Abstract
ASAP1 regulates F-actin-based structures and functions, including focal adhesions (FAs) and circular dorsal ruffles (CDRs), cell spreading and migration. ASAP1 function requires its N-terminal BAR domain. We discovered that nonmuscle myosin 2A (NM2A) directly bound the BAR-PH tandem of ASAP1 in vitro. ASAP1 and NM2A co-immunoprecipitated and colocalized in cells. Knockdown of ASAP1 reduced colocalization of NM2A and F-actin in cells. Knockdown of ASAP1 or NM2A recapitulated each other's effects on FAs, cell migration, cell spreading, and CDRs. The NM2A-interacting BAR domain contributed to ASAP1 control of cell spreading and CDRs. Exogenous expression of NM2A rescued the effect of ASAP1 knockdown on CDRs but ASAP1 did not rescue NM2A knockdown defect in CDRs. Our results support the hypothesis that ASAP1 is a positive regulator of NM2A. Given other binding partners of ASAP1, ASAP1 may directly link signaling and the mechanical machinery of cell migration.
Keywords: actin, ADP ribosylation factor (ARF), cell biology, cell migration, cytoskeleton, GTPase-activating protein (GAP), myosin
Introduction
Arf GTPase-activating proteins (Arf GAPs)3 are a structurally diverse family of proteins encoded by 31 genes in humans (1). The enzymatic function of Arf GAPs is to facilitate the hydrolysis of GTP on ADP-ribosylation factors (Arfs), which are members of the Ras superfamily. Both Arf GAPs and Arfs have been implicated as regulators of the actin cytoskeleton (2, 3). The role of Arf GAPs in actin-based structures has been primarily examined from the perspective of signaling. Two Arf GAPs are found to affect Arf6 and Rac1 activity (4, 5). The changes in Arf6 and Rac1, however, cannot completely account for the specific effects of particular Arf GAPs on the actin-based structures called focal adhesions (FAs) (4). Structural domains outside the catalytic Arf GAP domain may also contribute to the control of the actin cytoskeleton.
ASAP1 is an Arf GAP that associates with and regulates actin-based structures including FAs and circular dorsal ruffles (CDRs). It also affects cell spreading and cell migration, which involve actin remodeling (6–9). ASAP1 is composed of BAR, PH, Arf GAP, Ank repeats, Proline-rich, E/DLPPKP repeat, and SH3 domains (Fig. 1A). ASAP1 binds to Src and CrkL through the proline-rich domain and focal adhesion kinase (FAK) via its SH3 domain (6, 10, 11), which has been found to contribute to regulation of cell adhesions. The BAR domain of ASAP1 is also critical for its cellular function in regulation of actin-based structures (6). Specific mechanisms by which the BAR domain contributes to ASAP1 control of actin-based structures have not been elucidated.
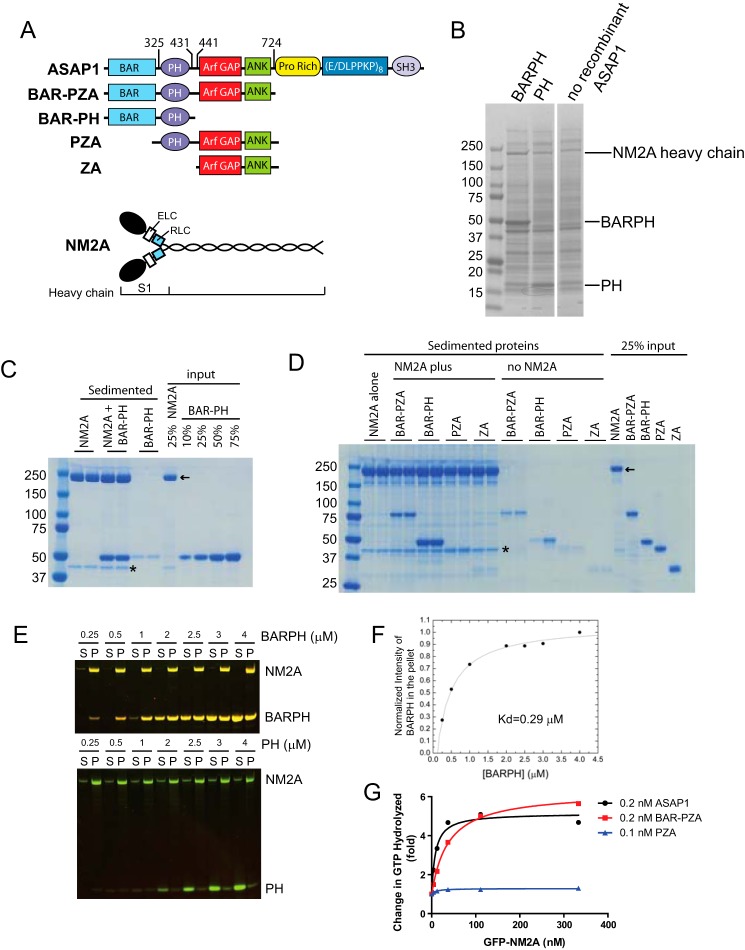
FIGURE 1.
ASAP1 directly interacts with NM2A. A, schematic representations of ASAP1 and NM2A domain structure and recombinant proteins used in this study. Targeting of ASAP1 to membranes is mediated primarily by the Pro-rich and SH3 domains. The PH domain regulates GAP activity of ASAP1. B, Coomassie Blue-stained SDS-PAGE gel of proteins sedimented with sucrose-loaded LUVs alone (no recombinant ASAP1) or coated with BAR-PH or PH. The image is a gel in which two central lanes were removed. The image of the gel is otherwise unaltered. C, co-sedimentation of ASAP1 and purified platelet NM2A. BAR-PH and NM2A were incubated either alone or together as indicated. Reaction mixtures were centrifuged and sedimented proteins were separated by SDS-PAGE and visualized with Coomassie Blue stain. Included in the gel were 25% of the NM2A in the reaction and the indicated percent of BAR-PH in the reaction mixture. Reactions were run in duplicate, and one experiment of three is shown. D, co-sedimentation of ASAP1 fragments (1 μm) with NM2A. The indicated fragments of ASAP1 were incubated with or without 0.6 μm NM2A and sedimented by centrifugation. Reactions were run in duplicate with the indicated proteins. The sedimented proteins and 25% input were separated by SDS-PAGE and visualized with Coomassie Blue stain. Asterisk indicates actin. Arrow indicates NM2A. Different fragments of ASAP1 were annotated in the schematic of ASAP1 domain structure. E and F, concentration-dependent ASAP1-NM2A association. NM2A was expressed in and purified from Sf9 cells. BAR-PH was titrated into a binding reaction containing 0.6 μm NM2A. At high concentrations of BAR-PH, some BAR-PH remains in the supernatant (S), indicating saturated binding. P, pellet. PH remains in the supernatant at all concentrations tested. The data of BAR-PH, after background corrections, were fit to an equation for one site binding. G, purified NM2A stimulates ASAP1 GAP activity dependent on BAR domain. GFP-NM2A purified from Sf9 cells was titrated into Arf GAP reactions containing the indicated recombinant ASAP1.
Cytoplasmic nonmuscle myosin 2A (NM2A) is an F-actin binding ATPase that functions as a molecular motor in cellular events that involve force or translocation (12). NM2A is comprised of 2 heavy chains, 2 essential light chains (ECL), and 2 regulatory light chains (RLC) (Fig. 1A) and self-associates to form bipolar minifilaments. NM2A cross-links F-actin and the complex of NM2A and F-actin generates contractility necessary for cellular functions including regulation of FAs, cell spreading, and cell migration.
Here, we set out to identify proteins that bind to and mediate the cellular functions of ASAP1. We found that NM2A directly binds to ASAP1 and associates with ASAP1 in cells. Knockdown of ASAP1 in cells reduced NM2A-F-actin colocalization. Knockdown of ASAP1 or NM2A had similar effects on FAs, cell spreading, cell migration, and CDRs. The effects of ASAP1 on CDRs require NM2A. Based on our results, we propose that ASAP1 positively regulates NM2A and speculate that ASAP1 integrates cell signals and links them to the machinery of actin remodeling.
Experimental Procedures
Plasmids, Antibodies, and Reagents
Mammalian expression vectors for N-terminal Flag-tagged ASAP1b and [ΔBAR]ASAP1b have been described (6). Rabbit anti-ASAP1 antisera (antisera 642) has been described (9). Anti-NM2A mouse monoclonal and rabbit polyclonal Abs were from Abcam (Cambridge, MA). Anti-NM2A polyclonal Ab (PRB-440P) was from Covance Research Products Inc. Recombinant human PDGF-BB, anti-vinculin monoclonal Ab (hVIN-1) and fibronectin were purchased from Sigma-Aldrich (St. Louis, MO). Anti-paxillin and β-actin monoclonal Abs were from BD Biosciences (San Jose, CA). Anti-Hsp70 was from Santa Cruz Biotechnology (Dallas, TX). Rhodamine- or Alexa Fluor-labeled phalloidin, Alexa Fluor-labeled secondary antibodies and Dynabeads were from Invitrogen (Carlsbad, CA). Horseradish peroxidase-conjugated anti-mouse and anti-rabbit IgG Abs were from Bio-Rad.
Cell Culture and Transfection
siRNA against mouse ASAP1 (5′-GCAUGCAUCUAGACGCUUAUU-3′), human ASAP1, mouse and human NM2A (MYH9) ON-TARGET smart pool and control siRNA (siCONTROL Non-Targeting siRNA #4) were purchased from Thermo Scientific (Lafayette, CO). Subconfluent cells were transfected with 100 nm siRNA using DharmaFECT transfection reagent 1 (Thermo Scientific, Lafayette, CO). Functional experiments with siRNA transfected cells were performed 72 h following transfection when ASAP1 and NM2A expression was reduced. For rescue experiments requiring exogenous protein expression, 48 h after siRNA transfection, cells were transfected with plasmids expressing Flag-ASAP1b, Flag-[ΔBAR]ASAP1b, GFP-NM2A, GFP or pCI vector using Lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA). Twenty-four hours later, effects on CDRs were assessed using confocal microscopy.
Protein Expression and Purification
Recombinant full-length mASAP1b was expressed in baculovirus-infected Sf9 cells and isolated using ion exchange and size exclusion columns. Bacterial expression vectors for [1–724]ASAP1-His6 (BAR-PZA), [1–431]ASAP1-His6 (BAR-PH), His10-[325–724]ASAP1 (PZA), His10-[325–441]ASAP1 (PH), His10-[441–724]ASAP1 (ZA), and myristoylated Arf1 (myrArf1) were described previously (13,14). Briefly, ASAP1 PZA and ZA were expressed in bacteria and purified using HiTrapQ followed by HisTrap columns (GE Healthcare) and then dialyzed. ASAP1 BAR-PZA and BAR-PH were expressed in bacteria and purified using HisTrap columns followed by hydroxylapatite and/or Sephacryl S100 columns. The purification of platelet NM2 was described (15). Flag-tagged or GFP-tagged cDNA construct of full-length (amino acids 1–1960) human NM2A were overproduced along with cDNA constructs encoding for myosin regulatory light chain and essential light chain in the baculovirus/Sf9 system.
Liposome Precipitation and Floatation Assays
All lipids were obtained from Avanti Polar Lipids. Large unilamellar vesicles (LUVs) were prepared by extruding the lipid mixture through 1-μm pore polycarbonate membrane (13, 14). In liposome precipitation assay, sucrose-loaded LUVs contained 40% phosphatidylcholine (PC), 25% phosphatidylethanolamine (PE), 15% phosphatidylserine (PS), 9% PtdIns (PI), 1% PtdIns (4,5)P2 (PIP2) and 10% cholesterol. In liposome floatation assays, LUVs contained 40% PC, 19.8% PE, 0.2% rhodamine-PE, 5% DGS-NTA (Ni2+), 15% PS, 9% PI, 1% PIP2, and 10% cholesterol. Two lysis buffers containing protease and phosphatase inhibitor mixture were used to prepare lysates of NIH3T3 fibroblasts: 25 mm Tris-HCl, pH 7.5, 100 mm NaCl, 0.5 mm MgCl2, 1 mm DTT, and 0.08% Triton X-100 (sucrose-loaded LUV precipitation); 50 mm HEPES pH 7.4, 120 mm Potassium acetate, 1 mm MgCl2 and 1 mm DTT (LUV floatation). Cells were lysed using isobiotec cell homogenizer with 4 μm clearance and sonication, followed by centrifugation at 20,800 × g for 15 min to obtain precleared lysates.
10 μm BARPH-His6 or His10-PH proteins were incubated with 1 mm LUVs either loaded with or without sucrose for 5 min at room temperature. Lysates (160∼175 μg) were added to the LUVs and incubated for 15 min at room temperature. The bound proteins were precipitated with sucrose-loaded LUVs by centrifugation at 253,800 × g for 15 min at 4 °C. In the floatation assay, lysis buffer containing 60% (w/v) sucrose was added in equal volume to the mixtures of LUVs and lysates to a final concentration of 30%, and then overlaid with 25% sucrose in lysis buffer. The bound proteins were floated with LUVs by centrifugation in a S55S swinging bucket rotor at 53000 rpm (240,000 × g) for 30 min at 4 °C. The bound proteins were dissolved in SDS sample buffer, separated by SDS-PAGE, visualized by Coomassie Blue staining, and identified by mass spectrometry.
Mass Spectrometry
The samples were separated by SDS-PAGE and each sample lane cut into 12 slices. The protein bands were reduced, alkylated with iodoacetamide, and in-gel trypsin digested for 16 h at 37 °C, as described (16). The digested peptides were extracted from the gel slices and lyophilized to dryness. The dried peptides were resuspended in water containing 2% acetonitrile, 0.5% acetic acid, and injected onto a 0.2 × 50 mm Magic C18AQ reverse phase column (Michrom Bioresources, Inc.) using the Paradigm MS4 HPLC (Michrom Bioresources, Inc.). Peptides were separated at a flow rate of 2 ml/min followed by online analysis by tandem mass spectrometry using an LTQ ion trap mass spectrometer (Thermo Scientific) equipped with an ADVANCE CaptiveSpray ion source (Michrom Bioresources, Inc.). Peptides were eluted into the mass spectrometer using a linear gradient from 95% mobile phase A (2% acetonitrile, 0.5% acetic acid, 97.5% water) to 65% mobile phase B (10% water, 0.5% formic acid, 89.5% acetonitrile) over 20 min followed by 95% mobile phase B over 5 min. Peptides were detected in positive ion mode using a data-dependent method in which the nine most abundant ions detected in an initial survey scan were selected for MS/MS analysis. The raw mass spectrometry data were searched against the IPI database using TurboSEQUEST in BioWorks v. 3.2 (ThermoElectron, San Jose, CA).
Immunoprecipitation and Western Blotting
NIH3T3 fibroblasts or HeLa cells after 72 h incubation with non-target or NM2A siRNA, were mechanically broken (passage through a 28-gauge needle) in ice-cold Nonidet P-40 buffer [20 mm Tris·HCl (pH 7.4), 50 mm NaCl, 0.1% Nonidet P-40, and EDTA-free protease inhibitor mixture]. Lysates (100 μg) were incubated (4 °C with mixing, overnight) with 2 μg of NM2A antibody (PRB-440P) or control IgG and 30 μl of Dynabeads Protein G (4 °C with mixing, 6 h). After incubation, beads were washed five times with Nonidet P-40 buffer. The bound proteins were eluted with SDS sample buffer and detected by Western blotting using standard methods and visualized with enhanced chemiluminescence.
Immunofluorescence and Confocal Microscopy
Cells were plated on 10 μg/ml fibronectin coated coverslips and fixed in 4% paraformaldehyde. For CDR experiments, cells plated in plain Opti-MEM for 5.5 h were treated with 20 ng/ml PDGF for the times indicated before fixation. Fixed cells were incubated in 15 mm glycine for 10 min, 50 mm NH4Cl for 2 × 10 min, then permeabilized and blocked with 0.2% saponin, 0.5% BSA, and 1% FBS in PBS for 20 min. Cells were incubated in primary antibody for 1 h, followed by secondary antibodies for 1h and mounted in DakoCytomation Fluorescent Mounting Medium (Dako, Carpinteria, CA). Images for fixed cells were taken on a Zeiss LSM 510 attached to a Zeiss Axiovert 100 m with a 63 × 1.4 numerical-aperture (NA) plan Neofluar oil immersion lens (Carl Zeiss, Thornwood, NY).
Co-sedimentation Assays
Two sources of full-length NM2A were used for binding to ASAP1 fragments. First, purified platelet NM2A at 0.13 mg/ml (0.6 μm) was incubated at 30 °C for 20 min with 1 μm of purified ASAP1 fragments (BAR-PZA, BAR-PH, PZA, or ZA) in the buffer containing 25 mm HEPES, pH7.4, 150 mm KCl, 1 mm MgCl2, and 1 mm DTT. The mixtures were sedimented by ultracentrifugation at 200,000 × g for 20 min at 4 °C. The sedimented proteins were separated by SDS-PAGE and visualized by Coomassie Blue staining. Second, 0.6 μm recombinant NM2A in 10 mm MOPS pH 7.2, 25 mm NaCl, 5 mm MgCl2, 0.1 mm EGTA, and 2 mm DTT was mixed with an equal volume of BAR-PH (0.25–4 μm) in PBS and incubated for 10 min at 22 °C. The samples were sedimented for 15 min at 100,000 × g in a Beckman TLA-100 rotor at 4 °C. The supernatant was removed and the pellet resuspended in an equal volume of assay buffer. Supernatant and pellet fractions were resolved on a 4–12% BIS-Tris gel (Invitrogen) and the gel stained with PageBlue (Fermentas). Gels were scanned with an Odyssey system (Li-Cor Biosciences). Image J was used to quantify the bands on the protein gel by densitometry.
Enzymatic Assays
The enzymatic activity of ASAP1 to convert myrArf1·GTP to myrArf1·GDP was determined as described (13). Full-length GFP-NM2A was titrated into the reaction containing myrArf1·GTP as the substrate and different concentrations of full-length ASAP1, BAR-PZA, or PZA as enzyme. Reactions were stopped after 3 min. Protein-bound nucleotide was trapped on nitrocellulose, eluted with formic acid, and separated by thin-layer chromatography. The hydrolysis of the Arf-bound GTP was determined by the relative mass of radiolabeled GDP and GTP.
Cell Spreading and Migration Assays
To measure the rate of cell spreading, NIH3T3 fibroblasts or HeLa cells were allowed to spread on 10 μg/ml fibronectin coated coverslips and fixed at various times. Cell area was measured based on F-actin staining by rhodamine-phalloidin. For migration assay, NIH3T3 fibroblasts were plated at 10,000 cell/well on μ-Slide 8 well (ibidi, Verona, WI) coated with 10 μg/ml fibronectin. The next day, cells were imaged in DMEM containing no phenol red and supplemented with penicillin/streptomycin using phase contrast microscopy (PMT 488 nm line of Argon laser at 0.2% power) on a Zeiss 710 NLO attached to a Axio Observer microscope with a 20×, 0.8 numerical-aperture (NA) Plan Apochromat lens (Carl Zeiss, Thornwood, NY). Cells were maintained at 37 °C and 5% CO2 in an environmental chamber during imaging. Images were acquired at 8-min intervals for at least 5.5∼6.5 h. Cell movement was analyzed by manually tracking the nucleus of each cell from the time-lapse images using the Manual Tracking plugin for ImageJ developed by F. Cordelières. Migration speed was calculated by dividing the total distance moved by the time. Plots of cell trajectories emanating from a common origin were generated by the DiPer program (17).
Image Analysis and Statistics
The number of FAs per cell (4, 18) and cell area were analyzed using the particle analysis tool in ImageJ (Rasband, W.S., ImageJ, U. S. National Institutes of Health, Bethesda, MD). For quantification of colocalization of NM2A with F-actin, z stack images of consecutive optical planes spaced by 0.3 μm were acquired to cover the whole cell volume using confocal microscopy. Pearson's coefficient was determined using Imaris 7.4.0. Differences between treatments were analyzed by one-way ANOVA using Bonferroni's multiple comparison test with p < 0.05 considered to be significant.
Results
ASAP1 Binds Directly to NM2A
To identify proteins that bind the BAR domain of ASAP1, BAR-PH and, as a control, the isolated PH domain of ASAP1 were expressed and purified from bacteria and adsorbed to sucrose-filled large unilamellar vesicles (LUVs) containing phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2), which specifically binds to the ASAP1 PH domain. The isolated BAR domain of ASAP1 is not stable as a recombinant protein, but the BAR-PH tandem is. LUVs with no ASAP1 fragments were another control. LUVs were mixed with lysates of NIH3T3 fibroblasts and rapidly sedimented by centrifugation. Sedimented proteins were separated by SDS-PAGE and stained with Coomassie blue (Fig. 1B). Proteins enriched in the samples containing BAR-PH, compared with the PH domain or LUV alone, were identified by mass spectrometric analysis. The major enriched protein was NM2A, represented by NM2A heavy chain that ran between the 150 and 250 kDa markers. NM2A was also observed in the LUVs alone lane and the PH plus LUVs lane at similar quantities in each. The band was of greater intensity in the BAR-PH plus LUVs lane. NM2A was also enriched in the BAR-PH-coated LUVs that lack sucrose and were floated through a sucrose gradient (data not shown).
To test for direct interaction between ASAP1 and NM2A, a co-sedimentation assay was performed using purified proteins in the absence of LUVs. NM2A forms filaments that sediment in a centrifugal field, which is the basis for our binding assays. First, enriched NM2A from platelets was incubated with BAR-PH of ASAP1. NM2A filaments formed during the incubation were sedimented by centrifugation (Fig. 1C). Duplicate samples from one experiment are shown. When incubated under these conditions, 45 ± 8% of the NM2A in the mixture sedimented in the absence of BAR-PH and 50 ± 8% in the presence of BAR-PH (mean ± S.D. of three experiments in duplicate). In the absence of NM2A, 5 ± 3% of BAR-PH sedimented (mean ± S.D. of four experiments in duplicate). In the presence of NM2A, 45 ± 5% of the BAR-PH sedimented (mean ± S.D. of three experiments in duplicate). To determine which domain of ASAP1 mediated the interaction, enriched NM2A from platelets was incubated with different fragments of ASAP1 (Fig. 1D). ASAP1 fragments sedimented to some extent in the absence of NM2A. BAR-PH and BAR-PZA sedimented more than other fragments, but the sedimented proteins did not exceed 10% of the total protein used in the assay. In contrast, ∼50% of BAR-PH co-sedimented with NM2A filaments, indicating association of BAR-PH with the sedimented NM2A (Fig. 1, C and D). BAR-PZA also co-sedimented with NM2A filaments but not as efficiently as BAR-PH. In contrast, ASAP1 fragments lacking the BAR domain, PZA and ZA did not sediment with NM2A (Fig. 1D). Direct interaction between ASAP1 BAR-PH domain and NM2A was further examined using a homogeneous preparation of recombinant NM2A expressed in Sf9 insect cells. BAR-PH, but not PH, co-sedimented with recombinant NM2A. The binding was concentration-dependent (Kd = 0.29 μm) (Fig. 1, E and F). As another means of testing for direct interaction, the effect of recombinant NM2A on Arf GAP activity of ASAP1 was examined (Fig. 1G). NM2A stimulated Arf GAP activity of full-length and BAR-PZA but had no effect on PZA, which lacks the BAR domain (Fig. 1G). Different than the binding studies, the GAP activity was measured in the presence of LUVs. ASAP1 is efficiently recruited to LUVs and NM2A also associates with LUVs to some extent (19), which may increase effective concentrations and explain why the EC50 for NM2A stimulation of GAP activity (10∼50 nm) is smaller than the Kd.
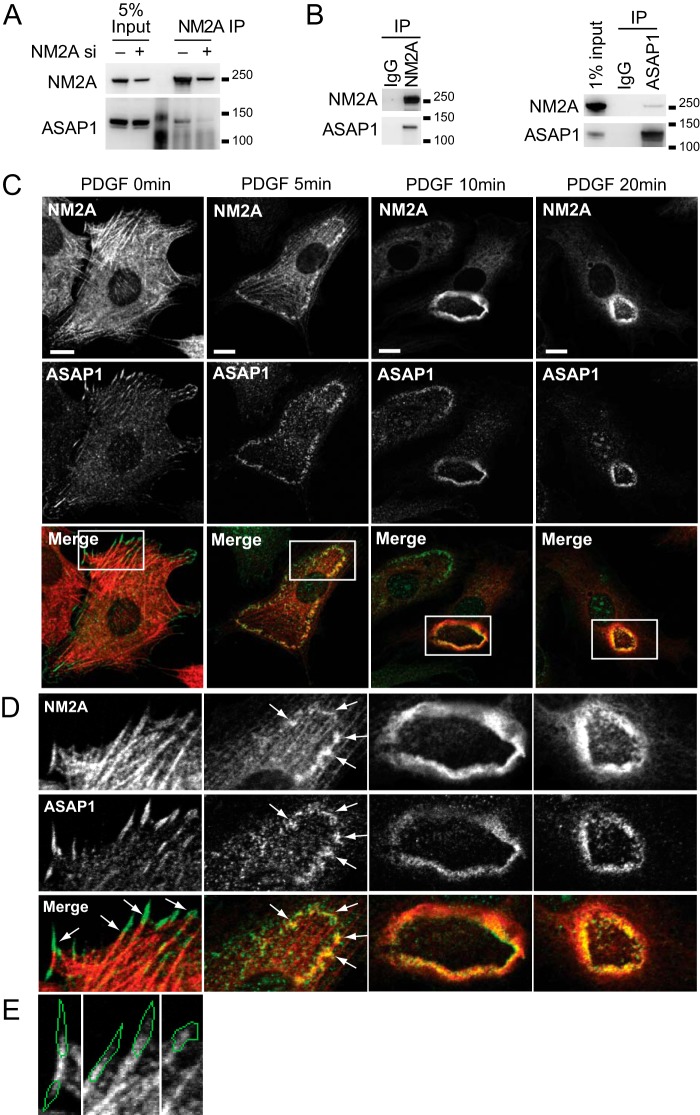
ASAP1-NM2A interaction was also detected in cells. Endogenous NM2A was immunoprecipitated from HeLa cell lysates. The amount of ASAP1 detected in the anti-NM2A precipitates from control cells was greater than that from NM2A knockdown cells (Fig. 2A). Co-immunoprecipitation of endogenous NM2A and ASAP1, precipitating with either antibody against NM2A (Fig. 2B, left panel) or ASAP1 (Fig. 2B, right panel) was also detected in NIH3T3 fibroblasts. We examined the colocalization of ASAP1 and NM2A in cells. ASAP1 localizes to FAs and translocates to CDRs upon PDGF treatment as previously described (Fig. 2C) (9). NM2A and ASAP1 colocalized at the junction of FAs and stress fibers and more extensively at CDRs (Fig. 2, C–E).
FIGURE 2.
ASAP1 associates with NM2A in cells. A and B, co-immunoprecipitation of ASAP1 with NM2A from HeLa cells (A) and NIH3T3 fibroblasts (B). Endogenous NM2A was immunoprecipitated from lysates of HeLa cells treated with siRNA (A) or NIH3T3 fibroblasts (left panel of B), followed by anti-ASAP1 immunoblots. In the right panel of B, ASAP1 was immunoprecipitated from lysates of NIH 3T3 fibroblasts followed by immunoblotting for NM2A. C–E, ASAP1 and NM2A colocalize in CDRs and at stress fiber/FA junctions. NIH3T3 fibroblasts plated on FN were treated with PDGF. Cells were fixed and co-stained for ASAP1 and NM2A. D, higher magnification of the boxed areas in C; arrows indicate colocalization. E, before PDGF treatment, NM2A was detected at the proximal region of ASAP1-positive FAs. The image for ASAP1 is not shown, but ASAP1-positive area is outlined in green. Bars, 10 μm.
ASAP1 Regulates NM2A-F-actin Interaction
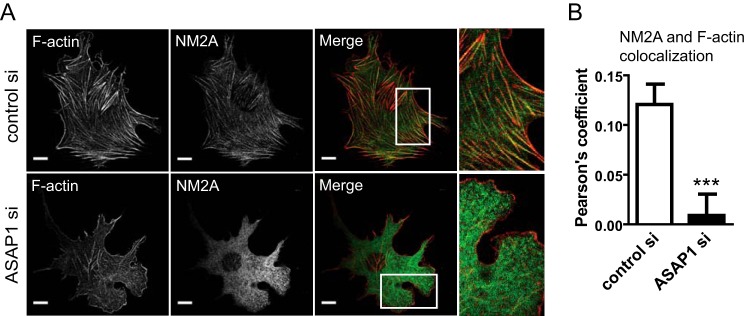
Function of NM2A depends on its interaction with F-actin. As a first test of the hypothesis that NM2A is an ASAP1 effector, we examined the colocalization of NM2A and F-actin as a measurement of actomyosin interaction in cells. Cells treated with control or ASAP1 siRNA were co-stained for NM2A and F-actin. Actin filaments, though present, were often thinner with less alignment than typical stress fibers (Fig. 3A). With these morphological changes, colocalization of NM2A and F-actin was reduced by ASAP1 knockdown (Fig. 3, A and B).
FIGURE 3.
ASAP1 affects cellular distribution of NM2A. siRNA-treated NIH3T3 fibroblasts were fixed and co-stained with rhodamine-phalloidin and anti-NM2A antibody. Representative images (A) and quantification (mean ± S.E., 30 cells from three experiments) (B) are shown. Boxed areas are shown in higher magnification. The magnification of the boxed area for ASAP1si is rotated 90°. Bars, 10 μm. ***, p < 0.001.
NM2A Mediates the Effect of ASAP1 on Actin-based Structures and Related Cellular Functions
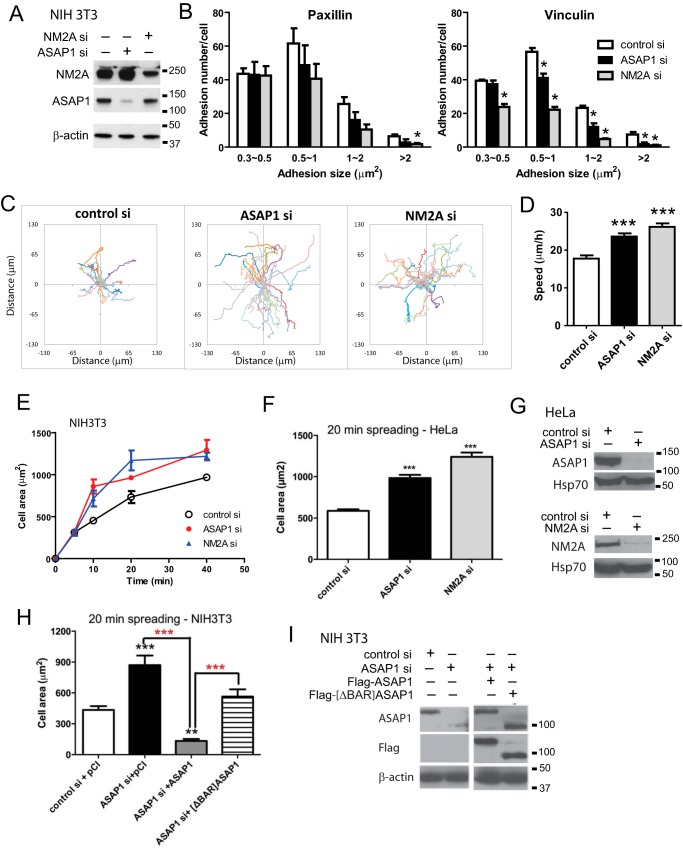
A prediction of the hypothesis that NM2A mediates the effects of ASAP1 on actin remodeling is that knockdown of either ASAP1 or NM2A recapitulates each other's cellular effects. We first examined cellular events regulated by NM2A. NM2-induced contractility is required for maturation of FAs (20, 21). Maturation of FAs from the precursor structures, nascent adhesions, is reflected on the growth in size from diffraction-limited 0.2 μm2 to more than 1 μm2. To examine effects of ASAP1 and NM2A knockdown on FAs, NIH3T3 fibroblasts, treated with control siRNA or siRNA targeting NM2A or ASAP1, were plated on fibronectin-coated coverslips, and FAs were detected by immunofluorescence staining using antibodies against the FA proteins paxillin and vinculin. We quantified the effect of ASAP1 or NM2A knockdown on the number and size distribution of the FAs. Consistent with NM2A's role in promoting FA maturation, NM2A knockdown decreased the number of mostly large adhesions (>1 μm2) and had less effect on the small adhesions (0.3∼0.5 μm2) (Fig. 4, A and B). Similarly, ASAP1 knockdown reduced the number of large adhesions but did not affect small adhesions. This result is consistent with the disorganized actomyosin stress fibers observed in ASAP1 knockdown cells because the size of FAs grow upon engagement with the actomyosin stress fibers. Cell movement, such as two-dimensional migration and cell spreading, also depends on actin and NM2A. NM2A activity has been reported to inhibit cell two-dimensional migration and spreading (22, 23). Consistent with these previous observations, we found that NM2A knockdown accelerated the speed of migration and rate of cell spreading in NIH3T3 fibroblasts (Fig. 4, C–E). ASAP1 knockdown also increased the speed of migration and rate of cell spreading. HeLa cell spreading was similarly affected by ASAP1 and NM2A knockdown (Fig. 4, F and G). The effect of replacing endogenous ASAP1 with Flag-ASAP1 was examined in NIH3T3 fibroblasts. The effect of ASAP1 knockdown on cell spreading was reversed by expression of wild type Flag-ASAP1 (Fig. 4H). Flag- [ΔBAR]ASAP1, expressed to the same level as Flag-ASAP1 (Fig. 4I) (∼100-fold greater than endogenous ASAP1), also slowed spreading of ASAP1 knockdown cells but less than did full-length ASAP1 (Fig. 4H).
FIGURE 4.
ASAP1 and NM2A have similar effects on cell adhesion, migration and spreading. A, efficiency of ASAP1 and NM2A knockdown in NIH3T3 fibroblasts was determined by immunoblotting. B, effect of ASAP1 and NM2A knockdown on FAs. siRNA-treated NIH3T3 fibroblasts plated on FN-coated coverslips were fixed and stained with anti-paxillin or vinculin antibody. The number and size of adhesions are the mean ± S.E. of three experiments. *, p < 0.05. C and D, effect of ASAP1 and NM2A knockdown on two-dimensional random migration. siRNA-treated NIH3T3 fibroblasts in FN-coated chambers were imaged using time-lapse phase contrast microscopy. Tracks of cells (C) and average migration speed (D) of 38 control si, 58 ASAP1 si, and 45 NM2A si-transfected cells are shown. ***, p < 0.0001. E, effect of ASAP1 and NM2A knockdown on cell spreading. NIH3T3 fibroblasts treated with siRNA were plated on FN for the times indicated. The area of rhodamine-phalloidin stained cells was determined for at least 30 cells per condition. Results shown are the combined mean ± S.E. from two experiments. F, effect of ASAP1 and NM2A knockdown on spreading of HeLa cells in 20 min was determined as described for NIH3T3 fibroblasts. Data are the mean ± range for two experiments. G, efficiency of ASAP1 and NM2A knockdown in HeLa cells determined by immunoblotting. Hsp70 was used as a loading control. H, effect of replacing endogenous ASAP1 with Flag-ASAP1 and Flag-[ΔBAR]ASAP1 on spreading of NIH 3T3 fibroblasts. NIH 3T3 fibroblasts, treated with siRNA and expressing pCI vector, Flag-tagged wild type ASAP1 or [ΔBAR]ASAP1, were plated on FN for 20 min. The results are from one experiment representative of three. I, efficiency of ASAP1 knockdown and relative expression of Flag-ASAP1 and Flag-[ΔBAR]ASAP1 determined by immunoblotting. Actin was used as a loading control. The recombinant ASAP1 were expressed to a level ∼100-fold that of the endogenous protein determined by titrating cell lysates (not shown). ***, p < 0.001 compared with control; **, p < 0.01 compared with control. In panel H, the red stars are p < 0.001 for the indicated comparisons.
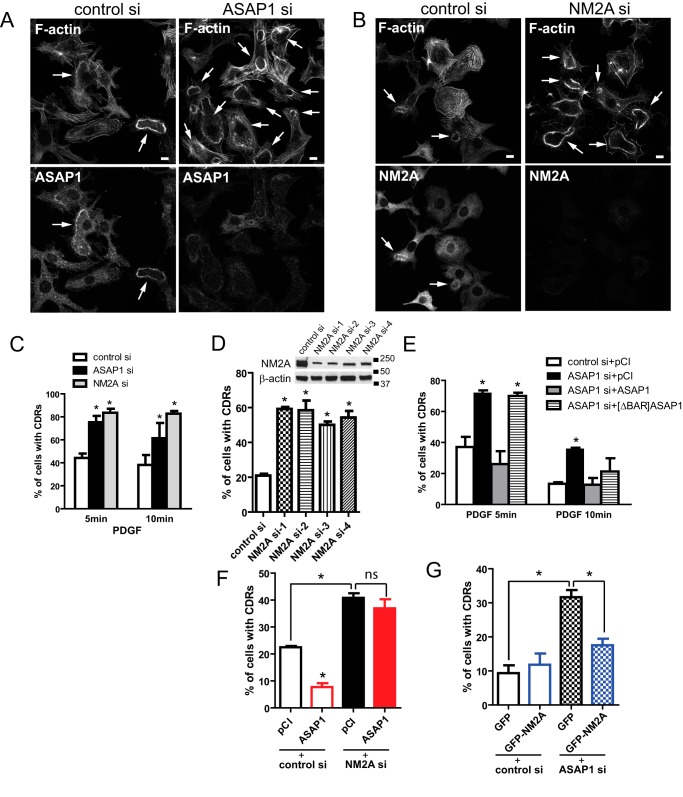
ASAP1 is known to reduce the appearance of CDRs but a role of NM2A in CDRs is not well characterized (9). CDRs have been implicated in receptor internalization and cell migration (24–28). The formation and dissolution of CDRs require extensive and dynamic actin remodeling, making it a useful system to study the role of ASAP1-NM2A interaction in actin remodeling. We found that ASAP1 knockdown increased the percentage of CDR-containing cells (Fig. 5, A and C). NM2A knockdown also increased the percentage of cells exhibiting CDRs (Fig. 5, B and C). Four different siRNAs targeting NM2A had the same phenotype, making an off-target effect unlikely (Fig. 5D). The effect of ASAP1 knockdown was reversed by overexpression of wild type Flag-ASAP1. In contrast, Flag-[ΔBAR]ASAP1, which cannot bind NM2A, did not rescue the defect at 5 min and had a partial effect at 10 min (Fig. 5E). If effects of ASAP1 on CDRs are mediated by NM2A, then one might predict that overexpression of ASAP1 would not reverse the effect of NM2A knockdown, but overexpression of NM2A could reverse the effect of ASAP1 knockdown. Consistent with predictions (Fig. 5, F and G), ASAP1 overexpression decreased CDRs in control cells but had no effect in NM2A knockdown cells. In contrast, NM2A overexpression had no effect on CDRs in control cells but reduced CDRs in ASAP1 knockdown cells. Collectively, these results support the hypothesis that ASAP1 regulates NM2A.
FIGURE 5.
Effect of ASAP1 and NM2A on circular dorsal ruffles. A–D, siRNA-treated NIH3T3 fibroblasts were treated with PDGF, fixed and co-stained for F-actin (rhodamine-phalloidin) and endogenous ASAP1 (A, C) or NM2A (B, C). D, effect of different siRNA targeting NM2A on CDR formation. Efficiency of NM2A knockdown determined by immunoblotting is shown in the inset. E–G, ASAP1 and NM2A regulate CDRs. NIH3T3 fibroblasts treated with the indicated siRNA and expressing Flag-ASAP1, Flag-[ΔBAR]ASAP1, GFP, GFP-NM2A, or pCI vector as indicated were treated with PDGF. Cells were fixed and stained for F-actin (rhodamine-phalloidin) alone (G) or co-stained for ASAP1 fragments with anti-Flag antibody (E, F). 50∼150 cells per treatment were scored in each experiment. Data presented are the mean ± S.E. from at least three experiments. *, p < 0.05.
Discussion
We set out to identify ASAP1-interacting protein(s) that mediate the effects of ASAP1 on actin-based structures. We found that ASAP1 directly bound to the actin-associated motor, NM2A. ASAP1 and NM2A affected a common set of actin structures and cell behaviors that depend on actin remodeling. The effect of ASAP1 on cell spreading and CDRs was dependent on the domain that bound directly to NM2A. The findings support the hypothesis that ASAP1 is a regulator of NM2A.
ASAP1, a multi-domain Arf GAP, may represent a new class of NM2A regulators. ASAP1 itself is regulated by a unique set of signals including phosphorylation, and by multiple interactions that are relevant to the control of the cytoskeleton (6, 29). Phosphoinositide binding to the PH domain may have a regulatory role, coordinating myosin activity with actin polymerization, which is also controlled by PI(4,5)P2 (30). Also plausible, Arf·GTP binding to the ASAP1 Arf GAP domain might regulate the interaction with NM2A. This hypothesis is in line with the diminished binding of NM2A to BAR-PZA containing the Arf GAP domain compared with the isolated BAR-PH domain and with the effect of NM2A on ASAP1 GAP activity. Thus, ASAP1 may mediate effects of Arf on the actin cytoskeleton (2). ASAP1 is regulated by proteins known to affect actin remodeling including FAK, which binds to the SH3 domain of ASAP1, and Src family proteins, which bind to the proline-rich domain (6, 10). The multiple binding partners regulate ASAP1 and target it to the plasma membrane, where it could localize NM2A function. Thus, ASAP1 could be the physical link between cellular signals and the mechanical machinery of actin remodeling in the context of cell movement and migration. Five other Arf GAPs may regulate NM2A. ASAP1–3 and ACAP1–3 all have an N-terminal BAR-PH tandem and the five tested affect the actin cytoskeleton (3). Like ASAP1, all interact with signaling proteins and phosphoinositides. With unique responses to combined signals, each of these Arf GAPs could provide regulation specific to actin remodeling in disparate cellular behaviors that depend on NM2A.
In testing for NM2A-ASAP1 association, we discovered NM2A stimulated ASAP1 GAP activity. Although NM2A was potent (half maximal effect at 10–50 nm NM2A), the effect was modest with a 2–5-fold stimulation, compared with the >10,000-fold stimulation observed with PI(4,5)P2 (31, 32). Although stimulated GAP activity may have a direct effector role, another plausible hypothesis, particularly given the modest effect, is that Arf·GTP binding to ASAP1 controls binding to NM2A. In this case, the GAP activity controls ASAP1-NM2A association.
In the course of these studies, it was discovered that ASAP1 binds actin filaments.4 The finding is consistent with the result presented in this report in Fig. 1B in which there was enrichment of actin in LUVs with BAR-PH. Though outside the scope of this current paper, we are investigating the possible mechanisms by which ASAP1, by binding both NM2A and actin, could regulate the dynamics of the actomyosin complex including stabilization by forming an ASAP1-actin-myosin ternary complex and by directly affecting the ATPase activity of myosin.
In summary, we have tested the hypothesis that ASAP1 directly binds to NM2A to regulate actin-based structures and related cellular functions. We speculate that the interaction of ASAP1 with NM2A is controlled by ligands of ASAP1. Thus, PI(4,5)P2 binding, Arf·GTP binding, GTP hydrolysis, and Arf·GDP dissociation each could affect a specific transition along the cycle of binding to and dissociation from NM2A. Given that ASAP1 also binds to a number of signaling molecules, including PI(4,5)P2 and oncoproteins, it may integrate cellular signals and physically link the signals and cellular membrane to the machinery of cell movement important to normal physiology and pathological processes such as tumor cell invasion and metastasis.
Author Contributions
P. W. C. conceived the study, designed, performed experiments presented in Fig. 1B, Fig 2, C–E, Fig. 3, Fig. 4, and Fig. 5, prepared reagents for Fig. 1D and wrote the manuscript. X. J. prepared reagents, designed and implemented experiment presented in Fig. 1, C and F and edited the manuscript. S. M. H. designed and performed experiments presented in Fig. 1, D and E and edited the manuscript. K. L. designed and performed experiments presented in Fig. 2, A and B and edited the manuscript. R. L. designed and performed experiments presented in Fig 4, F–I and edited the manuscript. L. M. J. designed and performed the mass spectrometric analysis. A. N. conceived of initial experiments testing the effects of ASAP1 on NM2A and compiled preliminary data. J. M. conceived and directed the experiments presented in Fig. 2, A and B and edited the manuscript. J. R. S. conceived and directed the experiments presented in Figs. 1 and 3 and wrote and edited the manuscript. P. A. R. conceived of the study, and wrote and edited the manuscript.
Acknowledgments
We thank Drs. Robert Adelstein and Mary Anne Conti for the GFP-NM2A plasmid and discussions and Dr. Kenneth Yamada for critical review of the manuscript.
This work was supported by the intramural program of the NCI, National Institutes of Health (to P. W. C., X. J., R. L., and P. R., Project BC 007365) and NHLBI, National Institutes of Health (to K. L., J. M., A. N., S. H., and J. S.). The authors declare that they have no conflicts of interest with the contents of this article.
S. M. Heissler, P. W. Chen, K. Chinthalapudi, J. R. Sellers, and P. A. Randazzo, manuscript in preparation.
- ArfGAP
- Arf GTPase-activating protein
- Arf
- ADP-ribosylation factor
- BAR
- Bin/Amphiphysin/Rvs
- PH
- pleckstrin-homology domain
- ANK
- ankyrin repeat
- Pro-rich
- proline-rich domain
- SH3
- Src homology3 domain
- CDR
- circular dorsal ruffle
- ELC
- essential light chain
- RLC
- regulatory light chain
- S1
- globular motor domain and lever arm of NM2A
- FN
- fibronectin
- FA
- focal adhesion
- PI
- PtdIns
- LUV
- large unilamellar vesicle.
References
- 1. Kahn R. A., Bruford E., Inoue H., Logsdon J. M., Nie Z. Z., Premont R. T., Randazzo P. A., Satake M., Theibert A. B., Zapp M. L., and Cassel D. (2008) Consensus nomenclature for the human ArfGAP domain-containing proteins. J. Cell Biol. 182, 1039–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Myers K. R., and Casanova J. E. (2008) Regulation of actin cytoskeleton dynamics by Arf-family GTPases. Trends Cell Biol. 18, 184–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Randazzo P. A., Inoue H., and Bharti S. (2007) Arf GAPs as regulators of the actin cytoskeleton. Biol. Cell 99, 583–600 [DOI] [PubMed] [Google Scholar]
- 4. Chen P. W., Jian X. Y., Yoon H. Y., and Randazzo P. A. (2013) ARAP2 signals through Arf6 and Rac1 to control focal adhesion morphology. J. Biol. Chem. 288, 5849–5860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nishiya N., Kiosses W. B., Han J. W., and Ginsberg M. H. (2005) An α(4) integrin-paxillin-Arf-GAP complex restricts Rac activation to the leading edge of migrating cells. Nat. Cell Biol. 7, 343–347 [DOI] [PubMed] [Google Scholar]
- 6. Bharti S., Inoue H., Bharti K., Hirsch D. S., Nie Z., Yoon H. Y., Artym V., Yamada K. M., Mueller S. C., Barr V. A., and Randazzo P. A. (2007) Src-dependent phosphorylation of ASAP1 regulates podosomes. Mol. Cell. Biol. 27, 8271–8283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu Y. H., Yerushalmi G. M., Grigera P. R., and Parsons J. T. (2005) Mislocalization or reduced expression of arf GTPase-activating protein ASAP1 inhibits cell spreading and migration by influencing Arf1 GTPase cycling. J. Biol. Chem. 280, 8884–8892 [DOI] [PubMed] [Google Scholar]
- 8. Onodera Y., Hashimoto S., Hashimoto A., Morishige M., Mazaki Y., Yamada A., Ogawa E., Adachi M., Sakurai T., Manabe T., Wada H., Matsuura N., and Sabe H. (2005) Expression of AMAP1, an ArfGAP, provides novel targets to inhibit breast cancer invasive activities. EMBO J. 24, 963–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Randazzo P. A., Andrade J., Miura K., Brown M. T., Long Y. Q., Stauffer S., Roller P., and Cooper J. A. (2000) The Arf GTPase-activating protein ASAP1 regulates the actin cytoskeleton. Proc. Natl. Acad. Sci. U.S.A. 97, 4011–4016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Y. H., Loijens J. C., Martin K. H., Karginov A. V., and Parsons J. T. (2002) The association of ASAP1, an ADP ribosylation factor-GTPase activating protein, with focal adhesion kinase contributes to the process of focal adhesion assembly. Mol. Biol. Cell 13, 2147–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oda A., Wada I., Miura K., Okawa K., Kadoya T., Kato T., Nishihara H., Maeda M., Tanaka S., Nagashima K., Nishitani C., Matsuno K., Ishino M., Machesky L. M., Fujita H., and Randazzo P. (2003) CrkL directs ASAP1 to peripheral focal adhesions. J. Biol. Chem. 278, 6456–6460 [DOI] [PubMed] [Google Scholar]
- 12. Vicente-Manzanares M., Ma X. F., Adelstein R. S., and Horwitz A. R. (2009) Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10, 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen P. W., Jian X., Luo R., and Randazzo P. A. (2012) Approaches to studying Arf GAPs in cells: in vitro assay with isolated focal adhesions. Curr. Protoc. Cell Biol., Chapter 17, Unit 17. 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jian X. Y., Brown P., Schuck P., Gruschus J. M., Balbo A., Hinshaw J. E., and Randazzo P. A. (2009) Autoinhibition of Arf GTPase-activating protein activity by the BAR domain in ASAP1. J. Biol. Chem. 284, 1652–1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Daniel J. L., and Sellers J. R. (1992) Purification and characterization Of platelet myosin. Methods Enzymol. 215, 78–88 [DOI] [PubMed] [Google Scholar]
- 16. Shevchenko A., Tomas H., Havlis J., Olsen J. V., and Mann M. (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 [DOI] [PubMed] [Google Scholar]
- 17. Gorelik R., and Gautreau A. (2014) Quantitative and unbiased analysis of directional persistence in cell migration. Nat. Protoc. 9, 1931–1943 [DOI] [PubMed] [Google Scholar]
- 18. Chen P. W., and Kroog G. S. (2010) Leupaxin is similar to paxillin in focal adhesion targeting and tyrosine phosphorylation but has distinct roles in cell adhesion and spreading. Cell Adh. Migr. 4, 527–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li D., Miller M., and Chantler P. D. (1994) Association of cellular myosin II with anionic phospholipids and the neuronal plasma membrane. Proc. Natl. Acad. Sci. U.S.A. 91, 853–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Choi C. K., Vicente-Manzanares M., Zareno J., Whitmore L. A., Mogilner A., and Horwitz A. R. (2008) Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10, 1039–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pasapera A. M., Schneider I. C., Rericha E., Schlaepfer D. D., and Waterman C. M. (2010) Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol. 188, 877–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cai Y. F., Rossier O., Gauthier N. C., Biais N., Fardin M. A., Zhang X., Miller L. W., Ladoux B., Cornish V. W., and Sheetz M. P. (2010) Cytoskeletal coherence requires myosin-IIA contractility. J. Cell Sci. 123, 413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Doyle A. D., Kutys M. L., Conti M. A., Matsumoto K., Adelstein R. S., and Yamada K. M. (2012) Micro-environmental control of cell migration - myosin IIA is required for efficient migration in fibrillar environments through control of cell adhesion dynamics. J. Cell Sci. 125, 2244–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Buccione R., Orth J. D., and McNiven M. A. (2004) Foot and mouth: Podosomes, invadopodia and circular dorsal ruffles. Nat. Rev. Mol. Cell Biol. 5, 647–657 [DOI] [PubMed] [Google Scholar]
- 25. Gu Z. Z., Noss E. H., Hsu V. W., and Brenner M. B. (2011) Integrins traffic rapidly via circular dorsal ruffles and macropinocytosis during stimulated cell migration. J. Cell Biol. 193, 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoon J. L., Wong W. K., and Koh C. G. (2012) Functions and regulation of circular dorsal ruffles. Mol. Cell. Biol. 32, 4246–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Itoh T., and Hasegawa J. (2013) Mechanistic insights into the regulation of circular dorsal ruffle formation. J. Biochem. 153, 21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Orth J. D., and McNiven M. A. (2006) Get off my back! Rapid receptor internalization through circular dorsal ruffles. Cancer Res. 66, 11094–11096 [DOI] [PubMed] [Google Scholar]
- 29. Inoue H., and Randazzo P. A. (2007) Arf GAPs and their interacting proteins. Traffic 8, 1465–1475 [DOI] [PubMed] [Google Scholar]
- 30. Campellone K. G., and Welch M. D. (2010) A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11, 237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kam J. L., Miura K., Jackson T. R., Gruschus J., Roller P., Stauffer S., Clark J., Aneja R., and Randazzo P. A. (2000) Phosphoinositide-dependent activation of the ADP-ribosylation factor GTPase-activating protein ASAP1 - Evidence for the pleckstrin homology domain functioning as an allosteric site. J. Biol. Chem. 275, 9653–9663 [DOI] [PubMed] [Google Scholar]
- 32. Jian X., Tang W. K., Zhai P., Roy N. S., Luo R., Gruschus J. M., Yohe M. E., Chen P. W., Li Y., Byrd R. A., Xia D., and Randazzo P. A. (2015) Molecular basis for cooperative binding of anionic phospholipids to the PH domain of the Arf GAP ASAP1. Structure 23, 1977–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]