Abstract
The immune system has evolved to protect multicellular organisms from the attack of a variety of pathogens. To exert this function efficiently, the system has developed the capacity to coordinate the function of different cell types and the ability to down-modulate the response when the foreign attack is over. For decades, immunologists believed that these two characteristics were primarily related to cytokine/chemokine-based communication and cell-to-cell direct contact. More recently, it has been shown that immune cells also communicate by transferring regulatory RNAs, microRNAs in particular, from one cell to the other. Several studies have suggested a functional role of extracellular regulatory RNAs in cell-to-cell communication in different cellular contexts. This minireview focuses on the potential role of extracellular RNA transfer in the regulation of adaptive immune response, also contextualizing it in a broader field of what is known of cell-free RNAs in communication among different organisms in the evolutionary scale.
Keywords: cellular immune response, extracellular vesicles, lymphocyte, nucleic acid, RNA, Cell-free RNAs, immune system, T cells, Treg cells
Cell-free Nucleic Acids: A Historical Perspective
The first description of extracellular nucleic acids in the blood of healthy and sick individuals goes back to observations published by Mandel and Metais in 1948 (1). After this first study, a series of research groups described both DNA and RNA circulating as extracellular molecules in the body. These molecules were considered as potential biomarkers mainly in cancer and for prenatal diagnosis (2, 3). More recently, microRNAs (miRNAs),3 small RNA molecules (18–25 nucleotides), which in their mature single-stranded forms act as post-transcriptional repressors through pairing with messenger RNAs (4), have been shown to circulate in human blood associated with either vesicles or protein complexes that protect them from degradation (5–7). In the last 7 years, blood-circulating miRNAs have become the most promising clinical biomarkers for the diagnosis, prognosis, and therapeutic options of a variety of pathological conditions such as cancer (8–10), cardiovascular disorders (11, 12), diabetes (13), and liver diseases (14, 15), among others (16). Although cell-free nucleic acids have the potential to revolutionize medical diagnostics, the biological significance of these molecules in the extracellular environment still remains elusive. RNA release can be a passive phenomenon that results from tissue damage, or an active secretory process of healthy cells. Hence, is RNA simply discarded by cells as waste, or does it have a role in cell-to-cell communication? Or both?
In immunology, transfer of immunity by RNA was first described more than 40 years ago (17). Moreover, RNA extracted from the thymus was shown to induce maturation and expression of a T cell-specific antigen in bone marrow lymphocytes (18) and to activate the proliferation of bone marrow plasma cells in vitro (19). Then, it was first proposed that an analogous mechanism in cell maturation in vivo might involve RNA release by thymus cells. In the mid-70s, lymphocytes were shown to release double-stranded DNA in complex with low molecular weight RNA. The information carried by extracellular nucleic acids could be transferred from one cell type to another in the course of an immune response (20–22). The capability to communicate through RNA molecules is possibly very old and universally distributed in living organisms.
Traveler RNA: A Universal Living Language
The hypothesis of the so-called “RNA world” suggests that RNA was the first nucleic acid in primordial cells ∼3.8 billion years ago (23, 24) and may be the answer to a famous “chicken and egg” problem: which came first, DNA or protein? RNA is the only molecule capable of both storing genetic information and catalyzing chemical reactions, being at the same time genotype and phenotype. About 3.6 billion years ago, the more stable DNA molecule almost completely replaced RNA for the storage of genetic information, and thus allowed the passage from an RNA world to a DNA world, although one in which RNA molecules still performed multiple functions, participating in cellular chemical reactions, regulating the transcription of genes, and modulating the activity of proteins. Although the last century of biological research was dominated by the idea that RNA is mostly devoted to the translation of genes into proteins, and basic biology textbooks focus on mRNAs, tRNAs, and rRNAs, today RNA is recognized as the most versatile biological macromolecule, with coding RNA being only a continent of the RNA planet.
One of the best characterized non-coding functions of RNA is interference (RNAi), originally defined as the capability of double-stranded RNA to induce sequence-specific degradation of messenger RNAs. RNAi was first described in plant biology in 1990 (25), and 8 years later, a double-stranded RNA was shown to trigger a powerful cellular silencing mechanism in Caenorhabditis elegans that could be transmitted in the germ line and pass through several generations, in addition to spreading from tissue to tissue in the same individual (26). RNA-induced gene-silencing mechanisms developed early at the basal eukaryotic lineage, possibly to control viruses and transposable elements (27). RNAi may have an even older origin, linked to the replication/transcription pathways present in the ancient RNA world (28). Importantly, RNA molecules regulate biological mechanisms across different species or even different kingdoms pertaining to the interconnected conditions of parasitism/infection/food intake. Endogenous non-coding RNAs from Escherichia coli interfere with gene expression and regulate physiological conditions in C. elegans (29, 30). A fungal pathogen that infects Arabidopsis thaliana uses extracellular small RNAs to hijack the host RNAi machinery by binding to plant AGO1 and selectively silencing host genes, with the effect of suppressing host immunity and achieving infection (31). The protozoan parasite Trypanosoma cruzi releases specific tRNA-derived small RNAs that are transferred between and from parasites to mammalian cells, eventually altering their infection susceptibility (32). Plant miRNAs present in food and acquired orally were shown to use mammalian AGO2 to form AGO2-associated RNA-induced silencing complex (RISC), thus regulating the expression of target genes in mammals (33), although later studies reported notes of caution, suggesting that plant miRNAs via dietary exposure may not be universal in animals (34).
In conclusion, RNA is not the labile molecule we used to think about at the end of the previous century. RNA molecules are involved in several cellular activities and are not even constrained inside single cells; instead they are mobile and travel between organisms of the same or different species, with increasingly recognized ecological roles. These RNA functions may be extremely ancient, as RNA molecules are involved in quorum sensing, a phenomenon of sociality among bacteria, that can reach a high level of regulation, including cheating and high competition (35, 36). If extracellular RNA-based communication arose even before the origin of the eukaryotic cell, then we can hypothesize that it is a widespread biological process, a powerful and universal code that allows one organism/cell to influence the behavior of others (37).
The need for a sophisticated type of containment brought about the use of phospholipidic bilayers with the formation of vesicles at a growing grade of complexity. Vesicles give two major advantages to RNA messages: protection from degradation and the possibility, through the presence of proteins, to confer a higher level of specificity to the message.
Extracellular Vesicles as Cell-to-Cell Words with a Huge Biological Impact: The Case of the Immune System
More recently, a growing body of evidence has unveiled that not all extracellular vesicles are created equal. There exist vesicles of nanometric size (20–100 nm, often referred to as exosomes) that are formed by the inward budding and subsequent fusion to the plasma membrane of multivesicular endosomes (38), and vesicles of larger size (0.2–1 μm) that bud directly from the plasma membrane are called microvesicles and also comprise apoptotic and senescent bodies. An exhaustive description of the biogenesis, trafficking, morphology, and isolation techniques of extracellular vesicles goes beyond the scope of the present review, but it can be found elsewhere (39).
The first accurate description of extracellular vesicles (EVs) goes back to 1987, when sheep reticulocytes cultured in vitro were shown to release vesicles containing a number of activities characteristic of the reticulocyte plasma membrane, including acetylcholinesterase and the transferrin receptor. Vesicle externalization was thus suggested to be a mechanism for shedding specific membrane functions (40). For quite some time after those first observations, it was believed that EVs are released by cells to eliminate molecules that need to be down-modulated during specific processes.
Starting from 1996, EVs started to be regarded as more than a mere garbage bin as it was discovered that both dendritic cells and B lymphocytes release vesicles containing MHC class II that are able to induce antigen-specific MHC class II-restricted T cell responses, and this was the first demonstration of a specific role for vesicles in antigen presentation in vivo (41, 42). Also, activated T lymphocyte-derived exosomes, bearing T cell receptor (TCR) from the pool of activated complexes, were shown to target cells bearing the right combination of peptide/MHC complexes (43). Since those first studies, many others have contributed to discovering a significant biological role of EVs in controlling the immune response. The general mechanism by which this control is exerted is through the exchange of vesicles as powerful vehicles to specifically deliver signals from cell to cell in an autocrine, paracrine, and also endocrine fashion. The picture is very complex if one thinks that most if not all cells of the immune system are known to release EVs; that vesicle content can change according to activation status and environmental cues of the releasing cells; and that the potential targets are both immune and non-immune cells. EVs can help amplify activation, as in the case of exosomes derived from stimulated T cells that cooperate with IL-2 to induce growth of resting T cells, by stimulating a proliferative response and the secretion of more cytokines and at higher levels than cells stimulated by IL-2 alone (44). In contrast, CD95 ligand-bearing EVs circulating in blood can deliver death signals and suppress the immune response in a Fas/FasL-dependent manner (45).
Although no systematic studies have been conducted yet, several cytokines and chemokines have been found in association with EVs. Examples are IL-1β (46, 47); IL-6 (48); IL-18 (49); and TNF-α (50). To date, it is not known whether EV association imprints a different specificity when compared with free cytokines in vivo and what is the effect of EV association on our conventional cytokine measurements. In 2001, EVs officially entered the immune regulatory arena when suppressing exosomes (e.g. “tolerosomes”) were first described as supramolecular structures assembled in and released from the small intestinal epithelial cell that carry MHC class II molecules with bound gut lumen antigenic peptides, fully capable of inducing antigen-specific tolerance (51). Tolerosomes were shown to induce donor-specific allograft tolerance characterized by strong inhibition of the anti-donor proliferative response and the delay of the appearance of chronic rejection, thus suggesting that these vesicles can induce regulatory responses able to modulate allograft rejection (52). EVs can also mediate regulatory T (Treg) cell differentiation: exosome-like particles released from thymic cells were shown to promote the conversion of naive T cells into forkhead-box-P (Foxp)3+ Treg cells, and may be one of the endogenous drivers for inducing Treg cells under physiological conditions via vesicle-associated TGF-β (53). It is conceivable that the thymus can also communicate with peripheral organs, including lymphoid and non-lymphoid sites, via thymic particles to regulate host immune tolerance. Importantly, tumor cells take advantage of EV-based immune-suppressive systems by secreting vesicles that can either induce apoptosis, impairment of T cell receptor signaling, and cytotoxicity on T effector and natural killer (NK) lymphocytes or promote lymphocyte-suppressive activity through the release of Treg-promoting exosomes (54). In particular, tumor-derived vesicles can induce, expand, and up-regulate biological activities of Treg cells through an EV-associated TGF-β-mediated mechanism (55). Furthermore, although studies are still in their infancy, it also seems that bacteria can produce vesicles able to cross the epithelial barrier and interact with cells of the host connective tissue, primarily cells of the immune system (56).
In conclusion, to date extracellular vesicles are fully recognized as communication modules between and toward cells of the immune system and are regarded by some authors as important as cytokines and chemokines in regulating not only microenvironment cues but also cell targets at a distance (57). Besides the immune system, specific biological roles of EVs have been described in pregnancy, embryonic development, tissue repair, vascular biology, nervous system, bone calcification, and liver homeostasis, and EVs have been constantly detected in all mammalian (in particular human) body fluids investigated: blood, urine, saliva, synovial, bile, cerebrospinal fluid, bronco-alveolar fluid, amniotic fluid, breast milk, and seminal plasma (58). When characterizing EV composition, with the aim of identifying the functional players associated with these tiny messages, a component is invariably found: microRNA.
Extracellular Vesicle-associated MicroRNAs: Novel Players of Innate and Adaptive Immune Cell Function
Although miRNAs at the intracellular level are recognized as key players of gene expression regulation in eukaryotic cells (59), and also in cells of the immune system specifically (60, 61), the biological role of miRNAs released at the extracellular level has just started to be explored (Fig. 1). Valadi et al. (62) were the first to show that exosomes contain messengers and regulatory RNAs (including miRNAs) inside their membranous structures when neither DNA nor ribosomal RNA (18S and 28S rRNA) is detectable. Moreover, vesicle transfer was described as specific, given that mast cell-derived EVs were internalized by other mast cells but not by CD4+ lymphocytes. Since that first study, RNA (as well as miRNAs for that matter) has been consistently found in vesicles released by most cells investigated, from stem cells to neurons, hepatocytes, and blood cells, among others. Most of the attention has been devoted to pathological conditions, but several recent studies also demonstrate the physiological relevance of miRNA release. Some examples of miRNAs in EVs as a cell-to-cell communication pathway are the regulation of hematopoiesis in the bone marrow (63), muscle cell differentiation (64), and the crosstalk between neurons and astrocytes (65).
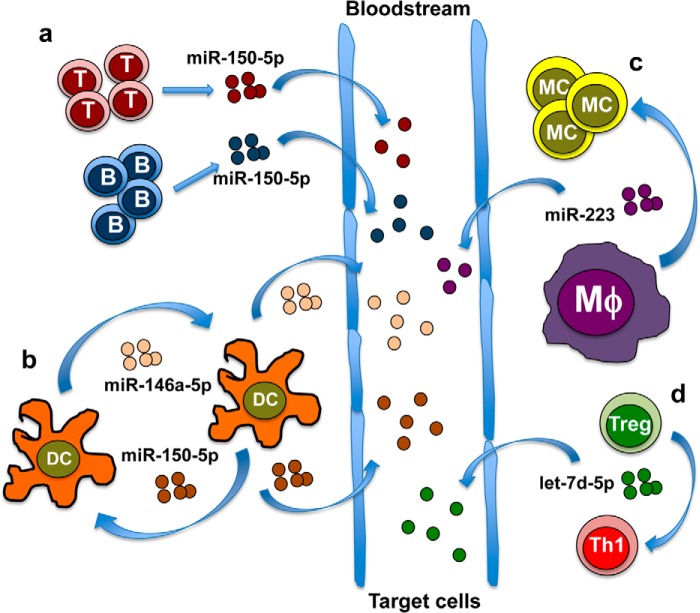
FIGURE 1.
Examples of EV-associated miRNAs relevant in the cross talk of immune cells. a, both B and T lymphocytes release EV-associated miR-150, which increases significantly in blood upon immune system activation. b, dendritic cell-derived miR-155 enhances whereas miR-146a reduces inflammatory response. c, mature macrophages release EV-associated miR-223, which induces the differentiation of recruited monocytes. d, regulatory T cells release Let-7d, which affects Th1 cells, halting their proliferation and IFNγ secretion. Whether EV-associated miRNAs functioning at a paracrine level may also be released in the bloodstream and which cells would be targeted is still largely unknown. Abbreviations: T = T cells; B = B cells; DC = dendritic cells; MC = monocytes; Mφ = macrophage.
An increasing number of studies have demonstrated that EV-associated miRNAs have a direct role in the immune system. During γ-herpesvirus EBV infection, the passage of functional viral miRNAs from infected to non-infected B cells via EVs is an additional mechanism adopted by the virus to control immune cells of the host and gain persistent infection (66). EBV may not be an isolated case. HIV-encoded trans-activating response element miRNA has been detected within the exosomes of HIV-infected T lymphocytes, together with the host miRNA machinery proteins Dicer and Drosha. Notably, exposure of naive T cells to exosomes from infected cells increased susceptibility of the recipient cells to HIV-1 infection and exosome-associated trans-activating response RNA down-regulated Bim and Cdk9 proteins in recipient cells, thus lowering apoptosis induction (67). In inflammatory conditions, mature macrophages produce and release EVs at high concentrations that are able to induce phenotypic differentiation and functional maturation of monocytes into other macrophages. Importantly, these EVs are enriched with miR-223, which is necessary for vesicle-induced monocyte maturation, and thus it is part of a feedback mechanism that works to induce the differentiation of recruited monocytes and the release of more vesicle as a local response activating the native immune system (68) (Fig. 1).
During the formation of an immune synapse, EVs released by T cells contain miRNAs that are transferred to the antigen-presenting cells in a unidirectional and antigen-driven fashion and are capable of modulating gene expression in recipient cells (69, 70). There are two important features of this type of communication: (i) the miRNA repertoire of vesicles differs significantly from that of parental cells, suggesting that the selection of secreted miRNAs may be highly regulated; and (ii) cell cognate interaction that forms upon the establishment of an immune synapse is necessary to promote the release of vesicles on one side, and to induce the fusion of these vesicles with the plasma membrane of the recipient cell on the other, demonstrating that the transfer is finely controlled.
EV-associated miRNAs have also been shown to be different in mature versus immature dendritic cells and to actively participate in the crosstalk of these cells for the fine-tuning of antigen-presenting cells and the immune response (71). Dendritic cells do communicate through vesicle exchange with different miRNAs having different and specific biological roles: in particular, EV-delivered miR-155 enhances whereas miR-146a reduces inflammatory response, by mediating target gene repression and reprogramming the cellular response to endotoxin (72) (Fig. 1).
This mechanism of cell communication is envisaged to occur in a paracrine environment, with vesicles traveling very short distances or even passing from one cell to the other upon cell-to-cell direct interaction. However, EV-associated miRNAs can also travel long distances and become endocrine messages (73). In accordance with this hypothesis, both B cells and T cells release EV-associated miR-150, which increases significantly in the blood of humans after vaccination with influenza virus and correlates with a higher immune response (74). In contrast, the level of extracellular miR-150 decreases in the blood of septic patients with an unfavorable outcome (75). Therefore, the modulation of circulating miR-150 may be an important regulatory loop aimed at down-modulating adaptive immune responses through the transmission of extracellular messages to other immune cells (also at distance) and the consequent regulation of miR-150 target genes (Fig. 1). Indeed, it has been demonstrated that vesicle-packaged miR-150 specifically regulates target gene expression and function in recipient cells (76). The described examples of communication via EV-associated miRNAs in the immune system are reported in Table 1.
TABLE 1.
Extracellular miRNAs with an identified role in immune cell communication
| Extracellular miRNA | Releasing cell | Target cell | Reference |
|---|---|---|---|
| miR-150–5p | T and B cells | Unknown | 74 |
| Let-7d-5p | Treg cell | Th1 cell | 88 |
| miR-150–5p | Monocyte | Endothelial cell | 76 |
| miR-223 | Macrophage | Monocyte | 68 |
| miR-146a-5p, miR-155–5p | Dendritic cell | Dendritic cell | 72 |
| HIV-encoded trans-activation response element miRNA | HIV-infected T cell | T cell | 67 |
| EBV-encoded miR-BART15 | EBV-infected B cell | Dendritic cell | 66 |
A long-distance miRNA-based communication can also occur between mother and child, with miRNAs traveling in milk during the months of lactation. Tolerosomes had already been described in breast milk, and shown to block allergic responses or prevent allergy development (77). More recently, human breast milk was found to be enriched with immune-related miRNAs (such as miR-17-92 cluster, known to be fundamental for the development of the immune system (78)) that may be transferred into the infant body via the digestive tract, and thus play a critical role in the development of the immune system in infants (79). It has been specifically hypothesized that milk miRNAs might induce long-term thymic Treg lineage commitment and down-regulate IL-4/Th2-mediated atopic sensitization by controlling pivotal target genes involved in the regulation of FoxP3 expression, IL-4 signaling, and immunoglobulin class switching (80).
Treg Cells and miRNAs: Fighting the Battle of Immune Tolerance at Both Intracellular and Extracellular Level
miRNA expression is necessary for the development of Treg cells in the thymus and the efficient induction of Foxp3 by TGF-β in a cell-autonomous fashion (81). Moreover, ectopic expression of the Treg cell signature transcription factor, FoxP3, in different cell lineages results in the acquisition of a Treg cell-specific miRNA profile, thus suggesting a key role for FoxP3 in the control of Treg cell intracellular miRNA content (81). Genome-wide analysis of its transcriptional binding sites revealed that Foxp3 directly activates several miRNAs, among which is miR-155, which is indispensable for Treg cells to normally respond to their key survival and growth factor, IL-2, but largely dispensable for Treg cell suppressor function (82). The expression of other miRNAs is necessary for Treg-mediated prevention of inflammation and autoimmunity, because the ablation of miRNA precursor-processing enzyme Dicer in Treg cells results in a fatal early-onset autoimmune pathology indistinguishable from that of Foxp3 KO mice (83, 84). In particular, Dicer-deficient Foxp3+ T cells completely lose their suppressive activity, even though their Foxp3 expression is comparable with that of control cells, thus suggesting that the impairment is secondary to a reduced expression of suppressor effector molecules rather than to an altered Foxp3 expression (83, 84). Strikingly, specific ablation of miR-146a alone in Treg cells is able to cause fatal lymphoproliferative diseases characterized by sharply augmented Th1 responses, secondary to an increased production of pro-inflammatory Th1 cytokines (IFNγ, above all) by both Treg and T conventional cells and to reduced suppressive capability of miR-146a-deficient Tregs (85).
miRNA expression is found altered in different autoimmune conditions. Divekar et al. (86) recently demonstrated that Tregs of systemic lupus erythematosus mice exhibit altered regulatory phenotype and reduced suppressive capacity and Dicer expression, together with a distinct miRNA profile. Moreover, miR-26a, an IL-6-associated miRNA that inhibits the generation of Th17 and promotes the generation of Tregs in vivo, thus functioning as a regulator of the Th17/Treg cell balance, has been found dramatically decreased both in experimental autoimmune encephalomyelitis mice and in subjects affected by multiple sclerosis (87).
Notably, a recent study on T cell subsets in mice demonstrated that Treg cells release EVs containing miRNAs that differ from those released by other effector cells and that the release is tightly regulated by distinct factors, such as IL-2, amphiregulin, the calcium ionophore, monensin, and hypoxia. More importantly, Treg cells deficient for Dicer (necessary for miRNA maturation) but also Treg cells deficient for Rab27 (necessary for vesicle release) are unable to suppress Th1, demonstrating that non-cell-autonomous gene silencing, mediated by miRNA-containing EVs, is indeed a required mechanism employed by Treg cells to suppress T-cell-mediated disease. In particular, the authors identified Let-7d as preferentially packaged and transferred to Th1 cells, halting Th1 cell proliferation and IFNγ secretion, and speculated that the local cytokine environment dictating the intracellular miRNA profile of Treg cells may also shape the extracellular EV-associated miRNA content (88). These findings suggest for the first time that non-cell-autonomous gene silencing, mediated by miRNA-containing EVs, is a mechanism that cooperates with other known Treg cell mechanisms to reinforce their suppressive function and halt T cell-mediated diseases.
The analysis of Treg-specific miR-146a KO mice described above has clearly suggested that one miRNA can be crucial in maintaining a certain optimal range of its targets, such as Stat1, whose deregulated activation leads to a severe failure of immunologic tolerance (85). However, whether miR-146a acts only at the intracellular or also at the extracellular level, through EV-associated passage from Treg cells to the Th1 effector counterpart, has not been investigated yet.
Moreover, it is important to underline that several cytokines and metabolic factors also regulate EV release by Tregs (88), but whether they can also shape their content is still unknown. It is possible that, together with a dominant cell-intrinsic role for miRNA secretion, the sensitivity of EV release to the microenvironment adds a further mechanism of plasticity to fine-tune specific T cell responses, thus ensuring immune tolerance.
Extracellular Vesicle-associated miRNAs: How Much Is Enough?
Purified populations of EVs usually contain many different miRNAs. Therefore, for the most part, EV-mediated miRNA transfer has the advantage over classical intercellular communication mediated by soluble factors wherein instead of one single messenger, a single vesicle may suppress functionally related genes in parallel, leading to a very effective paracrine control over neighboring cells (89). Unexpectedly, when a quantitative assessment of EV content of miRNAs was performed using a stoichiometric approach, it was found that, despite the biological source of the vesicles, the ratio between the number of miRNA molecules and the number of vesicles is lower than 1, even for the most abundant miRNAs, meaning that there are vesicles that do not contain any copy of them (90). This study may have discouraged people working in the field because the authors stated that miRNAs may be individually unlikely to be functional as communication vehicles. Instead, it is clear that the absolute number of miRNAs per vesicle is not the only parameter to be taken into account when evaluating the cell-to-cell communication potential of miRNA. If vesicle uptake is an infrequent but very selective event, the low-occupancy/high-concentration occupancy model (in which there are rare vesicles in the population carrying many copies of a given miRNA) may be consistent with vesicle-mediated communication through miRNA-mediated silencing of mRNAs. However, if cellular uptake of vesicles is rapid, miRNAs having a low-concentration/low-occupancy occupancy model (a small fraction of vesicles carrying a low concentration of miRNA) may also accumulate within the cell in physiologically relevant quantities, given that the minimum threshold in mammalian cells to repress a target mRNA is estimated to be 100 miRNA copies (91).
Recently, tumor-derived EVs were shown to contain pre-miRNAs along with Dicer and to possess cell-independent capacity to process precursors into mature miRNAs (92). If this capacity is not limited to tumor cell-derived vesicles but is a more general feature of EVs, then the quantification of mature miRNAs inside vesicles is not giving us the correct idea of the actual mRNA repression potential given by further miRNA maturation in vesicles themselves. Moreover, very low quantities of miRNAs may also be compatible with the recently proposed non-conventional activities, such as binding to Toll-like receptors as well as affecting DNA transcription and/or epigenetic states, that presumably require much lower concentrations of miRNA delivery than conventional mRNA targeting (98).
The majority of studies on the role of EVs in mediating RNA-based cell-to-cell communication (including the stoichiometric evaluation of miRNA quantity (90)) are based on the isolation of nanometric vesicles (50–200 nm in diameter), which excludes larger vesicles, such as microvesicles and oncosomes, that also contain miRNAs as well as longer molecules of RNAs. Therefore, it is conceivable that these larger EVs carry significantly higher numbers of miRNA molecules and function as an alternative vehicle for miRNA-based intercellular communication. Last, but not least, it is not clear what proportion of EV transcriptome consists of fragments of mRNAs, tRNAs, and other larger RNA molecules, which may play a regulatory function similarly to miRNAs (93), in addition to long interspersed elements and long terminal repeats (94). All these complex perspectives need to be further investigated.
The Shared Transcriptome: Single Cell Identity Goes Fuzzy
The most recent studies on symbiosis, defined as the mutually beneficial living arrangement of organisms belonging to different species, are shedding new lights on several aspects of these interactions and challenging the view of unitary individuals, genomes, and, at the end, life. Compartmentalization was an essential pillar in the evolutionary history of life as we know it (95), but living organisms have never forgotten the network environment in which they appeared. We are facing a paradigm shift in which our knowledge is progressing because we have abandoned the idea that biological systems have precise boundaries and embraced the concept of holobiont (96) and hologenome (97). Here we have seen that RNA is mobile and is exchanged among cells of the same organisms or of different organisms of the same species or of different species or even kingdoms. It is possible to envisage two main functions associated with sharing of the transcriptome: (i) regulation of target cell via non-coding regulatory RNAs (such as miRNAs); and ii) the buffering effect of transcriptome modulation to mount a coordinate response in the case of an initially de-synchronized cell population.
The growing knowledge on this ancient, universal, widespread, and possibly efficient mechanism of communication will eventually lead us to embrace the new concept of “holotranscriptome,” a sort of shared transcriptome with RNA molecules crossing the strict borders we impose to the biological unit of a cell.
Concluding Remarks
Networking has been crucial for life since the first steps ∼4 billion years ago, steps in which RNAs may have played and may still play a central role. A deeper analysis of RNA-based cell-to-cell communication will give us the tremendous opportunity to better understand relevant and broad biological issues, such as the ecological and physio-pathological relations between organisms and cells, with insight into infections, symbioses, nutrition, and diseases. Moreover, if human Treg cells will also be demonstrated to make use of EV-associated miRNAs to regulate immune tolerance, then the thorough analysis of this new potential pathogenic mechanism of immune system dysregulation may have an extraordinary impact on human health.
Acknowledgments
We thank Dolores Di Vizio, Fabio Grassi, and Nicola Manfrini for helpful discussions. We also thank Sara Ricciardi for the continuous support.
This work was supported by grants from the European Foundation for the Study of Diabetes/Juvenile Diabetes Research Foundation (EFSD/JDRF)/Lilly Programme 2015 and the Italian Space Agency (ASI) RadioEBV Grant 2014-033-RO (to G. M.). This work was also supported by the Ministero della Salute Grant GR-2010-2315414, the Fondo per gli Investimenti della Ricerca di Base (FIRB) Grant RBFR12I3UB_004, and the Fondazione Italiana Sclerosi Multipla (FISM) Grant 2014/R/21 (to V. D. R.). The authors declare that they have no conflicts of interest with the contents of this article.
- miRNA
- microRNA
- miR
- microRNA
- EV
- extracellular vesicle
- Treg
- regulatory T cell
- Th1
- T helper1 cells
- Foxp
- forkhead-box-P.
References
- 1. Mandel P., and Metais P. (1948) [The nucleic acids from blood plasma in humans]. C. R. Seances Soc. Biol. Fil. 142, 241–243 [PubMed] [Google Scholar]
- 2. Butt A. N., and Swaminathan R. (2008) Overview of circulating nucleic acids in plasma/serum. Ann. N.Y. Acad. Sci. 1137, 236–242 [DOI] [PubMed] [Google Scholar]
- 3. Lo Y. M. (2001) Circulating nucleic acids in plasma and serum: an overview. Ann. N.Y. Acad. Sci. 945, 1–7 [DOI] [PubMed] [Google Scholar]
- 4. Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K., Guo J., Zhang Y., Chen J., Guo X., Li Q., Li X., Wang W., Zhang Y., Wang J., et al. (2008) Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 18, 997–1006 [DOI] [PubMed] [Google Scholar]
- 6. Arroyo J. D., Chevillet J. R., Kroh E. M., Ruf I. K., Pritchard C. C., Gibson D. F., Mitchell P. S., Bennett C. F., Pogosova-Agadjanyan E. L., Stirewalt D. L., Tait J. F., and Tewari M. (2011) Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U.S.A. 108, 5003–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mitchell P. S., Parkin R. K., Kroh E. M., Fritz B. R., Wyman S. K., Pogosova-Agadjanyan E. L., Peterson A., Noteboom J., O'Briant K. C., Allen A., Lin D. W., Urban N., Drescher C. W., Knudsen B. S., Stirewalt D. L., et al. (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U.S.A. 105, 10513–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boeri M., Verri C., Conte D., Roz L., Modena P., Facchinetti F., Calabrò E., Croce C. M., Pastorino U., and Sozzi G. (2011) MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc. Natl. Acad. Sci. U.S.A. 108, 3713–3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moussay E., Wang K., Cho J. H., van Moer K., Pierson S., Paggetti J., Nazarov P. V., Palissot V., Hood L. E., Berchem G., and Galas D. J. (2011) MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U.S.A. 108, 6573–6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taylor D. D., and Gercel-Taylor C. (2008) MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 110, 13–21 [DOI] [PubMed] [Google Scholar]
- 11. D'Alessandra Y., Devanna P., Limana F., Straino S., Di Carlo A., Brambilla P. G., Rubino M., Carena M. C., Spazzafumo L., De Simone M., Micheli B., Biglioli P., Achilli F., Martelli F., Maggiolini S., et al. (2010) Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur. Heart J. 31, 2765–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goretti E., Vausort M., Wagner D. R., and Devaux Y. (2013) Association between circulating microRNAs, cardiovascular risk factors and outcome in patients with acute myocardial infarction. Int. J. Cardiol. 168, 4548–4550 [DOI] [PubMed] [Google Scholar]
- 13. Guay C., and Regazzi R. (2013) Circulating microRNAs as novel biomarkers for diabetes mellitus. Nat. Rev. Endocrinol. 9, 513–521 [DOI] [PubMed] [Google Scholar]
- 14. Starkey Lewis P. J., Dear J., Platt V., Simpson K. J., Craig D. G., Antoine D. J., French N. S., Dhaun N., Webb D. J., Costello E. M., Neoptolemos J. P., Moggs J., Goldring C. E., and Park B. K. (2011) Circulating microRNAs as potential markers of human drug-induced liver injury. Hepatology 54, 1767–1776 [DOI] [PubMed] [Google Scholar]
- 15. Chen Y. J., Zhu J. M., Wu H., Fan J., Zhou J., Hu J., Yu Q., Liu T. T., Yang L., Wu C. L., Guo X. L., Huang X. W., and Shen X. Z. (2013) Circulating microRNAs as a fingerprint for liver cirrhosis. PLoS One 8, e66577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Reid G., Kirschner M. B., and van Zandwijk N. (2011) Circulating microRNAs: association with disease and potential use as biomarkers. Crit. Rev. Oncol. Hematol. 80, 193–208 [DOI] [PubMed] [Google Scholar]
- 17. Friedman H. (1973) RNA in the immune response. Introductory remarks. Ann. N.Y. Acad. Sci. 207, 5–7 [DOI] [PubMed] [Google Scholar]
- 18. Archer S. J. (1978) Induction of a T-cell specific antigen on bone marrow lymphocytes with thymus RNA. Immunology 34, 123–129 [PMC free article] [PubMed] [Google Scholar]
- 19. Nakamura K., Nakamura Y., Kagawa H., and Kawahara M. (1979) The proliferation of plasma cells from mouse bone marrow in vitro. III. Primary and secondary immune responses associated with thymic RNA. Immunol Commun. 8, 511–529 [DOI] [PubMed] [Google Scholar]
- 20. Anker P., Stroun M., and Maurice P. A. (1975) Spontaneous release of DNA by human blood lymphocytes as shown in an in vitro system. Cancer Res. 35, 2375–2382 [PubMed] [Google Scholar]
- 21. Stroun M., Anker P., Beljanski M., Henri J., Lederrey C., Ojha M., and Maurice P. A. (1978) Presence of RNA in the nucleoprotein complex spontaneously released by human lymphocytes and frog auricles in culture. Cancer Res. 38, 3546–3554 [PubMed] [Google Scholar]
- 22. Anker P., Jachertz D., Stroun M., Brögger R., Lederrey C., Henri J., and Maurice P. A. (1980) The role of extracellular DNA in the transfer of information from T to B human lymphocytes in the course of an immune response. J. Immunogenet. 7, 475–481 [DOI] [PubMed] [Google Scholar]
- 23. Crick F. H. (1968) The origin of the genetic code. J. Mol. Biol. 38, 367–379 [DOI] [PubMed] [Google Scholar]
- 24. Orgel L. E. (1968) Evolution of the genetic apparatus. J. Mol. Biol. 38, 381–393 [DOI] [PubMed] [Google Scholar]
- 25. Napoli C., Lemieux C., and Jorgensen R. (1990) Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2, 279–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fire A., Xu S., Montgomery M. K., Kostas S. A., Driver S. E., and Mello C. C. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811 [DOI] [PubMed] [Google Scholar]
- 27. Obbard D. J., Gordon K. H., Buck A. H., and Jiggins F. M. (2009) The evolution of RNAi as a defence against viruses and transposable elements. Philos. Trans. R. Soc. Lond. B Biol. Sci. 364, 99–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Salgado P. S., Koivunen M. R., Makeyev E. V., Bamford D. H., Stuart D. I., and Grimes J. M. (2006) The structure of an RNAi polymerase links RNA silencing and transcription. PLoS Biol. 4, e434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liu H., Wang X., Wang H. D., Wu J., Ren J., Meng L., Wu Q., Dong H., Wu J., Kao T. Y., Ge Q., Wu Z. X., Yuh C. H., and Shan G. (2012) Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nat. Commun. 3, 1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Timmons L., and Fire A. (1998) Specific interference by ingested dsRNA. Nature 395, 854. [DOI] [PubMed] [Google Scholar]
- 31. Weiberg A., Wang M., Lin F. M., Zhao H., Zhang Z., Kaloshian I., Huang H. D., and Jin H. (2013) Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 342, 118–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garcia-Silva M. R., das Neves R. F., Cabrera-Cabrera F., Sanguinetti J., Medeiros L. C., Robello C., Naya H., Fernandez-Calero T., Souto-Padron T., de Souza W., and Cayota A. (2014) Extracellular vesicles shed by Trypanosoma cruzi are linked to small RNA pathways, life cycle regulation, and susceptibility to infection of mammalian cells. Parasitol. Res. 113, 285–304 [DOI] [PubMed] [Google Scholar]
- 33. Zhang L., Hou D., Chen X., Li D., Zhu L., Zhang Y., Li J., Bian Z., Liang X., Cai X., Yin Y., Wang C., Zhang T., Zhu D., Zhang D., et al. (2012) Exogenous plant MIR168a specifically targets mammalian LDLRAP1: evidence of cross-kingdom regulation by microRNA. Cell Res. 22, 107–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Y., Wiggins B. E., Lawrence C., Petrick J., Ivashuta S., and Heck G. (2012) Analysis of plant-derived miRNAs in animal small RNA datasets. BMC Genomics 13, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Svenningsen S. L., Tu K. C., and Bassler B. L. (2009) Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. The EMBO J. 28, 429–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tu K. C., Long T., Svenningsen S. L., Wingreen N. S., and Bassler B. L. (2010) Negative feedback loops involving small regulatory RNAs precisely control the Vibrio harveyi quorum-sensing response. Mol. Cell 37, 567–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Knip M., Constantin M. E., and Thordal-Christensen H. (2014) Trans-kingdom cross-talk: small RNAs on the move. PLoS Genet. 10, e1004602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stoorvogel W., Kleijmeer M. J., Geuze H. J., and Raposo G. (2002) The biogenesis and functions of exosomes. Traffic 3, 321–330 [DOI] [PubMed] [Google Scholar]
- 39. Raposo G., and Stoorvogel W. (2013) Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Johnstone R. M., Adam M., Hammond J. R., Orr L., and Turbide C. (1987) Vesicle formation during reticulocyte maturation: association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 262, 9412–9420 [PubMed] [Google Scholar]
- 41. Raposo G., Nijman H. W., Stoorvogel W., Liejendekker R., Harding C. V., Melief C. J., and Geuze H. J. (1996) B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 183, 1161–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Théry C., Duban L., Segura E., Véron P., Lantz O., and Amigorena S. (2002) Indirect activation of naive CD4+ T cells by dendritic cell-derived exosomes. Nat. Immunol. 3, 1156–1162 [DOI] [PubMed] [Google Scholar]
- 43. Blanchard N., Lankar D., Faure F., Regnault A., Dumont C., Raposo G., and Hivroz C. (2002) TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/ζ complex. J. Immunol. 168, 3235–3241 [DOI] [PubMed] [Google Scholar]
- 44. Wahlgren J., Karlson Tde L., Glader P., Telemo E., and Valadi H. (2012) Activated human T cells secrete exosomes that participate in IL-2 mediated immune response signaling. PLoS One 7, e49723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim S. H., Bianco N. R., Shufesky W. J., Morelli A. E., and Robbins P. D. (2007) MHC class II+ exosomes in plasma suppress inflammation in an antigen-specific and Fas ligand/Fas-dependent manner. J. Immunol. 179, 2235–2241 [DOI] [PubMed] [Google Scholar]
- 46. MacKenzie A., Wilson H. L., Kiss-Toth E., Dower S. K., North R. A., and Surprenant A. (2001) Rapid secretion of interleukin-1β by microvesicle shedding. Immunity 15, 825–835 [DOI] [PubMed] [Google Scholar]
- 47. Qu Y., Franchi L., Nunez G., and Dubyak G. R. (2007) Nonclassical IL-1β secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. J. Immunol. 179, 1913–1925 [DOI] [PubMed] [Google Scholar]
- 48. Kandere-Grzybowska K., Letourneau R., Kempuraj D., Donelan J., Poplawski S., Boucher W., Athanassiou A., and Theoharides T. C. (2003) IL-1 induces vesicular secretion of IL-6 without degranulation from human mast cells. J. Immunol. 171, 4830–4836 [DOI] [PubMed] [Google Scholar]
- 49. Gulinelli S., Salaro E., Vuerich M., Bozzato D., Pizzirani C., Bolognesi G., Idzko M., Di Virgilio F., and Ferrari D. (2012) IL-18 associates to microvesicles shed from human macrophages by a LPS/TLR-4 independent mechanism in response to P2X receptor stimulation. Eur. J. Immunol. 42, 3334–3345 [DOI] [PubMed] [Google Scholar]
- 50. Zhang H. G., Liu C., Su K., Yu S., Zhang L., Zhang S., Wang J., Cao X., Grizzle W., and Kimberly R. P. (2006) A membrane form of TNF-α presented by exosomes delays T cell activation-induced cell death. J. Immunol. 176, 7385–7393 [DOI] [PubMed] [Google Scholar]
- 51. Karlsson M., Lundin S., Dahlgren U., Kahu H., Pettersson I., and Telemo E. (2001) “Tolerosomes” are produced by intestinal epithelial cells. Eur. J. Immunol. 31, 2892–2900 [DOI] [PubMed] [Google Scholar]
- 52. Pêche H., Renaudin K., Beriou G., Merieau E., Amigorena S., and Cuturi M. C. (2006) Induction of tolerance by exosomes and short-term immunosuppression in a fully MHC-mismatched rat cardiac allograft model. Am. J. Transplant. 6, 1541–1550 [DOI] [PubMed] [Google Scholar]
- 53. Wang G. J., Liu Y., Qin A., Shah S. V., Deng Z. B., Xiang X., Cheng Z., Liu C., Wang J., Zhang L., Grizzle W. E., and Zhang H. G. (2008) Thymus exosomes-like particles induce regulatory T cells. J. Immunol. 181, 5242–5248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Filipazzi P., Bürdek M., Villa A., Rivoltini L., and Huber V. (2012) Recent advances on the role of tumor exosomes in immunosuppression and disease progression. Semin. Cancer Biol. 22, 342–349 [DOI] [PubMed] [Google Scholar]
- 55. Szajnik M., Czystowska M., Szczepanski M. J., Mandapathil M., and Whiteside T. L. (2010) Tumor-derived microvesicles induce, expand and up-regulate biological activities of human regulatory T cells (Treg). PLoS One 5, e11469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhang G., Ducatelle R., Pasmans F., D'Herde K., Huang L., Smet A., Haesebrouck F., and Flahou B. (2013) Effects of Helicobacter suis γ-glutamyl transpeptidase on lymphocytes: modulation by glutamine and glutathione supplementation and outer membrane vesicles as a putative delivery route of the enzyme. PLoS One 8, e77966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Simons M., and Raposo G. (2009) Exosomes: vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 21, 575–581 [DOI] [PubMed] [Google Scholar]
- 58. Yáñez-Mó M., Siljander P. R., Andreu Z., Zavec A. B., Borràs F. E., Buzas E. I., Buzas K., Casal E., Cappello F., Carvalho J., Colás E., Cordeiro-da Silva A., Fais S., Falcon-Perez J. M., Ghobrial I. M., et al. (2015) Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 4, 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. He L., and Hannon G. J. (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522–531 [DOI] [PubMed] [Google Scholar]
- 60. Xiao C., and Rajewsky K. (2009) MicroRNA control in the immune system: basic principles. Cell 136, 26–36 [DOI] [PubMed] [Google Scholar]
- 61. Bronevetsky Y., and Ansel K. M. (2013) Regulation of miRNA biogenesis and turnover in the immune system. Immunol. Rev. 253, 304–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J. J., and Lötvall J. O. (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 [DOI] [PubMed] [Google Scholar]
- 63. Salvucci O., Jiang K., Gasperini P., Maric D., Zhu J., Sakakibara S., Espigol-Frigole G., Wang S., and Tosato G. (2012) MicroRNA126 contributes to granulocyte colony-stimulating factor-induced hematopoietic progenitor cell mobilization by reducing the expression of vascular cell adhesion molecule 1. Haematologica 97, 818–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Forterre A., Jalabert A., Chikh K., Pesenti S., Euthine V., Granjon A., Errazuriz E., Lefai E., Vidal H., and Rome S. (2014) Myotube-derived exosomal miRNAs downregulate Sirtuin1 in myoblasts during muscle cell differentiation. Cell Cycle 13, 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Morel L., Regan M., Higashimori H., Ng S. K., Esau C., Vidensky S., Rothstein J., and Yang Y. (2013) Neuronal exosomal miRNA-dependent translational regulation of astroglial glutamate transporter GLT1. J. Biol. Chem. 288, 7105–7116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pegtel D. M., Cosmopoulos K., Thorley-Lawson D. A., van Eijndhoven M. A., Hopmans E. S., Lindenberg J. L., de Gruijl T. D., Würdinger T., and Middeldorp J. M. (2010) Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. U.S.A. 107, 6328–6333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Narayanan A., Iordanskiy S., Das R., Van Duyne R., Santos S., Jaworski E., Guendel I., Sampey G., Dalby E., Iglesias-Ussel M., Popratiloff A., Hakami R., Kehn-Hall K., Young M., Subra C., et al. (2013) Exosomes derived from HIV-1-infected cells contain trans-activation response element RNA. J. Biol. Chem. 288, 20014–20033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Ismail N., Wang Y., Dakhlallah D., Moldovan L., Agarwal K., Batte K., Shah P., Wisler J., Eubank T. D., Tridandapani S., Paulaitis M. E., Piper M. G., and Marsh C. B. (2013) Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood 121, 984–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Mittelbrunn M., Gutiérrez-Vázquez C., Villarroya-Beltri C., González S., Sánchez-Cabo F., González M. Á., Bernad A., and Sánchez-Madrid F. (2011) Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Gutiérrez-Vázquez C., Villarroya-Beltri C., Mittelbrunn M., and Sánchez-Madrid F. (2013) Transfer of extracellular vesicles during immune cell-cell interactions. Immunol. Rev. 251, 125–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Montecalvo A., Larregina A. T., Shufesky W. J., Stolz D. B., Sullivan M. L., Karlsson J. M., Baty C. J., Gibson G. A., Erdos G., Wang Z., Milosevic J., Tkacheva O. A., Divito S. J., Jordan R., Lyons-Weiler J., et al. (2012) Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 119, 756–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Alexander M., Hu R., Runtsch M. C., Kagele D. A., Mosbruger T. L., Tolmachova T., Seabra M. C., Round J. L., Ward D. M., and O'Connell R. M. (2015) Exosome-delivered microRNAs modulate the inflammatory response to endotoxin. Nat. Commun. 6, 7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cortez M. A., Bueso-Ramos C., Ferdin J., Lopez-Berestein G., Sood A. K., and Calin G. A. (2011) MicroRNAs in body fluids: the mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 8, 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. de Candia P., Torri A., Gorletta T., Fedeli M., Bulgheroni E., Cheroni C., Marabita F., Crosti M., Moro M., Pariani E., Romanò L., Esposito S., Mosca F., Rossetti G., Rossi R. L., et al. (2013) Intracellular modulation, extracellular disposal and serum increase of MiR-150 mark lymphocyte activation. PLoS One 8, e75348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Roderburg C., Luedde M., Vargas Cardenas D., Vucur M., Scholten D., Frey N., Koch A., Trautwein C., Tacke F., and Luedde T. (2013) Circulating microRNA-150 serum levels predict survival in patients with critical illness and sepsis. PLoS One 8, e54612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhang Y., Liu D., Chen X., Li J., Li L., Bian Z., Sun F., Lu J., Yin Y., Cai X., Sun Q., Wang K., Ba Y., Wang Q., Wang D., et al. (2010) Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol. Cell 39, 133–144 [DOI] [PubMed] [Google Scholar]
- 77. Admyre C., Telemo E., Almqvist N., Lötvall J., Lahesmaa R., Scheynius A., and Gabrielsson S. (2008) Exosomes: nanovesicles with possible roles in allergic inflammation. Allergy 63, 404–408 [DOI] [PubMed] [Google Scholar]
- 78. Mendell J. T. (2008) miRiad roles for the miR-17–92 cluster in development and disease. Cell 133, 217–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhou Q., Li M., Wang X., Li Q., Wang T., Zhu Q., Zhou X., Wang X., Gao X., and Li X. (2012) Immune-related microRNAs are abundant in breast milk exosomes. Int. J. Biol. Sci. 8, 118–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Melnik B. C., John S. M., and Schmitz G. (2014) Milk: an exosomal microRNA transmitter promoting thymic regulatory T cell maturation preventing the development of atopy? J. Transl. Med. 12, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Cobb B. S., Hertweck A., Smith J., O'Connor E., Graf D., Cook T., Smale S. T., Sakaguchi S., Livesey F. J., Fisher A. G., and Merkenschlager M. (2006) A role for Dicer in immune regulation. J. Exp. Med. 203, 2519–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lu L. F., Thai T. H., Calado D. P., Chaudhry A., Kubo M., Tanaka K., Loeb G. B., Lee H., Yoshimura A., Rajewsky K., and Rudensky A. Y. (2009) Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity 30, 80–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chong M. M., Rasmussen J. P., Rudensky A. Y., and Littman D. R. (2008) The RNAseIII enzyme Drosha is critical in T cells for preventing lethal inflammatory disease. J. Exp. Med. 205, 2005–2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Liston A., Lu L. F., O'Carroll D., Tarakhovsky A., and Rudensky A. Y. (2008) Dicer-dependent microRNA pathway safeguards regulatory T cell function. J. Exp. Med. 205, 1993–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Lu L. F., Boldin M. P., Chaudhry A., Lin L. L., Taganov K. D., Hanada T., Yoshimura A., Baltimore D., and Rudensky A. Y. (2010) Function of miR-146a in controlling Treg cell-mediated regulation of Th1 responses. Cell 142, 914–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Divekar A. A., Dubey S., Gangalum P. R., and Singh R. R. (2011) Dicer insufficiency and microRNA-155 overexpression in lupus regulatory T cells: an apparent paradox in the setting of an inflammatory milieu. J. Immunol. 186, 924–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhang R., Tian A., Wang J., Shen X., Qi G., and Tang Y. (2015) miR26a modulates Th17/T reg balance in the EAE model of multiple sclerosis by targeting IL6. Neuromolecular Med. 17, 24–34 [DOI] [PubMed] [Google Scholar]
- 88. Okoye I. S., Coomes S. M., Pelly V. S., Czieso S., Papayannopoulos V., Tolmachova T., Seabra M. C., and Wilson M. S. (2014) MicroRNA-containing T-regulatory-cell-derived exosomes suppress pathogenic T helper 1 cells. Immunity 41, 89–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lotvall J., and Valadi H. (2007) Cell to cell signalling via exosomes through esRNA. Cell Adh. Migr. 1, 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Chevillet J. R., Kang Q., Ruf I. K., Briggs H. A., Vojtech L. N., Hughes S. M., Cheng H. H., Arroyo J. D., Meredith E. K., Gallichotte E. N., Pogosova-Agadjanyan E. L., Morrissey C., Stirewalt D. L., Hladik F., Yu E. Y. (2014) Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc. Natl. Acad. Sci. U.S.A. 111, 14888–14893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Brown B. D., Gentner B., Cantore A., Colleoni S., Amendola M., Zingale A., Baccarini A., Lazzari G., Galli C., and Naldini L. (2007) Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 25, 1457–1467 [DOI] [PubMed] [Google Scholar]
- 92. Melo S. A., Sugimoto H., O'Connell J. T., Kato N., Villanueva A., Vidal A., Qiu L., Vitkin E., Perelman L. T., Melo C. A., Lucci A., Ivan C., Calin G. A., and Kalluri R. (2014) Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Batagov A. O., and Kurochkin I. V. (2013) Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3′-untranslated regions. Biol. Direct 8, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Nolte-'t Hoen E. N., Buermans H. P., Waasdorp M., Stoorvogel W., Wauben M. H., and 't Hoen P. A. (2012) Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 40, 9272–9285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Koshland D. E., Jr. (2002) The seven pillars of life. Science 295, 2215–2216 [DOI] [PubMed] [Google Scholar]
- 96. Mindell D. P. (1992) Phylogenetic consequences of symbioses: Eukarya and Eubacteria are not monophyletic taxa. Biosystems 27, 53–62 [DOI] [PubMed] [Google Scholar]
- 97. Zilber-Rosenberg I., and Rosenberg E. (2008) Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol. Rev. 32, 723–735 [DOI] [PubMed] [Google Scholar]
- 98. Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., Lovat F., Fadda P., Mao C., Nuovo G. J., Zanesi N., Crawford M., Ozer G. H., Wernicke D., Alder H., et al. (2012) MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 109, E2110–E2116 [DOI] [PMC free article] [PubMed] [Google Scholar]