Abstract
The transcription factor far upstream element binding protein (FBP) binds and activates the MYC promoter when far upstream element is via TFIIH helicase activity early in the transcription cycle. The fundamental biology and pathology of FBP are complex. In some tumors FBP seems pro-oncogenic, whereas in others it is a tumor suppressor. We generated an FBP knockout (Fubp1−/−) mouse to study FBP deficiency. FBP is embryo lethal from embryonic day 10.5 to birth. A spectrum of pathology is associated with FBP loss; besides cerebral hyperplasia and pulmonary hypoplasia, pale livers, hypoplastic spleen, thymus, and bone marrow, cardiac hypertrophy, placental distress, and small size were all indicative of anemia. Immunophenotyping of hematopoietic cells in wild-type versus knockout livers revealed irregular trilineage anemia, with deficits in colony formation. Despite normal numbers of hematopoietic stem cells, transplantation of Fubp1−/− hematopoietic stem cells into irradiated mice entirely failed to reconstitute hematopoiesis. In competitive transplantation assays against wild-type donor bone marrow, Fubp1−/− hematopoietic stem cells functioned only sporadically at a low level. Although cultures of wild-type mouse embryo fibroblasts set Myc levels precisely, Myc levels of mouse varied wildly between fibroblasts harvested from different Fubp1−/− embryos, suggesting that FBP contributes to Myc set point fixation. FBP helps to hold multiple physiologic processes to close tolerances, at least in part by constraining Myc expression.
The transcription factor Myc plays a decisive role in cell growth, differentiation, and senescence and so must be highly regulated. The far upstream element (FUSE) binding protein (FBP) binds single-stranded DNA of FUSE 1.7 kb upstream of the major P2 promoter of the human MYC gene.1, 2, 3 FUSE melting in response to dynamic supercoils propagated from the MYC promoters enables FBP binding. FBP activates TFIIH helicase activity to accelerate the transcription cycle from preinitiation complex assembly through promoter escape and onto pause release.4, 5, 6 Positive regulation of MYC is at least in part counteracted by FBP interacting repressor (FIR) that binds FBP and FUSE and opposes TFIIH helicase activity, retarding the transcription cycle.5, 7 Besides MYC, FBP regulates the expression of the cyclin-dependent kinase inhibitor p21, stathmin, and USP29, a powerful stabilizer of p53 induced by oxidative stress.8, 9, 10 Because the FBP/FIR system reads single-stranded DNA sequences exposed by transcriptionally generated dynamic supercoiling, it is proposed that the FBP/FIR/FUSE system serves as a molecular cruise control that monitors the mechanical output of promoters in real time while providing both positive and negative feedback.11, 12, 13
FBP (encoded by FUBP1) is the oldest member of a mammalian three gene family of single-strand nucleic acid binding proteins that includes FBP2 encoded by KHSRP and FBP3 encoded by FUBP3.14 The FBPs are reported to interact with a wide range of DNA or RNA targets and to participate in a variety of biological processes, including transcription, splicing, RNA transport, translational regulation, and mRNA degradation.15, 16, 17, 18, 19, 20, 21 The architecture of the FBPs include a central domain that comprises four regularly spaced K homology (KH) motifs that recognize sequences in single-stranded DNA or RNA with roughly equal (nanomolar) avidity.2, 3, 14, 15, 21, 22, 23 The carboxyl termini of the proteins include tyrosine-rich motifs that activate the TFIIH 3′ to 5′ helicase, promoting transcription.7, 24 FBP, FBP2, and FBP3 have three, four, and two tyrosine-rich motifs, respectively.14, 24 The amino termini of FBP and FBP2 include an α-helix that interacts with RNA recognition motif 2 domain of FIR,2, 4, 7, 16, 23 whereas an A→V substitution in FBP3 precludes this interaction.16 A variety of in vitro and tissue culture experiments have associated FBP binding with a complex set of DNA and RNA targets. The functional redundancy or compensation between the members of the FBP family is not known. FBP2 knockout (KO) mice are viable and fertile; they exhibit impaired 3′ untranslated region–directed degradation of several interferon-induced mRNAs, accounting for their resistance to vesicular stomatitis virus and herpes simplex 1.25 They also display diminished induction of some targets such as tumor necrosis factor at the transcriptional level, perhaps a manifestation of FBP-like transcription-regulatory activity.20 The biological role of FBP3 remains largely unexplored. (FUBP3 KO-first mice are in production; MGI:2443699). On the basis of their molecular relatedness, their DNA binding properties, and transcription regulatory capacity, it is suggested that the FBPs comprise a family of related factors that fine-tune gene expression.16 Alternatively, these factors may share little beyond their ability to engage single-stranded nucleic acids, and have diverged to participate in and regulate diverse processes.
Deregulation of FBP is documented in neoplastic disease. Although high levels of FBP expression are associated with tumor growth and a poor prognosis in hepatocellular, non-small cell lung carcinoma, and gliomas, loss of FBP expression occurs in a significant fraction of oligodendrogliomas where it is a tumor suppressor.3, 9, 10, 21, 26, 27, 28, 29, 30, 31, 32, 33, 34 The oncogenic and suppressor roles associated with FBP may be reconciled in light of its positive and negative transcription governor functions or, opposite roles for FBP family members across tissue types. However, the biology of FBP on an organismal scale remains largely unstudied. Here, we report the generation and first characterization of a germline Fubp1 null mutant allele in mice, which, when homozygous, is embryonic lethal and impairs embryonic hematopoiesis.
Materials and Methods
Flow Cytometry
Staining of cells for fluorescence-activated cell sorting analysis used the following conjugated antibodies obtained from BD Pharmingen (BD; Franklin Lakes, NJ) or eBioscience (eB; San Diego, CA). Cells were resuspended with Hank's Balanced Salt Solution (Ca2+, Mg2+ free; Invitrogen, Carlsbad, CA) that contained 2% fetal bovine serum and were incubated for 30 minutes on ice with one or more of the following antibodies: Mac-1phycoerythrin (PE; BD), Gr-1–fluorescein isothiocyanate (FITC; BD), CD45.2 (Ly5.2)-allophycocyanin (eB), B220-FITC (BD), CD3e-PE (eB), Ter119-FITC (BD), CD41-PE (eB), cKit-FITC (BD), Sca-1–PE (BD), CD48-PE (eB), and CD150-FITC (eB). To stain lineage-positive cells, biotinylated antibody cocktail (Stemcell Technologies, Vancouver, BC, Canada) and Streptavidin-allophycocyanin (BD) were used. After staining, cells were washed twice with phosphate-buffered saline and resuspended with HF2 that contained 1 μg/mL propidium iodide (Sigma-Aldrich, St. Louis, MO). The analyses were performed with a dual laser FACScan (BD Biosciences, Franklin Lakes, NJ).
Colony Forming Cell, Homing Assay, and Cell Proliferation Assay
Colony-forming cells were plated with 3 × 104 of fetal liver (FL) cells onto 35-mm petri dishes in Methocult M3434 methylcellulose medium (Stemcell Technologies) supplemented with cytokines (50 ng/mL recombinant mouse stem cell factor, 10 ng/mL recombinant mouse IL-3, 10 ng/mL recombinant human IL-6, and 3 U/mL recombinant human erythropoietin) and were incubated at 37°C in a 5% CO2 incubator. Number of colonies was counted at day 12 after plating the cells. Homing efficiency was evaluated by tracking donor stem/progenitor cells (CD45.2+, Sca-1+, and lin− cells) in bone marrow (BM) of recipients 18 hours after injecting 4 to 8 × 106 FL cells into the lethally irradiated B6 Ly5.2 (CD45.1) mouse. Recovery efficiency of donor cells was calculated with the method previously described by Pleett et al.35 For cell proliferation assessment, pregnant mice were euthanized, and embryonic day (E)13.5 FL cells were harvested from fetuses at 20 hours after injecting 1 mg of bromodeoxyuridine intraperitoneally. Lineage, CD48, CD150, and bromodeoxyuridine staining were performed with the manufacturer's suggested protocol and reagents (BrdU Flow Kit; BD).
Targeting the Fubp1 Allele
The Fubp1 targeting vector was generated according to well-described principles and methods.36 The targeting region of the recombination vector and its relation with the FUBP1 locus are shown in Supplemental Figure S1A. The targeting vector also included a herpes simplex thymidine kinase gene for negative counterselection. After transfection into 129 embryonic stem cells, selections for G418 resistance and against thymidine kinase activity, followed by screening with the use of Southern blots to assess homologous recombination, yielded three clones in which the targeting vector had recombined as expected. Injection of targeted embryonic stem cells into B6 blastocysts produced chimeras that, on crossing with B6 mice, showed germline transmission. Breeding of these mice revealed that the targeted Fubp1 allele behaved as a recessive embryonic lethal mutation, producing only stillborn homozygotes. Removal of the Neo cassette by crossing with B6 mice that expressed an actin-FLIP transgene did not reverse the embryonic lethality, suggesting that one or both of the remaining loxP cassette sites interfered with Fubp1 expression to prevent use as a conditional allele. Therefore, the Fubp1-floxed allele, deleted of the Neo cassette, was crossed with B6 mice that expressed an actin-Cre transgene that yielded germline deletion of exons 8 through 13, removing entirely KH motifs 2 and 3 and the amino terminal half of KH4, yielding an allele absolutely incapable of expressing a protein that binds properly with single-strand nucleic acids.2, 3, 15 This deletion was easily distinguished from the wild-type (WT) allele with the use of the primers and PCR screen in Supplemental Figure S1A and S2, as shown in Supplemental Figure S1B.
Mice
Mouse work was performed according to protocols by D.L. and P.D.A. approved by the Animal Care and Utilization Committee of the Center for Cancer Research, National Cancer Institute.
FL Transplantation and Radioprotection Assay
Radioprotection assays were performed as described.37 The B6 Ly5.2 recipient mice (Ly5.1) were purchased from Charles River Laboratories (Wilmington, MA) and maintained under microisolation conditions. Mice between 8 and 10 weeks of age were used as transplantation recipients. E13.5 FL cells (Ly5.2; 5 × 105) per mouse were injected into lethally irradiated recipient mice (1000 cGy) by tail vein injection along with 5 × 105 helper from BM nucleated cells from a nonirradiated healthy donor. Peripheral blood obtained from the tail vein of the recipients at 6, 16, and 24 weeks after transplantation and the red blood cell-lyzed peripheral blood samples were stained with anti-CD45.2 (Ly5.2) antibody for engraftment analysis. For radioprotection assay, 5 × 105 E13.5 FL cells per mouse were injected into lethally irradiated recipient mice (1000 cGy) by tail vein injection without helper BM nucleated cells, and the recipients were followed for >2 weeks after transplantation.
Immunoblotting
Mouse embryonic fibroblasts were cultured according to standard protocols. Cells were harvested in SDS lysis buffer and sonicated for clarification. Lysates were normalized by bicinchoninic acid assays and run on SDS-PAGE. Primary antibodies used for immunoblotting are listed in Table 1. Signals from the horseradish peroxidase-conjugated secondary antibodies were detected with enhanced chemiluminescence detection system and quantified with Kodak Image Station 4000 MM (Kodak, Rochester, NY).
Table 1.
Primary Antibodies Used in This Study
| Name | Source | Catalog no. |
|---|---|---|
| FBP | Custom | NA |
| Santa Cruz (Dallas, TX) | sc-11098 | |
| c-Myc | Epitomics (Burlingame, CA) | 1472-1 |
| Actin | Santa Cruz | sc-1616 |
| GAPDH | Santa Cruz | sc-47724 |
| Lamin A | Santa Cruz | sc-20680 |
| α-Tubulin | Sigma-Aldrich | T-9026 |
| pan-Histone H3 | Abcam (Cambridge, UK) | ab1791 |
| H3K9/14ac | Cell Signaling (Danvers, MA) | 9677 |
| H3K18ac | Abcam | ab1191 |
| H3K9me3 | Abcam | ab8898 |
| H3K27me3 | Millipore (Darmstadt, Germany) | 07-449 |
| H4K5/8/12/16ac | Millipore | 06-866 |
| H4K12ac | Millipore | 06-761 |
| H4K20me3 | Millipore | 07-463 |
FBP, far upstream element binding protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; NA, not applicable.
Primary Mouse Embryonic Fibroblast Cells
Pregnant female mice were sacrificed on gestation day 13.5. Embryos were surgically removed and separated from maternal tissues and yolk sac in phosphate-buffered saline (pH 7.2). Embryos were then decapitated and eviscerated (heads were used for genotyping). The bodies were minced finely in a minimal volume of phosphate-buffered saline and then incubated in 0.05% trypsin-5 HIM EDTA solution, with shaking at 37°C for 15 to 30 minutes. After settling for 2 minutes, the supernatant fluid was drawn off and centrifuged for 3 minutes at 1000 × g. The pellet was resuspended in culture medium, and cells were plated at approximately 104 cells/cm2. After 12 hours, the medium was changed to remove any nonadherent cells.
Quantitative Immunofluorescence and Data Analysis
Primary mouse embryonic fibroblasts (MEFs) from a litter of E13.5 embryos [three WT and three Fubp1−/− (KO)] were seeded in a fibronectin-coated glass-bottom 96-well plate overnight, fixed with 2% paraformaldehyde, stained with MYC antibody and Hoechst 33342, and imaged with a Nikon Eclipse Ti-inverted fluorescence microscope (Nikon, Tokyo, Japan). Nuclear Myc intensity of single MEFs was measured with NIS-Elements Advanced Research software version 4.30.02 (Nikon), and total nuclear Myc of approximately 2000 random cells of each sample were box plotted with ggplot2 package in R.38, 39
Results
FBP KO Is Embryonically Lethal
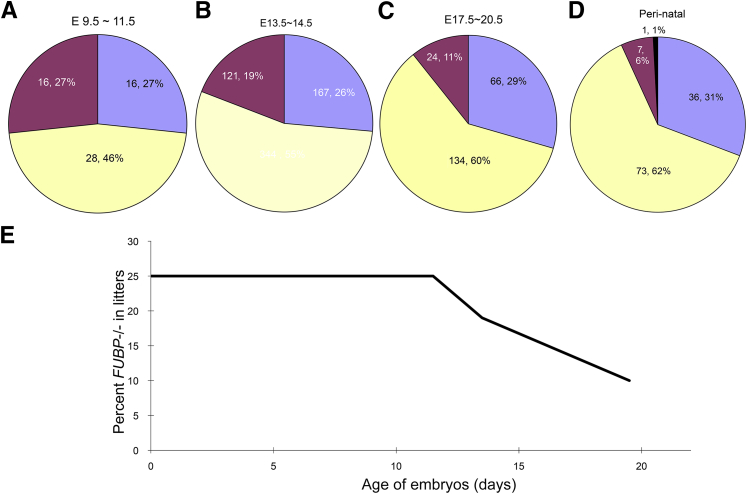
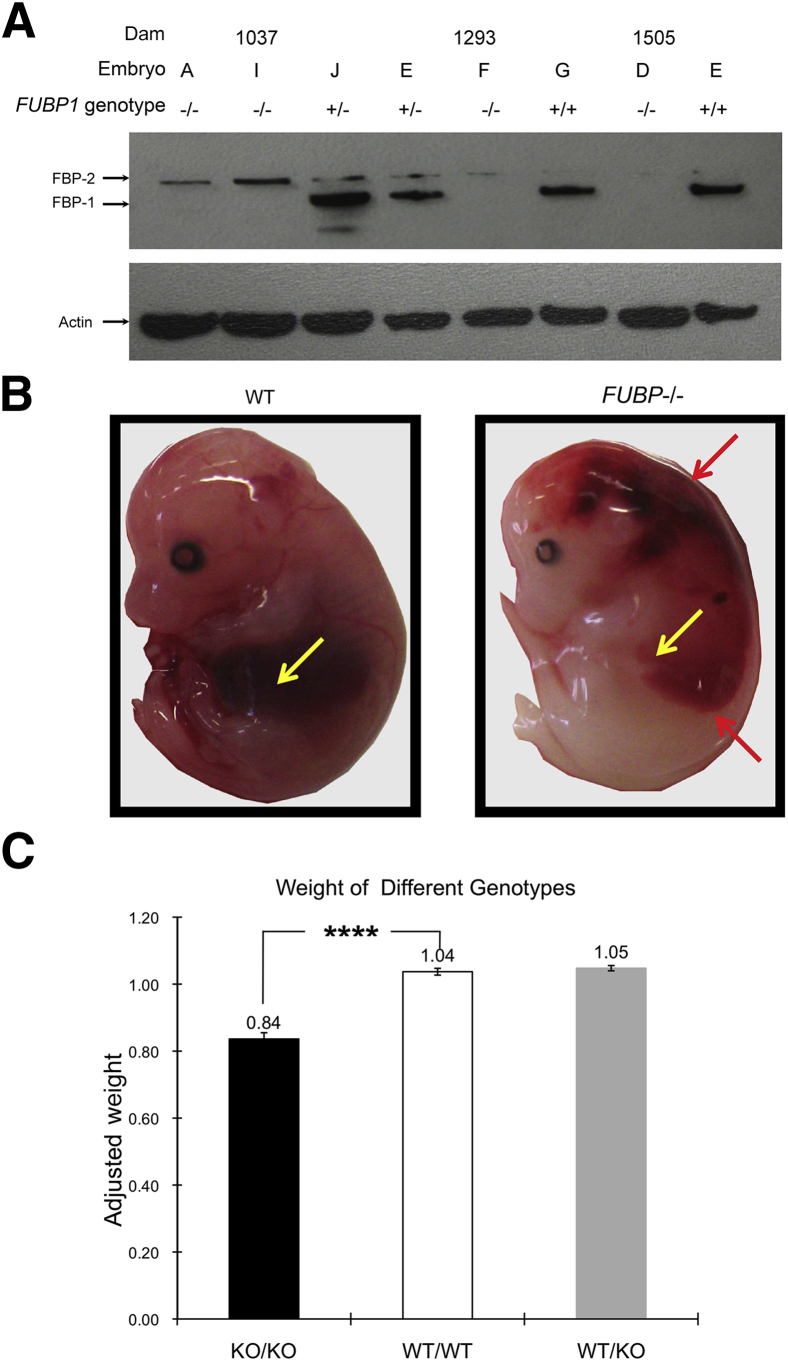
Crosses between mice heterozygous for the KO allele of FUBP delivered only heterozygous or WT live pups in the ratio of 2 to 1, consistent with prenatal lethality of Fubp1−/Fubp1− embryos. Stillborn pups were recovered at a rate of 6% (Figure 1) (a single live-born Fubp1−/− pup was pale, runt, in obvious respiratory distress, and died within 15 minutes of birth). Prenatally, Fubp1−/− embryos died steadily across a broad developmental span from approximately E10.5 until birth (Figure 1). To confirm the inability of the targeted Fubp1 allele to support FBP expression, mouse embryo fibroblasts from E13.5 WT, heterozygous, and KO embryos were prepared and analyzed by immunoblot and real-time quantitative PCR. No FBP was detected in Fubp1−/− mice with the use of several antibodies to FBP (Figure 2A), and Fubp1 RNA was diminished by at least 250-fold. No consistent compensatory or reactive change in the level of FBP2/KSRP was seen (Figure 2A).
Figure 1.
FBP knockout embryos die throughout the latter half of gestation. A–D: From E10.5 until birth there is continual attrition of Fubp1−/− embryos; relative distribution of viable KO (red), heterozygous (yellow), wild-type (blue) embryos at E9.5 to E11.5 (A), E13.5 to E14.5 (B), E17.5 to E20.5 (C), and perinatal (D) (note that with only a single exception all delivered KO embryos were stillborn, and that exception died within 15 minutes of birth). E: Graph of viability of FBP KO embryos versus developmental stage. E, embryonic day; FBP, far upstream element binding protein; KO, knockout.
Figure 2.
The absence of FBP is associated with small size and signs of defective hematopoiesis. A: FBP protein is absent in MEFs cultured from Fubp1−/− embryos. Immunoblot of FBP in MEFs cultured from single embryos with the genotype specified above the blot. B:Fubp1−/− embryos are smaller than their littermates. Data collected from Fubp1−/− (red), w/w (blue), and heterozygous (yellow) embryos. The average weight of each litter was set to 1. Only viable embryos were weighed to exclude the effects of embryonic death and resorption. C: An example of the gross anatomical pathology of Fubp1−/− embryos. Red arrows indicate hemorrhage; yellow arrows indicate the embryonic liver. n = 50 Fubp1−/− embryos (B); n = 75 wt/wt embryos (B), n = 131 heterozygous embryos (B). ∗∗∗∗P < 0.0001. FBP, far upstream element binding protein; KO, knockout; MEF, mouse embryonic fibroblast; WT, wild-type.
Morphology of Fubp1−/+ Heterozygotes
Fubp1+/− mice appeared to be grossly normal. These heterozygotes bred without difficulty and delivered normal-sized litters. No histologic deficits were appreciated. The absence of any apparent anatomic or microscopic features to reliably discriminate heterozygotes from their WT littermates prompted us to restrict our efforts to the comparison of the WT and Fubp1−/Fubp1− populations, to define rigorously the mutant phenotype without consideration of subtle or dose-dependent deficits.
Morphologic and Histologic Pathology of FBP KO Embryos
Fubp1−/− embryos appeared growth restricted and had an average gestational age-adjusted weight that was approximately 20% below their WT siblings (Figure 2B). Viable Fubp1−/− embryos after E10.5 displayed a spectrum of prenatal disorders compared with WT littermates. To assess the role of FBP throughout development, FBP WT and KO embryos were euthanized at E13.5 to E19.5. Necropsies thorough this interval revealed that, although some Fubp1−/− embryos appeared similar to their WT littermates, others were pale, runted, and hemorrhagic, suggesting defective hemostasis (Figure 2C). In contrast to WT embryos whose red-brown livers invariably filled a large portion of the abdomen, the livers of Fubp1−/− embryos were more often small and pale (Figure 2C). Because the embryonic liver is the main site of hematopoiesis in the second-half of murine gestation, it seemed that poor oxygenation secondary to anemia might contribute to embryonic demise.
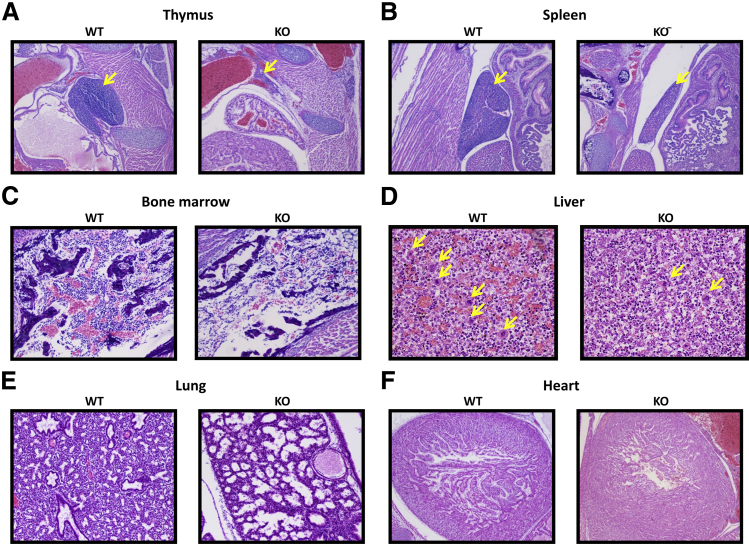
Whole-mount histologic sections of Fubp1−/− embryos showed striking underdevelopment of lymphoid tissues, often displaying a severely hypoplastic thymus (Figure 3A) and spleen (Figure 3B). Rarefaction of hepatic and BM hematopoietic precursors across all lineages (Figure 3, C and D) suggested dysfunctional hematopoiesis. Because smears of embryonic livers were technically unsatisfactory for the assessment of hematopoiesis, hepatic megakaryocytes were counted on histologic sections; Fubp1−/− mice had a significantly decreased number of hepatic megakaryocytes (15.57 ± 4.65 per 10 hpf; n = 7) compared with WT progeny (34.67 ± 5.85 per 10 hpf; n = 5) (P < 0.001) (Figure 3D).
Figure 3.
A–F: Histopathology of late-stage (E19.5) Fubp1−/− embryos compared with WT litter mates. Hematoxylin and eosin-stained sections of thymus(A), spleen (B), bone marrow (C), liver (D), lung (E), and heart (F). Arrows indicate magakaryocytes. Sections are from embryonic day 19.5 littermates. Original magnification: ×40 (A, B, and F); ×100 (C–E). KO, knockout; WT, wild-type.
The lungs of FBP KO fetuses were markedly small and hypoplastic (Figure 3E), whereas Fubp1 WT embryos presented higher-order branching of the bronchioalveolar tree, with progressive thinning of the alveolocapillary membrane and flattening of the terminal sac epithelium, and Fubp1−/− embryos presented reduced branching of air spaces and broad, thick-walled terminal sacs that were lined by predominantly bronchial-type cuboidal epithelium. Concentric hypertrophy of the cardiac ventricular wall was also present in FBP KO mice, suggesting increased vascular resistance (Figure 3F).
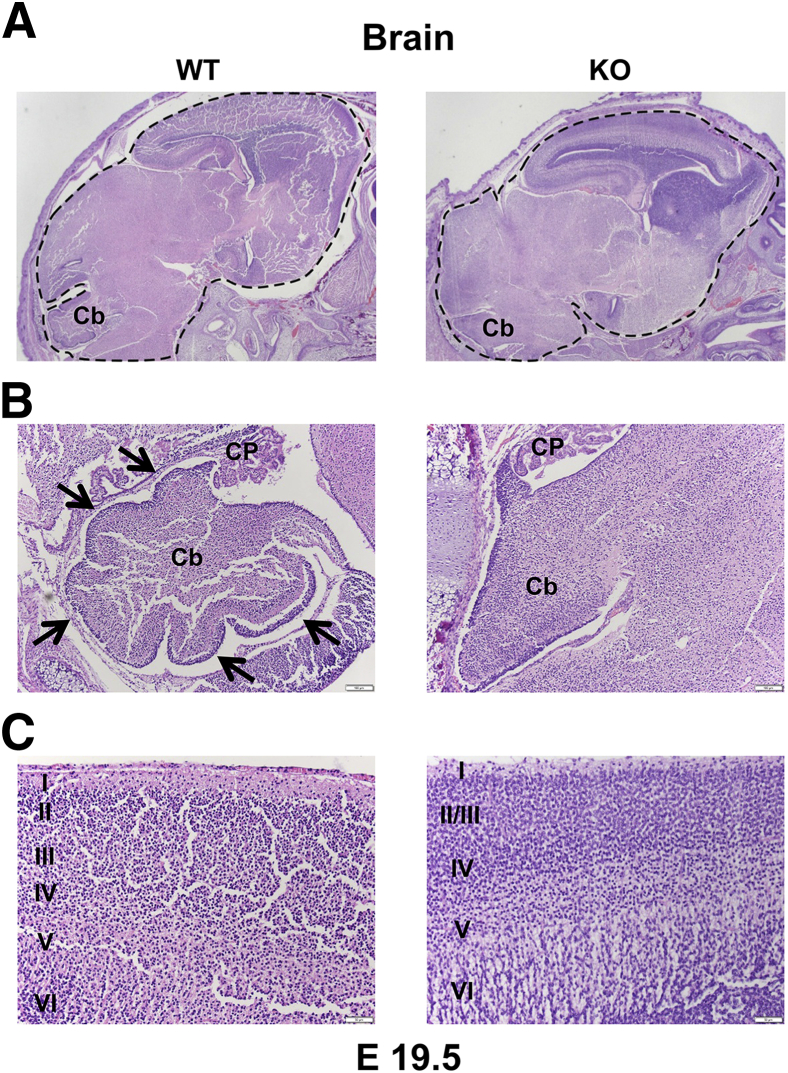
Central nervous system abnormalities in the Fubp1−/− embryos were evident in later stage embryos. Serial sagittal sections of WT and Fubp1−/− embryo brains at E19.5 showed that Fubp1−/− embryos presented notable overall brain parenchymal hypercellularity compared with their WT littermates, and none of the Fubp1 KO embryo sections showed the normal lobations of the cerebellum that were evident in WT embryos (Figure 4, A and B). At higher power, Fubp1−/− embryos showed less obvious organization of the diencephalic cortical layers (Figure 4C). Withstanding these seminal observations, further histologic characterization of the neurodevelopmental defect in Fubp1−/− embryos was deferred to a future study pending more detailed histologic sectioning and examination of additional planes of view.
Figure 4.
Brain development in Fubp1−/− mice. A and B: Sagittal sections of E19.5 Fubp1 WT and KO embryo brains (dashed outline) stained with hematoxylin and eosin show globally increased parenchymal cellularity in KO embryos compared with WT (A), and the normal Cb lobations (arrows) (B) of WT embryos present at the level of the (CP) in the IV ventricle are not seen in KO embryos. C: Decreased organization of layers I to VI of the diencephalic cortex is found with less distinct cortical layering in KO embryos compared with WT embryos. Original magnification: ×20 (A); ×100 (B); ×200 (C). Cb, cerebellar; CP, choroid plexus; E, embryonic day; KO, knockout; WT, wild-type.
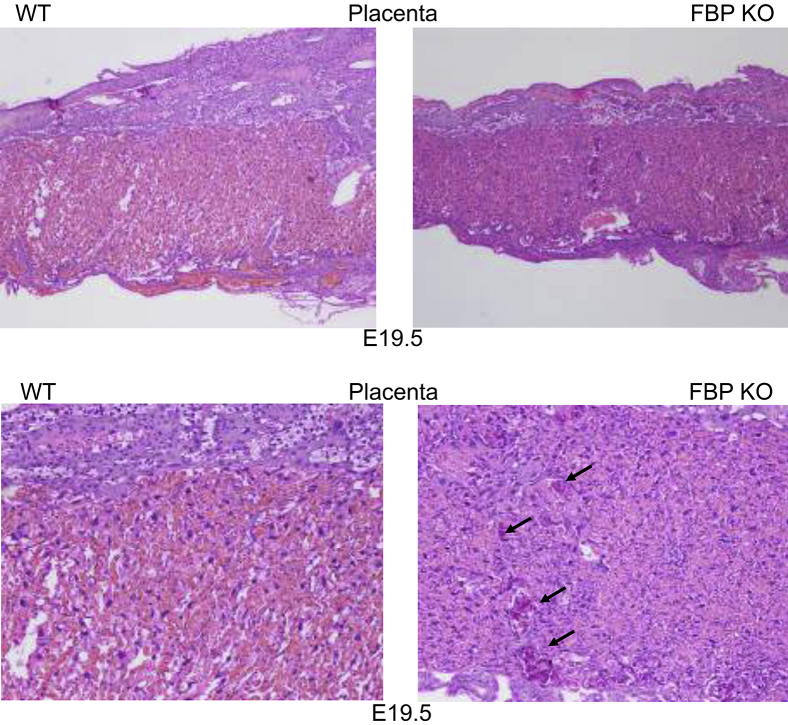
Because placental insufficiency would compound poor hematopoiesis and augment embryonic lethality, the placentas of KO and WT embryos were histologically examined to assess maternal-fetal perfusion. The placentas of Fubp1−/− embryos were thinner and poorly developed (Figure 5) and showed a prominence of labyrinth trophoblasts, which arranged into poorly formed maternal vascular spaces; however, the thin-walled capillary bed of fetal circulation was ill defined. Although the giant cell and spongiotrophoblastic layers appeared intact, a striking number of microcalcifications were found along these layers, suggesting placental stress (Figure 5).
Figure 5.
The placentas of Fubp1−/− embryos are stressed. Sections show placentas of WT or Fubp1−/− E19.5 littermates stained with hematoxylin and eosin (top). Collapse of the labyrinth reduces the thickness of the knockout placenta, and is accompanied (bottom) by calcifications (arrows) in the KO but not the WT placenta. Original magnification: ×40 (top); ×100 (bottom). E, embryonic day; FBP, far upstream element binding protein; KO, knockout; WT, wild-type.
It must be underscored that the morphologic variation among the KO embryos was surprisingly broad, ranging from subtle to fulminant pathology. Nevertheless, the apparent bleeding diathesis, diminution of lymphoid tissues, cardiac hypertrophy, and placental dysmorphism (the placental being a main hematopoietic organ in early mouse development) suggested that poor blood formation might be a central pathogenetic mechanism underlying these morphologic observations, contributing to fetal demise.
FBP Is Essential for Proper Hematopoiesis
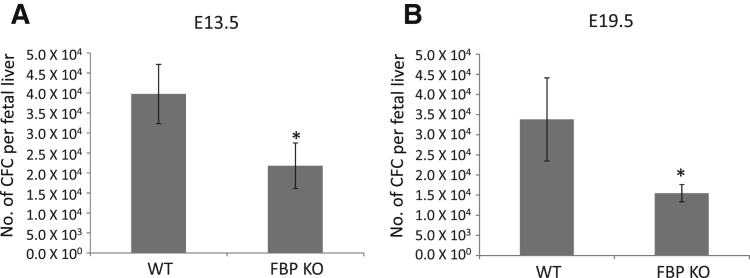
To compare hematopoiesis in KO embryos with WT embryos, livers were harvested at E13.5 or E19.5, and the various lineages were examined for surface markers with the use of flow cytometry and for colony formation in methylcellulose to assess the number and commitment of colony-forming units. At one or both stages, disorders were noted of erythroid, granulocyte/macrophage, and lymphoid lineages by immunophenotyping or colony-forming assays (Table 2 and Figure 6). Red cell anemia and B-cell lymphopenia were significant at both E13.5 and E19.5. When KO samples were compared with WT samples, decrements in all other lineages at E13.5 approached statistical significance in accord with the former's small, pale livers. Among individual KO embryos no consistent pattern of lineage deficiency was noted. (Because of ongoing embryonic attrition, it seemed likely that KO embryos surviving to E19.5 were likely to be less severely affected than those succumbing at earlier stages.) Combined with the histologic deficit of hepatic megarkaryocytes, the overall pattern suggested a complicated, variable, multilineage hematopoietic deficiency that paralleled the variation in size, morphology, and histology of the liver, spleen, and thymus in KO embryos relative to WT embryos. This trilineage deficit suggested a problem with hematopoietic stem cells (HSCs).
Table 2.
Immunophenotype of Fetal Liver Cells
| Fetal liver | Mac-1 (×104) | Gr-1 (×104) | M/G (×104) | CD3 (×104) | B220 (×104) | CD71 (×104) | Ter119 (×104) | Ter/CD71 (×104) | CD41 (×104) | No. cell/liver (×104) |
|---|---|---|---|---|---|---|---|---|---|---|
| E13.5 | ||||||||||
| WT (n = 7) | 20.6 ± 0.6 | 6.22 ± 0.22 | 22.4 ±2.4 | 8.8 ± 0.84 | 11.7 ± 1.7 | 175 ± 757 | 11.5 ± 1.5 | 1080 ± 080 | 17.9 ± 7.9 | 1300 ± 270 |
| KO (n = 7) | 13.6 ± 3.6 | 2.89 ± 0.89 | 17.5 ± 7.5 | 3.53∗ ± 0.53∗ | 5.75∗ ± 0.75∗ | 102 ± 025∗ | 3.32∗ ± 0.32∗ | 612 ± 122 | 8.30 ± 0.30∗ | 739 ± 281 |
| E19.5 | ||||||||||
| WT (n = 6) | 30.4 ± 0.4 | 4.11 ± 0.11 | 82.8 ± 2.8 | 16.2 ± 6.2 | 37.6 ± 7.6 | 129 ± 296 | 121 ± 216 | 1170 ± 170 | 47.4 ± 7.4 | 1520 ± 421 |
| KO (n = 14) | 27.6 ± 7.64 | 5.08 ± 0.084 | 82.0 ± 2.04 | 14.9 ± 4.94 | 17.8∗ ± 7.8∗ | 125 ± 258∗ | 60.9∗∗ ± 0.9∗ | 1100 ± 100 | 35.3 ± 5.3∗ | 1370 ± 162 |
Data are expressed as means ± SEM of cell number per fetal liver. Ter/CD71 represents double positive of Ter119 and CD71 antigen on the cell surface.
E, embryonic day; KO, knockout; M/G, macrophage/granulocyte; WT, wild-type.
P < 0.05.
P < 0.01 versus WT of the same immunophenotype.
Figure 6.
Fubp1−/− embryos have reduced numbers of hematopoietic progenitor development. A and B: Colony-forming assays were performed as described in Materials and Methods at E13.5 (A) and E19.5 (B). ∗P < 0.05. CFC, colony-forming cell; E, embryonic day; FBP, far upstream element binding protein; KO, knockout; WT, wild-type.
FBP Is Essential for HSC Function
To assess HSC numbers, lineage-negative embryonic liver cells were examined for signaling lymphocyte activation markers (SLAMs) CD48 and CD150.40 Because HSCs are CD48−/CD150+, these are among the most useful markers to quantify HSCs among other non-renewing multipotent hematopoietic progenitors. Although the relative proportion of the lin− CD48−/CD150+ cells in the embryonic livers was increased twofold at E13.5 and fivefold at E19.5 in KO relative to WT lin− cells, because the total yield of liver cells from the Fubp1−/− embryos was decreased, the HSC population was in fact only slightly, but not significantly, elevated (54,000 versus 43,000 at E13.5; 1900 versus 1400 at E19.5) in KO versus WT livers (Table 3). That equivalent (or even increased) numbers of HSCs were unable to support appropriate levels of hematopoiesis suggested a deficiency of HSC function and not of HSC production.
Table 3.
Hematopoietic Stem/Progenitor Cells in Fetal Liver
| Fetal liver | LNCD48+CD150− (×104) | LNCD48+CD150+ (×104) | LNCD48−CD150+ (×104) |
|---|---|---|---|
| E13.5 | |||
| WT (n = 7) | 18.7 ± 4.62 | 5.13 ± 1.30 | 4.31 ± 1.08 |
| KO (n = 7) | 11.6 ± 5.56 | 3.34 ± 0.97 | 5.44 ± 1.58 |
| E19.5 | |||
| WT (n = 2) | 64.1 | 0.50 | 0.14 |
| KO (n = 4) | 53.0 ± 5.90 | 0.41 ± 0.12 | 0.19 ± 0.055 |
Data are expressed as means ± SEM of total cell number per fetal liver.
E, embryonic day; KO, knockout; WT, wild-type.
To directly assess the function of hematopoietic progenitors, a radioprotectant assay was performed. These well-established assays found that 900 to 1000 rad of whole body irradiation is universally lethal to mice, due to BM failure, unless the mice are rescued by co-injection of HSCs shortly after irradiation.41, 42 Embryonic livers were harvested at E13.5 and transplanted into lethally irradiated mice. In contrast to WT mice whose livers were fully competent to protect the blood system, mice receiving cells from Fubp1−/− embryos invariably died within 2 weeks, suggesting that the FBP KO cells had insufficient hematopoietic progenitors to protect or prolong survival of the irradiated host for even a short period (Table 4). To assess longer-term HSC function and to unmask any lineage-specific deficits that might become manifest only after a prolonged interval, competitive transplantation assays were performed.
Table 4.
Radioprotectant Assay Summary
| Donor | No. of recipients | No. of survived recipients | Survival, % |
|---|---|---|---|
| WT | 8 | 6 | 75 |
| KO | 8 | 0 | 0∗∗ |
Survival mice counted on day 16 after transplantation. Fetal liver cells (5 × 105) were injected into lethally irradiated recipients.
∗∗P < 0.01.
KO, knockout; WT, wild-type.
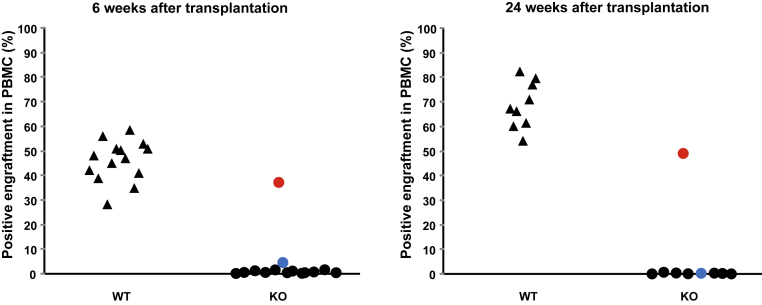
Embryonic liver cells from pooled WT or KO mice that carried the Ly5.2 allele of the Ptprc gene (which encodes the Ly5.2 antigen present on all of the nucleated hematopoietic cells) were mixed with BM from Ly5.1+ WT donors and transplanted into irradiated hosts. Although Ly5.2 WT FL cells efficiently competed with the Ly5.1 BM to repopulate the hematopoietic system, FL cells from the KO mice did not contribute to hematopoietic reconstitution in 16 of the 18 recipients of FL KO cells. In these cases, the recipient mice survived the lethal dose of irradiation due to hematopoietic reconstitution supplied by the Ly5.1 BM HSCs (Figure 7). However, 2 of 18 recipients of the FBP KO FL cells showed some evidence of HSC engraftment (Figure 7). In the first of these, engraftment increased progressively from weeks 6 through 24 weeks, stabilizing at approximately 50%. In the second mouse, low-level engraftment (4.6% at 6 weeks) dwindled further to 2.6% at 16 weeks (data not shown), becoming negative by 24 weeks, presumably reflecting extinction of short-term HSCs and/or multipotent hematopoietic progenitors and the absence of long-term HSC engraftment in this animal. (It must be stressed that these two animals were engrafted with the same pool of embryonic liver cells that completely failed to engraft in all other recipients, and therefore must represent a stochastic/idiosynchratic event.) Therefore, although present in at least normal numbers as assessed with SLAM, the FBP KO HSCs were functionally defective.
Figure 7.
Stochastic function of Fubp1−/− embryonic liver-derived HSCs in competitive transplantation assays versus WT bone marrow. Ly5.1 Fubp1−/− embryonic liver cells pooled from multiple Fubp1−/− (dots) or WT (triangles) embryos along with WT Ly5.2 bone marrow were co-transplanted into lethally irradiated mice, and the peripheral blood was monitored at intervals to assess short-term (6 weeks) and long-term (24 weeks) HSC function. In most recipients, Fubp1−/− Ly5.1-HSCs failed entirely to contribute to hematopoiesis. In one animal (blue) low levels of Ly5.1+ cells were detected early (6 and 12 weeks; not shown) but were lost by 24 weeks. In a single animal (red) Fubp1−/− Ly5.1 engraftment contributed to all lineages at all time points. (Because cells from several embryonic livers were pooled, the pattern supports sporadic engraftment by Fubp1−/− HSCs; normal HSC function of a single embryonic liver donor should have reconstituted all irradiated recipients.) HSC, hematopoietic stem cell; KO, knockout; PBMC, peripheral blood mononuclear cell; WT, wild-type.
Two aspects of normal HSC function were tested to assess the nature of the FBP KO HSC deficit. First, the capacity for WT and KO HSCs to home to the BM was compared. Ly5.2 embryonic liver cells were injected into lethally irradiated Ly5.1 recipients, and after 24 hours animals were sacrificed, and their BM was recovered and evaluated for Ly5.2+ Lin−/Sca-1+ cells. A tendency for KO cells to home less effectively than the WT cells approached but did not achieve significance at the 0.95% CI (Table 5). Even if the KO HSCs have a quantitative deficiency in homing, the magnitude would appear to be insufficient to account for the global failure of FBP KO HSCs to engraft. Second, because proper HSC function involves a choreographed program of cells dividing both symmetrically and asymmetrically to maintain stem cell pools, FBP KO HSCs were assessed for an abnormality in proliferation. Embryos were pulse-labeled with bromouridine in utero, then HSCs from WT and KO embryos were recovered and assessed for bromouridine incorporation as a measure of ongoing proliferation. KO HSCs incorporated significantly more bromouridine than their WT siblings (Table 6). Together with the other hematopoietic and HSC deficits, the image suggests that HSCs lacking FBP aberrantly partition between renewal and commitment. Although disturbances in the expression of Myc and Mycn alter the balance between HSC self-renewal and differentiation,43, 44 the scarcity of HSCs and their relatively low level of Myc expression (in comparison with high Myc expression states or tumors) thwarted several attempts to assess the levels and variation in Myc between individual WT and Fubp1−/− HSCs. Therefore, the influence of FBP on Myc expression was assessed in other embryonic cells.
Table 5.
Homing Assay Summary
| Donor FL | No. of input LS in recipients | No. of output LS in recipients | Homing (%) |
|---|---|---|---|
| WT (n = 5) | 2.04 × 104 ± 5.76 × 103 | 4.24 × 103 ± 1.17 × 103 | 26.8 ± 9.92 |
| KO (n = 6) | 1.93 × 104 ± 4.52 × 103 | 2.22 × 103 ± 8.21 × 102 | 10.9 ± 2.70 |
Data are expressed as means ± SEM.
FL, fetal liver cell; KO, knockout; LS, lineage− and Sca-1+ cells; WT, wild-type.
Table 6.
FBP Cell Proliferation in Lineage− CD48−CD150+ Cell
| BrdU− cells (%) |
BrdU+ cells (%) |
||
|---|---|---|---|
| WT (n = 6) | KO (n = 6) | WT (n = 6) | KO (n = 6) |
| 56.8 ± 2.3 | 48.3∗ ± 3.1 | 43.2 ± 2.3 | 51.7∗ ± 3.1 |
Data are expressed as means ± SEM.
BrdU, bromodeoxyuridine; FBP, far upstream element binding protein; KO, knockout; WT, wild-type.
P < 0.05 versus WT.
Lacking FBP, Myc Is Dysregulated
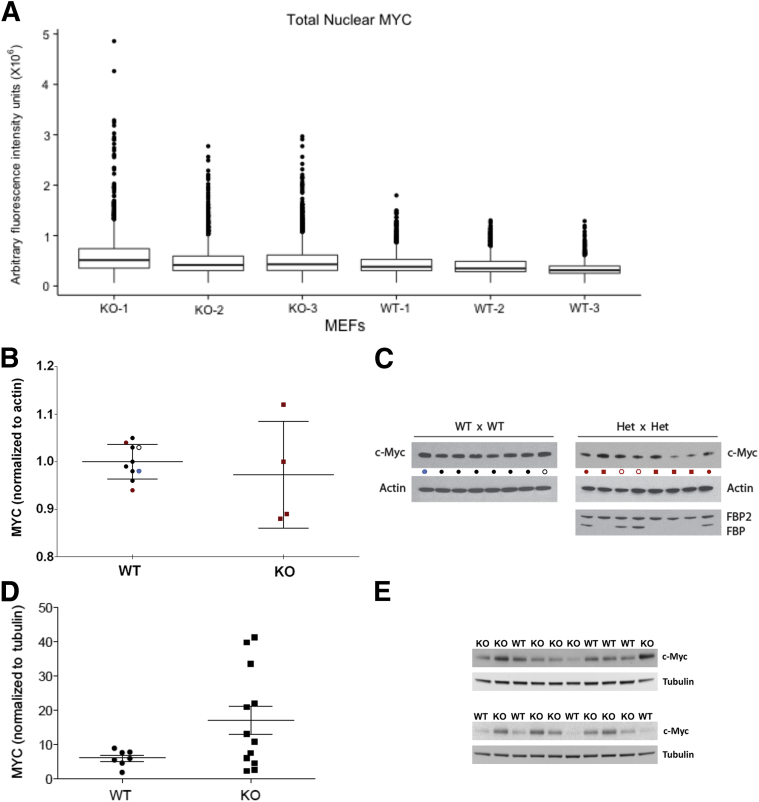
Mutations in the p89/xeroderma pigmentosum type B (XPB)/ERCC3 helicase subunit of TFIIH increase cell-to-cell variation of MYC.6 Similarly, the FBP/FIR/FUSE system acts through this subunit and is proposed to act as a molecular cruise control to dampen fluctuations in MYC expression.4, 5, 7, 10 If this proposal has merit, then Myc levels in Fubp1−/− cells should vary more than in WT cells. The relatively uniform expression of Myc in MEFs made it a good substrate to assess the variation elicited on loss of FBP. Myc levels were assessed in Fubp1 KO or WT MEFs at the single-cell level by quantitative immunofluorescent microscopy. WT MEFs growing at steady state express relatively uniform levels of Myc, whereas KO MEFs express a broader and more extreme range of Myc levels (Figure 8A). Comparison of the distributions across cultures of MEFs derived from several individual embryos showed that FBP indeed helps to stabilize Myc levels across a population. (The influence of FBP on the Myc distribution in hematopoietic cells was not studied because of the intrinsic heterogeneity and normal temporal variations in MYC levels during the development and differentiation of this tissue.)
Figure 8.
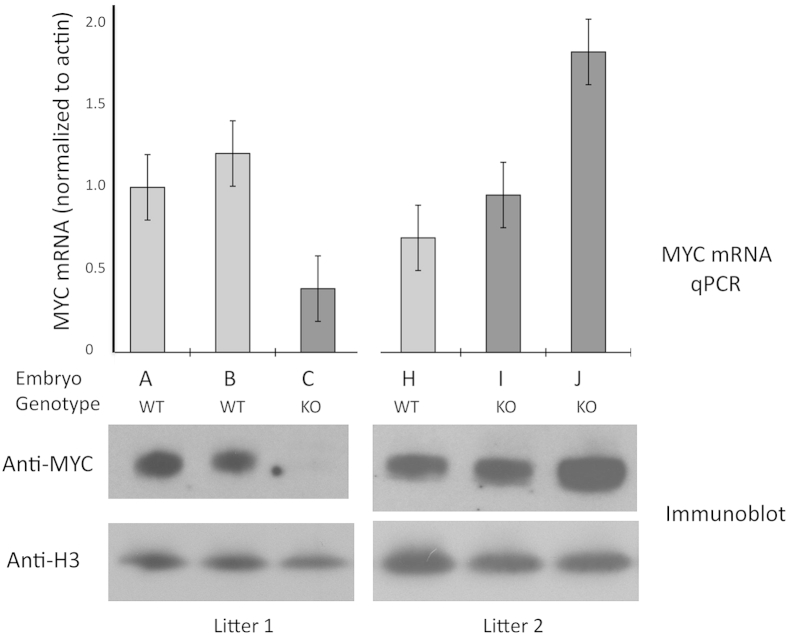
FBP helps to set and stabilize MYC levels. A: Loss of FBP is associated with increased cell-to-cell variation in MYC levels. MEFs were recovered at E13.5 from embryos with the indicated genotypes, cultured, stained for MYC, and quantified as described in Materials and Methods. B and C: Loss of FBP is associated with increased variability in MYC set points between individual embryos. Individual embryos (E14.5) from the litters of two concurrently sacrificed Fubp1+/− dams that had been mated with an Fubp1+/− male were genotyped with head tissue, while MEFs were prepared and cultured and individual extract was analyzed to reveal variable MYC set points. C: Immunoblots were analyzed for MYC and actin (or other housekeeping proteins, tubulin, glyceraldehyde-3-phosphate dehydrogenase, or histone; not shown). MEFs recovered from a WT × WT litter were similarly prepared and analyzed and shown in B and C as a reference. WT extracts are indicated with dots; the black, blue, and open dots show results derived from a single litter of WT embryos; the blue and open dots show technical duplicates of a single extract; only the blue dot was included in the statistical analysis with the other single embryo MEFs. The red dots indicate WT-MEFs recovered from two Fubp1+/−× Fubp1+/− litters. B and C: the results of immunoblots of MEFs from individual Fubp1−/− embryos are indicated with red squares. The MEFs from three of the four Fubp1−/− embryos (red squares) are outside the range of all WT MEFs. The open red dots in C show immunoblots of extracts from heterozygous Fubp1+/− single embryo MEFs that were not included in this analysis. D and E: A comparison is shown of the MYC levels normalized with tubulin from a collection of MEFs derived from either WT or Fubp1−/− single embryos. Although the difference between the means of the WT and Fubp1−/− is not significant at the 95% CI, the F test is positive at P < 0.0003, indicating that the Fubp1−/− MYC levels are significantly more variable than the WT. E, embryonic day; FBP, far upstream element binding protein; KO, knockout; MEF, mouse embryonic fibroblast; WT, wild-type.
Besides variation at the single-cell level, the set points of MYC seemed to vary more between single embryo cultures of Fubp1−/− than of WT MEFs. In some litters KO embryos seemed to express less Myc than their WT siblings, whereas in other litters the opposite seemed to occur. To test whether FBP contributed to the Myc set point fixation, Myc protein levels were compared between WT and FBP KO MEFs cultured from single embryos collected at E13.5. A quantitative comparison by immunoblot of WT and KO MEFs from two litters (E13.5), harvested at the same time, showed that the KO MEFs contributed both the high and low extremes of the range of MYC expression, whereas the Myc expression in their WT littermates clustered in the same narrow range with WT MEFs derived from a cross between WT parents (Figure 8, B and C). Mean Myc levels between cultured MEFs from different WT embryos varied on the same scale as the technical variation of duplicates from a single extract (Figure 8, B and C). The Myc variation among the FBP KO MEFs was significantly greater than among the WT MEFs (F test P = 0.011). Examination across a larger number of single embryo MEF samples revealed pronounced and even more statistically significant variation in MYC levels among the KO MEF cultures compared with the WT cultures (F test P < 0.003) (Figure 8, D and E). The results were equivalent whether Myc levels were normalized to actin, tubulin, glyceraldehyde-3-phosphate dehydrogenase (not shown) or histone H3 (not shown). The F test of all of the data aggregated was significant with P < 0.0003. The spectrum of Myc expression across all samples seemed not to resolve into discrete high versus low populations, arguing that feed-forward driven bistability blunted by FBP does not account for the broad range of Myc.45
Myc and actin mRNA levels were measured by real-time quantitative PCR in WT and Fubp1−/− MEFs from two representative litters. These same MEFS were evaluated for MYC and histone H3 (an excellent standard for normalization to cell number) by immunoblot and for Myc mRNA normalized to actin. Myc mRNA levels paralleled MYC protein levels, indicating that expression variation was occurring at the RNA level (Figure 9). To assess whether the variation in Myc output among Fubp1−/− MEFs was accompanied by global changes in activating or repressive chromatin marks, total histones were assayed for covalent modifications by immunoblot across a panel of extracts from KO or WT MEFs. No clear pattern related particular modifications with either the presence or absence of FBP or with the level of Myc (Supplemental Figure S2).
Figure 9.
FBP constrains Myc levels at the RNA level. MEFs from individual embryos in two litters were assayed for Myc mRNA levels normalized to actin mRNA by qPCR and Myc and histone H3 protein by immunoblots. FBP, far upstream element binding protein; KO, knockout; MEF, mouse embryonic fibroblast; qPCR, real-time quantitative PCR; WT, wild-type.
Discussion
The inevitable embryonic lethality of Fubp1−/− mice reveals that FBP2 and FBP3 are neither redundant nor compensatory. Because FBP1 and FBP2 are coexpressed in virtually all tissues, it is likely that they affect distinct processes or resolvable target sets. Notwithstanding almost ubiquitous expression, germline FBP KO proved especially deleterious to hematopoiesis both in embryos and in lethally irradiated hosts who received a transplant with embryonic Fubp1−/− HSCs. Because all blood lineages were affected, in the face of normal or even increased HSC numbers (as estimated by SLAM), we infer HSC dysfunction. Deficits were noted in erythroid, granulocyte-macrophage, and lymphoid lineages from FBP KO livers. Of note, on the basis of mRNA expression profiles and on the disposition of cis elements for known transcription factors, FBP had been predicted by Ebert and colleagues46 to regulate hematopoiesis, especially erythropoiesis and granulocytopoiesis, and was placed just below PBX1 and SOX4 in the hierarchy of hematopoietic master regulators; however direct evidence to confirm a hematopoietic role for FBP was lacking. A recent study also reported the involvement of FBP in embryonic and adult HSC function. Contrary to the results here, the investigators report that HSCs of FBP-deficient mice proliferate poorly but differentiate properly.47 Note that this study relied on gene-trap and shRNA targeting of the Fubp1 allele or its mRNA; neither of these methods assures the complete absence of functional FBP as guaranteed by the germline deletion of the FBP's nucleic acid binding domain in the current experiments. Hypomorphic but not null FBP activity in these systems is strongly suggested by comparison of the competitive transplantation assays. Although germline deletion HSCs were completely unable to contribute to the hematopoietic systems of 16 of 18 primary irradiated recipients, the HSCs from mice targeting FBP via genetrap or shRNA successfully engrafted into all primary irradiated hosts. Re-transplantation of FBP-targeted HSCs from these primary recipients into a second series of lethally irradiated mice was required to elicit HSC failure. The simplest explanation for this discrepancy is that the gene-trapped Fubp1 alleles are in fact not null but are hypomorphic, supporting a milder phenotype. Residual FBP activity in some HSCs may alter the kinetics and pattern of HSC cell division and differentiation.
Germline KOs of hematopoietic master regulators die at discrete points or within narrow windows of embryonic development, for example, GATA1 at E10.5 to E11.5,48 PBX1 at E15 to E16,49 EKLF at E15 to E16,50 and SOX4 at E14, although SOX4−/− embryos expire from failed cardiac development.51 In contrast Fubp1−/− embryonic lethality occurs across half of gestation, from E10.5 until birth, suggesting that FBP is supportive of, but not essential for, multiple hematopoietic stages and transitions. What determines whether a particular FBP KO embryo dies early or succumbs at later stages of development? The co-occurrence of viable and nonviable KO embryos within a single preterm litter suggests a nonmaternal origin to this variation. FBP has been proposed to be a molecular cruise control to constrain both upward and downward excursions of MYC and presumably other FBP targets. The passage of embryos through key developmental bottlenecks might be stochastic due to fluctuating FBP targets because the levels of one or more key players fall outside the envelope that supports development and/or viability. Although the probability that an embryo crosses any single developmental checkpoint may be high, the probability that it traverses all of them successfully may be negligible. Note that embryos and developmental processes are robust to elevated levels of background apoptosis, so random cell death is unlikely to explain this phenotype. Rather, FBP dysfunction at key hematopoietic and perhaps other transitions, may deprive embryos of a sufficient pool of the progenitors needed to populate subsequent developmental stages. Because total HSC numbers in the E13.5 embryo are not reduced, the initial colonization of the embryonic liver by HSCs derived from the aorta-gonad-mesenephros region of the aorta appears adequate. Subsequent multilineage anemia may reflect impaired recruitment and commitment of Fubp1−/− HSCs for productive hematopoiesis, perhaps via dysregulation of multiple FBP binding targets, including MYC, the cyclin-dependent kinase inhibitor p21, and the regulator of p53 levels JTV-1/AIMP2. Although beyond the scope of this article, investigation of conditional Fubp1−/− cells would help to define better the role of FBP in hematopoiesis, with the use of a strategy similar to that used for delineating MYC function in hematopoiesis as reviewed.52 A transcriptional map of human hematopoietic cells suggests that FBP is an important regulator of an expression module up-regulated in HSCs and primitive erythroid cells.46 Therefore, conditional deletion of FBP in lymphoid progenitors (with the use of RAG1-Cre, Lck-Cre, or CD4-Cre) may be expected to be less deleterious than deletion of FBP in HSC (through a Vav-Cre) or erythroid progenitors (GATA1-Cre).
Pulmonary hypoplasia in KO embryos most likely reflected a requirement for FBP during lung development unrelated to the hematopoietic defect. A deficiency of the FBP-interacting protein AIMP2/JTV1 that increases FBP levels elicits pulmonary hyperplasia and impedes terminal differentiation.53 Together these observations invite consideration of a primary role for FBP in lung development.
The hypercellularity noted in late stage Fubp1−/− brains contrasts with pulmonary hypoplasia and anemia and indicates that FBP does not universally support and, in some instances may check, proliferation. The hypercellularity of the brain in the absence of FBP is in accord with its identification as a tumor suppressor in oligodendrogliomas, but it contrasts with the association of FBP with higher levels of MYC, proliferation, and worse prognosis in gliomas.28 Across different embryonic tissues at different developmental stages, FBP function seems to be contextual, suggesting that it adjusts rather than defines biological state.
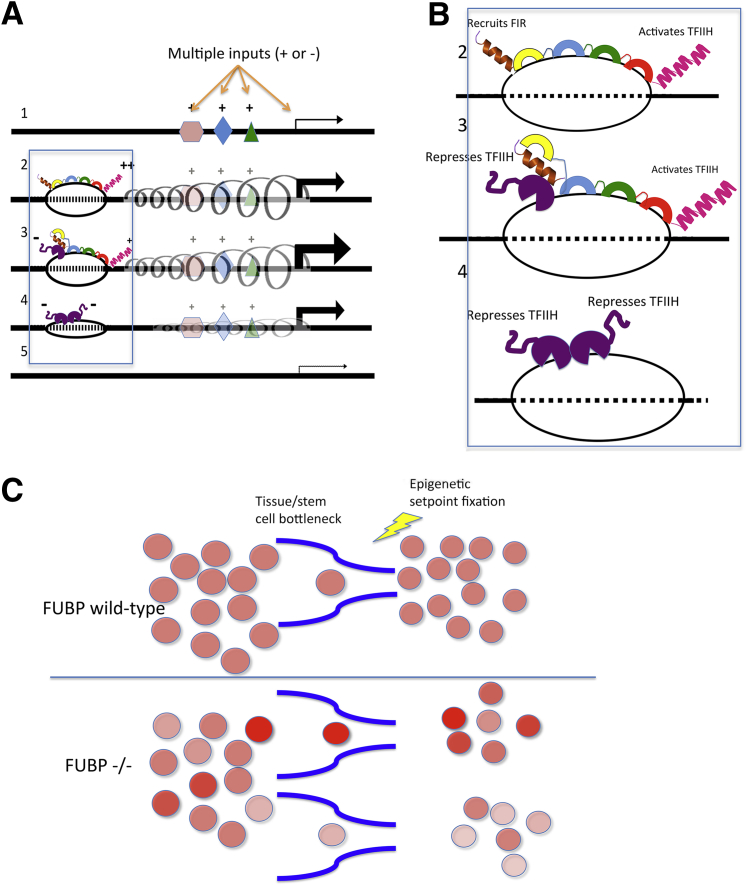
The results in this study provide direct in vivo support for the conjecture that FBP is the lynchpin of a molecular cruise control that coordinates positive and negative feedback onto the MYC promoter to help set and constrain MYC levels (Figure 10, A and B). As the FUSE first melts in response to the torque generated by ongoing transcription (Figure 10, A and B),5, 12, 13 FBP sequence selectively engages DNA via its KH motifs.2, 3, 15, 22 Then FBP loops and stimulates the p89/XPB/ERCC3 3′ to 5′ helicase/translocase of TFIIH at the promoter increasing transcription.4, 5, 6 As transcription intensifies, the melted DNA bubble at FUSE expands upstream, allowing the recruitment of FIR via DNA-protein interactions and protein-protein interaction between a shallow groove in FIR RNA recognition motif 2 and the most NH2 terminal α-helix of FBP (Figure 10, A and B).22, 23, 54, 55 FIR then restores TFIIH helicase activity to basal levels, depressing transcription.7 Falling levels of supercoiling combined with FIR's recruitment and dimerization with a second monomer evict FBP54 and down-regulate MYC (Figure 10, A and B). Note that the FBP/FIR/FUSE system depends on the action of other transcription factors to trigger MYC transcription and to generate dynamic supercoiling (Figure 10, A and B). Lacking FBP, the lynchpin of the cruise control, both activation by FBP and repression by FIR, fail, and MYC becomes coarsely regulated. Whether MYC was up- or down-regulated would depend on the prevailing driving or braking impulses provided to the MYC promoter by the ensemble of factors and pathways active in a given tissue or cell type. MYC output would be expected to fluctuate with the loss of positive and negative feedback from FBP, FIR, and FUSE.56
Figure 10.
Scheme for FBP operating as a molecular cruise control to help set MYC levels and to constrain fluctuations around the set point. A and B, 1:MYC transcription is initiated by a conventional transcription factor(s) represented by symbols. 2: As transcription intensifies, dynamic supercoiling melts the FUSE, allowing sequence-selective binding by FBP via KH motifs, especially KH3 and KH4.3, 4, 5, 11, 12, 13, 15, 22 The carboxyl terminal domain of FBP loops (not shown) to and activates TFIIH bound at the promoter. 3: FBP-augmented transcription expands the FUSE bubble, and FIR is recruited both by DNA binding with FUSE and by protein-protein interaction between the amino terminus of FBP and RNA recognition motif 2 of FIR.7, 15, 23, 54, 554: FIR-repressed transcription decreases supercoiling that in turn ejects FBP, enabling the recruitment and dimerization of a second FIR monomer.54, 55 Unless reactivated, eventually FIR dissociates, and the FUSE bubble relaxes to duplex DNA5; the blue box encompasses the region shown as an enlargement in that includes the FUSE region (B). C:Top: In wild-type mice, during development stem cells are selected to expand and populate tissue compartments. Under the supervision of FBP, MYC levels are uniform. Bottom: Lacking FBP, the positive and negative real-time feedback that constrains MYC levels fails, and MYC levels fluctuate. Tissues formed from stem cells drawn from a population in which MYC levels are broadly distributed and then epigenetically fixed may express higher or lower levels of the oncogene throughout the tissue. FBP, far upstream element binding protein; FIR, FBP interacting repressor; FUSE, far upstream element; KH, K homology; RRM2, RNA recognition motif 2.
The increased variation in Myc expression elicited by FBP KO mirrors the variation in MYC levels seen in XP.6 In XPB disease, the responses of the 3′ to 5′ p89/XPB helicase of TFIIH to effector domains of FBP and FIR are mutationally disabled, and the output of MYC is buffeted by the changes in the flux of the pathways that converge onto the MYC locus at the level of the chromatin and transcription machineries.
The variability in Myc levels between whole populations of MEFs derived from individual Fubp1−/− embryos cannot be explained only by increased cell-to-cell fluctuations. The large number of cells pooled for each sample analyzed by immuoblot renders single-cell fluctuation irrelevant on this scale. So beyond a potential role in constraining MYC fluctuations in single cells, FBP must also be constraining the set point for Myc expression across larger pools of cells (Figure 10C). Lacking FBP, upward (mostly) or downward deviations of MYC are projected across whole populations of MEFs from an individual embryo. What determines the magnitude and direction of this deviation is unknown. Most simply, in a small pool of progenitor cells (perhaps stem cells) lacking FBP, Myc levels at first fluctuate and then become epigenetically fixed as individual cells pass through developmental bottlenecks and expand, projecting these aberrant set points onto a larger population (Figure 10C). How these set points are remembered is unknown, but current theory would invite consideration of histone modifications and chromatin arrangement. It is also possible that noncell autonomous mechanisms instruct and enforce Myc levels through processes such as supercompetition.57 Much of the variation in the pathology of the Fubp1−/− embryos may directly relate to changes in the level of Myc expression. Because the protean influence of MYC as a transcription amplifier is projected across the transcriptome, deconvoluting the primary effects of FBP KO from its indirect effect via Myc on multiple targets and networks may prove challenging.58, 59 If abnormal Myc levels become epigenetically fixed, then Myc-driven pathology might ensue stochastically in the affected tissues.
Whereas the bi-allelic loss in oligodendrogliomas exposed FBP as a tumor suppressor, paradoxically, high levels of the WT mRNA and/or protein have repeatedly been associated with tumorigenesis, that is, liver, stomach, lung, and kidney cancers and gliomas other than those arising in oligodendrocytes. Acting as a molecular cruise control, FBP may moderate either the upward or downward excursions of MYC. In different cellular environments, the prevailing signaling to MYC may deliver a net positive or negative impulse to the promoter. Loss of moderation by FBP would exaggerate promoter output in either direction, explaining how context would determine whether FBP is pro-oncogenic or antioncogenic.
Acknowledgments
We thank Dr. Jessica Snyder (University of Washington School of Medicine) for her contributions in evaluating mouse embryonic neuropathology, James Soh for help with Fubp1 mouse breeding and genotyping, and Zuqin Nie for advice and help.
W.Z., Y.-J.C., E.R.P.C., Y.Z., H.-J.C., R.B., J.L., L.T., E.B., P.D.A., and D.L. conceived and designed experiments and acquired, analyzed, or interpreted data.
Footnotes
Supported by the NIH Intramural Research Program, Center for Cancer Research of the National Cancer Institute.
W.Z., Y.J.C., and E.R.P.C. contributed equally to this work.
Disclosures: None declared.
Current address of W.Z., Shanghai Institutes for Biomedicine Innovation Center for Pharmaceutical Translation, Shanghai, China; of E.R.P.C., Department of Pathology, Duke University School of Medicine, Duke University Medical Center, Durham, NC; of J.L., Center for Drug Evaluation and Research, Food and Drug Administration, Silver Spring, MD.
Supplemental material for this article can be found at http://dx.doi.org/10.1016/j.ajpath.2015.10.028.
Supplemental Data
Generation of the Fubp1−/− allele. A: Arrangement of loxP and FRT sites and the neomycin-resistance cassette and their alignment with the native Fubp1 allele. Crosses with actin-FLIP transgenic mice removed the neomycin cassette from the germline, and then crosses with actin-Cre transgenic mice deleted KH motifs 2, 3, and the amino terminal half of KH4. B: Primers and strategy for PCR genotyping. A sample genotyping gel is shown for three litter mate embryos (A, I, and J). KH, K homology; KO, knockout; WT, wild-type.
High or low Myc set point Fubp1−/− MEFs do not display consistent changes in global histone modification patterns. Extracts from wild-type or Fubp1−/− MEFs were subjected to electrophoresis and immunoblotted against the indicated panel of antibodies. MEF, mouse embryonic fibroblast.
References
- 1.Avigan M.I., Strober B., Levens D. A far upstream element stimulates c-myc expression in undifferentiated leukemia cells. J Biol Chem. 1990;265:18538–18545. [PubMed] [Google Scholar]
- 2.Duncan R., Bazar L., Michelotti G., Tomonaga T., Krutzsch H., Avigan M., Levens D. A sequence-specific, single-strand binding protein activates the far upstream element of c-myc and defines a new DNA-binding motif. Genes Dev. 1994;8:465–480. doi: 10.1101/gad.8.4.465. [DOI] [PubMed] [Google Scholar]
- 3.Michelotti G.A., Michelotti E.F., Pullner A., Duncan R.C., Eick D., Levens D. Multiple single-stranded cis elements are associated with activated chromatin of the human c-myc gene in vivo. Mol Cell Biol. 1996;16:2656–2669. doi: 10.1128/mcb.16.6.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu J., Akoulitchev S., Weber A., Ge H., Chuikov S., Libutti D., Wang X.W., Conaway J.W., Harris C.C., Conaway R.C., Reinberg D., Levens D. Defective interplay of activators and repressors with TFIH in xeroderma pigmentosum. Cell. 2001;104:353–363. doi: 10.1016/s0092-8674(01)00223-9. [DOI] [PubMed] [Google Scholar]
- 5.Liu J., Kouzine F., Nie Z., Chung H.J., Elisha-Feil Z., Weber A., Zhao K., Levens D. The FUSE/FBP/FIR/TFIIH system is a molecular machine programming a pulse of c-myc expression. EMBO J. 2006;25:2119–2130. doi: 10.1038/sj.emboj.7601101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weber A., Liu J., Collins I., Levens D. TFIIH operates through an expanded proximal promoter to fine-tune c-myc expression. Mol Cell Biol. 2005;25:147–161. doi: 10.1128/MCB.25.1.147-161.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J., He L., Collins I., Ge H., Libutti D., Li J., Egly J.M., Levens D. The FBP interacting repressor targets TFIIH to inhibit activated transcription. Mol Cell. 2000;5:331–341. doi: 10.1016/s1097-2765(00)80428-1. [DOI] [PubMed] [Google Scholar]
- 8.Liu J., Chung H.J., Vogt M., Jin Y., Malide D., He L., Dundr M., Levens D. JTV1 co-activates FBP to induce USP29 transcription and stabilize p53 in response to oxidative stress. EMBO J. 2011;30:846–858. doi: 10.1038/emboj.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabenhorst U., Beinoraviciute-Kellner R., Brezniceanu M.L., Joos S., Devens F., Lichter P., Rieker R.J., Trojan J., Chung H.J., Levens D.L., Zornig M. Overexpression of the far upstream element binding protein 1 in hepatocellular carcinoma is required for tumor growth. Hepatology. 2009;50:1121–1129. doi: 10.1002/hep.23098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singer S., Malz M., Herpel E., Warth A., Bissinger M., Keith M., Muley T., Meister M., Hoffmann H., Penzel R., Gdynia G., Ehemann V., Schnabel P.A., Kuner R., Huber P., Schirmacher P., Breuhahn K. Coordinated expression of stathmin family members by far upstream sequence element-binding protein-1 increases motility in non-small cell lung cancer. Cancer Res. 2009;69:2234–2243. doi: 10.1158/0008-5472.CAN-08-3338. [DOI] [PubMed] [Google Scholar]
- 11.Kouzine F., Gupta A., Baranello L., Wojtowicz D., Ben-Aissa K., Liu J., Przytycka T.M., Levens D. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kouzine F., Liu J., Sanford S., Chung H.J., Levens D. The dynamic response of upstream DNA to transcription-generated torsional stress. Nat Struct Mol Biol. 2004;11:1092–1100. doi: 10.1038/nsmb848. [DOI] [PubMed] [Google Scholar]
- 13.Kouzine F., Sanford S., Elisha-Feil Z., Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 14.Davis-Smyth T., Duncan R.C., Zheng T., Michelotti G., Levens D. The far upstream element-binding proteins comprise an ancient family of single-strand DNA-binding transactivators. J Biol Chem. 1996;271:31679–31687. doi: 10.1074/jbc.271.49.31679. [DOI] [PubMed] [Google Scholar]
- 15.Benjamin L.R., Chung H.J., Sanford S., Kouzine F., Liu J., Levens D. Hierarchical mechanisms build the DNA-binding specificity of FUSE binding protein. Proc Natl Acad Sci U S A. 2008;105:18296–18301. doi: 10.1073/pnas.0803279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chung H.J., Liu J., Dundr M., Nie Z., Sanford S., Levens D. FBPs are calibrated molecular tools to adjust gene expression. Mol Cell Biol. 2006;26:6584–6597. doi: 10.1128/MCB.00754-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gherzi R., Chen C.Y., Ramos A., Briata P. KSRP controls pleiotropic cellular functions. Semin Cell Dev Bio. 2014;34:2–8. doi: 10.1016/j.semcdb.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Jacob A.G., Singh R.K., Mohammad F., Bebee T.W., Chandler D.S. The splicing factor FUBP1 is required for the efficient splicing of oncogene MDM2 pre-mRNA. J Biol Chem. 2014;289:17350–17364. doi: 10.1074/jbc.M114.554717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Wang Z., Zhou X., Cheng Y., Xie Z., Manley J.L., Feng Y. Far upstream element-binding protein 1 and RNA secondary structure both mediate second-step splicing repression. Proc Natl Acad Sci U S A. 2013;110:E2687–E2695. doi: 10.1073/pnas.1310607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X., Lin W.J., Chen C.Y., Si Y., Zhang X., Lu L., Suswam E., Zheng L., King P.H. KSRP: a checkpoint for inflammatory cytokine production in astrocytes. Glia. 2012;60:1773–1784. doi: 10.1002/glia.22396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J., Chen Q.M. Far upstream element binding protein 1: a commander of transcription, translation and beyond. Oncogene. 2013;32:2907–2916. doi: 10.1038/onc.2012.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Braddock D.T., Louis J.M., Baber J.L., Levens D., Clore G.M. Structure and dynamics of KH domains from FBP bound to single-stranded DNA. Nature. 2002;415:1051–1056. doi: 10.1038/4151051a. [DOI] [PubMed] [Google Scholar]
- 23.Cukier C.D., Hollingworth D., Martin S.R., Kelly G., Diaz-Moreno I., Ramos A. Molecular basis of FIR-mediated c-myc transcriptional control. Nat Struct Mol Biol. 2010;17:1058–1064. doi: 10.1038/nsmb.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duncan R., Collins I., Tomonaga T., Zhang T., Levens D. A unique transactivation sequence motif is found in the carboxyl-terminal domain of the single-strand-binding protein FBP. Mol Cell Biol. 1996;16:2274–2282. doi: 10.1128/mcb.16.5.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin W.J., Zheng X., Lin C.C., Tsao J., Zhu X., Cody J.J., Coleman J.M., Gherzi R., Luo M., Townes T.M., Parker J.N., Chen C.Y. Posttranscriptional control of type I interferon genes by KSRP in the innate immune response against viral infection. Mol Cell Biol. 2011;31:3196–3207. doi: 10.1128/MCB.05073-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baumgarten P., Harter P.N., Tonjes M., Capper D., Blank A.E., Sahm F., von Deimling A., Kolluru V., Schwamb B., Rabenhorst U., Starzetz T., Kogel D., Rieker R.J., Plate K.H., Ohgaki H., Radlwimmer B., Zornig M., Mittelbronn M. Loss of FUBP1 expression in gliomas predicts FUBP1 mutation and is associated with oligodendroglial differentiation, IDH1 mutation and 1p/19q loss of heterozygosity. Neuropathol Appl Neurobiol. 2014;40:205–216. doi: 10.1111/nan.12088. [DOI] [PubMed] [Google Scholar]
- 27.Bettegowda C., Agrawal N., Jiao Y., Sausen M., Wood L.D., Hruban R.H., Rodriguez F.J., Cahill D.P., McLendon R., Riggins G., Velculescu V.E., Oba-Shinjo S.M., Marie S.K., Vogelstein B., Bigner D., Yan H., Papadopoulos N., Kinzler K.W. Mutations in CIC and FUBP1 contribute to human oligodendroglioma. Science. 2011;333:1453–1455. doi: 10.1126/science.1210557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding Z., Liu X., Liu Y., Zhang J., Huang X., Yang X., Yao L., Cui G., Wang D. Expression of far upstream element (FUSE) binding protein 1 in human glioma is correlated with c-Myc and cell proliferation. Mol Carcinog. 2015;54:405–415. doi: 10.1002/mc.22114. [DOI] [PubMed] [Google Scholar]
- 29.Jia M.Y., Wang Y.J. Far upstream element-binding protein 1(FUBP1) expression differs between human colorectal cancer and non-cancerous tissue. Neoplasma. 2014;61:533–540. doi: 10.4149/neo_2014_065. [DOI] [PubMed] [Google Scholar]
- 30.Ma J., Chen M., Xia S.K., Shu W., Guo Y., Wang Y.H., Xu Y., Bai X.M., Zhang L., Zhang H., Zhang M., Wang Y.P., Leng J. Prostaglandin E2 promotes liver cancer cell growth by the upregulation of FUSE-binding protein 1 expression. Int J Oncol. 2013;42:1093–1104. doi: 10.3892/ijo.2013.1782. [DOI] [PubMed] [Google Scholar]
- 31.Malz M., Bovet M., Samarin J., Rabenhorst U., Sticht C., Bissinger M., Roessler S., Bermejo J.L., Renner M., Calvisi D.F., Singer S., Ganzinger M., Weber A., Gretz N., Zornig M., Schirmacher P., Breuhahn K. Overexpression of far upstream element (FUSE) binding protein (FBP)-interacting repressor (FIR) supports growth of hepatocellular carcinoma. Hepatology. 2014;60:1241–1250. doi: 10.1002/hep.27218. [DOI] [PubMed] [Google Scholar]
- 32.Malz M., Weber A., Singer S., Riehmer V., Bissinger M., Riener M.O., Longerich T., Soll C., Vogel A., Angel P., Schirmacher P., Breuhahn K. Overexpression of far upstream element binding proteins: a mechanism regulating proliferation and migration in liver cancer cells. Hepatology. 2009;50:1130–1139. doi: 10.1002/hep.23051. [DOI] [PubMed] [Google Scholar]
- 33.Zhang F., Tian Q., Wang Y. Far upstream element-binding protein 1 (FUBP1) is overexpressed in human gastric cancer tissue compared to non-cancerous tissue. Onkologie. 2013;36:650–655. doi: 10.1159/000355659. [DOI] [PubMed] [Google Scholar]
- 34.Zubaidah R.M., Tan G.S., Tan S.B., Lim S.G., Lin Q., Chung M.C. 2-D DIGE profiling of hepatocellular carcinoma tissues identified isoforms of far upstream binding protein (FUBP) as novel candidates in liver carcinogenesis. Proteomics. 2008;8:5086–5096. doi: 10.1002/pmic.200800322. [DOI] [PubMed] [Google Scholar]
- 35.Plett P.A., Frankovitz S.M., Orschell C.M. Distribution of marrow repopulating cells between bone marrow and spleen early after transplantation. Blood. 2003;102:2285–2291. doi: 10.1182/blood-2002-12-3742. [DOI] [PubMed] [Google Scholar]
- 36.Tessarollo L., Palko M.E., Akagi K., Coppola V. Gene targeting in mouse embryonic stem cells. Methods Mol Biol. 2009;530:141–164. doi: 10.1007/978-1-59745-471-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morel F., Szilvassy S.J., Travis M., Chen B., Galy A. Primitive hematopoietic cells in murine bone marrow express the CD34 antigen. Blood. 1996;88:3774–3784. [PubMed] [Google Scholar]
- 38.Wickham H. Springer Science & Business Media; New York: 2009. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 39.R Development Core Team . Foundation for Statistical Computing; Vienna, Austria: 2008. R: A Language and Environment for Statistical Computing, Version 2.2. 1. [Google Scholar]
- 40.Kim I., He S., Yilmaz O.H., Kiel M.J., Morrison S.J. Enhanced purification of fetal liver hematopoietic stem cells using SLAM family receptors. Blood. 2006;108:737–744. doi: 10.1182/blood-2005-10-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorenz E., Congdon C., Uphoff D. Modification of acute irradiation injury in mice and guinea-pigs by bone marrow injections. Radiology. 1952;58:863–877. doi: 10.1148/58.6.863. [DOI] [PubMed] [Google Scholar]
- 42.Uchida N., Weissman I.L. Searching for hematopoietic stem cells: evidence that Thy-1.1lo Lin- Sca-1+ cells are the only stem cells in C57BL/Ka-Thy-1.1 bone marrow. J Exp Med. 1992;175:175–184. doi: 10.1084/jem.175.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laurenti E., Varnum-Finney B., Wilson A., Ferrero I., Blanco-Bose W.E., Ehninger A., Knoepfler P.S., Cheng P.F., MacDonald H.R., Eisenman R.N., Bernstein I.D., Trumpp A. Hematopoietic stem cell function and survival depend on c-Myc and N-Myc activity. Cell Stem Cell. 2008;3:611–624. doi: 10.1016/j.stem.2008.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilson A., Murphy M.J., Oskarsson T., Kaloulis K., Bettess M.D., Oser G.M., Pasche A.-C., Knabenhans C., MacDonald H.R., Trumpp A. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18:2747–2763. doi: 10.1101/gad.313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang H., Chen Y., Chen Y. Noise propagation in gene regulation networks involving interlinked positive and negative feedback loops. PLoS One. 2012;7:e51840. doi: 10.1371/journal.pone.0051840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Novershtern N., Subramanian A., Lawton L.N., Mak R.H., Haining W.N., McConkey M.E., Habib N., Yosef N., Chang C.Y., Shay T., Frampton G.M., Drake A.C.B., Leskov I., Nilsson B., Preffer F., Dombkowski D., Evans J.W., Liefeld T., Smutko J.S., Chen J., Friedman N., Young R.A., Golub T.R., Regev A., Ebert B.L. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011;144:296–309. doi: 10.1016/j.cell.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rabenhorst U., Thalheimer F.B., Gerlach K., Kijonka M., Böhm S., Krause Daniela S., Vauti F., Arnold H.H., Schroeder T., Schnütgen F., von Melchner H., Rieger M.A., Zörnig M. Single-stranded DNA-binding transcriptional regulator FUBP1 is essential for fetal and adult hematopoietic stem cell self-renewal. Cell Rep. 2015;11:1847–1855. doi: 10.1016/j.celrep.2015.05.038. [DOI] [PubMed] [Google Scholar]
- 48.Fujiwara Y., Browne C.P., Cunniff K., Goff S.C., Orkin S.H. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci U S A. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DiMartino J.F., Selleri L., Traver D., Firpo M.T., Rhee J., Warnke R., O'Gorman S., Weissman I.L., Cleary M.L. The Hox cofactor and proto-oncogene Pbx1 is required for maintenance of definitive hematopoiesis in the fetal liver. Blood. 2001;98:618–626. doi: 10.1182/blood.v98.3.618. [DOI] [PubMed] [Google Scholar]
- 50.Parkins A.C., Sharpe A.H., Orkin S.H. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature. 1995;375:318–322. doi: 10.1038/375318a0. [DOI] [PubMed] [Google Scholar]
- 51.Schilham M.W., Oosterwegel M.A., Moerer P., Ya J., de Boer P.A., van de Wetering M., Verbeek S., Lamers W.H., Kruisbeek A.M., Cumano A., Clevers H. Defects in cardiac outflow tract formation and pro-B-lymphocyte expansion in mice lacking Sox-4. Nature. 1996;380:711–714. doi: 10.1038/380711a0. [DOI] [PubMed] [Google Scholar]
- 52.Delgado M.D., León J. Myc roles in hematopoiesis and leukemia. Genes Cancer. 2010;1:605–616. doi: 10.1177/1947601910377495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim M.J., Park B.J., Kang Y.S., Kim H.J., Park J.H., Kang J.W., Lee S.W., Han J.M., Lee H.W., Kim S. Downregulation of FUSE-binding protein and c-myc by tRNA synthetase cofactor p38 is required for lung cell differentiation. Nat Genet. 2003;34:330–336. doi: 10.1038/ng1182. [DOI] [PubMed] [Google Scholar]
- 54.Crichlow G.V., Zhou H., Hsiao H.H., Frederick K.B., Debrosse M., Yang Y., Folta-Stogniew E.J., Chung H.J., Fan C., De la Cruz E.M., Levens D., Lolis E., Braddock D. Dimerization of FIR upon FUSE DNA binding suggests a mechanism of c-myc inhibition. EMBO J. 2008;27:277–289. doi: 10.1038/sj.emboj.7601936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsiao H.H., Nath A., Lin C.Y., Folta-Stogniew E.J., Rhoades E., Braddock D.T. Quantitative characterization of the interactions among c-myc transcriptional regulators FUSE, FBP, and FIR. Biochemistry. 2010;49:4620–4634. doi: 10.1021/bi9021445. [DOI] [PubMed] [Google Scholar]
- 56.Ji N., Middelkoop T.C., Mentink R.A., Betist M.C., Tonegawa S., Mooijman D., Korswagen H.C., van Oudenaarden A. Feedback control of gene expression variability in the Caenorhabditis elegans Wnt pathway. Cell. 2013;155:869–880. doi: 10.1016/j.cell.2013.09.060. [DOI] [PubMed] [Google Scholar]
- 57.Johnston L.A. Socializing with MYC: cell competition in development and as a model for premalignant cancer. Cold Spring Harb Perspect Med. 2014;4:a014274. doi: 10.1101/cshperspect.a014274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin C.Y., Loven J., Rahl P.B., Paranal R.M., Burge C.B., Bradner J.E., Lee T.I., Young R.A. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nie Z., Hu G., Wei G., Cui K., Yamane A., Resch W., Wang R., Green D.R., Tessarollo L., Casellas R., Zhao K., Levens D. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Generation of the Fubp1−/− allele. A: Arrangement of loxP and FRT sites and the neomycin-resistance cassette and their alignment with the native Fubp1 allele. Crosses with actin-FLIP transgenic mice removed the neomycin cassette from the germline, and then crosses with actin-Cre transgenic mice deleted KH motifs 2, 3, and the amino terminal half of KH4. B: Primers and strategy for PCR genotyping. A sample genotyping gel is shown for three litter mate embryos (A, I, and J). KH, K homology; KO, knockout; WT, wild-type.
High or low Myc set point Fubp1−/− MEFs do not display consistent changes in global histone modification patterns. Extracts from wild-type or Fubp1−/− MEFs were subjected to electrophoresis and immunoblotted against the indicated panel of antibodies. MEF, mouse embryonic fibroblast.