Abstract
The small GTPase Rap1 induces integrin activation via an inside-out signaling pathway mediated by the Rap1-interacting adaptor molecule (RIAM). Blocking this pathway may suppress tumor metastasis and other diseases that are related to hyperactive integrins. However, the molecular basis for the specific recognition of RIAM by Rap1 remains largely unknown. Herein we present the crystal structure of an active, GTP-bound GTPase domain of Rap1 in complex with the Ras association (RA)–pleckstrin homology (PH) structural module of RIAM at 1.65 Å. The structure reveals that the recognition of RIAM by Rap1 is governed by side-chain interactions. Several side chains are critical in determining specificity of this recognition, particularly the Lys31 residue in Rap1 that is oppositely charged compared with the Glu31/Asp31 residue in other Ras GTPases. Lys31 forms a salt bridge with RIAM residue Glu212, making it the key specificity determinant of the interaction. We also show that disruption of these interactions results in reduction of Rap1:RIAM association, leading to a loss of co-clustering and cell adhesion. Our findings elucidate the molecular mechanism by which RIAM mediates Rap1-induced integrin activation. The crystal structure also offers new insight into the structural basis for the specific recruitment of RA–PH module-containing effector proteins by their small GTPase partners.
Keywords: RIAM, Rap1, integrin signaling, inside-out signaling, crystal structure, RA–PH
Introduction
The small GTPase Rap1 is among the most closely related proteins to Ras in the Ras oncogene superfamily (Downward, 2003), and serves functions that are distinct from those of H-, K-, and N-Ras. Rap1 has been linked to cell proliferation, secretion, and migration (Altschuler and Ribeiro-Neto, 1998; D'Silva et al., 1998; Crittenden et al., 2004). Abnormal Rap1 activity often leads to integrin hyperactivity that is linked to tumor development and metastasis in many cancer types (Felding-Habermann et al., 2001; Hattori and Minato, 2003; Desgrosellier and Cheresh, 2010; Garmy-Susini et al., 2010). As adhesion receptors, integrins transduce signals in a bi-directional manner (Wickstrom and Fassler, 2011). In outside-in signaling, integrin binds to the extracellular matrix (ECM), forms highly organized clusters, and initiates downstream signaling cascades in the cytoplasm. Alternatively, integrin signaling can also be triggered by active Rap1 GTPase via an inside-out pathway, which requires the recruitment of a Rap1 effector protein known as Rap1-interacting adaptor molecule (RIAM) and talin to the plasma membrane (PM) (Ginsberg et al., 1992; Faull and Ginsberg, 1996; Lafuente et al., 2004; Lee et al., 2009).
RIAM functions downstream to Rap1 in inside-out integrin signaling. Upon Rap1 activation, RIAM translocates to the PM by interacting with Rap1 via a Ras association (RA) domain and by binding to phosphoinositide di-phosphate PI(4,5)P2 via a pleckstrin homology (PH) domain (Wynne et al., 2012). RIAM subsequently recruits talin via an N-terminal talin-binding (TB) sequence, leading to integrin activation (Wegener et al., 2007; Lee et al., 2009). The RA and PH domains of RIAM form an integrated RA–PH structural module that is only present in the Grb7/10/14 family and Mig10/RIAM/Lpd (MRL) family. In the latter, RIAM and Lamellipodin (Lpd) are the two mammalian orthologs (Holt and Daly, 2005). In addition to the RA–PH and TB regions, RIAM also contains two putative coiled-coil motifs (CC) and at least six proline-rich motifs (PP) that interact with cytoskeletal proteins Ena/VASP (Figure 1A) (Lafuente et al., 2004). Besides RIAM, Rap1 has another effector protein RAPL (Katagiri et al., 2003). However, RAPL binds to Ras more strongly than Rap1 in a splice variant form known as NORE1A, and exhibits tumor suppressive properties (Stieglitz et al., 2008). In contrast, RIAM binds to Rap1 specifically and shows little effect in signaling pathways of other Ras superfamily GTPases (Lafuente et al., 2004; Rodriguez-Viciana et al., 2004). Therefore, a high-resolution structure is needed to elucidate the molecular basis for this highly selective recognition of Rap1 and RIAM.
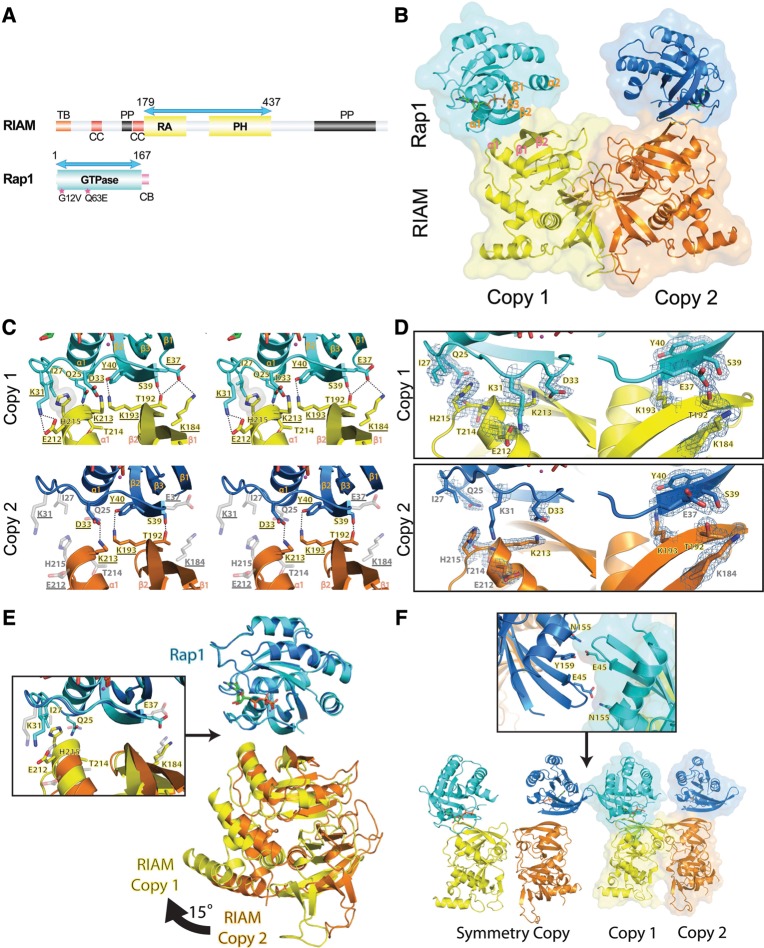
Figure 1.
Structural overview of the Rap1:RIAM complex and the intermolecular interface. (A) Schematic representation of the domain organization of RIAM and Rap1. RIAM (top): TB (talin-binding region) is in orange; CC (coiled-coil region) is in red; PP (poly-proline region) is in black; RA (Ras association) and PH (pleckstrin homology) domains are in yellow. Rap1 (bottom): GTPase domain is in cyan and the CAAX box (CB) is in pink. Double-headed arrows indicate the boundary of the constructs in the crystal structure. Constitutively active Rap1 mutations used in crystallization are denoted by asterisks. (B) Ribbon diagram and surface representation of Rap1:RIAM asymmetric units with copy 1 colored in cyan (Rap1) and yellow (RIAM), and copy 2 colored in blue (Rap1) and orange (RIAM); GTP and magnesium are illustrated in green stick and magenta sphere, respectively (same color scheme in C–F). Secondary structure elements at the Rap1:RIAM binding interface are labeled in orange and pink. (C) Stereo view of the Rap1:RIAM intermolecular interfaces in the two copies. H-bonds are denoted by dotted lines and hydrophobic interactions are represented by the gray surface. Side-chain interactions missing in copy 2 are shown in light gray color. Residues selected for mutagenesis analysis are underlined. (D) 2Fo-Fc electronic density map of the residues that mediate the Rap1:RIAM interaction at 1.0 σ contour level is shown in blue mesh. Residues lacking electronic density at 1.0 σ were built with a lower map contour. (E) Superposition of the two Rap1:RIAM complexes in the asymmetric unit. A 15° rotational shift of the RIAM molecule (right) leads to the disruption of several side-chain interactions in copy 2 (left panel). Interactions disrupted in copy 2 are indicated by gray-colored, translucent side chains. (F) Crystal contact of Rap1 molecules. Bottom panel: Rap1 of copy 1 in the original asymmetric unit (ribbon diagram with surface) makes contact with Rap1 of copy 2 from the neighboring unit (ribbon diagram). Top panel: residues that make crystal contacts are shown in stick.
We have determined the crystal structure of RIAM RA–PH in complex with the GTPase domain of Rap1 at 1.65 Å, and characterized the biochemical properties of the interaction. The structure of the Rap1:RIAM complex reveals that RIAM RA–PH interacts with Rap1 via a canonical binding mode, and the high specificity of this interaction is defined by their unique sequence characteristics. We also demonstrated that the determinant residues are essential for Rap1:RIAM association and integrin-mediated cell adhesion. This result affords detailed structural insight into how RIAM functions as a specific Rap1 effector in the inside-out integrin activation via a Rap1–RIAM–talin–integrin pathway. Our structure also provides a molecular model for the highly selective recognition of a small GTPase and an RA–PH structural module-containing protein.
Results
Crystal structure of RIAM RA–PH in complex with the Rap1 GTPase domain
We produced RIAM RA–PH of varying lengths as well as the constitutively active Rap1 GTPase domain (residues 1–167) bearing G12V or G12V/Q63E mutations for expression in Escherichia coli. GTP-loaded, active Rap1 was then mixed with RIAM RA–PH proteins at equal molar ratio for co-crystallization. Only the mixture of the RA–PH construct encoding residues 179–437 and Rap1-(G12V/Q63E) yielded crystals suitable for structure determination (Figure 1A). The 1.65-Å crystal structure, determined by molecular replacement, contains two Rap1:RIAM complexes in an asymmetric unit (Table 1).
Table 1.
Data collection and refinement statistics for Rap1:RIAM.
| Rap1:RIAM | |
|---|---|
| Crystal | |
| Space group | P212121 |
| a, b, c (Å) | 82.5, 85.8, 160.9 |
| α, β, γ (o) | 90.0, 90.0, 90.0 |
| Data collection | |
| Resolution (Å) | 50.0–1.65 |
| Completeness (%) | 99.4 (93.8) |
| Rsym (%) | 6.3 (44.8) |
| I/σ(I) | 29.2 (2.1) |
| Unique reflections | 137303 |
| Redundancy | 5.6 (4.0) |
| Refinement | |
| Resolution (Å) | 50.0–1.65 (1.69–1.65) |
| Rwork (%) | 19.7 (27.5) |
| Rfree (%) | 21.9 (28.5) |
| RMSD bonds (Å) | 0.005 |
| RMSD angle (o) | 1.070 |
| Protein atoms | 6923 |
| Solvent atoms | 734 |
| Total residues | 862 |
| Residue omit | |
| Chain A | 62–64 |
| Chain B | 279–292 |
| Chain C | 62–67, 167 |
| Chain D | 280–292 |
| Average B-factors (Å2) | |
| Main chain atoms | 13.2 |
| Side chain atoms | 13.8 |
| Solvent | 29.1 |
| Chain A (Rap1) | 14.6 |
| Chain B (RIAM) | 11.1 |
| Chain C (Rap1) | 18.9 |
| Chain D (RIAM) | 11.9 |
| Chain AB | 12.5 |
| Chain CD | 14.5 |
| Ramachandran | |
| Favored regions (%) | 98.0 |
| Allowed regions (%) | 100.0 |
Rsym = Σ|Iobs − Iavg|/ΣIavg; Rwork = Σ||Fobs − Fcalc||/ΣFobs; Rfree was calculated using 5% of data and the same sums.
The crystal structure reveals that Rap1 and RIAM bind canonically. The β2 of the RA domain in RIAM forms an antiparallel β-sheet interaction with the Rap1 β2 in the Switch I region (residues 25–40) (Figure 1B). In both copies, Rap1 interacts with the RA domain of RIAM via the Switch I region exclusively. In copy 1, side chains in the Switch I region of Rap1 make extensive interactions with side chains from the β1, β1–β2 loop, β2, and α1 of the RIAM RA domain. Gln25, Lys31, Asp33, Glu37, Ser39, and Tyr40 of Rap1 form H-bonds with Thr214, Glu212, Lys213, Lys184, Thr192, and Lys193 of RIAM, respectively. Furthermore, Ile27Rap1 makes van der Waals contact with His215RIAM (Figure 1C). In addition, a water-mediated H-bond network located between residues 25–41 of Rap1 and residues 189–213 of RIAM also strengthens this interface (Supplementary Figure S1). The well-defined electron density for these interacting residues is indicative of a stable binding interface (Figure 1D). Contrary to the ‘fully engaged’ copy 1, only three H-bonds remain in copy 2 (Figure 1C). A double mutation G12V/Q63E was introduced to Rap1 to prevent GTP hydrolysis. G12V is in the α1–β1 loop and makes little contact with the Switch regions. The Switch I region is essentially identical in all Ras:effector complexes. Q63E is in the disordered Switch II region, which is often highly dynamic in other Ras:effector complex structures as well. Thus, the G12V/Q63E double mutation does not appear to affect the Rap1 conformation in the Switch regions.
Compared with the structure of RIAM RA–PH alone (PDB ID 3TCA), the RA–PH molecule of RIAM in the complex remains virtually unchanged with an overall root-mean-square-deviation (rmsd) of 0.17 Å (246 Cα atoms). The two Rap1 molecules in the asymmetric unit are also mostly identical with an rmsd of 0.20 Å. Superposition of the Rap1 molecules from the two copies reveals an ∼15° rotation between the two RIAM RA–PH molecules (Figure 1E). This movement displaces the α1 helix of the RA domain away from the α1–β2 loop of Rap1 in copy 2, disrupting the Glu212Rap1:Lys31RIAM salt bridge and the interactions of Ile27Rap1:His215RIAM and Gln25Rap1:Thr214RIAM (Figure 1E). The loss of the Glu37Rap1:Lys184RIAM interaction in copy 2 is mainly due to a rotamer change in Glu37Rap1. We then analyzed crystal packing and B-factors of the two copies. The Rap1 molecule in copy 1 makes crystal contacts with the Rap1 molecule in copy 2 from a neighboring asymmetrical unit, primarily through weak H-bonding interactions formed by Glu45 and Asn155/Tyr159 (Figure 1F). Therefore, it is possible that the conformation of the Rap1:RIAM complex in copy 2 is due to crystal packing. Electron density and B-factor analyses also suggest that copy 1 (B-factor = 12.5 Å2) is more stable than copy 2 (B-factor = 14.5 Å2) (Figure 1D and Table 1). Thus, copy 1 reveals a stable configuration of the Rap1:RIAM complex.
Structural comparison with other Ras/Rap1:RA/RDB complexes
RIAM has been shown to participate in signaling pathways mediated by Rap1 but not other Ras family GTPases (Lafuente et al., 2004; Lee et al., 2009; Wynne et al., 2012). While Ras family GTPases share high sequence identity in the Switch I region, the interacting region sequence in Ras effector proteins is not well conserved despite the similar backbone conformation (Figure 2A). To elucidate the structural determinants for the highly specific Rap1:RIAM interaction, we compared the Rap1:RIAM complex with other Ras/Rap1:RA/RBD complexes with known structures, including Rap1:cRaf1, H-Ras:RalGDS, Rap1:KRIT1, and H-Ras:RAPL (Figure 2B, PDB ID 1GUA, 1LFD, 4DXA, and 3DDC, respectively) (Nassar et al., 1996; Huang et al., 1998; Stieglitz et al., 2008; Li et al., 2012). The Ras-binding domains in RalGDS, cRaf1, and RAPL exhibit high structural similarity to the RIAM RA domain, whereas KRIT1 interacts with the Rap1 GTPase domain with both F1 and F2 lobes. The GTPase domain of Rap1 binds to the RIAM RA domain with a moderate affinity (Kd = 0.7 μM) (Takala and Ylanne, 2012), similar to Rap1:RAPL (1.3 μM) (Stieglitz et al., 2008), Rap1:KRIT1 (1.8 μM) (Liu et al., 2011; Li et al., 2012), and Ras:RalGDS (1.3 μM) (Linnemann et al., 2002), but significantly lower than H-Ras:RAPL (0.08 μM) (Stieglitz et al., 2008), Ras:Raf1 (0.02 μM) (Herrmann et al., 1995), and Rap1:RalGDS (0.007 μM) (Linnemann et al., 2002). The buried surface in the Rap1:RIAM complex is 555 Å2, close to that of Rap1:cRaf-1 (673 Å2) and Ras:RalGDS (647 Å2). In contrast, H-Ras:RAPL (823 Å2) and Rap1:KRIT1 (921 Å2) complexes contain larger buried surfaces due to additional interactions outside of the Switch I region (Figure 2B).
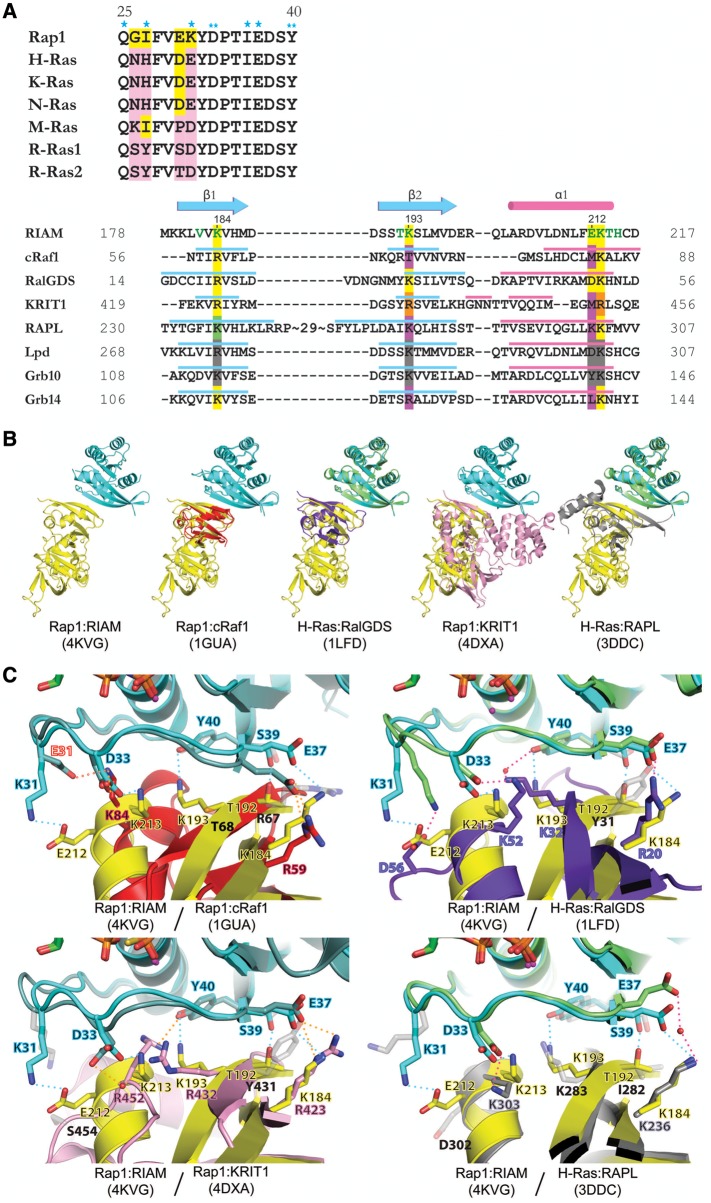
Figure 2.
Sequence and structural comparison of Rap1:RIAM and other Ras:effector complexes. (A) Structure-based sequence alignment of Switch I region of Ras family (upper panel) and Ras-binding domains (lower panel). Conserved residues are colored in yellow and non-conserved residues are colored in pink. Single asterisks indicate RIAM-interacting residues only seen in copy 1; double-asterisks denote RIAM-interacting residues seen in both copies. Secondary structure elements of the RA/RBD domain are shown in cyan (β-strand) and pink (α-helix). Green letters in the RIAM sequence indicate Rap1-interacting residues. Conserved Ras-interacting residues are highlighted in yellow. The RAPL residue highlighted in green indicates a water-mediated interaction with Ras. Non-conserved Ras-interacting residues are highlighted in purple. Arg432 and Arg452 of KRIT1 (highlighted in orange) interact with Rap1 Asp33 and Tyr40 reversely with respect to their sequence alignment. Conserved residues in Lpd and Grb10 are highlighted in gray, as no complex structure is available for comparison. (B) Superposition of Rap1:RIAM with Rap1:cRaf1 (PDB ID 1GUA, cRaf1 in red), H-Ras:RalGDS (PDB ID 1LFD, H-Ras in green, RalGDS in purple), Rap1:KRIT1 (PDB ID 4DXA, KRIT1 in pink), and H-Ras:RAPL (PDB ID 3DDC, H-Ras in green, RAPL in gray). (C) Superposition of Ras/Rap1:RA/RBD complexes for Rap1 residues Lys31, Asp33, Glu37, Ser39, and Tyr40, and corresponding interacting residues. The color scheme of the Rap1/Ras and RA/RBD ribbon models are the same as in B. The interaction formed by the mutated residue in Rap1:cRaf1 (K31E and K84, respectively) is labeled with red text and red dotted line. A water molecule is shown as a red sphere and H-bonds are denoted by dotted lines of cyan (Rap1:RIAM), orange (Rap1:cRaf1/KRIT1), or pink (H-Ras:RalGDS/RAPL). Contacting residues are labeled in colored text accordingly to the color scheme of the molecules and non-contacting residues are shown in light gray stick figures with black text.
Among the five complex structures, the Asp33Rap1:Lys213RIAM and Glu37Rap1:Lys184RIAM interactions are conserved (Asp33Rap1:Arg432KRIT1 and Glu37H-Ras:Lys236RAPL are mediated by water molecules), whereas the Tyr40Rap1:Lys193RIAM interaction is absent in Rap1:cRaf1 and H-Ras:RAPL complexes, suggesting that this interaction is specific for Rap1:RIAM, Rap1:KRIT1, and H-Ras:RalGDS (Figure 2C). A Lys31:Glu212 salt bridge further strengthens the Rap1:RIAM complex (Figure 1C). Interestingly, this salt bridge is not present in any other Rap1:RA/RBD complex structures. Within the Switch I region, four residues (Gly26, Ile27, Glu30, and Lys31) in Rap1 are different from other Ras GTPases (Figure 2A). Among these four residues, Lys31 is the most striking one, as the residues at this position in other Ras GTPases are all negatively charged (Glu or Asp). To date, no side-chain interaction mediated by the Lys31 of Rap1 is observed in any Rap1:RA/RBD complex. We now show that Rap1 Lys31 forms a salt bridge with a unique Glu212 residue in the RIAM RA domain. Although a conserved aspartate residue is present in Lpd and RalGDS at the corresponding position (Asp302Lpd and Asp51RalGDS) (Figure 2A), it appears that the aspartate residues may not be able to interact with the Lys31 of Rap1. Lpd has been reported to be unable to interact with Rap1 specifically (Krause et al., 2004), and Asp51RalGDS does not interact with the Lys31 residue in a Rap1-like Ras-(E31K) mutant (Huang et al., 1998). Thus, our structure, for the first time, reveals that Lys31 serves as a key specificity determinant of Rap1 in recruiting its effector protein by forming a unique salt bridge. Interestingly, in RalGDS, an Asp56RalGDS residue in an extended loop following α1 also forms a salt bridge with the Lys31 side chain of the Ras-(E31K) mutant (Figure 2C), indicating that Lys31 of Rap1 may also contribute to the tight association of RalGDS and Rap1.
Mutations of binding determinants in the RIAM:Rap1 complex diminish the interaction
To assess the contribution of the H-bonds observed in the Rap1:RIAM complex structure, we made point mutations K31A, K31E, D33A, E37A, and Y40A in the constitutively active Rap1-(G12V), and evaluated the binding of RIAM to Rap1 using in-cell co-immunoprecipitation (Co-IP) assays. All Rap1 mutants significantly diminished their association with full-length RIAM (Figure 3A). Conversely, we made K184A, K193A, E212A, and K213A mutations in RIAM for the Co-IP assays. These mutants also weakened the Rap1:RIAM interaction (Figure 3B). Thus, these results support that the electrostatic interactions between residues in the Switch I region of Rap1 and the β1–β2–α1 region of RIAM are essential for the association of Rap1 and RIAM. To further validate the specificity determinant role of the Lys31Rap1:Glu212RIAM salt bridge in Rap1:RIAM binding, we made complementary mutants of Rap1-(K31E) and RIAM-(E212K) and assessed their interaction using Co-IP. While single mutations of Rap1 (K31A or K31E) or RIAM (E212A or E212K) diminish the association, the complementarily mutated pair Rap1-(K31E) and RIAM-(E212K) successfully restores the binding (Figure 3C). These results were also confirmed by in vitro pull-down assays using purified RIAM RA–PH and Rap1 GTPase domain bearing these mutations (Supplementary Figure S2).
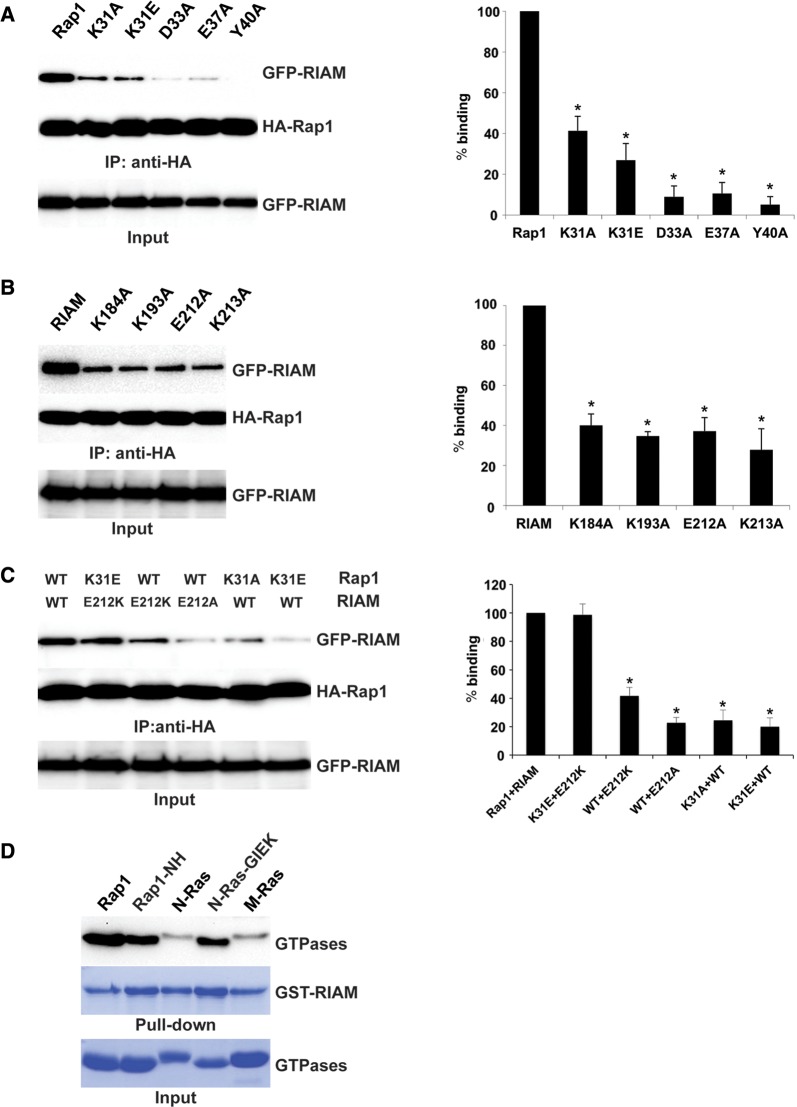
Figure 3.
Validation of the determinant residues for RIAM–Rap1 interaction. (A) Association of various Rap1 (WT, K31A, K31E, D33A, E37A, and Y40A) and WT RIAM measured by Co-IP. (B) Association of various RIAM forms (WT, K184A, K193A, E212A, and K213A) and constitutively active HA-Rap1-(G12V) measured by Co-IP. (C) Co-IP assays for complementarily mutated pair of RIAM-(E212K) and Rap1-(K31E). (D) In vitro pull-down assay of RIAM binding to Rap1, N-Ras, and M-Ras. All Rap1 and Ras proteins are constitutively active forms (Rap1-(G12V), N-Ras-(G12V), and M-Ras-(Q71L)). Rap1-NH and N-Ras-GIEK denote Rap1-(G12V) bearing mutations of G26N/I27H and N-Ras-(G12V) bearing mutations of N26G/H27I/D30E/E31K, respectively. *P < 0.01, n = 3.
To investigate the role of the remaining non-conserved residues in the Rap1 Switch I region (Gly26, Ile27, and Glu30) in Rap1:RIAM binding, we made mutations to swap these residues in N-Ras and Rap1, and assessed the binding of their GTPase domains with RIAM RA–PH. Rap1-NH (G26N/I27H) does not alter the binding significantly, whereas the Rap1-like N-Ras mutation N26G/H27I/D30E/E31K (GIEK) exhibits substantial gain of affinity towards RIAM RA–PH (Figure 3D). These results support that Lys31 is the key determinant residue that defines the binding specificity of Rap1 and RIAM, and the Lys31Rap1:Glu212RIAM salt bridge, along with other side-chain interactions, stabilizes the Rap1:RIAM complex.
Rap1:RIAM complex is required for the co-clustering of RIAM and Rap1 at the PM and integrin-mediated cell adhesion
It has been shown that RIAM functions as a scaffold that connects the membrane targeting sequence CAAX box of Rap1 to talin, promotes recruitment of talin to the PM, and activates integrin (Lee et al., 2009). Previously, a CHO-A5 cell line expressing αIIbβ3 integrin has been established to reconstitute Rap1-induced co-clustering of Rap1, RIAM, talin, and αIIbβ3 integrin (Han et al., 2006; Lee et al., 2009). To establish the biological relevance of the Rap1:RIAM interaction in subcellular localization, we co-transfected CHO-A5 cells with GFP-RIAM and HA-Rap1, and monitored co-clustering of RIAM and Rap1 by fluorescence microscopy. We first validated that wild-type (WT) RIAM and Rap1-(G12V) co-cluster at the PM with integrin αIIbβ3 in these cells (Figure 4A). In contrast, co-transfection of Rap1-(G12V) with RIAM mutants K193A, K213A, E212K, E212A, and K183A significantly inhibits co-clustering of RIAM and Rap1 with integrin at the PM (Figure 4B). Similarly, mutations of Rap1 in the corresponding residues (D33A, Y40A, E37A, K31A, and K31E) also diminish the co-clustering (Figure 4C). We then tested whether the complementarily mutated pair Rap1-(K31E) and RIAM-(E212K) can restore the co-clustering in CHO-A5 cells. When Rap1-(K31E) or RIAM-(E212K) is co-expressed with WT RIAM or Rap1, respectively, the co-clustering is inhibited, and the loss of co-clustering can be rescued by co-expressing the complementarily mutated pair (Figure 4D). Thus, mutations in the interface of the RIAM:Rap1 complex disrupt its co-clustering in CHO-A5 cells, and co-expression of RIAM-(E212K) and Rap1-(K31E) restores the co-clustering.
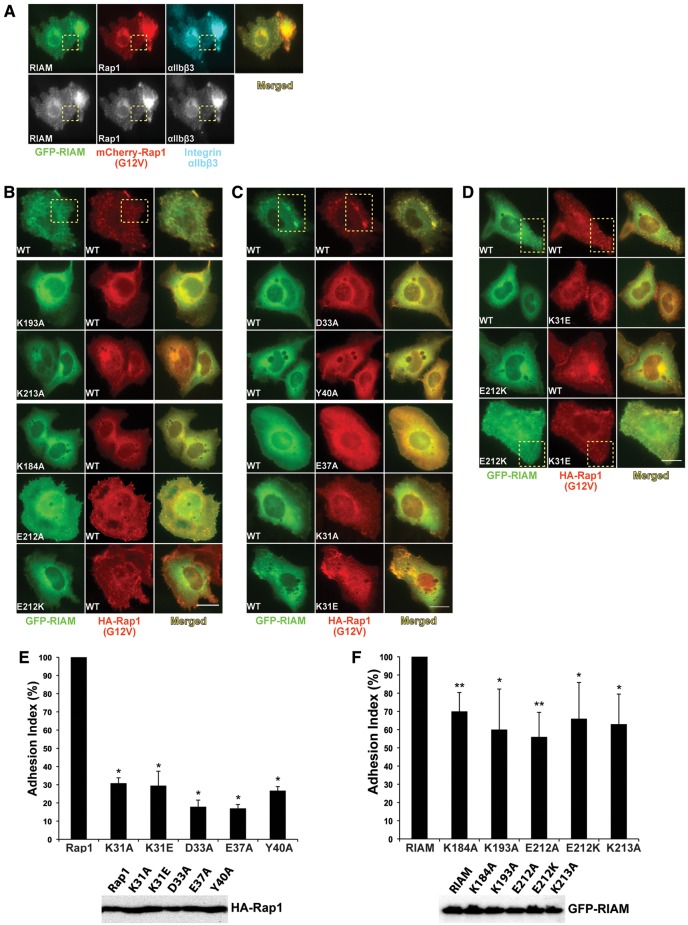
Figure 4.
RIAM:Rap1 complex is required for the co-clustering of RIAM and Rap1 at the PM and integrin-mediated cell adhesion. (A) Rap1 and RIAM co-cluster with integrin αIIbβ3. A5 cells co-expressing GFP-RIAM and mCherry-Rap1-(G12V) were stained with integrin αIIbβ3 antibody. Images in gray scale are shown in the lower row. (B) Fluorescent images of A5 cells co-expressing Rap1-(G12V) and RIAM mutants K184A, K193A, E212A, E212K, and K213A. (C) Images of A5 cells co-expressing GFP-RIAM and Rap1 mutants K31A, K31E, D33A, E37A, and Y40A. (D) Co-clustering of the complementarily mutated pair of GST-RIAM-(E212K) and HA-Rap1-(K31E). Representative co-clusters are indicated in the yellow dotted boxes. Scale bar, 10 µm. (E) The adhesion index of HEK293T cells transfected with the WT HA-Rap1-(G12V) and interface mutants K31A, K31E, D33A, E37A, and Y40A. The percentage of adhesion of each mutant was normalized to that of Rap1-(G12V) and presented in a bar graph (upper panel; *P < 0.001 compared with Rap1-(G12V), n = 3). HA-Rap1 expression was verified by western blotting (lower panel). (F) The adhesion index of HEK293T cells transfected with the WT RIAM and interface mutants K183A, K193A, E212A, E212K, and K213A (upper panel; *P < 0.05, **P < 0.01 compared with RIAM, n = 3). RIAM expression was verified by western blotting (lower panel).
Overexpression of Rap1 or RIAM induces integrin-mediated cell adhesion (Lafuente and Boussiotis, 2006). To assess the effect of Rap1 and RIAM interface mutations in cell adhesion, we transfected active Rap1 or interface mutations into HEK293T cells grown on fibronectin-coated microplates and quantified cell adhesion. Cells transfected with Rap1-(G12V) bearing Rap1:RIAM interface mutations show an ∼70%–80% decrease in cell adhesion (Figure 4E). We then transfected cells with WT RIAM or its mutations and measured the cell adhesion. Cells transfected with RIAM mutants also exhibit an ∼35%–50% decrease in cell adhesion compared with WT RIAM (Figure 4F). Co-transfection of Rap1 and RIAM results in highly elevated level of cell adhesion, which makes the effect of complementarily mutated pair in restoration of cell adhesion insignificant. Nevertheless, the reduction in cell adhesion caused by the single mutations of Rap1 or RIAM suggests that the integrity of the Rap1:RIAM complex is required for inducing integrin-mediated cell adhesion.
Discussion
In addition to RIAM, Lpd is the other mammalian ortholog in the MRL protein family. Although both Lpd and RIAM are ligands for Ena/VASP proteins and are capable of binding talin (Krause et al., 2004; Lafuente et al., 2004; Lee et al., 2009), the two RA–PH-containing adaptor proteins serve distinct functions in Ras signaling. It has been shown that RIAM, but not Lpd, interacts with Rap1 specifically (Krause et al., 2004). Our data also confirmed that Rap1 interacts with the RA–PH domains of RIAM much more strongly than that of Lpd, despite their high sequence identity in the Rap1/Ras-binding site (Supplementary Figure S3A). It is possible that the replacement of the RIAM Glu212 by a shorter Asp302 in Lpd destabilizes the Glu212RIAM:Lys31Rap1 salt bridge, which is supported by our data that a RIAM-(E212D) mutant exhibits >50% reduction in Rap1 binding (Supplementary Figure S3B). Previous structural studies have also shown that the Lys31 residue in a Ras-(E31K) mutant does not interact with the Asp302Lpd equivalent Asp51RalGDS or K302DRAPL (Nassar et al., 1996; Stieglitz et al., 2008). Furthermore, the PM-abundant PI(4,5)P2 plays important roles in RIAM-mediated integrin activation. The poor PI(4,5)P2-binding affinity of Lpd may also restrict its PM localization and thereby hinder its association with Rap1 (Chang et al., 2013). In contrast, RIAM and talin both preferentially interact with PI(4,5)P2 via the PH domain and the F2–F3 domains, respectively, leading to the release of talin autoinhibition and the initiation of integrin clustering (Goksoy et al., 2008; Saltel et al., 2009; Wynne et al., 2012). In addition, other proteins such as kindlin, migfilin, and ILK have also been suggested to enhance integrin activation (Honda et al., 2009; Moser et al., 2009; Das et al., 2011). Particularly, PtdInsP kinase Iγ has been shown to locally synthesize PI(4,5)P2 and is capable of modulating talin function and cell adhesion (Legate et al., 2011; Calderwood et al., 2013). Some of these protein components may also promote the association and the PM localization of Rap1:RIAM complex by unknown mechanisms. Thus, it appears that Lpd cannot replace RIAM in mediating inside-out integrin signaling.
Instead, Lpd has recently been shown to interact with M-Ras and participate in M-Ras-mediated dendrite development (Tasaka et al., 2012). Our data confirm that Lpd binds to M-Ras much more strongly than to Rap1 and N-Ras, whereas RIAM preferentially binds to Rap1 (Supplementary Figure S3C). In M-Ras (PDB ID 3KKO), Lys36 (Gly26Rap1 equivalent) forms a salt bridge with Asp165 in the C-terminal helix. It is possible that this interaction stabilizes the hydrophobic packing of Ile37M-Ras (Ile27Rap1 equivalent), Val39M-Ras (Val29Rap1 equivalent), and His305Lpd (Supplementary Figure S3D). Since the histidine residue is only conserved in RIAM (His215RIAM), M-Ras may also interact with RIAM but not with Grb14, which is supported by our in vitro binding result (Supplementary Figure S3E).
During our analysis of the Rap1:RIAM complex, a structure of the H-Ras:Grb14 complex was reported (Qamra and Hubbard, 2013). Grb14 belongs to the RA–PH-containing Grb7/10/14 adaptor protein family and inhibits insulin signaling upon Ras recruitment (Depetris et al., 2009). Both Rap1:RIAM and H-Ras:Grb14 complexes adopt the canonical Ras:effector binding mode. The overall B-factor of the Rap1:RIAM complex is much lower than that of the H-Ras:Grb14 complex (14 Å2 vs. 67 Å2). In both complexes, salt bridges are present at Asp33:Lys (RIAM Lys213 and Grb14 Lys140) and Asp37:Lys (RIAM Lys184 and Grb14 Lys111). The interactions of Lys31Rap1:Glu212RIAM, Ser39Rap1:Thr192RIAM, and Tyr40Rap1:Lys193RIAM are absent in H-Ras:Grb14 (Supplementary Figure S4). Instead, H-Ras makes additional contact with Grb14 through a Switch II region residue Tyr64.
Oligomerization is a common mechanism for regulating the activity and signaling of receptor molecules and related signaling proteins. Since the RA–PH structural module binds membrane phosphoinositides with moderate to low affinities compared with a conventional PH domain (Depetris et al., 2009; Wynne et al., 2012; Chang et al., 2013), dimerization of the RA–PH-containing proteins may serve to increase their avidity to the PM, thereby facilitating the association with membrane-anchored Ras GTPases. For example, Grb10 dimerizes via an SH2 domain and a helical extension of the PH domain (Stein et al., 2003; Depetris et al., 2009). Lpd also dimerizes via the coiled-coil region preceding the RA domain (Chang et al., 2013). Hence, it is possible that RIAM may also function as a dimer in integrin signaling. Intriguingly, RIAM is responsible for the PM translocation of talin, which functions as a dimer through a C-terminal helix as well (Gingras et al., 2008). While the conserved coiled-coil motif that mediates the dimerization of Lpd (PDB ID 4GMV) (Chang et al., 2013) is disordered in the crystal structure of RIAM (PDB ID 3TCA) (Wynne et al., 2012), we discovered a common interface mediated by residues His389 and Tyr398 of the two PH domains in the crystals of Rap1:RIAM complex and RIAM alone (PDB ID 3TCA) (Supplementary Figure S5). The two distinct crystals consist of different protein components (CC–RA–PH in 3TCA; RA–PH and Rap1 in the Rap1:RIAM crystal), indicating that this common interface may be biologically relevant. Thus, this finding may facilitate the efforts in further investigating the regulatory mechanism of the RIAM-mediated integrin activation.
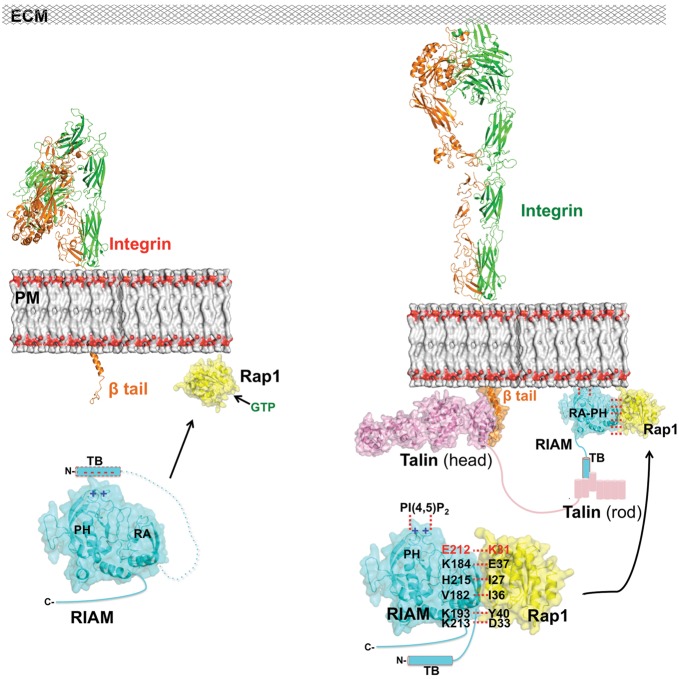
As a subgroup of the Ras superfamily, the Ras family GTPases include the Rat sarcoma (Ras) oncoproteins, Rap, R-Ras, Ral, and Rheb proteins (Wennerberg et al., 2005), which share high sequence identity and structural similarity in the GTPase domain. It is therefore important for Ras GTPases to recognize specific effector proteins for their distinct signaling functions. Previous in vitro data have suggested that RIAM interacts with GTP-bound Rap1, but not with GDP-bound Rap1, and retains weak, rather nonspecific binding capability with other Ras GTPases (Figure 3D) (Lafuente et al., 2004; Wynne et al., 2012). Nonetheless, this interaction is largely suppressed in cell signaling, likely by its effector proteins that interact with Ras with much better affinity (in low nanomolar range), such as Raf-1. In Rap1-induced integrin activation, Lys31 of Rap1 serves as the key specificity determinant that recognizes RIAM, and the binding of RIAM PH domain to PI(4,5)P2 facilitates the PM translocation of Rap1:RIAM complex. Afterwards, RIAM recruits talin via the TB region. Intriguingly, this region may also function in RIAM autoinhibition by blocking Rap1 binding (Wynne et al., 2012). Since the TB region is highly negatively charged, we hypothesize that TB may mask the positively charged PI(4,5)P2-binding surface in the PH domain and block PI(4,5)P2 binding. While it is unclear what triggers the release of the TB region, the PM translocation of the Rap1:RIAM complex or the presence of talin may initialize this event. Once talin is recruited to the PM by the TB region, the interaction of the talin head region and the cytoplasmic tail of integrin β subunit in turn induces integrin activation (Wegener et al., 2007; Elliott et al., 2010), by which integrin extends its extracellular domains and associates with the ECM (Zhu et al., 2008) (Figure 5).
Figure 5.
Working model for inside-out integrin activation mediated by Rap1:RIAM complex. Upper left: integrin (α subunit in green and β subunit in orange) is in a resting state prior to Rap1 (yellow) activation via GTP (green) exchange. Lower left: in a hypothetical model of the RIAM autoinhibition, the negatively charged TB region of RIAM (cyan cylinder with dashed outline) masks the positively charged PI(4,5)P2-binding site in the PH domain, blocking the PM translocation and talin association. Right: upon the binding of GTP, Rap1 switches to the active state and recruits RIAM via the backbone β-sheet interaction and a series of side-chain interactions (in black text) including the Lys31:Glu212 interaction unique to Rap1:RIAM (in red text); the PM localization of the Rap1:RIAM complex is facilitated by the association of PH and PM PI(4,5)P2, which also releases the TB region (cyan cylinder with solid outline) for talin (in pink) recruitment. Talin in turn activates integrin by a head-to-tail interaction. Activated integrin extends its extracellular domains to interact with ECM.
In summary, our results reveal new insight into the highly specific interaction between Rap1 and its effector protein RIAM through the RA–PH module. The high-resolution crystal structure of Rap1:RIAM complex will also facilitate the development of potential inhibitors targeting inside-out integrin activation by intervening in the specific binding of Rap1 and RIAM.
Materials and methods
Plasmid DNA constructs, mutagenesis, and antibodies
cDNAs encoding full-length mouse RIAM were cloned into pEGFP-C1 vector and the resulting plasmid was used as a PCR template to make RIAM RA–PH constructs. Full-length human Rap1A was cloned into mammalian expression vector pCGN with an N-terminal HA tag. Bacterial expression plasmids were constructed by subcloning gene fragments into a modified pET28a vector containing a TEV cleavage site for making 6× His-tagged protein and MCS, or a pGEX-5X-1 vector (GE Healthcare) for making GST-tagged proteins. Mutations were generated by using the QuikChange II XL site-directed mutagenesis kit (Stratagene). All constructs were confirmed by DNA sequencing. Anti-HA and anti-His mouse monoclonal antibodies were purchased from Sigma-Aldrich. Anti-GFP rabbit polyclonal antibody was purchased from Clontech Laboratories.
Protein expression and purification
The recombinant proteins were expressed in E. coli BL21 (DE3)-T1R strain (Sigma-Aldrich) in LB medium by induction with 0.4 mM IPTG overnight at 20°C. Bacterial cells were harvested, resuspended in lysis buffer (20 mM Tris, pH 7.5, 500 mM NaCl) and lysed by a high-pressure homogenizer (AVESTIN, Inc.). His-tagged proteins (Rap1, N-Ras, M-Ras, RIAM, Lpd, and Grb14) were purified using HisTrap Ni-affinity chromatography columns (GE Healthcare). The His tag was removed by recombinant TEV protease. Rap1, N-Ras, and M-Ras were further purified on an anion-exchange column (Resource Q, GE Healthcare). RIAM, Lpd, and Grb14 were purified on a cation-exchange column (Resource S, GE Healthcare). GST-tagged proteins (Rap1, M-Ras, RIAM, Lpd) were purified using GST-affinity chromatography columns (GSTrap, GE Healthcare) followed by a gel filtration chromatography column (Superdex-75, GE Healthcare). All purified proteins were quantified by measuring absorbance at 280 nm and confirmed by SDS–PAGE.
X-ray crystallography
To load GTP, purified Rap1 was first incubated with 10 mM GTP and 2 mM EDTA for 30 min on ice. MgCl2 was then added to the mixture to a final concentration of 5 mM. Both RIAM RA–PH and Rap1-GTP were concentrated to 5 mg/ml and mixed stoichiometrically on ice. The protein complex was mixed with an equal volume of reservoir solution to screen for crystallization conditions. Crystals of RIAM RA–PH in complex with Rap1 were grown by hanging drop vapor diffusion method at 4°C with a reservoir solution containing 0.1 M ammonium sulfate, 0.1 M Tris, pH 7.5, and 15% (w/v) PEG 1500. Cryoprotectant screening was performed on a Rigaku X-ray source. The crystals were flash frozen in liquid nitrogen with reservoir solution plus 20% (v/v) ethylene glycol. Final diffraction data were collected at beamline X25 of the National Synchrotron Light Source at Brookhaven National Laboratory (Upton, NY, USA), and processed using HKL2000 (Otwinowski and Minor, 1997). The complex structure of RIAM RA–PH and Rap1 GTPase domain was determined by molecular replacement using the RIAM RA–PH (PDB ID 3TCA) and Rap1 (PDB ID 1C1Y) structures as the search models. We performed model building with Coot (Emsley and Cowtan, 2004) and refined the structure using REFMAC (Vagin et al., 2004). The final atomic model contains residues 1–167 of chain A (Rap1), residues 178–437 of chain B (RIAM), residues 1–166 of chain C (Rap1), and 178–437 of chain D (RIAM), excluding residues 62–64 (chain A), residues 278–292 (chain B), residues 62–67 (chain C), and residues 280–292 (chain D). Atomic coordinates and structure factor have been deposited in the Protein Data Bank under the accession number 4KVG.
GST pull-down, immunoprecipitation, and western blotting
For in vitro pull-down assays, purified GST-RIAM RA–PH proteins immobilized on glutathione agarose were incubated with purified His-tagged Rap1 loaded with GTP in reaction buffer (20 mM Tris–HCl, pH 7.5, 100 mM NaCl, 2 mM DTT, 10 mM MgCl2) for 30 min at 4°C. After washing, bound proteins were eluted with elution buffer (100 mM Tris–HCl, pH 8.0, 20 mM reduced glutathione). The eluted samples were separated by SDS–PAGE and detected by western blotting with an anti-His antibody or Coomassie blue staining. For immunoprecipitation, HEK293T cells were co-transfected with HA-Rap1 and GFP-RIAM and cell lysates were prepared with lysis buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1% (v/v) Triton X-100, 1 mM DTT, and complete protease inhibitor cocktail (Roche)) at 24 h post-transfection. The cell lysates were clarified by centrifugation and immunoprecipitated with 5 μg of anti-HA antibody overnight at 4°C. Protein A/G PLUS-Agarose (Santa Cruz Biotechnology) was added to the reaction mixture and incubated for 1 h at 4°C with gentle rotating. Bound proteins were eluted with 1× sample buffer and resolved by SDS–PAGE. The fractions were transferred to Immobilon-P transfer membrane (EMD Millipore). The membranes were blocked with TBST buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl, 0.1% (v/v) Tween 20) containing 5% (w/v) BSA for 1 h and then incubated with anti-HA and anti-GFP antibodies for 1 h at room temperature. After incubation with HRP-conjugated secondary antibody, the blots were visualized by SuperSignal® West Pico Chemiluminescent Substrate (Thermo Scientific) and detected by a FluorChem E imager (ProteinSimple). Western blots were quantified by densitometric analysis using Image J (http://rsb.info.nih.gov/ij/). An unpaired t-test was performed to calculate the P-value using GraphPad Prism 5 (GraphPad software, Inc.).
Cell culture and transfection
HEK293T cells were maintained at 37°C with 5% CO2 in Dulbecco's Modified Eagle Medium (DMEM; Gibco) supplemented with 10% fetal bovine serum (FBS). CHO cells stably expressing integrin αIIbβ3 (A5) were grown in Ham's F-12 medium plus 10% FBS in the presence of G418 and Zeocin. For transient transfection, Lipofectamine™ 2000 (Invitrogen) was used according to manufacturer's recommendations.
Immunofluorescence staining and microscopy
αIIbβ3-expressing CHO cells (A5) were grown on glass cover slips and co-transfected with HA-Rap1-(G12V) and designated GFP-RIAM mutants, or with GFP-RIAM and HA-Rap1-(G12V) mutants. At 24 h post-transfection, the cells were fixed with 4% (w/v) paraformaldehyde for 30 min and permeabilized with 0.2% (v/v) Triton X-100 for 20 min at room temperature. The fixed cells were blocked with 1% BSA/10% normal goat serum (Invitrogen) and incubated with anti-HA antibody (Sigma-Aldrich) for 1 h followed by incubation with Alexa Fluor 555-conjugated goat anti-mouse antibody for 1 h at room temperature. The cover slips were washed with PBS buffer and mounted with ProLong Gold antifade reagent (Molecular Probes). To observe the co-clustering of RIAM and Rap1 with integrins, the A5 cells were co-transfected with GFP-RIAM and mCherry-Rap1, and immunostained with integrin αIIb/β3 antibody (P256, Santa Cruz Biotechnology) and Alexa Fluor 647-conjugated secondary antibody. The stained cells were imaged with a Nikon Eclipse TE2000-U microscope equipped with a Cascade 650 monochrome and MetaVue (Universal Imaging/Molecular Devices) software for acquisition using a 60× oil immersion objective (Nikon, Plan Apo, NA 1.40). The images were processed for presentation by Image J.
Cell adhesion assay
One day prior to cell adhesion assay, 96-well Maxisorp microplates (Nunc) were coated with human fibronectin (1 μg/ml, Sigma-Aldrich) in PBS buffer overnight at 4°C. The plates were then washed with PBS buffer and blocked with 1% (w/v) heat-denatured BSA in PBS buffer for 1 h at room temperature. HEK293T cells were transiently transfected with Rap1 or RIAM mutants, detached by trypsin at 48 h post-transfection, and resuspended in serum-free DMEM containing 25 mM HEPES, pH 7.5. The cell suspensions were incubated at 37°C, 5% CO2 for 10 min. Cells were plated onto 96-well microplate in triplicate and allowed to adhere to plate for 5 min at 37°C. Non-adherent cells were removed by gently washing with PBS buffer. The attached cells were fixed with 5% (w/v) glutaraldehyde (Molecular Probes) for 20 min at room temperature. Input cells were set up to determine 100% attachment by addition of cells in uncoated wells and fixing without washing. After washing with water, fixed cells were stained with 0.1% (w/v) crystal violet (Sigma-Aldrich) in 200 mM MES, pH 6.0 for 60 min at room temperature. The dyes absorbed by cells were solubilized in 10% (v/v) acetic acid for 5 min at room temperature. The absorbance was measured at 570 nm using an EnVision Multilabel plate reader (Perkin Elmer). The data were expressed in percentage as absorbance of cells bound to that of input cells. The cell adhesion percentages of Rap1 and RIAM mutants were normalized to that of Rap1-(G12V) and WT RIAM, respectively. An unpaired t-test was used to calculate the P-value.
Supplementary material
Supplementary material is available at Journal of Molecular Cell Biology online.
Funding
This work was supported by Fox Chase Cancer Center Startup Fund (to J.W.) and the Elizabeth Knight Patterson Fellowship (to Y.-C.C.).
Conflict of interest: none declared.
Supplementary Material
Acknowledgements
We thank S.R. Hubbard (NYU School of Medicine), J.R. Peterson and A.J. Andrews (Fox Chase Cancer Center) for manuscript comments, J.S. Bennett (University of Pennsylvania) for CHO-A5 cells, X. Zhang (UT Southwestern Medical Center) for the M-Ras plasmid, and the beamline staff of X4C and X25 at the National Synchrotron Light Source, Brookhaven National Laboratory for technical support.
References
- Altschuler D.L., Ribeiro-Neto F. Mitogenic and oncogenic properties of the small G protein Rap1b. Proc. Natl Acad. Sci. USA. 1998;95:7475–7479. doi: 10.1073/pnas.95.13.7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood D.A., Campbell I.D., Critchley D.R. Talins and kindlins: partners in integrin-mediated adhesion. Nat. Rev. Mol. Cell Biol. 2013;14:503–517. doi: 10.1038/nrm3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.C., Zhang H., Brennan M.L., et al. Crystal structure of Lamellipodin implicates diverse functions in actin polymerization and Ras signaling. Protein Cell. 2013;4:211–219. doi: 10.1007/s13238-013-2082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crittenden J.R., Bergmeier W., Zhang Y., et al. CalDAG-GEFI integrates signaling for platelet aggregation and thrombus formation. Nat. Med. 2004;10:982–986. doi: 10.1038/nm1098. [DOI] [PubMed] [Google Scholar]
- Das M., Ithychanda S.S., Qin J., et al. Migfilin and filamin as regulators of integrin activation in endothelial cells and neutrophils. PLoS One. 2011;6:e26355. doi: 10.1371/journal.pone.0026355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depetris R.S., Wu J., Hubbard S.R. Structural and functional studies of the Ras-associating and pleckstrin-homology domains of Grb10 and Grb14. Nat. Struct. Mol. Biol. 2009;16:833–839. doi: 10.1038/nsmb.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgrosellier J.S., Cheresh D.A. Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- D'Silva N.J., Jacobson K.L., Ott S.M., et al. Beta-adrenergic-induced cytosolic redistribution of Rap1 in rat parotid acini: role in secretion. Am. J. Physiol. 1998;274:C1667–C1673. doi: 10.1152/ajpcell.1998.274.6.C1667. [DOI] [PubMed] [Google Scholar]
- Elliott P.R., Goult B.T., Kopp P.M., et al. The structure of the talin head reveals a novel extended conformation of the FERM domain. Structure. 2010;18:1289–1299. doi: 10.1016/j.str.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Faull R.J., Ginsberg M.H. Inside-out signaling through integrins. J. Am. Soc. Nephrol. 1996;7:1091–1097. doi: 10.1681/ASN.V781091. [DOI] [PubMed] [Google Scholar]
- Felding-Habermann B., O'Toole T.E., Smith J.W., et al. Integrin activation controls metastasis in human breast cancer. Proc. Natl Acad. Sci. USA. 2001;98:1853–1858. doi: 10.1073/pnas.98.4.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garmy-Susini B., Avraamides C.J., Schmid M.C., et al. Integrin alpha4beta1 signaling is required for lymphangiogenesis and tumor metastasis. Cancer Res. 2010;70:3042–3051. doi: 10.1158/0008-5472.CAN-09-3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.R., Bate N., Goult B.T., et al. The structure of the C-terminal actin-binding domain of talin. EMBO J. 2008;27:458–469. doi: 10.1038/sj.emboj.7601965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg M.H., Du X., Plow E.F. Inside-out integrin signalling. Curr. Opin. Cell Biol. 1992;4:766–771. doi: 10.1016/0955-0674(92)90099-x. [DOI] [PubMed] [Google Scholar]
- Goksoy E., Ma Y.Q., Wang X., et al. Structural basis for the autoinhibition of talin in regulating integrin activation. Mol. Cell. 2008;31:124–133. doi: 10.1016/j.molcel.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J., Lim C.J., Watanabe N., et al. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr. Biol. 2006;16:1796–1806. doi: 10.1016/j.cub.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Hattori M., Minato N. Rap1 GTPase: functions, regulation, and malignancy. J. Biochem. 2003;134:479–484. doi: 10.1093/jb/mvg180. [DOI] [PubMed] [Google Scholar]
- Herrmann C., Martin G.A., Wittinghofer A. Quantitative analysis of the complex between p21ras and the Ras-binding domain of the human Raf-1 protein kinase. J. Biol. Chem. 1995;270:2901–2905. doi: 10.1074/jbc.270.7.2901. [DOI] [PubMed] [Google Scholar]
- Holt L.J., Daly R.J. Adapter protein connections: the MRL and Grb7 protein families. Growth Factors. 2005;23:193–201. doi: 10.1080/08977190500196267. [DOI] [PubMed] [Google Scholar]
- Honda S., Shirotani-Ikejima H., Tadokoro S., et al. Integrin-linked kinase associated with integrin activation. Blood. 2009;113:5304–5313. doi: 10.1182/blood-2008-07-169136. [DOI] [PubMed] [Google Scholar]
- Huang L., Hofer F., Martin G.S., et al. Structural basis for the interaction of Ras with RalGDS. Nat. Struct. Biol. 1998;5:422–426. doi: 10.1038/nsb0698-422. [DOI] [PubMed] [Google Scholar]
- Katagiri K., Maeda A., Shimonaka M., et al. RAPL, a Rap1-binding molecule that mediates Rap1-induced adhesion through spatial regulation of LFA-1. Nat. Immunol. 2003;4:741–748. doi: 10.1038/ni950. [DOI] [PubMed] [Google Scholar]
- Krause M., Leslie J.D., Stewart M., et al. Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev. Cell. 2004;7:571–583. doi: 10.1016/j.devcel.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Lafuente E., Boussiotis V.A. Rap1 regulation of RIAM and cell adhesion. Methods Enzymol. 2006;407:345–358. doi: 10.1016/S0076-6879(05)07029-1. [DOI] [PubMed] [Google Scholar]
- Lafuente E.M., van Puijenbroek A.A., Krause M., et al. RIAM, an Ena/VASP and Profilin ligand, interacts with Rap1-GTP and mediates Rap1-induced adhesion. Dev. Cell. 2004;7:585–595. doi: 10.1016/j.devcel.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Lampugnani M.G., Orsenigo F., Rudini N., et al. CCM1 regulates vascular-lumen organization by inducing endothelial polarity. J. Cell Sci. 2010;123:1073–1080. doi: 10.1242/jcs.059329. [DOI] [PubMed] [Google Scholar]
- Lee H.S., Lim C.J., Puzon-McLaughlin W., et al. RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J. Biol. Chem. 2009;284:5119–5127. doi: 10.1074/jbc.M807117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legate K.R., Takahashi S., Bonakdar N., et al. Integrin adhesion and force coupling are independently regulated by localized PtdIns(4,5)2 synthesis. EMBO J. 2011;30:4539–4553. doi: 10.1038/emboj.2011.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Zhang R., Draheim K.M., et al. Structural basis for small G protein effector interaction of Ras-related protein 1 (Rap1) and adaptor protein Krev interaction trapped 1 (KRIT1) J. Biol. Chem. 2012;287:22317–22327. doi: 10.1074/jbc.M112.361295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnemann T., Kiel C., Herter P., et al. The activation of RalGDS can be achieved independently of its Ras binding domain. Implications of an activation mechanism in Ras effector specificity and signal distribution. J. Biol. Chem. 2002;277:7831–7837. doi: 10.1074/jbc.M110800200. [DOI] [PubMed] [Google Scholar]
- Liu J.J., Stockton R.A., Gingras A.R., et al. A mechanism of Rap1-induced stabilization of endothelial cell–cell junctions. Mol. Biol. Cell. 2011;22:2509–2519. doi: 10.1091/mbc.E11-02-0157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser M., Legate K.R., Zent R., et al. The tail of integrins, talin, and kindlins. Science. 2009;324:895–899. doi: 10.1126/science.1163865. [DOI] [PubMed] [Google Scholar]
- Nassar N., Horn G., Herrmann C., et al. Ras/Rap effector specificity determined by charge reversal. Nat. Struct. Biol. 1996;3:723–729. doi: 10.1038/nsb0896-723. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Qamra R., Hubbard S.R. Structural basis for the interaction of the adaptor protein grb14 with activated ras. PLoS One. 2013;8:e72473. doi: 10.1371/journal.pone.0072473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P., Sabatier C., McCormick F. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol. Cell. Biol. 2004;24:4943–4954. doi: 10.1128/MCB.24.11.4943-4954.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saltel F., Mortier E., Hytonen V.P., et al. New PI(4,5)P2- and membrane proximal integrin-binding motifs in the talin head control beta3-integrin clustering. J. Cell Biol. 2009;187:715–731. doi: 10.1083/jcb.200908134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein E.G., Ghirlando R., Hubbard S.R. Structural basis for dimerization of the Grb10 Src homology 2 domain. Implications for ligand specificity. J. Biol. Chem. 2003;278:13257–13264. doi: 10.1074/jbc.M212026200. [DOI] [PubMed] [Google Scholar]
- Stieglitz B., Bee C., Schwarz D., et al. Novel type of Ras effector interaction established between tumour suppressor NORE1A and Ras switch II. EMBO J. 2008;27:1995–2005. doi: 10.1038/emboj.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala H., Ylanne J. Binding properties and stability of the Ras-association domain of Rap1-GTP interacting adapter molecule (RIAM) PLoS One. 2012;7:e31955. doi: 10.1371/journal.pone.0031955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaka G., Negishi M., Oinuma I. Semaphorin 4D/Plexin-B1-mediated M-Ras GAP activity regulates actin-based dendrite remodeling through Lamellipodin. J. Neurosci. 2012;32:8293–8305. doi: 10.1523/JNEUROSCI.0799-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A.A., Steiner R.A., Lebedev A.A., et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- Wegener K.L., Partridge A.W., Han J., et al. Structural basis of integrin activation by talin. Cell. 2007;128:171–182. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- Wennerberg K., Rossman K.L., Der C.J. The Ras superfamily at a glance. J. Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- Wickstrom S.A., Fassler R. Regulation of membrane traffic by integrin signaling. Trends Cell Biol. 2011;21:266–273. doi: 10.1016/j.tcb.2011.02.003. [DOI] [PubMed] [Google Scholar]
- Wynne J.P., Wu J., Su W., et al. Rap1-interacting adapter molecule (RIAM) associates with the plasma membrane via a proximity detector. J. Cell Biol. 2012;199:317–330. doi: 10.1083/jcb.201201157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Luo B.H., Xiao T., et al. Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell. 2008;32:849–861. doi: 10.1016/j.molcel.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.