Abstract
The incidence of chronic kidney disease (CKD) varies by ancestry, with African Americans (AA) having a threefold to fourfold higher rate than whites. Notably, two APOL1 alleles, termed G1 [c.(1072A>G; 1200T>G)] and G2 (c.1212_1217del6), are strongly associated with higher rates of nondiabetic CKD and an increased risk for hypertensive end-stage renal disease. This has prompted the opportunity to implement APOL1 testing to identify at-risk patients and modify other risk factors to reduce the progression of CKD to end-stage renal disease. We developed an APOL1 genotyping assay using multiplex allele-specific primer extension, and validated using 58 positive and negative controls. Genotyping results were completely concordant with Sanger sequencing, and both triplicate interrun and intrarun genotyping results were completely concordant. Multiethnic APOL1 allele frequencies were also determined by genotyping 7059 AA, Hispanic, and Asian individuals from the New York City metropolitan area. The AA, Hispanic, and Asian APOL1 G1 and G2 allele frequencies were 0.22 and 0.13, 0.037 and 0.025, and 0.013 and 0.004, respectively. Notably, approximately 14% of the AA population carried two risk alleles and are at increased risk for CKD, compared with <1% of the Hispanic and Asian populations. This novel APOL1 genotyping assay is robust and highly accurate, and represents one of the first personalized medicine clinical genetic tests for disease risk prediction.
Chronic kidney disease (CKD) is a progressive loss of renal function that can be defined by laboratory findings, such as pathological abnormalities, composition of blood or urine, and decreased glomerular filtration rates.1 CKD is classified into five different stages according to the presence or absence of kidney damage and the level of kidney function, and its symptoms include hypertension, anemia, hyperkalemia, metabolic acidosis, and bone disease.2 End-stage renal disease (ESRD) presents with anorexia, lethargy, nausea and vomiting, uremic pericarditis, seizures, muscle cramps, and coma. It has been reported that African Americans (AA) develop ESRD at rates four to five times higher than European Americans,3, 4 which is likely because of both environmental and genetic factors.5 Mapping by admixture linkage disequilibrium studies revealed that variant alleles in the apolipoprotein L1 (APOL1) gene on chromosome 22 were strongly associated with an increased risk of renal disease among AAs.6 Importantly, these APOL1 variants in the homozygous or compound heterozygous state increased the risk of hypertension-attributed ESRD and focal and segmental glomerulosclerosis by 10- to 17-fold and increased the risk of HIV-associated nephropathy by 29-fold.6, 7, 8
The two APOL1 risk alleles are termed G1, which is a haplotype with two missense variants in near-complete linkage disequilibrium [c.(1072A>G; 1200T>G); p.(S342G; I384M); rs73885319 and rs60910145], and G2, which is an in-frame two amino acid deletion (c.1212_1217del6; p.Asn388_Tyr389del; rs71785313).6, 9 Carriers of two G1 or G2 alleles are at significantly increased risks for developing CKD and ESRD compared with both heterozygous G1 or G2 carriers and noncarriers.6, 10 The overall incidence rate of CKD has been reported to be approximately 12 events/1000 person-years and approximately 8 events/1000 person-years for AA individuals at high and low APOL1 genetic risk, respectively.10 Absolute risks for progression to ESRD among AA individuals with CKD have been reported to be 58% and 37% for those at high and low APOL1 genetic risk, respectively.11 Taken together, these data provide a compelling rationale for individuals of African ancestry to determine their genetic risk for renal disease by APOL1 testing and subsequently promote better prevention and management for those identified to be at increased genetic risk.
Given the paucity of commercially available clinical APOL1 genotyping assays, we developed and validated a novel and cost-effective method to test APOL1 G1 and G2 using a multiplex allele-specific primer extension (ASPE) Luminex bead-based genotyping assay. In addition, 7059 DNA samples from adult AA, Hispanic, and Asian individuals were tested to determine the prevalence of the APOL1 G1 and G2 risk alleles in the multiethnic New York City metropolitan area.
Materials and Methods
Study Subjects
All self-reported AA DNA samples were obtained from the BioMe biobank of the Institute for Personalized Medicine at the Icahn School of Medicine at Mount Sinai (New York, NY) with Institutional Review Board approval.12 All self-reported Hispanic and Asian DNA samples were obtained from anonymous blood donors from the New York Blood Center with Institutional Review Board approval, as previously described.13, 14 All personal identifiers were removed before use, and isolated DNA samples were tested anonymously. Genomic DNA from the Institute for Personalized Medicine BioMe biobank and the New York Blood Center samples was isolated using the FlexiGene DNA kit and Puregene DNA Purification kit (Qiagen, Valencia, CA), respectively, according to the manufacturer's instructions.
APOL1 Genotyping
PCR was performed in 25 μL containing approximately 50 to 100 ng of DNA, 1.5× PCR buffer (Invitrogen, Carlsbad, CA), 5.0 mmol/L MgCl2, 0.2 mmol/L of each dNTP, 1.0 μmol/L of forward and reverse primers, and 1.0 U of Platinum Taq DNA Polymerase (Invitrogen). Primers were designed using the human hg19 reference genome and from GenBank (http://www.ncbi.nlm.nih.gov/genbank; accession number NM_145343.2), with forward primer, 5′-GAGCCAGAGCCAATCTTCAGTCAGT-3′ (chromosome 22:36661745-36661769), and reverse primer, 5′-GCCAGGCATATCTCTCCTGGTGGCT-3′ (chromosome 22:36662094-36662118) (Figure 1). The amplification consisted of an initial denaturation step at 94°C for 5 minutes, followed by 30 amplification cycles (94°C for 30 seconds, 66°C for 30 seconds, and 72°C for 1 minute), and a final incubation at 72°C for 5 minutes. After amplification, unincorporated nucleotides and PCR primers were degraded by treatment with 2.5 U of shrimp alkaline phosphatase and 8.0 U of exonuclease I (both from Affymetrix Inc., Santa Clara, CA) at 37°C for 30 minutes, followed by enzyme inactivation at 99°C for 15 minutes.
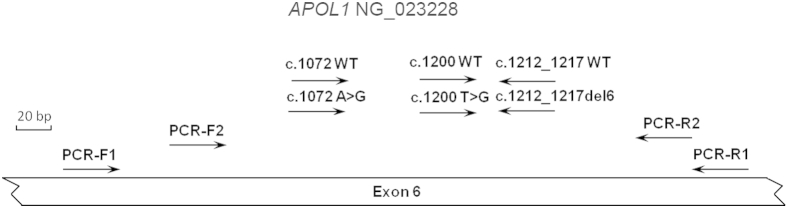
Figure 1.
PCR and multiplex allele-specific primer extension (ASPE) primer locations for APOL1 genotyping and Sanger sequencing. The NG_023228 genomic sequence of the APOL1 gene (http://www.ncbi.nlm.nih.gov/nuccore; accession number NG_023228.1) was used to design all PCR and ASPE primers for APOL1 genotyping and Sanger sequencing. The APOL1 genotyping and Sanger sequencing primer pairs used for amplification were PCR-F1/R1 and PCR-F2/R2, respectively. The six ASPE primers used to detect the WT, G1, or G2 alleles are illustrated with directional arrows and labeled as c.1072WT, c.1072A>G, c.1200WT, c.1200T>G, c.1212_1217WT, and c.1212_1217del6. Primer sequences are listed in Materials and Methods and Table 1.
Digested samples were subjected to multiplex ASPE with primers that were coupled with tags corresponding to complementary MicroPlex-xTAG antitags (Luminex, Austin, TX). The reaction was performed in 20 μL containing 5 μL of treated PCR product, 1.5× Takara PCR buffer, 5 μmol/L each of deoxy-ATP, dTTP, and deoxy-GTP, 5 μmol/L of biotin-14-dCTP (Invitrogen), 25 pmol/L each of tagged ASPE primers (Table 1 and Figure 1), and 1.25 U of Takara Taq HS polymerase (Takara Bio Inc., Otsu, Japan). The amplification consisted of an initial denaturation step at 96°C for 2 minutes, followed by 40 extension cycles (94°C for 30 seconds, 54°C for 30 seconds, and 74°C for 30 seconds).
Table 1.
Multiplex ASPE Primer Sequences and Their Corresponding Tags for Bead Identification
| Target name | Tag sequence | ASPE primer sequence | Chromosome 22 position of the ASPE primers (hg19) |
|---|---|---|---|
| c.1072A WT | 5′-ATCTCAATTACAATAACACACAAA-3′ | 5′-CATCCAGCACAAGAAAGAAGCT-3′ | 36661906-36661927 |
| c.1072A>G | 5′-TTAACAACTTATACAAACACAAAC-3′ | 5′-CATCCAGCACAAGAAAGAAGCC-3′ | 36661906-36661927 |
| c.1200T WT | 5′-CTAAATCACATACTTAACAACAAA-3′ | 5′-GAGCTGGAGGAGAAGCTAAACATT-3′ | 36662011-36662034 |
| c.1200T>G | 5′-ATACTTTACAAACAAATAACACAC-3′ | 5′-GAGCTGGAGGAGAAGCTAAACATG-3′ | 36662011-36662034 |
| c.1212_1217 WT | 5′-CAATTTACATTTCACTTTCTTATC-3′ | 5′-CCTGCAGAATCTTATAA-3′ | 36662046-36662062 |
| c.1212_1217del6 | 5′-AACTTTCTCTCTCTATTCTTATTT-3′ | 5′-CCTGCAGAATCTTATTG-3′ | 36662046-36662062 |
ASPE, allele-specific primer extension; WT, wild type.
ASPE products were hybridized to a population mix of MicroPlex-xTAG microsphere beads (Luminex) that were coupled with specific antitags (Table 1). The antitag for each bead population was complementary to the tag sequence attached to each ASPE primer. The hybridization reaction consisted of 5 μL of ASPE product and 45 μL of bead mix and was performed at 96°C for 2 minutes, followed by 37°C for 60 minutes. Hybridized samples were then washed twice with 150 μL of wash buffer [0.2 mol/L NaCl, 0.1 mol/L Tris, pH 8.0, and 0.08% (v/v) Triton X-100]. For detection, 100 μL of reporter solution [1 mg/L streptavidin-R-phycoerythrin (Molecular Probes Inc., Carlsbad, CA) in wash buffer] was added to each sample well and incubated at room temperature for 15 minutes in the dark. The samples were transferred to a Costar plate (Fisher Scientific, Pittsburgh, PA) and analyzed at ambient temperature on a Luminex 100 xMAP instrument that was set to read a minimum of 100 events per bead population. Dual lasers simultaneously identified color-coded beads and fluorescently labeled extension products to generate a signal for each variant.
Each Luminex run included at least one positive and negative control as well as a nontemplate control consisting of water. Positive and negative controls were confirmed by Sanger sequencing before use, with positive controls consisting of samples that were heterozygous, compound heterozygous, or homozygous for the G1 and/or G2 alleles, and negative controls consisting of samples that were wild-type for both the G1 and G2 alleles.
Genotyping Data Analysis
The output comma delimited data file generated by the Luminex system was analyzed using an Excel 2010 spreadsheet (Microsoft Corp., Redmond, WA). Using this program, all median fluorescence intensity (MFI) values for each allele of each mutation were evaluated, as well as the MFI values of a PCR-negative control sample. The maximum MFI allowed for the PCR-negative control was 300 U. If any signal on any bead exceeded 300 U for the PCR-negative control, all samples failed for the corresponding variant. Otherwise, net MFI values were calculated by subtracting the PCR-negative control MFI values from the respective MFI values of the sample being analyzed; net MFI values calculated to be negative were set to zero. As an acceptance criterion, the MFI value for at least one allele was required to be at least 300 MFI units for each variant. If this value was not met, the sample failed the variant in question and was assigned a no call. If a sample passed, the genotype for each variant was determined on the basis of allelic ratios, which equaled the net MFI for an allele divided by the sum of the net MFI for all alleles tested for that variant [eg, mutant/(wild-type plus mutant)]. Allelic ratios were set as follows: If the mutant allelic ratio was >0.85, the allele was called as mutant; if the mutant allelic ratio was 0.30 to 0.70, the call was heterozygous for the two alleles; if the mutant allelic ratio was <0.15, the call was wild-type; in all other cases, the call was no call and the sample was repeated and/or DNA reextracted.
Sanger Sequencing
The PCR conditions were the same as described above for the Luminex genotyping assay; however, APOL1 amplicons for Sanger sequencing were generated using independent M13 tagged forward (5′-CGACCTGGTCATCAAAAGCCTTGAC-3′) and reverse (5′-GGAGGCAGAGCTTGCAGTGAGCTG-3′) primers (Figure 1). PCR products were separated by agarose gel electrophoresis to ensure proper amplification, indicated by a single strong band at the expected amplicon size, and subsequently prepared for sequencing using Bigdye and ran on a 3730xl DNA Analyzer (SeqGen, Torrance, CA). Sequencing chromatograms had unique, nonoverlapping peaks for homozygous samples, overlapping peaks at the mutant position for heterozygous missense or nonsense mutations, and overlapping peaks at all positions after the change for deletions occurring in one strand. Sequencing results were required to show the variation in both the forward and reverse directions to be considered validated.
Results
APOL1 Genotyping Test Validation
To assess the analytical sensitivity and specificity of the APOL1 genotyping assay, we tested 48 positive and 10 negative control samples. The positive control DNAs were heterozygous or homozygous for the G1 and/or G2 alleles, and negative controls were wild-type at both loci (Table 2). G1GM has two missense alleles in cis (c.1072A>G and c.1200T>G), whereas G1G is defined by only one risk allele (c.1072A>G), and G2 is defined by the c.1212_1217del6 allele. Sanger sequencing was used to confirm all of the genotypes called by Luminex genotyping. The amplicons for Sanger sequencing were generated by primers that were independent of the genotyping PCR primer sequences to avoid rare variants that could cause allele dropout (Figure 1).
Table 2.
Comparison of APOL1 Genotyping Results with Sanger Sequencing
|
APOL1 genotyping | |||||
|---|---|---|---|---|---|
| c.1072A>G | c.1200T>G | c.1212_1217del6 | Diplotype | No. of samples | No. confirmed by Sanger sequencing |
| A/A | T/T | TTATAA/TTATAA | WT/WT | 10 | 10 |
| A/G | T/G | TTATAA/TTATAA | G1GM/WT | 6 | 6 |
| G/G | G/G | TTATAA/TTATAA | G1GM/G1GM | 6 | 6 |
| G/G | T/G | TTATAA/TTATAA | G1GM/G1G | 9 | 9 |
| A/A | T/T | TTATAA/del | G2/WT | 6 | 6 |
| A/A | T/T | del/del | G2/G2 | 8 | 8 |
| A/G | T/G | TTATAA/del | G1GM/G2 | 13 | 13 |
| Total | 58 | 58 | |||
G1GM, APOL1 c.(1072A>G; 1200T>G); G1G, APOL1 c.1072A>G; G2, APOL1 c.1212_1217del6; WT, wild type.
Among these 58 samples, which included all combinations of the G1 and G2 alleles, the Sanger sequencing results were completely concordant with the APOL1 genotyping results, indicating an analytical sensitivity of >99% (95% CI, 93%–100%) and an analytical specificity of >99% (95% CI, 72%–100%). To determine intrarun and interrun reproducibility, all of the 58 control samples were run on different days, which included three interrun repeated assays. These results had complete genotype concordance between each replicate.
Multiethnic APOL1 G1 and G2 Population Screening
The multiethnic APOL1 G1 (c.1072A>G, c.1200T>G) and G2 (c.1212_1217del6) allele and genotype frequency data are summarized in Tables 3 and 4, respectively. Genotyping 5453 AA individuals detected c.1072A>G, c.1200T>G, and c.1212_1217del6 allele frequencies of 0.22, 0.21, and 0.13, respectively. Importantly, 13.8% of all AA individuals carried two APOL1 G1 and/or G2 risk alleles, and are at increased risk for developing CKD and ESRD (Table 5). Genotyping 1146 Hispanic and 460 Asian individuals detected c.1072A>G, c.1200T>G, and c.1212_1217del6 allele frequencies of 0.037, 0.036, and 0.025, and 0.013, 0.013, and 0.004, respectively. These lower allele frequencies translated into only 0.3% of Hispanic and 0.4% of Asian individuals carrying two APOL1 risk alleles and being at increased risk for developing CKD and ESRD (Table 5).
Table 3.
Multiethnic APOL1 Allele Frequencies
| APOL1 genotype | African American (n = 10,906) |
Hispanic (n = 2292) |
Asian (n = 920) |
|||
|---|---|---|---|---|---|---|
| Frequency | 95% CI | Frequency | 95% CI | Frequency | 95% CI | |
| c.1072A>G | ||||||
| A | 0.784 | 0.777–0.792 | 0.963 | 0.956–0.971 | 0.987 | 0.980–0.994 |
| G | 0.216 | 0.208–0.223 | 0.037 | 0.029–0.044 | 0.013 | 0.006–0.020 |
| c.1200T>G | ||||||
| T | 0.789 | 0.781–0.796 | 0.964 | 0.956–0.971 | 0.987 | 0.980–0.994 |
| G | 0.211 | 0.204–0.219 | 0.036 | 0.029–0.044 | 0.013 | 0.006–0.020 |
| c.1212_1217del6 | ||||||
| TTATAA | 0.870 | 0.863–0.876 | 0.975 | 0.968–0.981 | 0.996 | 0.991–1.00 |
| del | 0.130 | 0.124–0.137 | 0.025 | 0.019–0.032 | 0.004 | 0.000–0.009 |
Table 4.
Multiethnic APOL1 Genotype Frequencies
| APOL1 genotype | African American (n = 5453) |
Hispanic (n = 1146) |
Asian (n = 460) |
|||
|---|---|---|---|---|---|---|
| Observed, % | Expected, %∗ | Observed, % | Expected, %∗ | Observed, % | Expected, %∗ | |
| c.1072A>G | ||||||
| A/A | 62.6 (n = 3411) | 61.5 | 92.8 (n = 1063) | 92.8 | 97.4 (n = 448) | 97.4 |
| A/G | 31.7 (n = 1732) | 33.8 | 7.2 (n = 82) | 7.1 | 2.6 (n = 12) | 2.6 |
| G/G | 5.7 (n = 310) | 4.7 | 0.1 (n = 1) | 0.1 | 0.0 (n = 0) | 0.0 |
| c.1200T>G | ||||||
| T/T | 63.2 (n = 3448) | 62.2 | 92.8 (n = 1064) | 92.9 | 97.4 (n = 448) | 97.4 |
| T/G | 31.3 (n = 1705) | 33.3 | 7.1 (n = 81) | 7.0 | 2.6 (n = 12) | 2.6 |
| G/G | 5.5 (n = 300) | 5.5 | 0.1 (n = 1) | 0.1 | 0.0 (n = 0) | 0.0 |
| c.1212_1217del6 | ||||||
| TTATAA/TTATAA | 75.9 (n = 4139) | 75.6 | 95.0 (n = 1089) | 95.0 | 99.1 (n = 456) | 99.1 |
| TTATAA/del | 22.1 (n = 1207) | 22.7 | 4.9 (n = 56) | 4.9 | 0.9 (n = 4) | 0.9 |
| del/del | 2.0 (n = 107) | 1.7 | 0.1 (n = 1) | 0.1 | 0.0 (n = 0) | 0.0 |
Predicted Hardy-Weinberg frequencies.
Table 5.
Multiethnic APOL1 G1 and G2 Diplotype Frequencies by Renal Disease Risk
| APOL1 diplotype∗ | African American, % (N = 5453) | Hispanic, % (N = 1146) | Asian, % (N = 460) |
|---|---|---|---|
| Normal risk | |||
| WT/WT | 44.6 (n = 2430) | 87.8 (n = 1006) | 97.0 (n = 446) |
| G1/WT | 25.6 (n = 1398) | 7.1 (n = 81) | 2.2 (n = 10) |
| G2/WT | 16.0 (n = 874) | 4.9 (n = 56) | 0.4 (n = 2) |
| Total | 86.2 (n = 4702) | 99.8 (n = 1143) | 99.6 (n = 458) |
| Increased risk | |||
| G1/G1 | 5.7 (n = 310) | 0.1 (n = 1) | 0.0 (n = 0) |
| G1/G2 | 6.1 (n = 334) | 0.1 (n = 1) | 0.4 (n = 2) |
| G2/G2 | 2.0 (n = 107) | 0.1 (n = 1) | 0.0 (n = 0) |
| Total | 13.8 (n = 751) | 0.3 (n = 3) | 0.4 (n = 2) |
The G1 risk haplotype includes both the G1GM and G1G allele configurations.
In total, 7059 DNA samples were used for the multiethnic APOL1 allele and genotype frequency screen; however, 11 additional samples were originally included (nine AA and two Hispanic) that resulted in a no call in one or more of the three tested variants (c.1072A>G, c.1200T>G, and/or c.1212_1217del6). These failures were determined to be because of poor DNA quality, and their data were excluded from the frequency calculations. However, the resulting APOL1 genotyping failure rate (approximately 0.16%) indicates that the reported assay is robust.
Discussion
To facilitate the implementation of clinical APOL1 genotyping to predict patient risk for developing CKD and ESRD, we developed and validated a novel and robust method to simultaneously interrogate the APOL1 G1 (c.1072A>G, c.1200T>G) and G2 (c.1212_1217del6) risk alleles using a multiplex bead-based genotyping assay on the Luminex platform. Our group has previously used this technology to develop multiplexed Ashkenazi Jewish carrier screening panels,15 and novel genotyping assays with this open platform have also been reported for hereditary hemochromatosis,16 HLA typing,17 human papillomavirus detection,18 and other applications.19 To the best of our knowledge, this APOL1 G1/G2 genotyping assay represents the first disease risk prediction genetic test validated for clinical use with this technology.
Clinical genetic testing has historically been centered on germ-line mutation detection for Mendelian diseases. However, candidate gene and genome-wide association studies have identified polymorphic DNA sequence variants that significantly contribute to common disease susceptibility, complex traits, and drug response phenotypes.20 In addition to advancing our scientific understanding of disease mechanisms and providing starting points for the development of medical treatments, the identification of susceptibility variants with significant disease associations also allows for the estimation of personal disease risks. As such, the personalized medicine paradigm now includes the use of individual genetic data in conjunction with other clinical, family, and demographic variables to inform decisions on disease prevention, diagnosis, treatment, and prognosis.21 Despite the ongoing debate regarding the clinical validity and utility of common disease risk genetic testing,22 public awareness of genetics and genome-guided medicine has undoubtedly increased during the past 5 years, which is likely, in part, because of the recent availability of direct-to-consumer genetic testing.
After decades of investigation on the epidemiology and genetics of common kidney diseases,23 the association between APOL1 and ESRD among nondiabetic kidney disease patients of African ancestry was elucidated by two independent groups in 2010.6, 8 Although most common disease susceptibility variants identified by genome-wide association studies have low odds ratios (ORs) typically <1.5,24 the APOL1 association OR is one of the highest reported for a common disease and ranges from 7 to 29 for the various etiologies of nondiabetic ESRD.23 This association is also notable because it is driven by relatively high-frequency variant alleles, which is most likely the result of powerful evolutionary selection pressure.25 Specifically, the higher frequency of APOL1 risk alleles observed in the AA population has been explained by the fact that APOL1 risk allele carriers are resistant to parasitic trypanosomal infection, giving them a selective heterozygote advantage over noncarriers in endemic regions of Africa.26 Similar to sickle cell disease, this advantage has allowed the APOL1 risk alleles to persist in the African population through positive selection for this gene region.6
According to the US Renal Data System, Medicare costs between 2008 and 2012 increased higher for patients with CKD than the total Medicare costs (53.6% versus 11.5%), and represented 19.6% of all Medicare parts A and B spending.27 Potential cost savings could be achieved through the prevention of disease progression to ESRD, and development of concurrent chronic conditions, such as diabetes and congestive heart failure. As such, a fast and reliable clinical APOL1 genotyping method has the potential to identify individuals at increased genetic risk for ESRD, allowing them to be monitored early, and potentially reduce their chances of disease progression. Our novel, high-throughput APOL1 genotyping method had an analytical sensitivity and specificity of >99% and was further evaluated by a large multiethnic population screen across the AA, Hispanic, and Asian populations of the New York City metropolitan area. Whites were not included in this screen because these risk alleles are rare in this population,6 with c.1072A>G and c.1200T>G allele frequencies of 0.0001051 and 7.65 × 10−5, respectively, in the Exome Aggregation Consortium European (non-Finnish) population (http://exac.broadinstitute.org; last accessed November 5, 2015). The G2 allele (c.1212_1217del6) is not currently reported in the Exome Aggregation Consortium database, likely because of the difficulties with detecting insertion/deletion variants by commonly used next-generation sequencing platforms.
As noted, approximately 14% of the AA population carried two APOL1 variant alleles and are at increased risk of developing CKD and ESRD, suggesting that targeted mutation testing with this validated assay could be effectively implemented for disease risk predication in this population. Despite the difficulties and challenges with disease risk prediction genetic testing,22, 28, 29, 30 the reported APOL1 genotyping assay represents one of the first analytically validated personalized medicine genetic tests for disease risk prediction. Although the clinical utility of APOL1 genetic testing is being evaluated by ongoing prospective Clinical Trials (http://www.clinicaltrials.gov; identifier NCT02234063), implementing APOL1 genetic testing for individuals of African descent has the potential to help prevent renal disease progression and minimize the continued increases in Medicare spending on CKD.
Footnotes
Supported, in part, by the Mount Sinai Genetic Testing Laboratory at the Icahn School of Medicine at Mount Sinai, and the Institute for Personalized Medicine at the Icahn School of Medicine at Mount Sinai, funded by the National Human Genome Research Institute of the NIH grant U01HG006380. The eMERGE Network was initiated and funded by NHGRI through the following grants: U01HG006828 (Cincinnati Children's Hospital Medical Center/Boston Children's Hospital); U01HG006830 (Children's Hospital of Philadelphia); U01HG006389 (Essentia Institute of Rural Health, Marshfield Clinic Research Foundation, and Pennsylvania State University); U01HG006382 (Geisinger Clinic); U01HG006375 (Group Health Cooperative/University of Washington); U01HG006379 (Mayo Clinic); U01HG006380 (Icahn School of Medicine at Mount Sinai); U01HG006388 (Northwestern University); U01HG006378 (Vanderbilt University Medical Center); and U01HG006385 (Vanderbilt University Medical Center serving as the Coordinating Center). Supported, in part, by the National Institute of General Medical Sciences of the NIH grant K23 GM104401 (S.A.S.).
J.Z. and A.F. contributed equally to this work.
Presented as a Reviewer's Choice Abstract at the 65th American Society of Human Genetics Annual Meeting, held October 6-10, 2015, in Baltimore, MD.
Disclosures: None declared.
Current address of J.Z., Baylor College of Medicine, Houston, TX.
Contributor Information
Jinglan Zhang, Email: jinglan.zhang@bcm.edu.
Stuart A. Scott, Email: stuart.scott@mssm.eddsu.
References
- 1.Kidney Disease Improving Global Outcomes KDIGO 2012 clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int Suppl. 2013;3:150. doi: 10.1038/ki.2013.243. [DOI] [PubMed] [Google Scholar]
- 2.National Kidney Foundation K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis. 2002;39(Suppl 2):S1–S266. [PubMed] [Google Scholar]
- 3.Byrne C., Nedelman J., Luke R.G. Race, socioeconomic status, and the development of end-stage renal disease. Am J Kidney Dis. 1994;23:16–22. doi: 10.1016/s0272-6386(12)80806-7. [DOI] [PubMed] [Google Scholar]
- 4.Satko S.G., Freedman B.I., Moossavi S. Genetic factors in end-stage renal disease. Kidney Int Suppl. 2005:S46–S49. doi: 10.1111/j.1523-1755.2005.09411.x. [DOI] [PubMed] [Google Scholar]
- 5.Patzer R.E., McClellan W.M. Influence of race, ethnicity and socioeconomic status on kidney disease. Nat Rev Nephrol. 2012;8:533–541. doi: 10.1038/nrneph.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Genovese G., Friedman D.J., Ross M.D., Lecordier L., Uzureau P., Freedman B.I., Bowden D.W., Langefeld C.D., Oleksyk T.K., Uscinski Knob A.L., Bernhardy A.J., Hicks P.J., Nelson G.W., Vanhollebeke B., Winkler C.A., Kopp J.B., Pays E., Pollak M.R. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fine D.M., Wasser W.G., Estrella M.M., Atta M.G., Kuperman M., Shemer R., Rajasekaran A., Tzur S., Racusen L.C., Skorecki K. APOL1 risk variants predict histopathology and progression to ESRD in HIV-related kidney disease. J Am Soc Nephrol. 2012;23:343–350. doi: 10.1681/ASN.2011060562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tzur S., Rosset S., Shemer R., Yudkovsky G., Selig S., Tarekegn A., Bekele E., Bradman N., Wasser W.G., Behar D.M., Skorecki K. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010;128:345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freedman B.I., Kopp J.B., Langefeld C.D., Genovese G., Friedman D.J., Nelson G.W., Winkler C.A., Bowden D.W., Pollak M.R. The apolipoprotein L1 (APOL1) gene and nondiabetic nephropathy in African Americans. J Am Soc Nephrol. 2010;21:1422–1426. doi: 10.1681/ASN.2010070730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Foster M.C., Coresh J., Fornage M., Astor B.C., Grams M., Franceschini N., Boerwinkle E., Parekh R.S., Kao W.H. APOL1 variants associate with increased risk of CKD among African Americans. J Am Soc Nephrol. 2013;24:1484–1491. doi: 10.1681/ASN.2013010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parsa A., Kao W.H., Xie D., Astor B.C., Li M., Hsu C.Y., Feldman H.I., Parekh R.S., Kusek J.W., Greene T.H., Fink J.C., Anderson A.H., Choi M.J., Wright J.T., Jr., Lash J.P., Freedman B.I., Ojo A., Winkler C.A., Raj D.S., Kopp J.B., He J., Jensvold N.G., Tao K., Lipkowitz M.S., Appel L.J., AASK Study Investigators; CRIC Study Investigators APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med. 2013;369:2183–2196. doi: 10.1056/NEJMoa1310345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tayo B.O., Teil M., Tong L., Qin H., Khitrov G., Zhang W., Song Q., Gottesman O., Zhu X., Pereira A.C., Cooper R.S., Bottinger E.P. Genetic background of patients from a university medical center in Manhattan: implications for personalized medicine. PLoS One. 2011;6:e19166. doi: 10.1371/journal.pone.0019166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott S.A., Jaremko M., Lubitz S.A., Kornreich R., Halperin J.L., Desnick R.J. CYP2C9∗8 is prevalent among African-Americans: implications for pharmacogenetic dosing. Pharmacogenomics. 2009;10:1243–1255. doi: 10.2217/pgs.09.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott S.A., Khasawneh R., Peter I., Kornreich R., Desnick R.J. Combined CYP2C9, VKORC1 and CYP4F2 frequencies among racial and ethnic groups. Pharmacogenomics. 2010;11:781–791. doi: 10.2217/pgs.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott S.A., Edelmann L., Liu L., Luo M., Desnick R.J., Kornreich R. Experience with carrier screening and prenatal diagnosis for 16 Ashkenazi Jewish genetic diseases. Hum Mutat. 2010;31:1240–1250. doi: 10.1002/humu.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardoso S.P., Patel R., Brown C., Navarrete C. Simultaneous detection of HFE C282Y, H63D and S65C mutations associated with type 1 haemochromatosis using a multiplex luminex bead assay. Tissue Antigens. 2011;78:171–177. doi: 10.1111/j.1399-0039.2011.01736.x. [DOI] [PubMed] [Google Scholar]
- 17.Heinemann F.M. HLA genotyping and antibody characterization using the luminex multiplex technology. Transfus Med Hemother. 2009;36:273–278. doi: 10.1159/000228834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozaki S., Kato K., Abe Y., Hara H., Kubota H., Kubushiro K., Kawahara E., Inoue M. Analytical performance of newly developed multiplex human papillomavirus genotyping assay using Luminex xMAP technology (Mebgen HPV Kit) J Virol Methods. 2014;204:73–80. doi: 10.1016/j.jviromet.2014.04.010. [DOI] [PubMed] [Google Scholar]
- 19.Dunbar S.A. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin Chim Acta. 2006;363:71–82. doi: 10.1016/j.cccn.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abul-Husn N.S., Owusu Obeng A., Sanderson S.C., Gottesman O., Scott S.A. Implementation and utilization of genetic testing in personalized medicine. Pharmacogenomics Pers Med. 2014;7:227–240. doi: 10.2147/PGPM.S48887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yim S.H., Chung Y.J. Reflections on the US FDA's warning on direct-to-consumer genetic testing. Genomics Inform. 2014;12:151–155. doi: 10.5808/GI.2014.12.4.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruzel-Davila E., Wasser W.G., Aviram S., Skorecki K. APOL1 nephropathy: from gene to mechanisms of kidney injury. Nephrol Dial Transplant. 2015 doi: 10.1093/ndt/gfu391. [Epub ahead of print] doi: 10.1093/ndt/gfu391. [DOI] [PubMed] [Google Scholar]
- 24.Manolio T.A. Bringing genome-wide association findings into clinical use. Nat Rev Genet. 2013;14:549–558. doi: 10.1038/nrg3523. [DOI] [PubMed] [Google Scholar]
- 25.Wasser W.G., Tzur S., Wolday D., Adu D., Baumstein D., Rosset S., Skorecki K. Population genetics of chronic kidney disease: the evolving story of APOL1. J Nephrol. 2012;25:603–618. doi: 10.5301/jn.5000179. [DOI] [PubMed] [Google Scholar]
- 26.Limou S., Nelson G.W., Kopp J.B., Winkler C.A. APOL1 kidney risk alleles: population genetics and disease associations. Adv Chronic Kidney Dis. 2014;21:426–433. doi: 10.1053/j.ackd.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.US Renal Data System . NIH, National Institute of Diabetes and Digestive and Kidney Diseases; Bethesda, MD: 2014. Chapter 6: Medicare expenditures for CKD. 2014 USRDS Annual Data Report: Epidemiology of Kidney Disease in the United States.http://www.usrds.org/2014/view/v1_06.aspx Available at. [Google Scholar]
- 28.American College of Medicine Genetics Board of Directors ACMG statement on direct-to-consumer genetic testing. Genet Med. 2004;6:60. doi: 10.109701.GIM.0000106164.59722.CE. [DOI] [PubMed] [Google Scholar]
- 29.Annas G.J., Elias S. 23andMe and the FDA. N Engl J Med. 2014;370:2248–2249. doi: 10.1056/NEJMc1404692. [DOI] [PubMed] [Google Scholar]
- 30.Hudson K., Javitt G., Burke W., Byers P., American Society of Human Genetics Social Issues Committee ASHG Statement∗ on direct-to-consumer genetic testing in the United States. Obstet Gynecol. 2007;110:1392–1395. doi: 10.1097/01.AOG.0000292086.98514.8b. [DOI] [PubMed] [Google Scholar]