Abstract
The biological activity of insulin and the insulin-like growth factor (IGF) ligands, IGF-I and IGF-II, is based in part on the relative abundance and distribution of their target receptors: the insulin receptor (IR) splice variants A (IR-A) and B (IR-B) and IGF 1 receptor (IGF-1R). However, the relative quantity of all three receptors in human tissues has never been measured together on the same scale. Due to the high homology between insulin receptor (IR)-A and IR-B proteins and lack of antibodies that discern the two IR splice variants, their mRNA sequence is the most reliable means of distinguishing between the receptors. Hence, highly specific primers for IR-A, IR-B, and IGF-1R mRNA were designed to accurately detect all three receptors by quantitative RT-PCR and enable direct quantification of relative receptor expression levels. A standard concentration curve of cDNA from each receptor was performed. Assay specificity was tested using competition assays and postamplification analysis by gel electrophoresis and cloning. Forward and reverse primer concentrations were optimized to ensure equal efficiencies across primer pairs. This assay enables a specific molecular signature of IGF/insulin signaling receptors to be assayed in different tissues, cell types, or cancers.
The complexities of insulin and insulin-like growth factor (IGF) signaling include the existence of multiple receptors through which the ligands, insulin, IGF-I, and IGF-II, can exert biological actions. These receptors, including the physiologically distinct insulin receptor (IR) splice variants IR-A (exon 11−) and IR-B (exon 11+), and the IGF 1 receptor (IGF-1R), are collectively ubiquitous, although they vary individually in their expression profiles. The three receptors share a large degree of sequence and structural homology and can form either homodimers or heterodimers (1). Upon ligand binding, downstream cellular responses include mitogenesis, survival, differentiation, and metabolic regulation (2).
The levels of the IR isoforms and IGF-1R vary across cell types, tissues, developmental stages, and pathological states (2). For example, developing tissues and certain cancers, including breast cancers, have proportionately higher levels of the IR-A isoform (3–8). Altered splicing patterns of IR are also described in skeletal muscle, liver, and adipose tissue of people with type 2 diabetes; however, reported levels vary, likely as a result of differences in measurement techniques as well as progression of diabetes within subjects (9–12). Hence, diabetes and other pathological states may alter the regulation of IR mRNA splicing and subsequently alter the cellular response to insulin as a result of an altered IR-A to IR-B ratio. Insulin analogs are used therapeutically for glycemic control in people with diabetes, and likely have different potency and kinetics of binding to each receptor than endogenous insulin (13, 14). Thus, determining relative levels of the three receptors, IGF-1R, IR-A, and IR-B, in a given tissue or disease state is likely to have predictive value as to potential biological outcomes.
In prior studies, we developed a sensitive and specific quantitative RT-PCR (qRT-PCR) assay to measure relative levels of the three receptors in mouse tissues and demonstrated the relative levels of the receptors were indeed predictive of physiological state and ligand function in normal mammary gland and brain (3, 15). In human tissues or cell lines, the relative quantity of the receptors has never been measured together on the same scale in a single assay. This capability would enable a specific molecular signature of IGF/insulin signaling receptors to be determined in different tissues, cell types, or cancers. The goal of this study was to develop a highly specific assay to determine relative expression levels of the three human receptors, exon 11− IR-A, exon 11+ IR-B and IGF-1R, on the same scale within a particular tissue or cell type.
Materials and Methods
Specific methods for assay development
qRT-PCR plasmid standards for IR and IGF-1R were synthesized using the same techniques described previously for the rodent assay (3) but with the use of human-specific primers. Plasmid DNAs were cloned and sequenced; linearized DNA used for standards was purified using the MinElute reaction cleanup kit, microcentrifuge protocol (QIAGEN) and quantified by spectrophotometry. Copy number was calculated using the following formula: (DNA concentration [grams per microliter]/[plasmid length [base pairs] * 660]) * 6.022 × 1023 = molecules per microliter.
qRT-PCR primer design and optimization
IR-A, IR-B, and IGF-1R qRT-PCR primers were designed against the human IR and IGF-1R sequences using Primer Premier version 5 software (Premier Biosoft International), with attention to the standard criteria for the qRT-PCR primer design. Several primer sets were developed and tested; the optimal primer sequences and product sizes are shown in Table 1.
Table 1.
Primer Sequences and Product Sizes
| Receptor | Primer Sequence (5′–3′) | Product Size, bp |
|---|---|---|
| IR-A and IR-B reversea,b | GTC ACA TTC CCA ACA TCG CC | |
| IR-A forwarda | TTT TCG TCC CCA GGC CAT C | 58 |
| IR-B forwardb | CCC CAG AAA AAC CTC TTC AGG | 87 |
| IGF-1R forwardc | GGC ACA ATT ACT GCT CCA AAG AC | |
| IGF-1R reversec | CAA GGC CCT TTC TCC CCA C | 121 |
| IGF-1R reverse-truncatedd | CAA GGC CCT TTC TCC C | 121 |
| β-Actin forwarde | AGC CAT GTA CGT TGC TAT CCA | |
| β-Actin reversee | ACC GGA GTC CAT CAC GAT G | 79 |
NCBI reference sequencing NM_001079817.2; forward, 2630-2648; reverse, 2687-2668 (IR-A).
NCBI reference sequencing NM_000208.3; forward, 2637-2657; reverse, 2723-2704 (IR-B).
NCBI reference sequencing NM_000875.4; forward, 3019-3041; reverse, 3139-3121.
NCBI reference sequencing NM_000875.4; reverse, 3139-3124; note: the truncated primer was validated for specificity and efficiency and used for data shown in Figures 3 and 4.
NCBI reference sequencing NM_001101.3; forward, 474-494; reverse, 552-534.
To test the detection limit and efficiency of each primer pair individually, 107 molecules/μL of each plasmid standard was serially diluted and used as template in qRT-PCR assays with a dissociation stage and reaction conditions as described below to generate a standard curve of threshold cycle (Ct) value vs log copy number.
For competition assays, 107 copies of a nonspecific plasmid DNA target was mixed with each standard and used as template in qRT-PCR assays (ie, to test IR-A primer specificity, 107 copies of IR-B standard, or IGF-1R standard was mixed with the IR-A standard). Standard curves were generated as described for the rodent assay (3).
Further tests of assay specificity were performed on postamplification qRT-PCR products. qRT-PCR products were taken directly from a postamplification assay plate and analyzed by gel electrophoresis. A single band of size consistent with dissociation curve analysis was present for each primer pair. Postamplification products were also cloned, and sequences were confirmed against National Center for Biotechnology Information (NCBI) published sequences.
Primer efficiency tests
Primer efficiency was assessed by two methods to determine whether the reaction efficiencies of the endogenous control primers and IR isoform and IGF-1R primers were suitably equivalent for the ΔΔCt method of relative quantification. First, optimal primer concentrations were determined by assessing efficiency of the primers at different forward and reverse concentration combinations. The nine concentration combinations, including 0.125, 0.25, and 0.5 μM for each forward and reverse primer, were based on the optimal activity of the DNA polymerase used in the qRT-PCR reaction (Bio-Rad Laboratories). Efficiencies were determined using a 2-fold serial dilution of 50–3.125 ng human universal RNA (QIAGEN) reverse transcribed to cDNA. Primer efficiency was calculated using the following equation: E = (10 − [1/slope]) − 1, where slope is the slope of the standard curve when the Ct value is plotted vs the known amount of template, and E = 1.0 when the slope is approximately −3.33 (ie, 100% reaction efficiency). The primer combination yielding an efficiency closest to 100% was further tested in a minimum of four different qRT-PCR experiments, with each primer concentration plated in triplicate. A mean efficiency value for each primer set was calculated. Primers were synthesized at the W. M. Keck Foundation Oligo Synthesis Resource (Yale University, New Haven, Connecticut).
An additional test to determine whether the primer efficiencies were suitably equivalent for the ΔΔCt method of relative quantification was performed by plotting the log cDNA dilution vs the ΔCt value (Ct of target − Ct of endogenous control) and fitting the data with a linear regression curve.
Comparison of the validated primer set and an unvalidated primer set
To test the value of rigorous primer validation for use in qRT-PCR experiments, a second set of primers for each receptor was designed and used according to common practices. Primer sequences were designed using Primer Premier version 5 software or Primer Blast, for optimal activity at a 60°C annealing temperature. Primers for IR-A and IR-B did not include a common reverse primer sequence, and no further validation or optimization was performed. The unvalidated set of primers was assayed at a common concentration of 0.5 μM for each receptor. Levels of receptors were quantified in two different tissues using the validated set of primers and the unvalidated set of primers. The same β-actin primer set was used in both assays to normalize gene expression.
RNA was extracted from deidentified adipose tissue (n = 3) and placenta (n = 4), collected per guidelines set forth by the Yale Human Investigations Committee. Tissue was homogenized in TRIzol (Life Technologies). RNA was isolated according to the standard protocol and purified via RNeasy spin columns (QIAGEN). RNA concentration and purity was determined via Nanodrop 2000 (Thermoscientific). Statistical analysis for interreceptor and intrareceptor comparisons in the validated-primer assays was assessed by a two-way ANOVA and multiple-comparisons correction with either a Tukey's test or a Sidak's test, after log transformation. For the intrareceptor comparisons in the unvalidated-primer assays, a two-tailed Student's t test was applied after log transformation using GraphPad Prism (GraphPad Software, Inc).
Quantitative RT-PCR
qRT-PCRs were performed on the Applied Biosystems 7900HT fast real-time PCR system using associated Sequence Detection Systems software, version 2.2.2 (Applied Biosystems), the Applied Biosystems 7300 real-time PCR system using SDS software version 1.2.3, or the Bio-Rad CFX96 detection system (Bio-Rad Laboratories). The plasmid concentration curve and competition assays were performed on the Applied Biosystems systems with the thermal profile as follows: 50°C for 2 minutes, 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds, and 60°C (annealing temperature) for 1 minute. IR-A, IR-B, IGF-1R, and β-actin reactions contained final concentrations of the following: 1× QuantiTect SYBR Green PCR master mix (QIAGEN), 0.25 μM reverse, and 0.25, 0.05, or 0.03 μM forward primers. qRT-PCRs performed to generate standard curves (ie, for primer sensitivity tests and competition assays that used plasmid standards as template) were run using the absolute quantification assay and included a dissociation stage. For primer efficiency testing and gene expression comparisons in human tissue, qRT-PCR was performed using assay-specific primer concentrations, SYBR Green containing deoxynucleotide triphosphates, fluorescein, and reverse transcriptase (Bio-Rad Laboratories) and amplified in the Bio-Rad CFX96 system under the following cycling conditions: 95°C for 3 minutes and 40 cycles of 95°C for 15 seconds followed by 60°C for 30 seconds and 72°C for 25 seconds, 95°C for 1 minute, 55°C for 1 minute, and an increase to 95°C at 0.5°C increments.
Results
Human IR isoform and IGF-1R qRT-PCR assay development
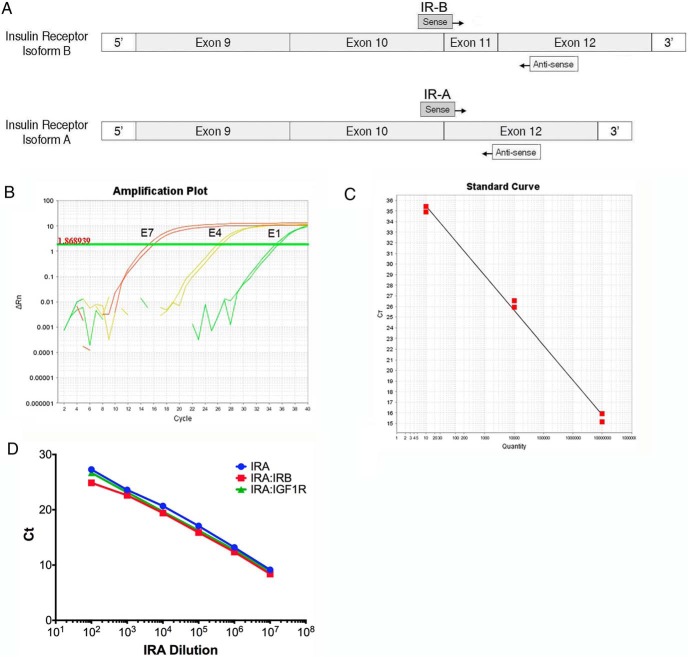
Due to the small sequence difference between IR-A and IR-B and the high degree of sequence homology between the IGF-1R and IR mRNAs, primer design is critical to developing a qRT-PCR assay to accurately detect all three receptors in human tissues. The validated mouse primers we developed previously were not valid for detection of human receptors due to sequence differences between species (3). Moreover, distinct primers in different regions of the receptors were necessary for obtaining high sensitivity and specificity for the human receptor sequences. As for the mouse assay, we applied an isoform-specific strategy identified by Vandenbroucke et al (16) using a primer that spans the exon junction specific to each isoform. For the human primers, we designed a common reverse primer that anneals within exon 12 to reduce interassay variability and obtain similar PCR kinetics. Specific forward primers were designed against exon/exon junctions unique to each IR isoform (Figure 1A). IGF-1R primers were targeted to a region of low IR homology.
Figure 1.
Human IR isoform-specific qRT-PCR primer design, and validation. A, Schematic representation of IR-B (exon 11+) and IR-A (exon 11−) primer design strategy. Both assays use a common reverse primer that anneals within exon 12 and isoform-specific forward primers that anneal across exon-exon junctions unique to each isoform. B, Sample amplification plot for IR-B assay using serial dilution of IR-B standard as template. Numbers to the left of each amplification curve indicate amount of standard used as template in each reaction. Horizontal line indicates threshold determined by Applied Biosystems sequence detection systems software. No template control reactions showed no amplification above baseline (not shown). C, A standard curve generated from the amplification plot of Ct vs log concentration of the plasmid standards for IR-B (slope = −3.33). D, Competition assay. A standard curve plot for IR-A (blue) is overlaid with standard curve plots using IR-A standards plus 107 copies of IR-B (red) or IGF-1R (green) standard at each data point.
For primer validation, 10-fold serial dilutions of plasmid standards were used as templates with each qRT-PCR primer pair. A sample amplification plot of IR-B standards demonstrates these primers are accurate over 6 orders of magnitude from 107 to 10 copies (Figure 1B). Figure 1C shows the standard curve from the IR-B amplification plot shown in Figure 1B. Similar results were obtained for the IR-A and IGF-1R standards and primer pairs (data not shown).
Competition assays were used to test the capability of the primers to specifically amplify their target in the presence of excess nonspecific, homologous target, as may be the case when analyzing total RNA from a biological sample. Serial dilution of standards was repeated in a fixed excess of 107 copies of competitor plasmid. Amplification plots were generated from serial dilution of IR-A standards in a fixed concentration of IR-B plasmid competitor or IGF-1R plasmid competitor (Figure 1D). There was no significant change in IR-A amplification curves down to 100 copies with addition of either competitor (slope = −3.32). Similarly, the efficiencies of IR-B and IGF-1R assays were not significantly changed in the presence of 107 copies of the other two plasmid competitors (data not shown). These data demonstrate that the primers can detect at least 100 copies of their target in the presence of 107 excess copies of highly homologous targets.
Assay optimization
The efficiency of all primer pairs must be approximately equivalent to allow accurate comparison of gene expression using the ΔΔCt method of relative quantification (Applied Biosystems user bulletin 2 P/N 4303859). Therefore, each primer pair was validated based on efficiency as well as sensitivity and specificity. The primer pairs of IR-A, IR-B, and IGF-1R were each tested for efficiency in relation to the endogenous control, β-actin. Primer efficiency (E) was calculated using the following equation: E = (10 − [1/slope]) − 1, where slope is the slope of the standard curve when the Ct value is plotted vs the known amount of template, and E = 1.0 when the slope is approximately −3.33 (ie, 100% reaction efficiency). Nine different combinations of primer concentrations were screened using an annealing temperature of 60°C (Supplemental Table 1). Because a common reverse primer for IR-A and IR-B was critical to the design of this assay, the same concentration of reverse primer was chosen for both receptors. Studies of the IGF-1R primer using low concentrations of target revealed primer dimerization. By truncating the reverse primer sequence for IGF-1R by 3 bases (Table 1), we were able to eliminate the formation of primer dimers and maintain specificity and high efficiency of amplification. The efficiency was consistently closest to 1.0 for the following primer pair concentrations: IR-A (F0.25, R0.25), IR-B (F0.5, R0.25), IGF-1R (F0.5, R0.25), and β-actin (F0.5, R0.5). The exact E values were 0.995, 1.004, 0.980, and 0.988, respectively. Because E is 1.0 when the reaction is 100% efficient, this assay enables accurate comparison of gene expression using the ΔΔCt method of relative quantification.
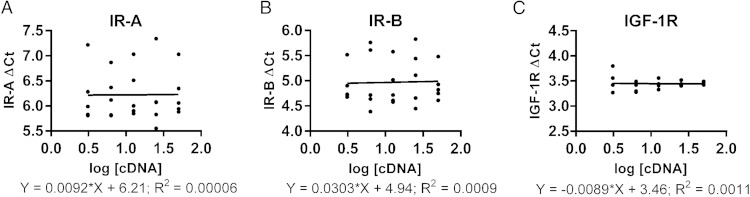
In a second assessment for efficiency equivalency, a linear regression curve was fit to the plot of the log cDNA dilution vs the ΔCt. According to Livak and Schmittgen (17), if the absolute value of the slope of the linear regression is close to zero, then the reaction efficiencies of the target and endogenous control genes are similar and the ΔΔCt method of calculation can be applied. The slope of the linear regression curve for IR-A and β-actin was 0.0092 (Figure 2A). The slope of the linear regression curve for IR-B and β-actin was 0.0303 (Figure 2B), and the slope of the linear regression curve for IGF-1R and β-actin was −0.0089 (Figure 2C). These values are below the value of 0.1 recommended in the Applied Biosystems user bulletin 2, indicating that the primers designed here for quantification of IR isoforms and IGF-1R mRNA are suitable for use in the ΔΔCt method of relative quantification.
Figure 2.
Determination of primer efficiency equivalency. A linear regression curve was fit to the plot of the log cDNA dilution vs the ΔCt, for IR-A (A), IR-B (B), and IGF1R (C). The data represent a minimum of four different qRT-PCR experiments, with each primer concentration plated in triplicate. Each point is the mean of the triplicate values from one qRT-PCR experiment. ΔCt was calculated as the Ct of the target gene minus the Ct of β-actin, the endogenous control. The y-axis of each graph is a close, compacted scale. The spread of values at each cDNA concentration is minimal and within the expected range. The slope of each curve is calculated to determine whether the reaction efficiencies of the target and endogenous control genes are similar. If the absolute value of the slope of the linear regression is close to zero, then the reaction efficiencies of the target and endogenous control genes are similar and the ΔΔCt method of calculation can be applied.
Gene expression outcomes using validated and unvalidated primer sets
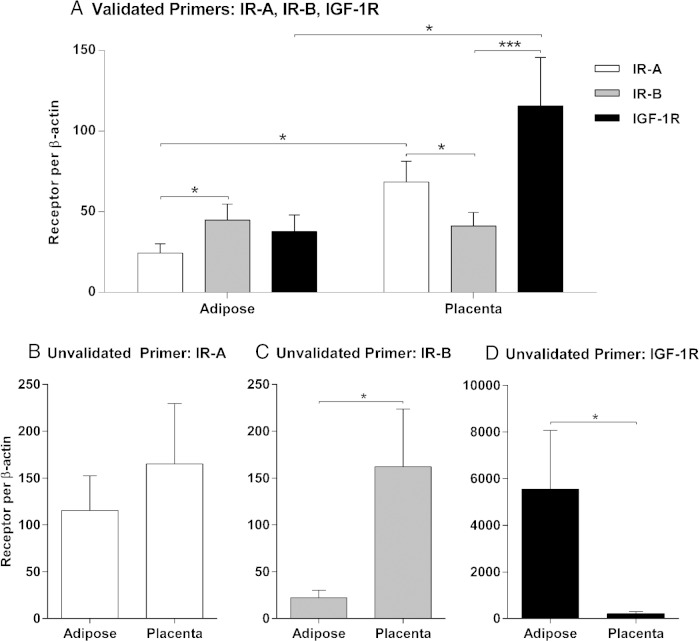
To demonstrate the value of rigorous primer validation in qRT-PCR experiments, IR-A, IR-B, and IGF-1R mRNAs were quantified in adipose tissue and placenta using the validated set of primers and a second, unvalidated set of primers (Figure 3 and Supplemental Data). Several discordant results are noted between the two assays, especially for IR-B and IGF-1R (Figure 3D and Supplemental Data). Detailed analyses support the conclusion that the unvalidated primers, particularly for IGF-1R, do not accurately detect differences in cDNA transcript concentration (Supplemental Figure 1). Moreover, the ability to quantify the three receptors on the same scale requires assay validation, thereby limiting the level of analysis possible with unvalidated primers.
Figure 3.
Comparison of two different primer sets used to quantify relative receptor expression in adipose and placenta tissue. Similarities and discrepancies in the expression of IR-A, IR-B, and IGF-1R in adipose and placenta using the validated primer set (A) or the unvalidated primer set (B–D) are shown. The qRT-PCR assay with the validated primers enables direct quantification of relative receptor expression levels due to assay validation and optimization. Thus, the mean ± SEM for each receptor is presented on the same scale as shown in panel A. The unvalidated primers were designed and used according to common practices; thus, they provide a quantification of individual receptor expression and cannot be used to compare levels of different receptors in a given tissue. The mean ± SEM for each receptor are shown in three separate graphs: IR-A (B), IR-B (C), and IGF-1R (D). *, P < .05; **, P < .001.
Conclusion
We developed a highly specific qRT-PCR assay to quantify levels of the human IR isoforms and IGF-1R on the same scale to provide a critical diagnostic and predictive tool for a variety of clinically relevant questions. The assay can be used to assess human cells or tissues in the following research applications: 1) to predict insulin and IGF sensitivity, 2) to predict the existence and levels of hybrid receptors, 3) to guide treatment strategies and predict potential treatment success for cancer, diabetes, and metabolic diseases, and 4) to screen new compounds and optimize development of new specific drugs for treatment of diabetes and cancer.
Acknowledgments
We acknowledge the Yale University Reproductive Sciences Biobank and Matthew Rodeheffer, PhD, for the collection of deidentified tissue.
This work was supported by National Institutes of Health Grant R01DK060612 from the National Institute of Diabetes and Digestive and Kidney Diseases (to T.L.W.) and by the Eunice Kennedy Shriver National Institutes of Child Health and Human Development Grant K08HD071010 (to C.A.F.).
Disclosure Summary: A.M.R. and T.L.W. are inventors on a patent application serial number 12/721,327 issued as a US Patent 8377655 on February 19, 2013, “Assay for the measurement of IGF type 1 receptor and insulin receptor expression.” The other authors have nothing to disclose.
Footnotes
- Ct
- threshold cycle
- E
- efficiency
- IGF-1R
- IGF type 1 receptor
- IR
- insulin receptor
- NCBI
- National Center for Biotechnology Information
- qRT-PCR
- quantitative RT-PCR.
References
- 1. Pandini G, Frasca F, Mineo R, Sciacca L, Vigneri R, Belfiore A. Insulin/insulin-like growth factor I hybrid receptors have different biological characteristics depending on the insulin receptor isoform involved. J Biol Chem. 2002;277:39684–39695. [DOI] [PubMed] [Google Scholar]
- 2. Belfiore A, Frasca F, Pandini G, Sciacca L, Vigneri R. Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease. Endocr Rev. 2009;30:586–623. [DOI] [PubMed] [Google Scholar]
- 3. Rowzee AM, Ludwig DL, Wood TL. Insulin-like growth factor type 1 receptor and insulin receptor isoform expression and signaling in mammary epithelial cells. Endocrinology. 2009;150:3611–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ziegler AN, Chidambaram S, Forbes BE, Wood TL, Levison SW. Insulin-like growth factor-II (IGF-II) and IGF-II analogs with enhanced insulin receptor-A binding affinity promote neural stem cell expansion. J Biol Chem. 2014;289:4626–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Frasca F, Pandini G, Scalia P, et al. Insulin receptor isoform A, a newly recognized, high-affinity insulin-like growth factor II receptor in fetal and cancer cells. Mol Cell Biol. 1999;19:3278–3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harrington SC, Weroha SJ, Reynolds C, Suman VJ, Lingle WL, Haluska P. Quantifying insulin receptor isoform expression in FFPE breast tumors. Growth Hormone, IGF Res. 2012;22:108–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vella V, Pandini G, Sciacca L, Mineo R, Vigneri R, Pezzino V, Belfiore A. A novel autocrine loop involving IGF-II and the insulin receptor isoform-A stimulates growth of thyroid cancer. J Clin Endocrinol, Metab. 2002;87:245–254. [DOI] [PubMed] [Google Scholar]
- 8. Chettouh H, Fartoux L, Aoudjehane L, et al. Mitogenic insulin receptor-A is overexpressed in human hepatocellular carcinoma due to EGFR-mediated dysregulation of RNA splicing factors. Cancer Res. 2013;73:3974–3986. [DOI] [PubMed] [Google Scholar]
- 9. Norgren S, Zierath J, Galuska D, Wallberg-Henriksson H, Luthman H. Differences in the ratio of RNA encoding two isoforms of the insulin receptor between control and NIDDM patients. The RNA variant without Exon 11 predominates in both groups. Diabetes. 1993;42:675–681. [DOI] [PubMed] [Google Scholar]
- 10. Anderson CM, Henry RR, Knudson PE, Olefsky JM, Webster NJ. Relative expression of insulin receptor isoforms does not differ in lean, obese, and noninsulin-dependent diabetes mellitus subjects. J Clin Endocrinol, Metab. 1993;76:1380–1382. [DOI] [PubMed] [Google Scholar]
- 11. Benecke H, Flier JS, Moller DE. Alternatively spliced variants of the insulin receptor protein. Expression in normal and diabetic human tissues. J Clin Invest. 1992;89:2066–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sesti G. Insulin receptor variant forms and type 2 diabetes mellitus. Pharmacogenomics. 2001;1:49–61. [DOI] [PubMed] [Google Scholar]
- 13. Trüb PK, Schäffer L, Sørensen A, Kristensen C, Jonassen I, Schmid C. Correlations of receptor binding and metabolic and mitogenic potencies of insulin analogs designed for clinical use. Diabetes. 2000;49:999–1005. [DOI] [PubMed] [Google Scholar]
- 14. Slieker LJ, Brooke GS, DiMarchi RD, et al. Modifications in the B10 and B26-30 regions of the B chain of human insulin alter affinity for the human IGF-I receptor more than for the insulin receptor. Diabetologia. 1997;40(suppl 2):S54–S61. [DOI] [PubMed] [Google Scholar]
- 15. Ziegler AN, Schneider JS, Qin M, et al. IGF-II promotes stemness of neural restricted precursors. Stem Cells. 2012;30:1265–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vandenbroucke II, Vandesompele J, Paepe AD, Messiaen L. Quantification of splice variants using real-time PCR. Nucleic Acids Res. 2001;29:E68–E68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-δδC[T]) method. Methods. 2002;25:402–408. [DOI] [PubMed] [Google Scholar]