Abstract
Comprehensive genomic analyses of common nervous system cancers provide new insights into their pathogenesis, diagnosis, and treatment. Although analogous studies of rare nervous system tumors are needed, there are major barriers to performing such studies. Cross-species comparative oncogenomics, identifying driver mutations in mouse cancer models and validating them in human tumors, is a promising alternative. Although still in its infancy, this approach is being applied to malignant peripheral nerve sheath tumors (MPNSTs), rare Schwann cell–derived malignancies that occur sporadically, after radiotherapy, and in neurofibromatosis type 1. Studies of human neurofibromatosis type 1–associated tumors suggest that NF1 tumor suppressor loss in Schwann cells triggers cell-autonomous and intercellular changes, resulting in development of benign neurofibromas; subsequent neurofibroma-MPNST progression is caused by aberrant growth factor signaling and mutations affecting the p16INK4A-cyclin D1-CDK4-Rb and p19ARF-Mdm2-p53 cell cycle pathways. Mice with Nf1, Trp53, and/or Cdkn2a mutations that overexpress the Schwann cell mitogen neuregulin-1 or overexpress the epidermal growth factor receptor validate observations in human tumors and, to various degrees, model human tumorigenesis. Genomic analyses of MPNSTs arising in neuregulin-1 and epidermal growth factor receptor-overexpressing mice and forward genetic screens with Sleeping Beauty transposons implicate additional signaling cascades in MPNST pathogenesis. These studies confirm the utility of mouse models for MPNST driver gene discovery and provide new insights into the complexity of MPNST pathogenesis.
Our understanding of the mechanisms underlying the development of common types of nervous system tumors has expanded exponentially over the past decade. In large part, this is because a cadre of investigators and consortia such as The Cancer Genome Atlas have comprehensively identified the genomic abnormalities in large cohorts of neoplasms such as glioblastomas,1, 2 ependymomas,3, 4, 5, 6, 7, 8 medulloblastomas,9, 10 and diffuse intrinsic pontine gliomas.11, 12, 13 These studies have provided key insights that are fundamentally altering both the diagnosis and treatment of central nervous system tumors. For diagnosis, it is now clear that for some tumor types, mutational status is more predictive of prognosis than classic pathologic criteria.14 As a result, the International Society of Neuropathology-Haarlem Working Group, which is developing the upcoming version of the Diagnostic Manual for Tumours of the Central Nervous System, has proposed a diagnostic approach that integrates pathologic and molecular findings to better predict patient outcomes.15 It is also now evident that there are several molecular subtypes of glioblastomas,16 ependymomas,5, 6, 7, 8 diffuse intrinsic pontine gliomas,11, 17, 18, 19 and medulloblastomas,20, 21, 22 each of which depends on distinct signaling cascades and thus potentially sensitive to different therapeutic agents. As a result, many academic centers are now performing oncogenomic testing to predict which drugs are potentially effective against a patient's tumor.
It is reasonable to expect that globally defining the genomic abnormalities that occur in large cohorts of rare nervous system cancers would similarly affect the care of patients with those neoplasms. However, there are important barriers to performing such analyses. The Cancer Genome Atlas typically tries to study 500 examples of each tumor type, using frozen tissue and a matched germline DNA; this cohort size will capture driver mutations present in at least 5% of that tumor type. In practice, the number of tumor samples that must be collected is much larger, because many samples will be rejected during quality assurance. In addition, The Cancer Genome Atlas prefers tumors from patients who have not yet received chemotherapy or radiotherapy, which reduces the pool of available tumor samples. Because large numbers of samples are needed and rare cancers that meet these criteria are encountered only a few times annually even in large medical centers, it is likely that investigators from different institutions will have to pool their tumor collections if these studies are to be performed. However, assembling even a small collection of rare tumor specimens requires several years, and many investigators are reluctant to donate their tumor collections to large consortia because of a concern that the effort invested in their collection will not be recognized. Consequently, human factors will also impede the collection of large numbers of rare human cancer specimens.
This should not dissuade us from performing comprehensive genomic analyses on all of the rare nervous system tumors we can collect; the information we will derive from these studies will undoubtedly be invaluable. However, to maximize the value of information derived from relatively small numbers of human tumors, we will need to partner these studies with other approaches. Recognizing this, many investigators are beginning to use genetically engineered mouse models to identify driver genes potentially relevant to the pathogenesis of rare tumor types. Here, I review how this approach has been used to probe the pathogenesis of malignant peripheral nerve sheath tumors (MPNSTs).
MPNSTs are rare Schwann cell–derived neoplasms that represent approximately 5% of the soft tissue sarcomas diagnosed annually.23 Although MPNSTs occur sporadically, after radiotherapy and in individuals with the autosomal dominant tumor susceptibility syndrome neurofibromatosis type 1 (NF1), our current understanding of the mechanisms responsible for MPNST pathogenesis in humans is derived almost exclusively from studies of NF1-associated MPNSTs. I begin by discussing the mechanisms responsible for the pathogenesis of plexiform neurofibromas, the benign precursors that give rise to MPNSTs in NF1 patients, and their transformation into MPNSTs. I then consider key genetically engineered mouse models that were created to validate observations in human MPNSTs, what we have learned from these models, and their relative suitability for MPNST driver gene discovery. I finish by reviewing the initial genomic analyses that have been performed with these mouse MPNST models.
Pathogenesis of Human Neurofibromas and MPNSTs
NF1 Tumor Suppressor Gene and Its Role in Plexiform Neurofibroma Pathogenesis
NF1 is the most common genetic disease that affects the human nervous system, occurring in 1 in every 3500 newborn infants.24 Although completely penetrant, the manifestations of NF1 are highly variable, even in the same family. NF1 patients commonly have learning disabilities, pigmentary lesions of the iris (Lisch nodules) and skin (axillary freckling, café-au-lait macules), bone dysplasias, and glial neoplasms in brain (optic gliomas, glioblastomas), large peripheral nerves (plexiform neurofibromas, MPNSTs), and skin (dermal neurofibromas). Less frequently, these individuals develop other tumors such as pheochromocytomas, rhabdomyosarcomas, leiomyosarcomas, and juvenile myelomonocytic leukemia. In NF1 patients, one allele of neurofibromin 1 (NF1), a tumor suppressor gene that contains 60 exons and spans 282,751 bp on chromosome 17 (17q11.2), is inactivated. Nonsense mutations,25 missense mutations,25 frameshift mutations,25 or mutations that affect RNA splicing26 are the usual means of NF1 inactivation; these mutations tend to cluster in exons 10a to 10c and 37, but they can be found anywhere in the NF1 coding sequence. Complete deletion of the NF1 gene also occurs, but is rare, being seen in only 5% of NF1 patients.27
Although the NF1 gene was cloned in 1990, the functions performed by neurofibromin, the 2818-amino acid polypeptide encoded by this gene, are still incompletely understood. We do know that neurofibromin inactivates members of the Ras family of small GTP-binding proteins.24 This function is mediated by a centrally located domain (amino acids 1203 to 1549) within neurofibromin that is homologous to the Saccharomyces cerevisiae GTPase-activating proteins (GAPs) IRA1 and IRA2. Ras proteins are activated on binding GTP; the neurofibromin Ras GAP domain stimulates an intrinsic GTPase activity in Ras proteins, causing them to cleave a phosphate from the bound GTP and inactivate themselves. Other domains in neurofibromin, namely the cysteine/serine-rich domain and a tubulin-binding domain located amino terminal to the Ras GAP domain, modulate its Ras GAP activity.24 The cysteine/serine-rich domain and TBD have opposing actions; phosphorylation of the cysteine/serine-rich domain by protein kinase Cα results in enhanced Ras GAP activity, whereas the interaction of tubulin with the tubulin-binding domain inhibits neurofibromin's Ras GAP function. A bipartite domain that contains both a region analogous to the yeast protein Sec14p and a pleckstrin homology domain is present carboxy terminal to the Ras GAP domain. Although it was suggested that this domain binds glycerophospholipids and interacts with other as yet unidentified proteins, the biological significance of these hypothetical interactions remains unknown. The carboxy terminus of neurofibromin also contains a focal adhesion kinase (FAK)–interacting domain that allows neurofibromin to modulate substrate interactions. Curiously, although neurofibromin has no known nuclear functions, a nuclear localization signal is present within the FAK-interacting domain. Conversely, some neurofibromin functions are not yet linked to specific domains in this protein. For example, although neurofibromin loss results in changes in cAMP and calcium signaling, it is not yet known which neurofibromin domains regulate these events.
Most investigators examining neurofibromin's tumor suppressor function have focused on the role of the Ras GAP domain. However, there is reason to think that other domains in neurofibromin also contribute to tumor suppression. For instance, neurofibromin is a member of a family of Ras GAPs that includes RASA1 (p120GAP), PLXNB2, RASA2 (Gap1m), RASA3 (GAP1IP4BP), RASA4 (CAPRI), RASAL1, SYNGAP (p135SynGAP), DAB2IP, RASAL2, and IQGAP1-3. Although several of these proteins are expressed ubiquitously, they do not compensate for neurofibromin loss in NF1-null tumors. Further, none of these other Ras GAPs produce a tumor susceptibility syndrome when mutated, indicating that loss of their Ras GAP activity is not sufficient for tumorigenesis. A comparison of the structure of neurofibromin to that of other Ras GAPs shows that, other than their Ras GAP domains, the domains present in other members of this protein family differ from those in neurofibromin. Given the structural and functional differences between neurofibromin and other Ras GAPs, it is thus possible that both the Ras GAP and other domains in neurofibromin are required for tumor suppression.
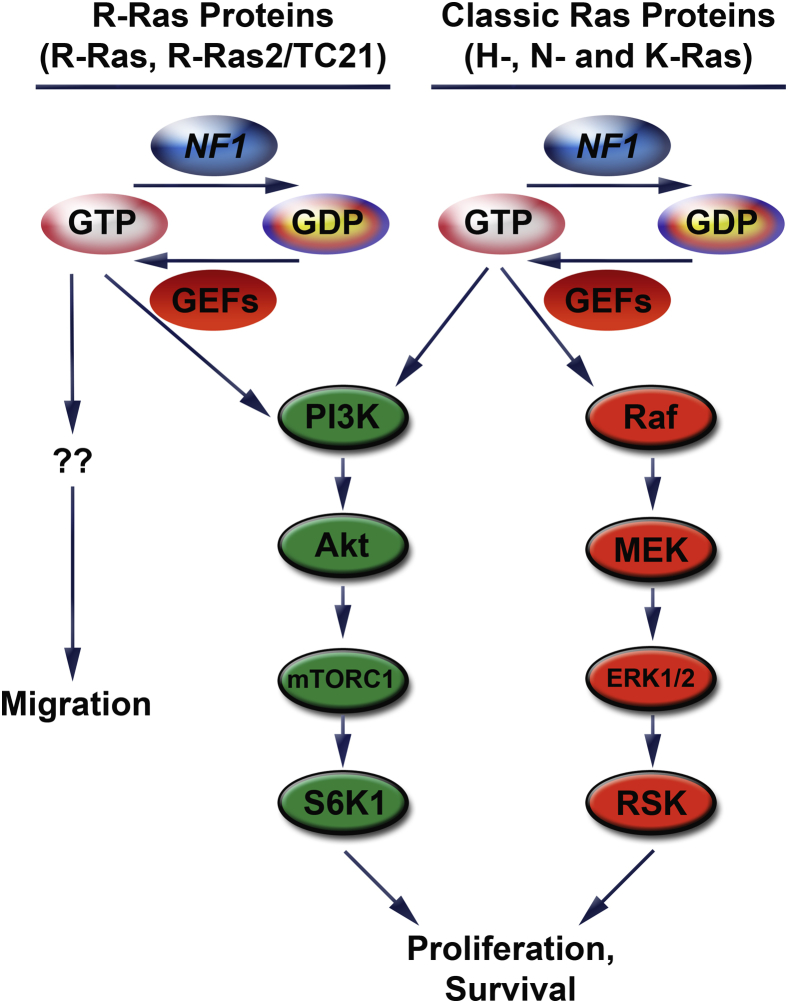
Shortly after the cloning of the NF1 gene, it was recognized that plexiform neurofibroma pathogenesis is initiated when a second-hit mutation inactivates the remaining functional NF1 gene in a cell within the Schwann cell lineage,24 resulting in a loss of neurofibromin expression and Ras hyperactivation.24 This led to the prediction that drugs targeting Ras signaling would be effective against NF1-null nerve sheath tumors. Unexpectedly, however, the farnesyltransferase inhibitor tipifarnib, which blocks a post-translational modification critical for Ras activity, did not inhibit the progression of plexiform neurofibromas.28 In retrospect, this clinical trial likely failed because the Ras activation that follows neurofibromin loss is more complex than that seen in tumors with an activating mutation of a single Ras protein. Neurofibromin inhibits both the classic Ras (H-, N-, and K-Ras) and R-Ras (R-Ras, R-Ras2/TC21, and M-Ras) subfamilies.24 As it happens, multiple members of both subfamilies (H-Ras, N-Ras, K-Ras, R-Ras, and R-Ras2) are simultaneously expressed and activated in MPNST cells. Further, these subfamilies have both overlapping and distinct functions in MPNST cells (Figure 1). Classic Ras and R-Ras proteins both contribute to MPNST proliferation. However, only classic Ras proteins promote the survival of MPNST cells, whereas R-Ras proteins drive their migration.29 Within the classic Ras subfamily, though, there appears to be functional redundancy; ablation of one subfamily member results in increased expression and activation of another subfamily member with little, if any, effect on proliferation. Tipifarnib inhibits only H-Ras, because other post-translational modifications such as geranylgeranylation allow K-Ras and N-Ras to be appropriately activated30 and thus circumvent inhibition by tipifarnib. Consequently, treatment with this drug likely resulted in compensatory activation of other, tipifarnib-resistant Ras proteins. Indeed, the simultaneous activation and intrafamily functional redundancy of Ras proteins in NF1-null MPNST cells suggests that it will be difficult to target these proteins therapeutically. Consequently, some laboratories have asked whether targeting signaling pathways downstream of Ras such as mitogen-activated protein extracellular signal-related kinase (ERK) kinase (MEK)31, 32 and the phosphatidylinositol 3-kinase (PI3K)-Akt-TSC2-mammalian target of rapamycin (mTOR)-S6 kinase33, 34 would be more effective therapeutically. Although initial results indicate that this is the case, note that the full repertoire of Ras-dependent signaling pathways activated in NF1-null peripheral nerve sheath tumors has not yet been defined. Indeed, a recent study that examined changes occurring in the phosphoproteome after inhibition of classic Ras signaling29 showed that classic Ras signaling controlled multiple additional signaling cascades in MPNSTs. It will be important to determine which of these signaling cascades contribute to tumorigenesis and whether they are further dysregulated by additional mutations arising during neurofibroma-MPNST progression.
Figure 1.
In plexiform neurofibromas and MPNSTs, neurofibromin loss deregulates multiple proteins from both the classic Ras and R-Ras subfamilies. NF1 stimulates an intrinsic GTPase activity in Ras proteins, causing them to cleave a phosphate from bound GTP and inactivate themselves. Ras is reactivated when guanine nucleotide exchange factors (eg, Sos), which themselves are activated by stimuli such as growth factors, cause Ras to release GDP and bind a new molecule of GTP. Despite their similar regulation, classic Ras and R-Ras proteins have overlapping (proliferation) and distinct (migration, survival) effects. This reflects the ability of different Ras families to differentially activate signaling pathways. Classic Ras proteins activate the Raf/MEK/ERK1/2/RSK and PI3K/Akt/mTORC1/S6K1 pathways, thereby enhancing proliferation and survival. It is known that R-Ras proteins can activate the PI3K pathway as well, but little is known about the cytoplasmic pathways they activate to promote migration. Note that, for the sake of simplicity, the illustration of the PI3K pathway was simplified, and some key regulators of this pathway are not shown (eg, Rheb, TSC1/2). ERK, extracellular signal-related kinase; GEF, guanine nucleotide exchange factor; MEK, mitogen-activated protein ERK kinase; MPNST, malignant peripheral nerve sheath tumor; mTOR, mammalian target of rapamycin; NF1, neurofibromin; PI3K, phosphatidylinositol 3-kinase; RSK, ribosomal S6 kinase.
Because NF1 loss occurs in Schwann cells, it might be expected that NF1-null nerve sheath tumors would be primarily composed of neoplastic Schwann cells. Paradoxically, however, neurofibromas are histologically complex lesions composed of NF1−/− Schwann cells intermingled with multiple other cell types, including NF1+/− Schwann cells, fibroblasts, perineurial-like cells, vasculature, mast cells, macrophages, and a poorly defined CD34-immunoreactive population. Further, animal models (see Currently Available Mouse Models of Neurofibroma and MPNST Pathogenesis) indicate that at least some of these other cell types are essential for neurofibroma pathogenesis. This admixture of cell types occurs, in part, because NF1-null Schwann cells demonstrate enhanced expression of paracrine signaling factors that recruit other cell types into the nascent neurofibroma. Although the nature of these paracrine signaling factors is incompletely understood, there is good evidence that at least one of them is stem cell factor (SCF, also known as Kit ligand), a protein that promotes the migration, survival, maturation, and activation of mast cells.35, 36 NF1 loss enhances SCF expression, with NF1−/− Schwann cells secreting sixfold higher levels of SCF than NF1+/− or wild-type Schwann cells.37 Intriguingly, changes in NF1 copy number also affects mast cell physiology; NF1+/− mast cells show enhanced proliferation, survival, and migration when challenged with SCF.38, 39 The hyperresponsiveness of NF1+/− mast cells also has cascading effects on other cell types in the developing neurofibroma. For instance, SCF-induced secretion of transforming growth factor-β is exaggerated in NF1+/− mast cells.40 When challenged with transforming growth factor-β, NF1+/− fibroblasts show increased production of collagen relative to wild-type fibroblasts, potentially explaining the dense collagenous deposits evident in human neurofibromas. Considered together, these observations suggest a model in which NF1 loss in Schwann cells results in both cell autonomous (eg, enhanced proliferation) and nonautonomous (recruitment of other NF1+/− cell types) effects that interact to drive neurofibroma pathogenesis. This hypothesis is consistent with observations from several laboratories showing that multiple NF1+/− cell types, including Schwann cells,41 fibroblasts,40, 42 and keratinocytes,43 show altered proliferative responses relative to their wild-type counterparts.
Genomic Abnormalities Mediating Neurofibroma-MPNST Progression
The transformation of a neurofibroma into a MPNST requires the accumulation of additional mutations, many of which affect tumor suppressor genes. Mutations that affect the p16INK4A-cyclin D1-cyclin-dependent kinase (CDK)4-Retinoblastoma (Rb) and p19ARF-Mdm2-p53 cell cycle regulatory pathways were among the first linked to MPNST pathogenesis. CDKN2A mutations occur in up to 50% of MPNSTs44, 45 and are especially deleterious because they dysregulate both of these cell cycle regulatory pathways. This is because CDKN2A encodes both p16INK4A, which inhibits CDK4 and CDK6, and p19ARF, a protein that inhibits Mdm2, an E3 ubiquitin-protein ligase which tags p53 for proteasomal degradation. However, these pathways can be disabled in other ways in MPNSTs. For instance, expression of Rb protein is lost in 25% of MPNSTs.46, 47 Deletions and other loss of function mutations of TP53 are also common, occurring in up to 75% of MPNSTs.24 Tumor suppressor mutations that affect other essential signaling cascades such as the PI3K-Akt-TSC2-mTOR-S6 kinase pathway have also been identified in MPNSTs. For instance, loss of the phosphate and tensin homologue tumor suppressor, an inhibitor of the PI3K-Akt-TSC2-mTOR-S6 pathway, is evident in approximately 60% of MPNSTs.48, 49 Epigenetic repression of tumor suppressors also plays a role in MPNST pathogenesis, as demonstrated by the recent finding that expression of Ras association domain family member 1, isoform A (RASSF1A) is silenced in MPNSTs by DNA methylation.50
The discovery of candidate oncogenes in MPNSTs was driven primarily by a search for therapeutic targets. As part of this effort, several laboratories have explored the possibility that aberrant growth factor signaling is a key upstream mediator of Ras activation in MPNSTs. These studies identified several growth factors and growth factor receptors that potentially contribute to neurofibroma and MPNST pathogenesis, including the epidermal growth factor (EGF) receptor,24 neuregulin-1 (NRG1),24 platelet-derived growth factor,24 hepatocyte growth factor, and its c-Met receptor,24 the insulin-like growth factor 1 receptor,51 transforming growth factor-β1,52 lysophosphatidic acid,53 midkine,52 Kit ligand and its c-Kit receptor,24 CXCR4 and its CXCL12 ligand,54 basic fibroblast growth factor, and vascular endothelial growth factor.55 However, the strength of the evidence supporting a role for each of these candidate molecules is highly variable, ranging from merely showing that a factor or receptor is present to showing strong genomic or functional evidence. As genomic evidence, genes encoding several receptor tyrosine kinases, including the EGF receptor, the NRG1 co-receptor erbB2, c-Kit, c-Met, and the platelet-derived growth factor receptor-α, are amplified in subsets of MPNSTs.24 Consistent with these genomic observations, pharmacologic inhibitors of the EGF receptor, NRG1 receptors, c-Kit, platelet-derived growth factor receptors, and c-Met inhibit the proliferation and/or invasion of MPNST cells.24 However, several important questions remain unanswered about the role these molecules play in MPNST pathogenesis. Of particular importance, it is not yet known whether there are distinct subsets of MPNSTs that depend on different growth factors for their growth or, alternatively, whether each of these growth factors play distinct, critically important roles in the same tumor.
Since multiple oncogenes and tumor suppressor genes have already been implicated in MPNST pathogenesis, it is reasonable to ask whether comprehensive genomic analyses of MPNSTs are likely to identify substantial numbers of currently unknown mutations that promote the pathogenesis of these tumors. A consideration of the chromosomal abnormalities in MPNSTs shows that the pathogenic abnormalities identified thus far represent only a fraction of the genomic changes in these tumors. Cytogenetic and array comparative genomic hybridization (aCGH) studies of human NF1-associated MPNSTs indicate that these neoplasms have highly complex hypodiploid or near-triploid genomes. This complexity has made it quite difficult to identify recurrent copy number changes that contain potential driver genes. Nonetheless, some such regions were identified; for instance, losses occur repeatedly in sites on chromosomes 1p, 9p, 11, 12p, 14q, 17q, 18, 22q, X, and Y, with focal gains commonly observed on chromosome 7, 8q, and 15q.56, 57, 58, 59, 60, 61, 62, 63, 64, 65 Consistent with the tumor suppressor mutations identified in MPNSTs, some repetitive losses, such as 17q (the NF1 gene) and 9p (CDKN2A), affect genes implicated in MPNST pathogenesis. However, for most of the regions of recurrent gain or loss, the driver gene or genes within these intervals have not been identified.
Only two studies have examined the genomes of human MPNSTs at nucleotide resolution. Zhang et al66 performed whole genome sequencing on four NF1-associated MPNSTs and matched buffy coats. They found that these tumors had a median of 60 tumor-specific small insertions-deletions and nonsynonymous single nucleotide polymorphisms that affect protein-coding sequences. Intriguingly, all four of the MPNSTs also had inactivating mutations of SUZ12, a gene encoding a chromatin-modifying protein that forms part of polycomb repressive complex 2 (PRC2). Targeted sequencing of an additional 42 formalin-fixed, paraffin-embedded MPNST specimens (31 NF1-associated, 11 sporadic) showed that both NF1-associated and sporadic MPNSTs had mutually exclusive mutations in SUZ12 or genes that encode other proteins that interact with PRC2 (EED, AEBP2, EPC1, EZH2, or RBBP7). SUZ12 mutations were not present in the neurofibromas from which some of these MPNSTs were derived, indicating that SUZ12 mutations occur during malignant progression. Lee et al67 performed whole exome sequencing, DNA copy number analyses, and whole transcriptome sequencing on 15 MPNSTs, including six NF1-associated, four sporadic, and four radiation-associated tumors. In these tumors and a subsequent validation cohort of 37 formalin-fixed, paraffin-embedded specimens, PRC2 mutations were evident in 70% of NF1-associated MPNSTs, 92% of sporadic MPNSTs, and 90% of radiation-induced MPNSTs. In keeping with the role of PRC2 in establishing and maintaining epigenetic regulation via methylation of histone H3, H3K27me3 immunoreactivity was lost in MPNSTs with mutation of PRC2 components and the transcriptomes of PRC2-wild type and -mutant MPNSTs were distinct.
At present, it is unclear whether the pathogenesis of sporadic, radiation-induced, and NF1-associated MPNSTs is driven by distinct mechanisms. Although NF1-associated and sporadic MPNSTs share some common pathogenic mechanisms (eg, EGF receptor amplification and activation of the PI3K-Akt-mTOR and MEK signaling cascades), a report that two sporadic MPNST cell lines (STS-26T and YST-1 cells) have intact NF1 genes68 has given the impression that NF1 mutation distinguishes between these two groups of neoplasms. An examination of currently available data shows that this conclusion is incorrect. Subsequent to the report that STS-26T cells have intact NF1 genes, it was found that these cells actually have a deletion within NF1.69 My laboratory has found that YST-1 cells have a genome quite distinct from that of MPNST cells, suggesting that they were incorrectly identified as an MPNST cell line (unpublished data). Further, one of the whole exome sequencing studies found NF1 mutations in 82% (18 of 22) of the sporadic and radiation-induced MPNSTs examined.67 Finally, it is not yet known whether sporadic MPNSTs with intact NF1 genes have other mutations that affect NF1-regulated signaling pathways; this possibility is consistent with observations from some genetically engineered mouse models (see Currently Available Mouse Models of Neurofibroma and MPNST Pathogenesis). Thus, we cannot yet conclude that sporadic and NF1-associated MPNSTs arise via distinct mechanisms. Indeed, the fact that transcriptome profiles distinguishing these tumor types have not yet been found (despite multiple attempts)70 is consistent with the possibility that the mechanisms driving the pathogenesis of sporadic and NF1-associated MPNSTs are quite similar.
Using Cross-Species Comparative Oncogenomics to Investigate MPNST Pathogenesis
Selecting an Animal Model That Maximizes the Advantages of Cross-Species Comparative Oncogenomics
Cross-species comparative oncogenomics is an approach in which a mouse cancer model is used to identify the somatic mutations that drive tumorigenesis, and the relevance of these mutations is then validated in the human counterpart of the mouse cancer. Because validation can be performed with formalin-fixed, paraffin-embedded tissue specimens, the size of the human tumor cohort available for validation is effectively amplified. The use of mouse cancer models also offers advantages that facilitate rapid identification of driver mutations. First, the genome of inbred mouse strains has less variability than human genomes. This means that somatic mutations in a mouse cancer are being identified against a relatively uniform genetic background rather than one with the extreme variability typical of humans. Second, there are intrinsic differences in the stability of human and mouse cancer genomes.71 Human cancer genomes are typically structurally complex, with numerous regions of focal chromosomal gain and loss; however, only a small subset of these copy number changes affect driver genes (Figure 2). In contrast, mouse cancer genomes tend to have whole chromosome gains or losses, with few focal copy number variations. Those focal gains or losses that do occur in mouse tumors arise under strong selective pressure and are more likely to contain a driver gene.72 Finally, because key driver genes and their associated pathways are evolutionarily conserved, driver mutations in mouse cancer models are usually relevant to the pathogenesis of their human counterpart. As evidence, cross-species comparative oncogenomic studies with the use of several genetically engineered mouse cancer models3, 73, 74, 75, 76, 77, 78, 79, 80 or murine cancers produced by insertional mutagenesis with the Sleeping Beauty transposon system64, 81, 82, 83 have proven highly effective for identifying novel oncogenes and tumor suppressor genes relevant to human tumor pathogenesis.
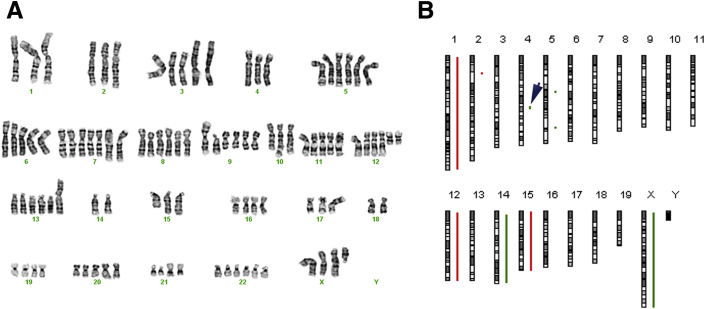
Figure 2.
Comparison of the genomes of a representative human MPNST and a mouse (P0-GGFβ3) MPNST. A: This human MPNST contained two major populations, one with 89 chromosomes and the other with 101 chromosomes. This highly complex karyotype was interpreted as 89 approximately 101<4n>,XXX,-X,-1,der(1)t(1;9)(q41;q21)x2,-2,der(2)(2pter->2p14::2q37->2p11.2::2q32->2qter),+der(3)(3qter->3p11::?::15q21->15qter),-4,+5,der(7)t(2;7)(q23;q22),+8,der(9)t(1;9)(q41;q13)x4,+i(9)(q10),-10,der(10)t(9;10)(q21;q25),+i(12)(p10),+add(13)(p11.2),-15,add(15)(q22)x2,-17,der(17)t(9;17)(q21;p11.1),add(17)(p11.2),-18,-18,add(20)(q12),der(21)t(19;21)(q13.3;p11.1)[cp15]. B: Array CGH analysis of an MPNST from P0-GGFβ3 mouse B233. In this ideogram, gains are indicated in red and losses in green. This genome is much simpler than that of the human MPNST shown in A, with only four whole chromosome gains or losses and four focal copy number variations. The arrowhead indicates a copy number loss that ablated the Cdkn2a/Cdkn2b gene. MPNST, malignant peripheral nerve sheath tumor.
To completely understand peripheral nerve sheath tumorigenesis, we need to identify the driver mutations responsible for neurofibroma pathogenesis, the transformation of a neurofibroma into a MPNST, and the progression from a low-grade MPNST to a high-grade malignancy. Ideally, cross-species comparative oncogenomics would thus be performed with animal models that recapitulate the process of tumorigenesis seen in humans and develop neurofibromas and MPNSTs that have genomic abnormalities mirroring those seen in their human counterparts. A variety of approaches were used to create genetically engineered mouse models that could be used for these studies (Table 1); these approaches included knocking out Nf1 and other tumor suppressor genes, overexpressing the growth factor NRG1, and overexpressing the EGF receptor. Given the central role that NF1 mutation plays in human neurofibroma and MPNST pathogenesis, one might expect that Nf1 knockout models would be the natural choice for cross-species comparative oncogenomics studies. However, there are important limitations to currently available Nf1 knockout models, and cancer genomics studies performed thus far have instead used the NRG1 and EGF receptor overexpression models. To explain these observations, it is useful to consider the strengths and weaknesses of these mouse models.
Table 1.
Genetically Engineered Mouse Models of Neurofibroma and MPNST Pathogenesis
| Model | Transgenic Mouse Model | Phenotype | Limitations |
|---|---|---|---|
| Nf1 knockout models | Nf1Δ31/Δ31 | Die of cardiac failure by embryonic day 13.5 | Early death prevents observation of tumorigenic effects of Nf1 loss |
| Nf1Δ31/+ | Develop pheochromocytomas (15% incidence); also show accelerated development of other non-NF1 tumors compared with wild-type mice | No neurofibromas or MPNSTs observed | |
| Krox20-Cre;Nf1flox/flox | Schwann cell hyperplasia | No neurofibromas or MPNSTs observed | |
| Krox20-Cre;Nf1flox/− | Plexiform neurofibroma development by 1 year of age (showing importance of both Nf1+/− and Nf1−/− cells in neurofibroma formation) | Neurofibromas in this model do not transform into MPNSTs | |
| Tumor microenvironment models | Krox2-Cre;Nf1flox/flox transplanted with Nf1+/− bone marrow | Developed plexiform neurofibromas infiltrated by donor mast cells (demonstrating importance of Nf1 haploinsufficiency in hematopoeitic lineage for neurofibroma formation) | Other Nf1+/− cell types within the hematopoietic lineage (eg, macrophages) may contribute to neurofibroma development |
| Nf1flox/+; Krox2-Cre transplanted with Nf1+/+ bone marrow | No neurofibromas developed (showing importance of Nf1 haploinsufficiency in hematopoeitic lineage for neurofibroma formation) | ||
| MPNST formation driven by dual tumor suppressor loss | trans-Linked Nf1+/−;p53+/− | Develop non-MPNST sarcomas characteristic of p53 LOH | No MPNST formation |
| cis-Linked Nf1+/−;p53+/− | Developed MPNSTs (∼30% incidence) | No neurofibroma precursor lesion | |
| Nf1+/−p16Ink4a−/− | Accelerated development of tumors characteristic of p16INK4a loss | No MPNST formation | |
| Nf1+/−p19Arf−/− | Accelerated development of tumors characteristic of p19Arf loss | No MPNST formation | |
| Nf1+/−p16Ink4a/p19Arf−/− | Developed MPNSTs (∼30% incidence) | No neurofibroma precursor lesion | |
| PNS tumor formation driven by dysregulated growth factor signaling | P0-GGFβ3 | Neurofibroma formation with progression to MPNSTs by 6 to 10 months | Although genetic complementation indicates that protumorigenic effect of NRG1 is in the Nf1 signaling cascade, tumors are not Nf1 null and other, as yet unknown, non-RasGAP neurofibromin functions that contribute to tumorigenesis may not be modeled |
| P0-GGFβ3;Trp53+/− | MPNSTs develop de novo and with complete penetrance by 6 to 9 months; also model MPNST progression from WHO grade II to IV | No neurofibroma precursor lesion | |
| CNPase-hEGFR | Schwann cell hyperplasia with mast cell recruitment and fibrosis | Neurofibroma formation exceedingly rare at advanced age; no MPNSTs observed | |
| Cnp-hEGFR;Trp53+/R270H | Develop both neurofibromas and MPNSTs | See discussion in text | |
| Cnp-hEGFR;Dhh-Cre;Ptenflox/+ | Develop neurofibromas | Neurofibromas do not transform into MPNSTs | |
| Cnp-hEGFR;Dhh-Cre;Ptenflox/flox | Develop MPNSTs de novo | No neurofibroma precursor lesion |
GAP, GTPase-activating protein; LOH, loss of heterozygosity; MPNST, malignant peripheral nerve sheath tumor; NF1, NF1, neurofibromatosis type 1; NRG, neuregulin; WHO, World Health Organization.
Currently Available Mouse Models of Neurofibroma and MPNST Pathogenesis
Mice with genetic ablation of the Nf1 gene were first created in 1994, when two laboratories independently created mice with deletion of exon 31 (Nf1Δ31/+ mice).84, 85 Surprisingly, although Nf1Δ31/+ mice were viable and fertile, they did not develop neurofibromas; these animals showed only some of the more rare manifestations of NF1 such as leukemias and pheochromocytomas. Mice homozygous for this mutation (Nf1Δ31/Δ31 mice) died at embryonic day 13.5 secondary to cardiac failure, and they displayed abnormally developed skeletal muscle, kidneys, and liver. However, nervous system pathology was limited to neuronal hyperplasia in sympathetic ganglia and, less commonly, exencephaly; neurofibromas were not evident.
One possible explanation for the absence of neurofibromas in Nf1Δ31/+ mice was that the frequency of second hit Nf1 mutations in the Schwann cells of these animals was rate limiting. Consequently, investigators next tried to create mice with a conditional knockout of Nf1 in Schwann cells (Krox20-Cre;Nf1flox/flox mice).86 These mice showed only mild Schwann cell hyperplasia, indicating that Nf1 loss in Schwann cells was not sufficient for neurofibroma pathogenesis. However, mice with complete Nf1 loss in Schwann cells and Nf1 haploinsufficiency in all other cell types (Krox20-Cre;Nf1flox/− mice) developed multiple neurofibromas in their cranial nerves and spinal nerve roots by 1 year of age. The contrasting phenotypes of Krox20-Cre;Nf1flox/flox and Krox20-Cre;Nf1flox/− mice provided the first experimental evidence that an Nf1 haploinsufficient tumor microenvironment was essential for neurofibroma pathogenesis. This concept (and the important role played by mast cells) was given further support by the subsequent demonstration that lethally irradiated Krox20-Cre;Nf1flox/flox mice grafted with Nf1+/− bone marrow developed multiple neurofibromas infiltrated by donor mast cells.87 Krox20-Cre;Nf1flox/− mice thus provided important new insights into the process of neurofibroma pathogenesis. Disappointingly, however, the neurofibromas in these animals only rarely transform and give rise to MPNSTs, and so Krox20-Cre;Nf1−/flox mice cannot be used to identify the driver genes responsible for the malignant transformation of neurofibromas and MPNST progression.
The lack of malignant transformation in the neurofibromas of Krox20-Cre;Nf1−/flox mice could, like the absence of neurofibromas in Nf1Δ31/+ mice, reflect a low rate of mutation in other key tumor suppressor genes involved in MPNST pathogenesis. To test this possibility, mice carrying null mutations of both Nf1 and Trp53 on the same (cis-linked Nf1+/−;Trp53+/− mice) and different (trans Nf1+/−;Trp53+/− mice) copies of chromosome 11 were created.88, 89 In mice, the most common mechanism of second hit mutation is whole chromosome loss. Consistent with this, 30% of cis-linked Nf1+/−;Trp53+/− mice developed MPNSTs by 5 months of age, whereas trans Nf1+/−;Trp53+/− mice typically died at 10 months of age with other tumor types characteristic of Trp53+/− mice. Likewise, 26% of Nf1+/− mice that also have homozygous mutations affecting both key Cdkn2a functions (Nf1+/−;p16Ink4a−/−;p19Arf+/− mice) also develop MPNSTs.90 Intriguingly, mice with mutations affecting only one of the two transcripts encoded by the Cdkn2a locus (Nf1+/−;p16Ink4a−/− or Nf1+/−;p19Arf−/− mice) do not develop MPNSTs,90, 91 indicating that both the p16INK4A-cyclin D1-CDK4-Rb and the p19ARF-Mdm2-p53 pathways must be dysregulated for MPNST pathogenesis to occur. These models provide strong experimental evidence that the Nf1, Trp53, and Cdkn2a genes play an important role in MPNST pathogenesis. However, in all of these models, MPNSTs arise de novo rather than from a neurofibroma precursor, and so they cannot be used to study transformation. Because a series of tumors from these animals have not been rigorously graded, it is also unclear whether they are appropriate models for identifying genes responsible for MPNST progression.
Although several growth factors and growth factor receptors contribute to neurofibroma and MPNST pathogenesis, only two of these molecules have been overexpressed in the Schwann cells of genetically engineered mice to directly examine their role in tumorigenesis. One of these is NRG1, a growth factor that regulates Schwann cell proliferation, survival, migration, and maturation during development. We have shown that NRG1 and all three of its receptors (the erbB2, erbB3, and erbB4 receptor tyrosine kinases) are co-expressed in human neurofibromas and MPNSTs; the resulting constitutive activation of these receptors drives the proliferation,92 survival,93 and migration94 of human MPNST cells. To determine whether inappropriate activation of the NRG1/erbB signaling is sufficient to induce tumorigenesis, we created transgenic mice in which a secreted NRG1 isoform is overexpressed in Schwann cells (P0-GGFβ3 mice). On both an outbred C57BL/6J × SJL/J background and when backcrossed onto a C57BL/6J background for five to eight generations, >90% of P0-GGFβ3 mice develop multiple neurofibromas in cranial, sympathetic, and spinal nerves that recapitulate the pathology of human plexiform neurofibromas. In 70% to 80% of P0-GGFβ3 mice, one or more neurofibromas transform into MPNSTs.95, 96 Further, P0-GGFβ3 MPNSTs have mutations in the p16INK4A-cyclin D/CDK4-Rb and p19ARF-Mdm-p53 pathways that parallel those seen in human MPNSTs. In addition, Ras is hyperactivated in P0-GGFβ3 MPNSTs in an erbB-dependent manner. However, immunoblot analyses96 and whole exome sequencing (unpublished data) showed that Nf1 is not mutated in P0-GGFβ3 MPNSTs, which raised the question of whether NRG1 promoted tumorigenesis via its effects on the Nf1 signaling cascade or instead activated another pathway that cooperated with the Nf1 signaling pathway to drive neoplasia. An opportunity to distinguish between these possibilities presented itself when it was noted that tumorigenesis was suppressed in P0-GGFβ3 mice bred onto a C57BL/6J background for ≥15 generations (inbred P0-GGFβ3 mice). To determine whether NRG1 interacts with Nf1 or Trp53 loss, cohorts of inbred P0-GGFβ3;Nf1+/− and P0-GGFβ3;Trp53+/− mice were established and examined for evidence of tumor formation.97 Although no tumors were seen in P0-GGFβ3;Nf1+/− mice, virtually all of the P0-GGFβ3;Trp53+/− mice developed MPNSTs. The MPNSTs in P0-GGFβ3;Trp53+/− mice, like those in cis-linked Nf1+/−;Trp53+/− mice, arose de novo, demonstrating that NRG1 overexpression could substitute for Nf1 loss and thus must promote tumorigenesis through the same signaling cascade affected by Nf1 loss. Further, P0-GGFβ3;Trp53+/− MPNSTs originated as lesions with the pathologic features of human World Health Organization grade II MPNSTs and subsequently progressed to become World Health Organization grade III and IV lesions. Thus, P0-GGFβ3 mice develop both neurofibromas and MPNSTs, recapitulate the process of neurofibroma-MPNST progression seen in human, and have tumor suppressor mutations analogous to those in human MPNSTs. In addition, P0-GGFβ3 and P0-GGFβ3;Trp53+/− mice model the evolution of MPNSTs as they progress from low to high grade.
Transgenic mice that overexpress the EGF receptor (erbB1) in Schwann cells [2′,3′-cyclic nucleotide 3′ phosphodiesterase (Cnp)-hEGFR mice) have also been generated.98 Although the EGF receptor is in the same family as the NRG1 receptors, the phenotype of Cnp-hEGFR mice is more subtle than that of P0-GGFβ3 mice. Cnp-hEGFR mice show Schwann cell hyperplasia, dissociation of Schwann cells from axons, increased numbers of intraneural mast cells, and fibrosis, but frank neurofibroma formation is rare, indicating that inappropriate expression of the EGF receptor is not sufficient for tumorigenesis. Nonetheless, the phenotype of Cnp-hEGFR mice does not exclude a role for the EGF receptor in neurofibroma and MPNST pathogenesis. As evidence, MPNST pathogenesis is impaired in cis-linked Nf1+/−;Trp53+/− mice that also carry a hypomorphic EGF receptor mutation (EGFRwa−2 mice). There is also evidence that inappropriate expression of the EGF receptor interacts with key tumor suppressor mutations to promote peripheral nervous system tumorigenesis. Crossing Cnp-hEGFR mice with Nf1+/− mice does not enhance tumorigenesis, much as was seen in inbred P0-GGFβ3;Nf1+/− mice. However, Cnp-hEGFR mice with partial (Cnp-hEGFR;Dhh-Cre;Ptenflox/+ mice) or complete (Cnp-hEGFR;Dhh-Cre;Ptenflox/flox mice) deletion of Pten in Schwann cells99 develop neurofibromas and MPNSTs, respectively. Likewise, Cnp-hEGFR mice that also express a dominant-negative Trp53 mutant (Cnp-hEGFR;Trp53+/R270H mice) develop neurofibromas and MPNSTs.100
Whole Genome aCGH and Forward Genetic Screens in Mouse MPNST Models
Two distinct approaches were used thus far to identify candidate driver genes in mouse MPNSTs, each of which is designed to take advantage of the distinct phenotypes of P0-GGFβ3 and Cnp-hEGFR mice. The first approach parallels that used by The Cancer Genome Atlas and uses whole genome aCGH, whole exome sequencing, and transcriptome sequencing to identify candidate driver mutations. Although findings from whole exome sequencing and transcriptome sequencing experiments have recently been presented at national meetings, only aCGH results are published,96, 97, 100 and so I focus on those experiments here.
Consistent with previous studies in other mouse tumors, whole genome aCGH in P0-GGFβ3 MPNSTs showed that the genomes of these mouse tumors were simpler than that of their human counterparts.96 Copy number variations that involve whole chromosomes or chromosome arms were common, occurring an average of 5.3 times (range, 3 to 10 times) per tumor genome. A smaller number of focal copy number gains or losses were also evident in each P0-GGFβ3 MPNST. An examination of the genes within 44 focal copy number gains or losses (26 gains, 18 losses) showed that 22 of these regions contained a total of 39 genes previously implicated in human neoplasia. One of the most common losses, which was present in >50% of the MPNSTs, was deletion of the Cdkn2a locus. The functions of the other genes were varied and included loci that encoded proteins regulating proliferation (Myc, Strn, Tpd52), chromatin remodeling (Ep400), progress through the cell cycle (Chfr, Chek2, Cdk2ap1, Cdk4), apoptosis (Bbc3/PUMA, XIAP, Ddit3/GADD153), transcriptional control (Ncoa3, Nr4a3, Zscan22), cell motility (Sept6, Vcl), and metabolism (Aldob, Mars, Alad). Genes encoding members of key cytoplasmic signaling pathways such as Hippo (Stk4), Notch (Dtx3), Hedgehog (Gli1), Arf (Agap2), Rho (Arhgap9), and PI3 kinase (Pten) were also repeatedly affected. Interestingly, several regions of recurrent copy number gain or loss did not contain any genes previously implicated in cancer pathogenesis, indicating that these regions may contain as yet unknown oncogenes or tumor suppressor genes.
MPNSTs from P0-GGFβ3;Trp53+/− and Cnp-EGFR;Trp53+/R270H mice have also been studied by whole genome aCGH.97, 100 Examination of a series of P0-GGFβ3;Trp53+/− MPNSTs that included World Health Organization grade II, III, and IV neoplasms showed that grade II tumors had fewer unbalanced copy number variations than grade IV lesions. Surprisingly, however, the number of copy number variations in grade III MPNSTs were not intermediate between grade II and IV neoplasms; instead, approximately one-half of the grade III MPNSTs had gains and losses that occurred with a frequency analogous to the grade IV tumors, with the remainder resembling the grade II MPNSTs. Although many of the candidate driver genes within the focal copy number variation in P0-GGFβ3;Trp53+/− MPNSTs were also seen in P0-GGFβ3 MPNSTs (eg, Myc), some genes encoding members of other signaling pathways (Prkcdbp, Mapk1, Rap1gds1, Arap, Crkl) were affected in P0-GGFβ3;Trp53+/− MPNSTs, raising the question of whether tumorigenesis in this mouse model involves distinct signaling cascades. The genomes of Cnp-EGFR;Trp53+/R270H MPNSTs had a complexity similar to that seen in P0-GGFβ3 MPNSTs. Again, Myc was among the genes recurrently amplified in these tumors; however, the signaling pathways affected in Cnp-EGFR;Trp53+/R270H MPNSTs included the Erk5, phospholipase C, tec kinase, and ephrin cascades.
The second approach that was used to identify driver genes in MPNSTs relies on the use of the Sleeping Beauty transposon system to accelerate tumorigenesis in Cnp-hEGFR and Cnp-EGFR;Trp53+/R270H mice.101 Examination of the common integration sites in 269 neurofibromas and 106 MPNSTs identified 695 and 87 genes, respectively, that were recurrently present in these tumors; 37 of these genes were detected in both neurofibromas and MPNSTs. Intriguingly, tumor suppressor genes predominated in these neoplasms, with 67 potential tumor suppressor genes and 20 candidate oncogenes identified. This collection of genes was then compared with archived aCGH, single nucleotide polymorphism array, RNA expression microarray, and methylome data from human Schwann cells, neurofibromas, and MPNSTs to identify a subset of genes potentially relevant to human neurofibroma and MPNST pathogenesis. In addition to driver genes known to be important for human MPNST pathogenesis (NF1, CDKN2A, PTEN), three signaling cascades (PI3K/AKT/mTOR, MAPK/ERK/JNK/p38, and Wnt/CTNNB1) were identified that were both repetitively targeted by Sleeping Beauty and potentially relevant to human tumorigenesis. These comparisons also identified the forkhead-box transcription factor FOXR2 as an oncogene potentially important for the development of MPNSTs. Thus, forward genetic screens with Sleeping Beauty transposons clearly identified genes relevant to the pathogenesis of human MPNSTs, underscoring the effectiveness of using mouse models to understand the pathogenesis of human peripheral nerve sheath tumors.
Conclusions and Future Directions
Although cross-species comparative oncogenomic studies of MPNSTs are still in their infancy, these analyses have already begun to provide new insights into the mechanisms responsible for the development of these deadly neoplasms (Figure 3). However, there are some important caveats that need to be considered as we move forward. First, the observation that different signaling pathways are being affected in different mouse MPNST models raises the question of whether particular mouse models are modeling specific MPNST subtypes. Consequently, it likely will be important to study multiple mouse models to assess this possibility. Second, it must be remembered that the identification of driver genes in mouse MPNSTs via whole exome sequencing, aCGH, and transcriptome sequencing is computational (as it is in comparable studies of human MPNSTs). Consequently, it is essential that candidate driver genes be functionally validated by a variety of approaches, including those described in the sections above. Third, it should be recognized that cross-species comparative oncogenomics will probably not effectively identify some important types of mutations such as fusion genes. This potential limitation reflects the fact that mouse MPNSTs do not demonstrate the extreme degree of chromothripsis102 and subsequent rejoining of chromosomal breaks seen in human MPNSTs. Nonetheless, it is increasingly evident that cross-species comparative oncogenomics is a promising approach to the challenges of performing genomics in rare tumor types and undoubtedly will be increasingly important as we turn our attention to these problematic neoplasms.
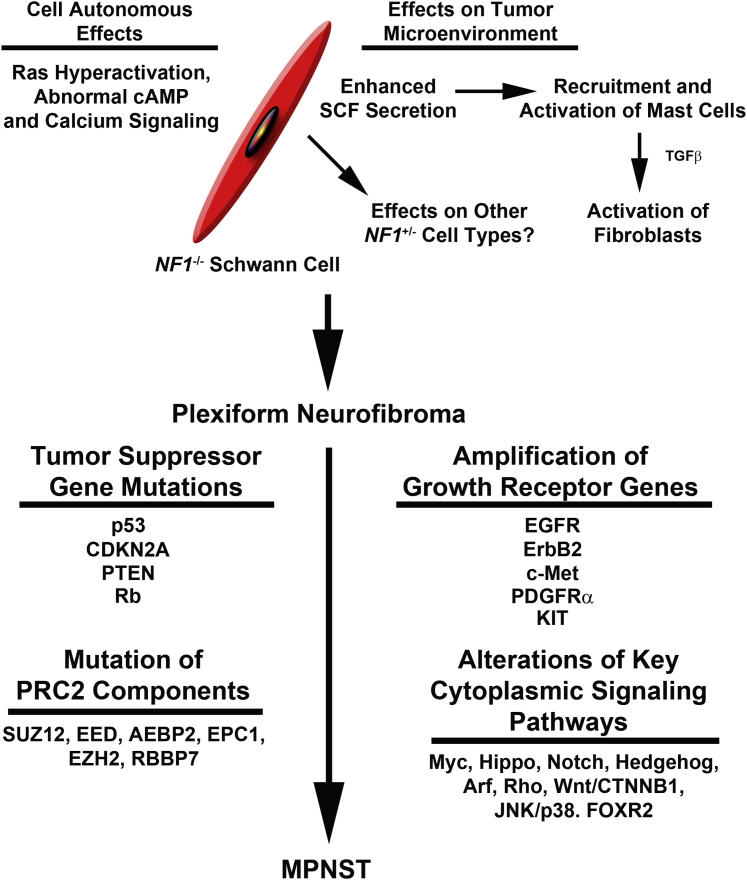
Figure 3.
Overview of our current understanding of neurofibroma- MPNST pathogenesis is based on both human and mouse studies. The upper half of the diagram indicates the cell autonomous and nonautonomous consequences of NF1 loss and how this leads to neurofibroma pathogenesis. Additional events involved in the transformation of a neurofibroma to an MPNST are indicated below. Although multiple events are indicated here, note that it is quite possible that many of these abnormalities may be specific to as yet undefined MPNST subtypes. Consistent with this, initial comparisons of PRC2-mutant and wild-type MPNSTs indicated that some of the indicated tumor suppressor mutations were specifically associated with PRC2 mutant tumors. EGFR, epidermal growth factor receptor; MPNST, malignant peripheral nerve sheath tumor; PDGFR, platelet-derived growth factor receptor; PRC2, Polycomb repressive complex 2; PTEN, phosphate and tensin homologue; TGF, transforming growth factor.
Acknowledgments
I thank Drs. Jody Fromm Longo and Amanda Prechtl for their helpful comments on this manuscript.
Neuropathology Theme Issue
Footnotes
Supported by NIH National Institute of Neurological Diseases and Stroke grant R01 NS048353; NIH National Cancer Institute grant R01 CA122804; the Department of Defense grants X81XWH-09-1-0086, W81XWH-11-1-0498, W81XWH-12-1-0164, W81XWH-14-1-0073, and W81XWH-15-1-0193; and The Children's Tumor Foundation grants 2014-04-001 and 2015-05-007.
Disclosures: None declared.
This article is part of a review series on neuropathology.
References
- 1.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parsons D.W., Jones S., Zhang X., Lin J.C., Leary R.J., Angenendt P., Mankoo P., Carter H., Siu I.M., Gallia G.L., Olivi A., McLendon R., Rasheed B.A., Keir S., Nikolskaya T., Nikolsky Y., Busam D.A., Tekleab H., Diaz L.A., Jr., Hartigan J., Smith D.R., Strausberg R.L., Marie S.K., Shinjo S.M., Yan H., Riggins G.J., Bigner D.D., Karchin R., Papadopoulos N., Parmigiani G., Vogelstein B., Velculescu V.E., Kinzler K.W. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson R.A., Wright K.D., Poppleton H., Mohankumar K.M., Finkelstein D., Pounds S.B., Rand V., Leary S.E., White E., Eden C., Hogg T., Northcott P., Mack S., Neale G., Wang Y.D., Coyle B., Atkinson J., DeWire M., Kranenburg T.A., Gillespie Y., Allen J.C., Merchant T., Boop F.A., Sanford R.A., Gajjar A., Ellison D.W., Taylor M.D., Grundy R.G., Gilbertson R.J. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466:632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Modena P., Lualdi E., Facchinetti F., Veltman J., Reid J.F., Minardi S., Janssen I., Giangaspero F., Forni M., Finocchiaro G., Genitori L., Giordano F., Riccardi R., Schoenmakers E.F., Massimino M., Sozzi G. Identification of tumor-specific molecular signatures in intracranial ependymoma and association with clinical characteristics. J Clin Oncol. 2006;24:5223–5233. doi: 10.1200/JCO.2006.06.3701. [DOI] [PubMed] [Google Scholar]
- 5.Witt H., Mack S.C., Ryzhova M., Bender S., Sill M., Isserlin R. Delineation of two clinically and molecularly distinct subgroups of posterior fossa ependymoma. Cancer Cell. 2011;20:143–157. doi: 10.1016/j.ccr.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wani K., Armstrong T.S., Vera-Bolanos E., Raghunathan A., Ellison D., Gilbertson R., Vaillant B., Goldman S., Packer R.J., Fouladi M., Pollack I., Mikkelsen T., Prados M., Omuro A., Soffietti R., Ledoux A., Wilson C., Long L., Gilbert M.R., Aldape K., Collaborative Ependymoma Research Network A prognostic gene expression signature in infratentorial ependymoma. Acta Neuropathol. 2012;123:727–738. doi: 10.1007/s00401-012-0941-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mack S.C., Witt H., Piro R.M., Gu L., Zuyderduyn S., Stutz A.M. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506:445–450. doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pajtler K.W., Witt H., Sill M., Jones D.T., Hovestadt V., Kratochwil F. Molecular classification of ependymal tumors across all CNS compartments, histopathological grades, and age groups. Cancer Cell. 2015;27:728–743. doi: 10.1016/j.ccell.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Parsons D.W., Li M., Zhang X., Jones S., Leary R.J., Lin J.C. The genetic landscape of the childhood cancer medulloblastoma. Science. 2011;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones D.T., Jager N., Kool M., Zichner T., Hutter B., Sultan M. Dissecting the genomic complexity underlying medulloblastoma. Nature. 2012;488:100–105. doi: 10.1038/nature11284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khuong-Quang D.A., Buczkowicz P., Rakopoulos P., Liu X.Y., Fontebasso A.M., Bouffet E., Bartels U., Albrecht S., Schwartzentruber J., Letourneau L., Bourgey M., Bourque G., Montpetit A., Bourret G., Lepage P., Fleming A., Lichter P., Kool M., von Deimling A., Sturm D., Korshunov A., Faury D., Jones D.T., Majewski J., Pfister S.M., Jabado N., Hawkins C. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta Neuropathol. 2012;124:439–447. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartzentruber J., Korshunov A., Liu X.Y., Jones D.T., Pfaff E., Jacob K. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 13.Wu G., Broniscer A., McEachron T.A., Lu C., Paugh B.S., Becksfort J., Qu C., Ding L., Huether R., Parker M., Zhang J., Gajjar A., Dyer M.A., Mullighan C.G., Gilbertson R.J., Mardis E.R., Wilson R.K., Downing J.R., Ellison D.W., Baker S.J., St. Jude Children's Research Hospital—Washington University Pediatric Cancer Genome Project Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44:251–253. doi: 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aldape K., Zadeh G., Mansouri S., Reifenberger G., von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129:829–848. doi: 10.1007/s00401-015-1432-1. [DOI] [PubMed] [Google Scholar]
- 15.Louis D.N., Perry A., Burger P., Ellison D.W., Reifenberger G., von Deimling A., Aldape K., Brat D., Collins V.P., Eberhart C., Figarella-Branger D., Fuller G.N., Giangaspero F., Giannini C., Hawkins C., Kleihues P., Korshunov A., Kros J.M., Beatriz Lopes M., Ng H.K., Ohgaki H., Paulus W., Pietsch T., Rosenblum M., Rushing E., Soylemezoglu F., Wiestler O., Wesseling P., International Society of Neuropathology--Haarlem International Society Of Neuropathology–Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol. 2014;24:429–435. doi: 10.1111/bpa.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verhaak R.G., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., Alexe G., Lawrence M., O'Kelly M., Tamayo P., Weir B.A., Gabriel S., Winckler W., Gupta S., Jakkula L., Feiler H.S., Hodgson J.G., James C.D., Sarkaria J.N., Brennan C., Kahn A., Spellman P.T., Wilson R.K., Speed T.P., Gray J.W., Meyerson M., Getz G., Perou C.M., Hayes D.N., Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paugh B.S., Broniscer A., Qu C., Miller C.P., Zhang J., Tatevossian R.G., Olson J.M., Geyer J.R., Chi S.N., da Silva N.S., Onar-Thomas A., Baker J.N., Gajjar A., Ellison D.W., Baker S.J. Genome-wide analyses identify recurrent amplifications of receptor tyrosine kinases and cell-cycle regulatory genes in diffuse intrinsic pontine glioma. J Clin Oncol. 2011;29:3999–4006. doi: 10.1200/JCO.2011.35.5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puget S., Philippe C., Bax D.A., Job B., Varlet P., Junier M.P., Andreiuolo F., Carvalho D., Reis R., Guerrini-Rousseau L., Roujeau T., Dessen P., Richon C., Lazar V., Le Teuff G., Sainte-Rose C., Geoerger B., Vassal G., Jones C., Grill J. Mesenchymal transition and PDGFRA amplification/mutation are key distinct oncogenic events in pediatric diffuse intrinsic pontine gliomas. PLoS One. 2012;7:e30313. doi: 10.1371/journal.pone.0030313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saratsis A.M., Kambhampati M., Snyder K., Yadavilli S., Devaney J.M., Harmon B., Hall J., Raabe E.H., An P., Weingart M., Rood B.R., Magge S.N., MacDonald T.J., Packer R.J., Nazarian J. Comparative multidimensional molecular analyses of pediatric diffuse intrinsic pontine glioma reveals distinct molecular subtypes. Acta Neuropathol. 2014;127:881–895. doi: 10.1007/s00401-013-1218-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pugh T.J., Weeraratne S.D., Archer T.C., Pomeranz Krummel D.A., Auclair D., Bochicchio J. Medulloblastoma exome sequencing uncovers subtype-specific somatic mutations. Nature. 2012;488:106–110. doi: 10.1038/nature11329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Northcott P.A., Shih D.J., Peacock J., Garzia L., Morrissy A.S., Zichner T. Subgroup-specific structural variation across 1,000 medulloblastoma genomes. Nature. 2012;488:49–56. doi: 10.1038/nature11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kool M., Korshunov A., Remke M., Jones D.T., Schlanstein M., Northcott P.A., Cho Y.J., Koster J., Schouten-van Meeteren A., van Vuurden D., Clifford S.C., Pietsch T., von Bueren A.O., Rutkowski S., McCabe M., Collins V.P., Backlund M.L., Haberler C., Bourdeaut F., Delattre O., Doz F., Ellison D.W., Gilbertson R.J., Pomeroy S.L., Taylor M.D., Lichter P., Pfister S.M. Molecular subgroups of medulloblastoma: an international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012;123:473–484. doi: 10.1007/s00401-012-0958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis J.J., Brennan M.F. Soft tissue sarcomas. Curr Probl Surg. 1996;33:817–872. [PubMed] [Google Scholar]
- 24.Carroll S.L. Molecular mechanisms promoting the pathogenesis of Schwann cell neoplasms. Acta Neuropathol. 2012;123:321–348. doi: 10.1007/s00401-011-0928-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Messiaen L.M., Callens T., Mortier G., Beysen D., Vandenbroucke I., Van Roy N., Speleman F., Paepe A.D. Exhaustive mutation analysis of the NF1 gene allows identification of 95% of mutations and reveals a high frequency of unusual splicing defects. Hum Mutat. 2000;15:541–555. doi: 10.1002/1098-1004(200006)15:6<541::AID-HUMU6>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 26.Wimmer K., Roca X., Beiglbock H., Callens T., Etzler J., Rao A.R., Krainer A.R., Fonatsch C., Messiaen L. Extensive in silico analysis of NF1 splicing defects uncovers determinants for splicing outcome upon 5' splice-site disruption. Hum Mutat. 2007;28:599–612. doi: 10.1002/humu.20493. [DOI] [PubMed] [Google Scholar]
- 27.Wimmer K., Yao S., Claes K., Kehrer-Sawatzki H., Tinschert S., De Raedt T., Legius E., Callens T., Beiglbock H., Maertens O., Messiaen L. Spectrum of single- and multiexon NF1 copy number changes in a cohort of 1,100 unselected NF1 patients. Genes Chromosomes Cancer. 2006;45:265–276. doi: 10.1002/gcc.20289. [DOI] [PubMed] [Google Scholar]
- 28.Widemann B.C., Dombi E., Gillespie A., Wolters P.L., Belasco J., Goldman S., Korf B.R., Solomon J., Martin S., Salzer W., Fox E., Patronas N., Kieran M.W., Perentesis J.P., Reddy A., Wright J.J., Kim A., Steinberg S.M., Balis F.M. Phase 2 randomized, flexible crossover, double-blinded, placebo-controlled trial of the farnesyltransferase inhibitor tipifarnib in children and young adults with neurofibromatosis type 1 and progressive plexiform neurofibromas. Neuro Oncol. 2014;16:707–718. doi: 10.1093/neuonc/nou004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brossier N.M., Prechtl A.M., Longo J.F., Barnes S., Wilson L.S., Byer S.J., Brosius S.N., Carroll S.L. Classic Ras proteins promote proliferation and survival via distinct phosphoproteome alterations in neurofibromin-null malignant peripheral nerve sheath tumor cells. J Neuropathol Exp Neurol. 2015;74:568–586. doi: 10.1097/NEN.0000000000000201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Appels N.M., Beijnen J.H., Schellens J.H. Development of farnesyl transferase inhibitors: a review. Oncologist. 2005;10:565–578. doi: 10.1634/theoncologist.10-8-565. [DOI] [PubMed] [Google Scholar]
- 31.Jessen W.J., Miller S.J., Jousma E., Wu J., Rizvi T.A., Brundage M.E., Eaves D., Widemann B., Kim M.O., Dombi E., Sabo J., Hardiman Dudley A., Niwa-Kawakita M., Page G.P., Giovannini M., Aronow B.J., Cripe T.P., Ratner N. MEK inhibition exhibits efficacy in human and mouse neurofibromatosis tumors. J Clin Invest. 2013;123:340–347. doi: 10.1172/JCI60578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watson A.L., Anderson L.K., Greeley A.D., Keng V.W., Rahrmann E.P., Halfond A.L., Powell N.M., Collins M.H., Rizvi T., Moertel C.L., Ratner N., Largaespada D.A. Co-targeting the MAPK and PI3K/AKT/mTOR pathways in two genetically engineered mouse models of schwann cell tumors reduces tumor grade and multiplicity. Oncotarget. 2014;5:1502–1514. doi: 10.18632/oncotarget.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johannessen C.M., Reczek E.E., James M.F., Brems H., Legius E., Cichowski K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc Natl Acad Sci U S A. 2005;102:8573–8578. doi: 10.1073/pnas.0503224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johannessen C.M., Johnson B.W., Williams S.M., Chan A.W., Reczek E.E., Lynch R.C., Rioth M.J., McClatchey A., Ryeom S., Cichowski K. TORC1 is essential for NF1-associated malignancies. Curr Biol. 2008;18:56–62. doi: 10.1016/j.cub.2007.11.066. [DOI] [PubMed] [Google Scholar]
- 35.Hirota S., Nomura S., Asada H., Ito A., Morii E., Kitamura Y. Possible involvement of c-kit receptor and its ligand in increase of mast cells in neurofibroma tissues. Arch Pathol Lab Med. 1993;117:996–999. [PubMed] [Google Scholar]
- 36.Ryan J.J., Klein K.A., Neuberger T.J., Leftwich J.A., Westin E.H., Kauma S., Fletcher J.A., DeVries G.H., Huff T.F. Role for the stem cell factor/KIT complex in Schwann cell neoplasia and mast cell proliferation associated with neurofibromatosis. J Neurosci Res. 1994;37:415–432. doi: 10.1002/jnr.490370314. [DOI] [PubMed] [Google Scholar]
- 37.Yang F.C., Ingram D.A., Chen S., Hingtgen C.M., Ratner N., Monk K.R., Clegg T., White H., Mead L., Wenning M.J., Williams D.A., Kapur R., Atkinson S.J., Clapp D.W. Neurofibromin-deficient Schwann cells secrete a potent migratory stimulus for Nf1+/- mast cells. J Clin Invest. 2003;112:1851–1861. doi: 10.1172/JCI19195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ingram D.A., Yang F.C., Travers J.B., Wenning M.J., Hiatt K., New S., Hood A., Shannon K., Williams D.A., Clapp D.W. Genetic and biochemical evidence that haploinsufficiency of the Nf1 tumor suppressor gene modulates melanocyte and mast cell fates in vivo. J Exp Med. 2000;191:181–188. doi: 10.1084/jem.191.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ingram D.A., Hiatt K., King A.J., Fisher L., Shivakumar R., Derstine C., Wenning M.J., Diaz B., Travers J.B., Hood A., Marshall M., Williams D.A., Clapp D.W. Hyperactivation of p21(ras) and the hematopoietic-specific Rho GTPase, Rac2, cooperate to alter the proliferation of neurofibromin-deficient mast cells in vivo and in vitro. J Exp Med. 2001;194:57–69. doi: 10.1084/jem.194.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang F.C., Chen S., Clegg T., Li X., Morgan T., Estwick S.A., Yuan J., Khalaf W., Burgin S., Travers J., Parada L.F., Ingram D.A., Clapp D.W. Nf1+/- mast cells induce neurofibroma like phenotypes through secreted TGF-beta signaling. Hum Mol Genet. 2006;15:2421–2437. doi: 10.1093/hmg/ddl165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim H.A., Rosenbaum T., Marchionni M.A., Ratner N., DeClue J.E. Schwann cells from neurofibromin deficient mice exhibit activation of p21ras, inhibition of cell proliferation and morphological changes. Oncogene. 1995;11:325–335. [PubMed] [Google Scholar]
- 42.Atit R.P., Crowe M.J., Greenhalgh D.G., Wenstrup R.J., Ratner N. The Nf1 tumor suppressor regulates mouse skin wound healing, fibroblast proliferation, and collagen deposited by fibroblasts. J Invest Dermatol. 1999;112:835–842. doi: 10.1046/j.1523-1747.1999.00609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korkiamaki T., Yla-Outinen H., Koivunen J., Karvonen S.L., Peltonen J. Altered calcium-mediated cell signaling in keratinocytes cultured from patients with neurofibromatosis type 1. Am J Pathol. 2002;160:1981–1990. doi: 10.1016/S0002-9440(10)61148-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kourea H.P., Orlow I., Scheithauer B.W., Cordon-Cardo C., Woodruff J.M. Deletions of the INK4A gene occur in malignant peripheral nerve sheath tumors but not in neurofibromas. Am J Pathol. 1999;155:1855–1860. doi: 10.1016/S0002-9440(10)65504-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nielsen G.P., Stemmer-Rachamimov A.O., Ino Y., Moller M.B., Rosenberg A.E., Louis D.N. Malignant transformation of neurofibromas in neurofibromatosis 1 is associated with CDKN2A/p16 inactivation. Am J Pathol. 1999;155:1879–1884. doi: 10.1016/S0002-9440(10)65507-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mawrin C., Kirches E., Boltze C., Dietzmann K., Roessner A., Schneider-Stock R. Immunohistochemical and molecular analysis of p53, RB, and PTEN in malignant peripheral nerve sheath tumors. Virchows Arch. 2002;440:610–615. doi: 10.1007/s00428-001-0550-4. [DOI] [PubMed] [Google Scholar]
- 47.Mantripragada K.K., Spurlock G., Kluwe L., Chuzhanova N., Ferner R.E., Frayling I.M., Dumanski J.P., Guha A., Mautner V., Upadhyaya M. High-resolution DNA copy number profiling of malignant peripheral nerve sheath tumors using targeted microarray-based comparative genomic hybridization. Clin Cancer Res. 2008;14:1015–1024. doi: 10.1158/1078-0432.CCR-07-1305. [DOI] [PubMed] [Google Scholar]
- 48.Gregorian C., Nakashima J., Dry S.M., Nghiemphu P.L., Smith K.B., Ao Y., Dang J., Lawson G., Mellinghoff I.K., Mischel P.S., Phelps M., Parada L.F., Liu X., Sofroniew M.V., Eilber F.C., Wu H. PTEN dosage is essential for neurofibroma development and malignant transformation. Proc Natl Acad Sci U S A. 2009;106:19479–19484. doi: 10.1073/pnas.0910398106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bradtmoller M., Hartmann C., Zietsch J., Jaschke S., Mautner V.F., Kurtz A., Park S.J., Baier M., Harder A., Reuss D., von Deimling A., Heppner F.L., Holtkamp N. Impaired Pten expression in human malignant peripheral nerve sheath tumours. PLoS One. 2012;7:e47595. doi: 10.1371/journal.pone.0047595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Danielsen S.A., Lind G.E., Kolberg M., Holand M., Bjerkehagen B., Sundby Hall K., van den Berg E., Mertens F., Smeland S., Picci P., Lothe R.A. Methylated RASSF1A in malignant peripheral nerve sheath tumors identifies neurofibromatosis type 1 patients with inferior prognosis. Neuro Oncol. 2015;17:63–69. doi: 10.1093/neuonc/nou140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang J., Ylipaa A., Sun Y., Zheng H., Chen K., Nykter M., Trent J., Ratner N., Lev D.C., Zhang W. Genomic and molecular characterization of malignant peripheral nerve sheath tumor identifies the IGF1R pathway as a primary target for treatment. Clin Cancer Res. 2011;17:7563–7573. doi: 10.1158/1078-0432.CCR-11-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carroll S.L., Stonecypher M.S. Tumor suppressor mutations and growth factor signaling in the pathogenesis of NF1-associated peripheral nerve sheath tumors: II. The role of dysregulated growth factor signaling. J Neuropathol Exp Neurol. 2005;64:1–9. doi: 10.1093/jnen/64.1.1. [DOI] [PubMed] [Google Scholar]
- 53.Nebesio T.D., Ming W., Chen S., Clegg T., Yuan J., Yang Y., Estwick S.A., Li Y., Li X., Hingtgen C.M., Yang F.C. Neurofibromin-deficient Schwann cells have increased lysophosphatidic acid dependent survival and migration-implications for increased neurofibroma formation during pregnancy. Glia. 2007;55:527–536. doi: 10.1002/glia.20482. [DOI] [PubMed] [Google Scholar]
- 54.Mo W., Chen J., Patel A., Zhang L., Chau V., Li Y., Cho W., Lim K., Xu J., Lazar A.J., Creighton C.J., Bolshakov S., McKay R.M., Lev D., Le L.Q., Parada L.F. CXCR4/CXCL12 mediate autocrine cell- cycle progression in NF1-associated malignant peripheral nerve sheath tumors. Cell. 2013;152:1077–1090. doi: 10.1016/j.cell.2013.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zietsch J., Ziegenhagen N., Heppner F.L., Reuss D., von Deimling A., Holtkamp N. The 4q12 amplicon in malignant peripheral nerve sheath tumors: consequences on gene expression and implications for sunitinib treatment. PLoS One. 2010;5:e11858. doi: 10.1371/journal.pone.0011858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Forus A., Weghuis D.O., Smeets D., Fodstad O., Myklebost O., van Kessel A.G. Comparative genomic hybridization analysis of human sarcomas: I. Occurrence of genomic imbalances and identification of a novel major amplicon at 1q21-q22 in soft tissue sarcomas. Genes Chromosomes Cancer. 1995;14:8–14. doi: 10.1002/gcc.2870140103. [DOI] [PubMed] [Google Scholar]
- 57.Lothe R.A., Karhu R., Mandahl N., Mertens F., Saeter G., Heim S., Borresen-Dale A.L., Kallioniemi O.P. Gain of 17q24-qter detected by comparative genomic hybridization in malignant tumors from patients with von Recklinghausen's neurofibromatosis. Cancer Res. 1996;56:4778–4781. [PubMed] [Google Scholar]
- 58.Mechtersheimer G., Otano-Joos M., Ohl S., Benner A., Lehnert T., Willeke F., Moller P., Otto H.F., Lichter P., Joos S. Analysis of chromosomal imbalances in sporadic and NF1-associated peripheral nerve sheath tumors by comparative genomic hybridization. Genes Chromosomes Cancer. 1999;25:362–369. [PubMed] [Google Scholar]
- 59.Mertens F., Rydholm A., Bauer H.F., Limon J., Nedoszytko B., Szadowska A., Willen H., Heim S., Mitelman F., Mandahl N. Cytogenetic findings in malignant peripheral nerve sheath tumors. Int J Cancer. 1995;61:793–798. doi: 10.1002/ijc.2910610609. [DOI] [PubMed] [Google Scholar]
- 60.Mertens F., Dal Cin P., De Wever I., Fletcher C.D., Mandahl N., Mitelman F., Rosai J., Rydholm A., Sciot R., Tallini G., van Den Berghe H., Vanni R., Willen H. Cytogenetic characterization of peripheral nerve sheath tumours: a report of the CHAMP study group. J Pathol. 2000;190:31–38. doi: 10.1002/(SICI)1096-9896(200001)190:1<31::AID-PATH505>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 61.Berner J.M., Sorlie T., Mertens F., Henriksen J., Saeter G., Mandahl N., Brogger A., Myklebost O., Lothe R.A. Chromosome band 9p21 is frequently altered in malignant peripheral nerve sheath tumors: studies of CDKN2A and other genes of the pRB pathway. Genes Chromosomes Cancer. 1999;26:151–160. [PubMed] [Google Scholar]
- 62.Schmidt H., Wurl P., Taubert H., Meye A., Bache M., Holzhausen H.J., Hinze R. Genomic imbalances of 7p and 17q in malignant peripheral nerve sheath tumors are clinically relevant. Genes Chromosomes Cancer. 1999;25:205–211. [PubMed] [Google Scholar]
- 63.Schmidt H., Taubert H., Meye A., Wurl P., Bache M., Bartel F., Holzhausen H.J., Hinze R. Gains in chromosomes 7, 8q, 15q and 17q are characteristic changes in malignant but not in benign peripheral nerve sheath tumors from patients with Recklinghausen's disease. Cancer Lett. 2000;155:181–190. doi: 10.1016/s0304-3835(00)00426-2. [DOI] [PubMed] [Google Scholar]
- 64.Schmidt H., Taubert H., Wurl P., Bache M., Bartel F., Holzhausen H.J., Hinze R. Cytogenetic characterization of six malignant peripheral nerve sheath tumors: comparison of karyotyping and comparative genomic hybridization. Cancer Genet Cytogenet. 2001;128:14–23. doi: 10.1016/s0165-4608(01)00393-4. [DOI] [PubMed] [Google Scholar]
- 65.Kresse S.H., Skarn M., Ohnstad H.O., Namlos H.M., Bjerkehagen B., Myklebost O., Meza-Zepeda L.A. DNA copy number changes in high-grade malignant peripheral nerve sheath tumors by array CGH. Mol Cancer. 2008;7:48. doi: 10.1186/1476-4598-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang M., Wang Y., Jones S., Sausen M., McMahon K., Sharma R., Wang Q., Belzberg A.J., Chaichana K., Gallia G.L., Gokaslan Z.L., Riggins G.J., Wolinksy J.P., Wood L.D., Montgomery E.A., Hruban R.H., Kinzler K.W., Papadopoulos N., Vogelstein B., Bettegowda C. Somatic mutations of SUZ12 in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46:1170–1172. doi: 10.1038/ng.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee W., Teckie S., Wiesner T., Ran L., Prieto Granada C.N., Lin M., Zhu S., Cao Z., Liang Y., Sboner A., Tap W.D., Fletcher J.A., Huberman K.H., Qin L.X., Viale A., Singer S., Zheng D., Berger M.F., Chen Y., Antonescu C.R., Chi P. PRC2 is recurrently inactivated through EED or SUZ12 loss in malignant peripheral nerve sheath tumors. Nat Genet. 2014;46:1227–1232. doi: 10.1038/ng.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Miller S.J., Rangwala F., Williams J., Ackerman P., Kong S., Jegga A.G., Kaiser S., Aronow B.J., Frahm S., Kluwe L., Mautner V., Upadhyaya M., Muir D., Wallace M., Hagen J., Quelle D.E., Watson M.A., Perry A., Gutmann D.H., Ratner N. Large-scale molecular comparison of human schwann cells to malignant peripheral nerve sheath tumor cell lines and tissues. Cancer Res. 2006;66:2584–2591. doi: 10.1158/0008-5472.CAN-05-3330. [DOI] [PubMed] [Google Scholar]
- 69.Pasmant E., Luscan A., Varin J., Laurendeau I., Parfait B., Vidaud D. Relevance of MPNST cell lines as models for NF1 associated-tumors. J Neurooncol. 2013;114:353–355. doi: 10.1007/s11060-013-1185-4. [DOI] [PubMed] [Google Scholar]
- 70.Carroll S.L., Ratner N. How does the Schwann cell lineage form tumors in NF1? Glia. 2008;56:1590–1605. doi: 10.1002/glia.20776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maser R.S., Choudhury B., Campbell P.J., Feng B., Wong K.K., Protopopov A. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Plaat B.E., Molenaar W.M., Mastik M.F., Hoekstra H.J., te Meerman G.J., van den Berg E. Computer-assisted cytogenetic analysis of 51 malignant peripheral-nerve-sheath tumors: sporadic vs. neurofibromatosis-type-1-associated malignant schwannomas. Int J Cancer. 1999;83:171–178. doi: 10.1002/(sici)1097-0215(19991008)83:2<171::aid-ijc5>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 73.Young N.P., Crowley D., Jacks T. Uncoupling cancer mutations reveals critical timing of p53 loss in sarcomagenesis. Cancer Res. 2011;71:4040–4047. doi: 10.1158/0008-5472.CAN-10-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu H., Tardivo L., Tam S., Weiner E., Gebreab F., Fan C., Svrzikapa N., Hirozane-Kishikawa T., Rietman E., Yang X., Sahalie J., Salehi-Ashtiani K., Hao T., Cusick M.E., Hill D.E., Roth F.P., Braun P., Vidal M. Next-generation sequencing to generate interactome datasets. Nat Methods. 2011;8:478–480. doi: 10.1038/nmeth.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maser R.S., Choudhury B., Campbell P.J., Feng B., Wong K.K., Protopopov A. Chromosomally unstable mouse tumours have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Garber M., Grabherr M.G., Guttman M., Trapnell C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods. 2011;8:469–477. doi: 10.1038/nmeth.1613. [DOI] [PubMed] [Google Scholar]
- 77.Endersby R., Zhu X., Hay N., Ellison D.W., Baker S.J. Nonredundant functions for Akt isoforms in astrocyte growth and gliomagenesis in an orthotopic transplantation model. Cancer Res. 2011;71:4106–4116. doi: 10.1158/0008-5472.CAN-10-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Varela I., Klijn C., Stephens P.J., Mudie L.J., Stebbings L., Galappaththige D., van der Gulden H., Schut E., Klarenbeek S., Campbell P.J., Wessels L.F., Stratton M.R., Jonkers J., Futreal P.A., Adams D.J. Somatic structural rearrangements in genetically engineered mouse mammary tumors. Genome Biol. 2010;11:R100. doi: 10.1186/gb-2010-11-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kim M., Gans J.D., Nogueira C., Wang A., Paik J.H., Feng B., Brennan C., Hahn W.C., Cordon-Cardo C., Wagner S.N., Flotte T.J., Duncan L.M., Granter S.R., Chin L. Comparative oncogenomics identifies NEDD9 as a melanoma metastasis gene. Cell. 2006;125:1269–1281. doi: 10.1016/j.cell.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 80.Zender L., Spector M.S., Xue W., Flemming P., Cordon-Cardo C., Silke J., Fan S.T., Luk J.M., Wigler M., Hannon G.J., Mu D., Lucito R., Powers S., Lowe S.W. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Uren A.G., Kool J., Matentzoglu K., de Ridder J., Mattison J., van Uitert M., Lagcher W., Sie D., Tanger E., Cox T., Reinders M., Hubbard T.J., Rogers J., Jonkers J., Wessels L., Adams D.J., van Lohuizen M., Berns A. Large-scale mutagenesis in p19(ARF)- and p53-deficient mice identifies cancer genes and their collaborative networks. Cell. 2008;133:727–741. doi: 10.1016/j.cell.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Starr T.K., Allaei R., Silverstein K.A., Staggs R.A., Sarver A.L., Bergemann T.L., Gupta M., O'Sullivan M.G., Matise I., Dupuy A.J., Collier L.S., Powers S., Oberg A.L., Asmann Y.W., Thibodeau S.N., Tessarollo L., Copeland N.G., Jenkins N.A., Cormier R.T., Largaespada D.A. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dupuy A.J., Rogers L.M., Kim J., Nannapaneni K., Starr T.K., Liu P., Largaespada D.A., Scheetz T.E., Jenkins N.A., Copeland N.G. A modified sleeping beauty transposon system that can be used to model a wide variety of human cancers in mice. Cancer Res. 2009;69:8150–8156. doi: 10.1158/0008-5472.CAN-09-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brannan C.I., Perkins A.S., Vogel K.S., Ratner N., Nordlund M.L., Reid S.W., Buchberg A.M., Jenkins N.A., Parada L.F., Copeland N.G. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- 85.Jacks T., Shih T.S., Schmitt E.M., Bronson R.T., Bernards A., Weinberg R.A. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat Genet. 1994;7:353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- 86.Zhu Y., Ghosh P., Charnay P., Burns D.K., Parada L.F. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science. 2002;296:920–922. doi: 10.1126/science.1068452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang F.C., Ingram D.A., Chen S., Zhu Y., Yuan J., Li X., Yang X., Knowles S., Horn W., Li Y., Zhang S., Yang Y., Vakili S.T., Yu M., Burns D., Robertson K., Hutchins G., Parada L.F., Clapp D.W. Nf1-dependent tumors require a microenvironment containing Nf1+/– and c-kit-dependent bone marrow. Cell. 2008;135:437–448. doi: 10.1016/j.cell.2008.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cichowski K., Shih T.S., Schmitt E., Santiago S., Reilly K., McLaughlin M.E., Bronson R.T., Jacks T. Mouse models of tumor development in neurofibromatosis type 1. Science. 1999;286:2172–2176. doi: 10.1126/science.286.5447.2172. [DOI] [PubMed] [Google Scholar]
- 89.Vogel K.S., Klesse L.J., Velasco-Miguel S., Meyers K., Rushing E.J., Parada L.F. Mouse tumor model for neurofibromatosis type 1. Science. 1999;286:2176–2179. doi: 10.1126/science.286.5447.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Joseph N.M., Mosher J.T., Buchstaller J., Snider P., McKeever P.E., Lim M., Conway S.J., Parada L.F., Zhu Y., Morrison S.J. The loss of Nf1 transiently promotes self-renewal but not tumorigenesis by neural crest stem cells. Cancer Cell. 2008;13:129–140. doi: 10.1016/j.ccr.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.King D., Yang G., Thompson M.A., Hiebert S.W. Loss of neurofibromatosis-1 and p19(ARF) cooperate to induce a multiple tumor phenotype. Oncogene. 2002;21:4978–4982. doi: 10.1038/sj.onc.1205632. [DOI] [PubMed] [Google Scholar]
- 92.Stonecypher M.S., Byer S.J., Grizzle W.E., Carroll S.L. Activation of the neuregulin-1/ErbB signaling pathway promotes the proliferation of neoplastic Schwann cells in human malignant peripheral nerve sheath tumors. Oncogene. 2005;24:5589–5605. doi: 10.1038/sj.onc.1208730. [DOI] [PubMed] [Google Scholar]
- 93.Kohli L., Kaza N., Lavalley N.J., Turner K.L., Byer S., Carroll S.L., Roth K.A. The pan erbB inhibitor PD168393 enhances lysosomal dysfunction-induced apoptotic death in malignant peripheral nerve sheath tumor cells. Neuro Oncol. 2012;14:266–277. doi: 10.1093/neuonc/nor226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Eckert J.M., Byer S.J., Clodfelder-Miller B.J., Carroll S.L. Neuregulin-1 beta and neuregulin-1 alpha differentially affect the migration and invasion of malignant peripheral nerve sheath tumor cells. Glia. 2009;57:1501–1520. doi: 10.1002/glia.20866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Huijbregts R.P., Roth K.A., Schmidt R.E., Carroll S.L. Hypertrophic neuropathies and malignant peripheral nerve sheath tumors in transgenic mice overexpressing glial growth factor beta3 in myelinating Schwann cells. J Neurosci. 2003;23:7269–7280. doi: 10.1523/JNEUROSCI.23-19-07269.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kazmi S.J., Byer S.J., Eckert J.M., Turk A.N., Huijbregts R.P., Brossier N.M., Grizzle W.E., Mikhail F.M., Roth K.A., Carroll S.L. Transgenic mice overexpressing neuregulin-1 model neurofibroma-malignant peripheral nerve sheath tumor progression and implicate specific chromosomal copy number variations in tumorigenesis. Am J Pathol. 2013;182:646–667. doi: 10.1016/j.ajpath.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brosius S.N., Turk A.N., Byer S.J., Brossier N.M., Kohli L., Whitmire A., Mikhail F.M., Roth K.A., Carroll S.L. Neuregulin-1 overexpression and Trp53 haploinsufficiency cooperatively promote de novo malignant peripheral nerve sheath tumor pathogenesis. Acta Neuropathol. 2014;127:573–591. doi: 10.1007/s00401-013-1209-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ling B.C., Wu J., Miller S.J., Monk K.R., Shamekh R., Rizvi T.A., Decourten-Myers G., Vogel K.S., DeClue J.E., Ratner N. Role for the epidermal growth factor receptor in neurofibromatosis-related peripheral nerve tumorigenesis. Cancer Cell. 2005;7:65–75. doi: 10.1016/j.ccr.2004.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Keng V.W., Watson A.L., Rahrmann E.P., Li H., Tschida B.R., Moriarity B.S., Choi K., Rizvi T.A., Collins M.H., Wallace M.R., Ratner N., Largaespada D.A. Conditional inactivation of Pten with EGFR overexpression in Schwann cells models sporadic MPNST. Sarcoma. 2012;2012:620834. doi: 10.1155/2012/620834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rahrmann E.P., Moriarity B.S., Otto G.M., Watson A.L., Choi K., Collins M.H., Wallace M., Webber B.R., Forster C.L., Rizzardi A.E., Schmechel S.C., Ratner N., Largaespada D.A. Trp53 haploinsufficiency modifies EGFR-driven peripheral nerve sheath tumorigenesis. Am J Pathol. 2014;184:2082–2098. doi: 10.1016/j.ajpath.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rahrmann E.P., Watson A.L., Keng V.W., Choi K., Moriarity B.S., Beckmann D.A., Wolf N.K., Sarver A., Collins M.H., Moertel C.L., Wallace M.R., Gel B., Serra E., Ratner N., Largaespada D.A. Forward genetic screen for malignant peripheral nerve sheath tumor formation identifies new genes and pathways driving tumorigenesis. Nat Genet. 2013;45:756–766. doi: 10.1038/ng.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kloosterman W.P., Koster J., Molenaar J.J. Prevalence and clinical implications of chromothripsis in cancer genomes. Curr Opin Oncol. 2014;26:64–72. doi: 10.1097/CCO.0000000000000038. [DOI] [PubMed] [Google Scholar]