Abstract
Newborn screening for cystic fibrosis enables early detection and management of this debilitating genetic disease. Implementing comprehensive CFTR analysis using Sanger sequencing as a component of confirmatory testing of all screen-positive newborns has remained impractical due to relatively lengthy turnaround times and high cost. Here, we describe CFseq, a highly sensitive, specific, rapid (<3 days), and cost-effective assay for comprehensive CFTR gene analysis from dried blood spots, the common newborn screening specimen. The unique design of CFseq integrates optimized dried blood spot sample processing, a novel multiplex amplification method from as little as 1 ng of genomic DNA, and multiplex next-generation sequencing of 96 samples in a single run to detect all relevant CFTR mutation types. Sequence data analysis utilizes publicly available software supplemented by an expert-curated compendium of >2000 CFTR variants. Validation studies across 190 dried blood spots demonstrated 100% sensitivity and a positive predictive value of 100% for single-nucleotide variants and insertions and deletions and complete concordance across the polymorphic poly-TG and consecutive poly-T tracts. Additionally, we accurately detected both a known exon 2,3 deletion and a previously undetected exon 22,23 deletion. CFseq is thus able to replace all existing CFTR molecular assays with a single robust, definitive assay at significant cost and time savings and could be adapted to high-throughput screening of other inherited conditions.
CME Accreditation Statement: This activity (“JMD 2016 CME Program in Molecular Diagnostics”) has been planned and implemented in accordance with the Essential Areas and policies of the Accreditation Council for Continuing Medical Education (ACCME) through the joint providership of the American Society for Clinical Pathology (ASCP) and the American Society for Investigative Pathology (ASIP). ASCP is accredited by the ACCME to provide continuing medical education for physicians.
The ASCP designates this journal-based CME activity (“JMD 2016 CME Program in Molecular Diagnostics”) for a maximum of 36 AMA PRA Category 1 Credit(s)™. Physicians should only claim credit commensurate with the extent of their participation in the activity.
CME Disclosures: The authors of this article and the planning committee members and staff have no relevant financial relationships with commercial interests to disclose.
With an overall incidence of 1 in approximately 3900 population in the United States, cystic fibrosis (CF; Online Mendelian Inheritance in Man no. 219700, http://www.ncbi.nlm.nih.gov/omim) is among the most common genetic disorders.1, 2 CF is an autosomal recessive disease caused by mutations in the CF transmembrane conductance regulator gene (CFTR). Whereas a single variant, the deletion of Phe508 (c.1521_1523delCTT, p.Phe508del), accounts for approximately 70% of CF chromosomes worldwide,3 nearly 2000 other single-nucleotide variants (SNVs), insertions and deletions (indels), and genomic copy number variations (CNVs) have been identified (Cystic Fibrosis Genetic Analysis Consortium, http://www.genet.sickkids.on.ca; CFTR2 database, http://www.cftr2.org, last accessed October 21, 2015). Moreover, these variants have diverse functional consequences and varying prevalences across ethnicities,1, 3, 4 emphasizing the need for comprehensive and definitive CFTR analysis in CF molecular testing.
Due to its high prevalence, devastating clinical sequelae, and responsiveness to early intervention, universal newborn screening (NBS) for CF has been implemented across the United States and in many countries worldwide, with substantial clinical effect. Indeed, NBS has been shown to accelerate the identification of children at risk for CF by approximately 1 year compared with symptomatic presentation.5 This early identification allows for early initiation of nutritional support, respiratory therapy, and prophylaxis against infectious complications, all of which have long-term benefits, including improved growth, reduced hospitalizations, and extended survival.5, 6
Although effective in identifying at-risk children, current CF NBS strategies vary widely.7 Most begin with immunoreactive trypsinogen testing of dried blood spots (DBSs) taken from newborn heel sticks. This assay is sensitive but has a marked lack of specificity. Indeed, the false-positive rate of immunoreactive trypsinogen testing is approximately 94%,8 generating a large number of newborns who require follow-up testing.7 To accommodate this lack of specificity, NBS programs rely on tiered screening strategies, which reflex hypertrypsinogenemic specimens to methods that interrogate a relatively small number of common mutations. These panels, however, have limited sensitivity, especially in nonwhite ethnic groups.1, 8 In California's diverse population, novel CFTR variations are being found in newborns at rates of 18 per year, in which 10% to 20% appear CF causing.8 Asymptomatic infants with one CF-causing mutation and a second mutation of variable clinical consequence and who have sweat chloride levels below the diagnostic range for CF in the first months of life are increasingly being reported to have CF as they age,9, 10 leaving NBS programs with limited CFTR testing misclassifying these infants as carriers.
These data support the need for comprehensive CFTR analysis as a component of CF NBS. Until recently, such comprehensive CFTR testing was performed exclusively through Sanger sequencing, which is costly, laborious, and time consuming and, therefore, impractical as a second-tier test for most NBS programs. The relatively recent advent of next-generation sequencing (NGS) technologies presents a solution to these limitations. Indeed, these approaches have the potential to provide clinical-grade sequence analysis of the entire CFTR gene at less cost than the currently used screening methods.11 Several studies have recently reported the development of NGS assays for CF testing,11, 12 one of which has been cleared by the US Food and Drug Administration.13 Despite this initial work, these assays remain limited in both cost-effectiveness and sample requirements. For example, current NGS assays often require relatively large quantities of high-quality genomic DNA derived from fresh peripheral blood samples, which requires an additional appointment and blood draw. Current screening programs, however, generally collect only DBSs from newborns and rely on 3.2-mm punches, which yield low quantities of DNA at variable quality,14 limiting currently used downstream NGS applications. To date, only a single NGS CF screening assay has been able to provide accurate results using a single 3.2-mm DBS punch; however, that assay is limited to approximately 170 pathogenic sequence variants, which, while an advance over currently used panel-based screening techniques, does not represent truly comprehensive CFTR testing.15 Additionally, existing NGS CF assays have reported multiplexing capabilities of 8 to 48 specimens per run,11, 12, 13, 15 which may be insufficient throughput for large screening programs that handle thousands of specimens per year and may lead to an unnecessarily high cost per specimen.
Here we describe the design, development, and validation of CFseq, a highly sensitive, specific, cost-effective, and rapid clinical NGS assay that reliably sequences all exons, flanking intronic regions, and key noncoding regions of the CFTR gene from a single 3.2-mm newborn DBS punch. This performance is achieved through optimized DBS sample processing and DNA extraction, a novel multiplex PCR (mPCR)-based target amplification strategy, and standard NGS using the MiSeq platform (Illumina, San Diego, CA) coupled with a customizable automated bioinformatics pipeline for data analysis and interpretation. Importantly, the CFseq design allows for 96 samples to be pooled and run simultaneously, reducing the per-sample reagent cost to as little as one-fifth that of traditional tiered NBS testing and the turnaround time to as little as 3 days. With these performance parameters, this single workflow may be able to completely replace existing algorithms for CF NBS and provide comprehensive analysis for all hypertrypsinogenemic infants, while reducing cost and turnaround time and providing superior molecular diagnosis.
Materials and Methods
Study Specimens
A total of 193 de-identified residual DBS specimens from California NBS were used for validating the performance of the assay. Two separate samples were selected to include a large variety of CFTR variant types: 111 unique known SNVs and indels, including both benign and pathogenic variants, and one known CNV (deletion of exons 2 and 3). The first sample was composed of 142 residual newborn DBS specimens undergoing Sanger sequencing at the Stanford Health Care Molecular Pathology Laboratory (Stanford, CA) between April 2013 and May 2014. The second sample included 51 archived specimens with known CF variants from the promoter region and all introns and exons, except introns 23 and 25 and exon 10. Residual DBS specimens were stored at 4°C or −20°C before testing. This study was exempt from written informed consent because the samples constituted nonidentifiable, residual clinical specimens used for clinical assay validation.
DNA Extraction
For each DBS, a 3.2-mm punch was obtained using a PE Wallac instrument (Perkin Elmer, Waltham, MA) and deposited into a 96-well plate. For consistency, the punch was made in the center of one blood spot on a five-spot NBS blood card. Three blank spots were punched between samples to prevent cross-contamination. DBS punch spots were washed twice with 180 μL of 10 mmol/L NaOH. Each punch spot was then suspended in 50 μL of 10 mmol/L NaOH solution and heated at 99°C for 15 minutes in an Applied Biosystems GeneAmp PCR System 9700 (Life Technologies, Grand Island, NY). The supernatant, containing eluted DNA, was mixed by pipetting and then transferred to a clean tube containing 50 μL of 20 mmol/L TrisCl pH 7.5.
Comparison of DNA Extraction Methods
We prepared simulated blood cards using standard NBS filter paper (provided by the California Department of Public Health) and 75 μL of residual whole blood in EDTA, so that multiple spots could be punched and used for parallel testing of different extraction methods. Specifically, we performed extractions using the crude sodium hydroxide–based method described under DNA Extraction, and two commercial reagents: the QIAamp DNA Micro Kit (Qiagen, Redwood City, CA), and the Generation Capture Column Kit without use of the presupplied columns (Gentra Systems, Minneapolis, MN). Extractions were performed according to the manufacturers' protocols. We tested eluates obtained directly from these methods and also after concentration, with an Amicon Ultra 30K filter (EMD Millipore, Billerica, MA). DNA yield was assessed using the Qubit HS assay (Life Technologies). We further tested the performance of the assay using one or two punch spots for extraction. We performed multiplex target capture, library preparation, and sequencing for eluates from all extraction in comparison.
Primer Design
We used a custom Perl script–integrating primer design code from Primer 316 to generate target-specific forward and reverse primers (Table 1) for 43 amplicons covering 16,513 bp of the CFTR region of interest (ROI), based on the hg19/GRCh37 human reference genome. The ROI was defined as all exons and 20 bp of flanking intronic sequence and selected portions of the 5′-untranslated region and introns 12 and 22 (legacy names, IVS11 and IVS19, respectively) known to contain pathogenic variants (Supplemental Table S1). Primer hybridization sites were selected to avoid common polymorphisms found in the National Center for Biotechnology Information's Single-Nucleotide Polymorphism Database (dbSNP) build 137, June 2012 release. Primers were designed to have similar length (mean, 23 bp; range, 21 to 27 bp), GC-content, and amplicon size (mean, 384 bp; range, 350 to 407 bp), matching the 2 × 250-bp paired-end sequencing chemistry of the MiSeq platform (Illumina). Exons larger than 350 bp were covered by overlapping amplicons. Adapter sequences (24 bp) were included at the 5′ end of each primer for postcapture amplification and sequence library construction.
Table 1.
CFseq Primers
| ID | Forward primer sequence | Reverse primer sequence | Length | Strand | Forward primer start | Forward primer end | Reverse primer start | Reverse primer end | Forward length | Reverse length |
|---|---|---|---|---|---|---|---|---|---|---|
| CFTR_promotor | 5′-TGCTTGGCTTCCTTTCGGTGGAT-3′ | 5′-CGGCAGTGTGGGTCTGATGCATT-3′ | 364 | F | 117119062 | 117119084 | 117119403 | 117119425 | 22 | 22 |
| CFTR_utr5_1 | 5′-TGGGCCGGTAATTACGCAAAGCA-3′ | 5′-CTTCCTAGACCCTCCTTCGCGTC-3′ | 400 | F | 117119232 | 117119254 | 117119609 | 117119631 | 22 | 22 |
| CFTR_utr5_2 | 5′-CGGAACTTTTCGGCTCTCTAAGGC-3′ | 5′-CGCACCTCCCTTTCCCGATTCTG-3′ | 397 | F | 117119426 | 117119449 | 117119800 | 117119822 | 23 | 22 |
| CFTR_rs139688774 | 5′-CCTAAAGAGAGGCCGCGACTGTC-3′ | 5′-ACCTACTACTCTGGGTGCCTGCC-3′ | 394 | F | 117119694 | 117119716 | 117120065 | 117120087 | 22 | 22 |
| CFTR_exon_1 | 5′-GTGGGTGGAGAAAGCCGCTAGAG-3′ | 5′-CAACCCATACACACGCCCTCCTC-3′ | 385 | F | 117119887 | 117119909 | 117120249 | 117120271 | 22 | 22 |
| CFTR_exon_2n1∗ | 5′-GTGACAGTCACATTAGTTCAGAGAT-3′ | 5′-ACTTATAATATGTTTGCTTTCTCTTCTC-3′ | 407 | F | 117144095 | 117144119 | 117144474 | 117144501 | 24 | 27 |
| CFTR_exon_3∗ | 5′-AGGACAACTAAAATATTTGCACATGC-3′ | 5′-AAATTGCCACCCGTGTTCCAGGA-3′ | 373 | F | 117149029 | 117149054 | 117149379 | 117149401 | 25 | 22 |
| CFTR_exon_4∗ | 5′-AGTCTTGTGTTGAAATTCTCAGGGT-3′ | 5′-TCCCTTACTTGTACCAGCTCACT-3′ | 389 | F | 117170876 | 117170900 | 117171242 | 117171264 | 24 | 22 |
| CFTR_exon_5∗ | 5′-TCTGCCTAGATGCTGGGAAATAA-3′ | 5′-CCCAGGAAAACTCCGCCTTTCCA-3′ | 400 | F | 117174096 | 117174118 | 117174473 | 117174495 | 22 | 22 |
| CFTR_exon_6n | 5′-TTAGTGTGCTCAGAACCACGAAG-3′ | 5′-TGACACTGAAGATCACTGTTCTATGCA-3′ | 405 | F | 117175170 | 117175192 | 117175548 | 117175574 | 22 | 26 |
| CFTR_exon_7 | 5′-GGTGGAAGTCTACCATGATAAACA-3′ | 5′-GCGTCTGGCACATAGGAGGCATT-3′ | 398 | R | 117176788 | 117176811 | 117176414 | 117176436 | 23 | 22 |
| CFTR_exon_8 | 5′-CATTAGAACTGATCTATTGACTGA-3′ | 5′-ACCATGCTCAGATCTTCCATTCCA-3′ | 400 | R | 117180444 | 117180467 | 117180068 | 117180091 | 23 | 23 |
| CFTR_exon_9 | 5′-AAGATGTAGCACAATGAGAGTAT-3′ | 5′-ACAACCATGAGCACCTGGCCATT-3′ | 368 | F | 117181977 | 117181999 | 117182322 | 117182344 | 22 | 22 |
| CFTR_exon_10 | 5′-TGGATCATGGGCCATGTGCTTTT-3′ | 5′-TCTCCAAAAATACCTTCCAGCACT-3′ | 399 | F | 117188572 | 117188594 | 117188947 | 117188970 | 22 | 23 |
| CFTR_exon_11n | 5′-TATACACTTCTGCTTAGGATGATAATTG-3′ | 5′-GGAAACATAAATATATGTAGACTAACCG-3′ | 393 | F | 117199428 | 117199455 | 117199793 | 117199820 | 27 | 27 |
| CFTR_processed_transcript_3 | 5′-GCCCCGATCACCAAATGCAAACA-3′ | 5′-ACCTGACTTCTCACTCATGGCTGT-3′ | 400 | F | 117204633 | 117204655 | 117205009 | 117205032 | 22 | 23 |
| CFTR_exon_12n | 5′-TGTGCCTTTCAAATTCAGATTGAGCA-3′ | 5′-AGGCAAACAAATACACTGACACCAAG-3′ | 407 | F | 117227727 | 117227752 | 117228108 | 117228133 | 25 | 25 |
| CFTR_intron_12∗ | 5′-ACAGAGTGTGGGGAAGAAACTGTGT-3′ | 5′-TGAAACCATAAGCAAGTAAAATCTACA-3′ | 391 | F | 117229276 | 117229300 | 117229640 | 117229666 | 24 | 26 |
| CFTR_exon_13† | 5′-TGCATGTAGTGAACTGTTTAAGGCA-3′ | 5′-AGCATGAGGCGGTGAGAAAAGGT-3′ | 391 | F | 117230316 | 117230340 | 117230684 | 117230706 | 24 | 22 |
| CFTR_exon_14_3a | 5′-ACAAAATGCTAAAATACGAGACA-3′ | 5′-GTCCAGGAGACAGGAGCATCTCC-3′ | 350 | F | 117231911 | 117231933 | 117232238 | 117232260 | 22 | 22 |
| CFTR_exon_14_3b | 5′-TCATGGGATGTGATTCTTTCGACCA-3′ | 5′-TCAGGACAGACTGCCTCCTTCGT-3′ | 381 | F | 117232152 | 117232176 | 117232510 | 117232532 | 24 | 22 |
| CFTR_exon_14_3c | 5′-ACTCAATTGCATTCTGTGGGGTGA-3′ | 5′-AGCCTTTAGAGAGAAGGCTGTCCT-3′ | 352 | R | 117232738 | 117232761 | 117232410 | 117232433 | 23 | 23 |
| CFTR_exon_15 | 5′-AAGCTGTGTTGCTCCAGTAGACA-3′ | 5′-TGTATACATCCCCAAACTATCTT-3′ | 395 | F | 117234787 | 117234809 | 117235159 | 117235181 | 22 | 22 |
| CFTR_exon_16† | 5′-TGTGGGCATGGGAGGAATAGGTG-3′ | 5′-GGAGTGCGGTGGCTACTCACAAT-3′ | 385 | F | 117242814 | 117242836 | 117243176 | 117243198 | 22 | 22 |
| CFTR_exon_17 | 5′-TCAGTAAGTAACTTTGGCTGCCA-3′ | 5′-ACCACAGGCCCTATTGATGGTGG-3′ | 359 | F | 117243536 | 117243558 | 117243872 | 117243894 | 22 | 22 |
| CFTR_exon_18n | 5′-GCTAATTCTTATTTGGGTTCTGAATGC-3′ | 5′-CAGGTTTGGGCCAGGTAAGCAGT-3′ | 407 | F | 117246609 | 117246635 | 117246993 | 117247015 | 26 | 22 |
| CFTR_exon_19n | 5′-ACACACTTTGTCCACTTTGCAATGT-3′ | 5′-AGTTTCCTTTTATATACACATGCATGT-3′ | 375 | F | 117250507 | 117250531 | 117250855 | 117250881 | 24 | 26 |
| CFTR_exon_20_2a1 | 5′-AGTTCCCATCTCTGGTAGCCAAGT-3′ | 5′-GTTGACAGGTACAAGAACCAGTTGG-3′ | 367 | F | 117251413 | 117251436 | 117251755 | 117251779 | 23 | 24 |
| CFTR_exon_20_2b1 | 5′-TCCGATTTCAAGGAAATTATTTGT-3′ | 5′-GGACGGCAGCCTTACTTTGAAAC-3′ | 362 | R | 117252038 | 117252061 | 117251700 | 117251722 | 23 | 22 |
| CFTR_exon_21 | 5′-AAGTCGTTCACAGAAGAGAGAAA-3′ | 5′-ACAGGTGAAAGAATGCTCACTGC-3′ | 391 | F | 117254606 | 117254628 | 117254974 | 117254996 | 22 | 22 |
| CFTR_exon_22_2a | 5′-AGCCCGACAAATAACCAAGTGACA-3′ | 5′-AATCTCACCCTCTGGCCAGGACT-3′ | 386 | F | 117267447 | 117267470 | 117267810 | 117267832 | 23 | 22 |
| CFTR_exon_22_2b | 5′-TGGCTTCTTTAGTTATTAACCTAGCA-3′ | 5′-CCTCAGGGGGCCAAATGACTGTC-3′ | 373 | R | 117268068 | 117268093 | 117267721 | 117267743 | 25 | 22 |
| CFTR_intron_target_27b | 5′-AGTAGTTGAATCATTCAGTGGGT-3′ | 5′-ACTTCAATGCACCTCCTCCCTGA-3′ | 353 | F | 117279805 | 117279827 | 117280135 | 117280157 | 22 | 22 |
| CFTR_exon_23 | 5′-TGAGTACAAGTATCAAATAGCAGT-3′ | 5′-TGGTCAGGATTGAAAGTGTGCAACA-3′ | 397 | R | 117282697 | 117282720 | 117282324 | 117282348 | 23 | 24 |
| CFTR_exon_24 | 5′-CCTGTTGCTCCAGGTATGTTAGGGT-3′ | 5′-ACTTGATGGTAAGTACATGGGTGT-3′ | 376 | R | 117293111 | 117293135 | 117292760 | 117292783 | 24 | 23 |
| CFTR_exon_25∗ | 5′-ATGTGTCACCATGAAGCAGGCAT-3′ | 5′-GCAGGTAGTGGGGGTAGAGGGAT-3′ | 377 | R | 117304966 | 117304988 | 117304612 | 117304634 | 22 | 22 |
| CFTR_exon_26∗ | 5′-TGCAGGAACTATCACATGTGAGA-3′ | 5′-CCCCATGGTTGAAAAGCTGATTGTGG-3′ | 355 | R | 117305675 | 117305697 | 117305343 | 117305368 | 22 | 25 |
| CFTR_exon_27_6a | 5′-TGTGCCAGTTTCTGTCCCTGCTC-3′ | 5′-AGGCAGAGGTAACTGTTCCACGA-3′ | 376 | F | 117306889 | 117306911 | 117307242 | 117307264 | 22 | 22 |
| CFTR_exon_27_6c | 5′-GAAACTCGTTAATTTGTAGTGTTG-3′ | 5′-ACCATCCTGTCCCCTGTGAAAGA-3′ | 400 | F | 117307518 | 117307541 | 117307895 | 117307917 | 23 | 22 |
| CFTR_exon_27_6b | 5′-ACCGCTGAAGTTTCCAGTTATCA-3′ | 5′-TCGTGGGACAGTCACCTCATGGA | 397 | R | 117307585 | 117307607 | 117307211 | 117307233 | 22 | 22 |
| CFTR_exon_27_6e1 | 5′-CCATGGGCACTGTGGGTAGACAC-3′ | 5′-TAGGTTCTCCCCTGTCCCAGTT-3′ | 372 | F | 117308082 | 117308104 | 117308432 | 117308453 | 22 | 21 |
| CFTR_exon_27_6d1 | 5′-ACATCTAGCCTGAAAACATACCA-3′ | 5′-TCCAGATCCTGGAAATCAGGGTT-3′ | 373 | R | 117308142 | 117308164 | 117307792 | 117307814 | 22 | 22 |
| CFTR_exon_27_6f | 5′-TGAAATATTGACTTTTTATGGCACT-3′ | 5′-TGGAGTGAGAGACTGATGAAACA-3′ | 393 | F | 117308381 | 117308405 | 117308751 | 117308773 | 24 | 22 |
F, forward; R, reverse.
These primers were also used for real-time quantitative PCR (qPCR) verification of the predicted CNVs in exon 2, 3, 4, 5, 25, and 26, and intron 12.
These primers were used in qPCR verification to target reference exons 13 and 16, which were used for normalize the amplification signal at the exons of interest.
Multiplex CFTR Target Capture
The 43 primer pairs were pooled together in a single tube for multiplex CFTR target capture. The 193 samples were tested on three runs: multiplexing 47, 95, or 51 samples per run; a no-template water control was also included on the first two runs. mPCR was performed in a Veriti 96-well thermal cycler (Applied Biosystems, Foster City, CA) using 4 μL of extracted DNA in a 20-μL final volume and the KAPA2G Fast Multiplex PCR Kit (Kapa Biosystems, Wilmington, MA) across the following thermal profile: 95°C for 3 minutes, 60°C for 3 minutes, 5 cycles at 95°C for 16 seconds and 60°C for 1 minute, 10 cycles at 95°C for 16 seconds and 72°C for 15 seconds, and 72°C for 2 minutes. After mPCR, each sample was treated with 8 units of exonuclease I (New England BioLabs, Ipswich, MA) at 37°C for 45 minutes, 80°C for 20 minutes, and 95°C for 5 minutes.
Sequence Library Construction and Sequencing
Sequencing library preparation for the MiSeq platform was performed according to the manufacturer's instructions using 4 μL of mPCR product (ie, captured DNA) per sample. PCR was set up in 20 μL–volume reactions, using common primers with sample-specific indices and Illumina's P5 and P7 adapter sequences attached at the 5′ end. Samples were barcoded with a single 6-bp index (up to 48 samples) or 8-bp dual indices (up to 96 samples) according to Illumina's index-sequencing protocol. The KAPA2G Fast Multiplex PCR Kit (Kapa Biosystems) was used for amplifying captured DNA samples with the following cycling conditions: 98°C for 50 seconds, 14 cycles at 98°C for 16 seconds and 72°C for 20 seconds, and a final extension at 72°C for 2 minutes. Four microliters of each sample was then pooled and purified using AMPure XP (Beckman Coulter, Brea, CA) using a bead/sample ratio of 0.7:1 and eluted in 30 μL (48 samples) or 60 μL (96 samples) in 10 mmol/L Tris-Cl buffer (pH 8.5). DNA concentration was measured by a 2100 Bioanalyzer instrument (Agilent Technologies, Santa Clara, CA) and sequenced on the MiSeq instrument using a 2 × 250-bp paired-end kit according to the manufacturer's protocol.
Sequencing Data Analysis
Image analysis and sample demultiplexing were performed with MiSeq Control Software version 2.4.1 and MiSeq Reporter version 2.5.1.3 (Illumina). Primer sequences were identified in reads using cross_match version 0.990329 (compiled with the “manyreads” option; http://www.phrap.org). Primers in regions where amplicons overlap were trimmed from read ends using Biopython17 in a customized Python script (available at https://github.com/eulaf/CFseq). The resulting processed fastq files were aligned to the hg19/GRCh37 human reference genome using Burrows-Wheeler Aligner–MEM version 0.7.12-r1039.18 Picard19 was used for sorting and converting files to BAM format. Customized Python scripts (available at https://github.com/eulaf/CFseq) using the pysam module were used for extracting the following quality-control (QC) metrics for each sample from the BAM file: total number of reads, percentage of reads that were properly paired and mapped to the reference genome, read depth of each amplicon, and read depths of individual base pairs within the CFTR ROI (Supplemental Table S1). At the amplicon level, coverage uniformity was calculated by obtaining the mean amplicon coverage of each sample and then calculating the percentage of amplicons in that sample that were covered by at least 20% (0.2 × mean) or 50% (0.5 × mean) of the mean amplicon coverage. At the base-pairs level, we assessed the percentage of bases within the ROI that were covered at least by 100 reads and the lowest read number per base pair of each sample. GC content of the CFTR gene was calculated for each position in the ROI using a sliding window of 21 bases. Sample coverage was extracted from pileups generated using pysam (parameters: stepper = “samtools”; max_depth = 99,999) on sample BAM files.
Variant Calling and Annotation
The clinical laboratory personnel performing the NGS assay and data analysis were blinded to the expected variants (E.F. and T.C.). Variant calling was accomplished using two publicly available tools: GATK version 3.3-0-g37228af20, 21 and FreeBayes version v0.9.21-7-g7dd41 db.22 Variant calls using GATK HaplotypeCaller were made in GVCF mode (parameters genotyping_mode = DISCOVERY, ERC = GVCF, variant_index_type = LINEAR, variant_index_parameter = 128000, maxReadsInRegionPerSample = 15,000) followed by joint genotyping using GATK GenotypeGVCFs. Customized scripts were used for computing actual read depths at variant positions and for performing variant quality filtering (QD <2.0 ‖ DP <5 ‖ FS >30 ‖ samtools mpileup read depth <20 ‖ alt depth/read depth <0.2). FreeBayes was run using default parameters, and variants were filtered using a customized script (QUAL <20 ‖ DP <20 ‖ QA/AO <20). Each variant was further annotated with the corresponding Human Genome Variation Society DNA and protein-level nomenclature and dbSNP rs number if available, using Annovar.23 Variants were also annotated with the corresponding coordinates in the hg19 reference assembly; legacy name if it existed; and whether the variant fell within the CFTR ROI (Supplemental Table S1) or was present in the literature, other public databases, and/or our clinical laboratory–curated CFTR database. This laboratory-curated CFTR database is a comprehensive machine-readable list of >2000 genomic variants that have been identified during clinical CF testing at the Stanford Health Care Molecular Pathology Laboratory, as well as variants reported in public databases, including the CF Mutation Database, the CFTR2 database, National Center for Biotechnology Information's dbSNP, and Ensembl (http://www.ensembl.org).
Variants are designated based on the following reference sequences: NM_000492.3 (cDNA; http://www.ncbi.nlm.nih.gov/nuccore/90421312) and NP_000483.3 (protein; http://www.ncbi.nlm.nih.gov/protein/90421313). Each variant in this laboratory-curated CFTR database is annotated with the source of information on the variant (public database versus internal finding), whether the variant is considered clinically reportable or benign, functional annotations and evolutionary conservation information derived from Annovar,23 and potential aliases of the variant based on empirical findings from automated variant callers. The latter is particularly relevant for the polymorphic poly-TG and poly-T tracts and small indels. For example, c.1521_1523delCTT, p.Phe508del is a deletion of the trinucleotide CTT but is identified as a deletion of TCT by the GATK variant caller. Similarly, each TG-polyT allele is associated with a specific GATK and FreeBayes alias (Supplemental Table S2). TG-polyT genotype calling was performed using a customized script to look for every possible TG-polyT combination in a sample, to count the number of reads supporting the call, and to report the allele frequency (ie, the proportion of reads supporting a given TG-polyT combination divided by the total number of reads covering the amplicon). We empirically found allele frequencies of 0.253 to 0.482 for known heterozygous samples and allele frequencies of 0.58 to 0.722 for known homozygous samples. Thus, we set the following allele-frequency thresholds for future applications: 25% to 55% to support a heterozygous call and >55% for a homozygous call; <25% was considered artefact. All customized scripts have been deposited to https://github.com/eulaf/CFseq.
CNV Analysis
Three different methods were applied to analyze CNV: ExomeDepth,24 CONTRA,25 and log2 coverage ratio plot. For each data set, we used the mean read coverage of all of the samples except the one under analysis as “reference.” In addition, samples with nonuniform read coverage were excluded from the reference sample set (specifically, samples 31 and 33 from the 51-sample set and samples 12, 48, 60, 92, and 93 from the 96-sample set; no samples were excluded from the 48-sample set). For ExomeDepth,24 the analysis was performed by calculating the reference for each sample individually (eg, for a 96-sample set, the coverage of all positions for each sample was calculated 95 times). Alternatively, the analysis can be easily parallelized by analyzing each sample on an independent central processing unit core in a computer server, thus reducing the overall computing time from roughly 15 hours to <1 hour. For the log2 coverage ratio plot method, the log2 coverage ratio for each position was calculated according to Li et al.25 The log2 ratio for each sample was smoothed with LOWESS26 and plotted for each data set, highlighting the exons of samples with significant differences in CNV from other samples in the same region.
CNV Confirmation by qPCR
We devised a method of quantitative assessment of CFseq-predicted CNVs by real-time quantitative PCR (qPCR). The primers used for targeting the predicted CNVs in exons 2, 3, 4, 5, 25, and 26 and intron 12, as well as two reference exons (13 and 16), are listed in Table 1. PCR was performed on a LightCycler 480 (Roche Diagnostics, Indianapolis, IN), using 10-μL reactions with 5 μL of 2× LightCycler 480 SYBR Green I Master mix, 1 or 3 μL of DNA template volume, and 0.1 to 0.8 μmol/L primer concentration using the following cycling parameters: 5 minutes at 95°C; 50 cycles of 10 seconds at 95°C, 10 seconds at 58°C, and 20 seconds at 72°C; single acquisition at 72°C; melting curve analysis from 60°C to 95°C, 10 acquisitions per second. We first used genomic DNA to validate the primers and to establish standard curves for each primer set. Empirical testing using DBS-derived DNA was used for optimizing primer concentrations and template amounts for each primer set. For each sample with a predicted CNV, we performed qPCR at the exon/intron predicted to be deleted or amplified, and normalized the signal to the two reference exons as follows. Crossing point values were calculated via standard curves, and CNVs were analyzed via Relative Quantification Analysis built into LC480 software release 1.5.0 SP3. Each target primer set T was paired with the two reference primer sets R to calculate the geometric mean of the resulting ratio [eg, T1/R (all) = (T1/R1 × T1/R2)1/2].
The same analysis was also performed in parallel on two control specimens (C1 and C2) from the same sample set. The T1/R (all) ratio of C1 was then used as a normalizer for the T1/R (all) ratios of C2 and the test sample. If no CNV was present, the normalized T1/R (all) ratio was expected to be approximately 1. If a heterozygous deletion was present, the normalized T1/R (all) ratio was expected to be approximately 0.5, whereas if a duplication was present, the normalized ratio was expected to be >1.5. In CNVs with normal spot experiments (CNV1 and CNV2), all normal spots were used for controlling and sampling DNA, so the expected normalized ratios should all have been close to 1.
Primer Binding-Site Assessment by Sanger Sequencing
To rule out the possibility that a predicted exon/intron deletion was due to allele dropout, we sequenced the binding regions of the forward and reverse primers in specimen S6 (intron_12) from the 48-sample set and in specimens S8 (exon_5) and S23 (exon_4) from the 51-sample set. Only specimens with single exon deletions were assessed because we reasoned that it was unlikely for two consecutive exons in the same sample to have had rare/private variants at primer-binding sites. The following primers were used for amplification and sequencing. To assess the forward NGS primer for exon_4: CFTR_exon_4-1_F (5′-AGCCTACTCTGATACTGAAAGTTGT-3′) and CFTR_exon_4-1_R (5′-GCGTTCCTCCTTGTTATCCGGGT-3′); the reverse NGS primer for exon_4: CFTR_exon_4-2_F (5′-ACCCGGATAACAAGGAGGAACGC-3′) and CFTR_exon_4-2_R (5′-AGGCTGTGTGAGTCATCTTAACAGGA-3′); the forward NGS primer for exon_5: CFTR_exon_5-1_F (5′-ACATGAAAAATTCAAGGCCAAGGCT-3′) and CFTR_exon_5-1_R (5′-TGTTCAGGTTGTTGGAAAGGAGAC-3′); the reverse NGS primer for exon_5: CFTR_exon_5-2_F (5′-GCTGTCAAGCCGTGTTCTAGATA-3′) and CFTR_exon_5-2_R (5′-AAACACATTATCTGTCCCAAGGA-3′); the forward NGS primer for intron 12: CFTR_intron_target_17bn-1_F (5′-TGGTTTTGCTGTAAGGTGCACACA-3′) and CFTR_intron_target_17bn-1_R (5′-TCATAACATTTAAATTTTTCAGGTGTGA-3′); and the reverse NGS primer for intron 12: CFTR_intron_target_17bn-2_F (5′-AAGGTTACTATCAATCACACCTGA-3′) and CFTR_intron_target_17bn-2_R (5′-TCCTGCCCTTGAAGATGTTGGGT-3′). Amplicons were purified with the ExoSAP-IT kit (Affymetrix, Santa Clara, CA). Bidirectional sequencing was performed as previously described,27 using the BigDye Terminator mix (Life Technologies) on an ABI 3730 genetic analyzer (Life Technologies). DNA sequences were analyzed using Mutation Surveyor software version 4.09 (SoftGenetics, State College, PA).
Statistical Analysis
All statistical analyses, including determinations of sensitivity, specificity, 95% CIs, and linear correlations, were performed using Prism software (GraphPad Software, La Jolla, CA) and VassarStats (http://vassarstats.net, last accessed October 21, 2015). Positive predictive value was calculated based on results from the 48-sample set. Sanger-sequencing results from the 47 NBS specimens (ie, excluding the nontemplate control) demonstrated a total of 287 variant positions (ie, true-positives) and 633,367 nonvariant positions (ie, true-negatives) within the ROI (13,482 bp).
Results
Assay Design
Current NGS diagnostics are suboptimal for NBS applications due to their inability to accommodate DBS-derived material. To overcome this limitation, we developed an optimized DBS-processing protocol by systematically evaluating several DBS-extraction procedures and assessing their performance as NGS analytes (Supplemental Table S3). The selection of the sodiam hydroxide–based method as optimal was based on its adaptability to 96-sample multiplexing, ease of performance, reagent cost, total NGS read coverage per sample, and amplicon coverage uniformity. The final protocol is described in Materials and Methods. Importantly, the CFseq workflow uses only a fraction of the DNA extracted and eluted from a single 3.2-mm DBS punch spot (4 μL of approximately 80 μL, with a DNA concentration of approximately 0.2 ng/μL, ie, approximately 0.8 ng/test) (Supplemental Table S3), reserving this valuable resource for other genetic testing.
The selection of CFTR regions to be targeted by the assay was based on the most comprehensive CF NBS algorithm currently in clinical use in the United States.8 We chose to use an mPCR strategy for amplification of the approximately 16-Kbp target genomic sequence because mPCR allows for greater sensitivity, specificity, and sequencing uniformity compared with hybrid capture methods.28 Additionally, the mPCR approach is cost-effective as it relies only on oligonucleotide primers, conventional thermal cyclers, and other equipment already available in conventional molecular diagnostic laboratories. The length of capture amplicons was restricted to approximately 400 bp to accommodate a 2 × 250-bp paired-end Illumina MiSeq sequencing chemistry, to ensure sequence quality and coverage uniformity by terminal overlap of the forward and reverse sequencing reads.29
The MiSeq platform was selected because it is a widely used benchtop NGS instrument that has already been adopted by many clinical laboratories; however, the workflow of CFseq is expected to accommodate other platforms of similar or higher capacity. We chose to optimize the procedure for multiplexing up to 96 samples with consideration of the existing requirements for volume and turnaround time in our laboratory in performing high-volume genetic screening for CF from DBS. However, we did not seek to determine the upper limit of multiplexing capabilities of the assay, which we estimate to be significantly higher based on the achieved depth of sample coverage.
Finally, NGS data analysis consists of several components, including sample demultiplexing, alignment to reference sequence, SNV and small indel variant calling, CNV analysis, and evaluation of QC metrics (Figure 1). For this assay, we chose to use primarily publicly available data-analytical tools that have been widely used and validated and are easily accessible and deployable in laboratories without dedicated bioinformaticists. We did, however, incorporate several customized scripts (see Materials and Methods), and the performance of each of these assay components.
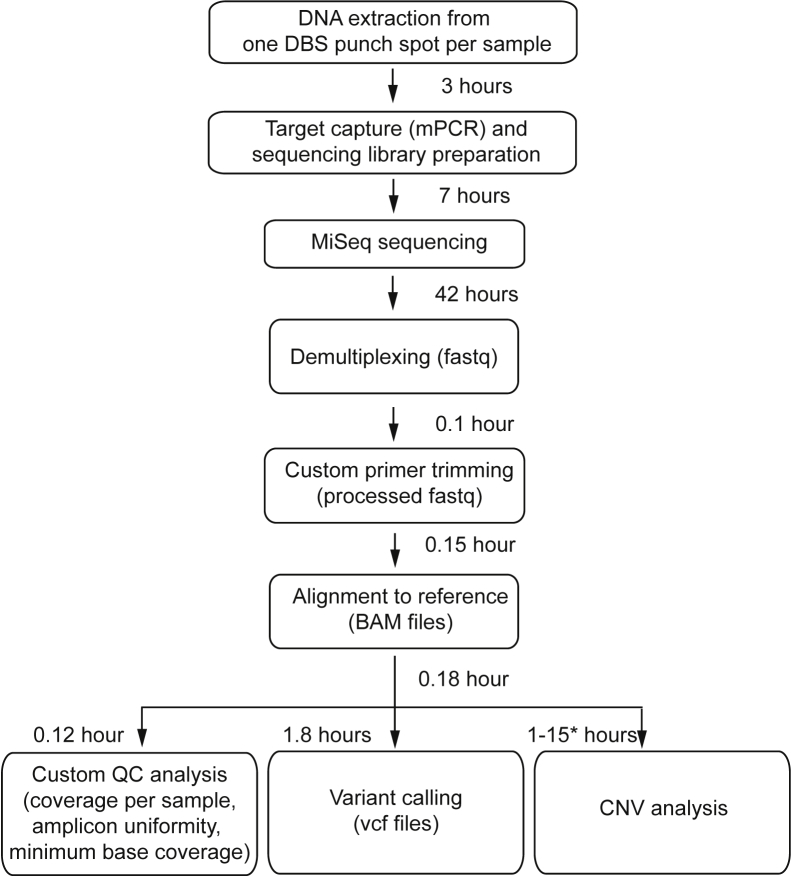
Figure 1.
Assay workflow for comprehensive CFTR sequencing using blood spots from newborns. Listed are the times for individual steps, when 96 specimens are multiplexed. Computation time for copy number variant (CNV) analysis can vary based on server cluster specifications, and therefore an estimated range is provided based on whether each sample is analyzed sequentially or in parallel (asterisk). DBS, dried blood spot; mPCR, multiplex PCR; MiSeq, MiSeq platform (Illumina, San Diego, CA); QC, quality control.
Quality Assurance and Assay Reliability
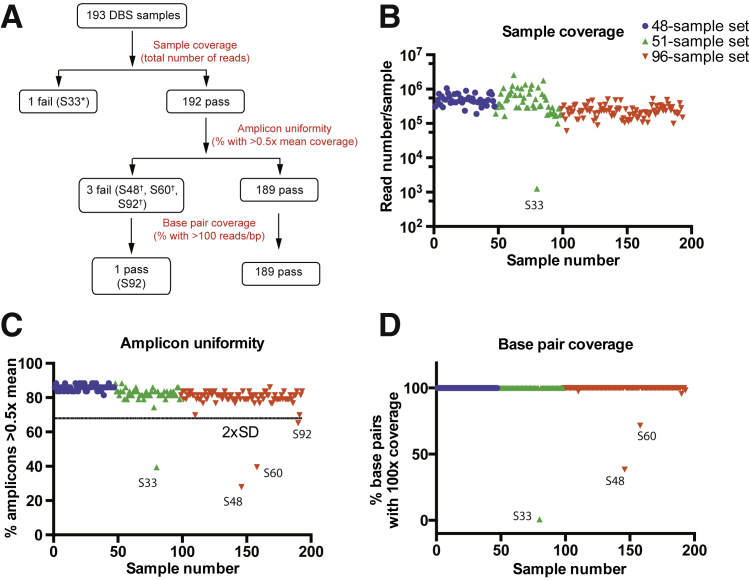
To assess assay performance, we implemented a QC algorithm whereby sequencing read coverage was examined on three different levels: individual samples, sample amplicons, and sequence bp (Figure 2A). The first QC metric, sample coverage, defined as the number of reads per sample, was used for detecting samples that completely failed multiplex amplification. In our validation experiments, this approach identified one sample (S33) with inadequate coverage (1200 total reads) (Figure 2B). The second QC metric, amplicon coverage uniformity, was used for identifying samples with partially failed amplification, such as individual amplicons that may have been insufficiently covered despite an overall normal read count for that sample. This metric is particularly important for CNV analysis since samples with nonuniform coverage cannot be analyzed accurately for CNVs. For each sample, we assessed the number of amplicons that had >0.5-fold the mean amplicon coverage and used a threshold of 2 SDs below the mean to flag samples for review. This approach identified three samples (S48, S60, and S92) with poor uniformity (Figure 2C). Lastly, we assessed base coverage for each sample, reasoning that if base coverage were sufficiently high, even samples with low amplicon uniformity could be analyzed further. As shown in Figure 2D, three samples flagged in the prior QC steps (S33, S48, and S60) also had base coverage of <100 reads/base and were excluded from further analysis and reflexed for repeated DNA extraction and sequencing; sample S92, in contrast, passed this coverage metric and yielded interpretable results in further analyses. Of the 190 samples that progressed to analysis, all target bases within the ROI were covered by at least 100 reads (Figure 2D and Table 2), providing a high degree of confidence for variant calling. Importantly, there was no correlation between GC content and bp coverage, indicating that even difficult-to-sequence areas of CFTR were appropriately covered (Supplemental Figure S1). Moreover, the QC performance metrics, including total number of reads per sample, amplicon coverage uniformity, and bp coverage, remained robust whether 48 or 96 samples were processed and tested simultaneously (Table 2).
Figure 2.
Quality-control (QC) algorithm for read coverage monitoring. A: Read coverage was examined on three different levels: individual samples, amplicons, and bp (red). The samples that failed to pass QC thresholds are indicated. B: Total reads per sample for all 193 samples that were sequenced. One sample (S33 from the 51-sample set) failed to amplify. C: Uniformity of amplicon coverage. Shown is the percentage of amplicons with read coverage >0.5-fold the mean amplicon coverage for each individual sample. Four samples did not pass the empirically established threshold of (mean – 2 × SD): S48, S60, and S92, all from the 96-sample set; in addition to S33 from the 51-sample set). D: Shown is the percentage of bp with coverage >100 reads per bp. Only three samples did not pass this threshold, indicating that the remaining 190 samples could be analyzed further. The asterisk indicates a 51-sample set; dagger, a 96-sample set. DBS, dried blood spot.
Table 2.
Coverage Metrics and Time Effort for Each Sequence Run
| Parameter | 48 samples | 96 samples | 51 samples |
|---|---|---|---|
| Mean reads passed filter/sample, n | 513,243 | 237,394 | 580,340 |
| Mean mapped reads/sample, % | 97.6 | 97.7 | 93.0 |
| Mean properly paired reads/sample, % | 89.9 | 89.6 | 81.2 |
| Amplicon coverage >0.2× mean, % | 97.6 | 97.0 | 99.6 |
| Amplicon coverage >0.5× mean, % | 85.8 | 79.3 | 81.6 |
| Lowest read coverage per base, n | 962 | 456 | 1440 |
| Target bases covered ≥100×,∗ % | 100.0 | 100.0† | 100.0† |
| Time effort, hours/run | |||
| Spot punch and DNA extraction | 2.1 | 3.3 | 2.1 |
| Target capture and library preparation | 5.8 | 7 | 5.8 |
| Sequencing time | 42 | 42 | 42 |
| Data analysis, QC and variant calling | 2.25 | 2.25 | 2.25 |
| CNV analysis‡ | 8 | 15 | 8 |
| Total time | 60.4 | 69.8 | 60.4 |
CNV, copy number variant; QC, quality control.
Target bases refers to the 13,482 bp that fall within the region of interest.
When the three samples that did not pass QC coverage metrics (S33 from the 51-sample set, S48 and S60 from the 96-sample set) are included, 98.9% (96-sample set) and 98.1% (51-sample set) of target bases are covered with >100 readsper bp.
The CNV analysis computing time is estimated based on calculating the reference for each sample individually in ExomeDepth.24 This analysis can also be parallelized by analyzing each sample on an independent central processing unit core in a computer server, thus reducing the overall computing time from roughly 15 hours to <1 hour.
Analytical Sensitivity and Specificity for SNVs and Small Indels
The 190 DBS samples that passed our QC filters contained 499 SNV and small indels that we had identified by Sanger sequencing and/or mutation panel testing (111 unique positions): 498 within the ROI and 1 well-known SNV outside the ROI (c.3272-26A>G). The 111 unique variants comprised 79 substitutions (including missense and intronic substitutions), 11 nonsense, 7 splice site, and 14 indel (<15 bp long) variants, which included 16 of the 23 mutations recommended by the American College of Medical Genetics and Genomics and American Congress of Obstetricians and Gynecologists and 37 of the 39 SNVs and small indels tested by the State of California (Supplemental Table S4). Variant calling was compared between two publicly available tools, GATK20, 21 and FreeBayes.22 FreeBayes detected all expected variants, whereas GATK failed to call 1 of the 499 variants (c.-288G>C, which is a variant of uncertain clinical significance located in the 5′-untranslated region). Thus, to increase sensitivity, our algorithm unified all variant calls from the two tools (GATK and FreeBayes). Indel realignment and base recalibration were found not to improve variant calling and in some cases reduced sensitivity (data not shown).
Relative to the applicable reference method, the analytical sensitivity of CFseq across the 499 variants was 100% (95% CI, 99.2% to 100.0%) (Table 3). There were 64 homozygous calls (allele frequency, >80%), all of which represented common benign polymorphisms that were concordant with initial results. The allele frequency of the 435 heterozygous calls ranged from 38% to 74%; therefore, the reference range of heterozygosity was defined as 35% to 80%. We also examined whether the assay detected any unexpected variants that were not previously reported for the 190 DBSs under study but that are classified as reportable in our laboratory-maintained CFTR variant database. CFseq identified only three unexpected variants, and on further analysis, all three were confirmed by Sanger sequencing: L997F (c.2991G>C, p.Leu997Phe), I1027T (c.3080T>C, p.Ile1027Thr), and c.1393-42G>A. These results demonstrate that CFseq is both highly sensitive and specific, with a positive predictive value of 100%. The positive predictive value of the assay was 100% (95% CI, 98.3% to 100%), based on the 48-sample set, for which NGS results at every ROI position could be correlated with Sanger sequencing.
Table 3.
Performance Specifications of the Assay in Clinical Specimens
| Specification | 48 samples | 96 samples | 51 samples |
|---|---|---|---|
| Analytical sensitivity (SNVs and small indels, %; n/N)∗ | 100; 287/287 | 100; 122/122 | 100; 89/89 |
| Analytical sensitivity (TG-polyT tracts, %; n) | 100; 47/47 | 100; 93/93 | 100; 29/29 |
| Sensitivity in reportable range† (%) | 100 | 100 | 100 |
| Analytical specificity (%) | 100 | 100 | 100 |
polyT, poly-thymidine; SNV, single-nucleotide variant; TG, thymidine-guanine.
Variants within the region of interest (ROI) called correctly by next-generation sequencing method/expected variants.
% bp within the 13,482-bp ROI that are covered by >100 reads per bp.
Intron 9 TG-PolyT Variant Calling
Intron 9 (legacy name, intron 8; IVS-8) of CFTR contains a polymorphic region consisting of a tract of 9 to 13 TG repeats followed by five to nine consecutive thymidines (polyT). The 5T allele, which is seen in approximately 5% of individuals,30 affects the splicing efficiency of exon 10 (legacy name, exon 9) in a context-dependent manner.3 When occurring together with 12 or 13 TGs and/or with other variants such as R117H (c.350G>A, p.Arg117His), a 5T allele can contribute to the clinical phenotypes associated with CF and CFTR-related disorders, such as congenital bilateral absence of the vas deferens.3 As such, detecting the exact TG-polyT genotype is critically important to any comprehensive CFTR testing platform; however, it poses a challenge for NGS alignment algorithms and variant callers due to its repetitive nature.11 Indeed, when default settings of the GATK variant caller were used, TG-polyT genotypes and allele frequencies were frequently miscalled. These miscalls necessitated a customized script for analysis of the region (see Materials and Methods), whereby the sequencing data from each sample were queried for all possible TG-polyT combinations (Supplemental Table S2) and allele frequencies were calculated for each possible combination. These computed allele frequencies were used for making the final TG-polyT allele calls based on empirically established thresholds, which differ from the thresholds for SNVs (25% to 55% for heterozygous, >55% for homozygous alleles). Using this approach, CFseq correctly assigned the TG-polyT genotype to all 169 samples for which the TG-polyT tract was known, including 27 disease-related genotypes (6 samples of 13TG-5T, 10 of 12TG-5T, and 11 of 11TG-5T) (Supplemental Table S5).
CNV Identification
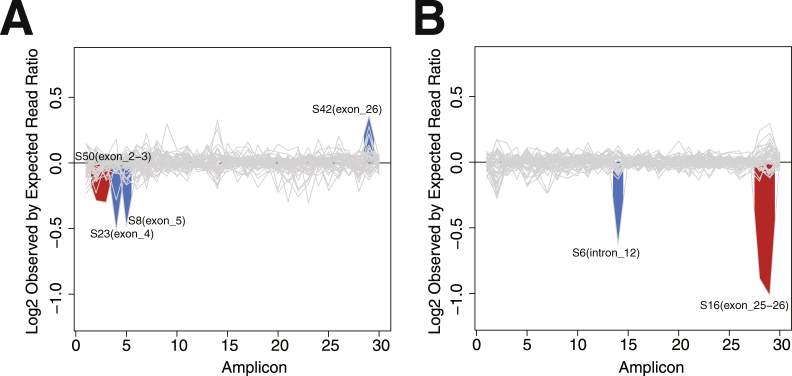
Large deletions or insertions in the CFTR gene are estimated to represent 1% to 2% of all CF-causing mutations31; however, NBS for CNVs is currently available for only one relatively common pathogenic variation (ex2,3del), using an oligonucleotide hybridization approach that targets the known breakpoints of the variant (Luminex Molecular Diagnostics, Inc., Toronto, ON, Canada). Other CNVs in CFTR are typically detected by multiplex ligation-dependent probe amplification.31, 32, 33 NGS platforms have also shown the ability to detect large exonic deletions or insertions11, 34, 35; however, to date, all of these approaches have utilized high-quality genomic DNA, and their compatibility with DBS-derived samples has not been assessed. Our sample set contained only a single known CNV, ex2,3del, which we were able to detect using a combination of publicly available tools described in Materials and Methods (Figure 3A). Interestingly, the CFseq CNV analysis also detected four other potential deletions and one potential duplication (Figure 3, A and B). Of these, the exon 25,26 deletion (legacy name: ex22,23del, S16) is of particular interest as it has been described previously.36
Figure 3.
CFTR copy number variant (CNV) detection in dried blood spot specimens. The results of three computational CNV prediction methods were integrated and plotted as the log2 ratio of observed versus expected read counts for each exon of each individual sample. Log2 ratios that significantly deviate from baseline are shaded in red if two consecutive exons show a difference in copy number, and in blue if a single exon/intron is affected. CNV results for the 51- and 48-sample sets are shown in A and B, respectively. No significant CNVs were detected in the 96-sample set.
As CNV determination is not a component of the standard NBS algorithm, these samples had not previously been assayed for CNVs, and given the inability of multiplex ligation-dependent probe amplification to assess DBS-derived material, we were unable to verify these findings with the current reference method. To increase confidence in the validity of the predicted CNVs, we analyzed each potential CNV sample with three distinct CNV detection algorithms, requiring concordance among all three methods. Four deletions, including the known ex2,3del, met this requirement (Table 4). Next, we utilized qPCR to quantitatively assess these CNV predictions and Sanger sequencing of the primer annealing regions to identify potential amplification failures in these samples. Of the three single-exon losses, qPCR confirmed one (S6, intron 12) but did not confirm another (S8, exon 5), whereas Sanger sequencing identified a rare variant (c.274-60C>T) under primer 4F as a possible cause of an amplification failure in sample S23, exon 4. qPCR confirmed the single-exon gain in sample S42, exon 26. For the two samples with losses of two consecutive exons, both exons were confirmed for sample S16 and only one for sample S50 (yes for exon 3, no for exon 2). Notably, the ex2,3del in S50 was previously detected by Luminex panel testing (see Study Specimens) and correctly confirmed by CFseq.
Table 4.
Comparison of CFTR Clinical Sequencing Assays
| Study | NGS assay | CFTR target size | Sample type (DNA amount) | DBS | Sensitivity, % | Specificity, % | Precision, % | Reagent cost | Effort | Limitations in NBS |
|---|---|---|---|---|---|---|---|---|---|---|
| Abou Tayoun et al12 | AmpliSeq custom panel with two separate PCR primer pools, Ion torrent sequencing | CDS, 10,343 bp | 79 peripheral blood samples and Coriell cell lines (20 ng) | No | 98.6 | 97 | 100 | $154 and $103 per sample when 12 and 35 samples, respectively, are pooled per run | NA | Homopolymer sequencing errors with 2184delA false-positive calls; poly-TG and polyT tract not reported |
| Trujillano et al11 | NimbleGen SeqCap EZ Choice array, Illumina HiSeq2000 | Entire CFTR genomic locus, 182 Kbp | 92 peripheral blood samples of CF patients and carriers (1.1 μg) | No | 100 | 100 | 91 | $200 per sample | ∼14 days' TAT | High DNA sample requirement, and limited multiplexing of 24 samples |
| Grosu et al13 | Illumina TruSeq Custom Amplicon, MiSeqDx Cystic Fibrosis Clinical Sequencing Assay | CDS, two deep intronic variants and two large deletions | 366 peripheral blood samples and cell lines (250 ng) | No | 99.68 | 100 | 99.7 | $220 per sample if 48 samples are pooled (∼$1320 per sample if 8 samples are pooled∗) | <3 hours; hands-on time based on Illumina Data sheet | High 250 ng DNA sample requirement; MiSeqDX special instrumentation |
| Bonini et al37 | NimbleGen SeqCap EZ kit, or long-range PCR, 454 GS Junior Sequencer | Entire CFTR genomic locus, 188 Kbp | 18 peripheral blood samples (500 ng) | No | NA | NA | High 500 ng DNA sample requirement, and high error rate in homopolymeric polypyrimidine tract base calling | |||
| Baker et al15 | Illumina TruSeq Custom Amplicon, MiSeqDx Cystic Fibrosis Clinical Sequencing Assay | CDS, two deep intronic variants and two large deletions | 232 DBS samples (1 × 3.2 mm DBS punch) | Yes | 100 | 100 | ∼220 per sample if 48 samples are pooled (∼$1320 per sample if eight samples are pooled∗) | ∼1 week TAT, <3 hours; hands-on time based on Illumina data sheet | Investigator-use-only mode limited to the sequence analysis of 162 CFTR mutations; MiSeqDx special instrumentation | |
| Loukas et al42 | Multiplicom MASTR version 2 multiplex PCR, Illumina MiSeq | CDS, selected intronic regions and promotor | 188 peripheral blood samples and 12 DBS | Yes | ∼$300 per sample when 10 samples are pooled per run | ∼1-week TAT | Limited multiplexing of up to 20 samples/run; no CNV analysis provided | |||
| Lefterova et al (this study) | Lab-developed multiplex PCR, Illumina MiSeq | CDS, two deep intronic variants, 16,513 bp | 193 DBS samples (1 × 3.2 mm punch, ∼1 ng/sample) | Yes | 100 | 100 | 100 | ∼$15 per sample when 95 samples are pooled per run | ∼7 hours; hands-on time, ∼3 days' TAT |
Characteristics of recently developed targeted NGS assays that enable CFTR sequence analysis and detection of common CNVs.
CDS, coding sequence; CF, cystic fibrosis; CNV, copy number variant; DBS, dried blood spot; NA, not available; NBS, newborn screening; NGS, next-generation sequencing; polyT, poly-thymidine; TAT, turnaround time; TG, thymidine-guanine.
Assay Reproducibility
To assess the reproducibility of CFseq, we first evaluated the consistency of amplicon coverage and found high degrees of correlation both within individual runs (Supplemental Figure S2A) and between runs (Supplemental Figure S2B and C). We also examined the reproducibility of variant calling within and between runs. For example, c.1521_1523delCTT, p.Phe508del, the most common pathogenic CFTR variant, was present in 29 samples in the 48-sample set, 56 samples in the 96-sample set, and 10 samples in the 51-sample set and was correctly identified in all samples at the expected heterozygous allele frequencies, indicating 100% reproducibility for the detection of this pathogenic variant. Similar results were obtained when comparing other variants that were present in multiple samples, such as G542X (c.1624G>T, p.Gly542X).
CFseq Cost and Time-of-Effort Assessments
Current NBS programs are effective in identifying the majority of newborns who will manifest CF symptoms; however, the labor, time, and expense of these programs are considerable due to the algorithmic approach. A great value of CFseq lies in the ability of the approach to collapse multiple workflows into a single robust pipeline while reducing cost and effort and streamlining laboratory operations. Currently, approximately 9000 newborns test positively in the immunoreactive trypsinogen screen in the California State program each year. Current algorithmic practice8 reflexes all of these samples to a 40-mutation panel (roughly 7 hours of hands-on effort per 39-sample run, typically around 2 days' turnaround time), followed by unidirectional Sanger sequencing for samples in which the panel approach detected only a single mutation (approximately 5 hours of hands-on time per 4-sample run and 12 hours' turnaround time, required for roughly 560 newborns per year in California). In contrast, CFseq can process 96 samples in approximately 7 hours of hands-on time with as little as 3 days' turnaround time (Table 2 and Figure 1). Using reagent list prices, we estimate that the reagent costs of CFseq represent approximately a fivefold cost savings over the conventional tiered NBS scheme, while providing comprehensive CFTR sequence and copy number analysis for every sample. We note that these estimates do not include labor costs or other laboratory costs, which are highly variable between laboratories and difficult to estimate. Additionally, the throughput of the CFseq workflow is much greater than those of existing assays; conventional Sanger sequencing, for example, can generally accommodate only three or four samples per run, even with the largest general-use sequencers available (27 amplicons for full CFTR coverage across 96 capillaries), whereas CFseq can easily accommodate 96 samples in each run, and potentially severalfold more with further sample multiplexing for even higher throughput.
Discussion
Current NBS strategies for CF are undeniably clinically useful; however, they have limitations due to their reliance on older molecular technology. We designed and validated CFseq, a highly sensitive, specific, rapid, and potentially cost-effective NGS assay for comprehensive clinical CFTR analysis from newborn DBSs. CFseq represents a major advance over currently used CF NBS techniques because it is the first to deliver comprehensive CFTR analysis for this sample type—including the detection of SNVs, indels, and large deletions—from a single 3.2-mm DBS punch. Moreover, its affordability, low hands-on time requirements, and ease of implementation make it generally accessible to clinical molecular laboratories equipped to perform NGS testing. In a single assay, CFseq has the potential to effectively replace all current CF genetic NBS assays (ie, mutation panels and Sanger sequencing), diagnostic assays (eg, bidirectional Sanger sequencing), and potentially assays for large deletions (eg, multiplex ligation-dependent probe amplification), although this latter functionality will require further validation.
NGS technologies have many advantages in molecular diagnostics, and other groups have already applied them to CF diagnosis, with varying degrees of success (Table 5). However, existing assays are generally impractical to implement in clinical NBS on a larger scale, such as the approximately 9000 DBS specimens processed annually in our laboratory. For example, the assays developed by Trujillano et al11 and Bonini et al,37 both of which use commercial hybrid-based target enrichment of the entire CFTR genomic locus, enable the detection of rare deep intronic variants; however, this approach markedly increases sample sequencing cost and requires DNA quantities that cannot be obtained from DBS without amplification (eg, whole genome amplification), which in turn may compromise copy-number detection.38 Moreover, the sequencing platforms used by Abou Tayoun et al12 and Bonini et al37 are error prone in homopolymer regions, which can affect base calling in the CFTR polymorphic TG-polyT tracts. Two recent studies (Grosu et al13 and Baker et al15) reported the implementation of the Cystic Fibrosis Clinical Sequencing Assay (Illumina), which has been configured by the manufacturer for a maximum of eight samples per run (Illumina technical note: MiSeqDx Cystic Fibrosis Clinical Sequencing Assay. San Diego, CA). Although both studies demonstrated multiplexing of 48 samples per run, the assay still results in higher per-sample sequencing costs, cannot easily be modified by end-users to include additional CFTR regions of interest, and can be run only on a dedicated instrument (MiSeqDx).
Table 5.
CNV Detection with CFseq
| Data set | Sample and region | CNV type | ExomeDepth24 | CONTRA25 | Coverage ratio plot | qPCR agreement | Primer region sequencing |
|---|---|---|---|---|---|---|---|
| 51 samples | S8 (exon_5) | Loss | N | Y | Y | N | N |
| S23 (exon_4)∗ | Loss | Y | Y | Y | N | c.274-60C>T† | |
| S42 (exon_26) | Gain | Y | Y | N | Y | NA | |
| S50 (exon_2-3)∗ | Loss | Y | Y | Y | Y-e3, N-e2 | NA | |
| 48 samples | S6 (intron_12)∗ | Loss | Y | Y | Y | Y | N |
| S16 (exon_25-26)∗ | Loss | Y | Y | Y | Y-e25, e26 | NA |
Analysis using three computational algorithms predicted CNVs in six samples, four of which were concordant among all three methods. qPCR quantitatively confirmed four of six CNV samples, with the exception of exon 2 in sample S50, which was a known exon 2,3 deletion sample based on Luminex panel testing. Sanger sequencing of primer annealing regions identified a rare variant under primer 4F in S23 (primer base position 18, marked with an underline below) as a possible cause of an allele-specific amplification failure. The CNV in sample 8 that was predicted by only two of the three methods was not confirmed.
CNV, copy number variant; NA, not available; qPCR, real-time quantitative PCR.
Predicted CNVs for which all three prediction algorithms agreed.
Located 8 bp from the 3′ end of primer 4F (5′-AGTCTTGTGTTGAAATTCTCAGGGT-3′).
CFseq was designed specifically to address the practical limitations of existing NGS assays for CF NBS applications and has four primary advantages. First, CFseq has very low input requirements of ≤1 ng and can accommodate DBS-derived samples without preamplification. Second, CFseq utilizes standard PCR instrumentation, thus avoiding more expensive capture technologies and specialized infrastructure that are typically required for commercial NGS assays. Third, CFseq enables testing at both high sample volumes and at very low cost, which are prerequisites for any assay to be implemented in NBS. Lastly, current NBS assays do not offer truly comprehensive CFTR analysis capable of detecting both rare sequence-based changes as well as larger deletions and/or duplications, whereas CFseq can detect all known CF-related CFTR variant types. Although the first three differentiators are largely logistical, the last may result in direct clinical care consequences. For example, whereas large deletions compose only 1% to 2% of all disease-causing CFTR mutations overall,31 large deletions can account for a much greater proportion of CF chromosomes in some populations.39, 40 The deletion of exons 2 and 3, ex2,3del, for example, accounts for up to 6% of all CF chromosomes in individuals of Eastern- and Western-Slavic descent.41 Routine CF screening tests do not detect most CNVs, and the specialized testing modalities available (eg, multiplex ligation-dependent probe amplification) are incompatible with DBS-derived samples. During its validation, CFseq, in contrast, detected multiple different deletions, including ex2,3del, ex25,26 (legacy name, ex22,23del), and three other putative single-exon or single-intron deletions and one putative single-exon gain. Using a combination of three CNV callers, normalized qPCR analysis, and Sanger sequencing to rule out allele dropout, we were able to confirm four of the six CNVs (Table 4). We found that the largest source of error in the qPCR analysis was the lack of reliable standard curves due to insufficient DNA amounts from DBS, which also limited the range of available DNA concentrations in this experiment. Other limitations of the method include an untested lower limit of size detection, laborious differentiation between allele dropout and true deletion (eg, by Sanger sequencing as in this study), and an unknown true rate of samples producing nonuniform coverage in routine clinical testing. Although these findings for CNV prediction based on CFseq are encouraging, they also demonstrate that clinical implementation will require further validation. Nonetheless, it is tempting to speculate that the widespread application of technologies such as CFseq might uncover additional CFTR structural variants and expand our understanding of the true frequency and diversity of these events.
Although CFseq provides comprehensive sequence analysis of the coding regions of CFTR and several key regulatory elements, it does not interrogate the entire intronic sequence of the gene such as the assays by Trujillano et al11 and Bonini et al.37 This design choice was made because of both the relative rarity of CF-associated variants in these regions [only two deeply intronic pathogenic variants (c.1679+1.6kbA>G and c.3717+12191C>T) have been described to occur with any appreciable frequency,4 and both are covered by CFseq], and the relatively high rate of intronic genetic variation relative to exonic. These two parameters make full intronic analysis of CF fraught with a high rate of variants of uncertain significance, which may cause parental anxiety and incur costly and unnecessary clinical follow-up.7 However, if additional novel, deeply intronic variants not currently covered by CFseq are characterized and validated as pathogenic,4 they can be easily incorporated into the existing assay through the addition of supplemental amplification primer sets.
Even within the CFTR coding sequence, many of the variants detected in comprehensive molecular assays are of unclear clinical significance. Therefore, although CF screening identifies newborns at risk for CF, it is typically followed by diagnostic testing, such as sweat chloride measurements and evaluation by CF specialist physicians, to reach a definitive clinical diagnosis. In that context, ongoing efforts to establish genotype–phenotype correlations are particularly important and can assist in the interpretation and classification of variants as pathogenic or benign.4 For example, the CFTR2 database (http://www.cftr2.org) characterizes the clinical and functional consequences of individual variants through clinical follow-up of a large number of CF patients.
Throughout the design and development of CFseq, every effort was made to facilitate the deployability of this workflow in the clinical molecular diagnostic laboratory. As with all NGS workflows, reproducibility requires duplication of both the technical assay performance and the bioinformatics interpretation. In our experience, most laboratories find the latter more difficult. As such, we used publicly available, free bioinformatics tools in our sequence data interpretation pipeline. Whereas these tools have excellent overall performance, we did note one specific weakness in this application: small indels, although reliably detected, were occasionally inaccurately named and designated in the .vcf output. For example, the c.2105-2117del13insAGAAA variant in our validation was called as two separate variants: an insertion of an A at chr7:117232191 and a deletion of TTCAATCCT at chr7:117232197. Similarly, the variants of the clinically relevant and highly polymorphic TG-polyT tract were called using nonstandard nomenclature. Despite the fact that the nomenclature for complex indels returned by these tools is occasionally inaccurate, variant detection is highly reliable and calls are reproducible (ie, any given variant will always be designated the same way). As such, we were able to implement a customized script in our analysis pipeline (see Materials and Methods) to match literature norms by using a simple alias-lookup table (Supplemental Table S2).
CFseq represents a major advance in CF diagnosis, but the technology underpinning the assay is fundamentally agnostic to the specific analytical target; that is, it may be easily adapted and applied to other NBS targets such as BTD (deficiencies in which cause biotinidase deficiency), PAH (mutations in which cause phenylketonuria), the hemoglobin genes (mutations in which cause various thalassemias and sickle cell disease), and many others (eg, the State of California currently screens for 80 genetic diseases). Moreover, given the low input requirements of CFseq, our optimized DBS sample–processing protocol yields sufficient material for many assays based on this technology platform (CFseq may be performed using as little as 0.8 ng of starting material, whereas our optimized DBS preparation may yield up to 20 ng per 3.2-mm DBS punch). Additionally, this approach is further amenable to the simultaneous interrogation of multiple targets in parallel; indeed, a comprehensive NBS panel comprising all routine genetic screening targets is well within the capabilities of this platform. It should be noted, however, that certain challenges will need to be addressed before NBS programs can broadly adopt comprehensive molecular assays. Those challenges include cost and maintenance of NGS instrumentation, and the level of technical expertise required for testing as well as for interpretation and reporting. Solutions to these challenges may involve either incorporating NGS-based testing into existing workflows, or outsourcing it to dedicated reference laboratories.
In summary, we describe the development of CFseq, the first truly comprehensive CFTR assay, and describe its advantages over existing assays in terms of analytical performance, sample requirements, turnaround time, hands-on labor, cost, and clinical yield. Moreover, this assay is specifically designed to be deployed in clinical molecular diagnostic laboratories, where it can consolidate CFTR testing, which now comprises at least two distinct sample types (DBSs for NBS and peripheral blood for testing beyond the newborn period) and at least three different technologies (mutation panel testing, Sanger sequencing, and multiplex ligation-dependent probe amplification) and their associated workflows. Importantly, this approach can accommodate DBS-derived samples and has the potential to incorporate multiple targets in addition to CFTR, for a truly comprehensive NBS platform.
Acknowledgments
We thank Dr. Juan Yang (Genetic Disease Screening Program) for assisting with the identification of DBS specimens with broad coverage across the CFTR gene, the laboratory staff at the Genetic Disease Laboratory, California Department of Public Health, for pulling archived specimens from the California Biobank Program (SIS Request 573) for development of the CFseq assay, and Kitchener Wilson, Wei Gu, Linda Gojenola, and the staff of the SHC Molecular Pathology Laboratory for technical assistance.
Footnotes
Supported by NIH grants R01HD081355 (C.S.) and P30CA016672 (W.W.) and CPRIT grant RP100030 (W.W.).
The California Department of Public Health is not responsible for the results or conclusions drawn by the authors of this publication.
M.I.L., P.S., and J.I.O. contributed equally to this work.
Disclosures: None declared.
Current address of C.S., Department of Genetics, Yale School of Medicine, New Haven, CT.
See related Commentary on page 163
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.jmoldx.2015.11.005.
Supplemental Data
Lack of correlation between GC content and next-generation sequencing read coverage for individual base pairs within the CFTR region of interest for a representative specimen. GC content was calculated in sliding 10-bp windows. The R2 for goodness-of-fit is indicated for the linear regression analysis.
Coverage reproducibility. A: Within-run reproducibility, comparing reads per amplicon between two randomly selected samples in one run (48-sample set). B and C: Between-run reproducibility, comparing reads per amplicon between two randomly selected samples in two different runs (B, 48- versus 51-sample set; C, 51- versus 96-sample set). The R2 for goodness-of-fit is indicated for each linear regression analysis.
References
- 1.Schrijver I. Mutation distribution in expanded screening for cystic fibrosis: making up the balance in a context of ethnic diversity. Clin Chem. 2011;57:799–801. doi: 10.1373/clinchem.2011.164673. [DOI] [PubMed] [Google Scholar]
- 2.Cutting G.R. Cystic fibrosis genetics: from molecular understanding to clinical application. Nat Rev Genet. 2015;16:45–56. doi: 10.1038/nrg3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zielenski J. Genotype and phenotype in cystic fibrosis. Respiration. 2000;67:117–133. doi: 10.1159/000029497. [DOI] [PubMed] [Google Scholar]
- 4.Sosnay P.R., Siklosi K.R., Van Goor F., Kaniecki K., Yu H., Sharma N., Ramalho A.S., Amaral M.D., Dorfman R., Zielenski J., Masica D.L., Karchin R., Millen L., Thomas P.J., Patrinos G.P., Corey M., Lewis M.H., Rommens J.M., Castellani C., Penland C.M., Cutting G.R. Defining the disease liability of variants in the cystic fibrosis transmembrane conductance regulator gene. Nat Genet. 2013;45:1160–1167. doi: 10.1038/ng.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grosse S.D., Boyle C.A., Botkin J.R., Comeau A.M., Kharrazi M., Rosenfeld M., Wilfond B.S., CDC Newborn screening for cystic fibrosis: evaluation of benefits and risks and recommendations for state newborn screening programs. MMWR Recomm Rep. 2004;53(RR-13):1–36. [PubMed] [Google Scholar]
- 6.Dijk F.N., Fitzgerald D.A. The impact of newborn screening and earlier intervention on the clinical course of cystic fibrosis. Paediatr Respir Rev. 2012;13:220–225. doi: 10.1016/j.prrv.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Wagener J.S., Zemanick E.T., Sontag M.K. Newborn screening for cystic fibrosis. Curr Opin Pediatr. 2012;24:329–335. doi: 10.1097/MOP.0b013e328353489a. [DOI] [PubMed] [Google Scholar]
- 8.Prach L., Koepke R., Kharrazi M., Keiles S., Salinas D.B., Reyes M.C., Pian M., Opsimos H., Otsuka K.N., Hardy K.A., Milla C.E., Zirbes J.M., Chipps B., O'Bra S., Saeed M.M., Sudhakar R., Lehto S., Nielson D., Shay G.F., Seastrand M., Jhawar S., Nickerson B., Landon C., Thompson A., Nussbaum E., Chin T., Wojtczak H., California Cystic Fibrosis Newborn Screening Consortium Novel CFTR variants identified during the first 3 years of cystic fibrosis newborn screening in California. J Mol Diagn. 2013;15:710–722. doi: 10.1016/j.jmoldx.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ren C.L., Fink A.K., Petren K., Borowitz D.S., McColley S.A., Sanders D.B., Rosenfeld M., Marshall B.C. Outcomes of infants with indeterminate diagnosis detected by cystic fibrosis newborn screening. Pediatrics. 2015;135:e1386–e1392. doi: 10.1542/peds.2014-3698. [DOI] [PubMed] [Google Scholar]
- 10.Kharrazi M., Yang J., Bishop T., Lessing S., Young S., Graham S., Pearl M., Chow H., Ho T., Currier R., Gaffney L., Feuchtbaum L. Newborn screening for cystic fibrosis in California. Pediatrics. 2015;136:1062–1072. doi: 10.1542/peds.2015-0811. [DOI] [PubMed] [Google Scholar]
- 11.Trujillano D., Ramos M.D., Gonzalez J., Tornador C., Sotillo F., Escaramis G., Ossowski S., Armengol L., Casals T., Estivill X. Next generation diagnostics of cystic fibrosis and CFTR-related disorders by targeted multiplex high-coverage resequencing of CFTR. J Med Genet. 2013;50:455–462. doi: 10.1136/jmedgenet-2013-101602. [DOI] [PubMed] [Google Scholar]
- 12.Abou Tayoun A.N., Tunkey C.D., Pugh T.J., Ross T., Shah M., Lee C.C., Harkins T.T., Wells W.A., Tafe L.J., Amos C.I., Tsongalis G.J. A comprehensive assay for CFTR mutational analysis using next-generation sequencing. Clin Chem. 2013;59:1481–1488. doi: 10.1373/clinchem.2013.206466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grosu D.S., Hague L., Chelliserry M., Kruglyak K.M., Lenta R., Klotzle B., San J., Goldstein W.M., Moturi S., Devers P., Woolworth J., Peters E., Elashoff B., Stoerker J., Wolff D.J., Friedman K.J., Highsmith W.E., Lin E., Ong F.S. Clinical investigational studies for validation of a next-generation sequencing in vitro diagnostic device for cystic fibrosis testing. Expert Rev Mol Diagn. 2014;14:605–622. doi: 10.1586/14737159.2014.916618. [DOI] [PubMed] [Google Scholar]
- 14.Saavedra-Matiz C.A., Isabelle J.T., Biski C.K., Duva S.J., Sweeney M.L., Parker A.L., Young A.J., Diantonio L.L., Krein L.M., Nichols M.J., Caggana M. Cost-effective and scalable DNA extraction method from dried blood spots. Clin Chem. 2013;59:1045–1051. doi: 10.1373/clinchem.2012.198945. [DOI] [PubMed] [Google Scholar]
- 15.Baker M.W., Atkins A.E., Cordovado S.K., Hendrix M., Earley M.C., Farrell P.M. Improving newborn screening for cystic fibrosis using next-generation sequencing technology: a technical feasibility study. Genet Med. 2015 doi: 10.1038/gim.2014.209. [Epub ahead of print] doi: 10.1038/gim.2014.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 17.Cock P.J., Antao T., Chang J.T., Chapman B.A., Cox C.J., Dalke A., Friedberg I., Hamelryck T., Kauff F., Wilczynski B., de Hoon M.J. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., McKenna A., Fennell T.J., Kernytsky A.M., Sivachenko A.Y., Cibulskis K., Gabriel S.B., Altshuler D., Daly M.J. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrison E, Marth G: Haplotype-based variant detection from short-read sequencing. arXiv preprint arXiv:1207.3907 [q-bio.GN] 2012
- 23.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Plagnol V., Curtis J., Epstein M., Mok K.Y., Stebbings E., Grigoriadou S., Wood N.W., Hambleton S., Burns S.O., Thrasher A.J., Kumararatne D., Doffinger R., Nejentsev S. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 2012;28:2747–2754. doi: 10.1093/bioinformatics/bts526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J., Lupat R., Amarasinghe K.C., Thompson E.R., Doyle M.A., Ryland G.L., Tothill R.W., Halgamuge S.K., Campbell I.G., Gorringe K.L. CONTRA: copy number analysis for targeted resequencing. Bioinformatics. 2012;28:1307–1313. doi: 10.1093/bioinformatics/bts146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cleveland W.S. LOWESS: a program for smoothing scatterplots by robust locally weighted regression. Am Stat. 1981;35 [Google Scholar]
- 27.Schrijver I., Pique L.M., Graham S., Pearl M., Cherry A., Kharrazi M. The spectrum of CFTR variants in nonwhite CF patients: implications for molecular diagnostic testing. J Mol Diagn. 2016;18:39–50. doi: 10.1016/j.jmoldx.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 28.Mamanova L., Coffey A.J., Scott C.E., Kozarewa I., Turner E.H., Kumar A., Howard E., Shendure J., Turner D.J. Target-enrichment strategies for next-generation sequencing. Nat Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 29.Shen P., Wang W., Chi A.K., Fan Y., Davis R.W., Scharfe C. Multiplex target capture with double-stranded DNA probes. Genome Med. 2013;5:50. doi: 10.1186/gm454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bombieri C., Claustres M., De Boeck K., Derichs N., Dodge J., Girodon E., Sermet I., Schwarz M., Tzetis M., Wilschanski M., Bareil C., Bilton D., Castellani C., Cuppens H., Cutting G.R., Drevinek P., Farrell P., Elborn J.S., Jarvi K., Kerem B., Kerem E., Knowles M., Macek M., Jr., Munck A., Radojkovic D., Seia M., Sheppard D.N., Southern K.W., Stuhrmann M., Tullis E., Zielenski J., Pignatti P.F., Ferec C. Recommendations for the classification of diseases as CFTR-related disorders. J Cyst Fibros. 2011;10(Suppl 2):S86–S102. doi: 10.1016/S1569-1993(11)60014-3. [DOI] [PubMed] [Google Scholar]
- 31.Svensson A.M., Chou L.S., Miller C.E., Robles J.A., Swensen J.J., Voelkerding K.V., Mao R., Lyon E. Detection of large rearrangements in the cystic fibrosis transmembrane conductance regulator gene by multiplex ligation-dependent probe amplification assay when sequencing fails to detect two disease-causing mutations. Genet Test Mol Biomarkers. 2010;14:171–174. doi: 10.1089/gtmb.2009.0099. [DOI] [PubMed] [Google Scholar]
- 32.Schouten J.P., McElgunn C.J., Waaijer R., Zwijnenburg D., Diepvens F., Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schrijver I., Rappahahn K., Pique L., Kharrazi M., Wong L.J. Multiplex ligation-dependent probe amplification identification of whole exon and single nucleotide deletions in the CFTR gene of Hispanic individuals with cystic fibrosis. J Mol Diagn. 2008;10:368–375. doi: 10.2353/jmoldx.2008.080004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng Y., Chen D., Wang G.L., Zhang V.W., Wong L.J. Improved molecular diagnosis by the detection of exonic deletions with target gene capture and deep sequencing. Genet Med. 2015;17:99–107. doi: 10.1038/gim.2014.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abel H.J., Duncavage E.J. Detection of structural DNA variation from next generation sequencing data: a review of informatic approaches. Cancer Genet. 2013;206:432–440. doi: 10.1016/j.cancergen.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hantash F.M., Milunsky A., Wang Z., Anderson B., Sun W., Anguiano A., Strom C.M. A large deletion in the CFTR gene in CBAVD. Genet Med. 2006;8:93–95. doi: 10.1097/01.gim.0000200945.54234.d7. [DOI] [PubMed] [Google Scholar]
- 37.Bonini J., Varilh J., Raynal C., Theze C., Beyne E., Audrezet M.P., Ferec C., Bienvenu T., Girodon E., Tuffery-Giraud S., Des Georges M., Claustres M., Taulan-Cadars M. Small-scale high-throughput sequencing-based identification of new therapeutic tools in cystic fibrosis. Genet Med. 2015;17:796–806. doi: 10.1038/gim.2014.194. [DOI] [PubMed] [Google Scholar]
- 38.Pugh T.J., Delaney A.D., Farnoud N., Flibotte S., Griffith M., Li H.I., Qian H., Farinha P., Gascoyne R.D., Marra M.A. Impact of whole genome amplification on analysis of copy number variants. Nucleic Acids Res. 2008;36:e80. doi: 10.1093/nar/gkn378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hantash F.M., Redman J.B., Starn K., Anderson B., Buller A., McGinniss M.J., Quan F., Peng M., Sun W., Strom C.M. Novel and recurrent rearrangements in the CFTR gene: clinical and laboratory implications for cystic fibrosis screening. Hum Genet. 2006;119:126–136. doi: 10.1007/s00439-005-0082-0. [DOI] [PubMed] [Google Scholar]
- 40.Audrezet M.P., Chen J.M., Raguenes O., Chuzhanova N., Giteau K., Le Marechal C., Quere I., Cooper D.N., Ferec C. Genomic rearrangements in the CFTR gene: extensive allelic heterogeneity and diverse mutational mechanisms. Hum Mutat. 2004;23:343–357. doi: 10.1002/humu.20009. [DOI] [PubMed] [Google Scholar]
- 41.Dork T., Macek M., Jr., Mekus F., Tummler B., Tzountzouris J., Casals T., Krebsova A., Koudova M., Sakmaryova I., Macek M., Sr., Vavrova V., Zemkova D., Ginter E., Petrova N.V., Ivaschenko T., Baranov V., Witt M., Pogorzelski A., Bal J., Zekanowsky C., Wagner K., Stuhrmann M., Bauer I., Seydewitz H.H., Neumann T., Jakubiczka S. Characterization of a novel 21-kb deletion, CFTRdele2,3(21 kb), in the CFTR gene: a cystic fibrosis mutation of Slavic origin common in Central and East Europe. Hum Genet. 2000;106:259–268. doi: 10.1007/s004390000246. [DOI] [PubMed] [Google Scholar]
- 42.Loukas Y.L., Thodi G., Molou E., Georgiou V., Dotsikas Y., Schulpis K.H. Clinical diagnostic next-generation sequencing: the case of CFTR carrier screening. Scand J Clin Lab Invest. 2015;75:374–381. doi: 10.3109/00365513.2015.1031689. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lack of correlation between GC content and next-generation sequencing read coverage for individual base pairs within the CFTR region of interest for a representative specimen. GC content was calculated in sliding 10-bp windows. The R2 for goodness-of-fit is indicated for the linear regression analysis.
Coverage reproducibility. A: Within-run reproducibility, comparing reads per amplicon between two randomly selected samples in one run (48-sample set). B and C: Between-run reproducibility, comparing reads per amplicon between two randomly selected samples in two different runs (B, 48- versus 51-sample set; C, 51- versus 96-sample set). The R2 for goodness-of-fit is indicated for each linear regression analysis.