisition per plane. Scale bar, 10 μm.
Same cell as in Movie S5. Volumetric image acquisition rate of 2.85 Hz, 1000 time points, 2.5 ms image acquisition per plane. Scale bar, 10 μm.
Yellow, clathrin-mediated endocytosis; gray, actin cytoskeleton. An XY maximum intensity projection of CLCa marks active sites of endocytosis at the plasma membrane and also labels intracellular structures associated with the Golgi apparatus, clearly visible in the perinuclear region of the cell. Color channels were acquired sequentially. Volumetric image acquisition rate of 0.20 Hz, 223 time points, 20 ms image acquisition per plane. Scale bar, 10 μm.
Gray, clathrin-mediated endocytosis; yellow, transferrin. Color channels were acquired simultaneously. Volumetric image acquisition rate of 0.36 Hz, 500 time points, 20 ms image acquisition per plane. Scale bar, 10 μm.
The vast majority of tracks show a confined displacement at the membrane and thus exhibit a low diffusion coefficient (red colored tracks). Rare events of active translocations can be detected with a maximum velocity 4.5 μm/s (shown in blue).
Upper left: 2.2 Hz sequence tracked with a maximum search radius of 1.8 μm/s. Upper right: 2.2 Hz sequence tracked with a maximum search radius of 2.8 μm/s. Lower left: 3.5 Hz sequence tracked with a maximum search radius of 2.8 μm/s.
Density is 0.72 object per μm3. Tracks are pseudocolored using their estimated diffusion coefficient (red tracks are below 0.5 μm3/s). The orange bounding box is 60 × 60 × 60 μm3.
XY maximum intensity projection of cells labeled with Tractin-GFP. Volumetric image acquisition rate of 0.17 Hz, 500 time points, 10 ms image acquisition per plane. Scale bar, 10 μm. The data were subjected to image registration to account for movement of the sample using the ImageJ function StackReg.
Volumetric image acquisition rate of 1.37 Hz, 3000 time points, 2 ms image acquisition per plane.