Abstract
SUMOylation and ubiquitination are two essential post translational modifications (PTMs) involved in the regulation of important biological processes in eukaryotic cells. Identification of ubiquitin (Ub) and small ubiquitin-related modifier (SUMO)-conjugated lysine residues in proteins is critical for understanding the role of ubiquitination and SUMOylation, but remains experimentally challenging. We have developed a powerful in vitro Ub/SUMO assay using a novel high density peptide array incorporated within a microfluidic device that allows rapid identification of ubiquitination and SUMOylation sites on target proteins. We performed the assay with a panel of human proteins and a microbial effector with known target sites for Ub or SUMO modifications, and determined that 80% of these proteins were modified by Ub or specific SUMO isoforms at the sites previously determined using conventional methods. Our results confirm the specificity for both SUMO isoform and individual target proteins at the peptide level. In summary, this microfluidic high density peptide array approach is a rapid screening assay to determine sites of Ub and SUMO modification of target substrates, which will provide new insights into the composition, selectivity and specificity of these PTM target sites.
Abbreviations: Ub, Ubiquitin; SUMO, Small ubiquitin-related modifier; PTM, post translational modification
Keywords: Ubiquitin, SUMO, High density peptide array, Ehrlichia, Effector
Highlights
-
•
A high throughput in vitro Ub/SUMO assay using microfluidic high density peptide array was developed.
-
•
The sensitivity and specificity of the assay was validated using human and bacterial proteins with known Ub/SUMO sites.
-
•
Specificity for SUMO isoforms on individual target proteins was determined at the peptide level.
-
•
This approach can rapidly identify specific Ub/SUMO sites in unknown substrates.
1. Introduction
Post translational modifications (PTMs) can dramatically affect protein function and interactions, ultimately influencing a plethora of eukaryotic cellular processes. PTMs include covalent linkage of a small protein, such as ubiquitin (Ub), the small ubiquitin-like modifier (SUMO), or other modifications such as phosphoryl, acetyl and methyl groups [1], [2]. Therefore, understanding the site and number of Ub and SUMO modifications on target proteins may provide valuable information that can lead to drug development or a better understanding of these PTMs with regard to protein function and role in various host cell processes and pathogen-host interactions [3], [4]. Recent proteomic screening approaches have identified new targets modified by Ub and SUMO, and the number of validated substrates is rapidly increasing [5], [6], [7]; however, the number of uncharacterized proteins that are targets of Ub/SUMO PTMs is vast.
Ub and SUMO (SUMO 1–4) are small proteins (76 and 100 amino acids, respectively) that are conjugated to substrates through a series of enzymatic reactions [8]. Ub-activating enzyme E1 forms a thiol-ester bond between its cysteine located in the active site and the carboxy terminal of Ub in an ATP-dependent manner. The ubiquitin molecule is then transferred to the second enzyme of the complex, E2 (Ub-conjugating enzyme), before reaching the final enzyme, an E3 Ub protein ligase, that is essential for binding the substrate [8]. The process can be repeated forming chains of poly-Ub. The human genome contains>600 annotated E3 Ub ligases, and more than 100 deubiquitinating enzymes (DUBs) that can remove Ub from the substrate [9]. Currently, experimental identification of Ub target sites is challenging due to rapid turnover of ubiquitinated proteins, the relatively large size of the Ub PTM, and an undefined consensus motif.
Similar to the process of ubiquitination, SUMOylation normally consists of three enzymatic steps: activation by enzyme E1 (SUMO-activating enzyme; SAE), SUMO-conjugation by enzyme E2 (UBC9), and SUMO-ligation by an E3 ligase. Sumoylation involves the covalent modification of target proteins at a lysine residue within the canonical consensus sequence JKxE/D (where J is a large hydrophobic amino acid and x any amino acid) [10]. However, recent proteomics studies have shown that a considerable proportion of SUMO-modified proteins do not contain consensus sites [11]. Furthermore, a model for non-consensus SUMO targeting has been proposed, where SUMO-Ubc9 thioester is recruited by a SUMO interaction motif (SIM: V/I-X-V/I-V/I), which interacts non-covalently with SUMO located on the substrate [12]. However, the non-consensus substrates remain largely unknown; therefore, novel tools that allow unbiased identification of SUMO target sites are needed.
The identification and validation of new Ub/SUMO substrates and deciphering of the Ub/SUMO acceptor site in a given protein is of considerable interest. A significant number of known SUMOylation sites in target proteins occur at the consensus motif. However, this motif also often occurs in non-SUMOylated proteins, and many functionally important SUMO sites are known to be in noncanonical sequences. SUMOylation sites are routinely determined by mass spectrometry (MS), which has been the primary approach for the characterization of Ub/SUMO modifications on substrate proteomes [5], [6], [7]. However, the application of MS to characterize Ub/SUMO sites can be hampered by low protein quantity and purity. Purification of intact modified species produces a peptide mixture which can be too complex for efficient detection of Ub/SUMO modified sites. Moreover, although MS is an accurate and reproducible approach, it is not suited for rapid analysis of lysine modification sites. In recent years, alternative approaches such as peptide and protein microarrays have been developed to study phosphorylation and SUMOylation [13], [14].
In this study, we developed a powerful in vitro approach to identify sites modified by Ub/SUMO that utilizes a high density peptide microfluidic chip. The approach was validated using a panel of human proteins and one bacterial effector protein with known Ub/SUMO sites, and the results demonstrated a high level of concordance with traditional MS analysis for identifying Ub/SUMO-modified sites.
2. Materials and methods
2.1. Microfluidic peptide microarray technology for analysis of ubiquitination and SUMOylation
The work flow for the Ub/SUMO peptide assay development was based on previous analysis of protein–protein interaction using a phosphopeptide microarray [15]. In this study, we detected Ub/SUMO PTMs on unmodified peptides that were synthesized on the chip. The various steps for this study included peptide design, peptide chip synthesis, ubiquitination and SUMOylation reaction, data processing and analysis. The peptides were organized into a layout file to direct chip synthesis and data analysis using PepArray pro software (http://www.pepcyber.org/PepArray/). After the chip synthesis, peptide based assays were conducted and the resulting image file (consisting of all the fluorescent spots of Ub/SUMOylation) was analyzed using previously described methods [16], [17]. The various steps in peptide design, detection and the process for identification of Ub/SUMOylation sites of selected proteins are provided in Fig. 1, Fig. 2.
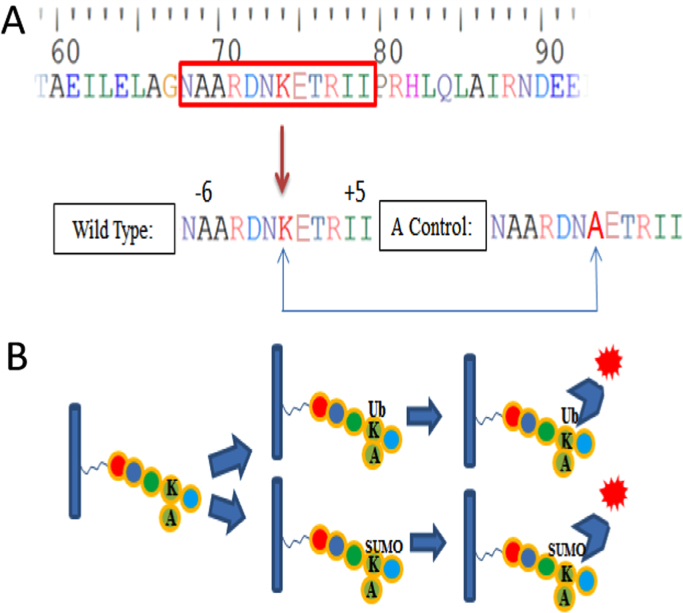
Fig. 1.
Overview of the peptide design and ubiquitination/SUMOylation detection strategy. (A) All wild type peptides are 12-mers by flanking a central lysine residue with 6 N-terminal amino acids and 5 C-terminal amino acids. For each peptide, a corresponding negative control sequence was included with alanine (A) substituted in place of K (A control). (B) The ubiquitination/SUMOylation status of lysine (K) in these peptides was detected using specific conjugated fluorescent anti-ubiquitin or anti-SUMO antibodies which recognize ubiquitinated or SUMOylated lysines and produce a fluorescent signal.
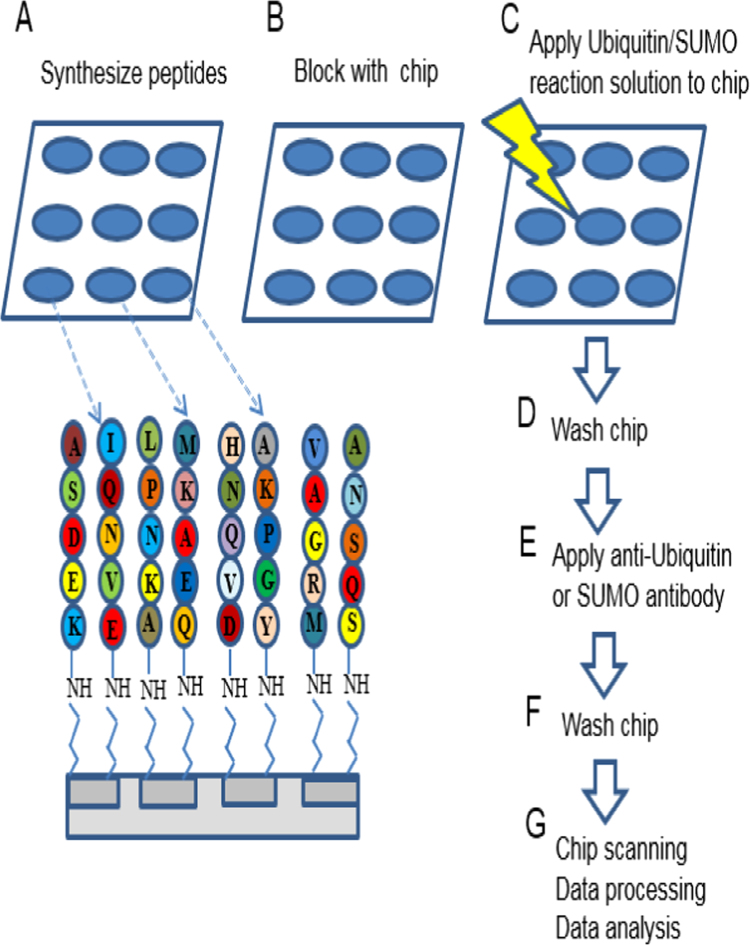
Fig. 2.
Experimental outline of ubiquitination and SUMOylation assay on peptide microarray. (A) All wild type peptides are synthesized onto microfluidic chip using standard t-Boc protecting amino acids and a photogenerated acid as deprotection reagent for the light-gated parallel synthesis. (B, C) The chip is blocked and incubated with ubiquitination or SUMOylation reaction solution and cell lysate. (D) The indirect and non-specific interactors are removed by denaturing washes and PBST washes. (E) The ubiquitination and SUMOylation status of peptides are detected using specific conjugated fluorescent anti-ubiquitin or anti-SUMO antibodies. (F, G) After washing, the chip is scanned using Anon GenePix 4400A (Molecular Devices) scanner using GenePix Pro 7 software. TIFF image files were further processed through Array-Pro Analyzer software and pixel density values were obtained as a text file (output data) for data analysis.
2.2. Source peptides
A peptide array was designed using an in-house developed PERL program [18]. A collection of 12-mer peptides from sequences of ubiquitinated or SUMOylated proteins confirmed experimentally by MS were selected for validation [6], [7]. Peptides (12-mer) from human and microbial target proteins (Ehrlichia) were synthesized by flanking a central lysine residue with 6 N-terminal amino acids and 5 C-terminal amino acids. For each peptide, a corresponding negative control sequence was included with alanine (A) substituted in place of K (A peptide). The sequence for all peptides used in this study, which includes the protein name, position of lysine, SUMO isoforms and sequence are presented in Table 1, Table 2. The designed peptides were organized into a layout file to direct chip synthesis and data analysis. All peptides including mutant controls with twelve residues were synthesized directly on chips in triplicate.
Table 1.
Peptides corresponding to ubiquitinated target sites.
| Protein | Modified K | Peptide sequence (wt) | Peptide sequence (ctrl) |
|---|---|---|---|
| NDUFS5 | 28 | SGEQPYKMAGRC | SGEQPYAMAGRC |
| NDFIP1 | 83 | DEAERTKAEATI | DEAERTAAEATI |
| EIF4A2 | 55 | YAYGFEKPSAIQ | YAYGFEAPSAIQ |
| SCAMP4 | 185 | AGGSFQKAQTEW | AGGSFQAAQTEW |
| DDIT4 | 129 | LVSQVGKELLRL | LVSQVGAELLRL |
| DIABLO | 147 | NHIQLVKLQVEE | NHIQLVALQVEE |
| EIF2S2 | 52 | PEPTEDKDLEAD | PEPTEDADLEAD |
| UBE2N | 82 | YHPNVDKLGRIC | YHPNVDALGRIC |
| Ubiquitin B-1 | 48 | RLIFAGKQLEDG | RLIFAGAQLEDG |
| BTF3L1P | 127 | LAEALPKQSVDG | LAEALPAQSVDG |
| TRAF2 | 27 | KTLLGTKLEAKY | KTLLGTALEAKY |
| ADRM1 | 21 | SRGASNKYLVEF | SRGASNAYLVEF |
| NAT13 | 37 | YNDKFYKDVLEV | YNDKFYADVLEV |
| SCAMP3 | 313 | TGASFQKAQQEF | TGASFQAAQQEF |
Amino acid residues in the peptides carrying a lys (K) in the ubiquitination sites are indicated in bold letters. For each peptide, a corresponding negative control sequence is included with alanine (A) substituted in place of K as indicated in bold letter. Wt (Wild-type), ctrl (control).
Table 2.
Peptides corresponding to SUMO target sites.
| Protein | Isoform specificity | Modified K | Peptide sequence (wt) | Peptide sequence (ctrl) |
|---|---|---|---|---|
| IkBa | S1 | 21 | GPRDGLKKERLL | GPRDGLAKERLL |
| P53 | S1 | 386 | HKKLMFKTEGPD | HKKLMFATEGPD |
| Elk-1 | S1 | 249 | ALPPEVKVEGPK | ALPPEVAVEGPK |
| HDAC4 | S1 | 559 | QAGVQVKQEPIE | QAGVQVAQEPIE |
| ANXA5 | S1 | 29 | TLRKAMKGLGTD | TLRKAMAGLGTD |
| RANBP2 | S1 | 1414 | FALVTPKKEGHW | FALVTPAKEGHW |
| PARP1 | S1 | 203 | KQLPGVKSEGKR | KQLPGVASEGKR |
| HSF1 | S1 | 298 | SPLVRVKEEPPS | SPLVRVAEEPPS |
| NCOA2 | S1 | 258 | GSEVTIKQEPVS | GSEVTIAQEPVS |
| RPS21 | S2 | 41 | NVAEVDKVTGRF | NVAEVDAVTGRF |
| ACIN1 | S2 | 532 | AQPLPLKIEELA | AQPLPLAIEELA |
| HNRNPD | S2 | 197 | PDTPEEKIREYF | PDTPEEAIREYF |
| ANAPC4 | S2 | 772 | GKPVKIKEEVLS | GKPVKIAEEVLS |
| HNRNPC | S2 | 229 | KQAVEMKNDKSE | KQAVEMANDKSE |
| CHD4 | S2 | 1304 | VEREIIKQEESV | VEREIIAQEESV |
| ZMYM4 | S2 | 273 | GLLDKIKDEPDN | GLLDKIADEPDN |
| PCNA | S2 | 164 | VVISCAKDGVKF | VVISCAADGVKF |
| PML | S1+S3 | 490 | CPRKVIKMESEE | CPRKVIAMESEE |
| RPS6 | S3 | 112 | LRASTSKSESSQ | LRASTSASESSQ |
| TRIM28 | S3 | 779 | NKLTEDKADVQS | NKLTEDAADVQS |
| UTP14A | S3 | 733 | HIINPIKAEDVG | HIINPIAAEDVG |
| ZFP106 | S3 | 1265 | EPSQELKFSVEQ | EPSQELAFSVEQ |
| Wiz | S3 | 666 | SPPGTVKAEEHQ | SPPGTVAAEEHQ |
| RPS3A | S3 | 249 | GDETGAKVERAD | GDETGAAVERAD |
| RPS11 | S3 | 249 | LLGETGKEKLPR | LLGETGAEKLPR |
| ZBTB21 | S1+S2 | 430 | VTEVRIKTEPSS | VTEVRIATEPSS |
| HNRNPM | S1+S2 | 698 | KGCGVVKFESPE | KGCGVVAFESPE |
| TRIM24 | S1+S2 | 741 | FPVVIVKQESDE | FPVVIVAQESDE |
| BEND3 | S1+S2 | 20 | LKSITVKVETEA | LKSITVAVETEA |
| NFRKB | S1+S2 | 488 | KDQAFCKQENED | KDQAFCAQENED |
| FOSL2 | S1+S2 | 222 | VGAVVVKQEPLE | VGAVVVAQEPLE |
| PTRF | S1+S2 | 161 | IYQDEVKLPAKL | IYQDEVALPAKL |
| SAFB2 | S1+S2 | 252 | SVGPDRKLAEEE | SVGPDRALAEEE |
| SNIP1 | S1+S2 | 30 | PAGVVVKQERLS | PAGVVVAQERLS |
| TRP120 | S1+S2 | 432 | FNPIVIKEEDKV | FNPIVIAEEDKV |
Amino acid residues in the peptides carrying a lys (K) in the consensus motif are indicated in bold letters. For each peptide, a corresponding negative control sequence is included with alanine (A) substituted in place of K as indicated in bold letter. Wt (Wild-type), ctrl (control).
2.3. Peptide chip and microfluidic device
The peptide chip (4k) contains 3968 pico-liter reaction chambers (128 rows and 31 columns) with a single peptide synthesized directly on the surfaces of the chamber, each reaction chamber can generate close to 1 fmol of peptide. The microfluidic device (µParaflo®; fabricated at University of Michigan) has one inlet and one outlet, and solution flows laterally through the column channels. The total reaction volume of the chip (1.2×62.0 cm2) is 10 µL and each cell can accommodate 0.2 nL of reaction volume. Fluid is introduced to the microfluidic device using a micro peristaltic pump delivering a defined volume (i.e. 50 µL/min).
2.4. Chip synthesis and post synthesis treatment
Peptide microarray synthesis was performed on the mParaflo® Microchip System as reported before [17], [19]. Briefly, a digital light gated microarray synthesizer consisting of a DNA synthesizer (Expedite 8909, PE Biosystems) as the automated reagent/solvent manifold and an optical unit with the same features as described previously [20]. The light source was a 500 W Hg lamp house (model 66033, Oriel Instruments) with a 405 nm filter. The array was placed in a holder that was connected to the synthesizer and peptide microarray synthesis was performed similarly to conventional peptide synthesis using Boc chemistry, except that a PGA (photo-generated acid) was formed at selected reaction sites for N-Boc deprotection instead of TFA, allowing subsequent selective coupling of a Boc amino acid monomer at designated sites on a chip. After de-protection of peptides anchored on the chip, the chip was washed with ethanol followed by 25% acetonitrile overnight to increase the solubility of the peptide. The residual acetonitrile was removed by washing with PBS for 2 h at room temperature. The surface blocking was conducted by incubation in blocking solution (1% BSA, 0.5% Gelatin, 0.05% TWEEN 20 in PBS; pH 6.8) at 4 °C, overnight. The Boc amino acids, N-hydroxybenzotriazole (HOBt), and 2-(1H-benzotriazole-1-yl)-1,1,3,3-tetramethyluroniumhexafluorophosphate (HBTU) were from GL Biochem (Shanghai Ltd) marketed by LC Sciences (Houston, TX). Other chemicals and organic solvents were ordered from Sigma-Aldrich (St. Louis, MO). All chemicals were used without further purification unless otherwise stated.
2.5. Cell lysate for Ub/SUMO assay
Human monocytic leukemia cells (THP-1) were propagated in RPMI 1640 medium with l-glutamine and 25 mM HEPES buffer (Invitrogen) supplemented with 1 mM sodium pyruvate (Sigma, St. Louis, MO), 2.5 g/liter d-glucose (Sigma), and 10% fetal bovine serum (HyClone). E. chaffeensis (Arkansas strain) was cultivated in THP-1 cells as previously described [21]. The whole cell lysates were extracted from E. chaffeensis-infected THP-1 cells using Abcam's Whole Cell Extraction Kit (ab113475) in the presence of 20 mM N-ethylmaleimide (NEM, covalent isopeptidase inhibitor, Sigma) and 20 mM Iodoacetamide (isopeptidase inhibitor, Sigma). Extractions were performed according to the manufacturer’s protocol. The cell lysate protein concentrations were determined using a protein assay (DC assay; BioRad).
2.6. Ubiquitination and SUMOylation reaction
Ubiquitination and SUMOylation reactions were performed using a SUMOylation kit (BML-UW8955, ENZO) or ubiquitination kit (BML-UW9920, ENZO) in presence of THP-1 cell lysates according to the manufacturer's protocol. Three microfluidic chips containing the peptide array were prepared for Ub, SUMO2/3 and SUMO1 analysis. For SUMO1 and SUMO2/3, the array was incubated with the SUMOylation Buffer, E1 enzyme, Ubc9, and SUMO1 or SUMO2 and SUMO3, ATP, Mg2+ and freshly prepared cell lysate. For the Ub assay, the array was incubated with E1, Ub, ATP, ubiquitinylation buffer, inorganic pyrophophatase, Mg2+ and cell lysate. Nonspecifically bound proteins were removed by washing with buffer containing 1% SDS, 0.1% β-mercaptoethanol and 100 mM Tris for 30 min, followed by a PBS wash containing 1% Tween-20 (PBST). SUMO2/3 conjugation was detected with SUMO3 Alexa Fluor 647 antibody (Novus, 1:500). The SUMO1 signal was detected using anti-SUMO1 (1:1,000), secondary anti-mouse IgG Alexa Fluor 594 (1:30,000) was used for final detection. The ubiquitin conjugation was detected by using anti-Ub Alexa Fluor 647 antibody (Santa Cruz Biotechnology, 1:300). The detailed procedure for SUMOylation and ubiquitination reactions is as follows: chip was washed with PBS containing 1% BSA and fluid circulated at 25 μL/min for overnight at 4 °C. The chip was washed with PBST at 50 μL/min for 30 minutes at RT, followed by SUMOylation or ubiquitination reaction solution circulated at 25 μL/min for 3 h at 37 °C. The chip was washed with 100 mM Tris containing 1% SDS, 0.1% β-mercaptoethanol solution at 50 μL/min for 30 min at RT, followed by PBST at 50 μL/min for 1 h at RT. Anti-Ub Alexa Fluor 647 antibody (1:300), SUMO3 Alexa Fluor 647 antibody (1:500), or anti-SUMO1 (1:1000) in PBST was circulated at 25 μL/min for overnight at 4 °C, then washed with PBST at 50 μL/min for 1 h at RT. For SUMO1 conjugation detection, secondary anti-mouse IgG Alexa Fluor 594 (1:30,000) was circulated at 25 μL/min for 1 h at RT, and washed with PBST at 50 μL/min for 1 h at RT.
2.7. Chip imaging
The peptide chip was removed from the microfluidic device and scanned using Anon GenePix 4400A (Molecular devices) scanner using GenePix Pro 7 software. TIFF image files were further processed through Array-Pro Analyzer software and pixel density values were obtained as a text file (output data).
2.8. Statistical analysis
The pixel data was merged with the layout file using an in-house micro-array analysis program (Excel macros) and the data was then processed in multiple stages to obtain the final data with background subtracted and replicates averaged using a local regression method which was used to remove system related variations, such as sample amount variations, dye labeling bias, and signal gain difference between scanners [16]. The significant positive signals were determined by comparison with the control peptide (A peptide) and negative controls for each assay. The data are presented as mean±SD. Differences between wild-type and control peptides were assessed with the two-tailed Student's t test, and significance was indicated by a P value of<0.05.
3. Results
3.1. Ub site validation
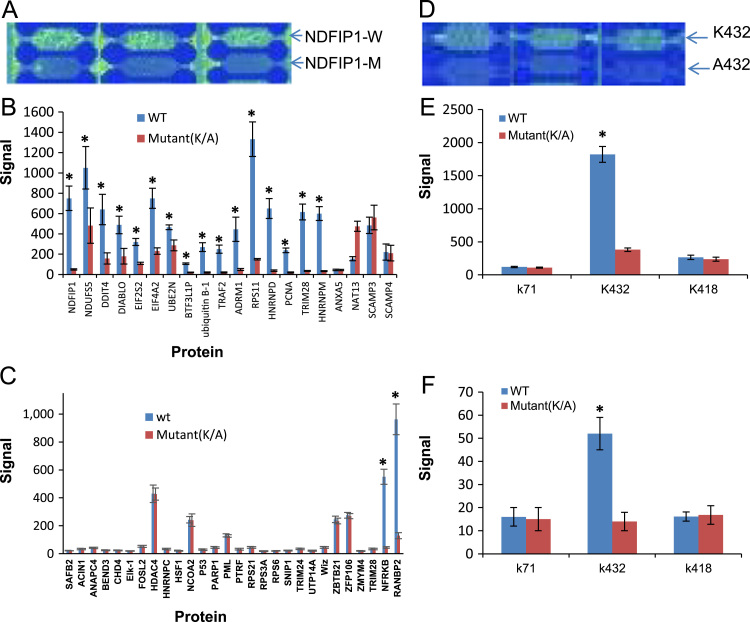
In previous studies, Qin's research group used MS with GST-qUBA reagent to isolate polyubiquitinated proteins and identify 294 endogenous ubiquitination sites on 223 proteins from human 293T cells [6]. Ubiquitination sites on both abundant and scarce regulatory proteins were identified. Although this ubiquitination site data set contains a significant number of substrates that are localized to the mitochondria, implicating ubiquitination in a wide range of mitochondrial functions, the highly conserved motif was not defined due to rapid turnover of ubiquitinated proteins and the relatively large size of the Ub PTM. In order to further validate these ubiquitination sites and evaluate the ability of our high density microfluidic chip platform to analyze these sites in vitro, 14 peptides containing ubiquitination sites corresponding to lysine (K) of these validated proteins were randomly selected and synthesized on the chip (Table 1). A microarray on a microfluidic chip containing these peptides was incubated with the E1, Ub, ATP, Mg2+ and freshly prepared cell lysate. A diagrammatic representation of the ubiquitination assay and representative image of a sample of NDFIP1 ubiquitination showing signal intensity compared to the control (alanine to lysine) peptide are shown in Fig. 3A. We analyzed fourteen peptides that were spotted on peptide microarray which exhibited fluorescent signals higher than the mutant peptides. Eleven peptides representing around 80% of the selected proteins had fluorescent signal that was significantly higher than the mutant peptides, demonstrating that the site corresponding to known ubiquitination sites was ubiquitinated in the in vitro assay (Fig. 3B). In addition, we analyzed ubiquitination signals for 34 SUMO conjugation peptides. Among these peptides, SUMOylation and ubiquitination targeted the same lysine residue in 6 peptides (RPS11, HNRNPD, PCNA, TRIM28, HNRNPM, ANXA5). The remaining 28 peptides (negative controls) did not contain predicted ubiquitination according to the mammalian Ubiquitination Site Database (mUbiSiDa) or based on experimental data [22], [23], [24], [25]. Significant signals for 5 peptides which contained known ubiquitination sites were detected (Fig. 3B). In addition, we detected significant signals for two peptides (NFRKB and RNABP2) from the negative control group (Fig. 3C), suggesting that these could be false positives. The remaining 26 peptides were confirmed as negative for PTMs in our ubiquitination assay (Fig. 3C). In addition, the 4 peptides (ANXA5, SCAMP3, SCAMP4 and NAT13) with known Ub sites were negative when compared to the mutant peptide.
Fig. 3.
Significant ubiquitination and SUMOylation signals (SUMO2/3 and SUMO1) were detected at the sites represented by peptides corresponding to lysine(K) of selected human and microbial (Ehrlichia) target proteins (A) Representative image of a small region from the peptide chip showing ubiquitination signal of wild type NDFIP1 (NDFIP1-W) compared with mutant control (NDFIP1-M). (B) Histogram plot of the ubiquitination signals to representing human proteins compared with mutant controls for 20 peptides containing ubiquitination sites. Data represent the means±S.D. of triplicate determinations (*, P<0.05). (C) Histogram plot of the ubiquitination signals to representing human proteins compared with mutant controls for 28 SUMO conjugation peptides that are not ubiquitinated. Data represent the means±S.D. of triplicate determinations (*, P<0.05). (D) Representative image of a small region from the peptide chip showing SUMO2/3 signals at K432 of TRP120 compared to the mutant control (A432). (E, F) Histogram plot of the SUMO2/3 and SUMO1 signals for K71, K432 and K418 of TRP120 compared to the corresponding mutant controls. Data represent the means±S.D. of triplicate determinations (*, P<0.05).
3.2. SUMO site validation
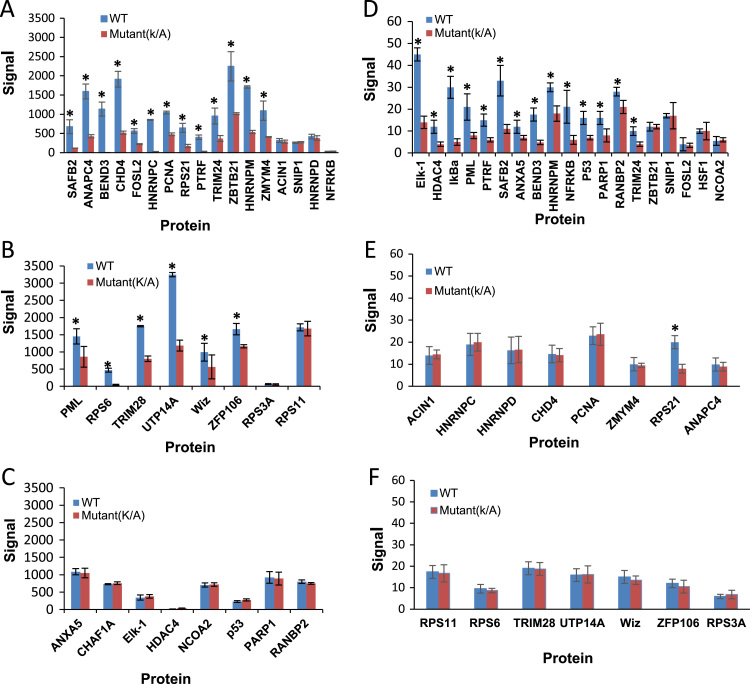
We previously reported that the E. chaffeensis T1S effector, TRP120, was modified by SUMO1 and SUMO2/3 isoforms in presence of E1, E2 ligase (Ubc9) at a carboxy-terminal canonical consensus SUMO conjugation motif (K432; KEDE) in vitro and in human cells [21]. In the present study, SUMO2/3 and SUMO1 were detected at the site corresponding to TRP120 K432 (Fig. 3D–F), which is consistent with our previous findings regarding TRP120 SUMOylation. Recently, Ribet’s group developed a powerful method for identifying SUMO sites by combining SILAC-based quantitative proteomics and immunocapture of SUMO-modified peptides, identifying 295 SUMO1 and 167 SUMO2 sites in endogenous human proteins [7], In addition, a significant number of SUMO3-modified Lys residues in target protein have been identified using novel proteomics approaches [26], [27], providing a useful database for which to validate our assay. In order to confirm the specificity for both SUMO isoform and individual target proteins at the peptide level and evaluate our high density microfluidic chip platform for in vitro SUMOylation assay, peptides (n=34) that contained scarce and abundant SUMOylation sites based on quantification of SUMOylation level were synthesized onto the chip (Table 2). Among 34 peptides, 26 contained a consensus motif (JKxE/D) and 8 contained non-canonical sites. SUMO2, SUMO3, SUMO1 and SUMO1+SUMO2/3 sites were detected on 8, 7, 9, and 10 peptides, respectively. The same analysis approach described for the ubiquitination assay was used for SUMO1 and SUMO2/3 conjugated sites mapping. As shown in Fig. 4A and B, of 17 SUMO2 conjugated sites selected, with a threshold of a P value of<0.05 indicating significance between each wild-type and mutant peptide for 13 SUMO2 conjugated peptides. Of the 8 selected SUMO3 targets, 6 had fluorescent signals that were significantly higher than the mutant peptide. 14/19 proteins with confirmed SUMO1 sites had signals that were significantly higher than the mutant peptide, indicating that these peptides were indeed modified with SUMO1 (Fig. 4D). We did not detect significant SUMO2/3 and SUMO1 signals for 8 known SUMO target proteins (HSF1, NCOA2, FOSL2: ZBTB21, SNIP, ACIN1, NFRKB, and HNRNPD) when compared to the mutant peptide. These peptides may represent false negatives for our SUMOylation assay. In addition, SUMO2/3 modifications were not detected on 8 SUMO1 specific targets (Fig. 4C). Therefore, these 8 peptides are true negatives for SUMO2/3 conjugation detection. We detected SUMO1 significant signal for one SUMO2 specific conjugation peptide (RPS21), suggesting RPS21 is either a false positive, or represents an uncharacterized SUMO1 site (Fig. 4E). The other SUMO2/3 specific conjugation peptides (n=14) were negative in SUMO1 assay (Fig. 4E and F). These results demonstrate the high isoform specificity of the in vitro SUMOylation assay corresponding to experimentally determined SUMO2/3 and SUMO1 sites.
Fig. 4.
Significant SUMOylation signals (SUMO2/3 and SUMO1) were detected at SUMOylation sites represented by peptides corresponding to lysine (K) of selected human proteins. SUMO2/3 assay: (A) SUMO2 targets (B) SUMO3 targets (C) SUMO1 targets. SUMO1 assay: (D) SUMO1 targets (E) SUMO2 targets (F) SUMO3 targets. Data represent the means±S.D. of triplicate determinations (*, P<0.05).
4. Discussion
We report herein the development of an in vitro Ub/SUMO assay with a peptide array housed within a microfluidic device, and demonstrate that this assay is useful for the identification of different Ub/SUMO target sites. Ubiquitination and SUMOylation are two important PTMs that play pivotal roles in in various host cell processes [3]. Both SUMOylation and ubiquitination are reversible, and in some cases, dynamic cycles of conjugation/deconjugation may be required for regulated activity. Because of its importance and complexity, identification of Ub/SUMO modified sites is one of the greatest challenges in gaining a full understanding of the regulatory roles of Ub and SUMO. However, it is both time-consuming and labor-intensive to use conventional experimental approaches to identify Ub/SUMO sites [5], [28], [29]. In addition, the core consensus motif in Ub substrates is not well defined. Most SUMO substrates contain the consensus motif JKxE/D, but 40% of SUMOylation sites are noncanonical [30].
There are two major considerations associated with detection of Ub and SUMO targets. First, only a small fraction of target protein is typically SUMOylated at steady state. Therefore, it is often hard to detect by direct immunoblotting. Second, SUMOylated targets are easily lost after nondenaturing lysis, due to highly active SUMO isopeptidases. In previous years, a variety of in vitro and in vivo approaches have been used to identify Ub/SUMO targets [5], [6], [7], [28], [29]. The most common strategy is to overexpress a tagged version of SUMO together with a putative target protein in cells and detect target modification by immunoblotting with antibodies specific to the tag [29]. Although this assay is limited to transfectable material, it is still a widely used assay. Mass spectrometry is also an important approach for confirmation of protein SUMOylation [5], [6], [7], but this method involves protein isolation, purification and enrichment and requires an antibody that recognizes diglycine. Moreover, it is not suitable for rapid analysis of lysine modification sites; however it does potentially enable quantification of large numbers of ubiquitination sites in a signal proteomic experiment. It is important to emphasize that only a small proportion of potential SUMO target proteins have been screened by mass spectrometry through identification of the SUMO-GG signature peptides. Therefore, it is crucial to identify potential SUMOylated substrates using more efficient and accurate in vitro assay in order to better understand in vivo identification of SUMOylation sites using overexpression system.
Compared with these conventional in vitro assay approaches, our microfluidic high density peptide array approach has several advantages in ubiquitin and SUMO target detection. First, unlike recombinant proteins, peptides are directly probed on the chip and enabling the large scale parallel synthesis of different peptides without the need for expensive, inconvenient purification step. The benefit of not using pre-synthesized peptides combined with the spotting process is highly uniform spots. The spot density obtained using this technology can be ten-fold higher than that of spotted chip [31]. In addition, labeling the peptides of interest with specific dye or radioactive material is avoided. This allows for simultaneous analysis of up to 4000 ubiquitin or SUMO targets in one experiment. The second advantage is a unique micro-fluidic device which provides picolitre scale reaction chambers (pico-array reactor). These types of peptide microarrays have the additional advantage of a controlled environment. Parameters such as time, temperature, flow rate, and flow direction can be preset and controlled. In addition, the assay is performed in a closed system (as opposed to open slides), which minimizes fluorescent antibody deterioration.
In mammals, three SUMO paralogues are commonly expressed: SUMO1 shares about 45% identity to SUMO2 and SUMO3, while SUMO2 and SUMO3 are 96% identical to each other [32]. The differences between SUMO1 and SUMO2/3 are mostly found in the second β-strand and the α-helix of both proteins [33]. In cells, different SUMO paralogues appear to share common properties but also have some distinct functions [34]. For example, SUMO-1 was more abundantly present on the mitotic spindle, and it was recruited very early to the reforming nuclear envelope, and later colocalized with chromosomes. SUMO-2 and SUMO-3 were not found on the reassembling nuclear envelope, but accrued on chromosomes earlier in the nuclear reformation process. Therefore, it is important to identify and distinguish target proteins conjugated preferentially to SUMO-1 and SUMO-2/3 in order to better elucidate their functions. In this studies, we did not detect SUMO2/3 and SUMO1 conjugates on selected SUMO1 and SUMO2/3 specific target peptides, respectively. This is consistent with previous results demonstrating the preferential conjugation of SUMO1 and SUMO2/3 by MS [7], [26], [27]. However, among those 11 selected peptides which contain overlapping SUMO1 and SUMO2/3 conjugation sites, SUMO1 and SUMO2 were not detected on one (SNIP1), and SUMO1 and SUMO2 were detected on seven others. In addition, one SUMO1 conjugates and two SUMO2 conjugates were specifically detected, respectively. It is known that proteins are conjugated in vitro to SUMO-1 and SUMO-2 by the E2 enzyme Ubc9 with similar efficiency [35]. This indicates that, in addition to the SUMO isoforms, target proteins, and the E1 and E2 enzymes that are used in SUMOylation reactions in vitro, additional factors may be present in cells that regulate the preferential usage of SUMO-1 or SUMO-2. E3 enzymes are likely candidates to fulfill this role in vivo. In addition to differences in target protein preferences for SUMO-1 and SUMO-2/3, the relative amount of conjugated SUMO compared with free SUMO is also different between these SUMO family members. It has been shown that a pools of free non-conjugated SUMO-2/3 and SUMO1 exists in different cells [36]. Thus, cell type-specific differences in conjugation efficiencies of SUMO-2/3 and SUMO1 appear to exist. Therefore, the elucidation of the cellular mechanism underlying target protein preferences for different SUMO family members is an important future objective.
The human genome encodes at least two ubiquitin E1, 53 E2 and ~500 E3 enzymes [6]. The appropriate pairing of E2 and E3 enzymes is an important determinant for specificity of target ubiquitination. Considering the numerous possible combinations of E2 and E3 enzymes, it remains challenging to identify the specific ubiquitin E2/E3 pair required for ubiquitination of a given substrate and to understand the molecular basis that control specific interaction. Therefore, in our in vitro ubiquitination and SUMOylation assays, we used cell lysates (THP-1) to provide all of necessary E2 and E3 enzymes for E. chaffeensis TRP120 and human protein ubiquitination site validation. In this study, we detected ubiquitination of 16 peptides representing around 80% of the selected proteins. However, we failed to detect ubiquitination on the other four proteins (SCAMP3, SCAMP4, ANXA5 and NAT13). Among these four proteins, SCAMP3 and SCAMP4 ubiquitination were not confirmed by SDS-PAGE gel in previous studies, and thus those findings may represent false positives. However, we cannot exclude that the difference observed is due to unbalanced expression level of E2 and E3 enzymes in different cells. It would be very interesting to further study the selected ubiquitinated proteins using the same human cell line used for ubiquitination site identification by MS. Moreover, we did not detect SUMO2 or SUMO1 on 8 known SUMO target proteins (HSF1, NCOA2, FOSL2: ZBTB21, SNIP, ACIN1, NFRKB, and HNRNPD) using our approach. In addition to the cell type-specific differences in conjugation efficiencies of SUMO-2 and SUMO1 that may exist among these targets, these findings are most likely related to the fact that six proteins contain phosphorylation dependent SUMO motifs (PDSM, ΨKxExxSP, in which K is the SUMO-conjugated lysine and S the phosphorylated serine). It is known that phosphorylation sites in close proximity to sumoylation sites directly affect the interaction between the substrate and the SUMO-conjugating machinery [37]. The PDSM motif was shown to significantly increase SUMOylation efficiency by mediating interactions between the target and a basic patch on Ubc9 [38]. These six proteins contain PDSMs and may not be phosphorylated in this in vitro sumoylation assay in the absence of phosphatase inhibitor, therefore, it is possible that our platform may not be suitable for sumoylation studies for those proteins which contain PDSMs.
Ubiquitination plays a fundamental role in the pathogenesis of many infectious diseases [4]. Recent studies have identified several bacterial effectors that interact with and modulate the ubiquitin-proteasome system (UPS) during infection by pathogenic bacteria. For example, Salmonella effectors SopE and SptP are targeted and degraded sequentially by ubiquitination to regulate bacterial invasion [4]. However, the host or bacterial E3 proteins that ubiquitinate these effectors are still largely unknown. In addition, some effectors that have E3 ligase activity are highly unstable, owing to their self-ubiquitylation activity within host cells, so they often undergo rapid proteasome-dependent degradation. Ehrlichia chaffeensis modulates numerous host cell processes, including gene transcription in order to survive within the macrophage. Using the current peptide microarray technology, we detected a significant SUMOylation signal at K432 in the C-terminus of TRP120. This finding was consistent with our previous results and reported TRP120 SUMOylated at K432 [21]. In addition, we have identified new SUMOylation sites for other Ehrlichia chaffeensis effector proteins with unknown sites by using this method. One of these novel SUMOylation sites was validated by other biological methods and was consistent with peptide array results (data not shown). Thus, this in vitro Ub/SUMO assay can be utilized not only to study eukaryotic protein modifications, but potentially to analyze modifications of pathogen associated proteins that exploit eukaryotic host cell Ub/SUMO pathways.
Our data set provides insight into the SUMO consensus motif and the functional groups of proteins being modified by SUMO under standard growth conditions. Regardless, the confirmed data set available from validation of this approach is fairly modest compared with the other PTMs. The MS-confirmed sites were validated in over 40 proteins using our approach with a single cell line. Utilizing the current approach to identify sites using multiple cell types and multiple cellular treatments will undoubtedly greatly increase global knowledge about ubiquitination and SUMOylation.
5. Conclusion
We report here in the development of an in vitro Ub/SUMO microfluidic peptide array assay that provides robust and sensitive method for the rapid screening of potential Ub/SUMO target sites. The sensitivity and specificity of the assay was validated using a panel of human proteins and a bacterial effector protein with known ubiquitin/SUMO sites. We also demonstrate a high level of concordance with traditional MS analysis for Ub/SUMO-modified sites identification. The results of the study revealed that this approach can be used for rapid screening of ubiquitination and SUMOylation sites on target proteins and confirm the specificity for both SUMO isoform and individual target proteins at the peptide level without laborious recombinant protein expression and protein purification. The current results suggest that the chip-based peptide microarray format could be useful for screening potential Ub/SUMO targets with unknown sites, and thereby contribute to our understanding the role of ubiquitin and SUMO enzymes within the Ubiquitin/SUMO network in normal physiology as well as disease pathogenesis.
Acknowledgments
This work was supported by National Institute of Allergy and Infectious Diseases (NIAID) Grants AI106859 and AI10553602 to JWM.
Footnotes
Transparency document associated with this article can be found in the online version at doi:10.1016/j.bbrep.2016.02.003.
Appendix A. Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
References
- 1.Krueger K.E., Srivastava S. Posttranslational protein modifications: current implications for cancer detection, prevention, and therapeutics. Mol. Cell. Proteom. 2006;5:1799–1810. doi: 10.1074/mcp.R600009-MCP200. [DOI] [PubMed] [Google Scholar]
- 2.Kim J.H., Choi H.J., Kim B., Kim M.H., Lee J.M., Kim I.S., Lee M.H., Choi S.J., Kim K.I., Kim S.I., Chung C.H., Baek S.H. Roles of sumoylation of a reptin chromatin-remodelling complex in cancer metastasis. Nat. Cell Biol. 2006;8:631–639. doi: 10.1038/ncb1415. [DOI] [PubMed] [Google Scholar]
- 3.Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 4.Kim M., Otsubo R., Morikawa H., Nishide A., Takagi K., Sasakawa C., Mizushima T. Bacterial effectors and their functions in the ubiquitin-proteasome system: insight from the modes of substrate recognition. Cells. 2014;3:848–864. doi: 10.3390/cells3030848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedrioli P.G., Raught B., Zhang X.D., Rogers R., Aitchison J., Matunis M., Aebersold R. Automated identification of SUMOylation sites using mass spectrometry and SUMmOn pattern recognition software. Nat. Methods. 2006;3:533–539. doi: 10.1038/nmeth891. [DOI] [PubMed] [Google Scholar]
- 6.Shi Y., Chan D.W., Jung S.Y., Malovannaya A., Wang Y., Qin J. A data set of human endogenous protein ubiquitination sites. Mol. Cell. Proteom. 2011;10 doi: 10.1074/mcp.M110.002089. M110.002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Impens F., Radoshevich L., Cossart P., Ribet D. Mapping of SUMO sites and analysis of SUMOylation changes induced by external stimuli. Proc. Natl. Acad. Sci. USA. 2014;111:12432–12437. doi: 10.1073/pnas.1413825111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson E.S. Protein modification by SUMO. Annu. Rev. Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 9.Grabbe C., Husnjak K., Dikic I. The spatial and temporal organization of ubiquitin networks. Nat. Rev. Mol. Cell Biol. 2011;12:295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denison C., Rudner A.D., Gerber S.A., Bakalarski C.E., Moazed D., Gygi S.P. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol. Cell. Proteom. 2005;4:246–254. doi: 10.1074/mcp.M400154-MCP200. [DOI] [PubMed] [Google Scholar]
- 11.Blomster H.A., Hietakangas V., Wu J., Kouvonen P., Hautaniemi S., Sistonen L. Novel proteomics strategy brings insight into the prevalence of SUMO-2 target sites. Mol. Cell. Proteom. 2009;8:1382–1390. doi: 10.1074/mcp.M800551-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu J., Zhu S., Guzzo C.M., Ellis N.A., Sung K.S., Choi C.Y., Matunis M.J. Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J. Biol. Chem. 2008;283:29405–29415. doi: 10.1074/jbc.M803632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., McCartney R.R., Schmidt M.C., Rachidi N., Lee S.J., Mah A.S., Meng L., Stark M.J., Stern D.F., De Virgilio C., Tyers M., Andrews B., Gerstein M., Schweitzer B., Predki P.F., Snyder M. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- 14.Schwamborn K., Knipscheer P., van Dijk E., van Dijk W.J., Sixma T.K., Meloen R.H., Langedijk J.P. SUMO assay with peptide arrays on solid support: insights into SUMO target sites. J. Biochem. 2008;144:39–49. doi: 10.1093/jb/mvn039. [DOI] [PubMed] [Google Scholar]
- 15.Krishnamoorthy S., Liu Z., Hong A., Zhu R., Chen H., Li T., Zhou X., Gao X., Novel Phosphopeptide A. Microarray based interactome map in breast cancer cells reveals phosphoprotein-GRB2 cell signaling networks. PLoS One. 2013;8 doi: 10.1371/journal.pone.0067634. e67634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou X., Zhu Q., Eicken C., Sheng N., Zhang X., Yan L., Gao X. MicroRNA profiling using µParaflo microfluidic array technology. Methods Mol. Biol. 2012;822:153–158. doi: 10.1007/978-1-61779-427-8_11. [DOI] [PubMed] [Google Scholar]
- 17.Gao X., Pellois J.P., Na Y., Kim Y., Gulari E., Zhou X. High density peptide microarrays. In situ synthesis and applications. Mol. Divers. 2004;8:177–187. doi: 10.1023/b:modi.0000036233.58271.25. [DOI] [PubMed] [Google Scholar]
- 18.Wall L., Christiansen T., Orwant J. 3rd ed. O’Reilly; Sebastopol: 2000. Programming Perl. [Google Scholar]
- 19.Gao X., Zhou X., Gulari E. Light directed massively parallel on-chip synthesis of peptide arrays with t-Boc chemistry. Proteomics. 2003;3:2135–2141. doi: 10.1002/pmic.200300597. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X.C., Cai S., Hong A.L., You Q.M., Yu P.L., Sheng N.J., Srivannavit O., Muranjan S., Rouillard J.M., Xia Y.M., Zhang X.L., Xiang Q., Ganesh R., Zhu Q., Matejko A., Gulari E., Gao X.L. Microfluidic PicoArray synthesis of oligodeoxynucleotides and simultaneous assembling of multiple DNA sequences, Nucleic Acids Res. 2004;32:5409–5417. doi: 10.1093/nar/gkh879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dunphy P.S., Luo T., McBride J.W. Ehrlichia chaffeensis exploits host SUMOylation pathways to mediate effector-host interactions and promote intracellular survival. Infect. Immun. 2014;82:4154–4168. doi: 10.1128/IAI.01984-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner S.A., Beli P., Weinert B.T., Nielsen M.L., Cox J., Mann M., Choudhary C. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol. Cell. Proteom. 2011;10 doi: 10.1074/mcp.M111.013284. M111.013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim W., Bennett E.J., Huttlin E.L., Guo A.L., Li J., Possemato A., Sowa M.E., Rad R., Rush J., Comb M.J., Harper J.W., Gygi S.P. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oshikawa K., matsumoto M., Oyamada K., Nakayama K.I. Proteome-wide identification of ubiquitylation sites by conjugation of engineered lysine-less ubiquitin. J. Proteom. Res. 2012;11:796–807. doi: 10.1021/pr200668y. [DOI] [PubMed] [Google Scholar]
- 25.Chen T., Zhou T., He B., Yu H.Y., Guo X.J., Song X.F., Sha J.H. mUbiSiDa: a comprehensive database for protein ubiquitination sites in mammals. PloS One. 2014;9 doi: 10.1371/journal.pone.0085744. e85744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamoliatte F., Bonneil E., Durette C., Caron-Lizotte O., Wildemann D., Zerweck J., Wenshuk H., Thibault P. Targeted identification of SUMOylation sites in human proteins using affinity enrichment and paralog-specific reporter ions. Mol. Cell. Proteom. 2013;12:2536–2550. doi: 10.1074/mcp.M112.025569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galisson F., Mahrouche L., Courcelles M., Bonneil E., Meloche S., Chelbi-Alix M.K., Thibault P. A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol. Cell. Proteom. 2011;10(2) doi: 10.1074/mcp.M110.004796. M110.004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bossis G., Chmielarska K., Gartner U., Pichler A., Stieger E., Melchior F. A fluorescence resonance energy transfer-based assay to study SUMO modification in solution. Methods Enzymol. 2005;398:20–32. doi: 10.1016/S0076-6879(05)98003-8. [DOI] [PubMed] [Google Scholar]
- 29.Hong Y., Xing X., Li S., Bi H., Yang C., Zhao F., Liu Y., Ao X., Chang A.K., Wu H. SUMOylation of DEC1 protein regulates its transcriptional activity and enhances its stability. PLoS One. 2011;6:e23046. doi: 10.1371/journal.pone.0023046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mazur M.J., van den Burg H.A. Global SUMO proteome responses guide gene regulation, mRNA biogenesis, and plant stress responses. Front. Plant Sci. 2012;3:215–225. doi: 10.3389/fpls.2012.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katz C., Levy-Beladev L., Rotem-bamberger S., Rito T., Rudiger S.G., Friedler A. Studying protein-protein interactions using peptide arrays. Chem. Soc. Rev. 2011;40:2131–2145. doi: 10.1039/c0cs00029a. [DOI] [PubMed] [Google Scholar]
- 32.Melchior F. SUMO—nonclassical ubiquitin. Annu. Rev. Cell Dev. Biol. 2000;16:591–626. doi: 10.1146/annurev.cellbio.16.1.591. [DOI] [PubMed] [Google Scholar]
- 33.Huang W.C., Ko T.P., Li S.S., Wang A.H. Crystal structures of the human SUMO-2 protein at 1.6 A and 1.2 A resolution: implication on the functional differences of SUMO proteins. Eur. J. Biochem. 2004;271:4114–4122. doi: 10.1111/j.1432-1033.2004.04349.x. [DOI] [PubMed] [Google Scholar]
- 34.Ayaydin F., Dasso M. Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol. Biol. Cell. 2004;15:5208–5218. doi: 10.1091/mbc.E04-07-0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tatham M.H., Kim S., Jaffray E., Song J., Chen Y., Hay R.T. Unique binding interactions among Ubc9, SUMO and RanBP2 reveal a mechanism for SUMO paralog selection. Nat. Struct. Mol. Biol. 2005;12:67–74. doi: 10.1038/nsmb878. [DOI] [PubMed] [Google Scholar]
- 36.Saitoh H., Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 37.Hietakangas V., Anckar J., Blomster H.A., Fujimoto M., Palvimo J.J., Nakai A., Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. USA. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohideen F., Capili A.D., Bilimoria P.M., Yamada T., Lima C.D. A molecular basis for phosphorylation-dependent SUMO conjugation by the E2 UBC9. Nat. Struct. Mol. Biol. 2009;16:945–952. doi: 10.1038/nsmb.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document
Transparency document