Abstract
Hexokinase 2 (Hxk2) from Saccharomyces cerevisiae is a bi-functional enzyme, being both a catalyst in the cytosol and an important regulator of the glucose repression signal in the nucleus. Despite considerable recent progress, little is known about the regulatory mechanism that controls nuclear Hxk2 association with the SUC2 promoter chromatin and how this association is necessary for SUC2 gene repression. Our data indicate that in the SUC2 promoter context, Hxk2 functions through a variety of structurally unrelated factors, mainly the DNA-binding Mig1 and Mig2 repressors and the regulatory Snf1 and Reg1 factors. Hxk2 sustains the repressor complex architecture maintaining transcriptional repression at the SUC2 gene. Using chromatin immunoprecipitation assays, we discovered that the Hxk2 in its open configuration, at low glucose conditions, leaves the repressor complex that induces its dissociation and promotes SUC2 gene expression. In high glucose conditions, Hxk2 adopts a close conformation that promotes Hxk2 binding to the Mig1 protein and the reassembly of the SUC2 repressor complex. Additional findings highlight the possibility that Hxk2 constitutes an intracellular glucose sensor that operates by changing its conformation in response to cytoplasmic glucose levels that regulate its interaction with Mig1 and thus its recruitment to the repressor complex of the SUC2 promoter. Thus, our data indicate that Hxk2 is more intimately involved in gene regulation than previously thought.
Keywords: gene expression, glucose, hexokinase, repressor protein, Saccharomyces cerevisiae, glucose signaling
Introduction
Transcriptional repressors bind to cis-acting elements of gene promoter regions to repress transcription by altering chromatin structure and regulating RNA polymerase II accumulation in the promoter (1). Thus, transcriptional repression involves the coordinated binding of several proteins in the promoter region at specific upstream repressing sequences (URS)4 to form a repressor complex. This repressor complex also interacts with other corepressors to regulate gene expression (2).
The Saccharomyces cerevisiae Tup1 protein is a global corepressor with WD40 repeats that can interact with Ssn6 (3, 4), and it has been suggested to be a potential yeast orthologue of the Groucho family of corepressors (5). The Tup family transcriptional corepressors are conserved between yeast and humans and regulate gene expression during stress response and cellular differentiation. S. cerevisiae Tup1 represses several genes regulated by glucose, oxidative stress, DNA damage, and other cellular stress responses (6). In the case of several genes regulated by glucose repression, Tup1 is recruited specifically to different promoters by the DNA-binding proteins Mig1 and Mig2 (7–9) to generate a glucose repression state of gene expression. Tup1 represses the expression of genes via distinct mechanisms as follows: by establishing a repressive chromatin structure around the target gene promoter, by recruiting histone deacetylases, and by directly interfering with the general transcription machinery (10, 11). However, the precise molecular mechanism of action of Tup1 family proteins in gene repression induced by high glucose levels has not been fully resolved. This is probably because the mechanism depends on the kinetics of formation and dissociation of the repressor complex, which in the case of glucose repression signaling is not well characterized.
Glucose is not only a fuel that serves as a preferential substrate for energy yielding metabolism but also functions as a signaling molecule that regulates the central pathways of carbohydrate metabolism. Although regulation of glucose repression is relatively well understood and the proteins that carry out this regulation have been studied extensively (12), the glucose signaling mechanism and the factors involved in the repressor complex structures at high and low glucose conditions are scarcely characterized. In high glucose conditions, the repressor protein Mig1 is the main transcription factor responsible for the repression of genes needed for utilization of alternative fermentable carbon sources (7, 12). Mig1 binds to DNA and inhibits transcription of SUC2, plus ∼350 other genes (13, 14), but it has been demonstrated that in several cases this process also requires the Hxk2 protein (15–17). Expression of the HXK2 gene is controlled by glucose availability and is mediated by the Rgt1 and Med8 transcription factors, which repress HXK2 expression in low-glucose-containing media (18, 19). Moreover, Hxk2 is also involved in the glucose-induced repression of the HXK1 gene, whereas the Hxk1 protein acts as a negative factor in the HXK2 gene expression (15). Transcriptional analysis of a Δhxk2 mutant strain showed significant up-regulation of genes with binding sites for Mig1 and/or Cat8 (15, 20, 21). In addition, Hxk2 also participates in the control of genes encoding sugar transporters (22) and genes modulating mitochondrial cytochrome content and respiratory activity (23). This is also supported by a diminished Crabtree effect in Δhxk2 mutants, resulting in a nearly complete respiratory metabolism at high glucose concentration (20). Thus, Hxk2 protein can be regarded as a global regulator of carbon metabolism that is essential for mediating the glucose repression signal.
In S. cerevisiae, Hxk2 is the predominant glucose kinase in cells growing in high glucose conditions and has dual functions (24, 25). It is a glycolytic enzyme, essential for cell energy metabolism in the cytoplasm, but also acts as a regulator of gene transcription in the nucleus (26, 27). To carry out their functions, Hxk2 has to shuttle in and out of the nucleus, but the protein is too large to translocate through the nuclear pore complex by diffusion. Thus, the Hxk2 transport across the nuclear envelope must be a mediated and regulated process. Recently, the carrier proteins involved in the Hxk2 transport across the nuclear envelope have been identified. The mechanisms by which Hxk2 enters and exits the nucleus are mediated by the α/β-importin (Kap60/Kap95) pathway (28) and the Xpo1 (Crm1) carrier protein (29), respectively. The directionality of transport is regulated by the guanine nucleotide-binding protein Gsp1 (28, 29), and the Snf1 kinase-Glc7 phosphatase protein pair works together to control the phosphorylation state of serine 14 of Hxk2 and thus its nucleocytoplasmic distribution (30).
Snf1 kinase, a homologue of mammalian AMP-activated kinase, also plays a central role in regulating the glucose repression signaling pathway. The Snf1 kinase, under low glucose conditions, is activated and modifies the phosphorylation state of Hxk2 (30) and Mig1 (31). Snf1 kinase forms a complex with an activating subunit Snf4 and one of the three Snf1-interacting proteins Sip1, Sip2, or Gal83, which target Snf1 to distinct subcellular locations (32). Regulation of Snf1 activity involves phosphorylation of the Snf1 catalytic subunit at threonine 210 by one of the three kinases, Sak1, Tos3, or Elm1 (33–35). ADP also appears to play a significant role in Snf1 activation in response to glucose limitation by protecting the enzyme against dephosphorylation by the Glc7-Reg1 phosphatase (36). In addition, high glucose levels inactivate Snf1 by stimulating the activity of the Glc7-Reg1 phosphatase, located in the cytosol (37–39). The activated Snf1 protein kinase phosphorylates at least four serine residues of the Mig1 protein in low glucose growth conditions (37). Nevertheless, only phosphorylation of the serine 311 in Mig1 has been shown to be enough to inhibit Mig1 repressor activity and to induce translocation of the protein from the nucleus to the cytoplasm (40). Thus, the activity of Mig1 is regulated by serine 311 phosphorylation and subcellular localization. In yeast, the regulatory subunit that targets the Glc7 protein phosphatase to Mig1 (41) and Hxk2 (30) in the glucose repression pathway is Reg1. In high glucose, Mig1 and Hxk2 are dephosphorylated by the Glc7-Reg1 protein phosphatase complex (30, 42) and are found in the nucleus, where they can repress transcription.
The SUC2 structural gene for invertase provides an attractive system to study glucose repression because glucose repression appears to be the only regulatory mechanism affecting the expression of SUC2 (43). Therefore, we used the invertase level as a marker for glucose repression and the SUC2 system as a good model to understand how Mig1 and Hxk2 proteins control gene expression and how these regulatory factors, together with others, participate in the structure and dynamics of the repressor complex.
Here, we demonstrate that under high glucose conditions Hxk2 is a critical factor for the stabilization of the SUC2 repressor complex. The presence of Hxk2 in the repressor complex is required to inhibit Mig1 phosphorylation by the Snf1 kinase (30). In low glucose conditions, the Hxk2 interaction with Mig1 is abolished, perhaps by Snf1-dependent Hxk2 phosphorylation (40), and a transient increment in Snf1 and Mig1 interaction is detected (37). These interaction patterns stimulate Mig1 and Hxk2 phosphorylation by Snf1 kinase at serine 311 (40) and serine 14 (30), respectively. Phosphorylation of Hxk2 and Mig1 results in the export of these proteins from nucleus to cytoplasm and the disassembly of the repressor complex at the SUC2 promoter. In this study, we also establish that besides Mig1 and Hxk2, the regulatory proteins Mig2, Snf1, Snf4, Gal83, and Reg1 are also part of the repressor complex at the SUC2 gene promoter both at high and low glucose conditions.
However, the following two questions have not been answered in an appropriate manner until now. How is the glucose level detected? How is this information transmitted to the nucleus to generate a coordinated cellular response? Recently, FRET sensor assays have proven to be useful in determining the cytosolic glucose levels in yeast. Quantitative in vivo measurement of cytosolic glucose shows a concentration of ∼10 mm in yeast cells growing in complex or synthetic media containing 50–100 mm glucose (44). Thus, in a high glucose-containing medium Hxk2 will be saturated with cytosolic glucose in a closed conformation (45). In contrast, in low glucose media Hxk2 has an open conformation (45). We describe here that the entry or exit of the Hxk2 protein to the SUC2 repressor complex is not regulated by phosphorylation but is regulated by the cytosolic glucose levels that ultimately determine the open or closed active site conformation of the Hxk2 protein. Thus, we present Hxk2 as an intracellular glucose sensor involved in signaling glucose levels to generate a coordinated transcriptional response.
Experimental Procedures
Strains and Growth Conditions
The S. cerevisiae strains used throughout this study were derived from W303-1A (46), DBY1315 (47), and BY4741 (Euroscarf) haploid wild-type strains and are listed in Table 1. Strains FMY350 and FMY351 expressing Snf1-HA were constructed, respectively, by homologous recombination in Y14403 and Y04620 strains using an HA-HIS3 tagging cassette obtained from the pFA6a-3HA-HIS3MX6 plasmid (48). Strains FMY403 and FMY833 expressing, respectively, Snf4-HA and Gal83-HA were constructed by homologous recombination in W303-1A strain using an HA-TRP1 tagging cassette obtained from pFA6a-3HA-HIS3MX6 plasmid (48). Strains FMY320, FMY321, and FMY322 expressing Mig1-GFP were constructed, respectively, by homologous recombination in W303-1A, Y04620 (Δhxk2), and DBY2052 (Δhxk1Δhxk2) strains using a GFP-HIS3 tagging cassette obtained from pFA6a-GFP-HIS3MX6 plasmid in the two first strains and a GFP-KanMX6 tagging cassette obtained from pFA6a-GFP-KanMX6 plasmid in the last strain (48). Strains FMY501, FMY507, and FMY509 expressing Mig2-GFP were constructed, respectively, by homologous recombination in W303-1A, H174 (Δmig1), and Y04620 (Δhxk2) strains using a GFP-HIS3 tagging cassette obtained from pFA6a-GFP-HIS3MX6 plasmid (48). Strains FMY901, FMY902, and FMY903 expressing Reg1-GFP were constructed, respectively, by homologous recombination in W303-1A, Y14403 (Δmig1), and Y04620 (Δhxk2) strains using a GFP-HIS3 tagging cassette obtained from pFA6a-GFP-HIS3MX6 plasmid (48).
TABLE 1.
S. cerevisiae strains used in this study
| Name | Relevant genotype | Source/Ref. |
|---|---|---|
| W303-1A | MATα ura3-52 trp1-289 leu2-3,112 his3-Δ1 ade2-1 can1-100 | 46 |
| DBY1315 | MATα ura3-52 leu2-3,2-112 lys2-801 gal2 | 47 |
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| DBY2052 | MATα ura3-52 leu2-3,2-112 lys2-801 gal2 hxk1::LEU2 hxk2-202 | 47 |
| DBY2053 | MATα ura3-52 leu2-3,2-112 lys2-801 gaI2 hxk1::LEU2 | 47 |
| MAP24 | MATα can1-100 his3-11,15 leu2-3,112 trp1-1 ura 3-1 mig1::loxp mig2::loxp-KAN-lox | 69 |
| H174 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 mig1–6J::LEU2 | 7 |
| FMY301 | MATα ura3-52 leu2-3,2-112 lys2-801 gal2 mig1::LEU2 hxk2-202 | 27 |
| FMY303 | MATα ura3-52 trp1-289 leu2-3,112 his3-Δ1 ade2-1 can1-100 SNF1-HA | 40 |
| FMY350 | MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 mig1::kanMX4 SNF1-HA | This work |
| FMY351 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxk2::kanMX4 SNF1-HA | This work |
| FMY403 | MATα ura3-52 trp1-289 leu2-3,112 his3-Δ1 ade2-1 can1-100 SNF4-HA | This work |
| FMY481 | MATα ura3-52 trp1-289 leu2-3,112 his3-Δ1 ade2-1 can1-100 snf1::kanMX4 SNF4-HA | This work |
| FMY833 | MATα ura3-52 trp1-289 leu2-3,112 his3-Δ1 ade2-1 can1-100 GAL83-HA | This work |
| FMY320 | MATα ura3-52 trp1-289 leu2-3,112 his3-Δ1 ade2-1 can1-100 MIG1-GFP | This work |
| FMY321 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxk2::kanMX4 MIG1-GFP | This work |
| FMY322 | MATα ura3-52 leu2-3,2-112 lys2-801 gal2 hxk1::LEU2 hxk2-202 MIG1-GFP | This work |
| FMY501 | MATα ura3-52 trp1-289 leu2-3,112 his3-Δ1 ade2-1 can1-100 MIG2-GFP | 70 |
| FMY507 | MATα ade2-1 canJ-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 mig1–6J::LEU2 MIG2-GFP | This work |
| FMY509 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxk2::kanMX4 MIG2-GFP | This work |
| FMY901 | MATα ura3-52 trp1-289 leu2-3,112 his3-Δ1 ade2-1 can1-100 REG1-GFP | This work |
| FMY902 | MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 mig1::kanMX4 REG1-GFP | This work |
| FMY903 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxk2::kanMX4 REG1-GFP | This work |
| Y04620 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hxk2::kanMX4 | Euroscarf |
| Y14403 | MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 mig1::kanMX4 | Euroscarf |
| Y14575 | MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 mig2::kanMX4 | Euroscarf |
| Y14311 | MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 snf1::kanMX4 | Euroscarf |
| Y14482 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 snf4::kanMX4 | Euroscarf |
| Y16694 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 gal83::kanMX4 | Euroscarf |
| Y13967 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 reg1::kanMX4 | Euroscarf |
Escherichia coli DH5α (F Ø80dlacZ ΔM15 recA1 endA1 gyrA96 thi-1 hsdR17(rk−rk−) supE44 relA1 deoRΔ 99U169) was the host bacterial strain for the recombinant plasmid constructions. Fusion protein expression was performed in E. coli BL21(DE3)pLysS (Promega).
Yeast cells were grown in the following media: YEPD, high glucose (2% glucose, 2% peptone, and 1% yeast extract); YEPE, low glucose (0.05% glucose, 3% ethanol, 2% peptone, and 1% yeast extract); YEPX, xylose medium (2% xylose, 2% peptone, and 1% yeast extract), and synthetic media containing the appropriate carbon source and lacking appropriate supplements to maintain selection for plasmids (2% glucose (S.D.) or 3% ethanol and 0.05% glucose (S.E.) and 0.67% yeast nitrogen base without amino acids). Amino acids and other growth requirements were added at a final concentration of 20–150 μg/ml. The solid media contained 2% agar in addition to the components described above.
Plasmids
GST fusion vectors pGEX/MIG1 and pGEX/HXK2 for expression, respectively, of GST-Mig1 and GST-Hxk2 were constructed as indicated (27). Plasmid pGEX-XPO1 for expression of GST-Xpo1 in E. coli was a gift from the C. N. Cole laboratory (49). Plasmids pWS93/Mig1-HA and pWS93/Snf1-HA were generated by cloning a 1.5- and 1.9-kb PCR product containing, respectively, the MIG1 and SNF1 genes into the BamHI site of vector pWS93, which expresses a triple HA epitope the from ADH1 promoter (a gift of P. Sanz, Valencia, Spain). The DNA sequence of all PCR-generated constructs was verified by sequencing.
Statistical Analysis
Data were obtained from at least three independent experiments. Results are shown as the means ± S.E.
Enzyme Assay
Cells were grown on 2% glucose (H-Glc) and then half of the cells were shifted to 0.05% glucose plus 3% ethanol (l-Glc) or to 2% xylose medium (Xyl). Samples were harvested by centrifugation and washed three times with ice-cold saline buffer. Invertase activity was assayed in whole cells, as described previously (50), and expressed as micromoles of glucose released per min per 100 mg of cells (dry weight).
Preparation of Crude Protein Extracts
Yeast protein extracts were prepared as follows: yeasts were grown in 10–20 ml of YEPD (H-Glc) at 28 °C to an absorbance at 600 nm of 0.8. Half of the culture was shifted to YEPE (l-Glc) or YEPX (Xyl) for 1 h. Cells were collected, washed twice with 1 ml of 1 m sorbitol and resuspended in 200 μl of PBS buffer (150 mm NaCl, 100 mm Na2HPO4, 18 mm NaH2PO4, pH 7.3) containing protease inhibitor (Roche Applied Science) plus 1 mm DTT and 0.1% Triton X-100. The cells were broken using a FastPrep homogenizer (Thermo Electron Co.). Two pulses of 20 s at 6.0 m/s were given in the presence of glass beads. Then, 200 μl of PBS buffer were added to the suspension. After centrifugation at 19,000 × g for 30 min at 4 °C, the supernatant was used as crude protein extract.
Immunoblot Analysis
Mutant or wild-type yeast cells were grown to an absorbance at 600 nm of 0.8–1.0 in selective medium containing high glucose conditions (2%) and shifted to low glucose conditions for 1 h. The cells were collected by centrifugation (3,000 × g, 4 °C, 2 min), and crude extracts were prepared as described above. For Western blotting, 20–40 μg of proteins were separated by SDS-12% PAGE and transferred to enhanced chemiluminescence PVDF transfer membrane (Amersham Biosciences HybondTM-P, GE Healthcare) by electroblotting, which was then incubated with anti-Hxk2, anti-GFP (Santa Cruz Biotechnology, Santa Cruz, CA), or anti-HA (Cell Signaling Tech.) as primary antibodies and then the appropriate secondary antibody. Horseradish peroxidase-conjugated protein-A was used as secondary reactant. West Pico Chemiluminescent system (Pierce) was used for detection.
Coimmunoprecipitation Assay
Immunoprecipitation experiments were performed using whole-cell extracts from different strains. The extracts were incubated with anti-HA, anti-Hxk2, anti-GFP, or anti-Pho4 polyclonal antibodies for 3 h at 4 °C. Protein A-Sepharose beads (GE Healthcare) were then added and incubated for 3 h at 4 °C in a spinning wheel. After extensive washing with PBS buffer, immunoprecipitated samples were boiled in SDS-loading buffer (50 mm Tris-HCl, pH 6.8, 100 mm DTT, 2% SDS, 0.1% bromphenol blue, 10% glycerol). The supernatant was subjected to SDS-12% PAGE. The proteins were transferred to an enhanced chemiluminescence PVDF membrane and immunoblotted as described above using anti-GFP or anti-Hxk2 polyclonal antibodies. Values shown are representative results from individual experiments.
GST Pulldown Experiments
E. coli cells from the BL21 (DE3) pLysS strain were transformed with the fusion protein expression vectors pGEX-MIG1, pGEX-HXK2, or pGEX-XPO1. Cells were grown to an absorbance at 600 nm of 0.6–0.8, induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 3 h, and collected by centrifugation. The pellet was resuspended in PBS buffer (150 mm NaCl, 100 mm Na2HPO4, 18 mm NaH2PO4, pH 7.3) and sonicated. Insoluble material was removed by centrifugation (17,000 × g for 20 min at 4 °C). The soluble extract was incubated with glutathione-Sepharose 4B (Amersham Biosciences) for 1 h at 4 °C, washed extensively with PBS buffer, and resuspended in the same buffer. The GST-Hxk2 fusion protein coupled to glutathione-Sepharose beads was incubated with 2.5 units of thrombin (2h at 4 °C) for site-specific separation of the GST affinity tag from Hxk2 protein. Identical amounts of Hxk2 affinity-purified protein were added to 20 μl of assay buffer (150 mm NaCl, 100 mm Na2HPO4, 18 mm NaH2PO4, pH 7.3, containing 8 mm magnesium acetate, 0.5 mm EDTA and 0.5 mm DTT) and incubated in the presence or absence of 1 mm ATP, 1 mm AMP-PNP, or different amounts of d-glucose and d-xylose for 30 min at 30 °C. The GST-Mig1 fusion protein coupled to glutathione-Sepharose was incubated with the assay mixture for 1 h at 4 °C in the assay buffer. The GST-Xpo1 fusion protein coupled to glutathione-Sepharose was incubated with whole-cell extracts from the FMY901 strain. The cell extracts were obtained from yeast cells grown in high glucose and shifted to low glucose medium for 60 min. Beads were gently washed five times with 2.5 ml of PBS buffer, boiled in 25 μl sample-loading buffer, and analyzed by SDS-PAGE followed by Western blot. Proteins were separated by SDS-12% PAGE and transferred to an enhanced chemiluminescence PVDF transfer membrane (Amersham Biosciences) by electroblotting. The membrane was then incubated with anti-Hxk2 antibody as primary antibody and anti-rabbit antibody (Santa Cruz Biotechnology) as secondary antibody. Horseradish peroxidase-conjugated protein-A was used as secondary reactant. West Pico chemiluminescent system (Pierce) was used for detection.
Chromatin Immunoprecipitation Assay
Chromatin immunoprecipitation (ChIP) assays were performed essentially as described previously (19). Cells were cultured in 100 ml of YEPD medium to reach an absorbance at 600 of 0.8. Cells from half of the culture were collected by centrifugation (3,000 × g, 4 °C, 2 min) at room temperature. Then, the cell pellet was washed two times with YEP medium, resuspended in YEPE or YEPX medium, and incubated for 1 h at 28 °C with shaking. 1 ml of 37% formaldehyde was added, and samples were incubated for an additional 30 min. Finally, cells were extensively washed, broken, and sonicated. The cross-linked chromatin solution was collected, and Hxk2 and GFP- or HA-tagged proteins were immunoprecipitated by incubating, respectively, the chromatin solution with anti-Hxk2 (26), anti-GFP, or anti-HA antibody (Santa Cruz Biotechnology) pre-bound to protein A-Sepharose beads (General Electric) for 4 h at 4 °C. Precipitated complexes were washed five times with PBS buffer; immunoprecipitants were eluted from the beads by heating to 65 °C for 10 min in elution buffer (50 mm Tris-HCl, pH 8.0, 10 mm EDTA, 1% SDS). Formaldehyde cross-link was reverted by heating to 65 °C overnight and incubation with proteinase K for 1 h at 37 °C. DNA was precipitated after phenol/chloroform extraction. Amounts of specific DNA target present in input and immunoprecipitated samples were analyzed by PCR using the following primers: OL-d, 5′-AGCTCGAGTTATTACTCTGAACAGGA-3′ (sense), and OL-r, 5′-TAGTCGACAAGTCGTCAAATCTTTCT-3′ (antisense), specific to the SUC2B-URS element of theSUC2 promoter. Quantitative PCR analysis was performed in triplicate with the same primers indicated above. ACT1 (OLACT1-d, GCCTTCTACGTTTCCATCCA, and OLACT1-r, GGCCAAATCGATTCTCAAAA, on the ORF of the ACT1 gene) and anti-rabbit antibody (Santa Cruz Biotechnology) were used as negative controls. The linearity of PCRs was shown by multiple template dilutions of input and immunoprecipitated DNA and varying the number of PCR cycles. Gene-specific PCR was usually 18–22 PCR cycles. In general, immunoprecipitated DNA samples were initially diluted 1:10, and inputs were diluted 1:50 with 10 mm Tris-HCl, pH 8.5. Control PCR for ACT1 (data not shown, expression was not influenced by glucose-induced nutritional stress), anti-rabbit antibody (unspecific antibody), and extracts prior to immunoprecipitation (input, whole-cell extract) were used as internal controls at 30–35 cycles to detect a background signal for normalization. Experiments were performed using three independent chromatin preparations, and quantitative PCR analysis was performed in real time using an Applied Biosystems 7300 sequence detector. Data are presented as fold immunoprecipitation over the unspecific antibody-precipitated DNA.
Results
Interdependent Binding of Mig1 and Hxk2 at the SUC2 Promoter Repressor Complex
Repression of the SUC2 gene is carried mainly by Mig1, a zinc finger DNA-binding protein (7). The Mig1 protein represses transcription by binding to two URS of the SUC2 promoter (SUC2A, 5′-−499AATAAAAATGCGGGGAA484-3′, and SUC2B, 5′- −449GGAAATTATCCGGGGGC−431-3′) and recruiting the general corepressors Tup1 and Ssn6 (13). Either SUC2A or SUC2B (Fig. 1A) is sufficient for Mig1-mediated repression, indicating that these two elements are functionally redundant with regard to Mig1 repressor function (51); hereafter, we refer to them as SUC2 elements. Previous experiments established that Mig1 makes a large contribution toward the glucose-induced repression of SUC2, but there is a lack of detailed information about the transmission mechanism of glucose signal from the environment to the Mig1 repressor complex. To answer this question, we employed a system that measures the extent to which the SUC2 elements can be cross-linked to a specific protein in vivo (52). We therefore investigated the interaction of Mig1, and several other factors significantly related to glucose repression signaling, with the SUC2 promoter by chromatin immunoprecipitation (ChIP). Cross-linked protein-DNA complexes were immunoabsorbed, and selected stretches of coprecipitated DNA were amplified by PCR and analyzed by gel electrophoresis. Moreover, to quantify the amount of each factor associated with the SUC2 element region, we used RT-PCR. As a control gene we used the constitutively expressed ACT1 gene (encoding for actin), and as a control antibody we used anti-rabbit. As can be seen in our results, there is little or no detectable DNA precipitation in the control lanes, demonstrating the specificity of our ChIP assay.
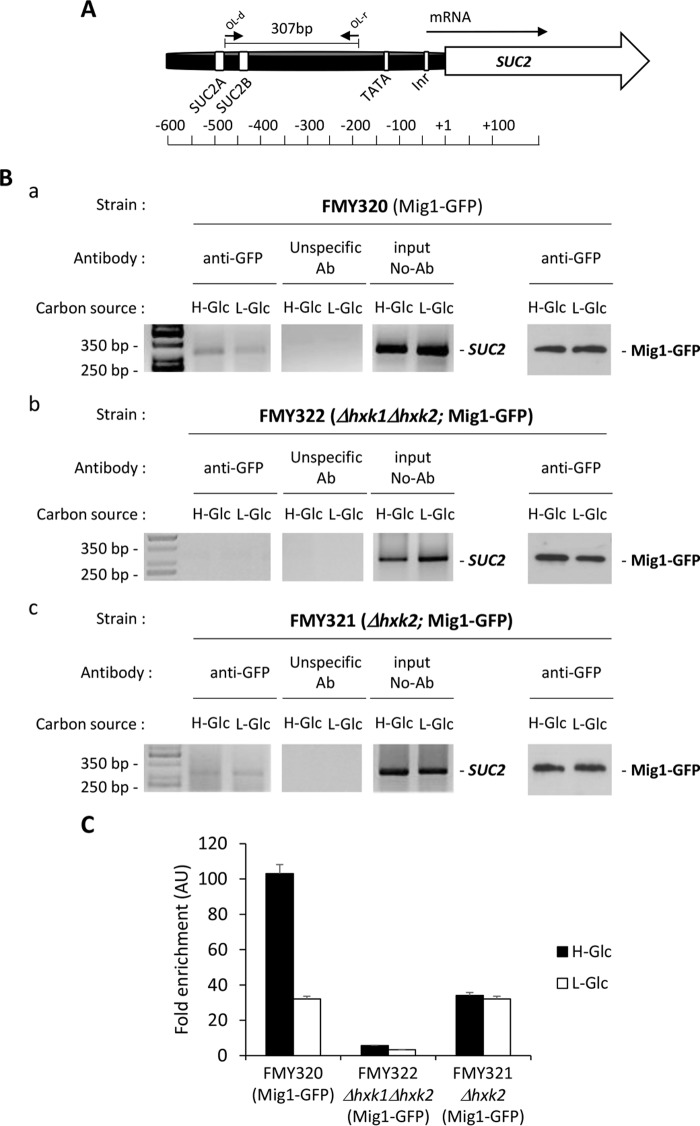
FIGURE 1.
Association of Mig1 repressor with the SUC2 promoter is glucose- and Hxk2-dependent. A, schematic diagram showing the location of primer pair at the SUC2 promoter used for the ChIP analysis. The numbers are presented with respect to the position of the first nucleotide of the initiation codon (+1). B, association of Mig1-GFP with the SUC2 promoter as measured by ChIPs. Strains FMY320 (panel a), FMY322 (Δhxk1Δhxk2) (panel b), and FMY321 (Δhxk2) (panel c) expressing a GFP-tagged Mig1 protein were grown in high glucose conditions (2% glucose, H-Glc) until an A600 of 0.8 was reached. Afterward, half of the culture was exposed to low glucose conditions (0.05% glucose plus 3% ethanol, l-Glc) for 60 min. Mig1 and the SUC2 promoter association was determined by ChIP assays. Results were analyzed by PCR. At least three independent experiments were performed with ACT1 (data not shown, expression was not influenced by glucose-induced nutritional stress), anti-rabbit antibody (Ab) (unspecific antibody), and extracts prior to immunoprecipitation (input, whole-cell extract) as internal controls. Last two lines in B, panels a–c, represent Western blot controls of the Mig1-GFP level. C, quantification of Mig1-GFP association in wild-type (wt), Δhxk1Δhxk2, and Δhxk2 mutant strains with the SUC2 promoter. Cells were treated as described for B, but ChIPs were analyzed by quantitative real time PCR. Data are expressed as signal normalized to the untreated sample. Error bars represent the standard error of the mean for three independent experiments.
Cells expressing a functional GFP-tagged form of the Mig1 protein at its chromosomal locus were exposed to high and low glucose conditions and used for ChIP assays. The results shown in Fig. 1, B and C, indicate that Mig1-GFP binding to SUC2 elements of the SUC2 promoter occurs in a glucose-dependent manner, with peak Mig1 binding activity in cells grown in high glucose conditions (Fig. 1, B, panel a, and C). Glucose starvation decreases the association of Mig1 with SUC2-URS region by 68% (Fig. 1, B, panel a, and C). Our finding is consistent with Mig1 subcellular localization in high glucose conditions but not in low glucose conditions (40, 53). It was previously reported that upon glucose removal, Mig1-GFP rapidly translocates to the cytoplasm (53). However, our results suggest that 32% of the protein remains associated with the SUC2 promoter after glucose starvation. This indicates that the interaction of Mig1 with the SUC2 promoter might be regulated by both the phosphorylation state of Mig1 (38) and an independent mechanism. The HXK2 gene is expressed in cells grown in high glucose (25), but after shifting the cells to a low glucose medium, the HXK2 gene is repressed, and the HXK1 gene is rapidly expressed (15). A possible explanation for the independent mechanism is that, in low glucose conditions, Hxk1 partially mimics the function performed by Hxk2 during glucose repression signaling in high glucose. To avoid the synthesis of Hxk1 in low glucose conditions, we used a Δhxk1Δhxk2 double mutant strain, with a functional GFP-tagged form of the Mig1 protein at its chromosomal locus, to perform ChIP assay. The results for Mig1-GFP SUC2 chromatin interactions are shown in Fig. 1, B, panel b, and C. In high and low glucose conditions, less than 5% of the protein became cross-linked with the SUC2-URS of the SUC2 promoter (Fig. 1C), a much lower degree of binding than found in a wild-type strain at low glucose conditions. Because the Hxk2 protein is necessary for glucose-induced repression of the HXK1 gene (15) and only 30% of SUC2 repression is lost in Δhxk2 mutant cells (Fig. 2), it seems likely that the Hxk1 protein could mimic Hxk2 function. To further address this possibility, we used a Δhxk2 mutant strain, with a functional GFP-tagged form of the Mig1 protein at its chromosomal locus, to perform the ChIP assay. We found that in the absence of Hxk2, the recruitment of Mig1 to the SUC2 promoter is significantly decreased in high glucose conditions (Fig. 1, B, panel c, and C). However, the recruitment of Mig1 to the SUC2 promoter in low glucose conditions is identical to that found in the wild-type strain in glucose-starved cells. Together, these results support that in high glucose conditions Mig1 recruitment to SUC2-URS of the SUC2 promoter is Hxk2-dependent, and the presence of Hxk1 is important to modulate the Mig1 SUC2-DNA binding in low glucose conditions.
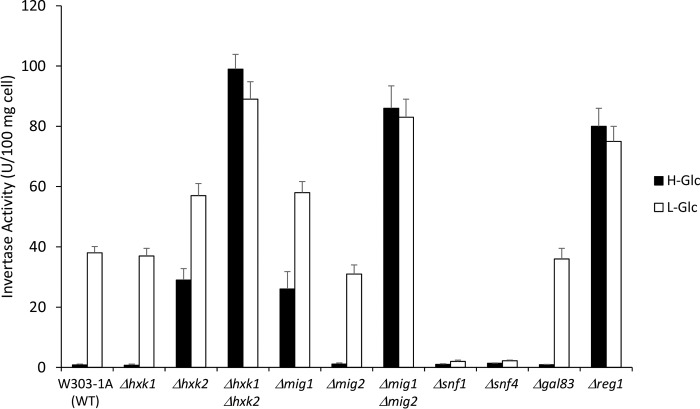
FIGURE 2.
Quantitative invertase assays were performed on cells with the indicated mutations. Whole cells from the wild-type strain, W303-1A, and the mutant strains Δhxk1, Δhxk2, Δhxk1Δhxk2, Δmig1, Δmig2, Δmig1Δmig2, Δsnf1, Δsnf4, Δgal83, and Δreg1 were used for invertase activity determination. Invertase was assayed using cells grown in high glucose medium (H-Glc, black bars) until an A600 nm of 0.8 was reached and then transferred to low glucose medium for 60 min (L-Glc, white bars). Error bars represent the standard error of the mean for three independent determinations using three colonies of each strain.
Because it is known that the Hxk2 protein interacts directly with Mig1 in vivo and in vitro (27), first we investigated the interaction of Hxk2 with the SUC2 promoter in high and low glucose conditions. In second place, we investigated how Mig1 affects this interaction. We tested direct interactions in vivo by ChIP experiments using a specific anti-Hxk2 antibody. A comparison between high and low glucose-growing cells revealed that nuclear Hxk2 was well engaged with the SUC2 elements of the SUC2 promoter in high glucose, but less than 8% of Hxk2 was bound to the SUC2 elements in low glucose conditions (Fig. 3, A, panel a, and B). The occupation rate of the SUC2 regions of the SUC2 promoter by Hxk2 in high glucose conditions is affected by the absence of the Mig1 repressor. In Δmig1 mutant cells growing in high or low glucose conditions, less than 2% of Hxk2 is associated with the SUC2 promoter, demonstrating that binding of Hxk2 to the SUC2 promoter is Mig1-dependent (Fig. 3, A, panel b, and B). These results indicate that Mig1 is required to capture the Hxk2 protein to the repressor complex of the SUC2 promoter and that Hxk2 does not interact directly with DNA.
FIGURE 3.
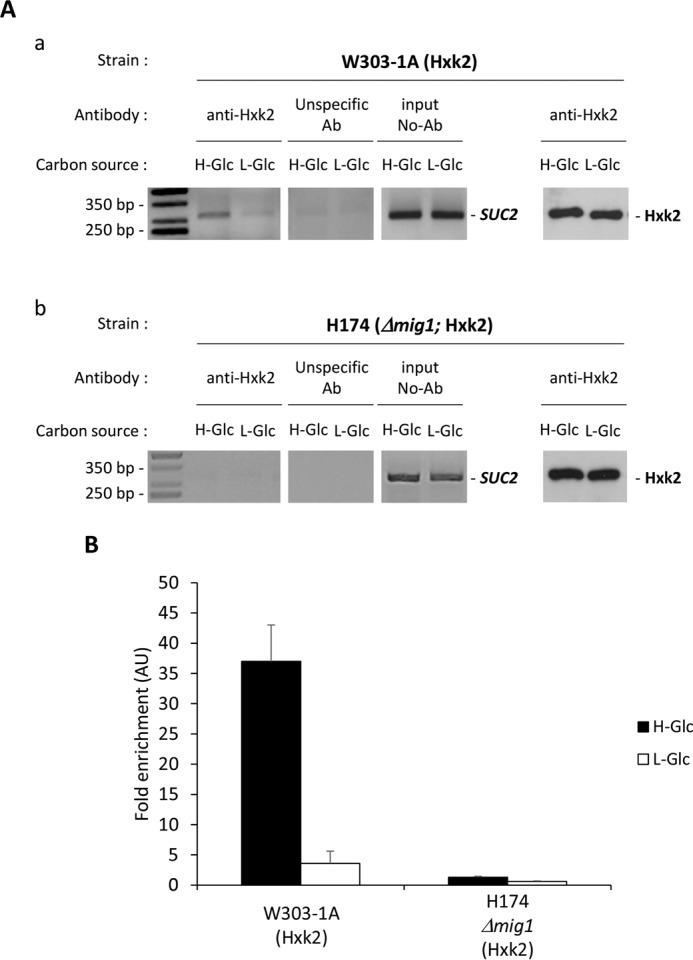
Association of Hxk2 with the SUC2 promoter is glucose- and Mig1-dependent. A, association of Hxk2 with the SUC2 promoter as measured by ChIPs. The wild-type strain W303-1A (panel a) and the mutant strain H174 (Δmig1) (panel b) were grown in high glucose conditions (2% glucose, H-Glc) until an A600 of 0.8 was reached. Afterward, half of the culture was exposed to low glucose conditions (0.05% glucose plus 3% ethanol, L-Glc) for 60 min. Hxk2 and the SUC2 promoter association was determined by ChIP. Results were analyzed by PCR. At least three independent experiments were performed with ACT1 (not shown, expression was not influenced by glucose induced nutritional stress), anti-rabbit antibody (Ab) (unspecific antibody), and extracts prior to immunoprecipitation (input, whole-cell extract) as internal controls. Last two lines in A, panels a–c, represent Western blot controls of the Hxk2 level. The agarose electrophoresis shown is representative of results obtained from three independent experiments. B, quantification of Hxk2 association in wild-type (wt) and Δmig1 mutant strain with the SUC2 promoter. Cells were treated as described for A (H-Glc, black bars; L-Glc, white bars), but ChIPs were analyzed by quantitative real time PCR. Data are expressed as signal normalized to the untreated sample. AU, arbitrary units. Error bars represent the standard error of the mean for three independent experiments.
In Δmig1 mutant cells growing in high glucose conditions, only 27% of SUC2 repression is lost (Fig. 2), so it seems likely that other DNA-binding proteins could mimic Mig1 function. Because like Mig1, Mig2 represses transcription in response to glucose through Tup1 and Ssn6 corepressors (9), we have investigated the role of Mig2 in the SUC2 repressor complex and how Mig1 and Hxk2 affect Mig2 SUC2 promoter interaction. A wild-type strain with a functional GFP-tagged form of the Mig2 protein at its chromosomal locus was used to determine by ChIP the SUC2-URS localization of Mig2. In wild-type cells grown in high and low glucose conditions, about 26–28% of Mig2 was recruited to the SUC2 elements of the SUC2 promoter (Fig. 4, A, panel a, and B). The level of Mig2 associated with the SUC2 promoter reaches a maximum at low glucose conditions in a Δhxk2 mutant strain (100%) (Fig. 4, A, panel c, and B). The level of Mig2 associated with the SUC2 promoter also increases at high and low glucose conditions in a Δmig1 mutant strain with respect to the wild type (Fig. 4, A, panel b, and B). Thus, in high glucose conditions in the absence of Mig1 protein, Mig2 is actively recruited to the SUC2 promoter (95%) contributing to maintain 70% of SUC2 gene repression. In low glucose conditions 45% of the Mig2 protein was associated with the SUC2 elements of the SUC2 promoter (Fig. 4, A, panel b, and B). Although Mig2 appears to play no role in SUC2 repression when Mig1 is present because its deletion has no effect on SUC2 expression in a MIG1 strain (Fig. 2), yeast cells are only fully derepressed in a Δmig1Δmig2 double mutant strain (Fig. 2). These data support the idea that Mig2 may bind to the SUC2 promoter mainly in the absence of Mig1 protein as happens in Δmig1 or Δhxk2 mutant strains. It also appears that the role of Mig2 in the yeast cell is as a redundant transcriptional repressor.
FIGURE 4.
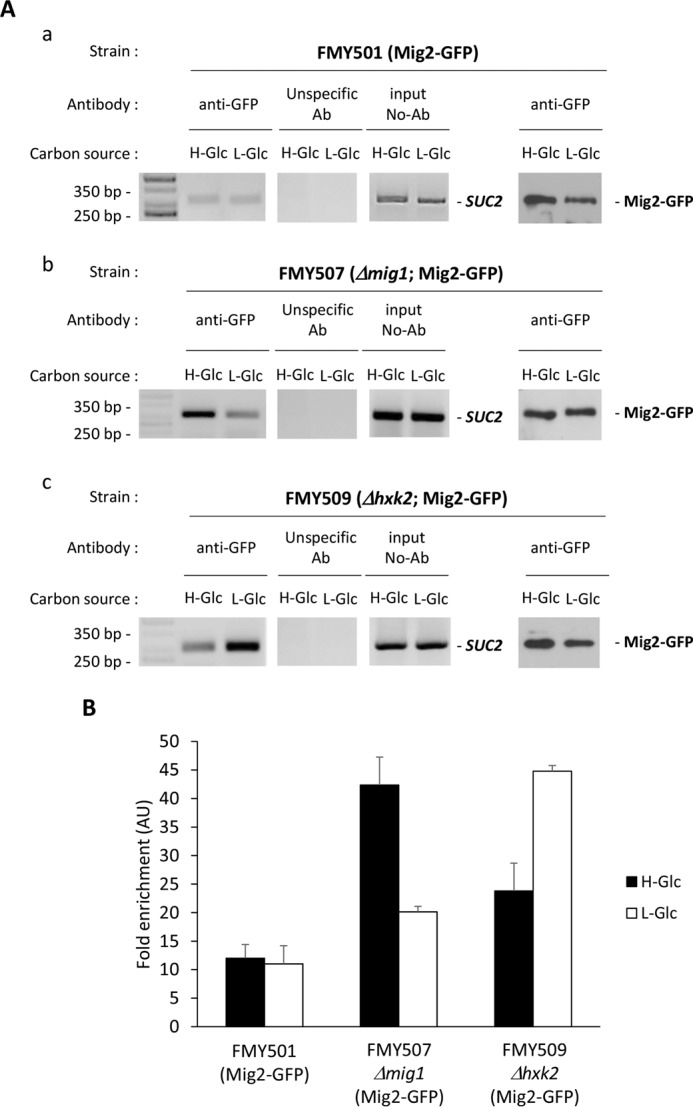
Association of Mig2-GFP with the SUC2 promoter is Mig1- and Hxk2-dependent. A, association of Mig2-GFP with the SUC2 promoter as measured by ChIPs. The FMY501 strain expressing a GFP-tagged Mig2 protein (panel a) and the mutant strains FMY507 (Δmig1) (panel b) and FMY509 (Δhxk2) (panel b), both expressing a GFP-tagged Mig2 protein, were grown in high glucose conditions (2% glucose, H-Glc) until an A600 of 0.8 was reached. Afterward, half of the culture was exposed to low glucose conditions (0.05% glucose plus 3% ethanol, L-Glc) for 60 min. Mig2 and the SUC2 promoter association were determined by ChIP. Results were analyzed by PCR. At least three independent experiments were performed with ACT1 (not shown, expression was not influenced by glucose-induced nutritional stress), anti-rabbit antibody (Ab) (unspecific antibody), and extracts prior to immunoprecipitation (input, whole-cell extract) as internal controls. Last two lines in A, panels a–c, represent Western blot controls of the Mig2-GFP level. The agarose electrophoresis shown are representative of results obtained from three independent experiments. B, quantification of Mig2 association in FMY501, FMY507 (Δmig1), and FMY509 (Δhxk2) strains with the SUC2 promoter. Cells were treated as described for A (H-Glc, black bars; L-Glc, white bars), but ChIPs were analyzed by quantitative real time PCR. Data are expressed as signal normalized to the untreated sample. Error bars represent the standard error of the mean for three independent experiments. AU, arbitrary units.
It is also tempting to speculate that Mig2 could function as a structural protein that, by interacting with other factors associated with the SUC2 promoter, could stabilize the repressor complex structure. To address this possibility, we examined the ability of Mig2 to interact with Mig1, Hxk2, and Snf1 by using an immunoprecipitation assay. Cell extracts from a strain with a modified MIG2 gene, which encodes a C-terminal Mig2 protein tagged with GFP, were immunoprecipitated with the antibodies indicated in Fig. 5. Following immunoprecipitation, the Mig2-GFP protein was detected in a complex with Mig1 (Fig. 5A), Hxk2 (Fig. 5B), and Snf1 (Fig. 5C), from both glucose-starved cells and glucose-rich medium-grown cells. When an anti-Pho4 antibody was used to detect unspecific immunoprecipitation, no signals were observed. Similar amounts of the Mig2-GFP protein were detected in the different protein extracts used by immunoblot analysis with an anti-GFP antibody. We conclude that Mig2 associates with Mig1, Hxk2, and Snf1 in vivo and that the association is independent of the glucose levels in the culture medium.
FIGURE 5.
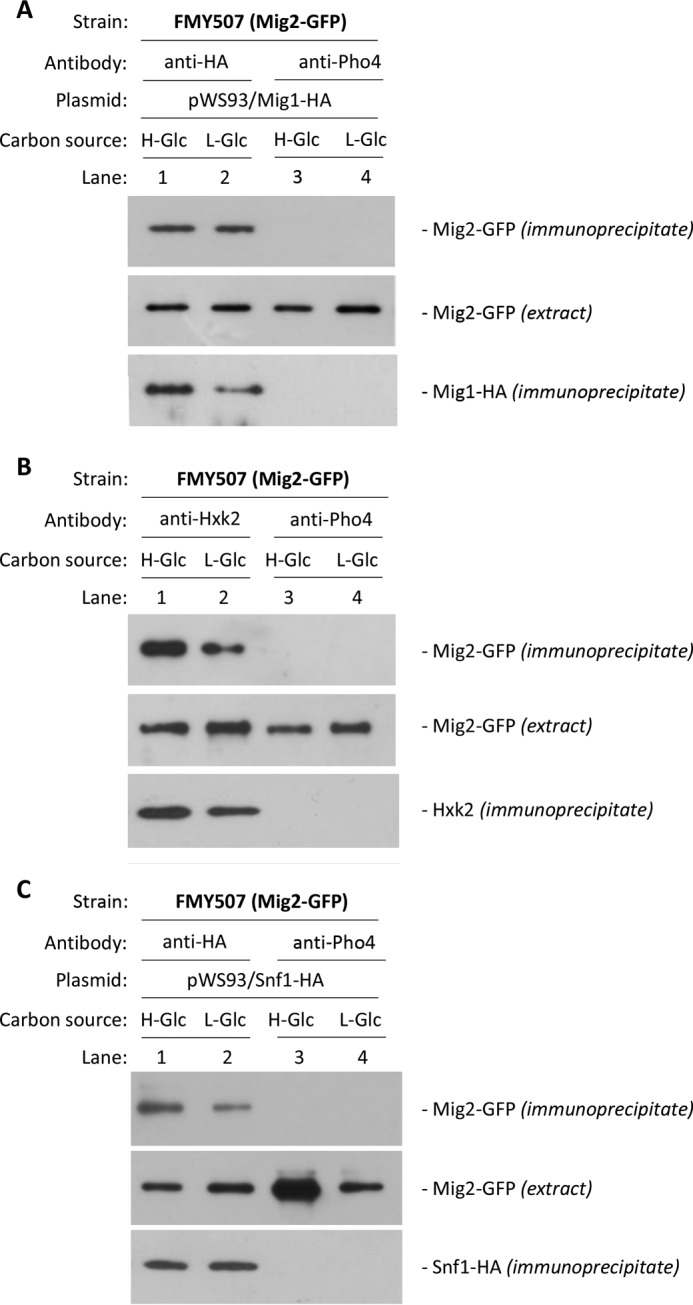
Interaction of Mig1, Hxk2, and Snf1 with Mig2. In vivo coimmunoprecipitation of Mig2-GFP with Mig1-HA (A), Hxk2 (B), and Snf1-HA (C) is shown. The FMY507 strain, expressing a Mig2 GFP-tagged fusion protein, was transformed with plasmids pWS93/Mig1 and pWS93/Snf1, which encode an HA-tagged Mig1 and Snf1 protein, respectively. The cells were grown in SG-media, lacking appropriate supplement to maintain selection for plasmid, until an A600 of 0.8 was reached and then shifted to high (H-Glc) and low (L-Glc) glucose conditions for 1 h. The cell extracts were immunoprecipitated with a monoclonal anti-HA, a polyclonal anti-Hxk2 antibodies, or a polyclonal antibody to Pho4 (lanes 3 and 4). Immunoprecipitates were separated by SDS-12% PAGE, and the level of immunoprecipitated Mig2-GFP in the blotted samples was determined by using anti-GFP antibody. The level of Mig2-GFP present in the different extracts was determined by Western blot using anti-GFP antibody. All Western blots shown are representative of results obtained from three independent experiments.
Hxk2 Recruits SNF1 Complex and Reg1-Glc7 Phosphatase to the Repressor Complex of the SUC2 Promoter
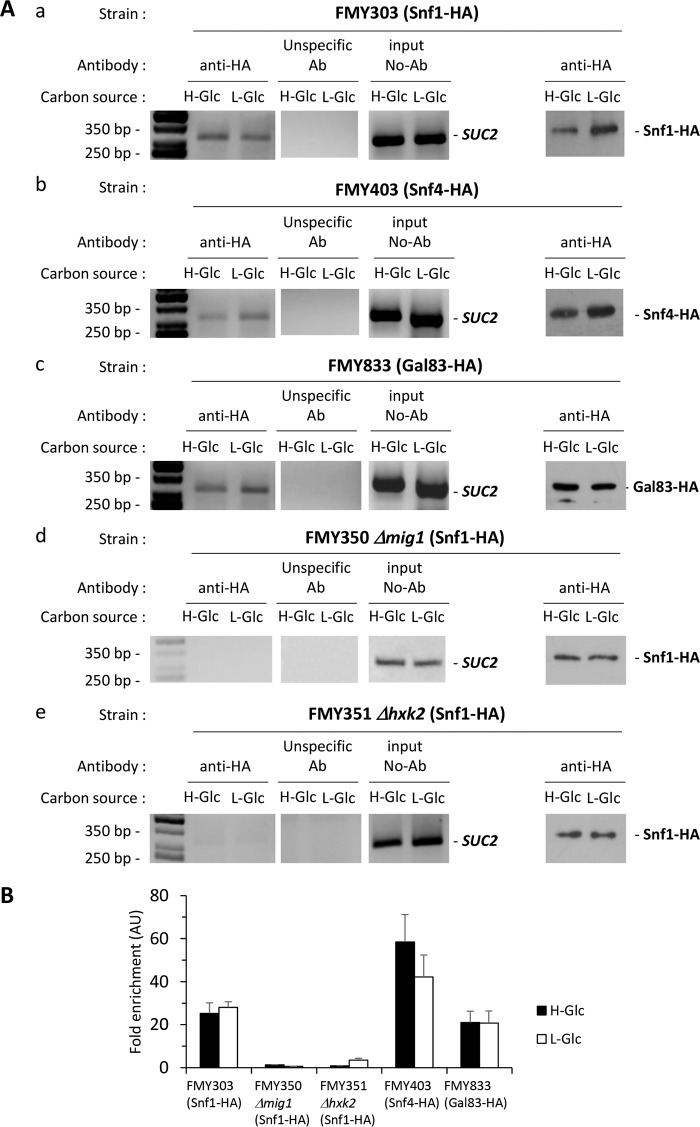
Since it has been previously described that the Snf1 kinase is constitutively associated with Hxk2 both at high and low glucose conditions and that Hxk2 accumulates in the nucleus upon SNF1 gene disruption, we considered a model in which the main regulatory proteins of the phosphorylation state of Mig1 and Hxk2 act by stable association with SUC2-URS on the SUC2 promoter. We used ChIP analysis to test this model. Chromatin binding experiments similar to that described above were pursued with Snf1, Snf4, Gal83, and Reg1 proteins, all of them characterized as important regulatory factors in glucose repression signaling. Thus, we investigated whether the SNF1 complex proteins interact with the SUC2-URS of the SUC2 promoter in high and low glucose conditions and how Mig1 and Hxk2 affect this interaction. Cells expressing functional HA-tagged forms of Snf1, Snf4, and Gal83 proteins at their chromosomal loci were first exposed to high glucose conditions until an A600 of 0.8 was reached and then shifted for 60 min to low glucose conditions. Proteins were then cross-linked to DNA with formaldehyde, followed by cell lysis and DNA fragmentation. Snf1-HA, Snf4-HA, and Gal83-HA were immunoprecipitated from extracts, and coprecipitating SUC2-URS of the SUC2 promoter sequence was detected by PCR and RT-PCR.
As shown in Fig. 6, A, panels a–c, and B, Snf1, Snf4, and Gal83 bind specifically to the SUC2 elements' sequence both at high and low glucose conditions. We find that Snf4 recruitment (100%) to the SUC2 promoter, at high glucose conditions, was more pronounced than Snf1 (43%) and Gal83 (36%). In low glucose conditions, 72% of Snf4 remains at the SUC2 promoter and a similar amount was found at high glucose conditions of Snf1 (48%) and Gal83 (35%). To determine whether Snf1-HA association with the SUC2 promoter was dependent on the Mig1 and Hxk2 proteins, the SNF1 gene was HA-tagged at its chromosomal locus in Δmig1 and Δhxk2 mutant cells. As can be seen in Fig. 6, A, panels d and e, and B, Snf1-HA was not detected in association with the SUC2 promoter both at high and low glucose conditions in Δmig1 and Δhxk2 mutant cells.
FIGURE 6.
Association of Snf1-HA, Snf4-HA, and Gal83-HA with the SUC2 promoter is Mig1- and Hxk2-dependent. A, association of Snf1, Snf4, and Gal83 with the SUC2 promoter as measured by ChIPs. The FMY303 (panel a), FMY403 (panel b), and FMY833 (panel c) strains expressing HA-tagged Snf1, Snf4, and Gal83 protein, respectively, and the mutant strains FMY350 (Δmig1; Snf1-HA) (panel d) and FMY351 (Δhxk2; Snf1-HA) (panel b), both expressing an HA-tagged Snf1 protein, were grown in high glucose conditions (2% glucose, H-Glc) until an A600 of 0.8 was reached. Afterward, half of the culture was exposed to low glucose conditions (0.05% glucose plus 3% ethanol, L-Glc) for 60 min. Snf1 and the SUC2 promoter association was determined by ChIP in the presence or absence of Mig1 (panel d) and Hxk2 (panel e) proteins. Results were analyzed by PCR. At least three independent experiments were performed with ACT1 (not shown, expression was not influenced by glucose-induced nutritional stress), anti-rabbit antibody (Ab) (unspecific antibody), and extracts prior to immunoprecipitation (input, whole-cell extract) as internal controls. Last two lines in A, panels a–e, represent Western blot controls of Snf1-HA (panels a, d, and e), Snf4-HA (panel b), and Gal83-HA (panel c) level. The agarose electrophoresis shown is representative of results obtained from three independent experiments. B, quantification of Snf1, Snf4, and Gal83 association in the presence of Mig1 and Hxk2 proteins with the SUC2 promoter was analyzed by quantitative real time PCR. Snf1 association in the absence of Mig1 and Hxk2 proteins with the SUC2 promoter was also analyzed by RT-PCR. Cells were treated as described for A (H-Glc, black bars; L-Glc, white bars). Data are expressed as signal normalized to the untreated sample. Error bars represent the standard error of the mean for three independent experiments. AU, arbitrary units.
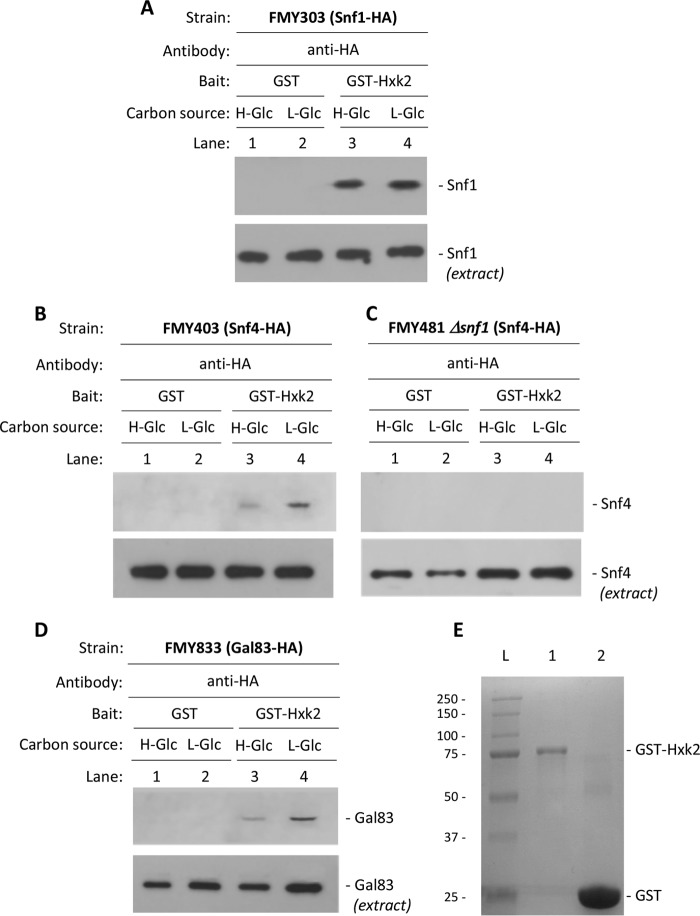
To analyze the specific requirements for incorporation of Snf1, Snf4, and Gal83 to the repressor complex at the SUC2 promoter, we conducted GST pulldown experiments to characterize physical interactions of these proteins with Hxk2. As was observed in Fig. 7A, and previously (30), a strong and specific signal of Snf1-HA was observed both with samples from high and low glucose-grown cells. We next tested the interactions of Snf4 and Gal83 with Hxk2 by GST pulldown experiments. We used extracts from the Snf4-HA- and Gal83-HA-producing strains and GST-Hxk2 expressed in bacteria. As shown in Fig. 7, B and D, weak signals of Snf4 and Gal83 proteins, retained by GST-Hxk2, were detected in extracts from either high or low glucose-grown cells. When a bacterially produced GST protein was used as bait in these experiments to detect nonspecific protein binding, no signals were observed. To confirm that the same amount of Snf1-HA, Snf4-HA, and Gal83-HA fusion proteins is present in each sample used in these experiments, cell extracts from high and low glucose-grown cells were subjected to Western blot analysis. Because in the extracts we keep the amount of each SNF1 complex factor constant, and the Snf4 and Gal83 signal intensity in Western blot experiments was greatly reduced in comparison with Snf1, we believe that these results suggest that the interaction takes place at the level of the Snf1 subunit and the weak detection of Gal83 and Snf4 proteins could be due to indirect Snf1-Snf4 and Gal83 interactions. To test this hypothesis, we used a GST pulldown assay in a Δsnf1 mutant strain with a functional HA-tagged form of the Snf4 protein at its chromosomal locus. Our results show that in cells grown in high and low glucose medium, Snf4 protein was not detected in complex with Hxk2 (Fig. 7C). We conclude that Snf4 association with Hxk2 is Snf1-dependent and that Snf4 does not interact directly with Hxk2.
FIGURE 7.
GST pulldown assays of the interaction of Snf1, Snf4, and Gal83 with Hxk2. A GST-Hxk2 fusion protein was purified on glutathione-Sepharose columns. Equal amounts of GST-Hxk2 were incubated with cell extracts from FMY303, FMY403, FMY481, and FMY833 strains expressing Snf1-HA (A), Snf4-HA (B), Snf4-HA in the absence of Snf1 (C), and Gal83-HA (D) fusion proteins, respectively. E, affinity purification of GST-Hxk2 fusion protein produced in bacteria. Each lane contains 2 μg of purified protein, and the molecular masses (kDa) of the ladder are depicted. Lanes: L, molecular mass ladder; 1, GST-Hxk2 SDS-12% PAGE; 2, GST 12% acrylamide SDS-PAGE. The yeast strains were grown in YEPD media until an A600 nm of 0.8 was reached and then shifted to high (H-Glc) and low (L-Glc) glucose conditions for 1 h. For the control samples, GST protein was also incubated with the high Glc and low Glc cell extracts, but no signals were detected (A–D, lanes 1 and 2). The level of Snf1, Snf4, and Gal83 present in the different extracts used were determined by Western blot using a monoclonal anti-HA antibody. The Western blots shown are representative of results obtained from three independent experiments.
Given these results, it could be tempting to propose that Hxk2 could act as an anchor point for the proteins that are part of the SUC2 gene repressor complex. To address this possibility, we examined the ability of Reg1 to interact in vivo with Hxk2 at the SUC2 promoter level by using a ChIP assay. The wild-type, Δmig1, and Δhxk2 strains with functional GFP-tagged forms of the Reg1 protein at its chromosomal locus were also used to determine by ChIP the SUC2-URS localization of Reg1 in the absence of Mig1 and Hxk2 proteins. The cells were first exposed to high glucose conditions until an A600 of 0.8 was reached and then shifted for 60 min to low glucose conditions. In FMY901 cells expressing the Reg1-GFP fusion protein, Reg1 was detected in a complex with SUC2-URS element of the SUC2 promoter both at high and low glucose conditions (Fig. 8, A, panel a, and B). However, less than 5% of the Reg1-GFP fusion protein bound to the SUC2 promoter in an FMY901 cell was detected in Δmig1 and Δhxk2 mutants, both in high and low glucose conditions (Fig. 8, A, panels b and c, and B). Thus, the repressor complex of the SUC2 promoter is an assembly of the proteins that directly bind to DNA (Mig1 and Mig2) as well as the machinery regulating the Mig1 repressing activity.
FIGURE 8.
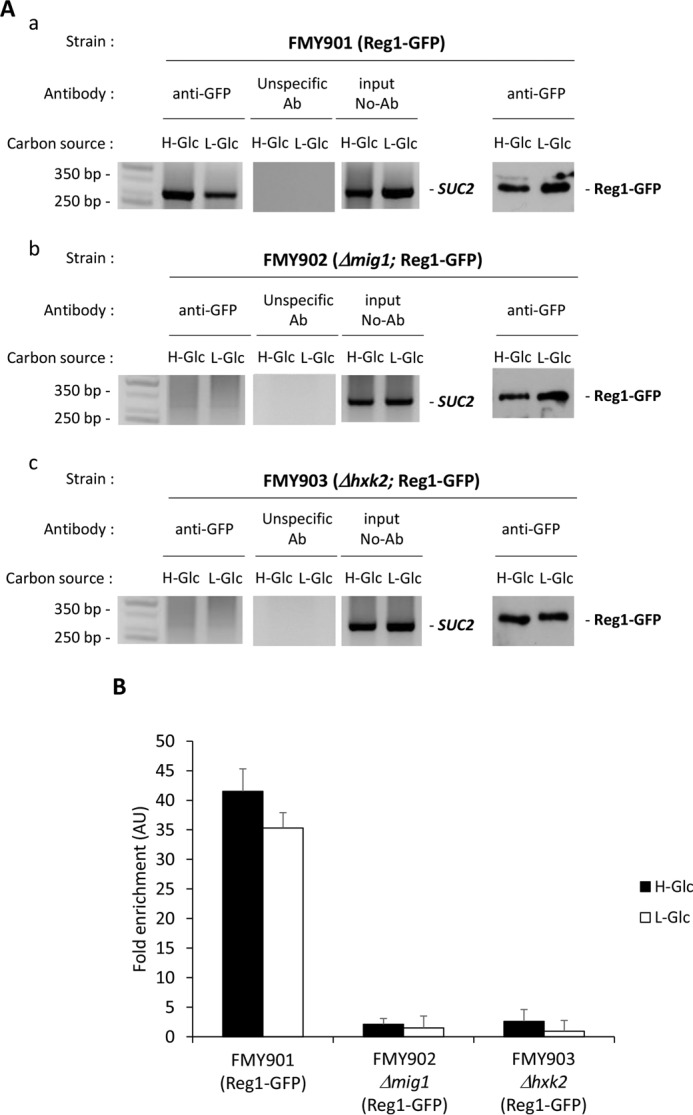
Association of Reg1-GFP with the SUC2 promoter is Mig1- and Hxk2-dependent. A, association of Reg1-GFP with the SUC2 promoter as measured by ChIPs. The FMY901 strain expressing a GFP-tagged Reg1 protein (panel a) and the mutant strains FMY902 (Δmig1) (panel b) and FMY903 (Δhxk2) (panel b), both also expressing a GFP-tagged Reg1 protein, were grown in high glucose conditions (2% glucose, H-Glc) until an A600 of 0.8 was reached. Afterward, half of the culture was exposed to low glucose conditions (0.05% glucose plus 3% ethanol, L-Glc) for 60 min. Reg1 and the SUC2 promoter association was determined by ChIP. Results were analyzed by PCR. At least three independent experiments were performed with ACT1 (not shown, expression was not influenced by glucose-induced nutritional stress), anti-rabbit antibody (Ab) (unspecific antibody), and extracts prior to immunoprecipitation (input, whole-cell extract) as internal controls. Last two lines in A, panels a–c, represent Western blot controls of the Reg1-GFP level. The agarose electrophoresis shown are representative of results obtained from three independent experiments. B, quantification of Reg1 association in FMY901, FMY902 (Δmig1), and FMY903 (Δhxk2) strains with the SUC2 promoter. Cells were treated as described for A (H-Glc, black bars; L-Glc, white bars), but ChIPs were analyzed by quantitative real time PCR. Data are expressed as signal normalized to the untreated sample. Error bars represent the standard error of the mean for three independent experiments. AU, arbitrary units.
To confirm these results, we also determined the ability of Reg1 to interact with Hxk2 by the coimmunoprecipitation assay. Cell extracts from a strain with a modified REG1 gene, which encodes for a C-terminal Reg1 protein tagged with GFP, were immunoprecipitated with anti-GFP antibody. Following immunoprecipitation, the Reg1-GFP protein was detected in a complex with Hxk2 (Fig. 9A), from both glucose-starved cells and glucose-rich medium cells. When an anti-Pho4 antibody was used to detect unspecific immunoprecipitation, no signals were observed. Similar amounts of the Hxk2 protein were detected in the different protein extracts used by immunoblot analysis with an anti-Hxk2 antibody. We conclude that Reg1 associates with Hxk2 in vivo and that the association is independent of the glucose levels in the culture medium.
FIGURE 9.
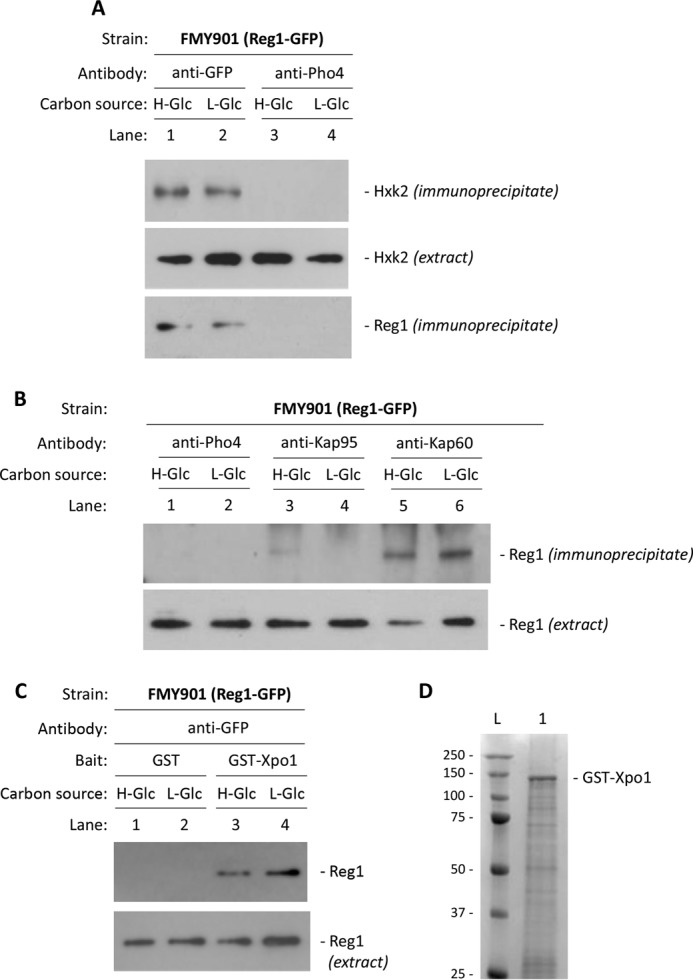
In vivo interaction of Reg1 with Hxk2, Kap60, Kap95, and Xpo1. The FMY901 strain, expressing a Reg1-GFP fusion protein, was grown in high glucose YEPD-medium (H-Glc) until an A600 of 0.8 was reached and then transferred to low glucose medium (L-Glc) for 60 min. A, cell extracts were immunoprecipitated with a polyclonal anti-GFP antibody (lanes 1 and 2) or a polyclonal antibody to Pho4 (lanes 3 and 4). B, cell extracts were immunoprecipitated with polyclonal anti-Kap95 and anti-Kap60 antibodies (lanes 3–6) or a polyclonal antibody to Pho4 (lanes 1 and 2). Immunoprecipitates were separated by SDS-12% PAGE, and coprecipitated Hxk2 or Reg1-GFP was visualized on a Western blot with polyclonal anti-Hxk2 or anti-GFP antibodies. The level of Hxk2 or Reg1 present in the different extracts was determined by Western blot using anti-Hxk2 or anti-GFP antibodies. C, GST-Xpo1 fusion protein was purified on glutathione-Sepharose columns. Equal amounts of GST-Xpo1 were incubated with cell extracts from FMY901 strain. D, affinity purification of GST-Xpo1 fusion protein produced in bacteria. The lane contains 4 μg of purified protein, and the molecular masses (kDa) of the ladder are depicted. Lanes: L, molecular mass ladder; 1, GST-Xpo1 10% acrylamide SDS-PAGE. The yeast strain was grown in YEPD media until an A600 of 0.8 was reached and then shifted to high (H-Glc) and low (L-Glc) glucose conditions for 1 h. For the control samples, GST protein was also incubated with the high Glc and low Glc cell extracts but no signals were detected (C, lanes 1 and 2). The level of Reg1 present in the different extracts used were determined by Western blot using an anti-GFP antibody. The Western blots shown are representative of results obtained from three independent experiments.
Because it has been described that Reg1 is predominantly cytoplasmic (54), although our results indicate that Reg1 also has nuclear localization, we examined the ability of the protein to interact with the import (Kap60-Kap95) and export (Xpo1) machinery used by Hxk2 to enter and exit the nucleus (28, 29). To test whether Reg1 binds to Kap60, Kap95, and Xpo1 in vivo, we used immunoprecipitation and GST pulldown assays in cells expressing Reg1-GFP. First, cell extracts from the FMY901 (Reg1-GFP) strain were immunoprecipitated with anti-Kap60 and anti-Kap95 antibodies. The resulting immunoprecipitates were assayed for the presence of Reg1-GFP by immunoblot analysis with anti-GFP antibodies. As shown in Fig. 9B, a strong and specific Reg1-GFP signal was observed with samples immunoprecipitated with an anti-Kap60 antibody in cells grown both in high and low glucose conditions. However, no interaction or a very weak interaction was observed when the experiment was done using samples immunoprecipitated with anti-Kap95 antibody, although similar amounts of Kap60 and Kap95 proteins were detected in the immunoprecipitates. Similarly, the same amount of the Reg1-GFP protein was detected by immunoblot analysis with anti-GFP antibody from the protein extract used for immunoprecipitation (Fig. 9B, lower panel). These results suggest that Reg1 enters the nucleus via the canonical route by directly binding to the α-importin Kap60, and the β-importin Kap95 is recruited to the ternary complex mainly during high glucose growth conditions. Then the tripartite protein complex could be recognized by the nuclear pore for transport into the nucleus.
To investigate the Reg1-Xpo1 interaction, we performed GST pulldown experiments with raw protein extracts from a strain expressing a functional Reg1-GFP protein at its chromosomal locus and purified GST-Xpo1 fusion protein produced from bacteria. As shown in Fig. 9C, a clear retention of Reg1 protein was observed in the sample containing GST-Xpo1 and raw extract from the FMY901 strain grown in high and low glucose conditions. However, the interaction between Reg1-GFP and Xpo1 was systematically stronger with samples from low glucose-grown cultures than from high glucose cultures, and our data suggest that the low glucose condition increases the affinity of Reg1 for the export receptor. When a control with GST protein alone in the reaction mixture was used, no signal was observed. These results suggest that Reg1 is a cargo of Xpo1 to exit the nucleus in S. cerevisiae. Because results from numerous studies support a direct substrate interaction mechanism to explain the ability of Reg1 to direct Glc7 to its substrates of interest, it is tempting to speculate that the Glc7 phosphatase could be recruited by Reg1 to the repressor complex of the SUC2 promoter to act over its substrates Snf1, Mig1, and Hxk2.
Regulation of the Hxk2 Incorporation to the Repressor Complex of the SUC2 Promoter
Hxk2 protein shuttles in and out the repressor complex of the SUC2 promoter in response to glucose availability. Because the shuttling back and forth between the nucleus and the cytoplasm of Hxk2 is also controlled by glucose availability and glucose determines the phosphorylation state of Hxk2 by controlling Snf1 kinase and Reg1-Glc7 phosphatase activities (30), an attractive hypothesis could be that the phosphorylation state of Hxk2 regulates its incorporation and dissociation from the repressor complex of the SUC2 promoter. To address this possibility, we used a Δsnf1 mutant strain to perform ChIP assays using a specific polyclonal antiserum against Hxk2. As shown in Fig. 10, A and B, ChIP analysis indicates that Hxk2 is able to interact with the SUC2 promoter in high glucose (95%), but less than 8% of Hxk2 was bound to the SUC2 elements in low glucose conditions, which is a similar result to that obtained using a wild-type strain. Thus, this result supports that phosphorylation of Hxk2 by the Snf1 kinase is not essential to regulate Hxk2-SUC2 promoter binding.
FIGURE 10.

Association of Hxk2 with the SUC2 promoter is not regulated by phosphorylation. A, association of Hxk2 with the SUC2 promoter as measured by ChIPs. The wild-type strain W303-1A (panel a) and the mutant strain Y14311 (Δsnf1) (panel b) were grown in high glucose conditions (2% glucose, H-Glc) until an A600 of 0.8 was reached; afterward, half of the culture was exposed to low glucose conditions (0.05% glucose plus 3% ethanol, L-Glc) for 60 min. Hxk2 and the SUC2 promoter association was determined by ChIP. Results were analyzed by PCR. At least three independent experiments were performed with ACT1 (not shown, expression was not influenced by glucose induced nutritional stress), anti-rabbit antibody (Ab) (unspecific antibody), and extracts prior to immunoprecipitation (input, whole-cell extract) as internal controls. Last two lines in A, panels a and b, represent Western blot controls of the Hxk2 level. The agarose electrophoresis shown is representative of results obtained from three independent experiments. B, quantification of Hxk2 association in wild-type (wt) and Δsnf1 mutant strain with the SUC2 promoter. Cells were treated as described for A (H-Glc, black bars; L-Glc, white bars), but ChIPs were analyzed by quantitative real time PCR. Data are expressed as signal normalized to the untreated sample. Error bars represent the standard error of the mean for three independent experiments. AU, arbitrary units.
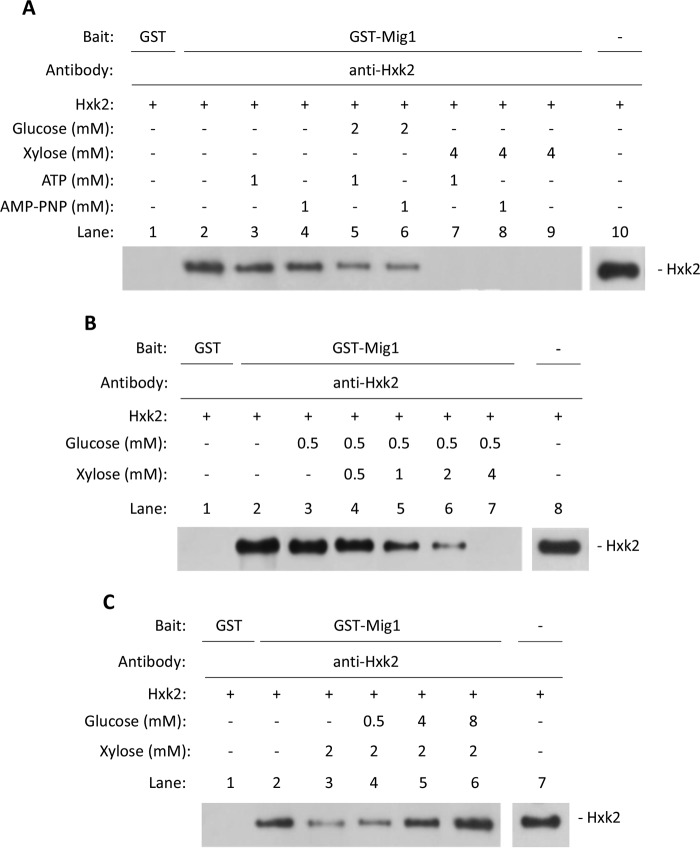
We then tested whether conformational changes in the Hxk2 protein affect the binding of Hxk2 to both Mig1 and the SUC2 promoter. d-Xylose binding in the presence of ATP promotes a conformational change in Hxk2 and its inactivation by autophosphorylation of serine 158 (55–58). Additionally, glucose protects against xylose-induced inactivation with efficiencies closely related to their respective glucose-Km or xylose-Ki values (58). Thus, it seems likely that xylose, a glucose non-phosphorylatable analogue, could induce an open conformation of the protein that mimics the low glucose conditions. To address this possibility, we first used GST pulldown assays to determine the effect of xylose on the interaction between Hxk2 and Mig1. GST and GST-Mig1 were purified from E. coli with the help of glutathione beads, and equal amounts of purified Hxk2 were incubated with the beads in the presence of different metabolites. After precipitation, the beads were washed, and bound Hxk2 was detected by Western blot with the help of a specific anti-Hxk2 antibody. Fig. 11A shows that GST-Mig1, but not GST alone, was able to precipitate the purified Hxk2 protein either in the absence or in the presence of substrates as glucose and ATP or the non-hydrolysable ATP analogue AMP-PNP. However, no signal was observed when the assay was done in the presence of 4 mm xylose (Fig. 11A, lanes 7–9). This experiment supports that the Hxk2-Mig1 interaction is abrogated by xylose and does not require either ATP or AMP-PNP. This result suggests that the efficiency of xylose as an inactivating agent of Hxk2-Mig1 interaction could be related to its ability to bind to Hxk2 and not to inhibit competitively the hexokinase reaction for which a metal-nucleotide complex is also required (58). To confirm that the xylose-Hxk2 binding is critical for suppression of Hxk2-Mig1 interaction, we measured it at a fixed glucose concentration (0.5 mm) and increasing xylose concentrations. Fig. 11B shows that increasing xylose concentration in the reaction mixture promotes the abrogation of Hxk2-Mig1 interaction, with total suppression at 4 mm xylose. A similar study was carried out with variable concentrations of glucose at a fixed (2 mm) concentration of xylose. The results shown in Fig. 11C support that glucose acts as a protecting agent with high efficiency to revert the changes induced by xylose. Taken together, these results show that the regulation of Hxk2 interaction with Mig1 involves a conformational change of the protein induced by the sugar substrate glucose and its analogue xylose. Thus, it could be assumed that the same flexibility of the Hxk2 active site that allows an “induced fit” (59) can promote an inactive conformation.
FIGURE 11.
In vitro conformational changes in Hxk2 affect its interaction with the Mig1 repressor. A, identical amounts of Hxk2 affinity-purified protein from bacteria were incubated in the presence of 2 mm glucose, 4 mm d-xylose, 1 mm MgATP2−, and 1 mm MgAMP-PNP for 30 min at 20 °C in PBS buffer. Total Hxk2 utilized in the experiment is shown (lane 10). B, time course of d-xylose inhibition of Hxk2-Mig1 interaction. Identical amounts of Hxk2 affinity-purified protein from bacteria were incubated in the presence of 0.5 mm glucose and increasing amounts of d-xylose, from 0.5 to 4 mm (lanes 4–7) for 30 min at 20 °C in PBS buffer. Total Hxk2 utilized in the experiment is shown (lane 8). C, time course of glucose activation of Hxk2-Mig1 interaction. Total Hxk2 utilized in the experiment is shown (lane 7). Identical amounts of Hxk2 affinity-purified protein from bacteria were incubated in the presence of 2 mm d-xylose and increasing amounts of glucose, from 0.5 to 8 mm (lanes 4–6) for 30 min at 20 °C in PBS buffer. Then, the GST-Mig1 fusion protein coupled to glutathione-Sepharose was incubated with the assay mixtures for 90 min at 4 °C in PBS buffer. Beads were gently washed five times with 2.5 ml of PBS buffer, boiled in 25 μl of sample-loading buffer, and analyzed by SDS-PAGE followed by Western blot using anti-Hxk2 antibodies. The Western blot shown is representative of results obtained from three independent experiments. In vitro kinase assay.
To confirm these results obtained in vitro with purified proteins, we performed ChIP assays in the presence of xylose to characterize in vivo Hxk2 binding to the SUC2 promoter. Fig. 12, A, panel a, and B, shows that the binding of Hxk2 to the SUC2 promoter was affected by the presence of xylose, and less than 6% of Hxk2 was bound to the SUC2-URS of the SUC2 promoter in these growth conditions, supporting that in the presence of xylose an inactivating Hxk2 conformational change is induced. As can be seen in Fig. 12, A, panel b, and B, the occupation rate of the SUC2 elements of the SUC2 promoter by Hxk2 in Δsnf1 mutant cells is also affected by the presence of xylose. A comparison between high glucose and xylose conditions revealed that nuclear Hxk2 was well engaged with the SUC2 elements of the SUC2 promoter in high glucose, but only 14% of Hxk2 was bound to the SUC2 elements in xylose conditions. In a Δsnf1 mutant strain Mig1 and Hxk2 are dephosphorylated, and cells are repressed both at high and low glucose conditions (Fig. 2). Although in this mutant strain an inactivating Hxk2 conformational change induced by xylose was observed (Fig. 12B), SUC2 gene expression was not promoted (Fig. 12C). A possible explanation is that although Hxk2 has adopted an inactive structure and only 14% of the protein remains associated with the SUC2 promoter (Fig. 12B), Mig1 is not phosphorylated and stays associated with the SUC2-URS element repressing gene transcription. Conversely, in a wild-type strain Hxk2 is removed from the repressor complex, favoring the phosphorylation of Mig1 by Snf1 kinase (30, 40), which promotes increased invertase activity (Fig. 12C). Therefore, Hxk2 phosphorylation does not affect the interaction of Hxk2 with Mig1, but it affects the regulation of Hxk2 nucleocytoplasmic shuttling between the nucleus and the cytoplasm (30). Taken together, these results strongly suggest that hexokinase 2 acts as an intracellular glucose sensor and that the structural conformation of Hxk2 regulates its interaction with Mig1 and it binding to the repressor complex of the SUC2 promoter.
FIGURE 12.
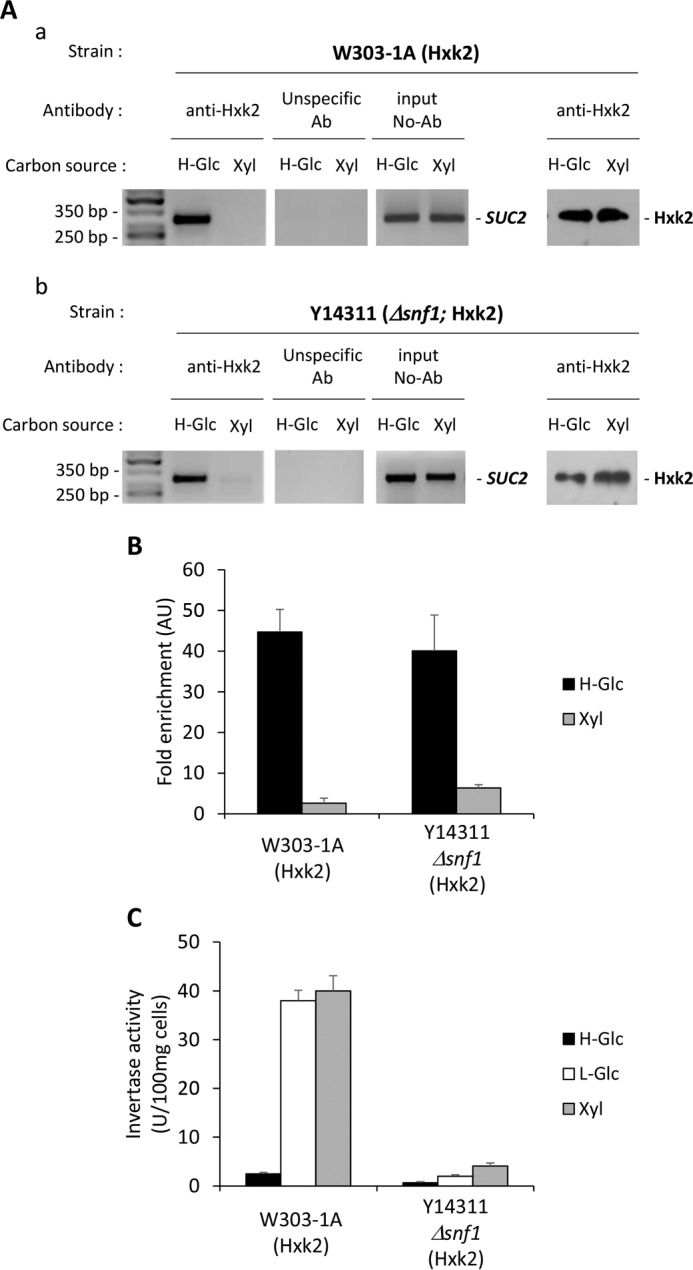
In vivo conformational changes of Hxk2 regulate its interaction with the Mig1 repressor. A, association of Hxk2 with the SUC2 promoter as measured by ChIPs. The wild-type strain W303-1A (panel a) and the mutant strain Y14311 (Δsnf1) (panel b) were grown in high glucose conditions (2% glucose, H-Glc) until an A600 of 0.8 was reached. Afterward, half of the culture was exposed to low glucose conditions (0.05% glucose plus 3% ethanol, L-Glc) for 60 min. Hxk2 and the SUC2 promoter association was determined by ChIP. Results were analyzed by PCR. At least three independent experiments were performed with ACT1 (not shown, expression was not influenced by glucose-induced nutritional stress), anti-rabbit antibody (Ab) (unspecific antibody), and extracts prior to immunoprecipitation (input, whole-cell extract) as internal controls. Last two lines in A, panel a and b, represent Western blot controls of the Hxk2 level. The agarose electrophoresis shown are representative of results obtained from three independent experiments. B, quantification of Hxk2 association in wild-type (wt) and Δsnf1 mutant strain with the SUC2 promoter. Cells were treated as described for A (H-Glc, black bars; L-Glc, white bars), but ChIPs were analyzed by quantitative real time PCR. Data are expressed as signal normalized to the untreated sample. Error bars represent the standard error of the mean for three independent experiments. AU, arbitrary units.
Discussion
Although previous studies (27, 30, 40) pointed to a role of Hxk2 in transcription, it was not clear how Hxk2 affects this process. Here, we show that Hxk2 is recruited to the SUC2-URS of the Mig1-dependent gene, SUC2, as a component of a repressor complex by direct physical interaction with Mig1. Subsequently, it promotes the formation of a functional repressor complex by recruiting the SNF1 complex proteins Snf1, Snf4, and Gal83, together with the Reg1 subunit of Glc7 protein phosphatase. This complex is disassembled during low glucose conditions and is reassembled in high glucose conditions. In this work, we tried to readdress two main questions. How does Hxk2 promote the formation and stabilization of a repressor complex at the SUC2 promoter? How does the glucose level regulate this process?
Mig1 and Mig2 have similar DNA-binding zinc fingers and recognize identical DNA-binding sequences (60). However, Mig1 protein is more important than Mig2 in SUC2 repression, because it is sufficient to achieve complete repression. To investigate the importance of Hxk2 in the Mig1 binding to the SUC2-URS, we used a ChIP assay and cells from both Δhxk1Δhxk2 double mutant and Δhxk2 single mutant strains expressing a Mig1 GFP-tagged protein. We found that in the absence of both Hxk1 and Hxk2 proteins, Mig1 cannot bind to the SUC2-URS region. In these mutant cells less than 5% of Mig1 was found associated with the SUC2 promoter both at high and low glucose conditions, a level as low as that measured in wild-type cells growing in low glucose. The most straightforward interpretation of these results could be that, in the absence of both Hxk1 and Hxk2 proteins, the Snf1 kinase is activated (39, 61), and Mig1 is phosphorylated and exported to the cytoplasm by binding to the Msn5 transporter (62). However, in the absence of Hxk2 protein, the recruitment of Mig1 to SUC2-URS is very similar to that found in a wild-type strain grown in low glucose conditions. A possible interpretation of these results could be that, because the HXK1 gene expression is highest during growth on low glucose and glucose-induced repression involves Hxk2, the Hxk1 protein could mimic, at least partially, the Hxk2 function at the SUC2 promoter repressor complex both in low glucose conditions and in Δhxk2 mutant cells. In this regard, Hxk1 and Hxk2 proteins have more than 77% identity, and the first 24 amino acids of the N-terminal region are identical in both. Because Hxk2-Mig1 interaction is mediated by a 10-amino acid motif in Hxk2 located between lysine 6 and methionine 15 (27, 63) (Hxk2 is numbered from residues 1 to 485; residue 1 is a valine because the initiator methionine is cleaved off from the primary translation product) and Hxk1 has in its N-terminal region the same 10-amino acid motif, we can supposed that Hxk1 could also interact with Mig1 protein in these metabolic conditions to prevent, at least in part, the Snf1 kinase-mediated phosphorylation of Mig1 at serine 311. Thus, in low glucose conditions 32% of the Mig1 protein is retained in the nucleus associated with the SUC2-URS, and 70% SUC2 repression is maintained. This behavior has not been previously described because the amount of Mig1-GFP retained in the nucleus is probably not detectable by conventional fluorescent microscopy, so in these conditions it has been claimed that Mig1 is localized exclusively in the cytoplasm (53). These results explain why full derepression of the SUC2 gene is only achieved in the absence of both Hxk1 and Hxk2 proteins (Fig. 2).
Moreover, the binding of the regulatory factors Snf1 and Reg1 to the SUC2 elements of the SUC2 promoter is Hxk2-dependent. In the absence of either Mig1 or Hxk2 proteins, recruitment of both Snf1 and Reg1 proteins to the SUC2 promoter is abolished. Because in a Δhxk2 mutant strain about 30% of Mig1 protein remains associated with the SUC2 promoter, we should detect a similar amount of Snf1 and Reg1 proteins associated with the SUC2-URS element if Mig1 was the critical factor for this association. However, less than 5% of these proteins detected was associated with the SUC2 promoter in a Δhxk2 strain, suggesting that the interaction of Snf1 and Reg1 proteins with the SUC2 repressor complex is Hxk2-dependent. These results are also supported by the fact that Hxk2 interacts with Snf1 and Reg1 proteins both at high and low glucose. Although Reg1 has been described as a predominantly cytoplasmic protein (54), our data for the first time support that a fraction of the Reg1 and Snf1 proteins present in the cell are constitutively associated with the SUC2-URS element of the SUC2 promoter, together with their substrates of interest, Hxk2 and Mig1, that exhibit the same nuclear localization.
Snf4 and Gal83 proteins, the regulatory units of the SNF1 complex, have also been detected to be associated with SUC2-URS elements at a similar level both in high and low glucose conditions. However, GST pulldown experiments show a weak interaction of Snf4 and Gal83 proteins with Hxk2, and in the absence of Snf1 no interaction of Snf4 with Hxk2 was observed. Thus, SNF1 complex regulatory proteins Snf4 and Gal83 binding to the SUC2 repressor complex appear to be Snf1-dependent.
Here, we show that Hxk2 is recruited to the SUC2-URS sequences of the SUC2 promoter by its interaction with the DNA-bound Mig1 protein, and subsequently it promotes the formation of a functional repressor complex. Snf1 and Reg1, whose interaction with Hxk2 was confirmed in vitro and in vivo, are also components of it. Mig2 appears to play no role in SUC2 repression when Mig1 is present, because in these conditions the absence of Mig2 has no effect on SUC2 expression. However, in wild-type cells grown in high and low glucose conditions, ∼25% of the Mig2 detected in the absence of Mig1 was recruited to the SUC2 promoter. The level of Mig2 associated with the SUC2 promoter increases in the absence of either Mig1 or Hxk2 proteins, which contributes to maintaining ∼70% of SUC2 repression, as observed in Δmig1 and Δhxk2 mutant cells (Fig. 2). Taking into account that Mig2 interacts with Mig1, Hxk2, and Snf1 proteins both in high and low glucose conditions, our results suggest that Mig2 could be a structural component of SUC2 repressor complex in high glucose. In the absence of Mig1 or Hxk2 proteins, an increased amount of Mig2 is dragged to the SUC2 promoter, and thus 70% of SUC2 repression is observed. In these conditions, SNF1 complex components would remain associated with the SUC2 promoter due to their interaction with Mig2. A full derepression of the SUC2 gene, in high and low glucose conditions, is only achieved in the absence of either Mig1-Mig2 or Hxk1-Hxk2 protein pairs, because in these conditions the SUC2 promoter repressor complex is completely disassembled.
A simplified diagram of how the glucose nutritional signal converge on Mig1 through the Hxk2, according to our results, is presented in Fig. 13. In high glucose conditions, the Gpr1 pathway triggers the activation of PKA, which activates the protein phosphatase Glc7-Reg1 (64, 65). This phosphatase maintains dephosphorylated Hxk2 and Mig1 proteins, which in this phosphorylation state have a nuclear localization (28, 30). Glc7-Reg1 also dephosphorylates and therefore maintains the inactivated kinase Snf1 (39, 66). Thus, under high glucose conditions it formed a repressor complex in the SUC2 promoter, which repress the gene expression. The repressor complex consists of different elements as follows: the transcriptional repressors Mig1 and Mig2 directly bound to the DNA, the protein Hxk2, the phosphatase Reg1-Glc7, and the three subunits of the SNF1 complex, Snf1, Snf4, and Gal83. Although Mig1 is the major transcriptional repressor, Mig2 is more a structural component that helps to maintain the repressor complex under high glucose conditions. The protein Hxk2 is a critical factor of this repressor complex. On the one hand, Hxk2 is required to stabilize the Mig1 association with the SUC2 promoter, and on the other hand, Snf1 and Reg1 association with the SUC2-URS is Hxk2-dependent. Snf4 and Gal83 proteins also take part in this repression complex, but its union with the SUC2 promoter is mediated by Snf1.
FIGURE 13.
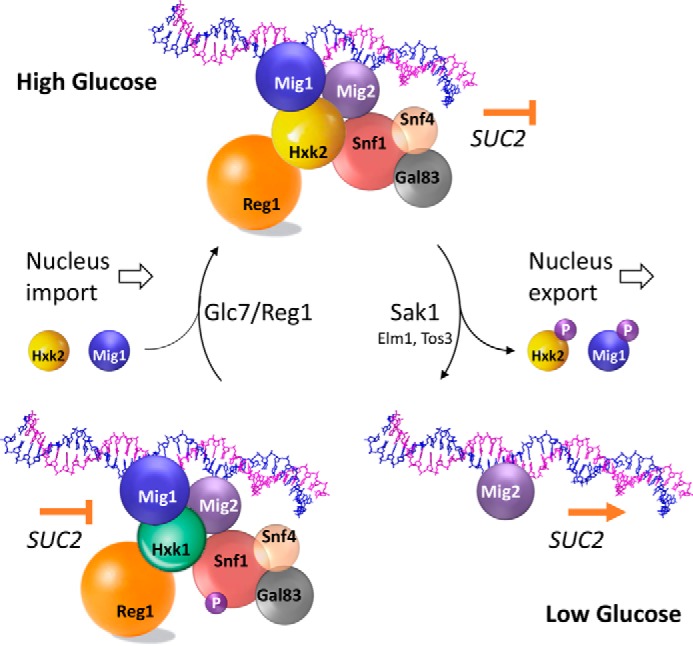
Model explaining how the SUC2 repressor complex dynamically disassembles and reassembles in a glucose-dependent manner.
When glucose becomes limiting, Sak1 kinase activates Snf1 by phosphorylation of threonine 210 (67). The activated Snf1 kinase phosphorylates Hxk2 and Mig1, and once they are phosphorylated they are exported to the cytoplasm (29, 30, 62), thus producing the disassembly of the repressor complex and the expression of the SUC2 gene. However, not all Mig1 protein is exported to the cytoplasm. Under low glucose conditions, about 30% of Mig1 remains associated with the SUC2 promoter due to the presence of Hxk1, which can mimic Hxk2 function sustaining the structure of the repressor complex. This model explains why the full expression of the SUC2 gene is only achieved in the double mutant strains Δmig1Δmig2 or Δhxk1Δhxk2.
Although our ChIP experiments support that the SUC2 repressor complex dynamically disassembles and reassembles in a glucose-dependent manner, they do not resolve the problem of how glucose signal is transmitted from the environment to the nucleus to regulate SUC2 expression. The results reported here provide for the first time a much clearer picture on this important point. The absence of Hxk2 impairs the recruitment of Mig1, Snf1, and Reg1 to the SUC2 promoter and hence the assembly of a functional repressor complex. Because Hxk2 seems to be a key factor for the assembly of SUC2 repressor complex, it could be a good candidate to operate as a glucose sensor that communicates glucose level information from the environment to the Mig1-regulated genes. The first unanswered question is as follows. How does phosphorylation of Hxk2 affect the Hxk2 recruitment to the SUC2 promoter? Our data clearly suggest that an impaired Hxk2 phosphorylation at serine 14, as happens in the absence of the Snf1 kinase, does not alter either repressor complex formation or its glucose-dependent regulation. Thus, Hxk2 promotes the assembly of the SUC2 repressor complex independently of its phosphorylation state. The phosphorylation state of Hxk2 regulates its nucleocytoplasmic shuttling between the nucleus and cytoplasm but not its assembly and disassembly to the repressor complex of the SUC2 promoter.
A second possibility is that Hxk2 assembly to the SUC2 promoter could be regulated by conformational changes induced by glucose levels (45). Because measuring dynamic changes of cytosolic glucose by FRET sensors demonstrated that cytosolic glucose accumulation is a direct function of glucose levels in the medium (44), it seems likely that an Hxk2 conformational change induced by glucose could be the signal that induces Mig1-Hxk2 interaction. The Hxk2 protein is folded into two domains of unequal size called the large and the small domain (68). These are separated by a deep cleft containing the residues making up the enzyme active site. In low glucose conditions, the Hxk2 structure adopts an open conformation of the cleft between the two domains. In high glucose conditions, a movement of about 8 Å of the domains closing the cleft was observed. The closed active site conformation is probably completed after additional conformational changes that accompany ATP binding. In addition to significant conformational changes of the loops involved in glucose and ATP binding, differences in the conformation of the external loops are also observed (45). Our in vivo and in vitro results show a striking correlation between a close conformation (high glucose conditions) of Hxk2 and its binding to the SUC2 repressor complex or an open conformation (low glucose conditions, xylose) of Hxk2 and its dissociation from the SUC2 promoter. Thus, here we present evidence that supports the hypothesis that Hxk2 acts as a glucose sensor in the signal transduction pathway mediating glucose-regulated gene expression in yeast; importantly, we document that the signaling activity of Hxk2 is linked to conformational changes induced by the glucose levels that promote its dissociation or reassociation to the SUC2 promoter to control gene expression.
Author Contributions
F. M. conceived and designed this work. P. H. and F. M. developed the methodology. M. V., A. F-C., A. R., and P. H. acquired the data. M. V., A. F-C., A. R., P. H., and F. M. analyzed and interpreted the data. A. F-C., A. R., P. H., and F. M. wrote and reviewed the manuscript. F. M. supervised and designed the study.
Acknowledgments
We are grateful to following people for generously providing yeast strains and plasmids: C.N. Cole for the pGEX-XPO1 plasmid and P. Sanz for the pWS93 plasmid.
This work was supported in whole or part by Ministerio de Ciencia e Innovación (Spain) Grants BFU2007-66063-C02-02 and BFU2010-19628-C02-01 (to F. M.). The authors declare that they have no conflicts of interest with the contents of this article.
- URS
- upstream repressing sequence
- AMP-PNP
- adenosine 5′-(β,γ-imino)triphosphate.
References
- 1. Struhl K. (1995) Yeast transcriptional regulatory mechanisms. Annu. Rev. Genet. 29, 651–674 [DOI] [PubMed] [Google Scholar]
- 2. Malavé T. M., and Dent S. Y. (2006) Transcriptional repression by Tup1-Ssn6. Biochem. Cell. Biol. 84, 437–443 [DOI] [PubMed] [Google Scholar]
- 3. Redd M. J., Arnaud M. B., and Johnson A. D. (1997) A complex composed of tup1 and ssn6 represses transcription in vitro. J. Biol. Chem. 272, 11193–11197 [DOI] [PubMed] [Google Scholar]
- 4. Varanasi U. S., Klis M., Mikesell P. B., and Trumbly R. J. (1996) The Cyc8 (Ssn6)-Tup1 corepressor complex is composed of one Cyc8 and four Tup1 subunits. Mol. Cell. Biol. 16, 6707–6714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Flores-Saaib R. D., and Courey A. J. (2000) Analysis of Groucho-histone interactions suggests mechanistic similarities between Groucho- and Tup1-mediated repression. Nucleic Acids Res. 28, 4189–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roth S. Y. (1995) Chromatin-mediated transcriptional repression in yeast. Curr. Opin. Genet. Dev. 5, 168–173 [DOI] [PubMed] [Google Scholar]
- 7. Nehlin J. O., and Ronne H. (1990) Yeast MIG1 repressor is related to the mammalian early growth response and Wilms' tumour finger proteins. EMBO J. 9, 2891–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lutfiyya L. L., Iyer V. R., DeRisi J., DeVit M. J., Brown P. O., and Johnston M. (1998) Characterization of three related glucose repressors and genes they regulate in Saccharomyces cerevisiae. Genetics 150, 1377–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lutfiyya L. L., and Johnston M. (1996) Two zinc-finger-containing repressors are responsible for glucose repression of SUC2 expression. Mol. Cell. Biol. 16, 4790–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang Z., and Reese J. C. (2004) Ssn6-Tup1 requires the ISW2 complex to position nucleosomes in Saccharomyces cerevisiae. EMBO J. 23, 2246–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Z., and Reese J. C. (2004) Redundant mechanisms are used by Ssn6-Tup1 in repressing chromosomal gene transcription in Saccharomyces cerevisiae. J. Biol. Chem. 279, 39240–39250 [DOI] [PubMed] [Google Scholar]
- 12. Gancedo J. M. (1998) Yeast carbon catabolite repression. Microbiol. Mol. Biol. Rev. 62, 334–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Treitel M. A., and Carlson M. (1995) Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc. Natl. Acad. Sci. U.S.A. 92, 3132–3136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gertz J., Siggia E. D., and Cohen B. A. (2009) Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature 457, 215–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rodríguez A., De La Cera T., Herrero P., and Moreno F. (2001) The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. Biochem. J. 355, 625–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moreno F., and Herrero P. (2002) The hexokinase 2-dependent glucose signal transduction pathway of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 26, 83–90 [DOI] [PubMed] [Google Scholar]
- 17. Moreno F., Ahuatzi D., Riera A., Palomino C. A., and Herrero P. (2005) Glucose sensing through the Hxk2-dependent signaling pathway. Biochem. Soc. Trans. 33, 265–268 [DOI] [PubMed] [Google Scholar]
- 18. Palomino A., Herrero P., and Moreno F. (2005) Rgt1, a glucose sensing transcription factor, is required for transcriptional repression of the HXK2 gene in Saccharomyces cerevisiae. Biochem. J. 388, 697–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Palomino A., Herrero P., and Moreno F. (2006) Tpk3 and Snf1 protein kinases regulate Rgt1 association with Saccharomyces cerevisiae HXK2 promoter. Nucleic Acids Res. 34, 1427–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schuurmans J. M., Boorsma A., Lascaris R., Hellingwerf K. J., and Teixeira de Mattos M. J. (2008) Physiological and transcriptional characterization of Saccharomyces cerevisiae strains with modified expression of catabolic regulators. FEMS Yeast Res. 8, 26–34 [DOI] [PubMed] [Google Scholar]
- 21. Schuurmans J. M., Rossell S. L., van Tuijl A., Bakker B. M., Hellingwerf K. J., and Teixeira de Mattos M. J. (2008) Effect of hxk2 deletion and HAP4 overexpression on fermentative capacity in Saccharomyces cerevisiae. FEMS Yeast Res. 8, 195–203 [DOI] [PubMed] [Google Scholar]
- 22. Ozcan S., and Johnston M. (1995) Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol. Cell. Biol. 15, 1564–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Noubhani A., Bunoust O., Bonini B. M., Thevelein J. M., Devin A., and Rigoulet M. (2009) The trehalose pathway regulates mitochondrial respiratory chain content through hexokinase 2 and cAMP in Saccharomyces cerevisiae. J. Biol. Chem. 284, 27229–27234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peláez R., Herrero P., and Moreno F. (2010) Functional domains of yeast hexokinase 2. Biochem. J. 432, 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Herrero P., Galíndez J., Ruiz N., Martínez-Campa C., and Moreno F. (1995) Transcriptional regulation of the Saccharomyces cerevisiae HXK1, HXK2 and GLK1 genes. Yeast 11, 137–144 [DOI] [PubMed] [Google Scholar]
- 26. Randez-Gil F., Herrero P., Sanz P., Prieto J. A., and Moreno F. (1998) Hexokinase PII has a double cytosolic-nuclear localisation in Saccharomyces cerevisiae. FEBS Lett. 425, 475–478 [DOI] [PubMed] [Google Scholar]
- 27. Ahuatzi D., Herrero P., de la Cera T., and Moreno F. (2004) The glucose-regulated nuclear localization of hexokinase 2 in Saccharomyces cerevisiae is Mig1-dependent. J. Biol. Chem. 279, 14440–14446 [DOI] [PubMed] [Google Scholar]
- 28. Peláez R., Fernández-García P., Herrero P., and Moreno F. (2012) Nuclear import of the yeast hexokinase 2 protein requires α/β-importin-dependent pathway. J. Biol. Chem. 287, 3518–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peláez R., Herrero P., and Moreno F. (2009) Nuclear export of the yeast hexokinase 2 protein requires the Xpo1 (Crm1)-dependent pathway. J. Biol. Chem. 284, 20548–20555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fernández-García P., Peláez R., Herrero P., and Moreno F. (2012) Phosphorylation of yeast hexokinase 2 regulates its nucleocytoplasmic shuttling. J. Biol. Chem. 287, 42151–42164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carlson M. (1999) Glucose repression in yeast. Curr. Opin. Microbiol. 2, 202–207 [DOI] [PubMed] [Google Scholar]
- 32. Vincent O., Townley R., Kuchin S., and Carlson M. (2001) Subcellular localization of the Snf1 kinase is regulated by specific β subunits and a novel glucose signaling mechanism. Genes Dev. 15, 1104–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nath N., McCartney R. R., and Schmidt M. C. (2003) Yeast Pak1 kinase associates with and activates Snf1. Mol. Cell. Biol. 23, 3909–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hong S. P., Leiper F. C., Woods A., Carling D., and Carlson M. (2003) Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. U.S.A. 100, 8839–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sutherland C. M., Hawley S. A., McCartney R. R., Leech A., Stark M. J., Schmidt M. C., and Hardie D. G. (2003) Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 13, 1299–1305 [DOI] [PubMed] [Google Scholar]
- 36. Mayer F. V., Heath R., Underwood E., Sanders M. J., Carmena D., McCartney R. R., Leiper F. C., Xiao B., Jing C., Walker P. A., Haire L. F., Ogrodowicz R., Martin S. R., Schmidt M. C., Gamblin S. J., et al. (2011) ADP regulates SNF1, the Saccharomyces cerevisiae homolog of AMP-activated protein kinase. Cell Metab. 14, 707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Treitel M. A., Kuchin S., and Carlson M. (1998) Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18, 6273–6280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McCartney R. R., and Schmidt M. C. (2001) Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276, 36460–36466 [DOI] [PubMed] [Google Scholar]
- 39. Sanz P., Alms G. R., Haystead T. A., and Carlson M. (2000) Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol. Cell. Biol. 20, 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ahuatzi D., Riera A., Peláez R., Herrero P., and Moreno F. (2007) Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J. Biol. Chem. 282, 4485–4493 [DOI] [PubMed] [Google Scholar]
- 41. Tu J., and Carlson M. (1995) REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 14, 5939–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Alms G. R., Sanz P., Carlson M., and Haystead T. A. (1999) Reg1p targets protein phosphatase 1 to dephosphorylate hexokinase II in Saccharomyces cerevisiae: characterizing the effects of a phosphatase subunit on the yeast proteome. EMBO J. 18, 4157–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Neigeborn L., and Carlson M. (1984) Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics 108, 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bermejo C., Haerizadeh F., Takanaga H., Chermak D., and Frommer W. B. (2011) Optical sensors for measuring dynamic changes of cytosolic metabolite levels in yeast. Nat. Protoc. 6, 1806–1817 [DOI] [PubMed] [Google Scholar]
- 45. Kuser P. R., Krauchenco S., Antunes O. A., and Polikarpov I. (2000) The high resolution crystal structure of yeast hexokinase PII with the correct primary sequence provides new insights into its mechanism of action. J. Biol. Chem. 275, 20814–20821 [DOI] [PubMed] [Google Scholar]
- 46. Wallis J. W., Chrebet G., Brodsky G., Rolfe M., and Rothstein R. (1989) A hyper-recombination mutation in S. cerevisiae identifies a novel eukaryotic topoisomerase. Cell 58, 409–419 [DOI] [PubMed] [Google Scholar]
- 47. Ma H., and Botstein D. (1986) Effects of null mutations in the hexokinase genes of Saccharomyces cerevisiae on catabolite repression. Mol. Cell. Biol. 6, 4046–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Longtine M. S., McKenzie A. 3rd, Demarini D. J., Shah N. G., Wach A., Brachat A., Philippsen P., and Pringle J. R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961 [DOI] [PubMed] [Google Scholar]
- 49. Hammell C. M., Gross S., Zenklusen D., Heath C. V., Stutz F., Moore C., and Cole C. N. (2002) Coupling of termination, 3′ processing, and mRNA export. Mol. Cell. Biol. 22, 6441–6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gascón S., and Lampen J. O. (1968) Purification of the internal invertase of yeast. J. Biol. Chem. 243, 1567–1572 [PubMed] [Google Scholar]
- 51. Bu Y., and Schmidt M. C. (1998) Identification of cis-acting elements in the SUC2 promoter of Saccharomyces cerevisiae required for activation of transcription. Nucleic Acids Res. 26, 1002–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hecht A., Strahl-Bolsinger S., and Grunstein M. (1999) Mapping DNA interaction sites of chromosomal proteins. Crosslinking studies in yeast. Methods Mol. Biol. 119, 469–479 [DOI] [PubMed] [Google Scholar]
- 53. De Vit M. J., Waddle J. A., and Johnston M. (1997) Regulated nuclear translocation of the Mig1 glucose repressor. Mol. Biol. Cell 8, 1603–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dombek K. M., Voronkova V., Raney A., and Young E. T. (1999) Functional analysis of the yeast Glc7-binding protein Reg1 identifies a protein phosphatase type 1-binding motif as essential for repression of ADH2 expression. Mol. Cell. Biol. 19, 6029–6040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fernández R., Herrero P., Fernández M. T., and Moreno F. (1986) Mechanism of inactivation of hexokinase PII of Saccharomyces cerevisiae by d-xylose. J. Gen. Microbiol. 132, 3467–3472 [DOI] [PubMed] [Google Scholar]
- 56. Fernández R., Herrero P., and Moreno F. (1985) Inhibition and inactivation of glucose-phosphorylating enzymes from Saccharomyces cerevisiae by d-xylose. J. Gen. Microbiol. 131, 2705–2709 [DOI] [PubMed] [Google Scholar]
- 57. Heidrich K., Otto A., Behlke J., Rush J., Wenzel K. W., and Kriegel T. (1997) Autophosphorylation-inactivation site of hexokinase 2 in Saccharomyces cerevisiae. Biochemistry 36, 1960–1964 [DOI] [PubMed] [Google Scholar]
- 58. DelaFuente G. (1970) Specific inactivation of yeast hexokinase induced by xylose in the presence of a phosphoryl donor substrate. Eur. J. Biochem. 16, 240–243 [DOI] [PubMed] [Google Scholar]
- 59. Koshland D. E. (1958) Application of a theory of enzyme specificity to protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 44, 98–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lundin M., Nehlin J. O., and Ronne H. (1994) Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol. Cell. Biol. 14, 1979–1985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Mayordomo I., Estruch F., and Sanz P. (2002) Convergence of the target of rapamycin and the Snf1 protein kinase pathways in the regulation of the subcellular localization of Msn2, a transcriptional activator of STRE (stress response element)-regulated genes. J. Biol. Chem. 277, 35650–35656 [DOI] [PubMed] [Google Scholar]
- 62. DeVit M. J., and Johnston M. (1999) The nuclear exportin Msn5 is required for nuclear export of the Mig1 glucose repressor of Saccharomyces cerevisiae. Curr. Biol. 9, 1231–1241 [DOI] [PubMed] [Google Scholar]
- 63. Herrero P., Martínez-Campa C., and Moreno F. (1998) The hexokinase 2 protein participates in regulatory DNA-protein complexes necessary for glucose repression of the SUC2 gene in Saccharomyces cerevisiae. FEBS Lett. 434, 71–76 [DOI] [PubMed] [Google Scholar]
- 64. Castermans D., Somers I., Kriel J., Louwet W., Wera S., Versele M., Janssens V., and Thevelein J. M. (2012) Glucose-induced posttranslational activation of protein phosphatases PP2A and PP1 in yeast. Cell Res. 22, 1058–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Barrett L., Orlova M., Maziarz M., and Kuchin S. (2012) Protein kinase A contributes to the negative control of Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot. Cell 11, 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Tabba S., Mangat S., McCartney R., and Schmidt M. C. (2010) PP1 phosphatase-binding motif in Reg1 protein of Saccharomyces cerevisiae is required for interaction with both the PP1 phosphatase Glc7 and the Snf1 protein kinase. Cell. Signal. 22, 1013–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu Y., Xu X., and Carlson M. (2011) Interaction of SNF1 protein kinase with its activating kinase Sak1. Eukaryot. Cell 10, 313–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Aleshin A. E., Zeng C., Bourenkov G. P., Bartunik H. D., Fromm H. J., and Honzatko R. B. (1998) The mechanism of regulation of hexokinase: new insights from the crystal structure of recombinant human brain hexokinase complexed with glucose and glucose-6-phosphate. Structure 6, 39–50 [DOI] [PubMed] [Google Scholar]
- 69. Mercado J. J., Vincent O., and Gancedo J. M. (1991) Regions in the promoter of the yeast FBP1 gene implicated in catabolite repression may bind the product of the regulatory gene MIG1. FEBS Lett. 291, 97–100 [DOI] [PubMed] [Google Scholar]
- 70. Fernández-Cid A., Vega M., Herrero P., and Moreno F. (2012) Yeast importin-β is required for nuclear import of the Mig2 repressor. BMC Cell Biol. 13, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]