Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that act as master regulators of many cellular processes. The expression of miRNAs is often deregulated in human tumors, causing the alteration of molecular mechanisms relevant for cancer progression. Importantly, miRNAs are detectable in the blood and their quantity fluctuations are the hallmark of pathogenic conditions, including cancer. Several groups reported the identification of circulating cell-free miRNAs (cf-miRNAs) in the human serum and plasma and demonstrated their diagnostic and prognostic utility. Other studies also shown that it may be feasible to apply such cf-miRNA signatures within screening programs in order to improve cancer early detection. Circulating cf-miRNAs therefore appear to be excellent candidates for blood-borne cancer biomarkers.
Keywords: MicroRNA, Liquid biopsy, Cancer, Cancer early detection
MicroRNAs (miRNAs) can be isolated from a wide range of different tissues and biological specimens, including biofluids. In 2008, pioneer studies demonstrated that miRNAs were detectable in the plasma and serum and that they were particular resistant to degradation by RNAses in the blood (Chen et al., 2008). Subsequent studies reported the presence of cell-free miRNAs (cf-miRNAs) in virtually all other body fluids (Weber et al., 2010), and raised several questions regarding the cf-miRNA stability in the body fluids, the mechanisms of release from cells, and their biological functions.
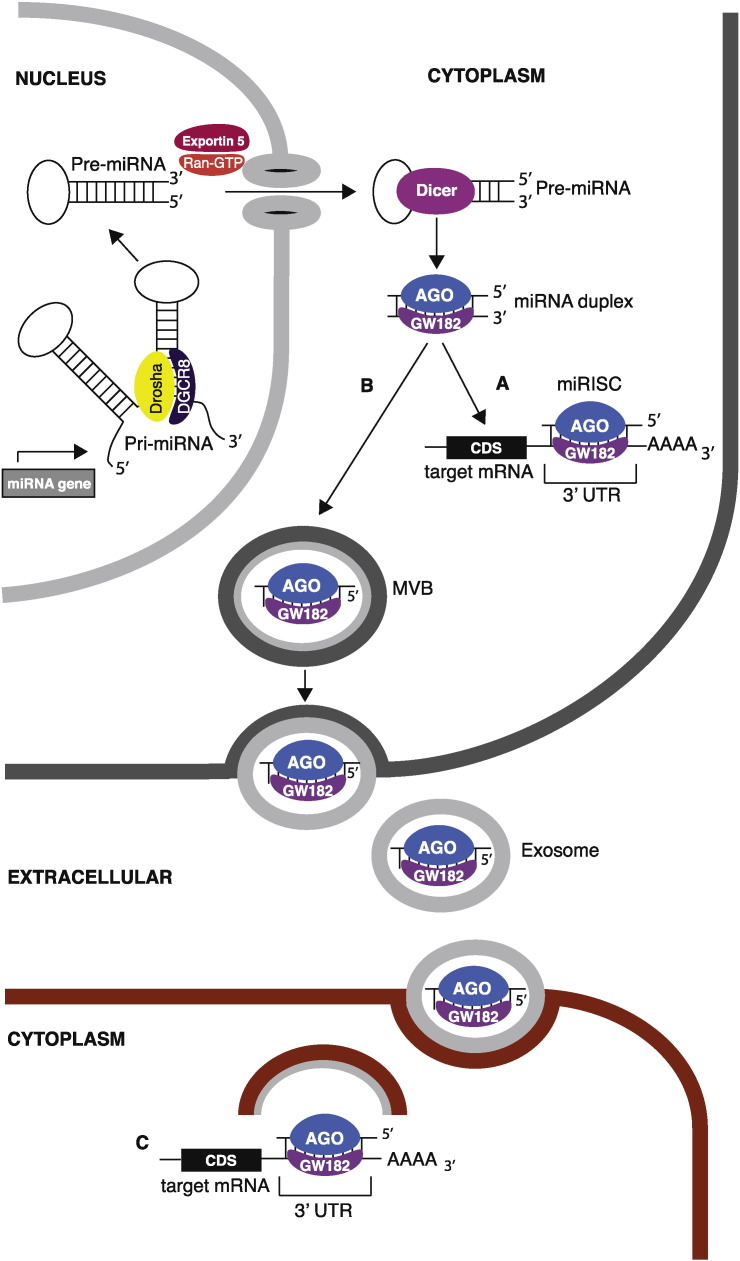
Cf-miRNAs were found within microvesicles/exosomes, apoptotic bodies (AB), HDL structures, or complexed with AGO proteins (that constitute the miRNA-induced silencing complex, miRISC) (Turchinovich and Burwinkel, 2012) (Fig. 1), which protect them by the action of RNAses. Of note, packaging of miRNAs in exosomes can be controlled by positive selection mechanisms, such as the ceramide-dependent secretory pathway controlled by the nSMase enzyme (e.g. the ceramide biosynthesis neutral sphingomyelinase) (Kosaka et al., 2010). However, how exactly miRNAs are selected and loaded to exosomes, and how the trafficking is regulated in physiological and pathological conditions, are not yet understood. Another unmet question is about the biological function of cf-miRNAs. In cancer cells, the extracellular release of miRNAs can be a way to reduce the intracellular level of miRNAs with tumor suppressor functions (Ohshima et al., 2010). On the other hand, cf-miRNAs can function as a paracrine signal to modify tumor microenvironment and support cancer progression; exosomal/MVB/AB-miRNAs may be indeed delivered to neighboring cells where, following uptake, they can modulate the transcription of target-mRNAs (Turchinovich and Burwinkel, 2012) (Fig. 1). An intriguingly alternative mechanism of action for cf-miRNAs was also proposed: the AGO2-complexed miR-21 and miR-29a may act as signaling molecules via binding to intracellular Toll-like receptors (murine TLR-7 and human TLR-8), which are a family of receptors characteristic of immune cells involved in the innate immune system (Fabbri et al., 2012). The activation of immune cells expressing TLRs stimulates secretion of inflammatory cytokines that ultimately induce tumor cell spread (Fabbri et al., 2012).
Fig. 1.
A) Intracellular miRNA processing and function. B) Extracellular release of mature miRNA. C) Modulation of transcription by exogenous miRNA.
Quantities and species of cf-miRNAs were shown to fluctuate in the presence of malignant and non-malignant disease (Chen et al., 2008). More recently, we and others proposed a reliable method to identify and quantify serum/plasma miRNAs starting from low amounts of serum/plasma (less than 300 μl) that could be easily implemented in the clinic for lung cancer early diagnosis. Using this method, two cf-miRNA signatures diagnostic for asymptomatic lung cancer were validated in high-risk individuals (> 55 years of age, smokers) enrolled in two large Italian lung screening trials (the COSMOS trial, n = 1115 and the MILD trial, n = 939) (Montani et al., 2015, Sozzi et al., 2014). Despite the fact that these two signatures were derived using different blood components (i.e., serum or plasma; see further below) and from different subjects, they have a substantial fraction of overlapping miRNAs (miR-92a-3p, miR-30c-5p, miR-30b-5p, miR-148a-3p, miR-140-5p), which further confirms the reliability of cf-miRNAs when used as cancer biomarkers. Of note, several other cf-miRNA signatures were recently proposed for the diagnosis of different cancer types (ovary, breast, prostate, liver, colorectal, brain, melanoma, pancreas, etc.). It would be interesting to perform a pan-cancer screening study to understand the ability of these signatures to discriminate among different types of cancer; this will be relevant for the application of these signatures in cancer screening programs.
There are a number of reasons that make the possibility to use blood-based miRNA signatures as diagnostic tools very attractive: i) cancer can be diagnosed in a minimally invasive way through a simple blood test; ii) a blood test does not require hospitalization and the access to complex and expensive technologies; and iii) blood-based miRNA tests can preselect high-risk individuals thus increasing the accuracy of cancer screening programs. Currently, selection criteria for high-risk individuals to be enrolled in cancer screening programs are principally based on epidemiological characteristics, such as age, smoking status, exposition to asbestos, obesity, presence of chronic diseases, familial history of cancer, and genetic predisposition to cancer (i.e. BRCA1/BRCA2 carriers). However, the incidence of cancer in these high-risk cohorts is dismal. For example, only ~ 1% of the high-risk individuals who underwent low-dose computed tomography (LDCT) screening were diagnosed with lung cancer (Montani et al., 2015), which limits the widespread application of such screening protocols. For this reason, the development of blood-based miRNA tests holds the potential to increase screening protocol sensitivity and specificity, while decreasing other unnecessary and potentially harmful diagnostic procedures (i.e., exposure to ionizing radiations during computed tomography, or mammography scans).
It is important to note, that almost half of the published cf-miRNA signatures were discovered analyzing serum samples, while the others were derived from plasma samples. Cf-miRNAs may vary in their quantities and species if measured in serum or in plasma samples (Wang et al., 2012). Therefore, when assessing the overlap among cf-miRNA signatures in different studies or planning validation–screening studies, the blood components used to derive these signatures should be taken into consideration.
It is foreseeable that the molecular analysis of liquid biopsies (blood, urine, etc.) will become the gold standard for developing non-invasive diagnostic and prognostic tests. Such tests will not require hospitalization and could be offered in peripheral health centers, therefore potentially augmenting the accrual of high-risk individuals to cancer screening programs. In addition, the possibility to collect multiple samples from the same patients will definitely help clinicians to better monitor therapy response and decide for an optimal treatment.
However, there are still some methodological limitations that need to be resolved before the large-scale application of such molecular tests. For example, the numbers of steps required to obtain and measure circulating nucleic acids are numerous and require expensive and complex technology. In such scenario, the development of point-of-care testing (POCT) to measure cf-miRNA directly in the blood (Jin et al., 2015) is overriding for the application of these next-generation blood tests in a population-scale. Furthermore, the sensitivity and specificity of cf-miRNA tests should be improved to augment the detection rate and avoid false positives and negative results, which are detrimental for cancer early detection screening programs.
Conflicts of Interest
FM and FB are coinventors on a patent application regarding a diagnostic cf-miRNA signature cited herein.
Acknowledgments
This work was supported by a grant from the Italian Association for Cancer Research (MFAG17568) to FB.
References
- Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X., Li Q., Li X., Wang W., Wang J., Jiang X., Xiang Y., Xu C., Zheng P., Zhang J., Li R., Zhang H., Shang X., Gong T., Ning G., Zen K., Zhang C.Y. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. (cr2008282 [pii]) [DOI] [PubMed] [Google Scholar]
- Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., Lovat F., Fadda P., Mao C., Nuovo G.J., Zanesi N., Crawford M., Ozer G.H., Wernicke D., Alder H., Caligiuri M.A., Nana-Sinkam P., Perrotti D., Croce C.M. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Geissler D., Qiu X., Wegner K.D., Hildebrandt N., Rapid A. Amplification-free, and sensitive diagnostic assay for single-step multiplexed fluorescence detection of microRNA. Angew. Chem. Int. Ed. Engl. 2015;54:10024–10029. doi: 10.1002/anie.201504887. [DOI] [PubMed] [Google Scholar]
- Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montani F., Marzi M.J., Dezi F., Dama E., Carletti R.M., Bonizzi G., Bertolotti R., Bellomi M., Rampinelli C., Maisonneuve P., Spaggiari L., Veronesi G., Nicassio F., Di Fiore P.P., Bianchi F. miR-test: a blood test for lung cancer early detection. J. Natl. Cancer Inst. 2015:107. doi: 10.1093/jnci/djv063. (djv063 [pii]) [DOI] [PubMed] [Google Scholar]
- Ohshima K., Inoue K., Fujiwara A., Hatakeyama K., Kanto K., Watanabe Y., Muramatsu K., Fukuda Y., Ogura S., Yamaguchi K., Mochizuki T. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sozzi G., Boeri M., Rossi M., Verri C., Suatoni P., Bravi F., Roz L., Conte D., Grassi M., Sverzellati N., Marchiano A., Negri E., La Vecchia C., Pastorino U. Clinical utility of a plasma-based miRNA signature classifier within computed tomography lung cancer screening: a correlative MILD trial study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014;32:768–773. doi: 10.1200/JCO.2013.50.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchinovich A., Burwinkel B. Distinct AGO1 and AGO2 associated miRNA profiles in human cells and blood plasma. RNA Biol. 2012;9:1066–1075. doi: 10.4161/rna.21083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Yuan Y., Cho J.H., McClarty S., Baxter D., Galas D.J. Comparing the MicroRNA spectrum between serum and plasma. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0041561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J.A., Baxter D.H., Zhang S., Huang D.Y., Huang K.H., Lee M.J., Galas D.J., Wang K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010;56:1733–1741. doi: 10.1373/clinchem.2010.147405. [DOI] [PMC free article] [PubMed] [Google Scholar]