Abstract
Kinesin-1 serves as a model for understanding fundamentals of motor protein mechanochemistry and for interpreting functional diversity across the kinesin superfamily. Despite sustained work over the last three decades, disagreements remain regarding the events that trigger the two key transitions in the stepping cycle: detachment of the trailing head from the microtubule and binding of the tethered head to the next tubulin binding site. This review describes the conflicting views of these events and highlights recent work that sheds light on these long-standing controversies. It concludes by presenting a consensus kinesin-1 chemomechanical that incorporates recent work, resolves discrepancies, and highlights key questions for future experimental work. It is hoped that this model provides a framework for understanding how diverse kinesins are tuned for their specific cellular roles.
Main Text
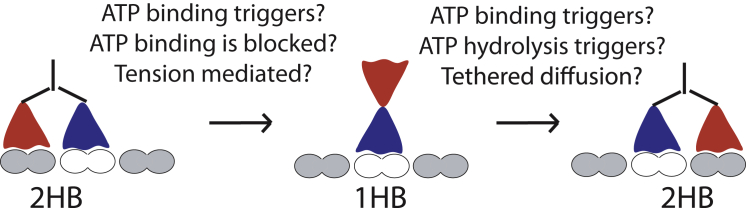
In the 30 years since the discovery of kinesin, there has been astonishing progress in understanding the inner workings of this protein machine and a vast appreciation of the role kinesin superfamily motors play in intracellular transport, cell division, and the regulation of microtubule dynamics. From an outsiders view, it may seem like the field has reached a consensus for the structural states and chemical transitions underlying kinesin-1 stepping. However, strong disagreements remain regarding the most fundamental aspects of the kinesin-1 stepping mechanism—namely, what processes trigger detachment of one head to initiate a step and what processes trigger reattachment of the tethered head to complete the step (Fig. 1) (1, 2). For instance, one class of hydrolysis cycle models holds that detachment of the trailing head from the two-heads-bound (2HB) state to a one-head-bound (1HB) intermediate is a prerequisite for ATP binding; this feature is termed front-head gating because activity in the front head is gated by the rear head (1, 3, 4, 5). A contrasting class of models (termed ATP-triggered detachment) flip this sequence and instead hold that ATP binding is the event that triggers detachment of the trailing head (6, 7). As a second example, the dominant paradigm for the nucleotide-dependent conformational changes underlying motor stepping holds that ATP binding drives docking of the neck linker domain in one head, positioning the partner head forward for proper binding to the microtubule (8). However, recent evidence (9, 10, 11) suggests that ATP binding alone is insufficient for this process and instead, ATP hydrolysis is the key chemical transition underlying full docking of the neck linker domain and stepping by the tethered head.
Figure 1.
Mechanochemical transitions that comprise one kinesin step. The starting 2HB state begins with no nucleotide in the front head. ATP-triggered step models (6, 7) hold that the transition to the 1HB state is triggered by ATP binding, whereas front-head gating models (3, 4, 30) hold that trailing head detachment precedes ATP binding. This rear head detachment may be accelerated by interhead tension, a mechanism termed rear-head gating because activity in the rear head is gated by the state of the front head (1, 18). In the 1HB state, the tethered head may reside near the previous binding site (45), nestled on the bound head (46), or diffuse freely (8). The second half of the step, which involves docking of the neck linker on the bound head, may be triggered by ATP binding and/or by ATP hydrolysis, and some component of the step involves tethered diffusion of the free head as it searches for its next binding site. The events that drive these two transitions are the subject of this review. To see this figure in color, go online.
The goal of this review is to answer three questions. 1) What events trigger the 2HB to 1HB transition that initiates the forward step? 2) What events trigger the 1HB to 2HB transition that completes the step? 3) What chemomechanical transitions determine processivity? The review begins with a discussion of the role of the neck linker domain in kinesin processivity and the role of gating between the kinesin heads. The heart of the review is an in-depth look at the two key transitions shown in Fig. 1 that comprise the kinesin stepping cycle. The review concludes with a consensus model for the kinesin-1 chemomechanical cycle that refines existing models, resolves a number of discrepancies in the literature, and sets the stage for understanding mechanisms that noncanonical kinesins have evolved to carry out their specific cellular functions.
The role of neck linker length in processive motor function
The kinesin motor can be divided into three domains. The catalytic core (or head) is the hallmark of the kinesin superfamily, and the 14 kinesin families are grouped based on conserved sequences in the head domain (12). Adjacent to the catalytic core is the neck linker, a sequence of length 14–18 residues in most N-terminal kinesins that undergoes structural rearrangements during stepping. Finally, the two heads in the dimer are connected by the neck-coil domain that connects to the distal coiled-coil and cargo-binding tail. Processivity, defined as the number of steps a motor takes before detaching from the microtubule, has generally been thought to result from tuning of rate constants inherent to the catalytic core of the motor, meaning that sequences in the catalytic core account for processivity differences between kinesin families. Hence, it was a surprise to find that when motors from diverse families were dimerized in a standard manner (using the neck-coil and coiled-coil of Drosophila kinesin-1) and their neck linkers shortened to the 14-residue length found in kinesin-1, they were all identically processive (13, 14). Furthermore, extending the neck linker led to a decrease in processivity of ∼threefold and a roughly 30% decrease in velocity. This dominant effect of neck linker length on motor function provides a unique perspective in which to understand interhead coordination in the kinesin hydrolysis cycle.
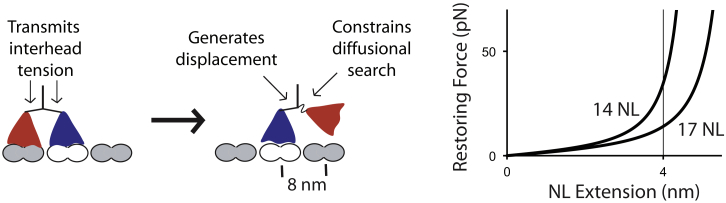
A proper understanding of mechanical coupling between the two motor domains requires understanding the mechanical properties of the neck linker domain that connects them. Based on probe mobility measurements and a lack of electron density in early x-ray and cryo-electron microscopy structures, the neck linker was interpreted to be unstructured in the apo and ADP states (8, 15). Unstructured polymers are generally modeled as worm-like chains (WLCs) with persistence lengths in the range of 0.5–2 nm (16). This entropic elasticity results in a highly nonlinear stiffness profile (Fig. 2). If both motor domains are bound to the microtubule 8 nm apart, the two neck linkers must then stretch to near their contour lengths and large interhead forces are predicted (16, 17). More importantly, lengthening the neck linker is expected to strongly reduce the force required to stretch the 8 nm spanning two tubulin subunits (16) (Fig. 2).
Figure 2.
Roles of the neck linker domain in kinesin stepping. When both heads are bound, the entropic elasticity of the neck linkers mediates interhead tension. Rear-head gating is defined as the accelerated detachment of the rear head due to this interhead tension. In the 1HB state, neck linker docking in the bound head causes a forward displacement of the tethered head. Following docking, entropic elasticity in the neck linker of the tethered head constrains its diffusional search for the next binding site. At right is a plot showing the nonlinear elasticity of a 14 or 17 amino acid polypeptide modeled as a WLC with 0.7 nm persistence length, showing different predicted forces at an extension of 4 nm, the distance needed to span the tubulin interdimer distance (16, 21). To see this figure in color, go online.
One interpretation of why extending the kinesin-1 neck linker reduces velocity is that interhead tension in wild-type (WT) motors accelerates the rate of trailing head detachment (defined as rear-head gating (1, 18, 19)), and lengthening the neck linker reduces interhead tension and slows the trailing head detachment rate. This hypothesis leads to the prediction that assisting loads imposed by an optical trap should restore this tension and recover WT stepping rates. Andreasson and colleagues found that, whereas assisting loads as large as 20 pN did not enhance the speed of WT kinesin-1, motors with extended neck linkers were indeed sped up by assisting loads (10). Consistent with predictions from a WLC model (16, 17), the extrapolated force necessary to fully restore WT speeds was ∼35 pN. Thus, this work supports a role for rear-head gating in kinesin-1 and provides a case where modeling the neck linkers as entropic springs is sufficient to explain the experimental results.
Unfortunately, this simple treatment of the neck linker as a purely mechanical element does not explain the effect of neck linker extension on processivity. Based on structural data (8, 20), neck linker docking by the bound head should provide a roughly 4 nm forward displacement, meaning that the tethered head must diffuse the remaining 4 nm to complete an 8 nm step. Achieving this displacement requires that Brownian forces on the head overcome entropic restoring forces of the neck linker that constrain this diffusive search. Brownian dynamics simulations predicted that diminishing these restoring forces by extending the neck linker should facilitate tethered head binding and enhance processivity (21, 22). However, precisely the opposite was found experimentally—neck linker extension diminished run length (13, 21).
Hence, in contrast to interhead tension in the 2HB state, the constrained diffusional search of the bound head for its next binding site is not well explained by entropic elasticity of the neck linker domain. One possible resolution is that the binding energy of the tethered head to the microtubule overcomes the entropic forces, and thus the binding rate is determined by the approximately microsecond first encounter rate (21). Alternatively, the neck linker in the tethered head may take on a relatively structured and rearward-pointing conformation (8, 15, 23, 24, 25), such that extending the neck linker orients the tethered head in a suboptimal position for binding to the next tubulin binding site.
Is gating the key to understanding processivity?
The molecular mechanisms underlying kinesin processivity have generally been discussed in the framework of gating, meaning that chemical or mechanical transitions occurring in one head are controlled, or gated, by the activity of the other head (1, 3, 4, 5, 10, 13, 14, 18, 19, 26, 27, 28, 29). In principle, one head could gate the other head by altering ATP binding, ATP hydrolysis, product release, or the microtubule attachment or detachment rates. However, the most common definition in the literature is that front-head gating is the inhibition of ATP binding by the front head when the motor in the 2HB state, whereas rear-head gating is the accelerated detachment of the trailing head in the 2HB state. The key is that both mechanisms prevent premature ATP binding to the front head before the rear head has time to detach. However, a closer analysis of the hydrolysis cycle along with more recent results from both kinesin-1 and kinesin-2 bring into question whether these gating mechanisms are the key features that control processivity.
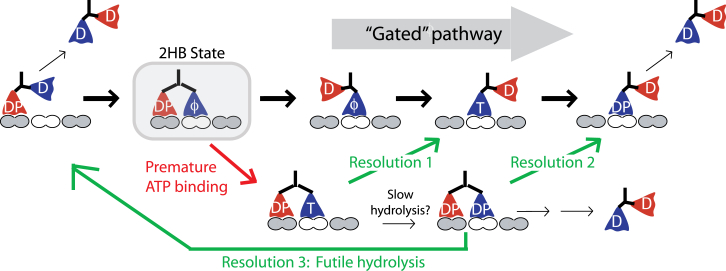
The first question is, does premature binding of ATP lead to termination of a processive run? We can analyze this by tracing the possible transitions that follow ATP binding and ask whether detachment is a likely result. In the model shown in Fig. 3, the 2HB state with an empty front head is highlighted. The gated pathway involves trailing head detachment, followed by ATP binding and completion of a step; failure of front-head gating is defined as ATP binding to the front head before the rear head has time to detach. The contention is that there is a high probability that this off-pathway event will result in motor detachment (1, 3, 4, 5, 6, 17, 29, 30). However, by laying out the sequence of possible transitions, it is not at all clear that motor detachment will be the end result. First, this 2HB ATP state can resolve by trailing head detachment (Resolution 1), and if ATP hydrolysis is slowed or blocked in the front head due to rearward neck linker tension or orientation (26), this resolution pathway will be preferred. If hydrolysis proceeds, the normal cycle can be resolved by detachment of either the rear head (Resolution 2) or the front head (Resolution 3, which constitutes a futile ATP hydrolysis cycle). Hence, even for a 2HB motor with ADP-Pi in both heads (DP/DP in Fig. 3), detachment of one head resolves to the normal hydrolysis cycle, although to a vulnerable 1HB ADP-Pi state. For detachment from the DP/DP state to occur, it must differ from these resolutions; for instance, by the front head rapidly releasing Pi, the trailing head detaching, and then the front head detaching in the ADP state, but this is speculation. Thus, the assumption that premature ATP binding leads to termination of a processive run should be questioned.
Figure 3.
Implications of premature ATP binding for kinesin processivity. Starting with the motor in a 2HB state with an empty front head, the gated pathway involves detachment of the trailing head followed by ATP binding, hydrolysis, and stepping. Alternatively, ATP can bind to the front head, defined as a breakdown of front-head gating. The first potential resolution is that the trailing head then detaches, and the cycle resumes the normal pathway (Resolution 1). Alternatively, the front head can hydrolyze ATP (which may be slowed due to rearward strain and/or the rearward position of the neck linker), putting both heads in a low affinity ADP-Pi state. From this DP/DP state, either the trailing head can detach (Resolution 2, which should be preferred based on the directional dependence of detachment (3, 5, 6, 10, 53)), or the front head can detach (Resolution 3, which results in a futile ATP hydrolysis cycle). Based on this scheme, for premature ATP binding to have a significant impact on processivity, the off-rate from the 2HB state having ADP-Pi in each head would have to be significantly faster than the transitions that resolve to the normal gated pathway. To see this figure in color, go online.
A recent study (26) provides an interesting perspective on this issue. A human Cys-lite motor with a 6 amino acid neck linker insert was studied by optical trapping and stopped flow to understand the effects of neck linker length on gating. Because of increased interhead flexibility, this motor was shown to release both bound ADP upon interaction with a microtubule. It was also shown to walk processively backward under hindering loads, a feature seen previously (6). Under no load, the walking speed was reduced by more than twofold, yet the ATPase rate was higher than WT, indicative of futile hydrolysis cycles. Interestingly, despite this uncoupling between the hydrolysis and stepping cycles, the run length was increased nearly twofold above WT. From this work, two important conclusions can be drawn. First, breakdown of interhead coordination need not alter processivity, and it can even enhance it. Second, seemingly subtle mutations can substantially alter a motor’s mechanochemistry—a similar six-residue neck linker insertion into a non-Cys-mutated Drosophila kinesin-1 resulted in shorter run lengths and no backstepping against hindering loads, and it displayed a qualitatively different force-velocity curve than its Cys-lite counterpart (10). The most plausible explanation is that Cys mutations in kinesin-1 enhance the ability of the trailing head to bind to the microtubule, which results in backstepping under load and enhanced processivity in the neck linker-extended construct (6, 26).
Finally, recent kinesin-1 and kinesin-2 nucleotide binding measurements bring into question whether front-head ATP binding necessarily follows trailing head detachment during processive stepping. The key experiment is to trap the motor on the microtubule in a 2HB state using AMPPNP, flush this complex against mant-labeled nucleotide, and measure the kinetics of nucleotide binding to the front head. The slow nucleotide binding rate measured previously for human Cys-lite kinesin-1 provided the strongest experimental evidence for front-head gating in kinesin-1 (3). Surprisingly, in kinesin-2 the kinetics of mantADP and mantATP binding in the 2HB state were not measurably different than in the control 1HB case (28). This lack of front-head gating in kinesin-2 also held for a motor with a shortened neck linker that is predicted to enhance interhead tension. Furthermore, in Drosophila kinesin-1 trapped in the 2HB state using AMPPNP, mantADP was found to bind to the front head with an off-rate of 357 s−1, and an on-rate in the range of 0.5 μM−1s−1 (11). Although this on-rate was not tightly specified by the data and more work is warranted here, it is notable that it represents a rate of 500 s−1 at 1 mM ATP, which is fast relative to the trailing head detachment rate discussed (see Fig. 5) below.
Figure 5.
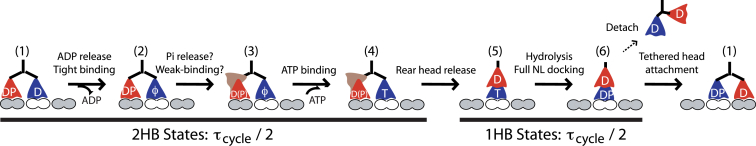
Consensus kinesin-1 chemomechanical cycle. Upon tethered head binding (State 1), ADP release is rapid, generating a 2HB state with an apo front head (State 2). The rear head then transitions into an ATP waiting state (State 3), a transition that may involve Pi release and may involve a different interaction with the microtubule but does not cause a displacement. Upon ATP binding (State 4), the rear head rapidly detaches and moves forward 8 nm (State 5). ATP hydrolysis results in a vulnerable 1HB state (State 6) in which the bound head can detach to terminate the run or the tethered head can attach to complete the step. At physiological ATP, the cycle duration (τcycle) is split evenly between 1HB and 2HB states; as ATP is reduced the 2HB duration increases (11). To see this figure in color, go online.
Hence, to summarize so far, 1) there is evidence in the literature that ATP binding is gated in the 2HB state (3, 4, 5, 29), but an analysis of the resolution pathways (Fig. 3) does not explain why this should substantially alter processivity; 2) a kinesin-1 mutant with disrupted chemomechanical coupling was actually more processive than WT (26); 3) for motors in the kinesin-2 family, ATP appears to readily bind in the 2HB state, yet the motors are still processive (28); 4) for kinesin-1, measured rates of nucleotide binding in the 2HB state are fast with respect to the overall stepping cycle. The second half of this review analyzes the two key transitions that make up the chemomechanical cycle and presents a consensus hydrolysis model in which processivity is predominantly controlled in the 1HB state by the race between detachment of the bound head and attachment of the tethered head to the next binding site.
The first half of the step: what triggers detachment of the trailing head?
A convenient way to understand the kinesin stepping mechanism is to consider one cycle that starts from the motor having both heads bound to the microtubule, proceeds to detachment of the trailing head to generate a one-head-bound intermediate state, and is completed by the tethered head binding to the next binding site 16 nm away (Fig. 1). Understanding the hydrolysis cycle then turns into answering the following question: what chemical events trigger these two key transitions? The question of what chemical event triggers detachment of the trailing head can be distilled to: at physiological ATP levels does the motor wait for ATP in a 1HB or 2HB state? There is substantial experimental evidence that at limiting nucleotide concentrations, kinesin-1 resides in a state with one bound and one tethered head (31, 32, 33, 34, 35). However, neither optical trapping studies using a bead attached to the kinesin tail (1, 36, 37) nor single-molecule tracking studies (7, 30, 38, 39) have been able to reliably detect a 1HB intermediate in the stepping cycle. Single-molecule fluorescence studies that track the position of one head and find 16 nm steps at both low and high ATP support a model in which the motor waits for ATP with both heads bound (6, 7, 30). Other studies with more limited temporal resolution detect 1HB waiting states at low ATP concentrations that extrapolate to an undetectable 1HB state at saturating ATP (33, 34). These latter studies argue that at limiting ATP, ATP binding is not necessary for trailing head detachment, but they do not resolve whether at saturating ATP, where nucleotide binding is very fast, ATP binding precedes trailing head detachment or vice versa. A study that analyzed the autocorrelation of fluorescence resonance energy transfer (FRET) intensities provided evidence for a 3 ms duration 1HB state, but it did not resolve the order of ATP binding and trailing head detachment at saturating ATP levels (40). Another study that used rhodamine quenching to analyze structural states concluded that the ATP waiting state is a rapid equilibrium between 2HB and 1HB states, with ATP only binding to the 1HB state (30).
My lab recently addressed this question by using interferometric scattering microscopy (41) to track the position of motors labeled with a single 30-nm gold nanoparticle on one head as the motors walked processively at saturating ATP (11). With the 1 ms and 2 nm spatiotemporal resolution achieved, we were able to detect a transient 1HB intermediate at saturating ATP that was localized midway between microtubule-bound positions 16.4 nm apart. Because only one head is labeled, alternating steps have different durations—the labeled head will be bound to the microtubule for the entire stepping cycle of the unlabeled head, and it will remain there for the 2HB portion of its cycle; it only resides at an intermediate position for the fraction of the cycle that it spends in the 1HB state. The threefold difference in long and short plateau durations and the nonexponential distribution of long plateaus were quantitatively described by the hydrolysis cycle having two rate-limiting steps—one in a 1HB state and one in a 2HB state. An attractive feature of this result is that it helps to explain previous optical trapping studies that found that the randomness of kinesin stepping is consistent with the cycle having two rate-limiting transitions (42, 43, 44).
To assess the ATP waiting state, the stepping rate was lowered by decreasing the ATP concentration (11). If the motor waits for ATP in a 1HB state, both the short and long duration plateaus will then be extended because both contain a 1HB phase of one of the heads. In contrast, if the ATP waiting state is a 2HB state, the transient 1HB state will then remain the same duration, whereas the longer plateau will be extended. The data matched the second prediction, providing strong evidence that the ATP waiting state is a 2HB state, or is at least a state in which the position of the trailing head is very close to its previous binding site if not necessarily tightly bound to it. This finding of a 2HB ATP waiting state is consistent with the previous fluorescence stepping data that detected only 16-nm head displacements (6, 7, 30), and it also supports the finding that reversal of ATP hydrolysis in the tethered head is considerably faster than a free motor in solution (45), which was interpreted as the tethered head in the ATP waiting state being able to interact with tubulin. This result is not consistent with an ATP waiting state at physiological ATP levels in which the tethered head freely diffuses around the mean position of the bound head (8), nor one in which the tethered head is nestled near the bound head (46). However, at limiting ATP, the data do not rule out slow transitions into these 1HB structural states.
One interesting feature of the data was the appearance of 1HB waiting states at very low ATP concentrations, a feature that could be quantitatively explained by an ATP-independent trailing head detachment pathway with a rate of 2 s−1. This result is consistent with a FRET study that observed transients consistent with alternating 1HB and 2HB states at limiting ATP but an apparently uniform 2HB population at saturating ATP (33). The conclusion that, given sufficient time at low ATP the trailing head will eventually detach to generate a 1HB state, provides an explanation for why 1HB states have been measured by FRET, stopped flow, and optical tweezers at limiting nucleotide (30, 31, 33, 34, 35). The finding that only 16 nm transitions were observed for the human Cys-lite dimer at limiting ATP (6, 7, 30) may be explained by this Cys-lite construct having a slower rate of trailing head detachment than WT, as discussed previously (10, 26), or it could result from the limiting signal/noise and the step-finding algorithm employed in those studies (discussed in (11)).
Thus, to answer the question: what events trigger the 2HB to 1HB transition that initiates the forward step? At physiological ATP levels, ATP binding to the front head in the 2HB state triggers the 2HB to 1HB transition. Aspects of this transition that require further work include full determination of the events immediately preceding this transition (which may include some combination of Pi release, a strong-to-weak transition, or trailing head detachment without substantial displacement), as well as a structural explanation for how ATP binding may trigger this transition.
The second half of the step: what drives forward binding of the tethered head?
For the last 15 years the paradigm used to describe nucleotide-driven conformational changes in kinesin has been the neck linker docking model, which holds that ATP binding leads to docking of the neck linker in a forward orientation that places the tethered in an optimal position for binding to the next binding site (8). This model can best be described as a ratchet-like mechanism in which thermal fluctuations drive movement of the tethered head, and chemical energy is used to trap the neck linker in one preferred orientation (1, 47, 48, 49, 50). Biochemical half-site experiments showed that microtubule binding by a dimeric motor triggers release of one bound ADP and retention of the second by the tethered head, whereas ATP binding causes rapid release of the second ADP (31). The finding that the nonhydrolyzable AMPPNP or the slowly hydrolyzable ATPγS, or ATP binding in a hydrolysis-compromised mutant also trigger release of the bound ADP (51, 52), although at a slower rate, led to a model in which ATP binding in the absence of hydrolysis is sufficient for docking of the neck linker and binding of the tethered head to the next binding site to complete a forward step. However, recent evidence calls into question whether ATP binding alone is the trigger for the forward step.
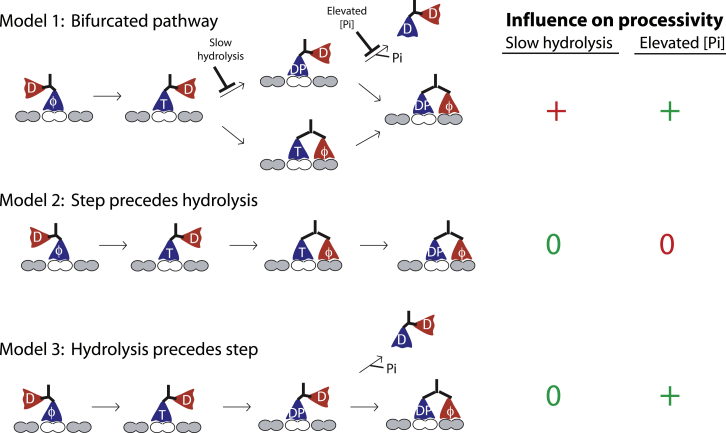
Defining the timing of hydrolysis and tethered head binding is of crucial importance because it is this transition that is thought to determine motor processivity. If ATP is hydrolyzed before the tethered head binds to its next binding site, it then leaves the motor with one weakly bound ADP-Pi head that is vulnerable to detachment. Instead, if ATP binding is sufficient to trigger the step to the next binding site, there is a short circuit by which the motor can then avoid this vulnerable state. This sequence of events can be divided into three possible models shown in Fig. 4. In the bifurcated pathway (Model 1 in Fig. 4), tethered head binding can occur either before or after ATP hydrolysis. Alternatively, the states may be sequential and involve either the tethered-head-binding step preceding hydrolysis (Model 2 in Fig. 4) or hydrolysis preceding tethered-head binding (Model 3 in Fig. 4).
Figure 4.
Unraveling the sequence of ATP hydrolysis and binding of the tethered head to complete the forward step. In experiments (9), slowing hydrolysis with ATPγS had no effect on processivity and increasing [Pi] enhanced processivity (denoted by 0 and +, respectively). Model predictions that agree and disagree with experiments are denoted in green and red, respectively. In the Bifurcated pathway (Model 1), tethered head binding and ADP release can occur either before or after ATP hydrolysis. If hydrolysis precedes tethered head binding (upper branch), the motor can potentially dissociate, ending a processive run. In this model, slowing ATP hydrolysis or elevating [Pi] are both predicted to enhance processivity. Alternatively, tethered head binding may precede ATP hydrolysis (Model 2), allowing the motor to avoid the vulnerable 1HB ADP-Pi state; in this case slowing hydrolysis or elevating [Pi] are predicted to have no effect on processivity. Experimental data from Milic et al. (9) are only consistent with Model 3, in which ATP hydrolysis precedes binding of the tethered head. To see this figure in color, go online.
This question was recently addressed by Milic and colleagues, who varied nucleotide conditions and measured run lengths under assisting loads (9). If ATP binding alone is sufficient for stepping, slowing ATP hydrolysis is then predicted to enhance processivity because the motor will reach a stable 2HB state before hydrolysis (Model 1 in Fig. 4). Instead, in the slowly hydrolysable nucleotide analog ATPγS, run lengths were unchanged, despite a more than 30-fold reduction in velocity. As a second experiment, run lengths were measured in varying concentrations of free Pi. If motor detachment involves Pi release, free Pi could then potentially stabilize the bound ADP-Pi state. Run lengths were indeed elevated at high [Pi], consistent with stabilization of the vulnerable ADP-Pi state from detachment in the presence of assisting loads. This result is inconsistent with a hydrolysis cycle in which binding by the tethered head precedes ATP hydrolysis (Model 2 in Fig. 4). The only model that accounts for the data is one in which ATP hydrolysis precedes the step that results in strong binding of the tethered head to its next binding site (Model 3 in Fig. 4).
These findings were extended by our recent work involving high resolution tracking of gold nanoparticle-labeled heads during kinesin-1 stepping (11). Under the unloaded conditions of these experiments, run length was also not elevated in ATPγS. Furthermore, if ATP binding alone is sufficient to complete the forward step, the prediction is that in ATPγS the motor will then overwhelmingly reside in a 2HB state limited by ATP hydrolysis in the trailing head. Instead, the opposite was found—stepping was highly variable in ATPγS and the predicted stable 2HB states were rare. These data are best explained by a model in which ATP binding does not dock the neck linker sufficiently for rapid stepping, and instead ATP hydrolysis is the key chemical transition that drives full neck linker docking and completion of the forward step (9). It is still difficult to explain why slowly and nonhydrolysable nucleotides trigger ADP release in the half-site assay, however. One possibility is that ADP in the tethered head is released without stable binding of the tethered head to the microtubule. Another possibility is that forward steps are not favored in the prehydrolysis state, but sidesteps are disinhibited by ATP binding, and ADP release by the tethered head is a result of sidesteps to adjacent protofilaments.
Thus, to answer the question: what events trigger the 1HB to 2HB transition that completes the step? ATP hydrolysis triggers the structural transition that results in microtubule binding and ADP release by the tethered head.
A consensus model for the kinesin-1 chemomechanical cycle
This recent work leads to an updated consensus model for the kinesin-1 chemomechanical cycle (Fig. 5) that helps to understand how the motor walks processively against loads and provides a framework for understanding noncanonical kinesins having different properties. The key constraints for the model are 1) at saturating ATP the total cycle duration is evenly split between 1HB and 2HB states (11), 2) ATP binding occurs in the 2HB state (7, 11, 30), and 3) ATP hydrolysis precedes the transition from the 1HB to the 2HB state (9, 10, 11).
The 2HB portion of the cycle begins with binding of the tethered head (State 1 in Fig. 5). Based on stopped flow measurements of mantADP binding in the 2HB state (11) and the finding that nucleotide affinity is diminished by rearward tension (53), ADP release is rapid, generating State 2. Transition to the 1HB state involves ATP binding, which is expected to be fast at saturating ATP, and trailing head detachment, which is expected to be fast based on the observation that assisting loads do not increase the stepping rate of WT kinesin-1 (10). To account for the 2HB states occupying half of the total cycle duration, a second transition is included that follows ADP release and precedes ATP binding. This State 2 → 3 transition may involve Pi release and/or conversion of the trailing head to a weak binding state (30, 34, 35) (denoted by multiple conformations of the trailing head in State 3). Experimental data (11) do not rule out ATP binding to State 2, but for simplicity the rate-limiting 2HB 2 → 3 transition and the rapid 3 → 4 ATP binding transition are shown sequentially. Trailing head detachment follows ATP binding to the lead head and completes the 2HB portion of the cycle. There are two 1HB states, one preceding ATP hydrolysis (State 5) and one following it (State 6). From the posthydrolysis State 6, the tethered head normally attaches to the next binding site to complete the step, but a small fraction of the time (<1% for kinesin-1), the motor detaches instead, terminating the run. In saturating ATP, the rate-limiting step in the 2HB phase of the cycle is the State 2 → 3 transition. Existing data cannot constrain the rate-limiting step in the 1HB phase, so it is either ATP hydrolysis or tethered head attachment.
Although a number of features in this chemomechanical model are well established, other features differ from models in the literature. The principle that ATP hydrolysis precedes tethered head attachment was established only recently (9, 10, 11), and it is not a component of most hydrolysis models (4, 26, 30, 33). A number of models include strict front-head gating—postulating that ATP binding by the front head necessarily follows detachment of the trailing head (3, 4, 30), whereas here ATP binding (State 3 → 4) occurs before measurable trailing head detachment. The lack of ATP binding in State 2 is a form of front-head gating; however, following the arguments from Fig. 3, as long as premature ATP binding does not lead to detachment of the front head, it can be permitted without consequence to the cycle.
Implicit in this model is that ATP binding to the front head triggers a powerstroke, meaning a conformational change that accelerates detachment of the trailing head. However, this transition is not clearly explained by existing structural data. There is strong evidence that ATP binding induces a rotation or seesaw motion of the catalytic domain in a direction perpendicular to the microtubule axis that opens a cleft where the neck linker docks (20, 24, 54, 55). However, in the 2HB State 4, the neck linker in the front head is expected to be pulled back into an undocked conformation, preventing the freedom of movement needed for such a ratchet-type mechanism. One possible resolution to this paradox is that the 2HB waiting state measured by gold nanoparticle tracking actually represents a dynamic equilibrium between 1HB and 2HB states that is strongly biased toward the 2HB state, an idea put forward previously (30, 33).
One clarifying aspect of this 2HB ATP waiting state is that it provides a simple explanation for backstepping. At its stall force, kinesin-1 continues to take forward and backward steps with a net rate of zero (37). If the motor waits for ATP in a 2HB state and the front head binds and hydrolyzes ATP and then detaches, the trailing head has a higher probability of being attached and ready to shoulder the load, rather than having to rebind to its previous binding site before the front head detaches.
The second clarifying principle for kinesin-1 is that during the forward step, ATP hydrolysis precedes attachment of the tethered head to the next binding site (9, 10, 11). This sequence simplifies the analysis of motor processivity by narrowing the focus to the posthydrolysis 1HB state having ADP-Pi in the bound head and ADP in the tethered head (State 6 in Fig. 5). Thus, to answer the question: what chemomechanical transitions determine processivity? The degree of processivity is determined by the relative rates of attachment and ADP release of the tethered head, versus microtubule detachment of the bound head. It follows that these parameters are tuned differently in motors that are more or less processive than kinesin-1.
This stepping model also provides an explanation for how motors step around microtubule-associated proteins and other roadblocks that decorate microtubules in vivo. In a recent high-resolution tracking study (56), kinesin-1 motors encountering roadblocks were found to pause with mean durations of 400 ms before detaching or taking a sidestep. Assuming the motor is waiting in a 1HB posthydrolysis state (State 6), this provides an estimate of 2.5 s−1 for the detachment rate from this vulnerable state. If the rate of posthydrolysis tethered head attachment were 100-fold faster than this (250 s−1 or 4 ms mean duration), the probability of detaching per step would be 1%, consistent with kinesin-1 processivity values. Interestingly, the processivity of kinesin-2, which has a longer neck linker than kinesin-1, is less affected by roadblocks, consistent with its greater interhead flexibility enabling it to more readily take sidesteps (57). Thus, a possible explanation for why some transport kinesins have evolved longer neck linkers despite the resulting decrease in processivity is that this adaptation enables sidestepping around obstacles (57).
How can we use this kinesin-1 hydrolysis cycle to interpret the properties of motors in other kinesin families? The strong load-dependence of kinesin-2 processivity (44) can be explained by the motor spending a large fraction of its time in a posthydrolysis 1HB state (State 6) that normally dissociates rather slowly but dissociates rapidly under load (28). Superprocessivity of kinesin-3 dimers (58) can be explained by either a slow detachment rate from the 1HB ADP-Pi state or a fast tethered head attachment rate. The fact that the motor is fast and K-loop modifications had little effect on dimer processivity (59) argues for fast attachment being key, whereas the finding that kinesin-3 detachment is highly load-dependent (60) argues that slow detachment from the ADP-Pi state is the underlying explanation. Kinesin-5 is an interesting case because it is slow and minimally processive, yet its detachment is relatively insensitive to load (61). This suggests that, rather than a long duration in a vulnerable 1HB state, kinesin-5 spends most of its cycle in a strongly bound state, perhaps with both heads bound (62). To validate the generality of the model, these hypotheses will need to be tested through high-resolution tracking experiments on these diverse kinesins.
The last three decades have witnessed amazing progress toward understanding these protein machines, and addressing these ongoing questions should make the next few years very exciting for understanding kinesin biophysics and the role of kinesins in cell biology.
Acknowledgments
I am grateful to members of the Hancock lab, particularly Keith Mickolajczyk, Geng-Yuan (Scott) Chen, and Shankar Shastry, and my collaborators in the Block lab, Johan Andreasson and Bojan Milic for their experimental and intellectual contributions. I also thank my many colleagues in the kinesin field whose work contributed directly and indirectly to this effort and apologize for work that could not be cited due to space constraints.
This work was supported by National Institutes of Health (NIH) grants R01GM076476 and R01GM100076.
Editor: Brian Salzberg.
Note Added in Proof
Following the acceptance of this article, a head-tracking study using a similar approach to (11) was published: Isojima H., R. Iino, …, M. Tomishige. 2016. Direct observation of intermediate states during the stepping motion of kinesin-1. Nat. Chem. Biol. Published online February 29, 2016. http://dx.doi.org/10.1038/nchembio.2028.
References
- 1.Block S.M. Kinesin motor mechanics: binding, stepping, tracking, gating, and limping. Biophys. J. 2007;92:2986–2995. doi: 10.1529/biophysj.106.100677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gennerich A., Vale R.D. Walking the walk: how kinesin and dynein coordinate their steps. Curr. Opin. Cell Biol. 2009;21:59–67. doi: 10.1016/j.ceb.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenfeld S.S., Fordyce P.M., Block S.M. Stepping and stretching. How kinesin uses internal strain to walk processively. J. Biol. Chem. 2003;278:18550–18556. doi: 10.1074/jbc.M300849200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klumpp L.M., Hoenger A., Gilbert S.P. Kinesin’s second step. Proc. Natl. Acad. Sci. USA. 2004;101:3444–3449. doi: 10.1073/pnas.0307691101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guydosh N.R., Block S.M. Backsteps induced by nucleotide analogs suggest the front head of kinesin is gated by strain. Proc. Natl. Acad. Sci. USA. 2006;103:8054–8059. doi: 10.1073/pnas.0600931103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yildiz A., Tomishige M., Vale R.D. Intramolecular strain coordinates kinesin stepping behavior along microtubules. Cell. 2008;134:1030–1041. doi: 10.1016/j.cell.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yildiz A., Tomishige M., Selvin P.R. Kinesin walks hand-over-hand. Science. 2004;303:676–678. doi: 10.1126/science.1093753. [DOI] [PubMed] [Google Scholar]
- 8.Rice S., Lin A.W., Vale R.D. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 9.Milic B., Andreasson J.O., Block S.M. Kinesin processivity is gated by phosphate release. Proc. Natl. Acad. Sci. USA. 2014;111:14136–14140. doi: 10.1073/pnas.1410943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andreasson J.O., Milic B., Block S.M. Examining kinesin processivity within a general gating framework. eLife. 2015;4:e07403. doi: 10.7554/eLife.07403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mickolajczyk K.J., Deffenbaugh N.C., Hancock W.O. Kinetics of nucleotide-dependent structural transitions in the kinesin-1 hydrolysis cycle. Proc. Natl. Acad. Sci. USA. 2015;112:E7186–E7193. doi: 10.1073/pnas.1517638112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence C.J., Dawe R.K., Wordeman L. A standardized kinesin nomenclature. J. Cell Biol. 2004;167:19–22. doi: 10.1083/jcb.200408113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shastry S., Hancock W.O. Neck linker length determines the degree of processivity in kinesin-1 and kinesin-2 motors. Curr. Biol. 2010;20:939–943. doi: 10.1016/j.cub.2010.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shastry S., Hancock W.O. Interhead tension determines processivity across diverse N-terminal kinesins. Proc. Natl. Acad. Sci. USA. 2011;108:16253–16258. doi: 10.1073/pnas.1102628108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rice S., Cui Y., Cooke R. Thermodynamic properties of the kinesin neck-region docking to the catalytic core. Biophys. J. 2003;84:1844–1854. doi: 10.1016/S0006-3495(03)74992-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hariharan V., Hancock W.O. Insights into the mechanical properties of the kinesin neck linker domain from sequence analysis and molecular dynamics simulations. Cell. Mol. Bioeng. 2009;2:177–189. doi: 10.1007/s12195-009-0059-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyeon C., Onuchic J.N. Internal strain regulates the nucleotide binding site of the kinesin leading head. Proc. Natl. Acad. Sci. USA. 2007;104:2175–2180. doi: 10.1073/pnas.0610939104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hancock W.O., Howard J. Kinesin’s processivity results from mechanical and chemical coordination between the ATP hydrolysis cycles of the two motor domains. Proc. Natl. Acad. Sci. USA. 1999;96:13147–13152. doi: 10.1073/pnas.96.23.13147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hancock W.O., Howard J. Processivity of the motor protein kinesin requires two heads. J. Cell Biol. 1998;140:1395–1405. doi: 10.1083/jcb.140.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sindelar C.V., Downing K.H. An atomic-level mechanism for activation of the kinesin molecular motors. Proc. Natl. Acad. Sci. USA. 2010;107:4111–4116. doi: 10.1073/pnas.0911208107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kutys M.L., Fricks J., Hancock W.O. Monte Carlo analysis of neck linker extension in kinesin molecular motors. PLOS Comput. Biol. 2010;6:e1000980. doi: 10.1371/journal.pcbi.1000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hancock W.O., Howard J. Kinesin: processivity and chemomechanical coupling. In: Schliwa M., editor. Molecular Motors. Wiley-VCH; Weinheim, Germany: 2003. pp. 243–269. [Google Scholar]
- 23.Muretta J.M., Jun Y., Rosenfeld S.S. The structural kinetics of switch-1 and the neck linker explain the functions of kinesin-1 and Eg5. Proc. Natl. Acad. Sci. USA. 2015;112:E6606–E6613. doi: 10.1073/pnas.1512305112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sindelar C.V. A seesaw model for intermolecular gating in the kinesin motor protein. Biophys. Rev. 2011;3:85–100. doi: 10.1007/s12551-011-0049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goulet A., Major J., Moores C.A. Comprehensive structural model of the mechanochemical cycle of a mitotic motor highlights molecular adaptations in the kinesin family. Proc. Natl. Acad. Sci. USA. 2014;111:1837–1842. doi: 10.1073/pnas.1319848111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clancy B.E., Behnke-Parks W.M., Block S.M. A universal pathway for kinesin stepping. Nat. Struct. Mol. Biol. 2011;18:1020–1027. doi: 10.1038/nsmb.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muthukrishnan G., Zhang Y., Hancock W.O. The processivity of kinesin-2 motors suggests diminished front-head gating. Curr. Biol. 2009;19:442–447. doi: 10.1016/j.cub.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen G.Y., Arginteanu D.F., Hancock W.O. Processivity of the kinesin-2 KIF3A results from rear head gating and not front head gating. J. Biol. Chem. 2015;290:10274–10294. doi: 10.1074/jbc.M114.628032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dogan M.Y., Can S., Yildiz A. Kinesin’s front head is gated by the backward orientation of its neck linker. Cell Reports. 2015;10:1967–1973. doi: 10.1016/j.celrep.2015.02.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Toprak E., Yildiz A., Selvin P.R. Why kinesin is so processive. Proc. Natl. Acad. Sci. USA. 2009;106:12717–12722. doi: 10.1073/pnas.0808396106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hackney D.D. Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc. Natl. Acad. Sci. USA. 1994;91:6865–6869. doi: 10.1073/pnas.91.15.6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirose K., Löwe J., Amos L.A. Congruent docking of dimeric kinesin and ncd into three-dimensional electron cryomicroscopy maps of microtubule-motor ADP complexes. Mol. Biol. Cell. 1999;10:2063–2074. doi: 10.1091/mbc.10.6.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mori T., Vale R.D., Tomishige M. How kinesin waits between steps. Nature. 2007;450:750–754. doi: 10.1038/nature06346. [DOI] [PubMed] [Google Scholar]
- 34.Asenjo A.B., Sosa H. A mobile kinesin-head intermediate during the ATP-waiting state. Proc. Natl. Acad. Sci. USA. 2009;106:5657–5662. doi: 10.1073/pnas.0808355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guydosh N.R., Block S.M. Direct observation of the binding state of the kinesin head to the microtubule. Nature. 2009;461:125–128. doi: 10.1038/nature08259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svoboda K., Schmidt C.F., Block S.M. Direct observation of kinesin stepping by optical trapping interferometry. Nature. 1993;365:721–727. doi: 10.1038/365721a0. [DOI] [PubMed] [Google Scholar]
- 37.Carter N.J., Cross R.A. Mechanics of the kinesin step. Nature. 2005;435:308–312. doi: 10.1038/nature03528. [DOI] [PubMed] [Google Scholar]
- 38.Nan X., Sims P.A., Xie X.S. Organelle tracking in a living cell with microsecond time resolution and nanometer spatial precision. ChemPhysChem. 2008;9:707–712. doi: 10.1002/cphc.200700839. [DOI] [PubMed] [Google Scholar]
- 39.Cappello G., Badoual M., Busoni L. Kinesin motion in the absence of external forces characterized by interference total internal reflection microscopy. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2003;68:021907. doi: 10.1103/PhysRevE.68.021907. [DOI] [PubMed] [Google Scholar]
- 40.Verbrugge S., Lansky Z., Peterman E.J. Kinesin’s step dissected with single-motor FRET. Proc. Natl. Acad. Sci. USA. 2009;106:17741–17746. doi: 10.1073/pnas.0905177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ortega-Arroyo J., Kukura P. Interferometric scattering microscopy (iSCAT): new frontiers in ultrafast and ultrasensitive optical microscopy. Phys. Chem. Chem. Phys. 2012;14:15625–15636. doi: 10.1039/c2cp41013c. [DOI] [PubMed] [Google Scholar]
- 42.Schnitzer M.J., Block S.M. Kinesin hydrolyses one ATP per 8-nm step. Nature. 1997;388:386–390. doi: 10.1038/41111. [DOI] [PubMed] [Google Scholar]
- 43.Svoboda K., Mitra P.P., Block S.M. Fluctuation analysis of motor protein movement and single enzyme kinetics. Proc. Natl. Acad. Sci. USA. 1994;91:11782–11786. doi: 10.1073/pnas.91.25.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andreasson J.O., Shastry S., Block S.M. The mechanochemical cycle of mammalian kinesin-2 KIF3A/B under load. Curr. Biol. 2015;25:1166–1175. doi: 10.1016/j.cub.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hackney D.D. The tethered motor domain of a kinesin-microtubule complex catalyzes reversible synthesis of bound ATP. Proc. Natl. Acad. Sci. USA. 2005;102:18338–18343. doi: 10.1073/pnas.0505288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alonso M.C., Drummond D.R., Cross R.A. An ATP gate controls tubulin binding by the tethered head of kinesin-1. Science. 2007;316:120–123. doi: 10.1126/science.1136985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fox R.F., Choi M.H. Rectified Brownian motion and kinesin motion along microtubules. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2001;63:051901–0519012. doi: 10.1103/PhysRevE.63.051901. [DOI] [PubMed] [Google Scholar]
- 48.Astumian R.D., Derényi I. A chemically reversible Brownian motor: application to kinesin and Ncd. Biophys. J. 1999;77:993–1002. doi: 10.1016/S0006-3495(99)76950-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taniguchi Y., Nishiyama M., Yanagida T. Entropy rectifies the Brownian steps of kinesin. Nat. Chem. Biol. 2005;1:342–347. doi: 10.1038/nchembio741. [DOI] [PubMed] [Google Scholar]
- 50.Hwang W., Lang M.J., Karplus M. Force generation in kinesin hinges on cover-neck bundle formation. Structure. 2008;16:62–71. doi: 10.1016/j.str.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 51.Ma Y.Z., Taylor E.W. Interacting head mechanism of microtubule-kinesin ATPase. J. Biol. Chem. 1997;272:724–730. doi: 10.1074/jbc.272.2.724. [DOI] [PubMed] [Google Scholar]
- 52.Klumpp L.M., Brendza K.M., Gilbert S.P. Motor domain mutation traps kinesin as a microtubule rigor complex. Biochemistry. 2003;42:2595–2606. doi: 10.1021/bi026715r. [DOI] [PubMed] [Google Scholar]
- 53.Uemura S., Ishiwata S. Loading direction regulates the affinity of ADP for kinesin. Nat. Struct. Biol. 2003;10:308–311. doi: 10.1038/nsb911. [DOI] [PubMed] [Google Scholar]
- 54.Gigant B., Wang W., Knossow M. Structure of a kinesin-tubulin complex and implications for kinesin motility. Nat. Struct. Mol. Biol. 2013;20:1001–1007. doi: 10.1038/nsmb.2624. [DOI] [PubMed] [Google Scholar]
- 55.Shang Z., Zhou K., Sindelar C.V. High-resolution structures of kinesin on microtubules provide a basis for nucleotide-gated force-generation. eLife. 2014;3:e04686. doi: 10.7554/eLife.04686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider R., Korten T., Diez S. Kinesin-1 motors can circumvent permanent roadblocks by side-shifting to neighboring protofilaments. Biophys. J. 2015;108:2249–2257. doi: 10.1016/j.bpj.2015.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hoeprich G.J., Thompson A.R., Berger C.L. Kinesin’s neck-linker determines its ability to navigate obstacles on the microtubule surface. Biophys. J. 2014;106:1691–1700. doi: 10.1016/j.bpj.2014.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Soppina V., Norris S.R., Verhey K.J. Dimerization of mammalian kinesin-3 motors results in superprocessive motion. Proc. Natl. Acad. Sci. USA. 2014;111:5562–5567. doi: 10.1073/pnas.1400759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Soppina V., Verhey K.J. The family-specific K-loop influences the microtubule on-rate but not the superprocessivity of kinesin-3 motors. Mol. Biol. Cell. 2014;25:2161–2170. doi: 10.1091/mbc.E14-01-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Arpağ G., Shastry S., Tüzel E. Transport by populations of fast and slow kinesins uncovers novel family-dependent motor characteristics important for in vivo function. Biophys. J. 2014;107:1896–1904. doi: 10.1016/j.bpj.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valentine M.T., Block S.M. Force and premature binding of ADP can regulate the processivity of individual Eg5 dimers. Biophys. J. 2009;97:1671–1677. doi: 10.1016/j.bpj.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krzysiak T.C., Grabe M., Gilbert S.P. Getting in sync with dimeric Eg5. Initiation and regulation of the processive run. J. Biol. Chem. 2008;283:2078–2087. doi: 10.1074/jbc.M708354200. [DOI] [PMC free article] [PubMed] [Google Scholar]