Abstract
The C57BLKS/J-Leprdb mouse has a point mutation in the leptin receptor gene and is one of the most useful animal model for non-insulin dependent diabetes mellitus in human. Since the homozygote of C57BLKS/J-Leprdb mouse is infertile, detection of point mutation in the leptin receptor gene is important for efficient maintaining strains as well as mass production of homozygotes. To develop a rapid and efficient genotyping method for C57BLKS/J-Leprdb mouse, the tetra-primer amplification-refractory mutation system polymerase chain reaction (ARMS-PCR) was used. The 407 and 199 bp PCR products were amplified from normal (+/+) mice; while the 407 and 268 bp PCR products were amplified from homozygotes (db/db) mice; and the 407, 268, and 199 bp PCR products were amplified from heterozygotes (db/+) mice. This result showed that the tetra-primer ARMS-PCR analysis by us is suitable to detect point mutation of the leptin receptor gene. Taken together, our method will dramatically reduce animal use for maintenance of strains as well as production of homozygote in the C57BLKS/J-Leprdb strains.
Keywords: Leptin receptor, genotyping, db mouse
The C57BLKS/J-Leprdb mouse, which has a mutation in the leptin receptor, is one of the most used animal model for non-insulin dependent diabetes mellitus (NIDDM). The C57BLKS/J-Leprdb mouse has a G→T point mutation in the leptin receptor gene [1]. Homozygotes of C57BLKS/J-Leprdb mice are infertile and can be easily distinguished from obese mice. Efficient maintenance of the C57BLKS/J-Leprdb strain requires an accurate identification method for heterozygotes. However, there is no phenotypic difference between normal and heterozygote C57BLKS/J-Leprdb mice. Progeny testing is used to identify heterozygotes of C57BLKS/J-Leprdb mice. For homozygote from a mating pair, both parents are required to be heterozygote, hence researchers spend much time and effort on progeny testing. The Coleman and Hummel incorporated misty gene (m) is closely linked to the leptin receptor and C57BLKS/J-Leprdb mice [2]. After misty gene incorporation, the heterozygote shows lean black mice and normal shows misty coat color after 3 to 4 weeks of age. The Coleman's method to identify heterozygote of C57BLKS/J-Leprdb mice has the disadvantage of inability to distinguish between heterozygote and normal offspring until 3 weeks of age. Horvat and Bünger developed a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay to overcome this problem [3]. The Horvet's method is also time consuming and expensive. The tetra-primer amplification-refractory mutation system-polymerase chain reaction (ARMS-PCR) analysis is used to identify known single nucleotide polymorphism in genomic DNA [4,5]. We developed a new method for easy identification of C57BLKS/J-Leprdb mice genotypes using ARMS-PCR.
We used C57BLKS/J-Leprdb mice that were supplied from Laboratory Animal Center, Hallym University. Animals were housed in a specific pathogen free room with controlled temperature (23±2℃), humidity (55±10%), and light (08:00 a.m~20:00 p.m.). The animal study was conducted in accordance with Institutional Animal Care and Use Committees of Hallym University (Hallym 2013-133). Genomic DNA was prepared from the mouse tail by standard SDS/proteinase K lysis and the phenol/chloroform extraction method [6]. The design of primers for amplification-refractory mutation system-polymerase chain reaction (ARMS-PCR) used the primer design web service for tetra-primer ARMS-PCR [7]. The PCR samples comprised 50 ng genomic DNA, 6 µL rTaq plus 5× PCR master mix (1 unit/4 µL thermostable DNA polymerases, 50 mM Tris-HCl (PH9.0), 16 mM (NH4)2SO4, 1.75 mM MgCl2, 2% DMSO, 0.1% Tween 20, 0.1 mg/mL BS, 0.05% bromophenol blue, 12% glycerol), and 2.5 p/mol primers. Target DNA was amplified in a Cyclogene Dri-Block® cycler (Techne Cambridge Ltd., U.K.), with initial denaturation at 94℃ for 5 min followed by 30 cycles of 94℃ for 30 sec, 49.1℃ 45 sec, and 72℃ for 45 sec and a final elongation period at 72℃ for 10 min. The PCR products were electrophoresed on a 3% agarose gel.
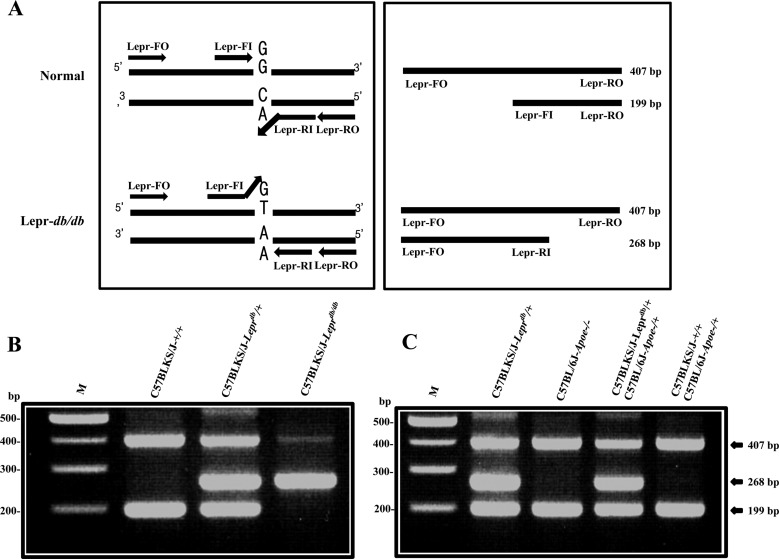
The mouse leptin receptor sequence (C57BL/6J) was retrieved from NCBI (Gene ID: 16847). The C57BLKS/J-Leprdb mice have transverse point mutation (G→T) in intron 18 of the leptin receptor [1]. This mutation creates a splicing donor site and produces exclusively short form mutant transcripts. We designed 4 primers for amplification of allele-specific PCR products from leptin receptor mutant and normal alleles using primer design web service for tetra-primer ARMS-PCR (Figure 1) i.e., Lepr-forward outer primer (Lepr-FO); AGGGTTAGA GATCTTTCATCTTTAGCT, Lepr-forward inner primer (Lepr-FI); TTAGAACATGTTTACATTTTGATGGAGGG, Lepr-reverse inner primer (Lepr-RI); ATTTGTTTGGAT TTGATACCAAACTTACTG, Lepr-reverse outer primer (Lepr-RO); AAATTTCAACTATAACTACCTGAGAC GTA. The 268 and 199 bp PCR products are amplified from the leptin receptor mutant allele (Lepr-FI and Lepr-RO primers) and normal allele (Lepr-FO and Lepr RI primers), respectively. A 407 bp PCR product is amplified from leptin receptor mutant and normal alleles by Lepr FO and Lepr-RO primers as an internal control (Figure 2A).
Figure 1. Sequences of the leptin receptor mutation and normal allele (NCBI, Gene ID: 16847). The primers for ARMS-PCR were designed using the primer design web service for tetra-primer ARMS-PCR. Underline indicates transverse point mutation (G→T) in intron 18 of the leptin receptor.
Figure 2. Detection of leptin receptor mutation by the tetra-primer ARMS-PCR analysis. A. Design of PCR products. Leptin receptor mutation (268 bp, Lepr-FO and Lepr-RI), wild type (199 bp, Lepr-FI and Lepr-RO), internal control from both alleles (407 bp, Lepr-FO and Lepr-RO). B. Genotyping of leptin receptor mutant mice by the tetra-primer ARMS-PCR. C. Genotyping of double knock-out mice by the tetra-primer ARMS-PCR. M: 100 bp ladder.
We tested our hypothesis by tetra-primer ARMS-PCR on the 3 genotypes i.e., db/db, db/+, +/+. The 407 and 199 bp PCR products were amplified from normal (+/+) mice; while the 407 and 268 bp PCR products were amplified from homozygote (db/db) mice; and the 407, 268, and 199 bp PCR products were amplified from heterozygote (db/+) mice (Figure 2B). Tetra-primer ARMS-PCR identification of heterozygotes was further confirmed by progeny testing. We showed that the tetraprimer ARMS-PCR analysis worked in other C57BL/6J background mice by genotyping progenies from C57BLKS/J-Leprdb/+ and C57BL/6J-Apoetm1Unc knock-out mating. The 407, 268, and 199 bp PCR products were amplified from double heterozygote (db/+, Apoe/+) mice (Figure 2C). This result indicated that the tetra-primer ARMS-PCR analysis was effective in mice with a C57BL/6J genetic background.
Currently, there are 3 methods to identify leptin receptor mutation. Genotyping by progeny testing is time consuming. The indirect genotyping method by misty gene cannot distinguish between heterozygote and normal mice from birth to 3 weeks of age [2]. Genotyping by PCR-RFLP is expensive and requires several steps such as PCR, restriction enzyme digestion, and electrophoresis [3]. The tetra-primer ARMS-PCR analysis has only 2 steps i.e., PCR and agarose gel electrophoresis. Thus, our tetra-primer ARMS-PCR analysis is the most efficient and rapid genotyping method to identify leptin receptor mutation on a C57BL/6J genetic background. Taken together, the tetra-primer ARMS-PCR analysis is useful for efficient maintenance of C57BLKS/J-Leprdb strain and high throughput production of double knock-out mice involving the leptin receptor mutation.
Acknowledgments
This work was supported by Basic Science Research Program (2011-0024062), and Priority Research Centers Program (NRF-2009-0094071) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology and Korea Mouse Phenotyping Project (NRF-2014M3A9D 5A01075129) of the Ministry of Science, ICT and Future Planning through the National Research Foundation.
Footnotes
Conflict of interests: The authors declare that there is no financial conflict of interests to publish these results.
References
- 1.Chen H, Charlat O, Tartaglia LA, Woolf EA, Weng X, Ellis SJ, Lakey ND, Culpepper J, Moore KJ, Breitbart RE, Duyk GM, Tepper RI, Morgenstern JP. Evidence that the diabetes gene encodes the leptin receptor: identification of a mutation in the leptin receptor gene in db/db mice. Cell. 1996;84(3):491–495. doi: 10.1016/s0092-8674(00)81294-5. [DOI] [PubMed] [Google Scholar]
- 2.Coleman DL, Hummel KP. Symposium IV: Diabetic syndrome in animals. Influence of genetic background on the expression of mutations at the diabetes locus in the mouse. II. Studies on background modifiers. Isr J Med Sci. 1975;11(7):708–713. [PubMed] [Google Scholar]
- 3.Horvat S, Bünger L. Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay for the mouse leptin receptor (Lepr(db)) mutation. Lab Anim. 1999;33(4):380–384. doi: 10.1258/002367799780487850. [DOI] [PubMed] [Google Scholar]
- 4.Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, Markham AF. Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS) Nucleic Acids Res. 1989;17(7):2503–2516. doi: 10.1093/nar/17.7.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ye S, Dhillon S, Ke X, Collins AR, Day IN. An efficient procedure for genotyping single nucleotide polymorphisms. Nucleic Acids Res. 2001;29(17):E88. doi: 10.1093/nar/29.17.e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oh SH, Nam H, Suh JG. A high resolution genetic mapping of the faded (fe) gene to a region between D10mit156 and D10mit193 on mouse chromosome 10. Lab Anim Res. 2013;29(1):33–38. doi: 10.5625/lar.2013.29.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins A, Ke X. Primer1: primer design web service for tetraprimer ARMS-PCR. Open Bioinforma J. 2012;6:55–58. [Google Scholar]