Abstract
DNA of malaria parasites, Plasmodium falciparum, is subjected to extraordinary high levels of genotoxic insults during its complex life cycle within both the mosquito and human host. Accordingly, most of the components of DNA repair machinery are conserved in the parasite genome. Here, we investigated the genome-wide responses of P. falciparum to DNA damaging agents and provided transcriptional evidence of the existence of the double strand break and excision repair system. We also showed that acetylation at H3K9, H4K8, and H3K56 play a role in the direct and indirect response to DNA damage induced by an alkylating agent, methyl methanesulphonate (MMS). Artemisinin, the first line antimalarial chemotherapeutics elicits a similar response compared to MMS which suggests its activity as a DNA damaging agent. Moreover, in contrast to the wild-type P. falciparum, two strains (Dd2 and W2) previously shown to exhibit a mutator phenotype, fail to induce their DNA repair upon MMS-induced DNA damage. Genome sequencing of the two mutator strains identified point mutations in 18 DNA repair genes which may contribute to this phenomenon.
Maintenance of genetic integrity is essential for proper functioning of a cell and is also vital for successful transmission of genetic information during the cell division. For this, eukaryotic cells developed an orchestrated DNA damage response encompassing distinct repair pathways for different types of DNA lesions1. These repair pathways are classified broadly into two categories: the excision repair pathways including nucleotide excision repair (NER), base excision repair (BER) and mismatch repair (MMR); and the double strand break repair pathways (DSBR) including homologous recombination repair (HR) and nonhomologous end joining (NHEJ)2. Given the complex life cycle of P. falciparum, the main causative agent of human malaria, which involves two hosts and numerous physiological/developmental stages, the parasite’s cellular DNA is under a constant exogenous and endogenous stress3. The major source of the genetic assaults in Plasmodium originates from replications errors, reactive oxygen species (ROS) generated during heme metabolism, and also free radicals generated by antimalarial drugs such as chloroquine and artemisinin3,4,5,6. Although, the majority of the genes of the DNA repair pathways are conserved in Plasmodium, until today, limited information exists about their regulation and specific function. Recently, it has been shown that Plasmodium parasites utilize predominantly the HR system to repair double strand breaks, which are the most deleterious forms of genetic aberrations and can lead to cell death7. On the other hand, the factors of canonical-NHEJ that are crucial for DSBR in most eukaryotes appear to be missing in the genome of the malaria parasites8. However, there is an experimental evidence for an alternative-NHEJ that has been suggested to repair double stranded breaks in the Plasmodium parasites9,10. This end joining process is believed to be guided by local microhomologies that are initially created by a non-processive DNA polymerase. This is followed by annealing, ligation and final repair in a process termed SD-MMEJ11. In P. falciparum, the DNA polymerase subunit β has been previously characterized12,13 and suggested to play the role in the SD-MMEJ presumably by creating the microhomologies9.
DNA damage recognition and repair occurs in the context of chromatin. In eukaryotes, chromatin plays a significant role particularly in regulating the DNA damage response14. During DNA damage, histone modifications and chromatin remodeling complexes alter the chromatin architecture to give access to the damaged site for the DNA repair enzymes15. In particular, H2A/H2A-X phosphorylation and H4 acetylations play significant roles in the recruitment of DNA repair factors such as ATM kinase, ATR kinase, Rad51, BRAC1, TP53, TIP60 HAT16,17. Most of the histone modifications such as acetylation, phoshorylation, methylation as well as several chromatin remodeling complexes, canonical in other eukaryotes, have been identified in P. falciparum18. However, the role of chromatin dynamics in the DNA damage response in P. falciparum remains unexplored.
Resistance of P. falciparum malaria to several classes of anti-malarial drugs such as chloroquine, pyrimethamine, sulfadoxine and more recently artemisinin had first emerged in Southeast Asia and South America and have (or it is expected to) subsequently spread around the world19,20,21,22,23. All drug resistance phenotypes have been associated with DNA polymorphisms; typically non-synonymous mutations in drug targets and/or drug transporters24,25,26. It has been suggested that a rapid emergence of the anti-malarial drug resistance phenotypes is linked with increased mutation capacity of some parasite strains (mutator phenotype), known as accelerated resistance to multiple drugs (ARMD)27. The ARMD parasites may harbor diverse genetic backgrounds which may provide additional advantage in developing the actual drug resistance mechanisms as well as alleviate a fitness cost27. Such mutator phenotypes were found in bacteria, yeast as well as mammalian cells28,29,30 and are typically associated with defects in the DNA replication machinery or DNA repair associated processes31. DNA mismatch repair deficient cells can be also unresponsive to several chemotherapeutic drugs such as etoposide, procarbazine, busulfan, cisplatin and doxorubicin32. Similarly, defective DNA repair machinery confers resistance to cisplatin and anti-parasitic drug monensin in other protozoa intracellular parasites Trypanosoma cruzii and Toxoplasma gondii.33,34. In malaria, it has been suggested that ARMD parasites have reduced DNA repair capacity as compared to the wild type (non-ARMD) parasites35. However, the underlying genetic and biochemical mechanisms responsible for the ARMD phonotype are still not understood.
Here, we characterized transcriptional responses of P. falciparum to DNA damaging agents in malaria parasite, to understand regulation of the parasite repair machinery. Using methyl methanesulphonate (MMS), we show that P. falciparum responds to DNA damage by upregulating specific genes of the DNA repair machinery and at the same time alters its chromatin structure. We also show that artemisinin, the primary anti-malarial drug, also induces DNA damage response (similar to MMS) involving both the transcriptional and epigenetic changes. Finally, P. falciparum ARMD strains seem to fail to induce these transcriptional and epigenetic responses to DNA damage which may underlay their increased mutational characteristics. Whole genome sequencing revealed several genetic polymorphisms in DNA repair genes that may contribute to the ARMD phenotype.
Results
Global transcriptional response to DNA damaging agents
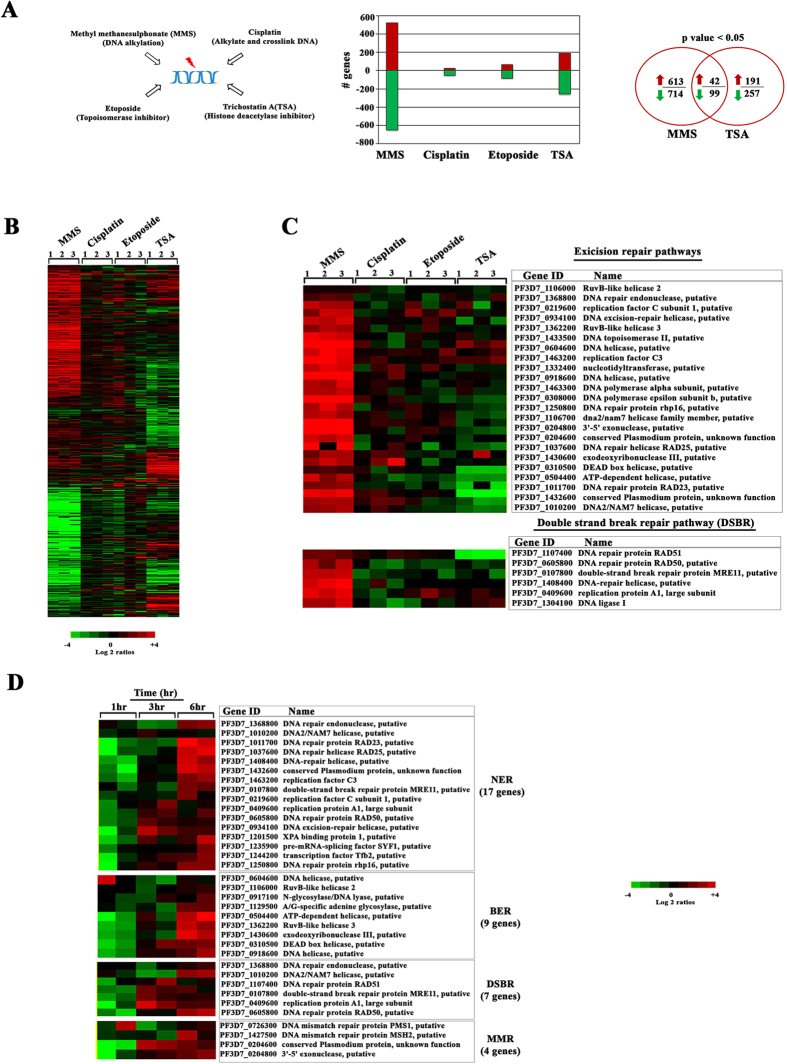
The main goal of this study was to elucidate regulation of gene expression in response to DNA damage in the human malaria parasites P. falciparum. As the first step, we carried out genome wide gene expression profiling of P. falciparum exposed to several classes of DNA damaging chemical agents. Briefly, synchronized P. falciparum cultures were treated with MMS (0.05%), cisplatin (100 μM), etoposide (100 μM) and TSA (50 μM) for 6 hours (hr) at the trophozoite stage (28–32 hr post invasion, hpi). At these concentrations these agents are known to cause either DNA damage or otherwise interact with DNA in P. falciparum. MMS alkylates DNA bases and by that induces single and double strand breaks36,37. Treatment of P. falciparum with MMS (0.05%) is known to cause DNA damage (validated by the comet assay) and also to induce two DSBR genes, PfRAD51, PfRAD5438. Cisplatin is a DNA intercalating agent that inhibits DNA replication and transcription39 while etoposide inhibits topoisomerase II, which subsequently induces DNA damage40. In P. falciparum, 100 μM of cisplatin treatment for 6 hr caused sequence specific cisplatin-DNA adduct formation, which subsequently led to DNA damage-mediated parasite death41. 100 μM treatment of P. falciparum by etoposide was shown to induce chromosomal cleavage and altered the genetic integrity which was confirmed by pulse field gel electrophoresis42. For comparison, we also used a histone deacetylase HDAC inhibitor, trichostatin A (TSA) that was previously shown to have a profound effect on the chromatin architecture and gene expression in P. falciparum43,44. To measure global transcriptional responses we utilized the genome-wide P. falciparum DNA microarray as previously described45. The perturbation analyses for these drugs were done in triplicates and the transcription results were highly reproducible (Pearson correlation among biological replicates were, 0.6–0.8, p value < 0.05). Interestingly, MMS and TSA affected transcription of a considerable number of genes while cisplatin and etoposide had only moderate-to-low effect (Fig. 1a,b). In particular, expression of 1170 genes was altered by MMS by 2-fold from which 523 and 647 genes were up- and down-regulated, respectively. In contrast, cisplatin and etoposide altered expression of 79 and 146 genes by 2-fold, respectively. The effect of TSA on gene expression was similar to previously published results with differential expression of 448 genes43. There was a small but statistically significant overlap between genes induced by MMS and TSA with 42 and 99 being commonly up and down regulated, respectively (Fig. 1a right panel). This suggests that MMS and TSA effects on gene expression are unique to each drug.
Figure 1. Global transcriptional response of P. falciparum to DNA damaging agents.
(A) Four DNA and/or chromatin perturbation agents MMS (0.05%), Cisplatin (100 μM), Etoposide (100 μM), and TSA (50 μM) were chosen to study the DNA damage response of P. falciparum parasites. The bar graph represents the number of genes differentially induced by >2 fold after synchronized parasites were treated at the trophozoite stage for 6 hr. The Venn diagram represents differentially expressed genes between the MMS or TSA treatments. (B) Heat map represents up (red) and down (green) regulated genes induced by the drug treatments by >2-fold. The results are mean value of a relative mRNA abundance in three experimental replicates. (C) The expression pattern of 29 excision and 6 DSB repair genes which were up-regulated by MMS are shown along with the effect of cisplatin, etoposide and TSA among three independent replicates (p-value < 0.05). (D) The heat map represents differential expression of DNA repair genes in a time course experiment with samples collected at 1, 3 and 6 hr of the MMS (0.05%) treatment. Only with the 6 hr of exposure to MMS, DNA repair machinery was up-regulated with 9 BER, 17 NER, 4 MMR and 7 DBSR genes significantly up-regulated in the two independent replicates (p- value 0.0002–0.02).
Amongst the 141 commonly co-regulated genes by MMS and TSA, there is a significant enrichment of factors of host parasite interactions such as “merozoite invasion” and “PfEMP1 domain architecture” (data not shown). This is consistent with previous suggestions that such genes facilitate a general stress response that is not directly linked to the perturbation/agent mode of action46,47. MMS alone induced an additional set of genes that can be linked with indirect responses such as protein biosynthesis, palmitome proteins, ribosomal assembly genes, molecular chaperones, nuclear genes destined for mitochondria/apicoplast and glycolysis (Supplementary Fig. S1). Most of these functionalities could also represent indirect responses to the stress conditions. Nonetheless, one of the most predominant functional group induced by MMS was a set of genes associated with the DNA repair machinery that reflects directly its presumed mode of action. This supports previous studies showing that treatment of P. falciparum parasites with MMS (0.05%) for 6 hr results in DNA double strands breaks and up-regulation of two factors of the HR pathway PfRAD51, PfRAD5438. Here, we show that MMS induces a broad DNA damage response including most of the factors of both the excision repair machinery and DBSR (Fig. 1c). After the 6 hr exposure, there were 9 genes of BER, 17 genes of NER, 4 genes of MMR and 7 genes of DBSR significantly up-regulated in the two independent replicates (p value < 0.05) (Fig. 1d). This transcriptional induction requires at least 6 hr exposure of P. falciparum cells to MMS as shorter time exposures did not exhibit statistically significant enrichment of the DNA repair genes/pathways amongst the upregulated genes. However, for all four pathways there was at least one gene whose transcriptional induction occurred as early as 3 hr; including XPA binding protein 1 (PF3D7_1201500) of NER; putative DEAD Box helicase (PF3D7_0310500) of BER; replication protein A1, large subunit (PF3D7_0409600) of DSBR; and the conserved Plasmodium protein (PF3D7_0204600) of MMR. In future experiments it will be interesting to investigate if these genes might represent rate limiting steps in DNA repair induction in their respective pathways.
DNA damage induces alterations in chromatin landscape
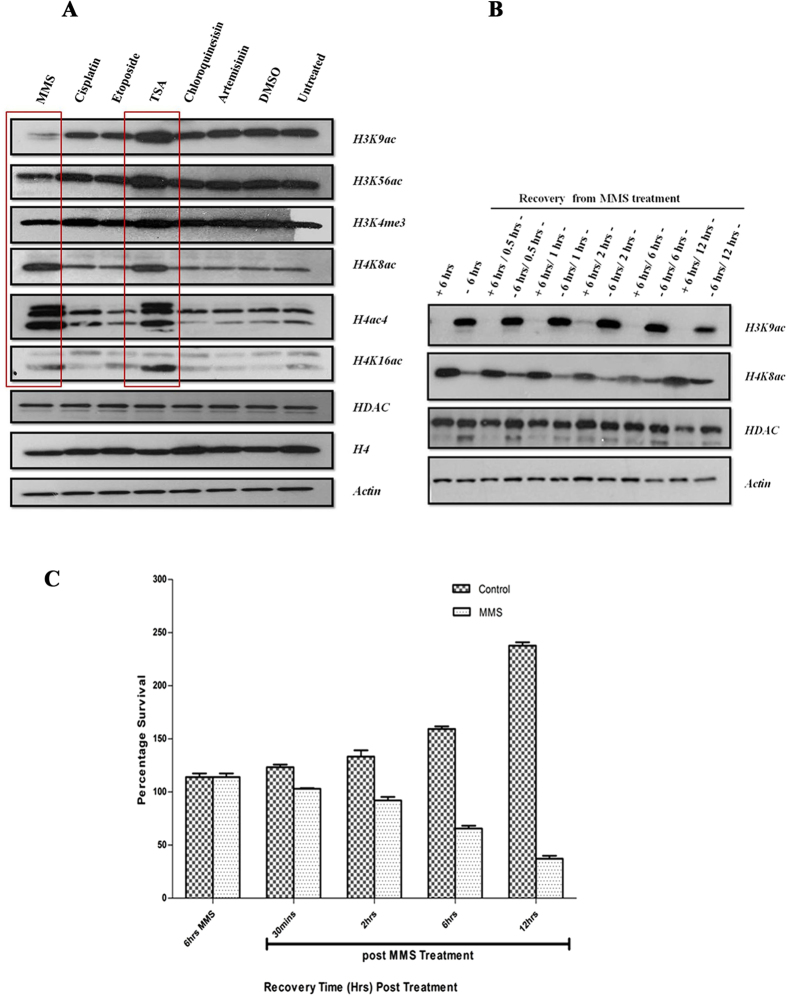
Chromatin alteration by histone modifications during DNA damage regulates the accessibility of DNA repair factors and other regulatory proteins16,48,49,50. The role of acetylation of histones 3 and 4 (H3 and H4) and phosphorylation of H2A/H2A-X in DNA damage response is well documented in yeast, mammals and more recently in Toxoplasma gondii51,52,53,54. Here, we wished to study the effect of DNA damage on histone modifications in P. falciparum. For that the trophozoite stage (28–32 hpi) parasites were treated with the same panel of drugs used for the transcriptional profiling with the same concentrations and duration of exposure (above). Simultaneously, trophozoite parasites were treated with inhibitory concentrations IC90 doses of artemisinin (50 nM) and chloroquine (70 nM). Subsequently, we utilized an immunoblotting analysis to study the abundance of several histone marks that were previously shown to play a role in the P. falciparum chromatin remodeling55. These included histone 3 Lysine 9 acetylation (H3K9ac), H3K56ac, H3K4me3 (lysine 4 trimethylation), H4K8ac, H4K16ac and H4Kac4 (K4, 8, 12, 16 acetylation) (Fig. 2a). Our results revealed that unlike cisplatin and etoposide, MMS, caused increases in acetylation at H4K8 and H4K16 and (likely) as a results of that at H4ac4. On the other hand, H3K9ac and H3K56ac were reduced after the 6hr MMS treatment. There were no detectable changes in the levels of the studied histone marks following treatment with IC90 doses of artemisinin and chloroquine. The increased levels of H4K8ac and the decreased levels of H3K9ac remained detectable even 12 hr after MMS was removed from the culture; past the 6 hr treatment (Fig. 2b). This may suggest that the repair in P. falciparum continues even after removal of MMS possibly in parasites in which the DNA damage was extensive that were beyond repair capacity. This is suggested by the fact that the majority of MMS treated parasite are dying gradually, diminishing the parasite population for as long as 12 hr post treatment (Fig. 2c). Interestingly, the studied histone modifications continue to function (possibly) as sensors/effectors of DNA repair, similar to other eukaryotes16. The MMS-induced effect on chromatin is distinct from TSA inhibition of HDAC that results in increases of H4K8ac, H4K16ac, H3K9ac and H3K56ac that was reproduced from our original study (Fig. 2a)43,56. The TSA-induced H4 and H3 hyperacetylations are reversible as early as 2 hr after drug removal43 which also contrasts the effect of MMS on chromatin remodeling.
Figure 2. DNA damage affects the overall abundance of histone modifications during the P. falciparum IDC.
(A) 3D7 parasites were treated with all drugs at the trophozoite stage for 6 hr and the overall abundance of individual histone modification was measured by western blotting. The MMS treatment resulted in reduction of H3K9ac and H3K56ac and increase of H4K8ac, H4K16ac and H4ac4. Conversely, TSA induced hyperacetylation of all studied H3 and H4 acetylations. Cisplatin and etoposide as well as artemisinin and chloroquine at IC-90 concentrations had no effect on the overall abundance on the histone modification compared to P. falciparum grown under normal condition or treated with the carrier solvent, DMSO. Specific antibody for HDAC enzyme, histone 4 and actin were used as loading controls. (B) Recovery dynamics of H3K9ac and H4K8ac were assessed by first treating with MMS for 6 hrs and subsequently drug was removed and samples were collected after 0.5 hrs, 1 hrs, 2 hrs, 6 hrs and 12 hrs. (C) Parasite survival after MMS (0.05%) treatment compared to untreated 3D7 (Control) parasites. Graphical representation shown is percentage survival in MMS treated and control parasites at 6hrs MMS and 30 mins, 2 hrs, 6 hrs and 12 hrs (post-MMS wash off treatment). The error bars represent SEM.
DNA damage induces genome wide spread of H4K8 acetylation whereas H3K9 acetylation preferentially marks the DNA repair genes
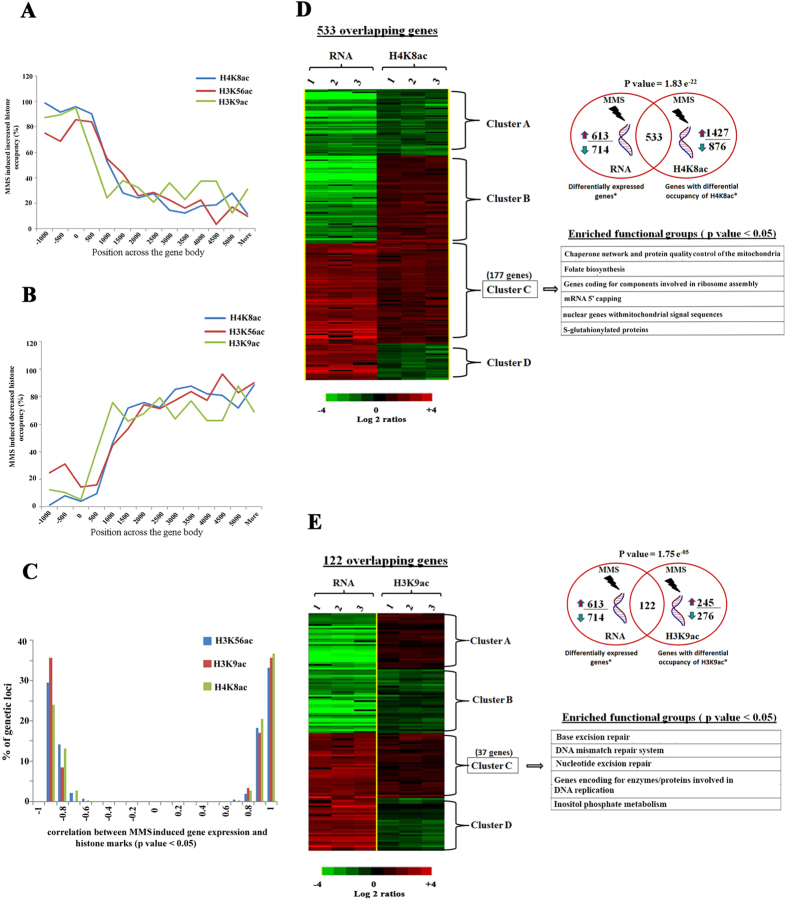
In the next step, we focused on the two predominant histone marks H3K9ac and H4K8ac, and carried out chromatin immunoprecipitations coupled with DNA microarray (ChIP-on-chip). The main objective was to identify chromosomal regions associated with H3K9ac, H4K8ac and H3K56ac that are responsive to DNA damage. Hence, trophozoite stage parasites (28–32 hpi) were treated with 0.05% MMS for 6 hr and ChIP-on-chip was performed as described55 (also see Materials and methods). The total number of genetic loci (defined by the microarray oligonucleotide probes) which were differentially acetylated as a result of MMS (induced/reduced) for H4K8ac, H3K9ac and H3K56ac were 2689, 528 and 2527, respectively (p value < 0.05). From these, 60% (1,592), 46% (243) and 52% (1,318) of genetic loci displayed a significant increase in H4K8ac, H3K9ac and H3K56ac occupancy in the presence of MMS. Crucially, MMS-induced H4K8ac, H3K9ac, H3K56ac appears to be enriched within intergenic regions; upstream of protein coding regions (Fig. 3a). In contrast, loss of acetylation was observed within the coding regions with progressively decreasing occupancy towards the 3′ termini of the genes (Fig. 3b). It has been previously shown that endogenous H4K8ac is predominantly enriched at the intergenic regions, whereas H3K9ac and H3K56ac are localized mainly within the 5′ regions of the coding regions55. Our result suggests that the MMS mediated DNA damage induces acetylations at regions that overlap with genetic loci that are already associated with these histone marks at putative promoter regions during the P. falciparum life cycle under normal growth conditions.
Figure 3. DNA damage induces genome wide spread of H4K8 acetylation whereas H3K9 acetylation preferentially marks the DNA repair genes.
(A,B) The horizontal axis is the microarray oligonucleotide probes (MOE) position relative to the ATG start codon. The vertical axis is the number of histone marks associated genetic loci normalized to the number of probes which represent that particular region on the genome and these genetic loci are those which have increased (A) or decreased (B) ChIP signal in presence of MMS (p value < 0.05). (C) The correlation between MMS induced gene expression and histone marks is shown. (D,E) Association between differential expression and altered histone marks occurrence during MMS treatment is shown. To study such association, we correlated ChIP-on-chip dataset and RNA expression dataset obtained from MMS treated trophozoite parasites and it results in significant overlap between up-regulated/down-regulated genes (>2 fold, p value < 0.05) due to MMS treatment and with those that had altered histone marks (p value < 0.05). For H4K8ac (D) and H3K9ac (E), we identified 533 and 122 overlapping genes. Hierarchical clustering and enriched functional groups of these 533 and 122 genes were also shown, which are divided into 4 clusters depending upon on correlation between ChIP-on-chip and transcriptome dataset.
To study the effect of the MMS induced histone modifications on transcription, we correlated the ChIP-on- chip and RNA expression results. Interestingly, there was a sharp bimodal distribution with positive and negative correlations between MMS induced gene expression and histone acetylations (p value < 0.05) (Fig. 3c). In particular, there were 533 genes whose altered transcription (>2 fold, p value < 0.05) correlated significantly with alterations in occupancy of H4K8ac (p value 1.83E-22, hypergeometric test). From these there were 177 genes in which the increase in H4K8ac coincided with transcriptional up-regulation. Pathway enrichment analyses identified that these genes belong to several cellular processes of protein metabolisms including ribosome assembly, chaperones/protein folding, as well as other biosynthetic pathways including mitochondria (“metabolic nuclear genes destined to mitochondria”) and folate biosynthesis (Fig. 3d). In yeast, MMS induces up-regulation of chaperones and also facilitates trafficking of proteins into mitochondria, which constitutes the indirect responses to DNA damage57,58. Our results suggest that this process is evolutionarily conserved and that some of the indirect transcriptional responses induced by MMS are linked with H4K8ac. For H3K9ac, we identified 122 genes (132 genetic loci, p value 1.75E-05, hypergeometric test) whose abundance correlated with expression. Out of the 122 overlapping genes, 37 show positive correlation (p value < 0.05). Strikingly, this gene set is enriched for factors of the DNA repair and DNA replication associated pathways (Fig. 3e). In mammals, it is the H3K56ac which typically regulates the expression of the DNA damage repair genes under stress conditions59. However, our results show that in P. falciparum, the occupancy of H3K56ac is not linked with DNA repair machinery but instead with genes encoding S-Glutathionylated and mitochondrial proteins (Supplementary Fig. S2). Hence, our results suggest that Plasmodium may have undergone an evolutionary diversion, where regulation of DNA repair was “taken over” by H3K9ac. Intriguingly, this occurs despite the fact that MMS causes an overall reduction of H3K9ac levels in P. falciparum. In future studies, it will be interesting to explore the molecular mechanisms that facilitate this chromatin-linked induction of the DNA damage repair genes.
Artemisinin induces DNA damage response in P. falciparum
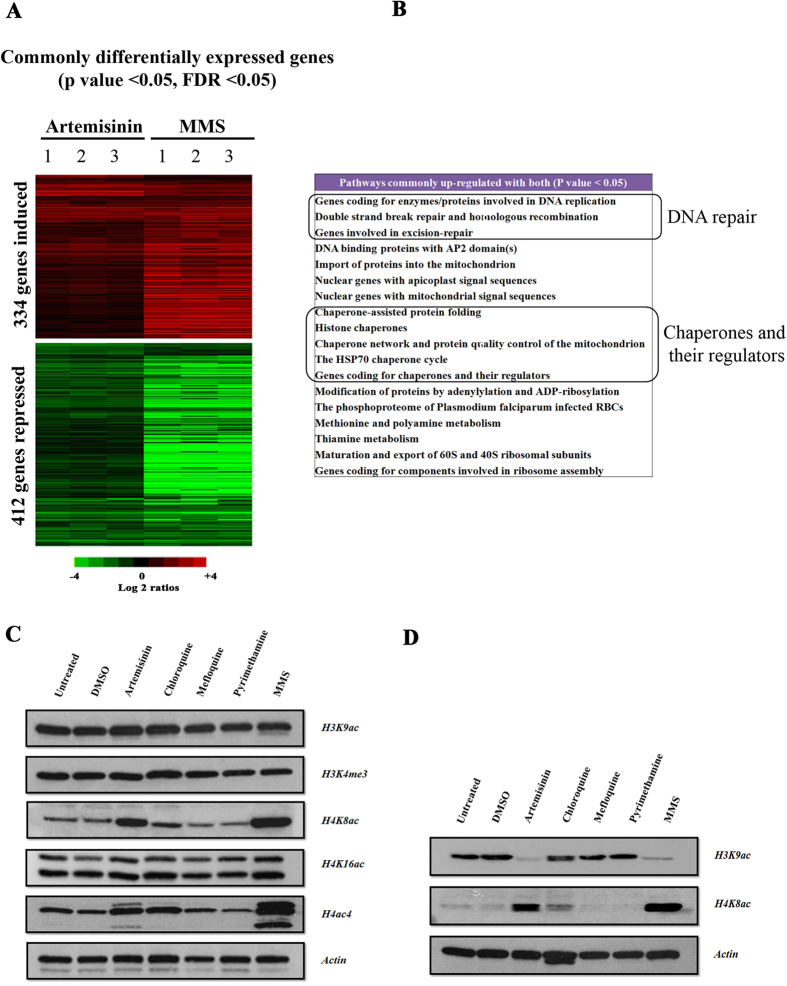
Artemisinin is the most potent antimalarial drug which has been established as the first line treatment of multiple drug resistant strains of P. falciparum60. Although, artemisinin have been shown to induce a DNA damage response in cancer cells, its effect on the Plasmodium genomic DNA remains debatable6. Artemisinin may induce DNA damage in malaria parasite giving positive results of an in vitro comet assay61. In order to provide evidence for an artemisinin role in DNA damage, we wished to investigate similarities between artemisinin and MMS in terms of their effect on the DNA repair machinery in P. falciparum. For this the 3D7 strain of P. falciparum was treated at the trophozoite stage (28–32 hpi) with 1 μM of artemisinin for 6 hr. We chose the physiologically relevant dose of 1 μM which corresponds to plasma concentrations of the drug when administered to patients62. Hierarchical clustering of RNA expression data identified 334 and 412 genes that were commonly up- and down-regulated, respectively, by both MMS and artemisinin (p value < 0.05, FDR < 0.05) (Fig. 4a). Functional enrichment analysis revealed that in addition to several biological processes associated with protein metabolism and DNA replication, at the given treatment conditions, artemisinin also induces genes of the DNA repair pathways (Fig. 4b). These include 8 genes of excision repair and 4 genes of DSBR machinery. Interestingly, Rad50 and MRE11 which constitute the MRN exonuclease complex which are regarded as DNA damage sensors of DSBR were also up-regulated. Moreover, Rad51, which is another important repair protein of DSBR pathway, was also up-regulated by artemisinin. Rad51 is a key nucleoprotein, which is required for strand invasion and strand annealing steps of DSBR38. It has been also reported that artemisinin inhibits topoisomerase enzymes in cancer cells, which leads to generation of single and double strand breaks and subsequently to cell death6. Interestingly, artemisinin treatment in the parasite also leads to under-expression of two DNA topoisomerases: PF3D7_1433500 and PF3D7_1365600. Moreover, artemisinin has an effect on the overall abundance of H3K9ac, H4K8ac and H4ac4; similar to MMS (Fig. 4c,d). This contrasts the lower dose of artemisinin IC90, which does not affect these histone modifications (see Fig. 2). Specifically, at 1 μM, artemisinin induced H4K8ac in the trophozoite and schizont stages and suppressed H3K9ac in the trophozoite stage but not at the schizont stage. In addition to artemisinin, we tested physiologically relevant concentrations of three antimalarial drugs including chloroquine, mefloquine and pyrimethamine and showed no effect on histone acetylation. The only exception is chloroquine that caused a moderate increase of H4K8ac in schizonts. Taken together, DNA damage is likely a part of artemisinin antimalarial mode of action given the fact that its effect on P. falciparum includes inductions of the DNA damage repair mechanisms and histone acetylations in the similar fashion to MMS; presumably as a result of DNA damage.
Figure 4. Artemisinin induces DNA damage response in P. falciparum.
(A) The synchronized P. falciparum cells were treated with clinical doses of artemisinin (1 μM) and MMS (0.05%) for 6 hr at the trophozoite stage. Genes with similar differential transcription for both MMS and artemisinin are shown in the heat map. The experiments were done in triplicates. (p value < 0.05, FDR < 0.05) (B) Pathways, which are commonly up-regulated by both artemisinin and MMS. Schizont (C) and Trophozoite stages (D) of parasites were treated with clinical doses of different anti-malarial drug for 6 hr: artemisinin (1 μM), chloroquine (30 μM), mefloquine (5 μM) and pyrimethamine (5 μM) and with MMS (0.05%). Immunoblot analyses were carried out using primary antibodies probed against the core histone modifications.
ARMD parasites displaying mutator phenotype have defective DNA repair
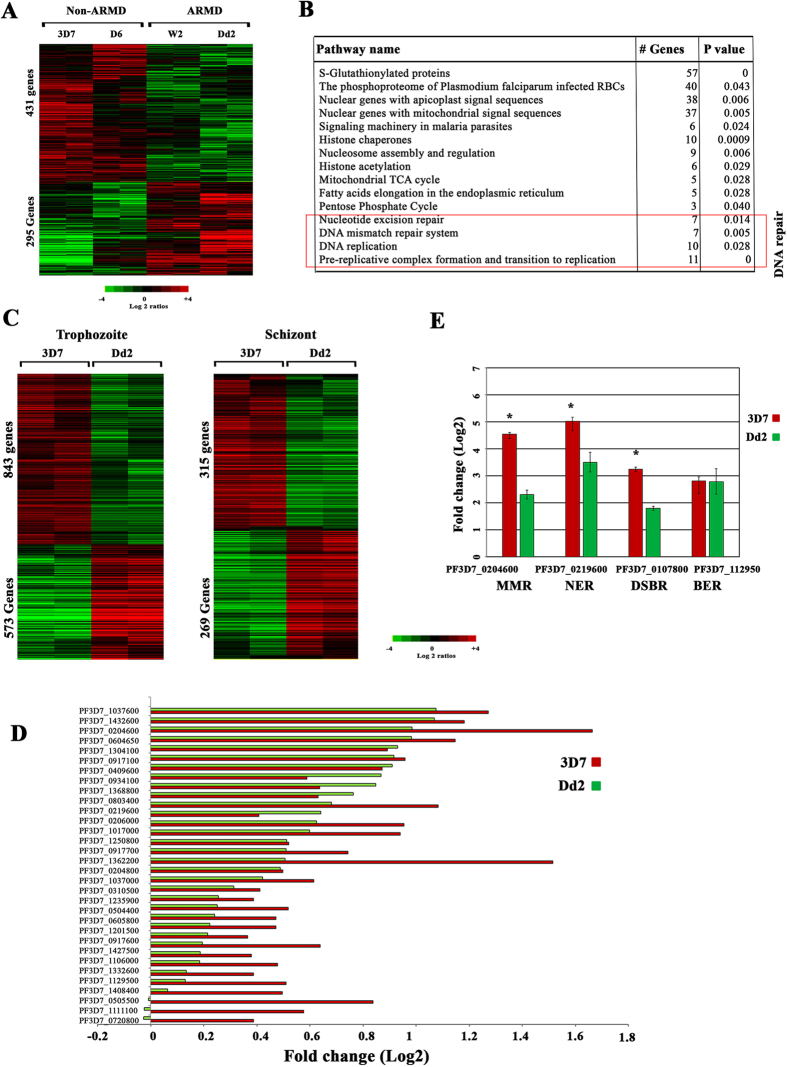
It had been previously suggested that defective DNA repair may contribute to the P. falciparum “mutator” phenotype termed ARMD (accelerated resistance to multiple drugs)35,63. To elucidate the underlying mechanism of the ARMD phenotype, we studied differences in transcription and chromatin structure between ARMD and non-ARMD parasites under DNA damaging conditions. For this, we treated two ARMD (Dd2 and W2) and two non-ARMD parasites strains (3D7 and D6) with MMS (0.05%) for 6 hr at the trophozoite stage (28–32 hpi). The two ARMD P. falciparum strains, Dd2 and W2, originate from Southeast-Asia (Indo-China) and display higher frequencies in acquiring drug resistance de novo under relatively short drug selection intervals27. In contrast, the non-ARMD strain 3D7 and D6 originate from “Netherland” and “Sierra Leone” and can remain drug susceptible even after prolonged pressure with anti-malarial drugs27. Overall, MMS was able to modulate expression of 1,065 1,018 1,093 and 1,014 genes in W2, Dd2, 3D7 and D6, respectively (>2 fold, p value < 0.05). From these, we identified 431 genes that were overexpressed in both non-ARMD and 295 genes overexpressed in both ARMD parasites simultaneously (>1.5 fold, p value < 0.05) (Fig. 5a). Results of a functional enrichment analysis of the 431 genes show a similar representation of physiological processes detected by our initial transcriptional perturbation study of the 3D7 strain (Fig. 5b). This suggests that the MMS effect on transcription is robust amongst nonisogenic strains of P. falciparum. As expected, genes of both MMR and NER repair machinery were found to be significantly up-regulated in these non-ARMD parasites. However, these genes were not induced by MMS in the ARMD parasite strains in both the trophozoite (24–28 hpi) and schizont (34–38 hpi) stages (Fig. 5c). Fig. 5d represents the microarray based signal for 34 DNA repair genes that were differentially induced by MMS in 3D7 but show no (statistically insignificant) changes in their expression in the Dd2, ARMD strain. Subsequently, differential expressions of 4 genes were validated by quantitative RT-PCR. These were: PF3D7_0204600 of MMR, PF3D7_0219600 of NER, PF3D7_0107800 of DSBR and PF3D7_112950 belongs to BER (Fig. 5e). These results suggest a failure to response to DNA damage by sufficient upregulation of DNA repair mechanisms may be one of the underlying mechanisms of the ARMD phenotype. This is however not linked with the changes in the histone modification that are responsive to MMS in all tested P. falciparum strains (Supplementary Fig. S3).
Figure 5. ARMD parasites displaying mutator phenotype have defective DNA repair.
(A) P. falciparum ARMD and non-ARMD parasites were treated with MMS (0.05%) at trophozoite stage for 6 hr. Heat map showing the MMS induced differentially expressed genes between the ARMD and non-ARMD parasites (p value < 0.05) at trophozoite stage. Shown are the mean-centered expression log2 ratios for these genes. (B) Functional enrichment analysis on the 431 up-regulated genes is shown. (C) The gene expression patterns of differentially expressed genes at trophozoite and schizont stage are shown between 3D7 and Dd2. (D) 34 repair genes, which were differentially expressed, were plotted for ARMD (Dd2) and non-ARMD (3D7). (E) Validation of differential expression of genes between 3D7 and Dd2 was done by Quantitative RT-PCR. Quantitative RT-PCR was done on four genes using RNA derived from samples treated with MMS for 6 hr. PF3D7_0204600 belongs to mismatch repair (MMR), PF3D7_0219600 belong to nucleotide excision repair (NER), PF3D7_0107800 belong to double strand break repair (DSBR) and PF3D7_112950 belongs to base excision repair (BER). Relative fold change in RNA expression was computed by 2−∆∆Ct method. The reference control gene used was PF3D7_1218600 (arginyl-tRNA synthetase). Error bars shows the standard deviation of the 2−∆∆Ct value over triplicates. (*represents genes which were differentially expressed between 3D7 and Dd2).
In yeast, bacteria as well as mammals, mutations in DNA repair genes underlay mutator phenotypes, which can subsequently result in rapid emergence of drug resistance28,29,30. Here, we carried out whole-genome sequencing using Illumina MiSeq platform of the four strains used in these experiments: ARMD (Dd2, W2) and non-ARMD (3D7, D6). For this, genomic DNA was harvested immediately after the transcriptomics study (less than 10 generation) in order to avoid accumulation of neutral mutations during normal grow in an in vitro culture as described64. Compared to the 3D7 reference strain, we identified 6,550, 7,231 and 7,253 non-synonymous SNPs within coding regions of 2099, 2,089 and 2,046 genes in the Dd2, W2 and D6 strains, respectively (Supplementary Table S1). Focusing on exonic non-synonymous SNPs in DNA repair genes, total 57 of these were polymorphic in at least one strain (Supplementary Table S2). From these, there are 18 DNA repair genes with SNPs found exclusively in the ARMD parasites (Table 1).
Table 1. List of SNP located in DNA repair genes of ARMD parasites (Dd2 and W2).
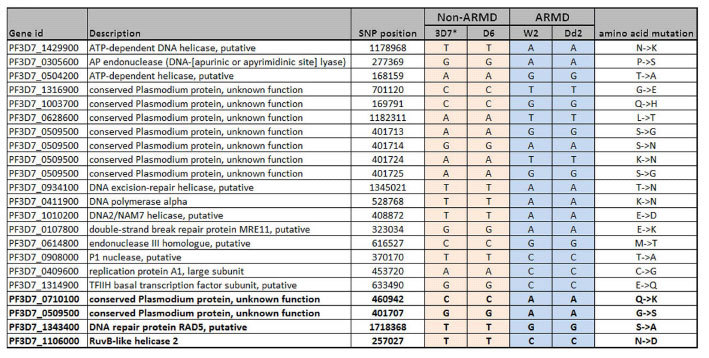
These are 18 DNA repair genes which have non-synonymous SNP within coding region of ARMD parasites. Only one repair gene (PF3D7_0509500) have 4 SNP’s whereas remaining 17 genes have only one SNP per gene. SNPs located in four genes previously identified to be associated with artemisinin resistance in South East Asia are highlighted in red.*3D7 is reference genome, Rows highlighted with red are those ns-SNPs previously identified in artemisinin resistant samples.
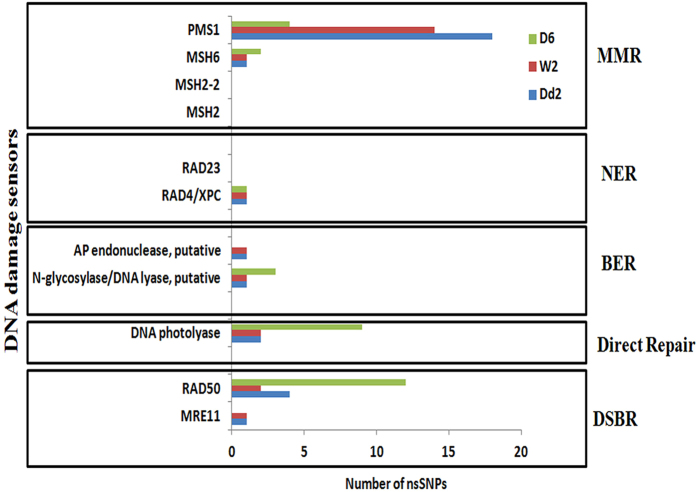
Four of the DNA repair genes have been previously reported to carry same mutations and were found to be associated with artemisinin resistance in Southeast Asia65. These are PF3D7_0509500 (conserved Plasmodium protein, unknown function), PF3D7_0710100 (conserved Plasmodium protein, unknown function), PF3D7_1106000 (RuvB-like helicase 2) and PF3D7_1343400 (DNA repair protein RAD5, putative)65,66. In addition, there were also ARMD-specific polymorphisms in genes encoding “sensors” of DNA damage (Fig. 6). This includes homologues of MRE11 (PF3D7_0107800) and AP lyase (PF3D7_0305600) belonging to DSBR and BER pathway, which showed polymorphisms in the ARMD strains compared to non-ARMD. Also, we identified extensive polymorphism in P. falciparum homologue of PMS1 protein (PF3D7_0726300) which potentially involved in MMR repair. Interestingly, artemisinin resistant parasites from Southeast Asia were found to carry nonsynonymous SNPs in the PMS1 gene65. Although, further experimental studies are needed to investigate the role of these mutations in the ARMD phenotype of P. falciparum, here we showed their occurrence in the parasites cells that failed to respond to DNA damage induced by MMS.
Figure 6. DNA polymorphism in DNA damage sensors between ARMD and non-ARMD parasites.
There are three different repair mechanisms: DNA excision repair (BER, MMR, NER), double strand break repair (DSBR) and direct repair (DNA photolyase). Each repair mechanism has different set of sensory proteins which detect DNA damage, for example RAD23 and XPC are the DNA damage recognition proteins for NER repair mechanism. Each box have the distribution of SNP in DNA damage sensory proteins for a specific repair pathway among D6, 3D7 (non-ARMD) and Dd2, W2 (ARMD). Also, included amino acid mutation which changed form reference.
Discussion
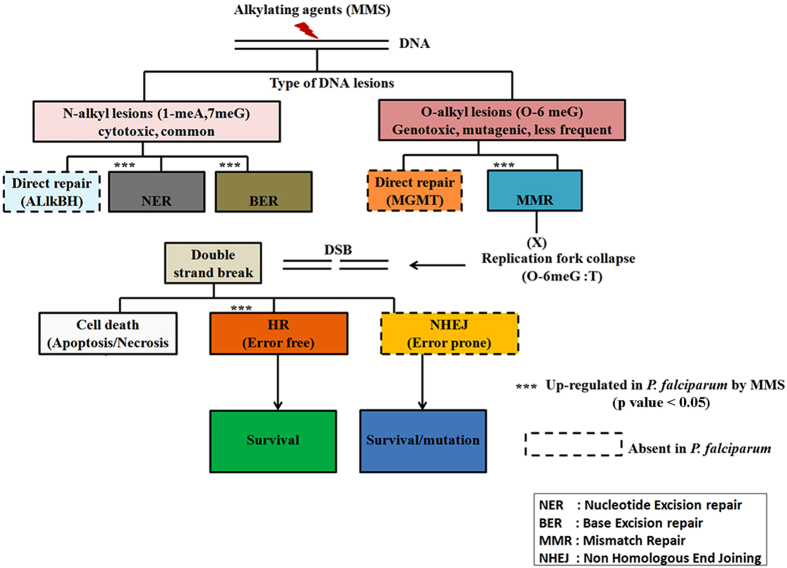
The DNA repair machinery as well as the overall DNA damage response in P. falciparum are believed to contribute to many crucial biological functions of malaria parasites including antigenic variation and copy number variation3,67. Hence, full characterizations of these processes will have a great value for understanding of general biology of the Plasmodium parasites. Here, we demonstrated that when challenged with a DNA damaging agent, P. falciparum cells respond by broad transcriptional changes (21% of the genome) which include up-regulation of both DSBR and excision repair pathways. Similar type of responses were reported in yeast, in which alkylating agents induce differential expression in 1/3 of the yeast genome68. Like in yeast, in P. falciparum, DNA damage leads to upregulation of genes involved in generic (indirect) stress responses such as protein synthesis and turnover as well as mitochondrial functions but also direct responses, involving genes of the DNA repair machinery. Many of the canonical DNA repair genes are conserved in the Plasmodium genome and in this study we are providing transcriptional evidence of their functional involvement. This include upregulation of enzymes that are believed to sense a DNA lesion and initiate specific repair pathways as a result69. In P. falciparum, these include the homologues of A/G specific adenine glycosylases, XPC-h23B, mutS heterodimer (MSH2-MSH6) and MRN exonuclease complex (MRE11-Rad50) which corresponds to DNA damage sensors for BER, NER, MMR and DSBR, respectively (Supplementary Table S3). Besides the sensors, MMS was previously shown to induce overexpression of three P. falciparum DSBR genes, PfRAD51, PfRAD54 and PfRPA138. Here, we show that the MMS-mediated DNA damage results in induction of the entire DNA repair machinery present in the Plasmodium genome (Fig. 7). Although, a single alkylating agent can generate different types of DNA lesions; each DNA lesion is recognized and repaired by a distinct pathway70. The two most common DNA adducts generated by MMS are: N7-methyl guanine (7 me G) and O6-methyl guanine (O6 me G)37. N-alkyl lesions are mostly repaired by BER, NER and direct repair enzyme AlkB homologue, whereas O-alkyl lesions are commonly repaired by MMR and a distinct direct repair enzyme, known as MGMT70,71. Particularly, O-6meG has higher tendency to form mispairs with thymine following replication71. The O-6 meG:T mispairs are recognized by mutS (MSH2-MSH6) which leads to activation of the MMR pathway72. If O6-meG:T mispairs, remains unrepaired, it leads to a futile cycle which results in the collapse of the replication fork and subsequently induction of double strand breaks37. This leads either to apoptosis (if the damage is too extensive) or the double strand breaks are repaired by either HR/NHEJ pathway37,70,71. Our results demonstrate that in Plasmodium, MMS treatment induces excision repair (NER, BER, and MMR) and DSBR (HR). As expected NHEJ and/or several unique direct repair enzymes (MGMT, AlkBH) do not play a role as most of their components are missing in the Plasmodium genome8,73. The DNA polymerase subunit β (PF3D7_0625300) the putative factor of SD-MMEJ, was not up-regulated by MMS which does not exclude this gene from DNA repair but indicate its distinct responsive properties to DNA damage.
Figure 7. Overview of the P. falciparum DNA damage pathways and their transcriptional properties under MMS-induced stress.
The initial N- and O-alkyl lesions of P. falciparum DNA caused by MMS leads initially to transcriptional induction of nucleotide excision repair (NER), base excision repair (BER) and mismatch repair (MMR). The subsequent induction of the double strand break repair (DSBR), namely the homologous recombination repair (HR), suggests that the MMS induced O-alkyl lesions was not be fully repaired by MMR. The transcriptional induction encompasses the majority (if not all) components of the P. falciparum DNA damage repair. Other components of eukaryotic DNA repair pathways such as nonhomologous end joining (NHEJ), or enzymes of direct repair (ALlkBH and MGMT) are not present in the P. falciparum genome (dashed line outline). Although extensive DSB could lead to cell death, 6 hr treatment with 0.05% MMS was likely counteracted by the DNA repair factors and did not lead to upregulation of putative factors of P. falciparum cell death. ***indicates statistically significant functional enrichment of genes of particular pathway amongst genes upregulated as a result of MMS-induced DNA damage. This outline is based on bioinformatics sequence analysis and the transcriptional results presented in this study.
In other eukaryotic cells, DNA damage can cause an arrest of the cell cycle and subsequently programmed cell death (reviewed in74,75,76). Analogously, exposure of P. falciparum to MMS leads to deceleration of the IDC (see below) and subsequent death even after MMS removal (Fig. 2c). In particular, after 6 hr exposure of P. falciparum schizonts to MMS, the overall transcriptome correlated with the 30 hr post invasion (hpi) rather than 38 hpi that was exhibited by the untreated cells (Supplementary Table S4) (for details about the correlation-based hpi estimation see77). This indicates that the IDC progression was arrested at the initial stage of the treatment and that thus the MMS induced transcriptional changes are related to DNA damage either as a direct response or as a consequence of this IDC slowdown. This is similar to transcriptional responses to other small molecular agents47. Nonetheless, there are two lines of evidence that the upregulation of DNA repair genes represents a direct response to the DNA damage; (i) the upregulation occurs at multiple IDC stages (trophozoites and schizonts) in which MMS otherwise deregulates different sets of genes; (ii) the DNA repair genes are not upregulated in the ARMD parasites that exhibit an IDC arrest identical to the Non-ARMD strains otherwise (for both see Fig. 5).
Our results also illustrate that MMS mediated DNA damage leads to considerable changes of histone modification, specifically H4K8ac, H3K9ac and H3K56ac. In yeast and mammals, DNA damage response is initiated by hyperacetylation of H4 by MYST histone acetyltransferase (MYST-HAT)49. It was also shown that mutations in any of the four H4 lysine residues impedes DNA repair in yeast78. In agreement with this, the Plasmodium, PfMYST-HAT acetylates H4K5, 8, 12 and 16 and its overexpression enhances protection from DNA damaging agents MMS and campthothecin79. This suggests that the histone 4 role in DNA damage is evolutionary conserved in the malaria parasites. The ChIP-on-chip results revealed that MMS induced H3K9ac and H4K8ac are predominantly enriched in the intergenic regions and thus are possibly associated with transcription (Fig. 3c). This is in contrast with histone deacetylase inhibitors such as apicidin and TSA that show very limited positive correlation between gene expression and altered acetylation induced by their effect in P. falciparum as well as mammals56,80. In particular, MMS-induced H4K8ac occurs at the transcriptionally up-regulated genes that constitute a putative generic response to DNA damage (protein turnover, mitochondria). Conversely, the increase of H3K9ac occurs preferentially at DNA repair genes that are also induced as a result of MMS-mediated DNA damage. Under normal growth condition, H3K9ac and H4K8ac are selectively enriched on actively transcribed genes belonging to metabolism and ribose assembly related functions in P. falciparum55. In the future, it will be important to investigate the molecular mechanism of H4K8ac and H3K9ac dynamics in transcriptional up-regulation during DNA repair.
Artemisinin appears to induce DNA damage in P. falciparum parasites presumably as a result of oxidative stress generated by this drug81. This is analogous to cancer cells where artemisinin engages DSBR82,83. Here, we observed that at a physiologically relevant dose (1 μM), artemisinin elicits similar transcriptional and epigenetic response that is induced by MMS. Importantly, both agents induced transcriptional up-regulation of parasite excision repair and DSBR. Additionally, we observed that both agents can induce expression of genes imported to the mitochondria/apicoplast. This is consistent with the results in yeast where DNA damage results in import of many proteins into mitochondria from nucleus, which includes pro-apoptotic proteins such as p53, as well as several DNA repair and cell cycle regulatory proteins84,85. Previously, Natalang et al. has shown that 3 hr exposure to 780 nM of artemisinin treatment at trophozoite stage altered transcript abundance of 398 genes none of which is involved in DNA repair46. Here, we showed that 6 hr treatment of P. falciparum cells by both MMS and artemisinin (1 μM) is needed before the gene expression and histone modification characteristics of DNA repair is detectable. Only then the DNA repair machinery is upregulated together with other ~1,500 genes (p value < 0.05). In natural infections, artemisinin reaches up to 1 uM concentrations in ~40 min and then it is rapidly eliminated with a half-life of ~20 min86. This pharmacokinetic profile may or may not provide sufficient time for artemisinin to cause DNA damage. Intriguingly, the kelch-13 protein (PF3D7_1343700) that has been associated with artemisinin resistance87 was upregulated in our artemisinin perturbation experiments (p value = 0.0004). Also, several factors of the DNA repair machinery have been linked with artemisinin resistance in vivo and in vitro65,66. The results here open a plausible possibility that DNA damage contributes to the “real-life” artemisinin mode of action and with that the artemisinin resistance phenotype that is presently spreading through Southeast Asia23,87,88.
Defects in DNA repair machinery were demonstrated to play significant roles in mutator phenotypes in various organisms such as yeast, mammals, bacteria, T. brucei and T. gondii 29,30,33,34,89. Here, we showed that the ARMD (mutator) phenotype of P. falciparum may be underlined by differential transcriptional regulation of genes involved in the DNA repair machinery when the parasites are under a DNA damaging stress. In particular, MMS was able to induce both MMR and NER repair machinery in the non-ARMD parasites but failed to up-regulate these genes in the ARMD parasites. This is consistent with the previous results, which suggested that defective DNA repair may contribute to the emergence of the ARMD parasites in Southeast Asia35,63. Although in our experiments, we did not link this phenomenon with (any) changes in histone modifications, further careful investigations are needed to see if the ARMD phenotype also involves epigenetic factors. Lastly, polymorphism in 18 DNA repair genes may lead to this impaired DNA repair functions. This suggestion is based on the fact that point mutations in mismatch repair genes leads to defective DNA repair which confers mutator phenotype in yeast and bacteria29,89. Further studies need to be done in order to understand the mechanism of how does mutation in DNA repair genes regulates the development of mutator phenotype in malaria parasites.
Material and Methods
Parasite culture and drug treatments
P. falciparum strains (3D7, Dd2, W2, and D6) were cultured using standard protocol for continuous culturing90. Drug treatments were carried out in sorbitol-synchronized trophozoite and schizont stage. For all drug treatment experiments, parasite culture was kept at 5% parasetemia and 2% hematocrit. Transcriptional profiling and immunodetection analysis were carried out at trophozoite stage in 3D7 under identical conditions with following drugs: MMS (0.05%), cisplatin (100 μM), etoposide (100 μM) and TSA (IC90/50nM). The four antimalarial drugs studied were artemisinin (1 μM), chloroquine (30 μM), mefloquine (5 μM) and pyrimethamine (5 μM).
RNA isolation and cDNA synthesis
For MMS (0.05%) induced transcriptional analysis, synchronized parasites were harvested after 1, 3 and 6 hr of treatment, whereas it was only one time point of 6 hr of treatment for cisplatin (100 μM), etoposide (100 μM) TSA (IC90/50 μM) and artemisinin (1 μM). Before RNA extraction, drug treated parasites were washed with PBS twice to obtain infected RBC pellets. Total RNA isolation and cDNA synthesis was performed as illustrated previosuly91. Prior to microarray, the integrity of each RNA samples was checked on 1% agarose gel electrophoresis. cDNA samples derived from drug treated parasites were labeled with Cy5 and hybridized against the Cy3 labeled 3D7 reference pool. This common reference pool was obtained by assembling equal amounts of RNA from 6 time points corresponding to 8 hr interval stages throughout the IDC of the 3D7 strain.
Microarray hybridization and data analysis
Identical amount of Cy5 and Cy3 labeled samples were hybridized to microarrays platform representing the P. falciparum genome. Our Plasmodium DNA array consists of 15,818 oligonucleotides probes (which includes 5,402 50-mer intergenic probes and 10,416 70-mer open reading frame probes) representing 5,343 protein coding genes45. The microarrays were scanned using GenePix scanner 400b and GenePix Pro 6.0 program (Axon Instruments). The raw microarray data obtained was subjected to lowess normalization as described55 and generates log transformed ratios of Cy5 intensity to Cy3 intensity. Hierarchical clustering was performed on these log-transformed ratios using Gene Cluster and results were visualized using Java Treeview.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was executed as explained previously91. Summarily, after 6 hr of MMS (0.05%) treatment at 3D7 trophozoite stage, parasites were harvested with 0.1% saponin. Isolated parasites were crosslinked with formaldehyde, lysed in 1% SDS buffer and homogenized using 200 strokes of dounce homogenizer. The derived nuclei pellet were sonicated with 8 pulses of 25% amplitude (10 sec pulse on, 50 sec off) in order to obtain DNA fragments in the range of 200–1000 bp. Sheared DNA was subjected to chromatin immunoprecipitation as described by Millipore ChIP Assay Kit (#17–295). Both immunoprecipitated DNA and input DNA (sonicated genomic DNA) from MMS treated and untreated samples was exponentially amplified using random primers 5′ GTTTCCCAGTCAGGAT-CNNNNNNNNN 3′ and 5′ GTTTCCCAGTCACGATC 3′ as described by91. For microarray, input was labeled with Cy 3 whereas immunoprecipitated DNA was labeled with Cy5. Input was made by mixing equal amount from MMS treated and untreated samples.
Immunoblot analysis
Identical amount of total protein lysates obtained from drug treated samples and control/DMSO treated samples were separated by 15% SDS-PAGE. Following separation, proteins were transferred onto nitrocellulose membrane. Immunoblot analyses were carried out using primary antibodies probed against the core histone modifications obtained from Millipore, Upstate and horseradish peroxidase conjugated secondary antibody was acquired from GE Healthcare. Additionally, polyclonal PfHDAC1anti-serum, histone 4 (Millipore) and actin (Millipore) were used as loading controls.
Quantitative real time PCR (RT-PCR)
Validations of microarray results for chosen genes were done by quantitative real time PCR (Roche) according to manufacturer instructions. Every cDNA sample was amplified in triplicates and to check specific amplification, no-template control was also included. Further, only those samples were included in data analysis, which shows single peak in melting curve. Relative fold change in RNA expression was computed by 2−∆∆Ct method92. The reference control gene used was PF3D7_1218600 (arginyl-tRNA synthetase).
High-throughput genome sequencing by Illumina MiSeq method
Genomic DNA was extracted from each of 3 parasites strains (Dd2, W2 and D6) and two microgram of genomic DNA was sequenced using whole-genome sequencing Illumina MiSeq method64. The sequenced data was aligned to the reference 3D7 Plasmodium genome. High quality SNP’s identified in the sequences using Genome Analysis Toolkit software (GATK) with stringent cut offs (minimum base quality score ≥ 30) as described by Mok et al.93. Using this cut-off, we identified 6,550, 7,231 and 7,253 non-synonymous SNP within coding regions in the Dd2, W2 and D6 strains, respectively.
Additional Information
Accession codes: We have submitted the microarray data to NCBI GEO, (Accession number GSE72580).
How to cite this article: Gupta, D. K. et al. DNA damage regulation and its role in drug-related phenotypes in the malaria parasites. Sci. Rep. 6, 23603; doi: 10.1038/srep23603 (2016).
Supplementary Material
Footnotes
Author Contributions D.K.G. and Z.B. designed the study and drafted the manuscript. D.K.G. performed the experiments and statistical analysis of the data. A.T.P contributed in the experiments and data anlyses. A.P.G. and L.Z. contributed intellectually. All authors read and approved the final draft of manuscript.
References
- Nowsheen S. & Yang E. S. The Intersection between DNA damage response and cell death pathways. Exp Oncol 34, 243–254 (2012). [PMC free article] [PubMed] [Google Scholar]
- Ataian Y. & Krebs J. E. Five repair pathways in one context: chromatin modification during DNA repairThis paper is one of a selection of papers published in this Special Issue, entitled 27th International West Coast Chromatin and Chromosome Conference, and has undergone the Journal’s usual peer review process. Biochem Cell Biol 84, 490–494 (2006). [DOI] [PubMed] [Google Scholar]
- Lee A. H., Symington L. S. & Fidock D. A. DNA Repair Mechanisms and Their Biological Roles in the Malaria Parasite Plasmodium falciparum. Microbiol Mol Biol Rev 78, 469–486 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percário S. et al. Oxidative Stress in Malaria. Int J Mol Sci 13, 16346–16372 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radfar A., Diez A. & Bautista J. M. Chloroquine mediates specific proteome oxidative damage across the erythrocytic cycle of resistant Plasmodium falciparum. Free Radic Biol Med 44, 2034–2042 (2008). [DOI] [PubMed] [Google Scholar]
- O’Neill P. M., Barton V. E. & Ward S. A. The Molecular Mechanism of Action of Artemisinin—The Debate Continues. Molecules 15, 1705–1721 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy N. et al. Dominant negative mutant of Plasmodium Rad51 causes reduced parasite burden in host by abrogating DNA double-strand break repair. Mol Microbiol 94, 353–366 (2014). [DOI] [PubMed] [Google Scholar]
- Gardner M. J. et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419, 498–511 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkman L. A., Lawrence E. A. & Deitsch K. W. Malaria parasites utilize both homologous recombination and alternative end joining pathways to maintain genome integrity. Nucleic Acids Res 42, 370–379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer M. et al. Zinc finger nuclease-based double-strand breaks attenuate malaria parasites and reveal rare microhomology-mediated end joining. Genome Biol 16, 249 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A. M. & McVey M. Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions. Nucleic Acids Res 38, 5706–17 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltiwanger B. M. et al. DNA base excision repair in human malaria parasites is predominantly by a long-patch pathway. Biochemistry 39, 763–72 (2000). [DOI] [PubMed] [Google Scholar]
- Nunthawarasilp P., Petmitr S. & Chavalitshewinkoon-Petmitr P. Partial purification and characterization of DNA polymerase beta-like enzyme from Plasmodium falciparum. Mol Biochem Parasitol 154, 141–7 (2007). [DOI] [PubMed] [Google Scholar]
- van Attikum H. & Gasser S. M. Crosstalk between histone modifications during the DNA damage response. Trends Cell Biol 19, 207–217. [DOI] [PubMed] [Google Scholar]
- Dinant C., Houtsmuller A. & Vermeulen W. Chromatin structure and DNA damage repair. Epigenetics Chromatin 1, 9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Jiang X. & Price B. D. Tip60: Connecting chromatin to DNA damage signaling. Cell cycle 9, 930–936 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo S. E. & Jackson S. P. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 25, 409–433 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy M. F., Selvarajah S. A., Josling G. A. & Petter M. Epigenetic regulation of the Plasmodium falciparum genome. Brief Funct Genomics (2013). [DOI] [PubMed] [Google Scholar]
- Payne D. Spread of chloroquine resistance in Plasmodium falciparum. Parasitol Today 3, 241–246 (1987). [DOI] [PubMed] [Google Scholar]
- Roper C. et al. Intercontinental Spread of Pyrimethamine-Resistant Malaria. Science 305, 1124 (2004). [DOI] [PubMed] [Google Scholar]
- Mita T. et al. Limited Geographical Origin and Global Spread of Sulfadoxine-Resistant dhps Alleles in Plasmodium falciparum Populations. J Infect Dis 204, 1980–1988 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noedl H. et al. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med 359, 2619–2620 (2008). [DOI] [PubMed] [Google Scholar]
- Ashley E. A. et al. Spread of Artemisinin Resistance in Plasmodium falciparum Malaria. N Engl J Med 371, 411–423 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayton K. & Su X.z. Genetic and Biochemical Aspects of Drug Resistance in Malaria Parasites. Curr Drug Targets Infect Disord 4, 1–10 (2004). [DOI] [PubMed] [Google Scholar]
- Anderson T., Nkhoma S., Ecker A. & Fidock D. How can we identify parasite genes that underlie antimalarial drug resistance? Pharmacogenomics 12, 59–85 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariey F. et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505, 50–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathod P. K., McErlean T. & Lee P.-C. Variations in frequencies of drug resistance in Plasmodium falciparum. Proc Natl Acad Sci USA 94, 9389–9393 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClerc J. E., Li B., Payne W. L. & Cebula T. A. High Mutation Frequencies Among Escherichia coli and Salmonella Pathogens. Science 274, 1208–1211 (1996). [DOI] [PubMed] [Google Scholar]
- Drotschmann K. et al. Mutator phenotypes of yeast strains heterozygous for mutations in the MSH2 gene. Proc Natl Acad Sci USA 96, 2970–2975 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. L. & Loeb L. A. The Mutation Rate and Cancer. Genetics 148, 1483–1490 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran P. Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis 22, 1931–1937 (2001). [DOI] [PubMed] [Google Scholar]
- Fink D., Aebi S. & Howell S. B. The role of DNA mismatch repair in drug resistance. Clin Cancer Res 4, 1–6 (1998). [PubMed] [Google Scholar]
- Augusto-Pinto L., Teixeira S. M. R., Pena S. D. J. & Machado C. R. Single-Nucleotide Polymorphisms of the Trypanosoma cruzi MSH2 Gene Support the Existence of Three Phylogenetic Lineages Presenting Differences in Mismatch-Repair Efficiency. Genetics 164, 117–126 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E. M. & Arrizabalaga G. Disruption of a mitochondrial MutS DNA repair enzyme homologue confers drug resistance in the parasite Toxoplasma gondii. Mol Microbiol 72, 425–441 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta R. F., Brown M. L., Terrell J. C. & Geyer J. A. Defective DNA Repair as a Potential Mechanism for the Rapid Development of Drug Resistance in Plasmodium falciparum. Biochemistry 43, 4885–4891 (2004). [DOI] [PubMed] [Google Scholar]
- Chlebowicz E. & Jachymczyk W. J. Repair of MMS-induced DNA double-strand breaks in haploid cells of Saccharomyces cerevisiae, which requires the presence of a duplicate genome. Mol Gen Genet 167, 279–286 (1979). [DOI] [PubMed] [Google Scholar]
- Kondo N., Takahashi A., Ono K. & Ohnishi T. DNA Damage Induced by Alkylating Agents and Repair Pathways. J Nucleic Acids 2010, 543531 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan A. M. & Kumar N. Opposing Roles for Two Molecular Forms of Replication Protein A in Rad51-Rad54-Mediated DNA Recombination in Plasmodium falciparum. mBio 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A. & Krishnamurthy S. Cellular Responses to Cisplatin-Induced DNA Damage. J Nucleic Acids 2010, 16 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinich N. O., Tafani M., Rothman R. J., Russo M. A. & Farber J. L. The Course of Etoposide-induced Apoptosis from Damage to DNA and p53 Activation to Mitochondrial Release of Cytochromec. J Biol Chem 277, 16547–16552 (2002). [DOI] [PubMed] [Google Scholar]
- Murray V., Campbell H. M. & Gero A. M. Plasmodium falciparum: DNA sequence specificity of cisplatin and cisplatin analogues. Exp Parasitol 128, 396–400 (2011). [DOI] [PubMed] [Google Scholar]
- Kelly J. M., McRobert L. & Baker D. A. Evidence on the chromosomal location of centromeric DNA in Plasmodium falciparum from etoposide-mediated topoisomerase-II cleavage. Proc Natl Acad Sci USA 103, 6706–6711 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews K. T. et al. Comparative Gene Expression Profiling of P. falciparum Malaria Parasites Exposed to Three Different Histone Deacetylase Inhibitors. PLoS ONE 7, e31847 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaal B., Gupta A., Wastuwidyaningtyas B., Luah Y. & Bozdech Z. Histone deacetylases play a major role in the transcriptional regulation of the Plasmodium falciparum life cycle. PLoS Pathog 6, e1000737 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Llinas M., Li J., Preiser P. & Bozdech Z. Selection of long oligonucleotides for gene expression microarrays using weighted rank-sum strategy. BMC Bioinformatics 8, 350 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natalang O. et al. Dynamic RNA profiling in Plasmodium falciparum synchronized blood stages exposed to lethal doses of artesunate. BMC Genomics 9, 388 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G. et al. Transcriptional profiling of growth perturbations of the human malaria parasite Plasmodium falciparum. Nat Biotechnol 28, 91–98 (2010). [DOI] [PubMed] [Google Scholar]
- Altaf M., Saksouk N. & Côté J. Histone modifications in response to DNA damage. Mutat Res 618, 81–90 (2007). [DOI] [PubMed] [Google Scholar]
- Xu Y. & Price B. D. Chromatin dynamics and the repair of DNA double strand breaks. Cell Cycle 10, 261–267 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossetto D., Truman A. W., Kron S. J. & Côté J. Epigenetic Modifications in Double-Strand Break DNA Damage Signaling and Repair. Clin Cancer Res 16, 4543–4552 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- House N. C. M., Koch M. R. & Freudenreich C. H. Chromatin modifications and DNA repair: beyond double-strand breaks. Front Genet 5, 296 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulli G., Di Micco R. & di Fagagna F. d. A. Crosstalk between chromatin state and DNA damage response in cellular senescence and cancer. Nat Rev Cancer 12, 709–720 (2012). [DOI] [PubMed] [Google Scholar]
- Dalmasso M. C., Onyango D. O., Naguleswaran A., Sullivan W. J. & Angel S. O. Toxoplasma H2A Variants Reveal Novel Insights into Nucleosome Composition and Functions for this Histone Family. J Mol Biol 392, 33–47 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonlaufen N., Naguleswaran A., Coppens I. & Sullivan W. J. MYST Family Lysine Acetyltransferase Facilitates Ataxia Telangiectasia Mutated (ATM) Kinase-mediated DNA Damage Response in Toxoplasma gondii. J Biol Chem 285, 11154–11161 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A. P. et al. Dynamic Epigenetic Regulation of Gene Expression during the Life Cycle of Malaria Parasite Plasmodium falciparum. PLoS Pathog 9, e1003170 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaal B. K., Gupta A. P., Wastuwidyaningtyas B. D., Luah Y.-H. & Bozdech Z. Histone Deacetylases Play a Major Role in the Transcriptional Regulation of the Plasmodium falciparum Life Cycle. PLoS Pathog 6, e1000737 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch A. P. et al. Genomic Expression Responses to DNA-damaging Agents and the Regulatory Role of the Yeast ATR Homolog Mec1p. Mol Biol Cell 12, 2987–3003 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkach J. M. et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol 14, 966–976 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vempati R. K. et al. p300-mediated Acetylation of Histone H3 Lysine 56 Functions in DNA Damage Response in Mammals. J Biol Chem 285, 28553–28564 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L. & Su X.-z. Discovery, mechanisms of action and combination therapy of artemisinin. Expert Rev Anti Infect Ther 7, 999–1013 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan A. M. & Kumar N. Anti-malarial action of Artesunate involves DNA damage mediated by Reactive Oxygen Species. Antimicrob Agents Chemother 59, 317–325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hien T. T. et al. Comparative Pharmacokinetics of Intramuscular Artesunate and Artemether in Patients with Severe Falciparum Malaria. Antimicrob Agents Chemother 48, 4234–4239 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellini M. A. et al. Malaria drug resistance is associated with defective DNA mismatch repair. Mol Biochem Parasitol 177, 143–147 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bopp S. E. R. et al. Mitotic Evolution of Plasmodium falciparum Shows a Stable Core Genome but Recombination in Antigen Families. PLoS Genetics 9, e1003293 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O. et al. Multiple populations of artemisinin-resistant Plasmodium falciparum in Cambodia. Nat Genet 45, 648–655 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala-Harrison S. et al. Genetic loci associated with delayed clearance of Plasmodium falciparum following artemisinin treatment in Southeast Asia. Proc Natl Acad Sci USA 110, 240–245 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanti Bhattacharyya M., Norris D. E. & Kumar N. Molecular players of homologous recombination in protozoan parasites: implications for generating antigenic variation. Infect Genet Evol 4, 91–98 (2004). [DOI] [PubMed] [Google Scholar]
- Jelinsky S. A., Estep P., Church G. M. & Samson L. D. Regulatory Networks Revealed by Transcriptional Profiling of Damaged Saccharomyces cerevisiae Cells: Rpn4 Links Base Excision Repair with Proteasomes. Mol Cell Biol 20, 8157–8167 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan C. H. & Russell P. The DNA damage response: sensing and signaling. Curr Opin Cell Biol 16, 629–633 (2004). [DOI] [PubMed] [Google Scholar]
- Drabløs F. et al. Alkylation damage in DNA and RNA—repair mechanisms and medical significance. DNA Repair 3, 1389–1407 (2004). [DOI] [PubMed] [Google Scholar]
- Fu D., Calvo J. A. & Samson L. D. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer 12, 104–120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett D. R. et al. Human MutSalpha recognizes damaged DNA base pairs containing O6-methylguanine, O4-methylthymine, or the cisplatin-d(GpG) adduct. Proc Natl Acad Sci USA 93, 6443–6447 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegg A. E. Repair of O6-alkylguanine by alkyltransferases. Mutat Res 462, 83–100 (2000). [DOI] [PubMed] [Google Scholar]
- Harper J. W. & Elledge S. J. The DNA damage response: ten years after. Mol Cell 28, 739–45 (2007). [DOI] [PubMed] [Google Scholar]
- Li Z., Pearlman A. H. & Hsieh P. DNA mismatch repair and the DNA damage response. DNA Repair (Amst) (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu B. M. & Cortez D. DNA damage response: three levels of DNA repair regulation. Cold Spring Harb Perspect Biol 5, a012724 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok S. et al. Artemisinin resistance in Plasmodium falciparum is associated with an altered temporal pattern of transcription. BMC Genomics 12, 391 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. W. et al. Acetylation of histone H4 by Esa1 is required for DNA double-strand break repair. Nature 419, 411–415 (2002). [DOI] [PubMed] [Google Scholar]
- Miao J. et al. The MYST Family Histone Acetyltransferase Regulates Gene Expression and Cell Cycle in Malaria Parasite Plasmodium falciparum. Mol Microbiol 78, 883–902 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Atalaya J. P., Ito S., Valor L. M., Benito E. & Barco A. Genomic targets, and histone acetylation and gene expression profiling of neural HDAC inhibition. Nucleic Acids Res 41, 8072–8084 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan A. M. & Kumar N. Antimalarial Action of Artesunate Involves DNA Damage Mediated by Reactive Oxygen Species. Antimicrob Agents Chemother 59, 317–325 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P. C. H. et al. Artesunate Derived from Traditional Chinese Medicine Induces DNA Damage and Repair. Cancer Res 68, 4347–4351 (2008). [DOI] [PubMed] [Google Scholar]
- Berdelle N., Nikolova T., Quiros S., Efferth T. & Kaina B. Artesunate Induces Oxidative DNA Damage, Sustained DNA Double-Strand Breaks, and the ATM/ATR Damage Response in Cancer Cells. Mol Cancer Ther 10, 2224–2233 (2011). [DOI] [PubMed] [Google Scholar]
- Griffiths L. M. et al. Dynamic Compartmentalization of Base Excision Repair Proteins in Response to Nuclear and Mitochondrial Oxidative Stress. Mol Cell Biol 29, 794–807 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tembe V. & Henderson B. R. Protein trafficking in response to DNA damage. Cell Signal 19, 1113–1120 (2007). [DOI] [PubMed] [Google Scholar]
- Dondorp A. et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361, 455–467 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto O. et al. Genetic architecture of artemisinin-resistant Plasmodium falciparum. Nat Genet 47, 226–234 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok S. et al. Drug resistance. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science 347, 431–5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra I., O’Neill A. J. & Miller K. The role of mutators in the emergence of antibiotic-resistant bacteria. Drug Resist Updat 6, 137–145. [DOI] [PubMed] [Google Scholar]
- Trager W. & Jensen J. Human malaria parasites in continuous culture. Science 193, 673–675 (1976). [DOI] [PubMed] [Google Scholar]
- Bozdech Z., Mok S. & Gupta A. DNA Microarray-Based Genome-Wide Analyses of Plasmodium Parasites. Methods Mol Biol 923, 189–211 (2013). [DOI] [PubMed] [Google Scholar]
- Livak K. J. & Schmittgen T. D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408 (2001). [DOI] [PubMed] [Google Scholar]
- Mok S. et al. Structural polymorphism in the promoter of pfmrp2 confers Plasmodium falciparum tolerance to quinoline drugs. Mol Microbiol 91, 918–934 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.