Abstract
Organ-on-a-chip devices have gained attention in the field of in vitro modeling due to their superior ability in recapitulating tissue environments compared to traditional multiwell methods. These constructed growth environments support tissue differentiation and mimic tissue–tissue, tissue–liquid, and tissue–air interfaces in a variety of conditions. By closely simulating the in vivo biochemical and biomechanical environment, it is possible to study human physiology in an organ-specific context and create more accurate models of healthy and diseased tissues, allowing for observations in disease progression and treatment. These chip devices have the ability to help direct, and perhaps in the distant future even replace animal-based drug efficacy and toxicity studies, which have questionable relevance to human physiology. Here, we review recent developments in the in vitro modeling of barrier tissue interfaces with a focus on the use of novel and complex microfluidic device platforms.
Keywords: Microfluidic technologies, Barrier tissues, In vitro modeling, Organ-on-a-chip, Drug discovery, Microphysiological systems
Highlights
-
•
Provide an introduction to chip technologies and their applications in drug discovery and disease modeling.
-
•
Review of current organ-on-a-chip technologies in the in vitro modeling of barrier tissues.
-
•
Highlight current chip technologies and address outstanding questions that need to be investigated prior to widespread use.
1. Introduction
Human organs are enormously complex, involving specialized structures, cells, and tissues that interact to carry out unique functions essential to survival. Due to this complexity, it's no wonder that there is currently a lack of reliable model systems to recapitulate tissue and organ level functions in the lab. In the past, researchers were left with two options: Static, in vitro cultures of human cells (primary or from cell lines), or the use of animal models; however, both of these options have flaws. In the case of human cell studies, primary cells are difficult to extract from tissues as a homogeneous population. They are also sensitive to passaging, resulting in early senescence, altered phenotype and metabolic capacities. The culture and use of these cells requires sophisticated techniques and advanced training. To overcome passaging and culturing issues, immortalized cells are often used. These cells are easy to grow, and can be expanded to many passages, however the immortalization procedure results in cells with significant changes that have not yet been studied with enough depth to truly identify differences from primary cell behavior (Astashkina et al., 2012). The most paramount issue with in vitro cell culture however, is the lack of physiological growth conditions that are found in vivo. To overcome this issue, animal models are often utilized, which leads to an entirely different set of limitations. The use of animal test subjects is expensive, lengthy, and at times controversial due to the question as to whether or not animal data can be extrapolated to human systems (Esch et al., 2011). Additionally, there is an innate lack of ability to study single factor changes in whole-body systems due to the sheer complexity of a living organism. It is clear that advances to in vitro culturing methods need to be made to bridge the gap between animal studies and cell culturing studies to address these issues.
Within the last few decades, major advances have been made in microfluidic technologies—specifically with their applications in organ-on-a-chip devices. The term microfluidics refers to a set of technologies that allow for the movement or manipulation of small volumes of liquid or gas. The synergy of microengineering and tissue engineering allows for the fabrication of growth environments that draw from the benefits of both human cell culture studies and animal models, supporting the growth of human cells in physiologically relevant conditions (Bhatia and Ingber, 2014, Schaffner et al., 1995). The term “organ-on-a-chip” has origins in the semiconductor industry where microfluidic technologies began, prior to being adapted and expanded by the micro-electromechanical systems (MEMS) field. Early devices were fabricated from glass (Harrison et al., 1992) and silicon (Van Lintel et al., 1988) in a modified form of photolithographic etching that was used in the manufacturing of computer chips, thus the “chip” in organ-on-a-chip. These materials are brittle, and require access to sophisticated fabrication tools. The development of easy and inexpensive prototyping techniques that utilized elastomer materials allowed researchers to explore the benefits of on-chip tissue culture for many different tissues (Sackmann et al., 2014).
Microfluidic technologies are able to exploit fundamental differences between the physical properties of fluids moving in macroscale systems and those in micrometer-scale channels. First, microfluidic channels can re-create fluidic characteristics that we find at the tissue level in vivo. Within microfluidic channels there is an immense difference in fluid turbulence compared to macroscale fluidic systems. In large scale systems, where inertia dominates viscous forces, fluids mix convectively. In contrast to this, microscale systems, like microcapillaries in tissues, have highly laminar flow. Second, the laminar flow allows for the generation of physical and chemical gradients and highly controlled fluid flow within microfluidic devices, making them attractive platforms for organ-on-a-chip devices. For example, when fluids come together in a channel, they flow in parallel and the only fluid mixing is due to diffusion across the fluid interface (Kamholz and Yager, 2001). This leads to very predictable and controllable fluid interactions through the addition of obstacles or mixing apparatuses into channels to cause perturbations in flow. Third, microfabricated tissue scaffolds can be incorporated into microfluidic systems, providing more in vivo-like microenvironments where tissues can replicate a more in vivo-like architecture. Fourth, within microfluidic systems, the liquid-to-cell ratios are much smaller than in typical well plates. This allows for growth factors to accumulate and affect tissue morphology and function. Additionally, capillary forces and surface and interfacial tension dominate over gravity in microscale systems, allowing for the passive driving of fluids through channels in opposition to gravity (Squires and Quake, 2005, Sung et al., 2010).
The goal of organ-on-a-chip devices is to recapitulate tissue- and organ-level functions in a simple, yet easily manipulated apparatus. The simplest systems involve the growth of one or more cell types in a single, perfused microfluidic chamber to exhibit the functions of one tissue type. More complex devices can involve multiple cell types that are separated by porous layers that mimic basal membranes of barrier tissues (Bhatia and Ingber, 2014). These multi-channel chip devices allow for models of tissue–tissue or tissue–blood interactions and barrier functions and will be the main focus of this review. Additionally, through the connection of these individual organ-on-a-chip devices, recent studies are showing the possibility to observe organ–organ crosstalk. Barrier tissues are of great interest to researchers because they are essential in understanding transport between tissues or tissue and blood, ADMET (absorption, distribution, metabolism, elimination and toxicology), and barrier integrity and function. Here, we will investigate the role of microfluidic technologies and other in vitro methods in the modeling of fluid-to-tissue interfaces from the last few years, specifically with examples in the modeling of skin, lung, gastrointestinal, kidney, endothelium and blood/brain interfaces. We will highlight some of the recent developments in the field, as well as the barriers and challenges faced with the adoption of these relatively new technologies. Additionally, we will discuss positive trends as well as lessons for future microfluidic and in vitro technologies in the modeling of barrier tissues.
2. Applications
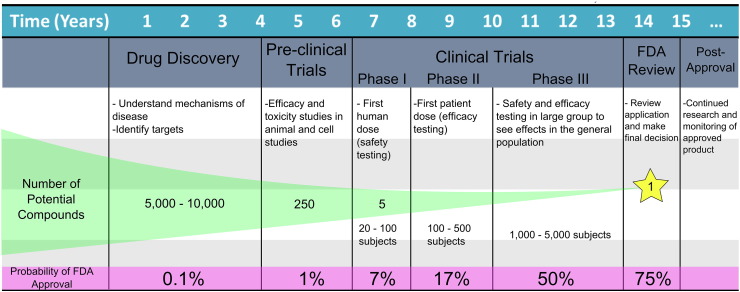
The applications and need for microphysiological devices are potentially far-reaching. These devices may offer improved predictive power for responses of human tissues compared to currently used test methods (both cell-based assays and animal testing). Tests with improved predictive power are desired because current attrition rates in late-stage pharmaceutical testing are high. Based on the results of pharmaceutical studies from 1960 to 2000, it was determined that 25% of drugs entering clinical development fail due to lack of efficacy, 20% from toxicology, and 12% due to clinical safety issues (Kola and Landis, 2004), and testing methods have not changed significantly since then. This year, it was reported by CMR International that even after the ‘First human dose’ (toxicity testing, phase I), there is only a 7% chance for a drug to make it to market, and this only slightly increases to 17% after the ‘First patient dose’ (efficacy testing, phase II) (Fig. 1) (CMR International, 2015). By this time, a large amount of time and money has been invested in the product. The cost to take a compound from concept to market is on average $2.5 billion as reported in the Tufts CSDD 2014 cost study, dwarfing the $802 million estimate in its last major study in 2003, even after being adjusted for post-approval expenses and other costs linked to approval outside of the US market (DiMasi et al., 2014). Additionally, the time required to bring a drug to market is substantial; 10–15 years on average. To reach phase I of clinical testing, it takes an average of 4–6 years (PhRMA, 2015) which goes against the ‘fail fast, fail cheaply’ goal of pharmaceutical companies in weeding out flawed products. No company wants to invest millions of dollars only to find that a drug falls flat in clinical trials, or worse yet, after the drug hits the market. When comparing financial investments to current attrition rates, it becomes clear that more predictive tests need to be implemented. To help direct these studies and possibly eliminate drugs showing toxicity and/or lack of efficacy early, organ-on-a-chip devices with human cells could be used in the evaluation of new drug therapies, vaccines and other biologic agents in a more cost effective, and time efficient manner during pre-clinical testing.
Fig. 1.
Current trends in pharmaceutical development. A model highlighting the time investment, number of potential compounds and probability of approval at each stage of pharmaceutical development from the discovery stage to post-approval (based on information from 2015 CMR International Pharmaceutical R&D Executive Summary and the IFPMA Facts and Figures 2014 report).
Additional applications for organ-on-a-chip devices are expansive, including the assessment of environmental toxin (National Academies, 2014), modeling of disease progression and treatment (Bhatia and Ingber, 2014, Huh et al., 2012), the advancement of personalized medicine through the improvement of screening and monitoring diagnostics (Neuzi et al., 2012, Schumacher et al., 2012), and even the development of vaccines to counter bioterrorism (Wang, 2004). The increased use of organ-on-a-chip devices in various fields can ultimately lead to improved pharmaceutical, health and regulatory decisions. The following section will discuss some of the existing technologies for the in vitro modeling of barrier tissues on chips.
3. Current Technologies in Chip Design/Culturing Methods
Recent advances in microfabrication techniques have made it possible to create more sophisticated cell culture environments, capturing more of the complex function and architecture of organs and tissues than traditional culturing methods. Often these advanced culturing methods rely on soft-lithography based PDMS replica molding, a fast prototyping method that is easily accessible. Many but not all of the technologies described in this review are based on the soft lithography of PDMS, however, novel hydrogel materials and plastic materials like polycarbonate are showing promise and will be discussed as well. The shape and dimensions of these devices can be easily manipulated to mimic a variety of organ/tissue microenvironments. The following section will review recent technologies in the modeling of human skin, lung, gastrointestinal tract, kidney, endothelium and blood–brain barrier tissues.
3.1. Skin
The skin is commonly referred to as our “largest organ”, serving as a protective barrier between the human body and surrounding environment. Understanding skin physiology is necessary in the development of safe topical products like transdermal drugs and cosmetics, but also in wound healing, skin disease, infection, scarring and a variety of other skin conditions. Most in vitro models of the skin have been static cultures that emulate the epidermis only, or combine epidermis with dermis, commonly called “full-thickness skin equivalents” (Black et al., 1999). Further functionality can be added to these basic models to include adipose tissue (lipid metabolism, fibroblast and keratinocyte proliferation), vasculature (oxygen and nutrient supply, removal of metabolites), and even hair follicles (skin metabolism, major penetration route for topically applied substances) (Atac et al., 2013).
Creating in vitro models of the skin is challenging, but advances have been made in reconstructing models in conventional, static growth environments. Full thickness in vitro skin was created by culturing human fibroblast cultures (taken from donor skin samples) in a collagen type I matrix, allowing for the growth of a dermis layer. Once the dermis had been established, human primary keratinocytes and melanocytes are then seeded on top of this layer and a differentiated epidermis is formed, leading to the creation of a human skin equivalent (HSE) composed of associated dermal and epidermal layers reconstructed in vitro. Results indicated that the epidermis contained 3–4 keratinocyte layers and that there was a clear distinction between dermis and epidermis (Souto et al., 2006). However, healthy donor skin samples can have poor donor variation and be difficult to obtain, so in an attempt to mitigate this issue, Reijnders et al. have proposed the creation of a HSE exclusively through the culturing of cell lines. In a manner similar to the previously described HSE creation, fibroblast cell lines were seeded onto a collagen matrix, and keratinocyte cell lines were seeded on top of these sheets once fibroblast proliferation had occurred. This artificial tissue was found to closely resemble the morphology of native tissues and primary cultures as confirmed through the formation of multiple keratinocyte layers and an increase in the expression of inflammatory cytokines and chemokines in response to burn and cold injuries. Additionally it was noted that re-epithelialization occurred after wounding events even in the cell line cultures (Reijnders et al., 2015).
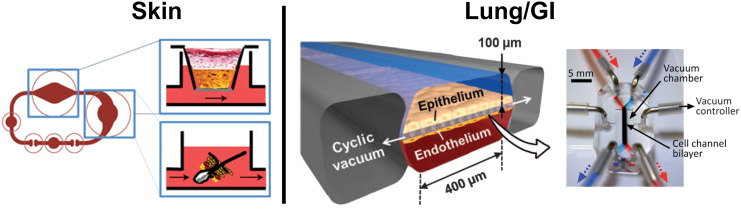
Recent developments in microfluidic technologies have made it possible to investigate the effects of shear stress on skin growth. The work of O'Neill et al. demonstrated that fluid perfusion over keratinocyte cultures increased overall cell viability and confluence compared to static cultures (O'Neill et al., 2008). A more advanced chip design by Atac et al. includes two fluidic chambers; the first containing skin biopsies grown over ex vivo subcutaneous tissue (adipocytes, fibroblasts, macrophages) in a liquid–air interface chamber, and in the second, submerged follicular hair extracts (FUEs) (Fig. 2). It was found that integration of SCT was successful in the chip, while in static cultures, these layers disintegrated. These results are indicative that dynamic perfusion influences cell growth and tissue integration. Additionally, hair shaft elongation was observed in FUEs indicating the possibility of postponing the catagen phase ex vivo (Atac et al., 2013).
Fig. 2.
Mimicking barrier tissues of organs in microdevices. SKIN CHIP: A series of microfluidic chambers were connected to model the skin and hair follicles. A Transwell chamber contained skin biopsies placed over ex vivo subcutaneous tissue with a fluidic basolateral chamber. A fluidic chamber containing follicular hair extracts (FUEs) followed the skin chamber in circuit. Reproduced from Atac et al. (2013) with permission of The Royal Society of Chemistry. LUNG/GI CHIP: This chip design was used to model both the lung and GI barrier tissues. Two fluidic channels run in parallel with a porous membrane to separate epithelium from endothelium. Additionally, empty chambers run along the sides of this device and pull a cyclic vacuum to mimic movement due to breathing or digestion respectively in lung or GI models. GI model reproduced from Kim et al. (2012) with permission of The Royal Society of Chemistry. Lung device from [Huh, D. et al. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci Transl Med, 4, 159ra147]. Reprinted with permission from AAAS.
In an attempt to model cross-talk between skin and liver, Wagner et al. designed a multi-chamber microfluidic device with a liquid–air interface for the growth of skin biopsies followed by a second chamber containing submerged human hepatocytes. All cell types were cultured within the multi-chamber device in HepaRG medium (supplemented Williams' medium E). The results showed that the liver chamber not only produced albumin which was subsequently consumed by the skin chamber, but equilibrium was reached with virtually no excess albumin in the system within a week of culturing (Wagner et al., 2013). This indicates crosstalk between organ chambers and opens up the possibility for a full body-on-a-chip device in the future.
These systems have been validated through testing with well characterized pharmaceutical compounds and shown in vivo-like reactions. For example, Abaci et al. created a fluidic HSE in a design similar to those previously described with an air interface and fluidic basolateral channel. To validate the capability of their platform to be used in drug toxicity studies, a clinically relevant dosing of doxorubicin (anti-cancer drug linked to vacuolar necrosis of keratinocytes) was added to perfusing culture media. Treated cultures displayed spatial detachment at the basal layer and epidermal-dermal interface. Additionally, no keratinocyte proliferation was observed after treatment (Abaci et al., 2015). Additional topical (at air interface) or media-dosing toxicity studies could further validate these models for pre-clinical testing.
3.2. Lung
The blood–air barrier in the lungs exists to prevent air bubbles from entering the blood, and blood and plasma from entering the alveoli. Additionally, this epithelial barrier serves to regulate tissue homeostasis in the lung, and stabilize the airways and keep them from drying through the secretion of surfactant films. This surfactant layer also protects lung tissue from harmful particulates. It is thought that abnormal responses of this epithelial barrier to the surrounding environment (air in the lungs) heavily contributes to the pathogenesis of chronic airway diseases (asthma, COPD, etc.). However, with a current lack of models due to differences in human lung morphology, physiology, and immunology from animal models, this barrier tissue is not yet fully understood.
The majority of in vitro pulmonary tissue models have mainly been created through the use of Transwells to mimic the air–liquid interface of lung barrier tissues (Blume and Davies, 2013). However, the use of 2-layered PDMS microfluidic devices is increasing. Typical design includes a top chamber that is air-filled and permits gas exchange (parenchymal chamber) and an underlying vascular chamber designed to mimic the capillary network, allowing for perfusion underneath the cultured tissues (Long et al., 2012). This liquid–air interface is separated by a porous membrane that allows for cell seeding and growth (Takayama et al., 2014). The work of Fritsche et al. demonstrated that immortalized mouse lung epithelial cells could be cultured on the surface of polystyrene microcarrier beads and injected into the parenchymal chamber, allowing for an even distribution of cells on the compartment surface. Additionally, cell specificity and functional capacity were demonstrated through positive staining for alveolar epithelial markers (Fritsche et al., 2009). Blume et al. used a similar design in their work to model inflammatory reactions of primary bronchial epithelial cells to pollen (characterized by IL-8 and CXCI release), which had not previously been observable in static cultures, indicating a higher sensitivity to environmental factors in chip devices with incorporated physiological shear stress (Blume et al., 2015). Liquid plug propagation was also investigated in the presence or absence of secreted and added surfactants using this same design (Tavana et al., 2011). The work of Huh et al. advanced this 2 channel chip design a step further and cultured vascular endothelial cells in the fluidic chamber on the bottom of the membrane, and alveolar epithelial cells on the membrane in the top channel. This co-culture allowed for a better formation and mimicking of barrier function. Additionally, they investigated the role of mechanical strain on the barrier tissue through the addition of air filled chambers on the sides of the channel with vacuum suction to cause slight deformations, mimicking the stretching of alveolar tissues during normal breathing (Fig. 2) (Huh et al., 2010). With this unique design, this group also mimicked a drug toxicity-induced pulmonary edema with interleukin-2, indicating the influence of both fluid flow and mechanical strain on vascular leakage (Huh et al., 2012).
3.3. GI
The gastrointestinal tract has the largest surface of any tissue within the human body exposed to the external environment. Its main function is selective absorption of nutrients from orally consumed matter, while maintaining a barrier between this foreign matter and tissues that may have an inflammatory reaction. Low bioavailability (fraction of an administered dose of unchanged drug that actually reaches circulation) of drugs has been one of the major reasons cited for the failure of novel pharmaceutical compounds entering clinical trials. Unfortunately, most in vitro drug/toxicity studies do not address bioavailability of orally administered drugs because the drug is added directly to tissue, when by contrast, oral administration is the preferred route. Therefore the pre-digestion, uptake and transformation of these compounds by the GI are rarely considered. Another limitation of previous models is the lack of living microbes on the luminal surface of cultured intestinal epithelium. Intestinal microbial symbionts contribute significantly to intestinal barrier function, metabolism, disease, and absorption of compounds (Round and Mazmanian, 2009, Turner, 2009). Therefore, testing systems that contain models of these barrier tissues with symbionts are invaluable for the correct prediction of drug bioavailability as early as possible in the drug development process.
In an effort to match the unique 3D shapes of the gut epithelium, Esch et al. have developed a protocol for the fabrication of a microporous polymeric membrane from partially polymerized SU8. They demonstrated that when cultured upon substrates with these unique shapes, Caco-2 cells mimicked key aspects of the gut epithelium. This technique can create highly controlled shapes allowing for the mimicking of a variety of different barrier tissues (Esch et al., 2012). This 3D geometry is relevant in the GI tract because cells can localize in different areas based on scaffold shape. Studies have shown that both protein expression and differentiation gradients are present in cultures grown on 3D, villus-like geometries, with more columnar and polarized cells near the top of the villus than in the crypt regions. This follows the observation that enterocytes become more differentiated as they migrate to the tip of the villus (Yu et al., 2012). This localization is also observed in gut bacteria, as many strains preferentially adhere to epithelial cells at various stages of differentiation (Costello et al., 2014). Works by Kim et al. have involved the use of 2 channel devices, very similar to those used in the modeling of lung barrier function, fabricated from PDMS. However, with this GI barrier, there is medium perfusing in both the top and bottom channels to mimic the barrier between gut epithelium and the bloodstream. Vacuum chambers were also present along the sides of these channels to allow for peristaltic-like movement that occurs naturally during digestion (Fig. 2). Co-cultures of gut epithelium with intestinal microbe Lactobacillus rhamnosus GG were viable and increased barrier function as indicated by heightened transepithelial electrical resistance (TEER) in fluidic cultures (static cultures experiences epithelial detachment). Additionally, TEER was 3–4 fold higher within the chip device compared to static cultures, nearing in vivo values, and there was evidence that peristaltic motions increased the permeability of cell layers without altering TEER, indicating a direct effect on paracellular mechanisms of transport (Kim et al., 2012). After long-term culturing within this device, macrovilli-like folds formed in epithelial monolayers increasing surface area by over 1.7-fold. Cells expressed microvilli forming brush borders, and well defined junctions. Cell reorganization also took place, with ratios and localization of absorptive, enteroendocrine, goblet and Paneth cells similar to those found in vivo (Kim and Ingber, 2013). These macrovilli and cell organizations have not been observed under traditional static culturing methods. Additionally, Mahler et al. created a static, Transwell tri-culture of Caco-2, HT29-MTX and Raji B cells that mimics the enterocytes, goblet cells, and M (microfold) cells that populate the healthy intestinal lining. Caco-2 and HT29-MTX cells were seeded in the apical chamber of a Transwell, and Raji B in the basolateral chamber. This tri-culturing method resulted in a physiologically relevant number of differentiated M cells, which play major roles in transport and barrier function (Mahler et al., 2012).
Finally, multi-organ systems incorporating the liver have recently come into focus. Work by Mahler et al. involving a multi-organ system with gut epithelium and a liver compartment demonstrated a “first pass” metabolism of acetaminophen (APAP) in a gut and liver chamber, highlighting the importance of pre-digestion and metabolism in the gut that occurs during oral administration of a drug. It was observed that within this system, APAP was absorbed and metabolized by the gut epithelium, then went on to be further broken down in the liver compartment, where toxic metabolites formed in a dose-dependent manner similar to in vivo results (Mahler et al., 2009). In similar work, Esch et al. coupled a fluidic GI (caco-2/HT29-MTX) and liver (HepG2/C3A) tissue microphysiological system to investigate the effects of ingested nanoparticles (50 nm) on the liver. Nanoparticles were either added to the multi-organ system, where they would pass through the GI module prior to reaching the liver, or to a liver-only control device. The GI module prevented 90% of nanoparticles from crossing the epithelial barrier, and the remaining nanoparticles reached the liver module, causing the release of aspartate aminotransferase (AST, an injury marker). This injury was observable at lower concentrations than was observed in liver-only models, indicating that compounding effects, or alteration of nanoparticle properties occurred in the GI module (Esch et al., 2014). In both of these multi-organ studies, HepG2/C3A and Caco-2/HT29-MTX cells were initially cultured in MEM (Minimum Essential Media) and DMEM (Dulbecco's Modified Eagle Medium), respectively. However, when cell types were combined within the multi-organ system, MEM was used throughout. These couplings of organ-on-a-chip devices allow for more in vivo-like cell behaviors and properties, but raise questions as to the need of a universal medium to support cell function as well as emulate blood's carrying capacity for drugs and metabolites.
3.4. Kidney
The kidney filters compounds from the bloodstream and is one of the main sites of elimination for drugs circulating in the body. Understanding the barrier functions of the kidney is essential in the development of drugs for the lengthening of residence time in the bloodstream and the subsequent removal of these compounds after they have taken effect on their target tissues. Additionally, the renal epithelial barrier is heavily involved in the reabsorption of useful compounds like sugars, salts, small proteins and water. As these compounds are removed from urine precursor, any drugs or compounds left behind are effectively concentrated, leading to many unexpected toxic effects. Kidney toxicity is one of the most frequently reported toxic effects during drug development, and renal toxicity is often only detected late into clinical trials (Bonventre et al., 2010, Laverty et al., 2011). Therefore an in vitro model that takes into effect reuptake in the kidney is essential in predicting renal toxicity early in the drug development process.
Shear stress plays a major role in renal barrier function as demonstrated through transcriptomic (gene) and proteomic (protein) analysis in MDCK (Madin-Darby canine kidney, proximal and distal tubule) cells. When cells were grown in a fluidic channel, a significant upregulation of ion transporters involved in calcium, phosphate and sodium homeostasis was observed, as well as genes involved in H+ transport and pH regulation of the urine. Additionally, there was a noted upregulation in phase I and II enzymes, multi-drug resistance genes and phase III transporters, allowing for a more in vivo-like metabolism of drugs (Snouber et al., 2012). Shear stress also plays major roles in protein handling, as demonstrated by Ferrell et al., who created a multi-layered device (fluidic top chamber, media reservoir on bottom, separated by a porous membrane) to demonstrate albumin handling of renal epithelial cells under fluidic stress. It was determined that physiological levels of shear stress significantly increased uptake and degradation (metabolism) capacity of cells (Ferrell et al., 2012). Work by Jang et al. using a similar chip design (Fig. 3) while culturing collecting duct (Jang and Suh, 2010), distal tubule (Jang et al., 2011), and proximal tubule (Jang et al., 2013) cells also reported increased effects on the translocation of aquaporin-2 (AQP2) and reorganization of F-actin in the presence of shear stress, hormonal stimulation (arginine vasopressin), and an osmotic gradient compared to static controls. In their studies with proximal tubule cells, it was also demonstrated that fluid shear stress can have a protective effect against cisplatin toxicity with a much smaller degree of LDH release and apoptosis observed in fluidic cultures compared to static controls, resulting in a higher fidelity toxicity response. In an effort to model proteinuria (high amount of serum in the urine, normally blocked by the glomerulus), Zhou et al. created a 3D chip that incorporates layers of BME (basement membrane extract) in a fluidic chamber, making it possible to observe EMT (epithelial to mesenchymal transition) and the subsequent migration of these cells into the BME. Cells that were exposed to human serum became apoptotic, or developed a mesenchymal phenotype, however cells exposed to heat inactivated serum were unaffected (Zhou et al., 2014). This device can be used to investigate the roles of EMT in the pathogenesis of renal fibrosis and loss of barrier function. Finally, in a move away from PDMS devices, fibrillogenesis of collagen in a liquid molding technique has recently been investigated for applications in nephron modeling. Mu et al. have developed a collagen and alginate device that contains two parallel tubes (vascular and nephron) that allow for passive diffusion through the device material (Mu et al., 2013). Additionally, renal epithelial cells were able to seed on all internal surfaces of the channel, whereas cells do not naturally “stick” to PDMS. This device design shows much promise for future applications in the modeling of barrier tissues due to the more realistic ECM-like material and possibility for passive diffusion between parallel channels, while still maintaining the separation of tissues.
Fig. 3.
Mimicking barrier tissues of organs in microdevices. KIDNEY CHIP: Kidney tubular epithelial cells were seeded onto an ECM-treated membrane in a fluidic, top channel. Underneath the membrane was a media reservoir to provide nutrients to the basolateral surface of the cell monolayer. Reproduced from Jang and Suh (2010) with permission of The Royal Society of Chemistry. ENDOTHELIUM CHIP: Fully enclosed, perfusable vessels formed in a collagen hydrogel. This hydrogel allows for endothelial growth on all internal surfaces of the vessel, allowing for a close recapitulation of blood vessel formation. Adapted by permission from Macmillan Publishers Ltd.: [NATURE PROTOCOLS] (Morgan et al., 2013), copyright (2013). BLOOD–BRAIN CHIP: A multi-layered microfluidic device with incorporated electrodes for internal TEER measurements. Astrocytes and brain endothelial cells were seeded on opposite sides of the membrane in a back-to-back co-culture resulting in higher resistance to permeability and barrier function than endothelial monocultures. Reproduced from Booth and Kim (2012) with permission of The Royal Society of Chemistry.
Finally, a microfluidic model of the kidney (MDCK) has been combined with a model of the liver (HepG2/C3a or HepRG) in order to observe the synergistic interaction between these organs in the presence of ifosfamide (an anticancerous drug), which, when metabolized, forms a nephrotoxic compound, chloroacetaldehyde. Fluidic cultures of HepaRG, and HepaRG/MDCK co-cultures were grown in HepaRG medium (without DMSO), and HepaG2/C3a and co-cultures with MDCK in MEM. Ifosfamide (50 μM) was either added to the fluidic multi-organ device, or to a kidney only device to validate this metabolic model. After a 72 h exposure, no nephrotoxic effects were observed in the kidney-only devices, however the kidney cells in the multi-organ device exhibited cell death, and an increased release of intracellular calcium indicating cell distress caused by the formation of toxic metabolites (Choucha-Snouber et al., 2013). This study highlights the need for multi-organ systems that address systemic organ-organ interactions, which may potentially allow for more effective predictive toxicology models than single-tissue systems.
3.5. Endothelium
The vascular system provides the perfusion of blood, containing nutrients and drugs to tissues and organs, while also removing waste and metabolites. Blood vessels are an integral part of the body and serve as a connection for all tissues and organs, with a low and selective permeability to fluid and solutes under normal physiological conditions. Additionally, the endothelial barrier in these vessels mediates the transfer of solutes and cells between the bloodstream and surrounding tissues, playing a central role in the regulation of metabolic activity, wound healing, immune response and disease progression. However, the breakdown of this barrier can lead to edema (leaking of watery fluids into surrounding tissues) and inappropriate interstitial forces. The endothelium is also of interest in the study of tumor metastasis, due to tumor cell interactions with endothelial barriers during organ invasion. It is therefore important to understand the functions of the endothelium to maximize selective drug delivery and the understanding of disease mechanisms.
Van der Meer et al. demonstrated that it is possible to control the shapes of vessel networks in addition to shear stress through their growth in PDMS channels within a microfluidic device. HUVECs and pericytes (10:1) were seeded into PDMS channels with collagen type I and within 12 h, formed tubular-like structures, following the contours of the microfluidic channels (van der Meer et al., 2013). There has also been evidence of cross-talk between endothelial cells and their surrounding tissues in unique cell–cell interactions. Chen et al. fabricated a bi-layer PDMS microfluidic device for the co-culture of porcine valvular endothelial cells (VECs) and valvular interstitial cells (VICs). The fluidic chamber on top contained VECs seeded on a fibronectin treated membrane, and the bottom chamber (static) contained VICs encapsulated in a methacrylated gelatin hydrogel. Utilizing this device, it was possible to investigate the roles of both shear stress and VEC co-culture on the differentiation of VICs to myofibroblast phenotypes. It was observed that the presence of endothelial cells significantly decreased VIC differentiation (78% VICs alone, 29% co-culture) and that the introduction of relevant shear stress to the fluidic chamber enhanced this effect (17% differentiation of VICs) (Chen et al., 2013). Therefore, both bio-mechanical and -chemical signaling factors must be considered in the design of healthy microvessels. In addition, Douville et al. have described a method for creating a 2-layer PDMS microfluidic device with embedded electrodes for the purpose of evaluating endothelial barrier function without disrupting cell growth. This device was tested with brain and kidney endothelial cells, as well as myoblast cells. Clear differences in epithelial resistance were observed in these various cell types, indicating proper cell growth and electrode function. Additionally, readings responded in real-time to changes in TEER as indicated by significant drop-offs when TritonX100 permeabilizing agent was added to culture media (Douville et al., 2010).
In an attempt to replace PDMS, a number of endothelial barrier models were created in collagen hydrogel channels. Morgan et al. developed a procedure for creating 3D cell cultures in collagen hydrogel, leading to fully enclosed perfusable vessels in an easily manipulated matrix (Fig. 3) (Morgan et al., 2013). HUVECs were successfully grown within these enclosed channels and displayed appropriate morphology, barrier function, angiogenic remodeling, and cell–cell junctions. In addition to modeling a relevant ECM, Price et al. investigated the effects of mechanical signals (shear stress, transmural pressure, and luminal pressure) on barrier function and microvessel stability with BECs (endothelial cells from human dermal blood microvessels). Results of these studies indicated that high shear stress (15–20 dyn/cm2) lead to barrier function comparable to that observed in vivo, and a 2-fold increase in longevity over studies with low shear stress. Additionally, positive transmural pressure was associated with vessel stability, resulting in a much lower degree of endothelial delamination than controls (Price et al., 2010). These porous materials also allow for investigations into angiogenesis and endothelial sprouting. Verbridge et al. created 3 parallel channels in collagen hydrogel. The central channel contained endothelial cells, with the neighboring channels serving as a source and sink for a VEGF biochemical gradient. This gradient promoted endothelial sprouting and invasion into the surrounding collagen matrix which would not be possible with less porous materials like PDMS (Verbridge et al., 2013). These in vitro models demonstrate controlled vessel formation, cell reactions to both mechanical and chemical stimulations, and a viable method for studying barrier function without disrupting cell layers. Additionally, the move to more porous materials like hydrogels allows for more accurate drug transport and diffusion across endothelial barriers and ECM.
3.6. Blood–Brain
The blood–brain barrier is a highly selective (metabolic and biochemical) blockade against the entry of substances into the brain tissue. This makes drug development targeting brain disease extremely difficult because passive diffusion across this membrane is almost non-existent with the exception of small molecules (< 500 Da), which have had little/poor response in the treatment of most diseases (Pardridge, 2003). Additionally, this barrier protects the brain against fluctuations in plasma composition, maintains parenchymal homeostasis and transport through unique, asymmetrically expressed carrier-mediated trans-membrane transport systems. Due to the fact that there are a lack of fenestrations and a very small amount of pinocytosis in the endothelial cells lining this barrier, in vitro modeling could allow for a greater understanding and testing of innovative methods for drug delivery that exploit existing carrier-mediated transport systems.
Considering that the majority of in vivo drug transport studies are on mice (current in vitro models use rat, bovine or porcine endothelial cells), Shayan et al. developed a murine model of the blood–brain barrier. This co-culture involved murine BMECs (brain microvascular endothelial cells) cultured with rat primary astrocytes. These static, Transwell models involved either a back-to-back co-culture of cells (endothelial cells on top, and astrocytes on the bottom of the membrane) or control cultures with astrocytes grown in a separate well. It was determined that back-to-back cultures displayed higher TEER and lower permeability to sodium fluorescein. Additionally, an overall 2.5-fold increase in functional proteins and transporters was observed by western blotting, and in a series of tests with hydrophobic and hydrophilic drugs, an overall correlation of 0.98 was determined in comparison to in vivo permeability values (Shayan et al., 2011a). In later work, it was also demonstrated with the same Transwell culturing method that back to back culturing of astrocytes with BMECs can increase TEER of the endothelial cells by up to 167%, and that this increase is specific to BMECs only, as the addition of astrocytes to aortic endothelial cells had no effect on TEER (Shayan et al., 2011b). In similar work, Booth et al. cultured brain endothelial cells in the presence and absence of astrocytes in a fluidic multi-layered PDMS device with integrated TEER electrodes to measure barrier function (Fig. 3). Dynamic flow was introduced in both the top and bottom fluidic channels, and astrocytes and endothelial cells were seeded on opposite sides of the membrane between the fluidic channels, resulting in a back to back co-culture. Epithelial resistance of the blood–brain barrier within the fluidic device was much higher than values observed in back-to-back static control Transwell cultures (25 vs 250 Ω ∗ cm2 in controls against chip devices respectively), indicating a strong influence of shear stress on epithelial barrier function. Additionally, these TEER results were confirmed through permeability studies with fluorescent dextrans, resulting in a significantly higher permeability in static controls and fluidic endothelial mono-cultures compared against fluidic co-cultures (Booth and Kim, 2012). Griep et al. also confirmed these modulations in epithelial resistance caused by the introduction of a dynamic fluidic environment through the use of an immortalized human brain cell line (hCMEC/D3) in a similar device design (Griep et al., 2013). Finally, work by Achyuta et al. combined neural cells obtained from primary rat cortical tissue (4% neurons, 95% astrocytes, and 1% microglia) with neurovascular cells (RBE4) in a multi-layered PDMS microfluidic device. Vascular cells were cultured in the fluidic top chamber, and neural cells in the bottom, static reservoir of the device. After 10 days of culture, neural cells showed evidence of firing inhibitory and excitatory potentials, and endothelial cells expressed von Willebrand factor (vWF), ZO-1 tight junctions, and diluted-acetylated low density lipoprotein (dil-a-LDL) uptake. These results, in combination with a reduced permeability of dextran across the endothelial barrier indicate recapitulation of the functions of “Neurovascular Units” (Achyuta et al., 2013). The results of these studies are promising, providing for unique opportunities in the in vitro study of nutrient, therapeutic agent, and nanomaterial delivery across this highly selective barrier tissue. However, although the co-culture of brain endothelial cells with astrocytes has a synergistic effect on the overall TEER and decreased permeability of this barrier tissue, limitations still remain with these models. Due to the low availability of human brain cells, and the low yield of BMEC isolations from animal subjects, many singular cell isolations must be performed for model scalability. Additionally, many of the models described above involve mixed species co-cultures of BMECs and brain cells, which is suboptimal compared against syngeneic co-culturing. With recent advances in the use of neural progenitor cells (NPCs) and human pluripotent stem cells (hPSCs), an all-human in vitro BBB model might soon be possible (Lippmann et al., 2011).
4. Advantages Over Current Methods
The use of microfluidic-based in vitro modeling has many advantages over traditional testing methods for clinical studies. In comparison to static in vitro culture, microfluidics allow for dynamic fluid flow through cultures. This highly controlled dynamic cell growth environment has been linked to increased cell growth, differentiation, and polarization, allowing for more in vivo-like tissue growth and behavior (Jang et al., 2011). Additionally, fluid flow enables ADMET properties, allowing for more accurate predictions of physiologically based pharmacokinetic (PBPK) properties than with static systems. The combination of microfluidics with this in silico modeling has the possibility for more accurate efficacy and toxicity predictions (Fig. 4). Fluid flow has also been linked with longer culture periods, up to a month as shown in microfluidic modeling of lung tissue (Huh et al., 2010). This allows for more clinically relevant time scales as many diseases develop over chronic exposure, or can take time to express symptoms. Microfluidics by definition also utilize smaller media volumes than static cultures in wells, using only a fraction of the volume of traditional cultures over the same area of cells. This ratio of fluid to tissue is a closer match to that found in the body, and leads to a smaller reagent requirement, reducing cost and maximizing information obtained from valuable samples early on in the drug development process. Finally, microfluidic chambers allow for the possibility of tissue/organ interconnection. Fluidic chambers can be connected to one another, allowing for a degree of fluidic interaction and cell signaling while maintaining physical separation, an application that is not currently possible in static cultures. Microfluidic in vitro modeling also has many advantages over in vivo animal studies.
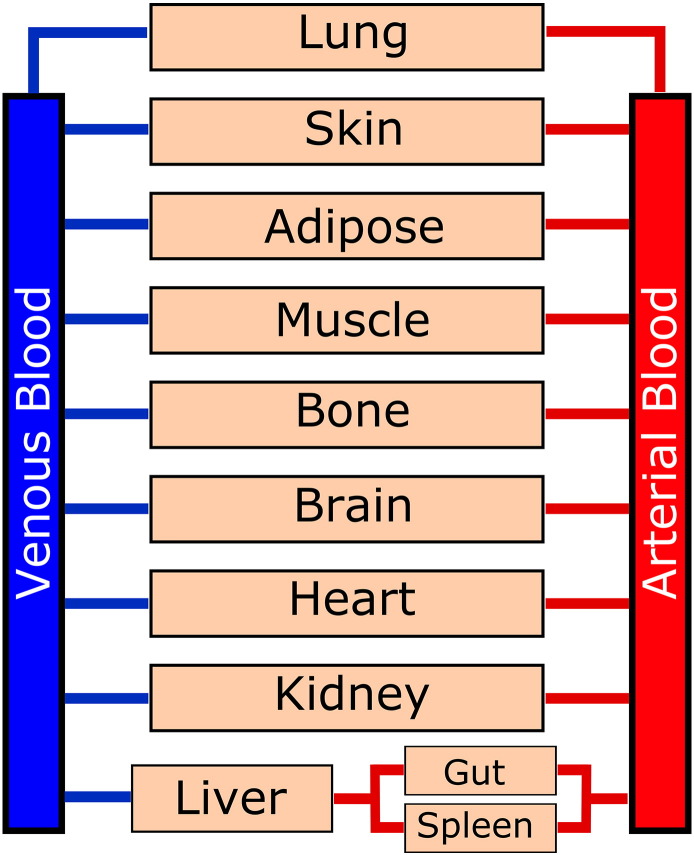
Fig. 4.
Physiologically-based pharmacokinetic (PBPK) model of a human system. This model shows the blood circulation throughout the human body through various tissues. Future multi-organ microfluidics will likely be designed following this type of schematic, with various residence times and permeability in each organ chamber.
One of the greatest flaws in animal testing is the questionable correlation of animal data to human systems. This often results in either a lack of efficacy or unexpected toxicity when transitioning from animal studies to human clinical trials. The use of in vitro microfluidic devices has the potential to overcome this issue through the use of human cells or tissues (in the case of tissue explants) in physiologically relevant microenvironments. By matching the growth environment of these tissues within a microfluidic device, it is possible to observe potential drug interactions as they would translate to human systems, potentially allowing for much better drug efficacy and toxicity predictions. Additionally, imaging is much simpler in microfluidics due to the transparent nature of many fabrication materials, and the control of tissue and cell positioning which is not possible in living systems. This allows for real time imaging under flow without disrupting cell growth. Microfluidics also allow for high throughput sample processing and more realistic sample sizes than with animal studies, where it can be time consuming and expensive to obtain a large sample of animal specimens. This throughput is perhaps one of the greatest advantages of the use of microfluidic technologies. Through the use of multi-organ systems like those described in this paper, it is possible to simultaneously assess the effects or toxicity of drugs and their metabolites on multiple tissue types, further reducing testing times. This type of high throughput analysis can allow for rapid, inexpensive and highly efficient testing. Additionally, it is possible to incorporate systems to monitor tissue and barrier health (TEER, automatic sampling, etc.) within these engineered devices, streamlining the testing process even further. Finally, many pathologies of toxicity and disease are poorly understood on molecular and biochemical levels which are challenging if not impossible to observe at the macro scale within a living system. Microfluidics offer an alternative at a small enough scale, where cell–cell or cell–tissue interactions can be simplified and accurately emulated, allowing for easily observable interactions.
5. Limitations
Though the use of organ- or tissue-on-a-chip devices shows a great deal of promise for future applications, it is important to remember that these are relatively new technologies and require further advancements prior to widespread use. It is first necessary to characterize these devices with drugs that have well studies ADMET properties, and to validate the relevance to clinical efficacy and toxicity. Additionally, the devices that are currently being used in the lab are labor intensive to create, and require specialized training to maintain. There are many technical challenges with fabrication processes, issues with bubbles and flow perturbations that can destroy cell cultures, and issues with cell contamination in porous microfluidic devices. The use of a rocker platform or a gravity-driven flow can aid in the elimination of bubbles for some microfluidic platforms, however this solution does not address unidirectional shear flow (Sung et al., 2010, Esch et al., 2015). Additionally, consistent cell seeding can be difficult to achieve within complex channel designs, and the simplified ECMs commonly used in culturing can lead to matrix degradation or contraction. To create a more complex ECM, some groups have used Matrigel, however this ECM is derived from tumor cells and is highly dissimilar to normal cell ECM and can have a high batch-to-batch variance. In addition to ECM, cell media does not reflect in vivo context, and can also impact cell phenotype (Kolbe et al., 2011). In order for these technologies to transition to industrial and clinical use, it is necessary to create “user friendly”, robust and scalable testing systems.
In addition to concerns with fabrication and cell maintenance, there is a consensus among all researchers that no in vitro culture will ever completely represent the complexity of whole animal systems. The feedback mechanisms of extensive interrelationships and crosstalk from multiple cell types and dynamics that modulate physiological processes are currently very difficult to recapitulate in vitro. Additionally, adaptive immune responses, and complex system-level behaviors of the endocrine, skeletal and nervous system have not yet been investigated (Astashkina et al., 2012). Finally, there will always be an issue of systematic, “off target” toxicity. In vitro studies often involve just a few cell or tissue types at most, but often toxicity can crop up in areas of the body that weren't necessarily a “target” of the drug being introduced. As a result of these limitations, in vivo animal studies retain the advantage of possibly uncovering such off target responses. However, with differences in animal and human physiology, such results are often not predictive of human response. Using such animal studies to suggest which organ compartments need to be added to the in vitro human model should be valuable. Chip devices need to make many advancements before modeling full tissue or organ level functions, but will have many exciting applications for future drug discovery, and disease modeling and treatment.
6. Outstanding Questions
In addition to the limitations mentioned above, there are a few outstanding questions that must be addressed prior to the widespread use of lab-on-a-chip devices. The first, and most concerning is: Can these devices really mimic organ level functions and interactions? Macro-scale architecture, and micro-scale spatial heterogeneity found in organoids and tissue sections can be very difficult to recapitulate in microfluidic channels, but play large roles in many physiological functions. Additionally, how can we account for the function of organs regulated by humoral, neurogenic, and metabolic factors? With existing organ on a chip technology, these factors can only be fully accounted for in whole-body, in vivo animal models.
We must also consider the availability of relevant human cells. It can be difficult to obtain healthy primary cells from a diverse population, so an alternative must be considered. Recently, it has been suggested that human induced pluripotent stem cell technologies could be utilized to provide relevant cell models, additionally establishing the possibility for personalized medicine (Williamson et al., 2013, Bellin et al., 2012). Cell availability is not only an issue with healthy tissue modeling, but must also be considered in disease modeling. Can a cell phenotype be discerned that represents the pathophysiology of underlying diseases? Further along this avenue, can a phenotype be altered in vitro in such a way that potential therapies might emerge? Another imperative question is whether it will be possible to develop a universal blood substitute for microfluidic culturing (Schaffner et al., 1995, Guo et al., 2011). Currently used media have been optimized for specific cell types, but in order to see tissue–tissue interactions in more complex multi-organ chamber devices, it will be necessary to have a universal solution that will equally supplement multiple cell types. The papers referenced earlier in this review that incorporated multiple cell/tissue types into multi-chamber devices faced this very issue and offer potential models to solve these problems. Questions pertaining to device fabrication materials have also been of recent concern. There has recently been a push toward thermoplastic materials (polystyrene, cyclic olefin copolymer, polymethyl methacrylate, polycarbonate) over elastomers (PDMS), which are used in the majority of organ-on-a-chip devices because of their low cost, high availability and ease of use in soft lithography. Recent studies have determined that elastomer materials can leach uncrosslinked oligomers into solution, absorb small molecules (affecting cell signaling, and pharmaceutical dosage), and have higher vapor permeability (evaporation can lead to detrimental effects on micro- and nanoliter fluid volumes) (Toepke and Beebe, 2006, Regehr et al., 2009). Finally, when considering device design, it is important to ask: What types of real-time detection and analysis can be incorporated into chip devices? Many of the works described in this review have incorporated TEER, and fluorescent or optical based measurements, however many recent advances in label-free protein assays have been reported. Electrical biosensors and automated sampling for label-free detection of protein and disease biomarkers have proven to be useful, however their incorporation into chip systems has not yet been investigated (Luo and Davis, 2013). Additionally, the use of universal assays will need to be incorporated at a larger scale. Current assays generally focus on cell function and behavior, but organs and tissues also need to be evaluated at a functional level. As an example, Stancesu et al. monitored the contractile stress of human cardiomyocytes and used Multi-electrode arrays (MEAs) to measure electrical activity in an in vitro model of the heart. This data, coupled with cell-based assays could provide relevant predictions for drug toxicity and efficacy ranging from cell–cell to systemic scales (Stancescu et al., 2015). It is also important to consider, when designing these monitoring and testing methods that with low cell culture volume and cell count compared to traditional culturing, it might be difficult to detect biological signals at this scale. These sensors will be required to have high sensitivity to detect changes in biological responses. Ultimately, the main question to be asked is: which is more important, complexity or practicality? It may, in the future, be possible to design whole-body microfluidics that can account for systemic organ interactions and signaling, and parallel the function of the human body, but would this sort of system be practical? The more intricate the device becomes, the more difficult it becomes to manufacture, distribute, and train operators. A reason balance between these two opposing forces must be reached in order for the creation of a practical, yet insightful technology to be widely utilized.
Once these questions have been addressed, microfluidic-based chip technologies have the potential for extensive breakthroughs in drug discovery, disease modeling and furthering our understanding of organ and tissue interactions within the human body. This novel modeling technology has many interesting applications that are just beginning to be explored. For example, disease modeling has already been highlighted, but further along that route is understanding cancer formation and metastasis. It is currently understood that cancer growth and spread is not only affected by the surrounding chemical environment, but also the surrounding ECM and mechanical factors such as shear stress which can easily be manipulated in microfluidics (Chivukula et al., 2015). Additionally, the study of many rare diseases may be possible. These diseases often have small sample populations for study and therefore suffer from a low availability for in vivo studies. If replicated in vitro, it may be possible to study a much larger population size and gain insight into some of the mechanisms of these diseases. Another interesting application is for stratified medicine—developing a drug for a particular set of the population, rather than a “one size fits all” approach to medical treatments (Trusheim et al., 2007). For example, treatments could be developed based on the presence or absence of particular biomarkers. High throughput testing methods, like the use of microfluidic in vitro models, will assist greatly in the development of these stratified treatments at the pharmaceutical development and testing stages. Additionally, these technologies have applications in predicting appropriate drug doses or administration regimens to achieve desired effects prior to clinical dosing tests. Finally, the study of nervous, endocrine, sensory, and reproductive systems for which we currently lack dynamic models will likely be of focus in the near future.
Understanding how barrier tissues function is essential for the design of effective pharmaceutical compounds. Once these devices are transitioned from research to clinical use, and we have more accurate predictive models early on in the development process, we will likely see an end to the recent stagnation in pharmaceutical development. Additionally, once pluripotent stem cells are incorporated into these technologies, there will presumably be a huge push for use of organ- or body-on-a-chip devices in personalized medicine, allowing for patient-tailored disease treatments and diagnostic testing. Overall, the field of in vitro modeling, and many others will greatly benefit from the widespread use of organ-on-a-chip devices as we move into the future.
7. Search Strategy and Selection Criteria
Material for this review was selected through searches on PubMed, MEDLINE, and from references contained in relevant articles, using the search terms: “microfluidics”, “microphysiological”, “lab-on-a-chip”, “in vitro modeling”, and “barrier tissues” in their applications for the modeling of the tissues discussed in this review. Only articles published in English were selected, with a focus on articles published after 2010.
Acknowledgments
We gratefully acknowledge support, in part, from the National Center for Advancing Translational Sciences at the National Institute of Health (UH2TR000156-01), the Clifford D. Clark Graduate Fellowship, NSF CMMI 1436173, NIH 1R15ES022828, and the Research Foundation for the State University of New York. These funding resources did not play any role in the writing of this manuscript.
References
- Abaci H.E., Gledhill K., Guo Z., Christiano A.M., Shuler M.L. Pumpless microfluidic platform for drug testing on human skin equivalents. Lab Chip. 2015;15:882–888. doi: 10.1039/c4lc00999a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achyuta A.K., Conway A.J., Crouse R.B., Bannister E.C., Lee R.N., Katnik C.P., Behensky A.A., Cuevas J., Sundaram S.S. A modular approach to create a neurovascular unit-on-a-chip. Lab Chip. 2013;13:542–553. doi: 10.1039/c2lc41033h. [DOI] [PubMed] [Google Scholar]
- Astashkina A., Mann B., Grainger D.W. A critical evaluation of in vitro cell culture models for high-throughput drug screening and toxicity. Pharmacol. Ther. 2012;134:82–106. doi: 10.1016/j.pharmthera.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Atac B., Wagner I., Horland R., Lauster R., Marx U., Tonevitsky A.G., Azar R.P., Lindner G. Skin and hair on-a-chip: in vitro skin models versus ex vivo tissue maintenance with dynamic perfusion. Lab Chip. 2013;13:3555–3561. doi: 10.1039/c3lc50227a. [DOI] [PubMed] [Google Scholar]
- Bellin M., Marchetto M.C., Gage F.H., Mummery C.L. Induced pluripotent stem cells: the new patient? Nat. Rev. Mol. Cell Biol. 2012;13:713–726. doi: 10.1038/nrm3448. [DOI] [PubMed] [Google Scholar]
- Bhatia S.N., Ingber D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014;32:760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- Black A.F., Hudon V., Damour O., Germain L., Auger F.A. A novel approach for studying angiogenesis: a human skin equivalent with a capillary-like network. Cell Biol. Toxicol. 1999;15:81–90. doi: 10.1023/a:1007541713398. [DOI] [PubMed] [Google Scholar]
- Blume C., Davies D.E. In vitro and ex vivo models of human asthma. Eur. J. Pharm. Biopharm. 2013;84:394–400. doi: 10.1016/j.ejpb.2012.12.014. [DOI] [PubMed] [Google Scholar]
- Blume C., Reale R., Held M., Millar T.M., Collins J.E., Davies D.E., Morgan H., Swindle E.J. Temporal monitoring of differentiated human airway epithelial cells using microfluidics. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonventre J.V., Vaidya V.S., Schmouder R., Feig P., Dieterle F. Next-generation biomarkers for detecting kidney toxicity. Nat. Biotechnol. 2010;28:436–440. doi: 10.1038/nbt0510-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth R., Kim H. Characterization of a microfluidic in vitro model of the blood–brain barrier (muBBB) Lab Chip. 2012;12:1784–1792. doi: 10.1039/c2lc40094d. [DOI] [PubMed] [Google Scholar]
- Chen M.B., Srigunapalan S., Wheeler A.R., Simmons C.A. A 3D microfluidic platform incorporating methacrylated gelatin hydrogels to study physiological cardiovascular cell–cell interactions. Lab Chip. 2013;13:2591–2598. doi: 10.1039/c3lc00051f. [DOI] [PubMed] [Google Scholar]
- Chivukula V.K., Krog B.L., Nauseef J.T., Henry M.D., Vigmostad S.C. Alterations in cancer cell mechanical properties after fluid shear stress exposure: a micropipette aspiration study. Cell Health Cytoskeleton. 2015;7:25–35. doi: 10.2147/CHC.S71852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choucha-Snouber L., Aninat C., Grsicom L., Madalinski G., Brochot C., Poleni P.E., Razan F., Guillouzo C.G., Legallais C., Corlu A., Leclerc E. Investigation of ifosfamide nephrotoxicity induced in a liver-kidney co-culture biochip. Biotechnol. Bioeng. 2013;110:597–608. doi: 10.1002/bit.24707. [DOI] [PubMed] [Google Scholar]
- CMR International . Thomson Reuters; 2015. Pharmaceutical R&D Executive Summary. August 2015. Available from: http://cmr.thomsonreuters.com/pdf/Executive_Summary_Final.pdf. [7 December 2015] [Google Scholar]
- Costello C.M., Sorna R.M., Goh Y.L., Cengic I., Jain N.K., March J.C. 3-D intestinal scaffolds for evaluating the therapeutic potential of probiotics. Mol. Pharm. 2014;11:2030–2039. doi: 10.1021/mp5001422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMasi J.A., Grabowski H.G., Hansen R.W. 2014. Cost to develop and win marketing approval for a new drug is $2.6 billion. Available from: http://csdd.tufts.edu/news/complete_story/pr_tufts_csdd_2014_cost_study. [7 December 2015] [Google Scholar]
- Douville N.J., Tung Y.C., Li R., Wang J.D., El-Sayed M.E., Takayama S. Fabrication of two-layered channel system with embedded electrodes to measure resistance across epithelial and endothelial barriers. Anal. Chem. 2010;82:2505–2511. doi: 10.1021/ac9029345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch M.B., King T.L., Shuler M.L. The role of body-on-a-chip devices in drug and toxicity studies. Annu. Rev. Biomed. Eng. 2011;13:55–72. doi: 10.1146/annurev-bioeng-071910-124629. [DOI] [PubMed] [Google Scholar]
- Esch M.B., Mahler G.J., Stokol T., Shuler M.L. Body-on-a-chip simulation with gastrointestinal tract and liver tissues suggests that ingested nanoparticles have the potential to cause liver injury. Lab Chip. 2014;14:3081–3092. doi: 10.1039/c4lc00371c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch M.B., Prot J.M., Wang Y.I., Miller P., Llamas-Vidales J.R., Naughton B.A., Applegate D.R., Shuler M.L. Multi-cellular 3D human primary liver cell culture elevates metabolic activity under fluidic flow. Lab Chip. 2015;15:2269–2277. doi: 10.1039/c5lc00237k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esch M.B., Sung J.H., Yang J., Yu C., Yu J., March J.C., Shuler M.L. On chip porous polymer membranes for integration of gastrointestinal tract epithelium with microfluidic ‘body-on-a-chip’ devices. Biomed. Microdevices. 2012;14:895–906. doi: 10.1007/s10544-012-9669-0. [DOI] [PubMed] [Google Scholar]
- Ferrell N., Ricci K.B., Groszek J., Marmerstein J.T., Fissell W.H. Albumin handling by renal tubular epithelial cells in a microfluidic bioreactor. Biotechnol. Bioeng. 2012;109:797–803. doi: 10.1002/bit.24339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche C.S., Simsch O., Weinberg E.J., Orrick B., Stamm C., Kaazempur-Mofrad M.R., Borenstein J.T., Hetzer R., Vacanti J.P. Pulmonary tissue engineering using dual-compartment polymer scaffolds with integrated vascular tree. Int. J. Artif. Organs. 2009;32:701–710. doi: 10.1177/039139880903201001. [DOI] [PubMed] [Google Scholar]
- Griep L.M., Wolbers F., De Wagenaar B., Ter Braak P.M., Weksler B.B., Romero I.A., Couraud P.O., Vermes I., Van Der Meer A.D., Van Den Berg A. BBB on chip: microfluidic platform to mechanically and biochemically modulate blood–brain barrier function. Biomed. Microdevices. 2013;15:145–150. doi: 10.1007/s10544-012-9699-7. [DOI] [PubMed] [Google Scholar]
- Guo X., Gonzalez M., Stancescu M., Vandenburgh H.H., Hickman J.J. Neuromuscular junction formation between human stem cell-derived motoneurons and human skeletal muscle in a defined system. Biomaterials. 2011;32:9602–9611. doi: 10.1016/j.biomaterials.2011.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison D.J., Manz A., Fan Z.H., Ludi H., Widmer H.M. Capillary electrophoresis and sample injection systemsintegrated on a planar glass chip. Anal. Chem. 1992;64:1926–1932. [Google Scholar]
- Huh D., Leslie D.C., Matthews B.D., Fraser J.P., Jurek S., Hamilton G.A., Thorneloe K.S., Mcalexander M.A., Ingber D.E. A human disease model of drug toxicity-induced pulmonary edema in a lung-on-a-chip microdevice. Sci. Transl. Med. 2012;4:159ra147. doi: 10.1126/scitranslmed.3004249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh D., Matthews B.D., Mammoto A., Montoya-Zavala M., Hsin H.Y., Ingber D.E. Reconstituting organ-level lung functions on a chip. Science. 2010;328:1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang K.J., Cho H.S., Kang Do H., Bae W.G., Kwon T.H., Suh K.Y. Fluid-shear-stress-induced translocation of aquaporin-2 and reorganization of actin cytoskeleton in renal tubular epithelial cells. Integr. Biol. (Camb.) 2011;3:134–141. doi: 10.1039/c0ib00018c. [DOI] [PubMed] [Google Scholar]
- Jang K.J., Mehr A.P., Hamilton G.A., Mcpartlin L.A., Chung S., Suh K.Y., Ingber D.E. Human kidney proximal tubule-on-a-chip for drug transport and nephrotoxicity assessment. Integr. Biol. (Camb.) 2013;5:1119–1129. doi: 10.1039/c3ib40049b. [DOI] [PubMed] [Google Scholar]
- Jang K.J., Suh K.Y. A multi-layer microfluidic device for efficient culture and analysis of renal tubular cells. Lab Chip. 2010;10:36–42. doi: 10.1039/b907515a. [DOI] [PubMed] [Google Scholar]
- Kamholz A.E., Yager P. Theoretical analysis of molecular diffusion in pressure-driven laminar flow in microfluidic channels. Biophys. J. 2001;80:155–160. doi: 10.1016/S0006-3495(01)76003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.J., Huh D., Hamilton G., Ingber D.E. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–2174. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- Kim H.J., Ingber D.E. Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation. Integr. Biol. (Camb.) 2013;5:1130–1140. doi: 10.1039/c3ib40126j. [DOI] [PubMed] [Google Scholar]
- Kola I., Landis J. Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- Kolbe M., Xiang Z., Dohle E., Tonak M., Kirkpatrick C.J., Fuchs S. Paracrine effects influenced by cell culture medium and consequences on microvessel-like structures in cocultures of mesenchymal stem cells and outgrowth endothelial cells. Tissue Eng. A. 2011;17:2199–2212. doi: 10.1089/ten.TEA.2010.0474. [DOI] [PubMed] [Google Scholar]
- Laverty H., Benson C., Cartwright E., Cross M., Garland C., Hammond T., Holloway C., Mcmahon N., Milligan J., Park B., Pirmohamed M., Pollard C., Radford J., Roome N., Sager P., Singh S., Suter T., Suter W., Trafford A., Volders P., Wallis R., Weaver R., York M., Valentin J. How can we improve our understanding of cardiovascular safety liabilities to develop safer medicines? Br. J. Pharmacol. 2011;163:675–693. doi: 10.1111/j.1476-5381.2011.01255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann E.S., Weidenfeller C., Svendsen C.N., Shusta E.V. Blood–brain barrier modeling with co-cultured neural progenitor cell-derived astrocytes and neurons. J. Neurochem. 2011;119:507–520. doi: 10.1111/j.1471-4159.2011.07434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C., Finch C., Esch M., Anderson W., Shuler M., Hickman J. Design optimization of liquid-phase flow patterns for microfabricated lung on a chip. Ann. Biomed. Eng. 2012;40:1255–1267. doi: 10.1007/s10439-012-0513-8. [DOI] [PubMed] [Google Scholar]
- Luo X., Davis J.J. Electrical biosensors and the label free detection of protein disease biomarkers. Chem. Soc. Rev. 2013;42:5944–5962. doi: 10.1039/c3cs60077g. [DOI] [PubMed] [Google Scholar]
- Mahler G.J., Esch M.B., Glahn R.P., Shuler M.L. Characterization of a gastrointestinal tract microscale cell culture analog used to predict drug toxicity. Biotechnol. Bioeng. 2009;104:193–205. doi: 10.1002/bit.22366. [DOI] [PubMed] [Google Scholar]
- Mahler G.J., Esch M.B., Tako E., Southard T.L., Archer S.D., Glahn R.P., Shuler M.L. Oral exposure to polystyrene nanoparticles affects iron absorption. Nat. Nanotechnol. 2012;7:264–271. doi: 10.1038/nnano.2012.3. [DOI] [PubMed] [Google Scholar]
- Morgan J.P., Delnero P.F., Zheng Y., Verbridge S.S., Chen J., Craven M., Choi N.W., Diaz-Santana A., Kermani P., Hempstead B., Lopez J.A., Corso T.N., Fischbach C., Stroock A.D. Formation of microvascular networks in vitro. Nat. Protoc. 2013;8:1820–1836. doi: 10.1038/nprot.2013.110. [DOI] [PubMed] [Google Scholar]
- Mu X., Zheng W., Xiao L., Zhang W., Jiang X. Engineering a 3D vascular network in hydrogel for mimicking a nephron. Lab Chip. 2013;13:1612–1618. doi: 10.1039/c3lc41342j. [DOI] [PubMed] [Google Scholar]
- Neuzi P., Giselbrecht S., Lange K., Huang T.J., Manz A. Revisiting lab-on-a-chip technology for drug discovery. Nat. Rev. Drug Discov. 2012;11:620–632. doi: 10.1038/nrd3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill A.T., Monteiro-Riviere N.A., Walker G.M. Characterization of microfluidic human epidermal keratinocyte culture. Cytotechnology. 2008;56:197–207. doi: 10.1007/s10616-008-9149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardridge W.M. Blood–brain barrier drug targeting: the future of brain drug development. Mol. Interv. 2003;3(90–105):51. doi: 10.1124/mi.3.2.90. [DOI] [PubMed] [Google Scholar]
- Pharmaceutical Research and Manufacturers of America (PhRMA) 2015. 2015 Profile Biopharmaceutical Research Industry. Available from: http://www.phrma.org/sites/default/files/pdf/2015_phrma_profile.pdf. [7 December 2015] [Google Scholar]
- Price G.M., Wong K.H., Truslow J.G., Leung A.D., Acharya C., Tien J. Effect of mechanical factors on the function of engineered human blood microvessels in microfluidic collagen gels. Biomaterials. 2010;31:6182–6189. doi: 10.1016/j.biomaterials.2010.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regehr K.J., Domenech M., Koepsel J.T., Carver K.C., Ellison-Zelski S.J., Murphy W.L., Schuler L.A., Alarid E.T., Beebe D.J. Biological implications of polydimethylsiloxane-based microfluidic cell culture. Lab Chip. 2009;9:2132–2139. doi: 10.1039/b903043c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijnders C.M., Van Lier A., Roffel S., Kramer D., Scheper R.J., Gibbs S. Development of a full-thickness human skin equivalent in vitro model derived from TERT-immortalized keratinocytes and fibroblasts. Tissue Eng. A. 2015;21:2448–2459. doi: 10.1089/ten.tea.2015.0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round J.L., Mazmanian S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackmann E.K., Fulton A.L., Beebe D.J. The present and future role of microfluidics in biomedical research. Nature. 2014;507:181–189. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- Schaffner A.E., Barker J.L., Stenger D.A., Hickman J.J. Investigation of the factors necessary for growth of hippocampal neurons in a defined system. J. Neurosci. Methods. 1995;62:111–119. doi: 10.1016/0165-0270(95)00063-1. [DOI] [PubMed] [Google Scholar]
- Schumacher S., Nestler J., Otto T., Wegener M., Ehrentreich-Forster E., Michel D., Wunderlich K., Palzer S., Sohn K., Weber A., Burgard M., Grzesiak A., Teichert A., Brandenburg A., Koger B., Albers J., Nebling E., Bier F.F. Highly-integrated lab-on-chip system for point-of-care multiparameter analysis. Lab Chip. 2012;12:464–473. doi: 10.1039/c1lc20693a. [DOI] [PubMed] [Google Scholar]
- Shayan G., Choi Y.S., Shusta E.V., Shuler M.L., Lee K.H. Murine in vitro model of the blood–brain barrier for evaluating drug transport. Eur. J. Pharm. Sci. 2011;42:148–155. doi: 10.1016/j.ejps.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Shayan G., Shuler M.L., Lee K.H. The effect of astrocytes on the induction of barrier properties in aortic endothelial cells. Biotechnol. Prog. 2011;27:1137–1145. doi: 10.1002/btpr.620. [DOI] [PubMed] [Google Scholar]
- Snouber L.C., Letourneur F., Chafey P., Broussard C., Monge M., Legallais C., Leclerc E. Analysis of transcriptomic and proteomic profiles demonstrates improved Madin-Darby canine kidney cell function in a renal microfluidic biochip. Biotechnol. Prog. 2012;28:474–484. doi: 10.1002/btpr.743. [DOI] [PubMed] [Google Scholar]
- Souto L.R., Rehder J., Vassallo J., Cintra M.L., Kraemer M.H., Puzzi M.B. Model for human skin reconstructed in vitro composed of associated dermis and epidermis. Sao Paulo Med. J. 2006;124:71–76. doi: 10.1590/S1516-31802006000200005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires T.M., Quake S.R. Microfluidics: fluid physics at the nanoliter scale. Rev. Mod. Phys. 2005;77:977–1026. [Google Scholar]
- Stancescu M., Molnar P., Mcaleer C.W., Mclamb W., Long C.J., Oleaga C., Prot J.M., Hickman J.J. A phenotypic in vitro model for the main determinants of human whole heart function. Biomaterials. 2015;60:20–30. doi: 10.1016/j.biomaterials.2015.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung J.H., Kam C., Shuler M.L. A microfluidic device for a pharmacokinetic-pharmacodynamic (PK-PD) model on a chip. Lab Chip. 2010;10:446–455. doi: 10.1039/b917763a. [DOI] [PubMed] [Google Scholar]
- Takayama S., White J., Zhang C. Inhalation Toxicology. third ed. CRC Press; 2014. Lungs-on-a-chip. [Google Scholar]
- Tavana H., Zamankhan P., Christensen P.J., Grotberg J.B., Takayama S. Epithelium damage and protection during reopening of occluded airways in a physiologic microfluidic pulmonary airway model. Biomed. Microdevices. 2011;13:731–742. doi: 10.1007/s10544-011-9543-5. [DOI] [PubMed] [Google Scholar]
- The National Academies (NAS) 2014. Emerging Science for Environmental Health Decisions Newsletter. Available from: http://nas-sites.org/emergingscience/files/2013/05/tissue-chips-final.pdf. [7 December 2015] [Google Scholar]
- Toepke M.W., Beebe D.J. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip. 2006;6:1484–1486. doi: 10.1039/b612140c. [DOI] [PubMed] [Google Scholar]
- Trusheim M.R., Berndt E.R., Douglas F.L. Stratified medicine: strategic and economic implications of combining drugs and clinical biomarkers. Nat. Rev. Drug Discov. 2007;6:287–293. doi: 10.1038/nrd2251. [DOI] [PubMed] [Google Scholar]
- Turner J.R. Intestinal mucosal barrier function in health and disease. Nat. Rev. Immunol. 2009;9:799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- Van Der Meer A.D., Orlova V.V., Ten Dijke P., Van Den Berg A., Mummery C.L. Three-dimensional co-cultures of human endothelial cells and embryonic stem cell-derived pericytes inside a microfluidic device. Lab Chip. 2013;13:3562–3568. doi: 10.1039/c3lc50435b. [DOI] [PubMed] [Google Scholar]
- Van Lintel H., Vandepol F., Bouwstra S. A piezoelectric micropump based on micromachining of silicon. Sensors Actuators. 1988;153-167 [Google Scholar]
- Verbridge S.S., Chakrabarti A., Delnero P., Kwee B., Varner J.D., Stroock A.D., Fischbach C. Physicochemical regulation of endothelial sprouting in a 3D microfluidic angiogenesis model. J. Biomed. Mater. Res. A. 2013;101:2948–2956. doi: 10.1002/jbm.a.34587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner I., Materne E.M., Brincker S., Sussbier U., Fradrich C., Busek M., Sonntag F., Sakharov D.A., Trushkin E.V., Tonevitsky A.G., Lauster R., Marx U. A dynamic multi-organ-chip for long-term cultivation and substance testing proven by 3D human liver and skin tissue co-culture. Lab Chip. 2013;13:3538–3547. doi: 10.1039/c3lc50234a. [DOI] [PubMed] [Google Scholar]
- Wang J. Microchip devices for detecting terrorist weapons. Anal. Chim. Acta. 2004;507:3–10. [Google Scholar]
- Williamson A., Singh S., Fernekorn U., Schober A. The future of the patient-specific body-on-a-chip. Lab Chip. 2013;13:3471–3480. doi: 10.1039/c3lc50237f. [DOI] [PubMed] [Google Scholar]
- Yu J., Peng S., Luo D., March J.C. In vitro 3D human small intestinal villous model for drug permeability determination. Biotechnol. Bioeng. 2012;109:2173–2178. doi: 10.1002/bit.24518. [DOI] [PubMed] [Google Scholar]
- Zhou M., Ma H., Lin H., Qin J. Induction of epithelial-to-mesenchymal transition in proximal tubular epithelial cells on microfluidic devices. Biomaterials. 2014;35:1390–1401. doi: 10.1016/j.biomaterials.2013.10.070. [DOI] [PubMed] [Google Scholar]