Abstract
Background
There are conflicting reports on the impact of soy on breast carcinogenesis. This study examines the effects of soy supplementation on breast cancer-related genes and pathways.
Methods
Women (n = 140) with early-stage breast cancer were randomly assigned to soy protein supplementation (n = 70) or placebo (n = 70) for 7 to 30 days, from diagnosis until surgery. Adherence was determined by plasma isoflavones: genistein and daidzein. Gene expression changes were evaluated by NanoString in pre- and posttreatment tumor tissue. Genome-wide expression analysis was performed on posttreatment tissue. Proliferation (Ki67) and apoptosis (Cas3) were assessed by immunohistochemistry.
Results
Plasma isoflavones rose in the soy group (two-sided Wilcoxon rank-sum test, P < .001) and did not change in the placebo group. In paired analysis of pre- and posttreatment samples, 21 genes (out of 202) showed altered expression (two-sided Student’s t-test, P < .05). Several genes including FANCC and UGT2A1 revealed different magnitude and direction of expression changes between the two groups (two-sided Student’s t-test, P < .05). A high-genistein signature consisting of 126 differentially expressed genes was identified from microarray analysis of tumors. This signature was characterized by overexpression (>2-fold) of cell cycle transcripts, including those that promote cell proliferation, such as FGFR2, E2F5, BUB1, CCNB2, MYBL2, CDK1, and CDC20 (P < .01). Soy intake did not result in statistically significant changes in Ki67 or Cas3.
Conclusions
Gene expression associated with soy intake and high plasma genistein defines a signature characterized by overexpression of FGFR2 and genes that drive cell cycle and proliferation pathways. These findings raise the concerns that in a subset of women soy could adversely affect gene expression in breast cancer.
Many women consume soy in the belief that it prevents breast cancer, or treats the disease. This practice is based primarily on results of epidemiological studies, yet the impact of soy on breast cancer (BC) is not clearly understood. Soy can exert either pro- or antiestrogenic effects and may have other effects on cellular events. It is not clear if soy is protective or harmful in some circumstances (1,2). The effect of soy intake on critical signaling molecules, cellular markers, and gene products associated with BC remains unknown. In prospective observational studies in Asian populations, soy intake was associated with reduced risk of BC incidence and recurrence (3–5). When stratified by amount of soy consumed, a dose-response relationship has been reported with a statistically significant trend of decreasing risk with increasing soy food intake, translating to a 16% risk reduction per 10mg of daily isoflavone consumed (4). Yet soy intake was unrelated to BC risk in multiple prospective studies in western populations, and patients with BC are frequently advised to avoid soy foods (4,6). The tumorigenic properties of soy isoflavones are well documented in BC cell lines, and animal models and are largely associated with their estrogenic properties (1,2,7).
Soybeans contain the isoflavones genistein and daidzein. Genistein stimulates growth of estrogen-sensitive BC cells through transactivation of the estrogen receptor (ER), and can block the inhibitory effects of tamoxifen (7–9). However, isoflavones have also been reported to decrease BC cell growth through ER-independent inhibition of tyrosine kinases and DNA topoisomerases (10–15). Additionally, genistein exerts anti-inflammatory and anti-angiogenic effects through the regulation of VEGF and VEGFR-2 expression (16–18).
Human intervention studies have not led to conclusive results regarding soy effects on biomarkers of mammary tumorigenesis. Gene expression profiling using microarray technologies has provided critical insights into the molecular classification of BC, improved our understanding of BC biology, and generated clinically useful information about prognosis and response to therapy (19). Given the presumed importance of soy in modulating BC risk, we aimed to identify its effects on the expression of genes and pathways in BC.
Methods
Study Design
The objective of this randomized, placebo-controlled study was to investigate the effects of soy supplementation on the molecular features of BC, including gene expression profiles and markers of BC risk (Clinicaltrials.gov identifier: NCT00597532; http://clinicaltrials.gov/show/NCT00597532). The primary endpoint of the study was comparison of changes in proliferation (Ki67) and apoptosis (Cas3) between the two groups. Secondary outcomes were changes in gene expression by NanoString and expression by microarrays and qPCR. Women with invasive BC scheduled for resection were randomly assigned to receive supplements of soy protein (intervention) or milk protein (placebo). Supplementation lasted from the initial surgical consultation to the day before surgery (minimum 7 days, maximum 30 days). Tissue from the diagnostic core biopsy was analyzed for gene expression by NanoString and for markers of cell proliferation and apoptosis by immunohistochemistry (IHC). Results were compared with those in the posttreatment tissue obtained at the time of surgery. Expression analysis by oligonucleotide microarray and qPCR were performed using total RNA isolated from frozen tissue. Microarray, qPCR, and NanoString gene expression studies were performed whenever tissue was available for research purposes. For inclusion and exclusion criteria, see the Supplementary Methods (available online). The study was approved by MSKCC’s Institutional Review Board, and all patients signed informed consent. All patients completed a modified dietary intake questionnaire including soy foods (20,21).
Soy/Placebo
Soy and placebo were dispensed by the hospital pharmacist in identically appearing packets containing 25.8g soy protein powder or 25.8g milk protein. All patients were counseled to consume two packets/day, mixed with water or juice from the day of consent through the day before surgery. Research staff and participants were blinded to group assignments. Full contents of soy and placebo are listed in the Supplementary Methods (available online).
Plasma Isoflavones
Blood samples were obtained at time of consent and day of surgery to measure plasma genistein and daidzein by high performance liquid chromatography (HPLC) (22,23).
NanoString and qPCR
NanoString and qPCR analyses are described in the Supplementary Methods (available online).
Immunohistochemistry and Pathology
Routine pathologic assessment of the initial diagnostic and subsequent surgical specimen was performed on all specimens. Tissues were fixed in 10% buffered formalin for 6 to 48 hours, routinely processed and embedded in paraffin. Formalin-fixed, paraffin-embedded (FFPE) tissue sections were used for the study when available, following completion of routine clinical histopathologic examination and sign out.
Immunohistochemical detection was performed using streptavidin-biotin-peroxidase and microwave antigen retrieval methodology (24). Human epidermal growth factor receptor 2 (HER2) positivity was defined as 3+ by IHC, or 2+ by IHC with gene amplification of 2.0 or greater. Amplification was measured by fluorescence in situ hybridization (FISH) (25). ER status was determined by IHC, and samples were considered positive if greater than 1% of cell nuclei were immunoreactive.
IHC for Ki67 and Cas3 was performed on representative FFPE tissue sections identified by the study pathologist in the Research Immunohistochemistry Core Laboratory of MSKCC on a Discovery XT instrument (Ventana). The Cas-3 (Asp175) antibody was from Cell Signaling (catalog #9661, rabbit polyclonal), and the dilution was 1:400 for 60 minutes. The Ki-67 antibody was clone MIB-1 (Dako Cytomation, Catalog# M7240, mouse monoclonal) and dilution was 1:400 for 60 minutes. Cell lines and tissue samples known to express the antigen under study were used as positive controls.
IHC scoring was performed using deidentified samples, without any information on clinical characteristics or study group assignment. Cells with positive Cas3 and Ki67 staining were counted in 10 high-power (40x objective) fields selected to represent the spectrum of staining seen on review of the whole section (26). IHC score was expressed as the percentage of positively staining tumor cells among the total number of tumor cells counted. At least 1000 malignant cells were evaluated for each specimen, and only nuclear staining was considered positive.
Statistical Analysis
The clinicopathological and demographic characteristics were compared between soy and placebo groups using the Fisher’s exact test for categorical variables and the Wilcoxon rank-sum test for continuous variables. Plasma isoflavones, and Ki-67 and Cas3 indices (% of positive cells) were assessed within groups (patient matched post/pre) using the Wilcoxon matched-pairs, signed-rank test, and between groups (soy-treated difference vs placebo-treated difference) using the Wilcoxon rank-sum test. The effect of treatment on NanoString gene expression was evaluated within groups using the paired t test. To compare NanoString gene expression between groups, the fold change (post/preratio) for each sample was compared using the Student’s t-test. For qPCR data, the average normalized qPCR value for a gene was used to compare gene expression between groups, and statistical significance was determined by unpaired t test with Welch’s correction (assume unequal variance). Correlation between genistein and daidzen and association between paired values were computed using the Pearson method.
All statistical tests were two sided, and P values of less than .05 for NanosString and less than .01 for microarrays were considered statistically significant. Analyses and data visualization were performed using GraphPad Prism for Mac OSX v. 6.0b (GraphPad Software, www.graphpad.com), Partek Genomics Suite 6.6, R version 3.0.1 (http://www.r-project.org), and Bioconductor version 2.13 (27).
Microarray and Bioinformatics Analysis
Affymetrix Human U133 Plus 2.0 microarray gene expression was analyzed using tools in Partek Genomic Suite 6.6 software (Partek Incorporated, www.partek.com). The Robust Multiarray Analysis (RMA) algorithm was used for global normalization and probeset summarization. Differentially expressed (DE) genes were determined using Student’s t test (unpaired equal variance) at a statistical significance level 0.01 and absolute fold-change of 2 or greater. Hierarchical clustering was performed by Euclidian distance and average linkage method in Partek Genomics Suite 6.64. Ingenuity Pathways Analysis (www.ingenuity.com) was performed with default settings. DAVID Functional Gene Classification Tool with default settings was used for pathway analysis with FDR = 0.01.
Gene Set Analysis (GSA) was performed using the Bioconductor package piano (www.bioconductor.org) in R statistical language (www.r-project.org) (28). The main function runGSA was applied with default parameters using fold changes as gene-level statistics and gene-set collections from the Broad Molecular signatures database (MSigDB: www.broadinstitute.org/gsea/msigdb/collections.jsp). Only gene-sets with adjusted P values less than .01 were reported. An Additional filtering step was applied that limited gene sets to those in which at least half of the genes in the gene set showed fold changes of at least 50%.
To predict molecular subtype of samples measured by microarrays, we obtained an independent set of 204 BCs with known molecular class assignments (data set GSE12276) (29). PAM50 genes were mapped to 139 probe sets according to gene symbol using the online NetAfxx portal (www.affymetrix.com). Molecular class was predicted for each BC sample of the training and test sets using a nearest centroid model based on the expression of the PAM50 genes and using the Partek Genomic Suite 6.6 software (30,31). A leave-one-out cross validation was used to estimate prediction accuracy in the training set.
Results
Patients
A total of 140 women with invasive breast adenocarcinoma (stage T1, T2, or T3) were randomized to participate in this study from 2003 to 2007 (32). Eight women dropped out (three elected to have surgery elsewhere, two withdrew, and three refused surgery). When available, blood and tumor tissues were analyzed form 132 remaining women (Figure 1). Median durations of soy or placebo supplementation were 14 and 15 days, respectively (P = .70). There were no side effects or complications related to the intervention or placebo. Measurements consisted of: plasma isoflavones (n = 125), tumor IHC (n = 104), NanoString (n = 14), gene expression profiling (n = 51), and qPCR (n = 46) (Figure 1).
Figure 1.
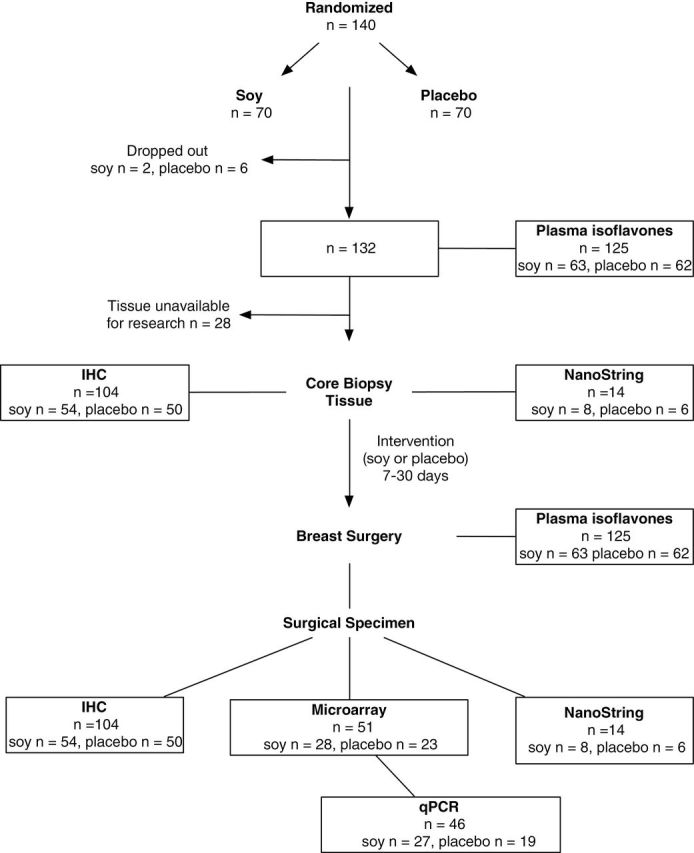
CONSORT diagram for study design and availability of samples for analysis.
Demographics and clinicopathological characteristics including age, race, menopausal status, TNM stage, tumor estrogen receptor (ER) status, HER2 status (by IHC and FISH) showed no differences between the two groups (Table 1). There were no statistically significant differences in baseline weight, BMI, dietary components, or alcohol consumption (Supplementary Table 1, available online).
Table 1.
Baseline patient and tumor characteristics
| Characteristic | Soy (n = 54) | Placebo (n = 50) | Total (n = 104) | P* |
|---|---|---|---|---|
| Age, y | .91 | |||
| Mean (SD) | 56.3 (11.3) | 56.1 (12.6) | 56.2 (11.9) | |
| Weight, kg | ||||
| Median (range) | 66 (43–123) | 68 (46–121) | 68 (43–123) | .43 |
| BMI, kg/m2 | ||||
| Median (range) | 25.5 (18.0–45.0) | 25.7 (17.7–44.4) | 25.7 (17.7–44.9) | .41 |
| Race, No. (%) | ||||
| White, non-Hispanic | 46 (85.2) | 42 (84) | 88 (84.6) | .92 |
| Black, non-Hispanic | 4 (7.4) | 5 (10) | 9 (8.7) | |
| White, Hispanic | 4 (7.4) | 3 (6) | 7 (6.7) | |
| Menopausal status, No. (%) | ||||
| Postmenopausal | 35 (64.8) | 28 (56) | 63 (60.6) | .44 |
| Premenopausal | 19 (35.2) | 22 (44) | 41 (39.4) | |
| Estrogen receptor status, No. (%) | ||||
| Positive | 43 (80) | 42 (84) | 85 (82) | .62 |
| Negative | 11 (20) | 8 (16) | 19 (18) | |
| HER2 status, No. (%) | ||||
| Positive | 7 (13) | 4 (8) | 11 (10.6) | .53 |
| Negative | 47 (87) | 46 (92) | 93 (89.4) | |
| Tumor stage, No. (%) | ||||
| T1 | 37 (68.5) | 36 (72) | 73 (70.2) | .94 |
| T2 | 15 (27.8) | 12 (24) | 27 (27) | |
| T3 | 2 (4.7) | 2 (4) | 4 (3.8) | |
| Nodal status, No. (%) | ||||
| 0 | 29 (53.7) | 31 (62) | 60 (57.7) | .53 |
| 1–3 | 24 (44.4) | 17 (34) | 41 (39.4) | |
| >3 | 1 (1.9) | 2 (4) | 3 (2.9) | |
* Fisher’s exact test was used for categorical variables and the Wilcoxon two-sided test for continuous variables. BMI = body mass index; Nodal status = number of metastatic axillary lymph nodes.
Plasma Isoflavones
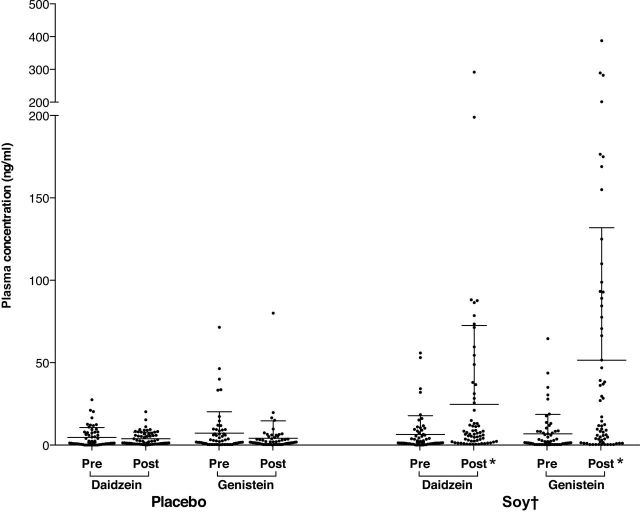
To assess adherence to soy consumption, paired plasma samples were compared before and after treatment for all participants (n = 125). The soy group had a seven-fold increase in plasma genistein (from median 1.6 [range = 0.4 to 64.6] to 11.6 [0.3 to 387.9] ng/mL, P < .001) and a four-fold increase in plasma daidzein (from median 1.5, [range = 0.1 to 55.9] to 6.7 [range = 0.5 to 291.5] ng/ml, P < .001). No statistically significant changes in isoflavone levels were observed in the placebo group (Figure 2). A strong positive correlation was observed between genistein and daidzein levels in the soy group (r = 0.94, P < .001). These results indicated a strong adherence to the assigned treatment.
Figure 2.
Plasma genistein and daidzein following soy intake. Post-treatment isoflavone levels increased compared with pretreatment levels following intake of soy (P < .001) but not placebo. The changes (post-pre) in plasma isoflavones were statistically significantly greater for women receiving soy compared with those receiving placebo (P < .001). * Indicates within-group statistical significance by the Wilcoxon matched-pairs signed rank test, P < .001. † Indicates statistical significance for the comparison of the fold-change between treatment groups by the Wilcoxon rank-sum test (P < .001).
The dispersion in posttreatment isoflavone levels in the soy group was large. While in most patients there was a marked increase, in a few levels changed minimally (Figure 2). This may be explained by differences in adherence to treatment, absorption, metabolism, and clearance of soy and its metabolites (22,32).
NanoString Analysis of Gene Expression Before and After Soy or Placebo
We measured expression of 202 BC-related genes by NanoString analysis in matched tumor samples obtained before and after intervention from 14 BCs. The availability of pretreatment core biopsy tissue limited the sample size to eight patients in the soy, and six in the placebo group. There were no statistically significant between-group differences in patient or tumor characteristics, including ER status (Supplementary Table 2, available online). We identified genes that were statistically significantly changed post-intervention and compared the magnitude and direction of gene expression changes between the two groups (Table 2). Fourteen genes changed in the soy group: 10 increased, and four decreased expression. In the placebo group, 10 genes changed, five increased, and five decreased. Three of these 10 genes were among those that changed in the soy group in the same direction. Thus a total of 21 (out of 202) genes in both groups demonstrated changes. The expression of these genes in pre- and posttreatment tumor samples from both groups is represented in Supplementary Figure 1 (available online).
Table 2.
Differentially expressed genes in pre- and posttreatment tumor samples as assessed by NanoString analysis*
| Gene | refseq | Paired analysis | Between-group analysis | |||
|---|---|---|---|---|---|---|
| Posttreatment / pretreatment | ||||||
| n = 14 patients | ||||||
| Placebo paired | Soy paired | Soy FC vs placebo FC | ||||
| FC | P | FC | P | P | ||
| CCNA2 | NM_001237.2 | -1.61 | .03 | 1.05 | .87 | -- |
| CCNE2 | NM_057735.1 | -2.33 | .05 | 1.49 | .35 | -- |
| CDKN1A | NM_000389.2 | 1.51 | .02 | 1.57 | .01 | -- |
| CDKN1B | NM_004064.2 | 1.07 | .83 | 1.59 | .00 | -- |
| DLC1 | NM_006094.3 | 1.40 | .25 | 1.58 | .05 | -- |
| EGFR | NM_005228.3 | 2.57 | .24 | 1.62 | .03 | -- |
| FANCC | NM_000136.2 | -1.27 | .18 | 1.27 | .04 | .04 |
| HDAC1 | NM_004964.2 | -1.01 | .96 | 1.20 | .05 | -- |
| HIST2H3C | NM_021059.2 | -1.05 | .85 | 1.72 | .03 | -- |
| JUN | NM_002228.3 | 3.96 | .04 | 2.58 | .09 | -- |
| LCMT2 | NM_014793.3 | -1.25 | .15 | -1.43 | .03 | -- |
| NFYB | NM_006166.3 | 1.04 | .83 | -1.64 | .04 | -- |
| PRMT6 | NM_018137.1 | 1.21 | .02 | 1.13 | .39 | -- |
| RAD1 | NM_133377.2 | -1.43 | .04 | 1.01 | .87 | -- |
| RPL27 | NM_000988.3 | 1.25 | .03 | 1.30 | .03 | -- |
| SERPINE1 | NM_000602.2 | 1.35 | .65 | 2.72 | .01 | -- |
| TBP | NM_003194.3 | -1.30 | .04 | -1.37 | .02 | -- |
| TGFA | NM_001099691.1 | 1.68 | .02 | 1.45 | .17 | -- |
| TP53 | NM_000546.2 | -1.19 | .02 | 1.00 | 1.00 | -- |
| UGT1A4 | NM_007120.2 | 1.69 | .45 | 0.43 | .01 | -- |
| UGT2A1 | NM_006798.2 | -1.33 | .26 | 1.57 | .01 | .03 |
* Fold change (FC) and two-sided P value from paired t test (P < .05) is shown for paired analysis within each group, and two-sided P value from unpaired t test (P < .05) is shown for the comparison of FC between groups.
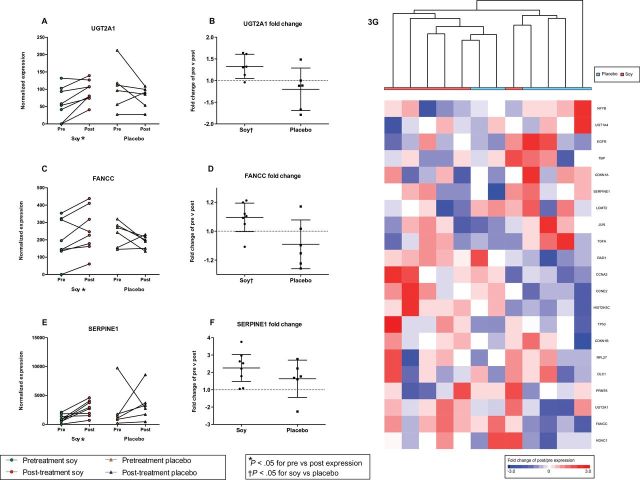
To determine gene expression changes, we focused on fold change (posttreatment/pretreatment ratio) for each of the 21 genes, and compared this value between treatment groups. Expression of FANCC and UGT2A1 increased in 87.5% of tumors following soy intake (mean FC = 1.27 and 1.57, P < .05) and decreased (mean FC = -1.26 and -1.33, P value not statistically significant) in the placebo group (Figure 3, A-D). While these fold changes were modest, they were consistent and in opposing direction in the two groups (P < .05). Genes with altered expression in the soy group also included SERPINE1 (mean FC = 2.7, P = .006) (Figure 3E). However, this increase was not statistically significant between soy and placebo groups (P = .26) (Figure 3F).
Figure 3.
Expression of FANCC and UGT2A1. Gene expression was measured in paired samples (pre/post) using NanoString. Expression levels of FANCC (A), UGT2A1 (C), and SERPINE1 (E) were increased following consumption of soy (P < .05), but not placebo. The expression fold changes (FC) were statistically significantly greater in tumors exposed to soy compared with placebo for FANCC (B) and UGT2A1 (D) (P < .05), but not SERPINE1 (P = .26) (F). A heat map and hierarchical clustering of the post/pre FC of the 21 genes DE between paired samples (P < .05) (G). Positive FC are colored red, negative FC are colored blue, and treatment group is indicated below the sample dendogram. *Indicates within-group statistical significance by paired t test P < .05, †Indicates statistical significance for the comparison of fold change between treatment groups by unpaired t test (P < .05).
To evaluate the patterns of gene expression changes in the matched tumors, we performed hierarchical clustering of the paired samples using the pre/postexpression fold change of the 21 differentially expressed (DE) genes (Figure 3G). Clustering showed a tendency to organize samples by soy or placebo group, and the heat map showed groups of genes correlated by expression fold change in response to soy or placebo. Genes related to cell cycle functions, including CCNA2, CCNE2, and CDKN1B, were closely grouped by cluster analysis, and demonstrated a pattern of expression with increases in soy and decreases in placebo group samples. These data suggest an effect of soy intake characterized by subtle yet consistent and statistically significant alterations in BC gene expression.
Genome-Wide Expression Analysis in Posttreatment Specimens
We performed genome-wide expression analysis of 51 specimens (39 ER+ and 12 ER-) from surgically resected tumors, 28 from soy group, and 23 from placebo. There were no statistically significant between-group differences in demographics or clinicopathological criteria (Supplementary Tables 3 and 4, available online). We identified 131 differently expressed (DE) genes between the two groups (absolute fold change ≥2 and P < .01). Of these, 11 were overexpressed and 120 were underexpressed in tumors of the soy relative to the placebo group.
We next considered the possibility that genistein plasma levels, rather than assignment to the soy group per se, may be a more relevant marker of genistein effects on BC gene expression. Therefore, we examined differential gene expression as a function of plasma genistein. Median genistein concentration in the soy group was 6.3ng/mL, and 25% demonstrated very low levels (<0.5ng/mL). We therefore limited tumors of the soy group to those from patients with serum genistein greater than 16ng/mL, which corresponded to the 95th percentile concentration of the placebo group. The resulting analysis consisted of posttreatment expression profiles of 11 tumors of the soy group with elevated plasma genistein, and 23 tumors from the placebo group with low plasma genistein, referred to as high and low-genistein subsets, respectively. Tumor characteristics, including ER status, were similar in high- and low-genistein subsets (Supplementary Figure 3, available online).
One hundred and twenty-six genes were differentially expressed in the high-genistein vs low-genistein subsets and defined a high-genistein expression signature (P < .01, FC ≥2; 47 overexpressed and 79 underexpressed) (Supplementary Table 5, available online). Hierarchical clustering of the DE genes was performed to investigate patterns of relative expression among tumor subsets (Figure 4). Tumors clustered in the sample dendogram according to plasma genistein levels as expected, reflecting the selection of DE genes by genistein subset.
Figure 4.
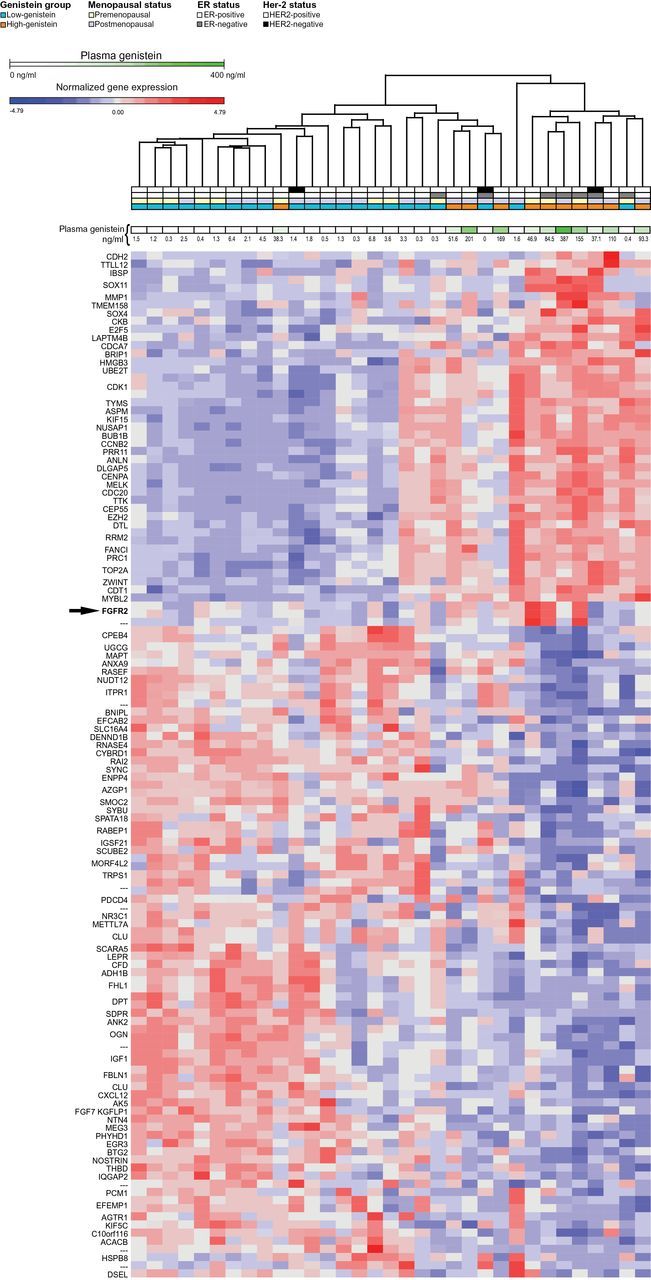
Hierarchical clustering of DE genes in high versus low- genistein subsets. Clustering of samples and 126 DE genes between high- and low-genistein samples (fold change > 2 and t-test P < .1) as identified by microarrays. Clustering was performed using Euclidian distance. Gene expression values are log2 transformed and standardized. ER status, menopausal status, and genistein plasma concentrations for each sample are indicated. Twelve tumors of the soy group with high (>16ng/mL) plasma genistein defined the high-genistein subset, and 22 tumors of the placebo group with low (<6.8ng/mL) genistein defined the low-genistein subset. FGFR2 was overexpressed in three of 12 tumors of the high-genistein group (arrow, P < .01).
Pathway analysis of the high-genistein signature revealed over-representation of pathways that regulate cell growth and proliferation in tumors of the high-genistein group (P < .001). DAVID analysis of overexpressed genes in the high-genistein group revealed that 18 of 23 categories represented cell cycle functions (Bonferroni P < .001, FDR < 0.01) (Supplementary Table 6, available online) (33). Similarly, IPA revealed that the Top Biological Functions and Network Modules of the 126 DE genes were cellular growth and proliferation, cell cycle, cell death and survival, cell development, and nucleic acid metabolism (P < .001) (Supplementary Figure 2, A and B, and Supplementary Table 7, available online). Results of a “downstream effect analysis,” which focuses on a gene’s function, were concordant with the network analysis, indicating enrichment of genes that regulate cell proliferation (Supplementary Table 8, available online). Specifically, genes in the top-ranked IPA network that were overexpressed in the high-genistein tumors included those which coordinately regulate G1/S and G2/M cell cycle processes, such as E2F5, BUB1, CCNB2 (Cyclin B2), MYBL2, CDK1, and CDC20 (Supplementary Figure 2B and Supplementary Table 8, available online). The receptor tyrosine kinase FGFR2, a known regulator of the cell cycle, was found to be overexpressed in the downstream effect analysis (Supplementary Table 8, available online).
Gene Set Analysis performed on fold changes of all genes between high- and low-genistein groups revealed a higher level of expression of numerous cell cycle gene sets, including RB1 cell cycle targets and E2F-family target genes (Supplementary Table 9, available online). Similar results were obtained when ER(-) samples were excluded from the analysis (Supplementary Table 10, available online).
To assess whether increased expression of cell cycle–related genes in the soy group was associated with BC molecular subtypes, we evaluated the distribution of the PAM50 subtypes in soy and placebo groups (34,35). We implemented a nearest centroid molecular classification model based on the expression of the PAM50 genes to predict a breast cancer’s intrinsic subtype as luminal A, luminal B, HER2-enriched, or Basal (35). Although all intrinsic subtypes were represented in the high and low-genistein groups, there was a trend for luminal A in the low-genistein group and luminal B in the high-genistein group (P = .06) (Supplementary Figure 3, available online). These data demonstrate enrichment for luminal A BCs in the placebo group and for luminal B tumors in the soy group. Although this may constitute a selection bias despite randomization, we cannot exclude the possibility that soy might increase expression of genes associated with luminal B, including proliferation and cell cycle–related genes.
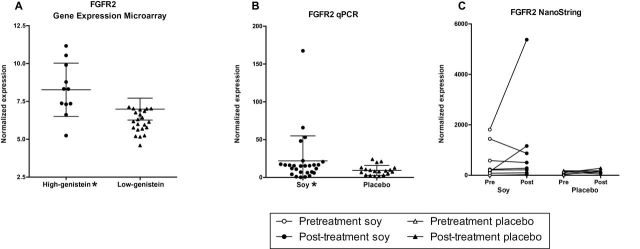
Expression of the protumorigenic growth factor receptor FGFR2 was elevated in the high- compared with the low-genistein group (FC = 2.4, P = .006) (Figure 5A; Supplementary Table 5, available online), with much higher expression in three out of 12 tumors with very high genistein (46.9, 84.5, 155ng/ml). To confirm overexpression of FGFR2, we performed quantitative real-time PCR (qPCR) in 27 tumors of the soy and 19 of the placebo group, revealing its overexpression by 2.3-fold in tumors of the soy vs placebo group (P = .03) (Figure 5B). The three cases with FGFR2 overexpression by microarray also demonstrated an increase by qPCR. Two of the three samples with FGFR2 overexpression were included in the NanoString paired analysis; one tumor demonstrated a three-fold, and the second a 7.7-fold increase in FGFR2 following soy treatment (Figure 5C). Taken together, these data raise the concern that FGFR2 expression was increased by soy in a subset of BCs.
Figure 5.
Differential expression of FGFR2. FGFR2 expression was increased 2.4 fold in the high-genistein subset of the soy group by microarray (P = .006) (A). FGFR2 overexpression by qPCR in soy vs placebo (P = .03) (B). Nanostring analysis of pre/post samples. Although there were no statistically significant differences for the group as a whole, in two samples from the soy group there was a marked increase in FGFR2 expression (C). *Indicates statistically significant overexpression (P < .05).
Soy Effects on Tumor Proliferation (Ki67) and Apoptosis (Cas3)
Markers of apoptosis (Cas3) and proliferation (Ki67) were examined in paired pre- and posttreatment tumor samples from 54 patients from the soy and 50 from the placebo group, and the percentage of positive staining tumor cells was assessed (Table 3). A comparison of changes (between pre- and posttreatment) in the placebo and soy groups showed no statistically significant differences (P = .2 for Ki67 and P = .3 for Cas3) (Table 3).
Table 3.
Measurements (median and range) of proliferation (Ki67) and apoptosis (Cas3) indices as determined by immunohistochemical analysis of pre- and posttreatment tumors in the soy and placebo groups*
| Index | Median (range) | Changes | |||||
|---|---|---|---|---|---|---|---|
| Soy (n = 54) | Placebo (n = 50) | Soy vs placebo | |||||
| Pre | Post | P | Pre | Post | P | P | |
| Ki67 | 15.5 (1.6–80) | 21 (4.0–80) | .087 | 16.5 (0–80) | 20 (1–72) | .71 | .21 |
| Cas3 | 1 (0–25) | 1.55 (0–31) | .007 | 1 (0–10) | 1.25 (0–31) | .53 | .35 |
* P-values are from the two-sided Wilcoxon matched-pairs signed rank test. Two-sided Wilcoxon rank-sum test was used to calculate the P values for the changes (post–pre) in the soy group.
NanoString Analysis of Gene Expression Before and After Soy or Placebo
We measured expression of 202 BC-related genes by NanoString analysis in matched tumor samples obtained before and after intervention from 14 BCs. The availability of pretreatment core biopsy tissue limited the sample size to eight patients in the soy, and six in the placebo group. There were no statistically significant between-group differences in patient or tumor characteristics, including ER status (Supplementary Table 2, available online). We identified genes that were changed postintervention, and compared the magnitude and direction of gene expression changes between the two groups (Table 2). Fourteen genes changed in the soy group: 10 increased, and four decreased expression. In the placebo group, 10 genes changed, five increased, and five decreased. Three of these 10 genes were among those that changed in the soy group in the same direction. Thus a total of 21 genes in both groups demonstrated changes. The expression of these genes in pre- and posttreatment tumor samples from both groups is represented in Supplementary Figure 1 (available online).
To determine gene expression changes, we focused on fold change (posttreatment/pretreatment ratio) for each of the 21 genes, and compared this value between treatment groups. Expression of FANCC and UGT2A1 increased in 87.5% of tumors following soy intake (mean FC = 1.27 and 1.57, P < .05), and decreased (mean FC = -1.26 and -1.33, P value not statistically significant) in the placebo group (Figure 3, A-D). While these fold changes were modest, they were consistent and in opposing direction in the two groups (P < .05). Genes with altered expression in the soy group also included SERPINE1 (mean FC = 2.7, P = .006) (Figure 3E). However, this increase was not significant between soy and placebo groups (P = .26) (Figure 3F).
To evaluate the patterns of gene expression changes in the matched tumors, we performed hierarchical clustering of the paired samples using the pre/postexpression fold change of the 21 differentially expressed genes (Figure 3G). Clustering showed a tendency to organize samples by soy or placebo group, and the heat map showed groups of genes correlated by expression fold change in response to soy or placebo. Genes related to cell cycle functions, including CCNA2, CCNE2, and CDKN1B, were closely grouped by cluster analysis, and demonstrated a pattern of expression with increases in soy and decreases in placebo group samples. These data suggest an effect of soy intake characterized by subtle, yet consistent alterations in BC gene expression.
Genome-Wide Expression Analysis in Posttreatment Specimens
We performed genome-wide expression analysis of 51 specimens (39 ER+ and 12 ER-) from surgically resected tumors, 28 from soy group and 23 from placebo. There were no statistically significant differences between group demographics or clinicopathological criteria (Supplementary Tables 3 and 4, available online). We identified 131 differently expressed (DE) genes between the two groups (absolute fold change ≥2, P < .01) (Supplementary Table 5, available online). Of these, 11 were overexpressed, and 120 were underexpressed in tumors of the soy relative to the placebo group.
We next considered the possibility that genistein plasma levels, rather than assignment to the soy group per se, may be a more relevant marker of genistein effects on BC genes expression. Therefore, we examined differential gene expression as a function of plasma genistein. Median genistein concentration in the soy group was 6.3ng/mL, and 25% demonstrated very low levels (<0.5ng/mL). We therefore limited tumors of the soy group to those from patients with serum genistein greater than 16ng/mL, which corresponded to the 95th percentile concentration of the placebo group. The resulting analysis consisted of posttreatment expression profiles of 11 tumors of soy group with elevated plasma genistein, and 23 tumors from the placebo group with low plasma genistein, referred to as high- and low-genistein subsets, respectively. Tumor characteristics including ER status were similar in high- and low-genistein subsets.
One hundred and twenty-six genes were differentially expressed in the high-genistein vs low-genistein subsets and defined a high-genistein expression signature (FC ≥2, P < .01; 47 overexpressed and 79 underexpressed). Hierarchical clustering of the DE genes was performed to investigate patterns of relative expression among tumor subsets (Figure 4). Tumors clustered in the sample dendogram according to plasma genistein levels as expected, reflecting the selection of DE genes by genistein subset.
Pathway analysis of the high-genistein signature revealed overrepresentation of pathways that regulate cell growth and proliferation in tumors of the high-genistein group (P < .001). DAVID analysis of overexpressed genes in the high-genistein group revealed that 18 of 23 categories represented cell cycle functions (Bonferroni FDR < 0.01, P < .001) (Supplementary Table 6, available online) (33). Similarly, IPA revealed that the Top Biological Functions and Network Modules of the 126 DE genes were cellular growth and proliferation, cell cycle, cell death and survival, cell development, and nucleic acid metabolism (P < .001) (Supplementary Figure 2, A and B, and Supplementary Table 7, available online). Results of a “downstream effect analysis,” which focuses on a gene’s function, were concordant with the network analysis, indicating enrichment of genes that regulate cell proliferation (Supplementary Table 8, available online). Specifically, genes in the top-ranked IPA network that were overexpressed in the high-genistein tumors included those which coordinately regulate G1/S and G2/M cell cycle processes, such as E2F5, BUB1, CCNB2 (Cyclin B2), MYBL2, CDK1, and CDC20 (Supplementary Figure 2B and Supplementary Table 8, available online). The receptor tyrosine kinase FGFR2, a known regulator of the cell cycle, was found to be overexpressed in the downstream effect analysis (Supplementary Table 8, available online).
Gene Set Analysis performed on fold changes of all genes between high- and low-genistein groups revealed higher level of expression of numerous cell cycle gene-sets, including RB1 cell cycle targets and E2F-family target genes (Supplementary Table 9, available online). Similar results were obtained when ER(-) samples were excluded from the analysis (Supplementary Table 10, available online).
To assess whether increased expression of cell cycle–related genes in the soy group was associated with BC molecular subtypes, we evaluated the distribution of the PAM50 subtypes in soy and placebo groups (34,35). We implemented a nearest centroid molecular classification model based on the expression of the PAM50 genes to predict a breast cancer’s intrinsic subtype as luminal A, luminal B, HER2-enriched, or Basal (35). Although all intrinsic subtypes were represented in the high- and low-genistein groups, there was a trend for luminal A in the low-genistein group and luminal B in the high-genistein group (P = .06) (Supplementary Figure 3, available online). These data demonstrate enrichment for luminal A BCs in the placebo group and for luminal B tumors in the soy group. Although this may constitute a selection bias despite randomization, we cannot exclude the possibility that soy might increase expression of genes associated with luminal B, including proliferation and cell cycle–related genes.
Protumorigenic growth factor receptor FGFR2 expression was elevated in the high- compared with the low-genistein group (FC = 2.4, P = .006) (Figure 5A; Supplementary Table 5, available online), with much higher expression in three out of 12 tumors with very high genistein (46.9, 84.5, 155ng/mL). To confirm overexpression of FGFR2, we performed quantitative real-time PCR (qPCR) in 27 tumors of the soy and 19 of the placebo group, revealing its overexpression by 2.3-fold in tumors of the soy vs placebo group (P = .03) (Figure 5B). The three cases with FGFR2 overexpression by microarray also demonstrated an increase by qPCR. Two of the three samples with FGFR2 overexpression were included in the NanoString paired analysis; one tumor demonstrated a three-fold, and the second a 7.7-fold increase in FGFR2 following soy treatment (Figure 5C). Taken together, these data raise the concern that FGFR2 expression was increased by soy in a subset of BCs.
Soy Effects on Tumor Proliferation (Ki67) and Apoptosis (Cas3)
Markers of apoptosis (Cas3) and proliferation (Ki67) were examined in paired pre- and posttreatment tumor samples from 54 patients from the soy and 50 from the placebo group, and the percentage of positive staining tumor cells was assessed (Table 3). A comparison of changes (between pre- and posttreatment) in the placebo and soy groups showed no statistically significant differences (P = .21 for Ki67 and P = .35 for Cas3) (Table 3).
Discussion
This study demonstrates that soy supplementation alters BC-related gene expression. Using multiple molecular approaches and bioinformatics techniques, we identified a large number of cell-cycle and proliferation-associated genes that were overexpressed in BCs from the soy group in patients with elevated plasma genistein. Expression of 21 genes measured by NanoString was altered from pretreatment levels as a consequence of treatment with soy or placebo. Two of these genes, UGT2A1 and FANCC, were upregulated in the soy group, suggesting a treatment effect. DE genes that increased in the soy group included the protumorigenic growth factor receptor FGFR2. To our knowledge, this is the first report to analyze gene expression in patient-matched tumors before and after soy intake.
We found FANCC and UGT2A1 to be altered by soy consumption. While the consequences of their increased expression in human BC are unknown, both genes have the potential to influence BC biology. FANCC encodes a DNA repair protein, and mutations are responsible for the autosomal recessive disorder Fanconi Anemia and may be implicated in BC development (36). UGT2A1 functions in metabolism, including 17β-estradiol and its enantiomers, and has been implicated in tobacco-related carcinogenesis (37,38).
Computational analysis of genome-wide microarray data from the patients with high and low levels of genistein (subsets of the soy and placebo groups) revealed overrepresentation of several cell cycle gene categories in the high-genistein gene signature and higher levels of expression of cell cycle gene sets, including targets of E2F-family transcription factors. Gene Set Analysis limited to ER(+) samples yielded similar results as above, suggesting that enrichment of gene sets in tumors of the high-genistein subset could not be explained solely on the basis of differences associated with ER status (Supplementary Table 10, available online). Analysis of publically available gene expression data in MCF-7 BC cells exposed to genistein (GSE5705) revealed upregulation of many of the same top-ranked gene sets, as identified in our data (cell cycle categories, targets of E2F and RB1; unpublished observations), which supports our findings (39).
A subset of tumors from the soy group was notable for increased FGFR2 expression as demonstrated by multiple gene expression techniques. There is extensive evidence that FGFR2 drives cancer growth through its role as a potent oncogene, and increased FGFR2 expression is a marker of poor prognosis in BC (40–42). We found statistically significant overexpression of FGFR2 by microarray and qPCR in tumors of patients taking soy compared with placebo and increased expression in two paired tumor samples before and after soy supplementation. In one sample, expression was increased from already elevated pretreatment levels, and one could speculate that the initial molecular alteration was reinforced by soy.
Soy intake did not result in statistically significant changes in cell proliferation and apoptosis indices compared with the placebo group. A similar observation was made in healthy breast tissue (43). In BCs from patients with elevated serum genistein, we observed increased expression of genes and gene sets associated with increased cell proliferation and cell cycle progression. A potential explanation for the discrepancy between Ki67 and gene expression results is that a nutritional intervention such as soy intake may take longer periods of time to influence a phenotype measured by immunohistochemical analysis. A second possibility is related to limitations of the Ki67 IHC. As a consequence of tissue heterogeneity and small amounts of available tissue, the Ki67 assessments in pretreatment core biopsies may not have represented the whole tumor.
Identifying gene expression effects associated with a nutritional intervention such as soy presents numerous challenges. Expression changes from diet intervention are expected to be subtle, and detection of alterations is complicated by the molecular heterogeneity of BCs and the need for large data sets. One solution, as implemented in this study, was to take an exploratory approach in which false discovery rate and corrections for multiple hypothesis testing are often withheld in favor of limiting type-II errors, but with the possibility of increasing type-I errors. Additionally, Gene Set Analysis provides an analytical strategy to detect modest but coordinated changes in the expression of biological pathways and sets of functionally related genes. A second possible solution is analysis of paired samples before and after soy intake. Until recently, gene expression analysis required use of snap-frozen tissue, rarely available from diagnostic core biopsies. NanoString technology allows measurement of gene expression from limited FFPE tissue samples, such as those obtained from diagnostic core biopsies. This technology allowed measurement of gene expression from the same tumor before and after soy consumption. Despite the small sample size, this approach identified consistent yet subtle patterns of altered gene expression associated with soy consumption.
This study has a number of limitations. This was a short-term study utilizing a large daily supplement of soy. The implications of our findings in patients consuming smaller amounts of soy over prolonged periods are unclear. Also, the clinical impact of the subtle changes in gene expression has not been examined. Nevertheless, these data raise concern that soy may exert a stimulating effect on BC in a subset of women.
Funding
This work was supported by a grant from the Breast Cancer Research Foundation (ClinicalTrials.gov number, NCT 00597532).
Supplementary Material
Notes
The microarray data from this study are available from the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession GSE58792.
The soy and placebo protein were provided by DuPont Nutrition & Health, St Louis, MO.
We would like to acknowledge the use of the Genomics Core Laboratory, partially funded by NCI Cancer Center Support Grant P30 CA008748-47. The authors would also like to thank and acknowledge Dr. Katherine Panageas of the Memorial Sloan-Kettering Cancer Center’s Department of Epidemiology and Biostatistics for her assistance in the study design and statistics and Margot Weissman for her assistance in the study analysis.
The study sponsors had no role in the design of the study, the collection, analysis, or interpretation of the data, the writing of the manuscript, nor the decision to submit the manuscript for publication.
Authors disclose no potential conflicts of interest related to this study.
References
- 1. Messina M, McCaskill-Stevens W, Lampe JW. Addressing the soy and breast cancer relationship: review, commentary, and workshop proceedings. J Natl Cancer Inst. 2006;98(18):1275–1284. [DOI] [PubMed] [Google Scholar]
- 2. Messina MJ, Loprinzi CL. Soy for breast cancer survivors: a critical review of the literature. J Nutr. 2001;131(11 Suppl):3095S–3108S. [DOI] [PubMed] [Google Scholar]
- 3. Shu XO, Zheng Y, Cai H, et al. Soy food intake and breast cancer survival. JAMA. 2009;302(22):2437–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu AH, Yu MC, Tseng CC, et al. Epidemiology of soy exposures and breast cancer risk. Br J Cancer. 2008;98(1):9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wu AH, Koh WP, Wang R, et al. Soy intake and breast cancer risk in Singapore Chinese Health Study. Br J Cancer. 2008;99(1):196–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dong JY, Qin LQ. Soy isoflavones consumption and risk of breast cancer incidence or recurrence: a meta-analysis of prospective studies. Breast Cancer Res Treat. 2011;125(2):315–323. [DOI] [PubMed] [Google Scholar]
- 7. Hsieh CY, Santell RC, Haslam SZ, et al. Estrogenic effects of genistein on the growth of estrogen receptor-positive human breast cancer (MCF-7) cells in vitro and in vivo. Cancer Res. 1998;58(17):3833–3838. [PubMed] [Google Scholar]
- 8. Allred CD, Allred KF, Ju YH, et al. Soy diets containing varying amounts of genistein stimulate growth of estrogen-dependent (MCF-7) tumors in a dose-dependent manner. Cancer Res. 2001;61(13):5045–5050. [PubMed] [Google Scholar]
- 9. Ju YH, Doerge DR, Allred KF, et al. Dietary genistein negates the inhibitory effect of tamoxifen on growth of estrogen-dependent human breast cancer (MCF-7) cells implanted in athymic mice. Cancer Res. 2002;62(9):2474–2477. [PubMed] [Google Scholar]
- 10. Hirano T, Oka K, Akiba M. Antiproliferative effects of synthetic and naturally occurring flavonoids on tumor cells of the human breast carcinoma cell line, ZR-75-1. Res Commun Chem Pathol Pharmacol. 1989;64(1):69–78. [PubMed] [Google Scholar]
- 11. Pagliacci MC, Smacchia M, Migliorati G, et al. Growth-inhibitory effects of the natural phyto-oestrogen genistein in MCF-7 human breast cancer cells. Eur J Cancer. 1994;30A(11):1675–1682. [DOI] [PubMed] [Google Scholar]
- 12. Kuriu A, Ikeda H, Kanakura Y, et al. Proliferation of human myeloid leukemia cell line associated with the tyrosine-phosphorylation and activation of the proto-oncogene c-kit product. Blood. 1991;78(11):2834–2840. [PubMed] [Google Scholar]
- 13. Cunningham BD, Threadgill MD, Groundwater PW, et al. Synthesis and biological evaluation of a series of flavones designed as inhibitors of protein tyrosine kinases. Anticancer Drug Des. 1992;7(5):365–384. [PubMed] [Google Scholar]
- 14. Okura A, Arakawa H, Oka H, et al. Effect of genistein on topoisomerase activity and on the growth of [Val 12]Ha-ras-transformed NIH 3T3 cells. Biochem Biophys Res Commun. 1988;157(1):183–189. [DOI] [PubMed] [Google Scholar]
- 15. Markovits J, Linassier C, Fosse P, et al. Inhibitory effects of the tyrosine kinase inhibitor genistein on mammalian DNA topoisomerase II. Cancer Res. 1989;49(18):5111–5117. [PubMed] [Google Scholar]
- 16. Buteau-Lozano H, Velasco G, Cristofari M, et al. Xenoestrogens modulate vascular endothelial growth factor secretion in breast cancer cells through an estrogen receptor-dependent mechanism. J Endocrinol. 2008;196(2):399–412. [DOI] [PubMed] [Google Scholar]
- 17. Guo Y, Wang S, Hoot DR, et al. Suppression of VEGF-mediated autocrine and paracrine interactions between prostate cancer cells and vascular endothelial cells by soy isoflavones. J Nutr Biochem. 2007;18(6):408–417. [DOI] [PubMed] [Google Scholar]
- 18. Farina HG, Pomies M, Alonso DF, et al. Antitumor and antiangiogenic activity of soy isoflavone genistein in mouse models of melanoma and breast cancer. Oncol Rep. 2006;16(4):885–891. [PubMed] [Google Scholar]
- 19. Reis-Filho JS, Pusztai L. Gene expression profiling in breast cancer: classification, prognostication, and prediction. Lancet. 2011;378(9805):1812–1823. [DOI] [PubMed] [Google Scholar]
- 20. Block G, Woods M, Potosky A, et al. Validation of a self-administered diet history questionnaire using multiple diet records. J Clin Epidemiol. 1990;43(12):1327–1335. [DOI] [PubMed] [Google Scholar]
- 21. Lanza E, Schatzkin A, Ballard-Barbash R, et al. The polyp prevention trial II: dietary intervention program and participant baseline dietary characteristics. Cancer Epidemiol Biomarkers Prev. 1996;5(5):385–392. [PubMed] [Google Scholar]
- 22. King RA, Bursill DB. Plasma and urinary kinetics of the isoflavones daidzein and genistein after a single soy meal in humans. Am J Clin Nutr. 1998;67(5):867–872. [DOI] [PubMed] [Google Scholar]
- 23. Xu X, Wang HJ, Murphy PA, et al. Neither background diet nor type of soy food affects short-term isoflavone bioavailability in women. J Nutr. 2000;130(4):798–801. [DOI] [PubMed] [Google Scholar]
- 24. Holzbeierlein J, Lal P, LaTulippe E, et al. Gene expression analysis of human prostate carcinoma during hormonal therapy identifies androgen-responsive genes and mechanisms of therapy resistance. Am J Pathol. 2004;164(1):217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lal P, Tan LK, Chen B. Correlation of HER-2 status with estrogen and progesterone receptors and histologic features in 3,655 invasive breast carcinomas. Am J Clin Pathol. 2005;123(4):541–546. [DOI] [PubMed] [Google Scholar]
- 26. Dowsett M, Nielsen TO, A’Hern R, et al. Assessment of Ki67 in breast cancer: recommendations from the International Ki67 in Breast Cancer working group. J Natl Cancer Inst. 2011;103(22):1656–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Varemo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids. Res 2013;41(8):4378–4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bos PD, Zhang XH, Nadal C, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haibe-Kains B, Desmedt C, Loi S, et al. A three-gene model to robustly identify breast cancer molecular subtypes. J Natl Cancer Inst. 2012;104(4):311–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Doane AS, Danso M, Lal P, et al. An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene. 2006;25(28):3994–4008. [DOI] [PubMed] [Google Scholar]
- 32. Edge SB, American Joint Committee on Cancer. AJCC cancer staging manual. 7th ed. New York: Springer; 2010. [DOI] [PubMed] [Google Scholar]
- 33. Stubert J, Gerber B. Isoflavones - Mechanism of Action and Impact on Breast Cancer Risk. Breast Care (Basel). 2009;4(1):22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. [DOI] [PubMed] [Google Scholar]
- 35. Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thompson ER, Doyle MA, Ryland GL, et al. Exome sequencing identifies rare deleterious mutations in DNA repair genes FANCC and BLM as potential breast cancer susceptibility alleles. PLoS Genet. 2012;8(9):e1002894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sneitz N, Krishnan K, Covey DF, et al. Glucuronidation of the steroid enantiomers ent-17beta-estradiol, ent-androsterone and ent-etiocholanolone by the human UDP-glucuronosyltransferases. J Steroid Biochem Mol Biol. 2011;127(3–5):282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bushey RT, Chen G, Blevins-Primeau AS, et al. Characterization of UDP-glucuronosyltransferase 2A1 (UGT2A1) variants and their potential role in tobacco carcinogenesis. Pharmacogenet Genomics. 2011;21(2):55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Shioda T, Rosenthal NF, Coser KR, et al. Expressomal approach for comprehensive analysis and visualization of ligand sensitivities of xenoestrogen responsive genes. Proc Natl Acad Sci U S A. 2013;110(41):16508–16513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim S, Dubrovska A, Salamone RJ, et al. FGFR2 promotes breast tumorigenicity through maintenance of breast tumor-initiating cells. PLoS One. 2013;8(1):e51671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sun S, Jiang Y, Zhang G, et al. Increased expression of fibroblastic growth factor receptor 2 is correlated with poor prognosis in patients with breast cancer. J Surg Oncol. 2012;105(8):773–779. [DOI] [PubMed] [Google Scholar]
- 43. Fletcher MN, Castro MA, Wang X, et al. Master regulators of FGFR2 signalling and breast cancer risk. Nat Commun. 2013;4:2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Khan SA, Chatterton RT, Michel N, et al. Soy isoflavone supplementation for breast cancer risk reduction: a randomized phase II trial. Cancer Prev Res (Phila). 2012;5(2):309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.