Abstract
Mutations in PROP1, the most common known cause of combined pituitary hormone deficiency in humans, can result in the progressive loss of all hormones of the pituitary anterior lobe. In mice, Prop1 mutations result in the failure to initiate transcription of Pou1f1 (also known as Pit1) and lack somatotropins, lactotropins, and thyrotropins. The basis for this species difference is unknown. We hypothesized that Prop1 is expressed in a progenitor cell that can develop into all anterior lobe cell types, and not just the somatotropes, thyrotropes, and lactotropes, which are collectively known as the PIT1 lineage. To test this idea, we produced a transgenic Prop1-cre mouse line and conducted lineage-tracing experiments of Prop1-expressing cells. The results reveal that all hormone-secreting cell types of both the anterior and intermediate lobes are descended from Prop1-expressing progenitors. The Prop1-cre mice also provide a valuable genetic reagent with a unique spatial and temporal expression for generating tissue-specific gene rearrangements early in pituitary gland development. We also determined that the minimal essential sequences for reliable Prop1 expression lie within 10 kilobases of the mouse gene and demonstrated that human PROP1 can substitute functionally for mouse Prop1. These studies enhance our understanding of the pathophysiology of disease in patients with PROP1 mutations.
The pituitary gland plays a critical role in vertebrate physiology through the secretion of hormones that regulate reproduction, stress management, metabolism, lactation, and water balance. Pituitary dysfunction can result from congenital malformation, pituitary adenoma, or head trauma. Although some cases are managed with pharmacotherapy and/or surgery, not all individuals respond, resulting in impaired quality of life or morbidity. Congenital pituitary hormone deficiency is defined as GH deficiency and loss of at least one other pituitary hormone. It is generally caused by mutations in transcription factors necessary for pituitary organogenesis, and the severity of the corresponding disease is related to the molecular function of the transcription factor during development of the pituitary and other craniofacial tissues (1, 2). For example, patients with mutations in transcription factors that act during the earliest stages of head development, such as HESX1 and SOX2, can have a syndromic presentation, including the lack of multiple pituitary hormones and septo-optic dysplasia (1). People with nonsyndromic pituitary deficiencies tend to have mutations in pituitary lineage-specific transcription factors such as PROP1 and PIT1 (POU1F1).
Mutations in PROP1 and PIT1 typically cause deficiency of GH, prolactin (PRL), and TSH, which are produced by 3 separate cell types known as the PIT1 lineage (3–6). PIT1 stimulates differentiation of progenitor cells to these 3 cell fates by organizing target gene enhancer regions in the nuclear matrix, using a mechanism involving MATRIN-3, β-CATENIN, and the DNA-binding protein, SATB1 (7). PIT1 also acts as a direct transcriptional activator of the GH, PRL, and TSH genes (8–12). PROP1, in turn, is a direct transcriptional activator of PIT1 (13). PROP1 partners with β-CATENIN and binds an early-acting enhancer of PIT1 to initiate its transcription (13). Prop1df/df (Ser83Pro) and Prop1−/− mice have a congenital deficiency in the PIT1 lineage cell types, but gonadotropes and corticotropes develop normally (14–17). In contrast, humans with PROP1 mutations often exhibit progressive loss of the PIT1 lineage, gonadotropin deficiency, and ultimately loss of corticotrope function (18). The basis for the phenotypic differences between Prop1−/− mice and humans are unknown, but it could be caused by temporal or spatial differences in PROP1 expression between the 2 species or differences in downstream targets due to evolutionary changes in the proteins.
PROP1 expression has been detected in SOX2-expressing pituitary progenitors (19, 20), which supports the hypothesis that the progressive loss of the hormone-producing lineages in humans could be due to depletion of progenitors marked by PROP1. An alternative theory is that Rathke's pouch harbors 2 separate populations of progenitor cells, one of which forms corticotropes and gonadotropes, and the other forms the PIT1 lineage of somatotropes, lactotropes, and thyrotropes (21). In this scenario, Prop1 expression is only required in the PIT1 lineage, and is not necessary for differentiation of corticotropes and gonadotropes. More complex models are also possible because there is evidence that antagonism between lineage-specific transcription factors plays a role in some cell fate decisions. For example, TBX19 (also known as TPIT) is an early determinate of corticotrope formation, and it inhibits the activity of STEROIDOGENIC FACTOR 1 (SF1, gene symbol Nr5a1), a key transcription factor for gonadotrope differentiation (21). SF1 also inhibits the activity of TBX19, generating a mechanism where the expression of either TBX19 or SF1 in a common progenitor leads to lineage specification and inhibition of the opposite cell type (21). GATA2 synergizes with PIT1 to promote thyrotrope fate and antagonize the gonadotrope fate (22–24). It is important to determine the lineages that derive from cells expressing PROP1 in order to distinguish between these possibilities.
The discovery of pituitary stem cells has focused much attention on the generation and function of this pituitary cell population (25). SOX2 is expressed in the pituitary progenitors of Rathke's pouch, and it is maintained in adult pituitaries in the stem cells that reside near the residual cleft of Rathke's pouch, a region known as the marginal zone (26). The pituitary progenitors arise during mouse embryogenesis at embryonic day of development 9.5 (e9.5) as Rathke's pouch is induced from the oral ectoderm (27). These progenitors continue to proliferate, and they maintain an epithelial structure. As pituitary progenitors exit the cell cycle between e11.5 and e13.5, they leave the epithelia and adopt a more rounded shape characteristic of hormone secreting cells of the anterior lobe (28–30). This transition is similar in appearance to an epithelial to mesenchymal transition (EMT), where cells down-regulate cell adhesion molecules characteristic of epithelial cells and become migratory in nature. However, the pituitary hormone-expressing cells are not truly mesenchymal; therefore, this transition in the pituitary is EMT like. Prop1 is expressed in Rathke's pouch between e10.5 and e14.5, a time when the pituitary progenitors proliferate, exit the cell cycle, and migrate into the anterior lobe (5). Prop1 plays a critical role in this transition, as Prop1−/− and Prop1df/df mice have dysmorphic pituitaries with an increase in epithelial-like cells and few anterior lobe cells (31). In addition, Prop1df/df embryos have persistent expression of N-CADHERIN in the progenitor cells at the ventral aspect of Rathke's pouch, and they fail to express SNAI2 (also known as SLUG), both of which are indicative of failed EMT (32). These results support the idea that Prop1 is expressed in progenitors of all pituitary cell types, not just in the PIT1 lineage.
We used transgenic mice to determine the minimal essential sequences necessary for mouse Prop1 expression and to assess whether human PROP1 is sufficiently conserved to functionally compensate for mouse Prop1. We also performed a lineage tracing experiment of the Prop1-expressing cell population to determine whether Prop1 is expressed in all pituitary progenitors or whether its expression is limited to the PIT1 lineage. We generated and characterized a Prop1-cre transgenic mouse line for the lineage tracing experiment, and this line provides a valuable tool for tissue-specific gene rearrangements in the pituitary progenitors. These studies provide a greater understanding of PROP1 function in pituitary organogenesis.
Materials and Methods
Transgene construct generation
A 25-kb rescue plasmid was subcloned from mouse bacterial artificial chromosome (BAC) RP23-250I22 using gap repair (33). Regions of homology to mouse chromosome 11 were generated by PCR, amplifying nucleotide 50 943 223 to 50 943 595 (using mouse genome reference assembly GRCm38.p2) at the 3′ end of Prop1 genomic locus and nucleotide 50 968 513 to 50 968 857 at the 5′ end. The amplified regions were cloned into pBluescript II (Aligent Technologies). The linearized plasmid was transformed into RP23-250I22 containing bacteria, and ampicillin-resistant colonies selected. The rescued plasmid was completely sequenced to confirm identity to the reference region of mouse chromosome 11. The 10-kb rescue plasmid was generated previously (34).
The Prop1-cre construct was generated using materials and methodology similar to that reported in the construction of the Cga-cre (35). We used a cre cassette (icre) with codons optimized for mammalian cells (36). The cre-FRT-gb2-kan-FRT containing plasmid was further modified to introduce the bovine GH polyadenylation signal (bGH-pA) downstream of the cre coding sequence. BAC clone RP23-479M11, obtained from the BACPAC Resource Center, was modified by inserting a cre expression cassette into Prop1 exon 2, in frame. An oligonucleotide with homology to Prop1 exon 2 (nucleotides 50 952 311 to 50 952 236 and 50 952 155 to 50 952 233) was used in combination with a 100-bp oligonucleotide containing 24 bp of homology to cre to amplify the cre-bGH-pA-FRT-gb2-kan-FRT cassette (Table 1). The PCR product was inserted into the BAC using homologous recombination, and kanamycin-resistant colonies were selected. The FRT-gb2-kan-FRT drug selection cassette was removed using enhanced flippase, leaving behind a single FRT site (37). Successfully targeted BACs were identified by pulsed field gel restriction enzyme mapping (38) and sequenced over the 10-kb critical region of Prop1 (Table 1).
Table 1.
Primers Used in Generating and Genotyping Transgenic Mice
| Prop1 Homology Domain | cre Cassette Homology Domain | |
|---|---|---|
| Prop1-cre transgene construction | ||
| Exon 2 A | GTGAGGATGGGCTGGTGGTCCCATTAATAAGGGGGTCTCCCTCCGTTTTTCTCCCTCCATAGACAGGAGCTCTGAG | ATGGTGCCCAAGAAGAAGAGGAAA |
| Exon 2 B | GGCGGCGCCGGGAGTGGGGACGGCCTCTCTGTGGGCAAAGCTTAGGTCTCCCCAGCTCTGTACCAGAGAGCCTCCTATAA | CTATACGAAGTTATAAGCTT |
| Prop1-cre transgene genotyping | ||
| A | CTGCACACAGACAGGAGCAT | |
| B | GGTCTCCCTCCGTTTTTCTC | |
| 25-kb Prop1 BAC transgene genotyping | ||
| A | AATTAACCCTCACTAAAGGG | |
| B | AAGGGCAGGTAAGATGCTGA | |
| 10-kb Prop1 BAC transgene genotyping | ||
| A | AAACCCCCAGAGAAGAGGAA | |
| B | GACCATGATTACGCCAAGCTA | |
| HsPROP1 BAC transgene genotyping | ||
| A | GGAGGGAACAGGTGTGGAG | |
| B | GGTGTCTCTCAGGCAACAGG | |
Each primer is listed 5′–3′, and A and B indicate forward and reverse primers, respectively.
Generation and genotyping of transgenic mice
All experiments using mice were approved by the Institutional Committee on the Use and Care of Animals for the University of Michigan (protocol number PR00004640 to S.A.C.) and the University of South Carolina (protocol number 2106-100665-012213 to S.W.D.). Mice were housed in specific pathogen-free conditions with automatic watering and ventilated cages. Mice were fed ad libitum irradiated 5080 chow with 21.5% of the calories from fat. Weaning was delayed to permit mutants to nurse longer, and after weaning, mutants were housed with normal animals to provide warmth.
Prop1df/df mice were provided by A. Bartke in 1988 and maintained as a stock. Prop1−/− mice (officially Prop1tm1Sac) were generated from mouse embryonic stem cells engineered to contain a deletion in Prop1 beginning with the initiator ATG in the first exon and extending through the first intron and a portion of exon 2 (17).
Linearized and purified 25- and 10-kb Prop1 rescue plasmids were injected separately into the pronuclei of fertilized eggs generated from a cross of DF/B-Prop1df/+ males of mixed genetic background (15) and (SJL/J X C57BL/6J) F1 females. Injected zygotes were transferred into pseudopregnant foster mothers. Prop1 plasmid transgenic; Prop1df/+ mice were crossed to N6-B6-Prop1+/− mice (17) to generate Prop1 plasmid transgenic; Prop1df/− offspring. These mice and all littermates were weighed, photographed, and genotyped at weaning. Genotyping for the Prop1 rescue plasmids was performed by PCR using the primers listed in Table 1, and the Prop1df allele and Prop1− allele were genotyped using previously described primers (17, 39). BAC RP11-452O4 containing 175 kb spanning the human PROP1 genomic locus was obtained from BACPAC Resources, expanded, and purified. Purified BAC (HsPROP1) was injected into the pronuclei of fertilized eggs from a cross of DF/B-Prop1df/+ males and (SJL/J X C57BL/6J) F1 females, as detailed above. HsPROP1; Prop1df/+ mice were crossed with Prop1df/+ mice and all offspring were weighed, photographed, and genotyped at weaning. HsPROP1 transgenic mice were genotyped by PCR using primers listed in Table 1 and primers for the chloramphenicol resistance gene in the BAC backbone as previously described (35).
The purified Prop1-cre modified BAC was injected into the pronuclei of fertilized eggs from a cross between C57BL/6J and B6D2F1 mice, and the fertilized eggs were transferred into pseudopregnant foster mothers. Transgenic cre positive mice were identified by PCR genotyping using primers listed in Table 1 and primers for the chloramphenicol resistance gene.
C57BL/6J, B6.129S4-Gt(ROSA)26Sortm1Sor/J (Rosastop-Lacz), and B6.Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo(RosamT/mG) mice were obtained from The Jackson Laboratory. Each line was crossed separately with hemizygous Prop1-cre mice to produce experimental embryos and pups.
X-gal staining and immunohistochemistry
Prop1-cre; Rosastop-LacZ embryos ranging from e9.5 to e12.5 for whole mount 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-gal) staining, along with Prop1-cre negative control tissues, were isolated in 1× PBS (pH 7.2), fixed for 15 minutes at 4°C in 4% paraformaldehyde in LacZ Fix, washed 3 times for 15 minutes at 4°C in LacZ wash buffer, before immersion in X-gal stain overnight at 37°C (40). Prop1-cre; Rosastop-LacZ e14.5 and e16.5 embryos were also collected in 1× PBS, fixed for 15 minutes at 4°C in LacZ Fix, washed 3 times for 15 minutes at 4°C in LacZ wash buffer, equilibrated in 30% sucrose at 4°C, and frozen for sagittal sections in Tissue-Tek Optimum Cutting Temperature Compound (Sakura) in a dry ice/ethanol bath. Tissues were cryosectioned at 10-μm thickness and mounted on Superfrost Plus slides (VWR). After sectioning, tissues were thawed at room temperature for 5 minutes, briefly fixed in LacZ Fix for 5 minutes at room temperature, washed 3 times in LacZ wash buffer, and incubated in X-gal stain overnight at 37°C. X-gal stained sections were counterstained in 2% neutral red for 2 minutes. Prop1-cre;Rosastop-LacZ 3-week-old pituitaries used for X-gal staining and immunohistochemistry were processed similarly, except that incubation in X-gal stain at 37°C occurred for 2.5 hours, instead of overnight. Immunohistochemistry for pituitary hormones was performed as previously described using antibodies from the National Hormone and Pituitary Program, National Institute of Diabetes and Digestive and Kidney Diseases (41).
Prop1+/+, Prop1+/−, and Prop1−/− e12.5 embryos were dissected from pregnant mothers and fixed in 3.7% formaldehyde in PBS overnight at 4°C. The embryos were processed for paraffin sectioning, and sectioned at 6-μm thickness (40). Select sections of each genotype were treated with xylene to remove paraffin and hydrated through 100% and 95% ethanol and into PBS. Slides were treated in 1.5% H2O2 and 50% methanol for 20 minutes to inactivate endogenous peroxidases, boiled in 10mM citrate (pH 6.2), for 10 minutes to unmask antigens, and blocked in 0.5% blocking reagent provided in the tyramide signal amplification (TSA) fluorescein kit (PerkinElmer) in 0.1M Tris-HCl (pH 7.5), and 0.15M NaCl for 10 minutes. Tissue sections were incubated with guinea pig anti-PROP1 antibody (Aimee Ryan) diluted 1:100 in block overnight at 4°C. After removing unbound primary antibody with PBS wash, sections were incubated with 1:100 biotin-conjugated antiguinea pig antibody (Jackson ImmunoResearch) for 30 minutes at room temperature, followed with 1:100 horseradish peroxidase-conjugated streptavidin (TSA fluorescein kit; PerkinElmer) for 30 minutes at room temperature. Slides were counterstained in 4′, 6-diamidino-2-phenylindole (DAPI) for 5 minutes before mounting in fluorescence mounting medium.
The PROP1 immunostaining protocol was modified for analysis of cryosections of Prop1-cre; RosamT/mG embryos. Embryos were dissected from pregnant mothers and fixed, embedded in Optimum Cutting Temperature, and cryosectioned as above for Prop1-cre; Rosastop-LacZ embryos. Select sections were thawed at room temperature and photographed on a Leica DMI3000 B microscope with fluorescence capabilities, using a Leica DFC345 FX camera, to capture the tandem dimeric Tomato (tdTOMATO) and enhanced green fluorescent protein (eGFP) fluorescence (Leica Microsystems). Images were processed with Leica Application Suite Core software to produce red and green overlay images. The tissue sections were fixed in 4% paraformaldehyde in PBS for 1 hour at room temperature, before incubation in 0.1% Triton X-100 in PBS for 15 minutes. Boiling in 10mM citrate for 10 minutes retrieved antigens and eliminated fluorescence from tdTOMATO and eGFP. Sections were incubated in 1% blocking reagent in 1× PBS (Molecular Probes Tyramide Signal Amplification kits; Thermo Fisher Scientific) and then incubated with guinea pig anti-PROP1 antibody diluted 1:500 in block overnight at 4°C. Biotin-conjugated antiguinea pig secondary antibody was used as above, followed by Dylight 488-conjugated streptavidin (Thermo Fisher Scientific) diluted 1:100 in block for 30 minutes at room temperature. Tissues were counterstained with DAPI before mounting in fluorescence mounting media. The green fluorescent signal was captured with the same microscope and camera, but pseudocolored purple instead of green. Fluorescent emission from DAPI was also photographed, but not used in the final overlay images. The purple image was overlaid on the tdTOMATO and/or eGFP images using Adobe Photoshop CS6 (Adobe Systems).
Pituitaries from 3-week-old Prop1-cre;RosamT/mG mice used for hormone and PECAM1 (also known as CD31) immunohistochemistry were collected and fixed, washed, frozen, and sectioned as above for Prop1-cre; Rosastop-LacZ embryos. Selected sections were thawed at room temperature for 5 minutes, fixed in 4% paraformaldehyde in 1× PBS for 15 minutes at room temperature, and incubated in 0.1% Triton X-100 in 1× PBS for 15 minutes. The sections were treated in 1.5% H2O2 in 50% methanol for 20 minutes before blocking in 1% blocking reagent in 1× PBS (Molecular Probes Tyramide Signal Amplification kits; Thermo Fisher Scientific) for 10 minutes. Guinea pig primary antibodies for LHβ (1:500), TSHβ (1:1000), proopiomelanocortin (POMC) (1:1000), and PRL (1:1000) and monkey primary antibody for GH (1:1000) (National Hormone and Pituitary Program, National Institute of Diabetes and Digestive and Kidney Diseases) (41) were diluted into block and incubated overnight at 4°C. PECAM1 rabbit primary antibody (Thermo Fisher Scientific) was diluted 1:100. Biotin-conjugated species appropriate secondary antibodies were diluted 1:100 and incubated for 30 minutes at room temperature, followed by streptavidin-conjugated horseradish peroxidase (1:100) for 30 minutes at room temperature, and then TSA with Alexa Fluor 350 (Thermo Fisher Scientific) for 3 minutes. Sections were mounted in fluorescence mounting media and then imaged as above for endogenous fluorescence from tdTOMATO, eGFP, and Alexa Fluor 350. Overlay images of eGFP and Alexa Fluor 350 (pseudocolored red) were produced with Leica Application Suite Core software. Under these conditions, background staining in the intermediate lobe prevented analysis of POMC expression in the melanotropes. For intermediate lobe POMC expression, an e18.5 Prop1-cre; RosamT/mG embryo was fixed, embedded in paraffin, and sectioned as above. Select sections were processed as above to remove paraffin and hydrate the tissues, followed by a 5-minute citrate boil to retrieve antigens and blocking in 1% blocking reagent in PBS (Molecular Probes Tyramide Signal Amplification kits; Thermo Fisher Scientific). Detection of eGFP was performed with a 1:500 dilution of rabbit anti-eGFP (Abcam), 1:200 dilution of biotin-conjugated antirabbit secondary (Thermo Fisher Scientific), and 1:100 dilution of streptavidin-conjugated Dylight 488 (Thermo Fisher Scientific). POMC detection utilized the guinea pig anti-POMC as above; followed by Dylight 594-conjugated antiguinea pig secondary (Thermo Fisher Scientific) diluted 1:100. Images were processed to match images produced for the other pituitary hormones.
All animals and tissues used for these experiments were chosen without regard to sex.
Results
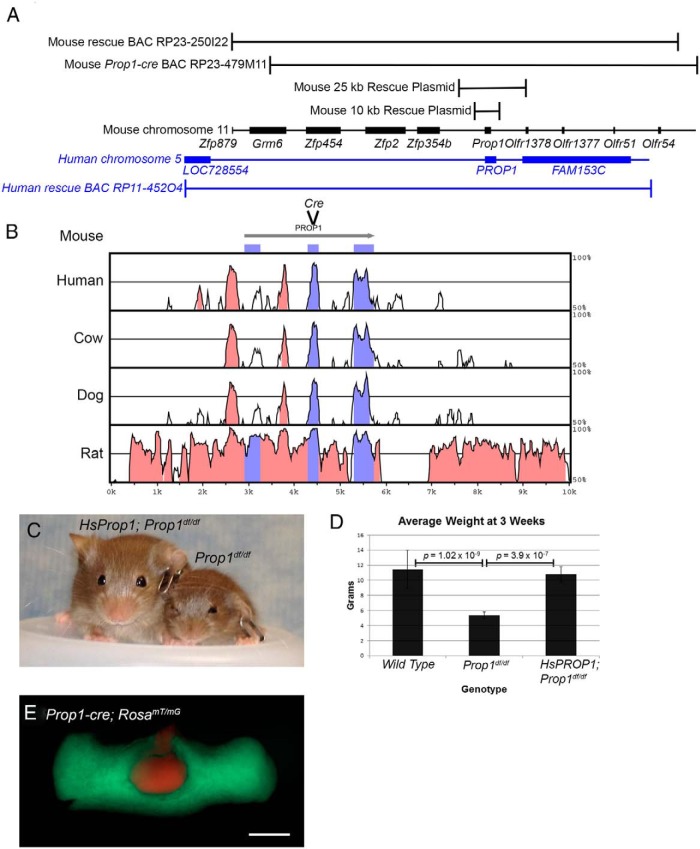
The order of genes flanking Prop1 differs in rodents and primates, suggesting that genomic rearrangements probably occurred during mammalian radiation (Figure 1A). This implies that conserved cis-regulatory elements necessary for Prop1 transcription are located in the area of homology between human and mouse. Previously, we demonstrated that a mouse line transgenic for a BAC containing the mouse Prop1 gene and the surrounding approximately 195-kb genomic region (BAC RP23-250I22) was sufficient to rescue the Prop1df/− dwarf phenotype (42). This result indicates that the BAC contains the cis-regulatory elements necessary for Prop1 transcription. We hypothesized that a considerably smaller region might be sufficient for correction because of the lack of synteny homology surrounding the human and mouse Prop1 genes (Figure 1A) and the clustering of conserved noncoding sequences (CNSs) within 10 kb of the gene (Figure 1B) (42). To identify the minimal essential sequences for Prop1 expression, we generated transgenic lines with either 25 or 10 kb surrounding the Prop1 gene and tested at least 4 lines from each construct for the ability to correct the dwarf phenotype (Table 2 and Figure 1A). Four different lines with the 25-kb BAC were able to correct the dwarf phenotype with 100% penetrance (n = 29 informative progeny total). Four out of 5 transgenic lines carrying the 10-kb Prop1 genomic region were sufficient for rescuing the Prop1df/− dwarf phenotype, but the penetrance was incomplete in 3 of the 4 lines (Table 2). The 10-kb transgene contains all of the obvious CNSs, but the incomplete penetrance of transgene correction relative to the larger constructs reveals a lack of insulation from insertion site effects in this transgene relative to the 25-kb construct.
Figure 1.
Prop1 genomic regions and conservation, Prop1-cre generation, and HsPROP1 transgenic rescue. A, Schematic diagram of mouse Prop1 genomic region on chromosome 11 (black) and human PROP1 on chromosome 5 (blue) as visualized using the UCSC Genome Browser (http://genome.ucsc.edu). Black brackets above the genomic schematic indicate the portion of mouse chromosome 11 contained in the original rescue BAC, RP23-250I22, the BAC used for generating the Prop1-cre, RP23-479M11, and 2 rescue plasmids of 25 and 10 kb. The blue bracket below the genomic schematic indicates the portion of human chromosome 5 contained in HsPROP1 BAC RP11-452O4. B, VISTA plot (http://genome.lbl.gov/vista/mvista/submit.shtml) comparing conserved genomic regions between mouse, human, cow, dog, and rat genomes in the region of the 10-kb rescue plasmid. Pink peaks indicate CNSs and purple peaks represent conserved coding sequences. Also indicated is the location where the cre cassette was inserted into BAC RP23-479M11 to generate Prop1-cre. C, Three-week-old HsPROP1; Prop1df/df transgenic and Prop1df/df mice. D, Graph of average weight at 3 weeks for wild-type, Propdf/df, and HsProp1; PROP1df/df mice. Brackets indicate P values for 2-tailed, t tests between wild-type (n = 37) and Prop1df/df (n = 11) and between Prop1df/df and HsPROP1; Prop1df/df (n = 18) mice. E, Pituitary from a 3-week-old Prop1-cre; RosamT/mG mouse. Green fluorescence indicates recombination from Prop1-cre, and red fluorescence indicates no recombination. Scale bar, 500 μm.
Table 2.
Transgenic Rescue of Prop1 Mutant Mice
| Transgenic Construct | Line | Full Sized Tg; Prop1df/− Micea | Dwarf Tg; Prop1df/− Micea | Penetrant Correction |
|---|---|---|---|---|
| 25-kb Prop1 Tg | 103 | 5 | 0 | 100 |
| 114 | 6 | 0 | 100 | |
| 450 | 12 | 0 | 100 | |
| 453 | 6 | 0 | 100 | |
| 231 | 2 | 5 | 29 | |
| 241 | 1 | 0 | 100 | |
| 10-kb Prop1 Tg | 244 | 0 | 2 | 0 |
| 248 | 6 | 1 | 86 | |
| 463 | 11 | 0 | 100 | |
| 853 | 5 | 0 | 100 | |
| HsPROP1 BAC Tg | 862 | 4 | 1 | 80 |
| 868 | 4 | 0 | 100 | |
| 884 | 4 | 0 | 100 |
Each line number represents the ear tag of the founder transgenic mouse.
The number represents the total number of dwarf mice or transgenic dwarf mice collected over multiple litters. Only 1/8 of the mice are expected to be transgenic and compound heterozygotes for Prop1 loss of function mutations (Prop1df/−).
To test the functional conservation of the PROP1 protein and the cis-regulatory elements that regulate its expression, we tested the ability of the human PROP1 gene to rescue the dwarf mouse phenotype. We generated 4 transgenic mouse lines carrying an approximately 196-kb BAC containing human PROP1 (Figure 1A) and performed the necessary crosses to generate mice that were Prop1df/df and harbored the human PROP1 BAC transgene (HsPROP1). All 4 HsPROP1 BAC transgenic lines rescued the mouse dwarf phenotype, and only one of the 3 lines exhibited incomplete penetrance (Table 2 and Figure 1, C and D). This indicates that human PROP1 can compensate functionally for the mouse gene, despite the 51.5% divergence in amino acids N-terminal to the homeodomain and an overall amino acid identity of 73%. It also demonstrates that transcriptional regulation is sufficiently conserved between mouse and man for rescue of the mouse genetic defect with mouse transactivating factors recognizing human cis-acting sequences and driving PROP1 expression.
BAC recombineering has simplified the ability to engineer large DNA constructs (33); therefore, we chose to generate a mouse Prop1-cre transgene by inserting the coding sequence for the cre recombinase into a mouse Prop1 BAC (36). We used a Prop1 containing BAC, RP23-479M11, which contains mouse genomic DNA similar to that shown to be sufficient for correcting the mutant phenotype (Figure 1A). This BAC was used for generating all Prop1-cre transgenes. The CNS in intron 1 of mouse Prop1 is known to mediate spatial expression within the pituitary gland and response to Notch signaling (42, 43), and it is more conserved than exon 1 (Figure 1B). Therefore, we attempted to preserve its function in the Prop1-cre construct. Using BAC recombineering, the cre coding sequence was inserted in-frame into exon 2, thus preserving the integrity of intron 1 completely, as well as the spatial arrangement relative to the promoter (Figure 1B). Three transgenic lines were generated and tested for cre activity: Tg(Prop1-cre)432Sac, Tg(Prop1-cre)506Sac, Tg(Prop1-cre)517Sac. All 3 lines exhibit cre activity throughout the pituitary anterior lobe at 3 weeks, when combined with cre reporter line RosamT/mG, where a membrane bound red fluorescent tdTOMATO protein is expressed before recombination, and a membrane bound green fluorescent eGFP protein is expressed following recombination (Figure 1E and data not shown) (44).
To determine whether Prop1-cre expression is properly restricted to the pituitary gland, we performed whole mount X-gal staining on Tg(Prop1-cre)517Sac; R26Rstop-Lacz embryos at e11.5 (Figure 2, A and B) (45). Robust X-gal staining was observed in the pituitary gland with no detectable ectopic activity in other tissues in 75% of the embryos analyzed (n = 18). A portion of the embryos (25%) displayed variable, ectopic expression in the brain and peripheral tissues in addition to the expression in Rathke's pouch (n = 6, data not shown). Individual mice and fetuses with ectopic cre activity were excluded from further analyses. We selected Tg(Prop1-cre)517Sac for our studies because it best reflected endogenous Prop1 expression. Tg(Prop1-cre)432Sac displayed a similar degree of ectopic cre activity, and cre reporter expression was detectable before endogenous Prop1 expression. Tg(Prop1-cre)506Sac displayed a higher rate of ectopic expression (data not shown). All data for the exon 2 Prop1-cre presented here was generated using Tg(Prop1-cre)517Sac and will be referred to from here on as Prop1-cre.
Figure 2.
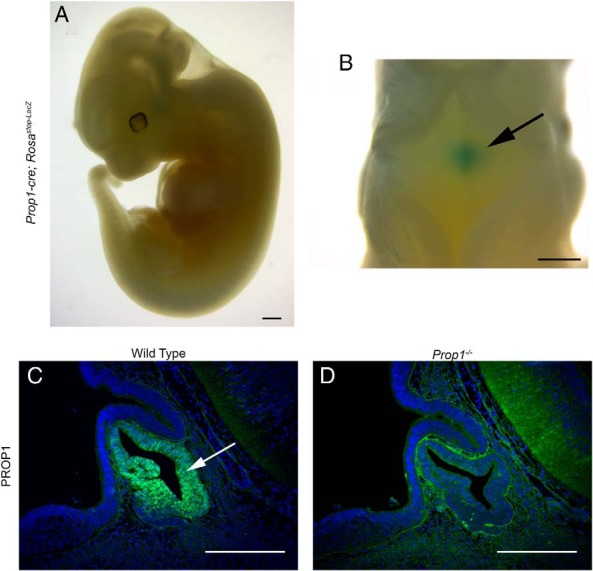
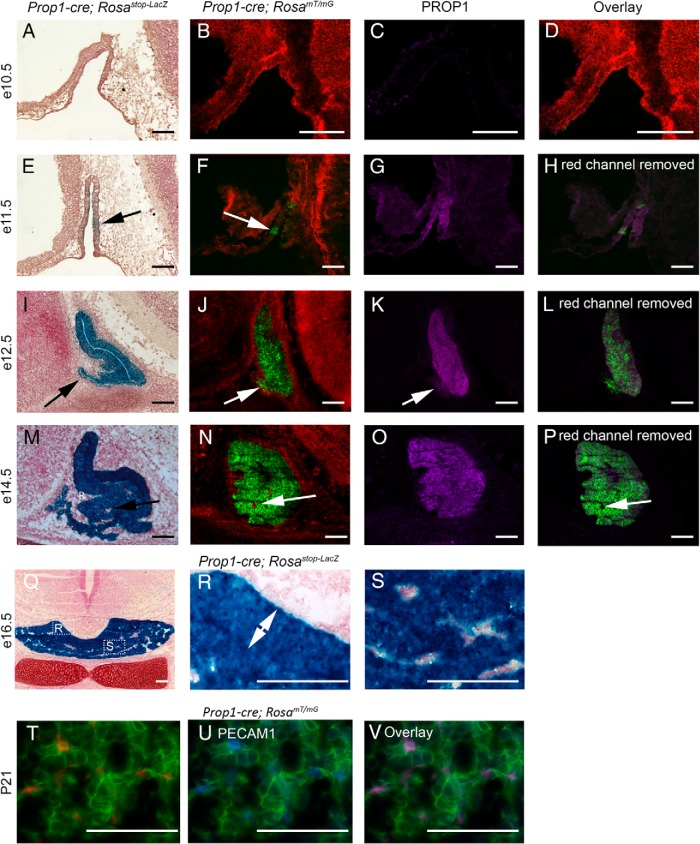
Pituitary-specific Prop1-cre activity and PROP1 immunostaining. A and B, Whole mount X-gal-stained e11.5 Prop1-cre; Rosastop-Lacz embryo. Blue indicates LacZ activity and cre expression. A, Lateral view. B, Dorsal view. Arrow indicates the developing pituitary gland as seen through the hindbrain. C and D, PROP1 expression (green) assayed by immunostaining, and counterstained with DAPI (blue), on e12.5 sagittal sections. C, Wild type. Arrow indicates PROP1 expression in Rathke's pouch. D, Prop1−/−. Scale bars, 500 μm (A and B) and 100 μm (C and D).
PROP1 antibodies have been used to detect mouse and rat PROP1 in pituitary sections (20, 46). We characterized the specificity of the PROP1 antibody B directed against the N-terminus by performing immunofluorescence for PROP1 on e12.5 wild-type and Prop1−/− tissues. As expected, strong, nuclear PROP1 immunoreactivity is detected throughout the middle zone of Rathke's pouch in wild-type embryos at e12.5, with lighter staining in the dorsal aspect of the pouch, and no staining in the rostral tip (Figure 2C). In contrast, no PROP1 staining was detected in the pituitaries of Prop1−/− embryos at e12.5, confirming the specificity of the antibody (Figure 2D). There is background staining in nonpituitary tissues, but it is not nuclear.
To determine whether the Prop1-cre transgene accurately reflects the endogenous PROP1 expression pattern, we used the Rosastop-LacZ and RosamT/mG reporter lines to characterize the timing of Prop1-cre activity in comparison with PROP1 expression by immunostaining. At e10.5, no cre activity is observed in Rathke's pouch of Prop1-cre; Rosastop-LacZ or Prop1-cre; RosamT/mG embryos (Figure 3, A and B). After imaging Prop1-cre; RosamT/mG cryosections for fluorescence generated from the RosamT/mG allele, the sections were processed to quench the endogenous fluorescence and then subjected to PROP1 immunostaining. Overlaying the initial Prop1-cre; RosamT/mG fluorescence image with the PROP1 immunostaining allows for direct comparison of Prop1-cre activity and PROP1 expression. Nuclear PROP1 is not detected in Rathke's pouch at e10.5 (Figure 3, C and D).
Figure 3.
Prop1-cre activity compared with PROP1 expression. A–D, e10.5 sagittal sections. E–H, e11.5 sagittal sections. I–L, e12.5 sagittal sections. M–P, e14.5 sagittal sections. Q–S, e16.5 coronal sections. T–V, Postnatal day 21 (P21) pituitary coronal sections. A, E, I, M, Q, R, and S, Prop1-cre; Rosastop-LacZ embryos X-gal stained (blue) to indicate LacZ activity caused by cre recombination, and counterstained with neutral red. Arrow in E indicates small region of LacZ activity. Arrow in I indicates LacZ activity in the rostral tip. B, F, J, and N, Prop1-cre; RosamT/mG embryos where red fluorescence indicates no cre activity and green fluorescence indicates cre recombination. Arrow in F indicates a small region of cre activity. Arrow in J indicates cre activity in the rostral tip. Arrow in N indicates a region with no cre activity (red). C, G, K, and O, PROP1 immunostaining (purple) on the same sections as B, F, J, and N. Arrow in K indicates no expression of PROP1 in the rostral tip. D, Overlay of Prop1-cre; RosamT/mG and PROP1 immunostaining images. H, L, and P, Overlay of eGFP fluorescence and PROP1 immunostaining. Arrow in P indicates a small region with no cre activity and no PROP1 expression in the pituitary anterior lobe. R and S, Boxed regions in Q, presented at higher magnification. Double arrow in R indicates the intermediate lobe. T, Fluorescence from Prop1-cre; RosamT/mG tdTOMATO (red) and eGFP (green). U, Same image as T, with the red channel removed and immunostained for PECAM1 (blue). V, Same image as T with all 3 channels, demonstrating colocalization of PECAM1 with tdTOMATO. Sale bars, 100 μm.
As expected, Prop1-cre mediated recombination is detected in Rathke's pouch at e11.5, using both reporter lines (Figure 3, E–G). Some cells in Rathke's pouch are negative for PROP1 immunoreactivity at e11.5, demonstrating that the initial expression of PROP1 does not occur simultaneously in all pituitary progenitor cells. The overlay of PROP1 immunostaining and RosamT/mG fluorescence indicates that PROP1 expression is detectable before Prop1-cre activity (Figure 3H). At e12.5, Prop1-cre activity detected by Rosastop-LacZ and RosamT/mG appears uniform throughout Rathke's pouch and extends to the rostral tip (Figure 3, I and J). In contrast, PROP1 protein is not expressed in the forming rostral tip (Figure 3K). Therefore, cells in the rostral tip must have expressed PROP1 before their migration into the forming anterior lobe.
By e14.5, Prop1-cre activity as assayed by both Rosastop-LacZ and RosamT/mG is uniform throughout the luminal area and the forming anterior lobe (Figure 3, M and N). PROP1 expression is still observed in the luminal area, but is down-regulated in the differentiating cells of the anterior lobe (Figure 3O). At e14.5, mesenchymal cells, which are Prop1-cre and PROP1 negative (Figure 3, M–P) appear to be entering the developing anterior lobe. Both the intermediate (Figure 3, Q and R) and anterior lobes (Figure 3, Q and S) are almost completely comprised of LacZ positive cells at e16.5. Those cells that do not express LacZ at e16.5 are likely part of the forming pituitary vasculature (Figure 3, Q and S). Immunostaining for PECAM1 on Prop1-cre; RosamT/mG pituitaries at 3 weeks of age confirm that the nonrecombined, red fluorescent cells in the anterior lobe are endothelial cells (Figure 3, T–V).
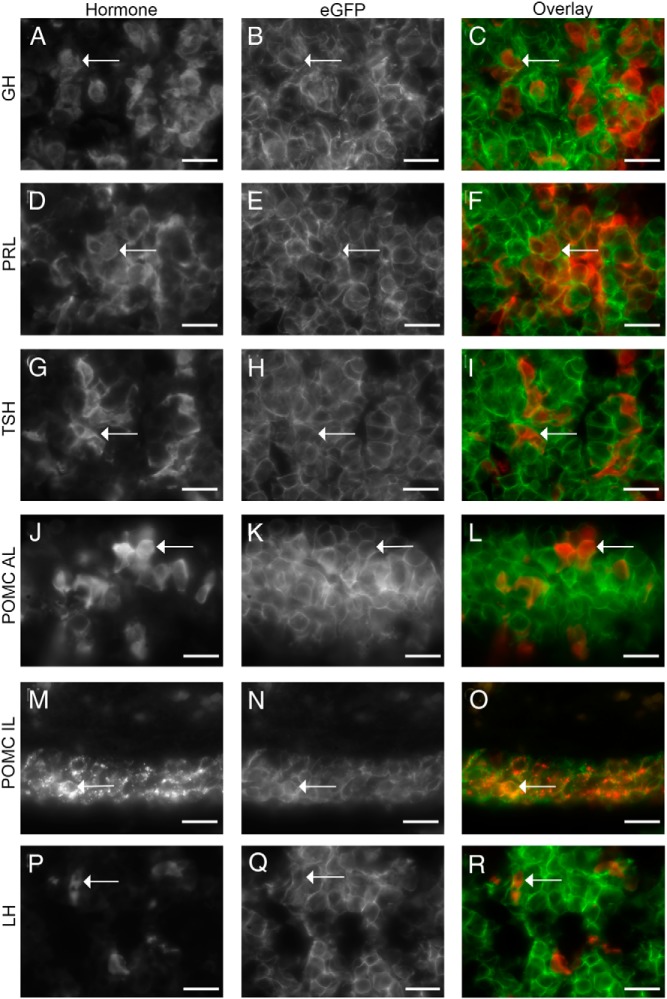
The descendants of the Prop1-cre-expressing progenitor cells are permanently marked by the recombination event, as evidenced by the green fluorescence in the anterior lobe of a postnatal day 21 Prop1-cre; RosamT/mG pituitary (Figure 1E). Similar results were obtained with the 2 other Prop1-cre transgenic lines. To confirm that the lineage-tracing experiments indicate that all hormone-expressing cell types are descended from a Prop1 progenitor, we assessed the colocalization of eGFP and individual hormones in Prop1-cre; RosamT/mG mice. As expected, we observed PIT1 lineage cells that were eGFP positive and expressed the hormones GH, TSH, or PRL (Figure 4, A–I). We also observed eGFP positive cells that expressed POMC in the anterior lobe (Figure 4, J–L) and intermediate lobe (Figure 4, M–O), indicating that Prop1-expressing progenitors gave rise to both corticotropes and melanotropes. LHβ immunostaining was also observed in eGFP positive anterior lobe cells (Figure 4, P–R). Therefore, all hormone-expressing cell types of the pituitary anterior and intermediates lobe arise from Prop1-expressing progenitors.
Figure 4.
All anterior and intermediate lobe hormone-expressing cell types are derived from a Prop1-cre progenitor. A, D, G, J, M, and P, Gray scale images of immunostaining for hormones from Prop1-cre; RosamT/mG pituitaries. GH (A), PRL (D), TSHβ (G), POMC in the anterior lobe (J), POMC in the intermediate lobe (M), and LHβ (P). (B, E, H, K, N, and Q) Gray scale images of eGFP detection of same images as A, D, G, J, M, and P. C, F, I, L, O, and R, Overlay of hormone (red) and eGFP (green) images. Arrows indicate the same cell in each image. Immunostainings for A–L and P–R were performed on P21 pituitaries, while immunostainings for M–O were performed on e18.5 pituitaries. Scale bars, 20 μm.
Discussion
We have generated Prop1-cre mouse strains that deliver efficient and highly penetrant cre activity early in pituitary gland development, in Rathke's pouch. Our lineage tracing experiments demonstrate that Prop1-cre marked progenitors give rise to all hormone secreting cell types of the anterior and intermediate lobes of the pituitary. This result suggests that there is a common Prop1-expressing progenitor pool in Rathke's pouch that gives rise to all hormone-producing cells of the anterior and intermediate lobes. Prop1 mutant mice are able to produce corticotropes, melanotropes, and gonadotropes, indicating that Prop1 is not necessary for the formation of these cell types in mice, despite being expressed in a common progenitor (14–16).
Prop1 mutant mice have an increase in the epithelial-like cells that surround Rathke's cleft, and the developing mutant pituitaries fail to express Slug, a transcription factor involved in EMTs (32, 47). The essential function of Prop1 is likely to be in regulating the release of progenitor cells from the marginal zone and promoting differentiation via an EMT-like mechanism.
PROP1 forms a complex with β-CATENIN to activate Pou1f1 transcription (13). However, Prop1 may not be absolutely required to activate Pou1f1. Small clonal populations of Pou1f1-expressing thyrotropes, somatotropes, and lactotropes form in the anterior lobe of Prop1df/df mice (48). This observation is in alignment with the fact that patients with PROP1 loss of function mutations typically present with short stature and low but detectable GH production, suggesting the initial presence of functional somatotropes (18, 49). Patients tend to subsequently lose production of other hormones, including reproductive hormones and ACTH. For example, patients may not present with short stature and begin GH replacement therapy until age 6, sex steroid replacement at age 15, and hydrocortisone replacement at age 18 or even later (18). This contrasts with the typical presentation of patients with PIT1 loss of function mutations, with onset of short stature and other hormone deficiencies before the age of 2 and no progressive features (49). The progressive loss of hormonal production by all anterior pituitary cell types may occur because PROP1 regulates a common pool of pituitary progenitors, not just the PIT1 lineage, and the progenitor pool may be exhausted over the life of the patient. It is also possible that activation of PIT1 is less dependent upon PROP1 in humans than in mice, resulting in an early expansion of the PIT1 lineage which cannot be sustained.
The expression of Prop1 in a common pituitary progenitor pool helps to explain the progressive loss of all hormone cell types in humans, but does not adequately explain why mice do not evolve ACTH deficiency. Both Prop1df/df and Prop1−/− mice on mixed genetic backgrounds or backcrossed onto a C57BL/6 background for 4 generations maintained a functional pituitary adrenal axis throughout life, with no loss of corticotropes in aged mice (50). Although we cannot rule out the possibility that some genetic background might provoke ACTH deficiency in mice, it seems that there are species differences in the requirement for PROP1 to initiate and maintain pituitary function. Longitudinal studies in humans and analysis of very large human pedigrees with PROP1 mutations reveals that the age of onset and degree of hormone insufficiency can vary even in individuals with the same mutation (18, 51). Mouse Prop1 and Hes1, a mediator of the Notch signaling pathway, regulate the pool of common pituitary progenitor cells such that loss of both Prop1 and Hes1 leads to premature differentiation of corticotropes and a smaller pituitary gland (32). Perhaps mice maintain sufficient Notch signaling in the absence of Prop1 to maintain a progenitor pool sufficient for continual corticotrope production, whereas humans do not.
PROP1 patients sometimes present with a “waxing and waning” pituitary phenotype, where sequential magnetic resonance imaging scans reveal that the pituitary undergoes periods characterized by hyperplasia and hypoplasia (52, 53). This phenotype has not been observed in mice. Instead, the mouse embryonic pituitary is dysmorphic, but has a normal volume, and the postnatal pituitary is hypoplastic due to reduced proliferation and increased apoptosis (31). The rescue of the mouse Prop1 deficiency with an HsPROP1 BAC demonstrates that the cis-regulatory elements and trans acting factors are sufficiently conserved for HsPROP1 to be expressed in mice and for it to activate the necessary downstream targets in mice. These experiments do not address the possibility of a temporal difference in PROP1 expression between mice and humans, which could contribute to the differential phenotypes.
The generation of a well-characterized Prop1-cre strain expands the available repertoire of genetic reagents available for tissue-specific conditional inactivation in the pituitary gland. There are several existing lines that target the oral ectoderm before Rathke's pouch formation, including Pitx1-cre, Hesx1-cre, and Foxg1-cre, but these promoters also drive cre expression in other tissues besides the pituitary gland (54–56). The Prop1-cre strains reported here offer pan-pituitary deletion from e11.5-e12.5 onward in most embryos. This complements other cre strains that are available for cell-specific deletion, such as the improved Cga-cre that is activated at e11.5 and promotes deletion in the thyrotropes and gonadotropes (35). The Pou1f1-cre, Sf1-cre, and GnRHR-cre (GRIC) mediate recombination during the differentiation process, beginning at e14.5, in a subset of cell types in the anterior lobe (13, 57, 58). The Gh-cre, Tshb-cre, Lhb-cre, Prl-cre, and Pomc-cre all mediate recombination in the specific pituitary hormone cell types, after terminal differentiation (59–64). The Prop1-cre complements these genetic resources by expressing cre after progenitor cell formation from e9.5 to e10.5, but before cell specification at e13.5–e14.5, and it targets all hormone secreting cell types of the pituitary gland.
Acknowledgments
We thank Thom Saunders, Wanda Filipiak, Maggie Van Keuren, and Michael Zeidler of the University of Michigan Transgenic Animal Model Core for preparation of BAC constructs and transgenic mice. The PROP1 antibody was a generous gift of Aimee Ryan of the Departments of Pediatrics and Human Genetics, McGill University, Montréal, Québec.
This work was supported by Eunice Kennedy Shriver National Institute of Child Health and Human Development Grant R0130428 (to S.A.C.), International Endocrine Scholar Program (to M.I.P.-M.), the Transgenic Animal Model Core (National Institutes of Health Grants CA46592, AR20557, and DK34933), and the University of South Carolina (start-up funding to S.W.D.).
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- BAC
- bacterial artificial chromosome
- CNS
- conserved noncoding sequence
- DAPI
- 4′, 6-diamidino-2-phenylindole
- e
- embryonic day of development
- eGFP
- enhanced green fluorescent protein
- EMT
- epithelial to mesenchymal transition
- kb
- kilobases
- POMC
- proopiomelanocortin
- PRL
- prolactin
- tdTOMATO
- tandem dimeric Tomato
- TSA
- tyramide signal amplification
- X-gal
- 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside.
References
- 1. Alatzoglou KS, Dattani MT. Genetic forms of hypopituitarism and their manifestation in the neonatal period. Early Hum Dev. 2009;85:705–712. [DOI] [PubMed] [Google Scholar]
- 2. Vallette-Kasic S, Brue T, Pulichino AM, et al. Congenital isolated adrenocorticotropin deficiency: an underestimated cause of neonatal death, explained by TPIT gene mutations. J Clin Endocrinol Metab. 2005;90:1323–1331. [DOI] [PubMed] [Google Scholar]
- 3. Li S, Crenshaw EB, 3rd, Rawson EJ, Simmons DM, Swanson LW, Rosenfeld MG. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene pit-1. Nature. 1990;347:528–533. [DOI] [PubMed] [Google Scholar]
- 4. Pfäffle RW, DiMattia GE, Parks JS, et al. Mutation of the POU-specific domain of Pit-1 and hypopituitarism without pituitary hypoplasia. Science. 1992;257:1118–1121. [DOI] [PubMed] [Google Scholar]
- 5. Sornson MW, Wu W, Dasen JS, et al. Pituitary lineage determination by the Prophet of Pit-1 homeodomain factor defective in Ames dwarfism. Nature. 1996;384:327–333. [DOI] [PubMed] [Google Scholar]
- 6. Wu W, Cogan JD, Pfäffle RW, et al. Mutations in PROP1 cause familial combined pituitary hormone deficiency. Nat Genet. 1998;18:147–149. [DOI] [PubMed] [Google Scholar]
- 7. Skowronska-Krawczyk D, Ma Q, Schwartz M, et al. Required enhancer-matrin-3 network interactions for a homeodomain transcription program. Nature. 2014;514:257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bodner M, Castrillo JL, Theill LE, Deerinck T, Ellisman M, Karin M. The pituitary-specific transcription factor GHF-1 is a homeobox-containing protein. Cell. 1988;55:505–518. [DOI] [PubMed] [Google Scholar]
- 9. Gordon DF, Haugen BR, Sarapura VD, Nelson AR, Wood WM, Ridgway EC. Analysis of Pit-1 in regulating mouse TSH β promoter activity in thyrotropes. Mol Cell Endocrinol. 1993;96:75–84. [DOI] [PubMed] [Google Scholar]
- 10. Ingraham HA, Chen RP, Mangalam HJ, et al. A tissue-specific transcription factor containing a homeodomain specifies a pituitary phenotype. Cell. 1988;55:519–529. [DOI] [PubMed] [Google Scholar]
- 11. Lin SC, Li S, Drolet DW, Rosenfeld MG. Pituitary ontogeny of the Snell dwarf mouse reveals Pit-1-independent and Pit-1-dependent origins of the thyrotrope. Development. 1994;120:515–522. [DOI] [PubMed] [Google Scholar]
- 12. Steinfelder HJ, Hauser P, Nakayama Y, et al. Thyrotropin-releasing hormone regulation of human TSHB expression: role of a pituitary-specific transcription factor (Pit-1/GHF-1) and potential interaction with a thyroid hormone-inhibitory element. Proc Natl Acad Sci USA. 1991;88:3130–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Olson LE, Tollkuhn J, Scafoglio C, et al. Homeodomain-mediated β-catenin-dependent switching events dictate cell-lineage determination. Cell. 2006;125:593–605. [DOI] [PubMed] [Google Scholar]
- 14. Bartke A, Goldman BD, Bex F, Dalterio S. Effects of prolactin (PRL) on pituitary and testicular function in mice with hereditary PRL deficiency. Endocrinology. 1977;101:1760–1766. [DOI] [PubMed] [Google Scholar]
- 15. Buckwalter MS, Katz RW, Camper SA. Localization of the panhypopituitary dwarf mutation (df) on mouse chromosome 11 in an intersubspecific backcross. Genomics. 1991;10:515–526. [DOI] [PubMed] [Google Scholar]
- 16. Tang K, Bartke A, Gardiner CS, Wagner TE, Yun JS. Gonadotropin secretion, synthesis, and gene expression in human growth hormone transgenic mice and in Ames dwarf mice. Endocrinology. 1993;132:2518–2524. [DOI] [PubMed] [Google Scholar]
- 17. Nasonkin IO, Ward RD, Raetzman LT, et al. Pituitary hypoplasia and respiratory distress syndrome in Prop1 knockout mice. Hum Mol Genet. 2004;13:2727–2735. [DOI] [PubMed] [Google Scholar]
- 18. Böttner A, Keller E, Kratzsch J, et al. PROP1 mutations cause progressive deterioration of anterior pituitary function including adrenal insufficiency: a longitudinal analysis. J Clin Endocrinol Metab. 2004;89:5256–5265. [DOI] [PubMed] [Google Scholar]
- 19. Chen J, Gremeaux L, Fu Q, Liekens D, Van Laere S, Vankelecom H. Pituitary progenitor cells tracked down by side population dissection. Stem Cells. 2009;27:1182–1195. [DOI] [PubMed] [Google Scholar]
- 20. Garcia-Lavandeira M, Quereda V, Flores I, et al. A GRFa2/Prop1/stem (GPS) cell niche in the pituitary. PLoS One. 2009;4:e4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pulichino AM, Vallette-Kasic S, Tsai JP, Couture C, Gauthier Y, Drouin J. Tpit determines alternate fates during pituitary cell differentiation. Genes Dev. 2003;17:738–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dasen JS, O'Connell SM, Flynn SE, et al. Reciprocal interactions of Pit1 and GATA2 mediate signaling gradient-induced determination of pituitary cell types. Cell. 1999;97:587–598. [DOI] [PubMed] [Google Scholar]
- 23. Gordon DF, Lewis SR, Haugen BR, et al. Pit-1 and GATA-2 interact and functionally cooperate to activate the thyrotropin β-subunit promoter. J Biol Chem. 1997;272:24339–24347. [DOI] [PubMed] [Google Scholar]
- 24. Gordon DF, Woodmansee WW, Black JN, et al. Domains of Pit-1 required for transcriptional synergy with GATA-2 on the TSH β gene. Mol Cell Endocrinol. 2002;196:53–66. [DOI] [PubMed] [Google Scholar]
- 25. Castinetti F, Davis SW, Brue T, Camper SA. Pituitary stem cell update and potential implications for treating hypopituitarism. Endocr Rev. 2011;32:453–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fauquier T, Rizzoti K, Dattani M, Lovell-Badge R, Robinson IC. SOX2-expressing progenitor cells generate all of the major cell types in the adult mouse pituitary gland. Proc Natl Acad Sci USA. 2008;105:2907–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davis SW, Ellsworth BS, Peréz Millan MI, et al. Pituitary gland development and disease: from stem cell to hormone production. Curr Top Dev Biol. 2013;106:1–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bilodeau S, Roussel-Gervais A, Drouin J. Distinct developmental roles of cell cycle inhibitors p57Kip2 and p27Kip1 distinguish pituitary progenitor cell cycle exit from cell cycle reentry of differentiated cells. Mol Cell Biol. 2009;29:1895–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Davis SW, Mortensen AH, Camper SA. Birthdating studies reshape models for pituitary gland cell specification. Dev Biol. 2011;352:215–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Seuntjens E, Denef C. Progenitor cells in the embryonic anterior pituitary abruptly and concurrently depress mitotic rate before progressing to terminal differentiation. Mol Cell Endocrinol. 1999;150:57–63. [DOI] [PubMed] [Google Scholar]
- 31. Ward RD, Stone BM, Raetzman LT, Camper SA. Cell proliferation and vascularization in mouse models of pituitary hormone deficiency. Mol Endocrinol. 2006;20:1378–1390. [DOI] [PubMed] [Google Scholar]
- 32. Himes AD, Raetzman LT. Premature differentiation and aberrant movement of pituitary cells lacking both Hes1 and Prop1. Dev Biol. 2009;325:151–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet. 2001;2:769–779. [DOI] [PubMed] [Google Scholar]
- 34. Watkins-Chow DE, Buckwalter MS, Newhouse MM, Lossie AC, Brinkmeier ML, Camper SA. Genetic mapping of 21 genes on mouse chromosome 11 reveals disruptions in linkage conservation with human chromosome 5. Genomics. 1997;40:114–122. [DOI] [PubMed] [Google Scholar]
- 35. Pérez-Millán MI, Zeidler MG, Saunders TL, Camper SA, Davis SW. Efficient, specific, developmentally appropriate cre-mediated recombination in anterior pituitary gonadotropes and thyrotropes. Genesis. 2013;51:785–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shimshek DR, Kim J, Hübner MR, et al. Codon-improved Cre recombinase (iCre) expression in the mouse. Genesis. 2002;32:19–26. [DOI] [PubMed] [Google Scholar]
- 37. Lee EC, Yu D, Martinez de Velasco J, et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. [DOI] [PubMed] [Google Scholar]
- 38. Van Keuren ML, Gavrilina GB, Filipiak WE, Zeidler MG, Saunders TL. Generating transgenic mice from bacterial artificial chromosomes: transgenesis efficiency, integration and expression outcomes. Transgenic Res. 2009;18:769–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cushman LJ, Watkins-Chow DE, Brinkmeier ML, et al. Persistent Prop1 expression delays gonadotrope differentiation and enhances pituitary tumor susceptibility. Hum Mol Genet. 2001;10:1141–1153. [DOI] [PubMed] [Google Scholar]
- 40. Hogan B, Beddington R, Costantini F, Lacey E. Manipulating the Mouse Embryo: A Laboratory Manual. 2nd Edition Plainview, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 41. Kendall SK, Gordon DF, Birkmeier TS, et al. Enhancer-mediated high level expression of mouse pituitary glycoprotein hormone α-subunit transgene in thyrotropes, gonadotropes, and developing pituitary gland. Mol Endocrinol. 1994;8:1420–1433. [DOI] [PubMed] [Google Scholar]
- 42. Ward RD, Davis SW, Cho M, et al. Comparative genomics reveals functional transcriptional control sequences in the Prop1 gene. Mamm Genome. 2007;18:521–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhu X, Zhang J, Tollkuhn J, et al. Sustained Notch signaling in progenitors is required for sequential emergence of distinct cell lineages during organogenesis. Genes Dev. 2006;20:2739–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. [DOI] [PubMed] [Google Scholar]
- 45. Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. [DOI] [PubMed] [Google Scholar]
- 46. Suh H, Gage PJ, Drouin J, Camper SA. Pitx2 is required at multiple stages of pituitary organogenesis: pituitary primordium formation and cell specification. Development. 2002;129:329–337. [DOI] [PubMed] [Google Scholar]
- 47. Ward RD, Raetzman LT, Suh H, Stone BM, Nasonkin IO, Camper SA. Role of PROP1 in pituitary gland growth. Mol Endocrinol. 2005;19:698–710. [DOI] [PubMed] [Google Scholar]
- 48. Gage PJ, Roller ML, Saunders TL, Scarlett LM, Camper SA. Anterior pituitary cells defective in the cell-autonomous factor, df, undergo cell lineage specification but not expansion. Development. 1996;122:151–160. [DOI] [PubMed] [Google Scholar]
- 49. Turton JP, Reynaud R, Mehta A, et al. Novel mutations within the POU1F1 gene associated with variable combined pituitary hormone deficiency. J Clin Endocrinol Metab. 2005;90:4762–4770. [DOI] [PubMed] [Google Scholar]
- 50. Nasonkin IO, Ward RD, Bavers DL, et al. Aged PROP1 deficient dwarf mice maintain ACTH production. PLoS One. 2011;6:e28355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Reynaud R, Chadli-Chaieb M, Vallette-Kasic S, et al. A familial form of congenital hypopituitarism due to a PROP1 mutation in a large kindred: phenotypic and in vitro functional studies. J Clin Endocrinol Metab. 2004;89:5779–5786. [DOI] [PubMed] [Google Scholar]
- 52. Nascif SO, Vieira TC, Ramos-Dias JC, Lengyel AM, Abucham J. Waxing and waning of a pituitary mass in a young woman with combined pituitary hormone deficiency (CPHD) due to a PROP-1 mutation. Pituitary. 2006;9:47–52. [DOI] [PubMed] [Google Scholar]
- 53. Turton JP, Mehta A, Raza J, et al. Mutations within the transcription factor PROP1 are rare in a cohort of patients with sporadic combined pituitary hormone deficiency (CPHD). Clin Endocrinol (Oxf). 2005;63:10–18. [DOI] [PubMed] [Google Scholar]
- 54. Andoniadou CL, Signore M, Sajedi E, et al. Lack of the murine homeobox gene Hesx1 leads to a posterior transformation of the anterior forebrain. Development. 2007;134:1499–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hébert JM, McConnell SK. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev Biol. 2000;222:296–306. [DOI] [PubMed] [Google Scholar]
- 56. Treier M, Gleiberman AS, O'Connell SM, et al. Multistep signaling requirements for pituitary organogenesis in vivo. Genes Dev. 1998;12:1691–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bingham NC, Verma-Kurvari S, Parada LF, Parker KL. Development of a steroidogenic factor 1/Cre transgenic mouse line. Genesis. 2006;44:419–424. [DOI] [PubMed] [Google Scholar]
- 58. Wen S, Schwarz JR, Niculescu D, et al. Functional characterization of genetically labeled gonadotropes. Endocrinology. 2008;149:2701–2711. [DOI] [PubMed] [Google Scholar]
- 59. Balthasar N, Coppari R, McMinn J, et al. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron. 2004;42:983–991. [DOI] [PubMed] [Google Scholar]
- 60. Castinetti F, Brinkmeier ML, Gordon DF, et al. PITX2 AND PITX1 regulate thyrotroph function and response to hypothyroidism. Mol Endocrinol. 2011;25:1950–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Castrique E, Fernandez-Fuente M, Le Tissier P, Herman A, Levy A. Use of a prolactin-Cre/ROSA-YFP transgenic mouse provides no evidence for lactotroph transdifferentiation after weaning, or increase in lactotroph/somatotroph proportion in lactation. J Endocrinol. 2010;205:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Charles MA, Mortensen AH, Potok MA, Camper SA. Pitx2 deletion in pituitary gonadotropes is compatible with gonadal development, puberty, and fertility. Genesis. 2008;46:507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nasonkin IO, Potok MA, Camper SA. Cre-mediated recombination in pituitary somatotropes. Genesis. 2009;47:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Luque RM, Amargo G, Ishii S, et al. Reporter expression, induced by a growth hormone promoter-driven Cre recombinase (rGHp-Cre) transgene, questions the developmental relationship between somatotropes and lactotropes in the adult mouse pituitary gland. Endocrinology. 2007;148:1946–1953. [DOI] [PubMed] [Google Scholar]