Abstract
Srs2 plays many roles in DNA repair, the proper regulation and coordination of which is essential. Post-translational modification by small ubiquitin-like modifier (SUMO) is one such possible mechanism. Here, we investigate the role of SUMO in Srs2 regulation and show that the SUMO-interacting motif (SIM) of Srs2 is important for the interaction with several recombination factors. Lack of SIM, but not proliferating cell nuclear antigen (PCNA)-interacting motif (PIM), leads to increased cell death under circumstances requiring homologous recombination for DNA repair. Simultaneous mutation of SIM in a srs2ΔPIM strain leads to a decrease in recombination, indicating a pro-recombination role of SUMO. Thus SIM has an ambivalent function in Srs2 regulation; it not only mediates interaction with SUMO-PCNA to promote the anti-recombination function but it also plays a PCNA-independent pro-recombination role, probably by stimulating the formation of recombination complexes. The fact that deletion of PIM suppresses the phenotypes of Srs2 lacking SIM suggests that proper balance between the anti-recombination PCNA-bound and pro-recombination pools of Srs2 is crucial. Notably, sumoylation of Srs2 itself specifically stimulates recombination at the rDNA locus.
Keywords: DNA repair, homologous recombination, proliferating cell nuclear antigen (PCNA), protein-protein interaction, small ubiquitin-like modifier (SUMO)
Introduction
Homologous recombination (HR)3 is a key DNA repair pathway with particular importance in the repair of DNA double-strand breaks and stalled replication forks. HR needs to be tightly regulated, however, because uncontrolled recombination can lead to genome rearrangements and cell death (1, 2). Srs2, the Saccharomyces cerevisiae DNA helicase and single-stranded DNA translocase (3, 4), constitutes an important means of HR regulation, affecting it both negatively as well as positively.
Srs2 was originally identified as a suppressor of sensitivity to DNA-damaging agents of post-replication repair mutants (rad6, rad18, rad5) (5–8). Because the suppression is dependent on the RAD52 pathway genes, Srs2 is thought to inhibit HR and channel the lesions into post-replication repair (7, 9). The role of Srs2 as an anti-recombinase is further supported by findings that inactivation of Srs2 causes spontaneous mitotic hyper-recombination (6, 9–11). The recombination that is up-regulated in the absence of Srs2 is dependent on the RAD52 group and is believed to cause accumulation of toxic recombination intermediates, resulting in sensitivity of srs2Δ cells to DNA-damaging agents (UV light, x-rays, methyl methanesulfonate) and synthetic lethality of srs2 with various gene deletions (6, 9–16). The possible mechanism of Srs2 anti-recombination function is based on the ability of Srs2 to dismantle Rad51 recombinase from the presynaptic filaments in vitro (17, 18). For this process, the ability of Srs2 to translocate on single-stranded DNA and interact with Rad51 is necessary, as the ATPase or Rad51 interaction-deficient Srs2 mutants are unable to dissociate Rad51 from the DNA in vitro and to counteract recombination in vivo (19–21). The mechanism of recombination regulation by Srs2 may also include direct protein interaction with SUMO-PCNA that results in limiting the extent of DNA repair synthesis (22).
Evidence of Srs2 pro-recombination function has also accumulated over the years. Srs2 has been shown to stimulate single-strand annealing and non-homologous end joining, where the ability of Srs2 to dissociate Rad51 from DNA probably stimulates these Rad51-independent pathways (23–29). More recent studies, however, have shown the importance of Srs2 in Rad51-dependent ectopic gene conversion between non-homologous chromosomes (28, 30, 31). Observations that srs2Δ cells exhibit extensive end resection and severely reduced strand invasion intermediates have suggested that Srs2 plays an early role in the process (31). On the other hand, specific decrease in the faster HR sub-pathway leading to noncross-overs in srs2Δ indicates that Srs2 promotes synthesis-dependent strand annealing (SDSA), whereas it does not affect the DSBR sub-pathway (28, 31). It has been proposed that Srs2 facilitates strand displacement, which may utilize its ability to dissociate Rad51 from recombination intermediates, its helicase activity, or both (28, 32). However, we were not able to observe such activity in vitro (33). Recently, Miura et al. (34) suggested that the role of Srs2 in SDSA is dependent on its Rad51 and SUMO-PCNA interactions. Srs2 also stimulates unequal recombination between sister chromatids (35), and it has been suggested that Srs2 promotes break-induced replication (27, 36). Moreover, deletions of SRS2 and most of the other HR factors, including RAD51, are lethal in the rad27Δ background (16, 37). This indicates that HR is essential for DNA repair in such cells and that Srs2 may either promote or regulate its proper execution (16, 37).
Apart from HR regulation, Srs2 plays a major role in the recovery from DNA damage checkpoint-mediated arrest (28, 30). In the absence of Srs2, Vaze et al. (30) found that cells were not able to recover from DNA damage checkpoint even if the DNA had been repaired, thus suggesting that Srs2 is needed to turn off the checkpoint.
Later studies demonstrated that Srs2 is recruited to the replication forks by the sumoylated form of PCNA, as blocking of PCNA sumoylation or deletion of C-terminal SIM in Srs2 suppresses rad6 and rad18 sensitivity equally to srs2 deletion (38–41). Accordingly, deletion of Srs2 SIM strongly reduces the interaction with SUMO and PCNA in the yeast two-hybrid assay (38, 42) as well as the interaction with SUMO-PCNA in vivo and in vitro (38, 41–43). Srs2 recently was shown also to contain a PCNA-specific interaction site, which together with the SIM is necessary for efficient interaction with SUMO-PCNA (42, 44, 45).
The multiple roles of Srs2 in DNA repair argue for the means to tightly and properly regulate its activity. Srs2 phosphorylation is induced by DNA damage and promotes the SDSA branch of HR (46, 47). We have previously shown that DNA damage-induced Srs2 sumoylation depends on the Srs2 SIM motif and targets three lysines in its C-terminal part (Lys-1081, -1089, -1142) (42, 46). Nevertheless, the role of Srs2 sumoylation remains unexplained.
In the present work we analyzed the role of SUMO and PCNA in regulating the multiple functions of Srs2. We show that the roles of SUMO in Srs2 regulation are ambivalent, as the SIM of Srs2 not only mediates interaction with SUMO-PCNA and thus promotes the anti-recombination function, but it also plays a PCNA-independent pro-recombination role. Accordingly, sumoylation of Srs2 promotes recombination at the rDNA locus. SUMO facilitates the formation of protein complexes required for proper repair, as suggested by the importance of Srs2 sumoylation and particularly its SIM for interactions with Mre11, Rad51, and Rad52. We also show that PCNA interaction is necessary for Srs2 inhibitory effect in direct-repeat recombination, which includes single-strand annealing and gene conversion events, and it plays a limited role in inhibiting recombination between homologous chromosomes. This study improves our understanding as to the roles of SUMO and PCNA in Srs2 regulation, and it shows that stimulation of SUMO-SIM interactions of a protein can, depending on the circumstances, target protein to diverse protein complexes and lead to different and even opposing outcomes.
Experimental Procedures
Yeast Strains and Plasmids
The S. cerevisiae strains used in this study are all RAD5 derivatives of strain W303-1A (48, 49) and are listed in Table 1. Yeast strains and media were prepared using standard techniques, as previously described (50). Strains SS149-15D, PK1-A, PK2-A, PK7-A, and PK8-A were generated by the PCR-based allele replacement method (51) using oligonucleotides described in Table 2. Correct integration was verified by sequencing. The srs2ΔPIM mutant contains deletion of amino acids 1159–1163 and in the srs2-SIM* mutant amino acids 1170–1173 (IIVI) were exchanged for alanines.
TABLE 1.
Yeast strains used in this study
Strains are derivatives of W1588–4C (MATa ade2-1 can1-100 ura3-1 his3-11,15 leu2-3,112 trp1-1) (49), a RAD5 derivative of W303-1A (48).
| Strain | Genotype | Source |
|---|---|---|
| ML8-9A | MATa ADE2 ura3-1 his3-11,15 leu2-3,112 trp1-1 | M. Lisby |
| SS149-15D | MATa ADE2 srs2-K1081/K1089R/K1142R | This study |
| PK1-A | MATa ADE2 srs2-SIM* | This study |
| PK2-A | MATa ADE2 srs2ΔPIM | This study |
| PK7-A | MATa ADE2 srs2ΔPIM-SIM* | This study |
| PK64-2D | MATa ADE2 srs2::HIS3 | This study |
| PK78--3C | MATa ADE2 siz1::KAN | This study |
| PK79-5C | MATa ADE2 siz2::KAN | This study |
| PK81-10A | MATa ADE2 siz1::KAN siz2::KAN | This study |
| PK100-2D | MATa ADE2 siz1::KAN siz2::KAN srs2-SIM* | This study |
| PK80-3C | MATa ADE2 mms21-CH::HIS3 | This study |
| PK77-5D | MATa ADE2 rad51Δ | This study |
| PK77-6D | MATa ADE2 srs2-SIM* rad51Δ | This study |
| PK82-6A | MATa ADE2 srs2::HIS3 rad51Δ | This study |
| yLK69 | MATa ADE2 rad18::LEU2 | H. Klein |
| PK05-16D | MATa ADE2 rad18::LEU2 srs2-K1081/K1089R/K1142R | This study |
| PK15-1B | MATa ADE2 rad18::LEU2 srs2-SIM* | This study |
| PK25-6D | MATa ADE2 rad18::LEU2 srs2ΔPIM | This study |
| PK83-3C | MATa ADE2 rad18::LEU2 srs2ΔPIM-SIM* | This study |
| PK47-1A | MATa ADE2 rad18::LEU2 srs2::HIS3 | This study |
| PK06-15C | MATα ADE2 rad27::URA3 | This study |
| PK70-10B | MATa ADE2 srs2-K41R | This study |
| PK8-A | MATa ADE2 srs2-K41R-SIM* | This study |
| ML144-8C | MATa ADE2 leu2-ΔEcoRI::URA3-HO::leu2-ΔBstEII | M. Lisby |
| PK02-2B | MATa ADE2 leu2-ΔEcoRI::URA3-HO::leu2-ΔBstEII srs2-K1081/K1089R/K1142R | This study |
| PK12-4C | MATa ADE2 leu2-ΔEcoRI::URA3-HO::leu2-ΔBstEII srs2-SIM* | This study |
| PK22-8C | MATa ADE2 leu2-ΔEcoRI::URA3-HO::leu2-ΔBstEII srs2ΔPIM | This study |
| PK84-1B | MATa ADE2 leu2-ΔEcoRI::URA3-HO::leu2-ΔBstEII srs2ΔPIM+SIM* | This study |
| PK46-6B | MATa ADE2 leu2-ΔEcoRI::URA3-HO::leu2-ΔBstEII srs2::HIS3 | This study |
| PK89-2A | MATa ADE2 leu2-ΔEcoRI::URA3-HO::leu2-ΔBstEII srs2-K41R | This study |
| PK88-13C | MATa ADE2 leu2-ΔEcoRI::URA3-HO::leu2-ΔBstEII srs2-K41R-SIM* | This study |
| ML412 | MATa ADE2 leu2-ΔBstEII lys2Δ TRP1 | M. Lisby |
| MATα ADE2 leu2-ΔEcoRI LYS2 trp1-1 | ||
| PK09 | MATa ADE2 leu2-ΔBstEII lys2Δ TRP1 srs2-K1081/K1089R/K1142R | This study |
| MATα ADE2 leu2-ΔEcoRI LYS2 trp1-1 srs2-K1081/K1089R/K1142R | ||
| PK19 | MATa ADE2 leu2-ΔBstEII lys2Δ TRP1 srs2-SIM* | This study |
| MATα ADE2 leu2-ΔEcoRI LYS2 trp1-1 srs2-SIM* | ||
| PK29 | MATa ADE2 leu2-ΔBstEII lys2Δ TRP1 srs2ΔPIM | This study |
| MATα ADE2 leu2-ΔEcoRI LYS2 trp1-1 srs2ΔPIM | ||
| PK87 | MATa ADE2 leu2-ΔBstEII lys2Δ TRP1 srs2ΔPIM-SIM* | This study |
| MATα ADE2 leu2-ΔEcoRI LYS2 trp1-1 srs2ΔPIM-SIM* | ||
| ML478 | MATa ADE2 leu2-ΔBstEII lys2Δ TRP1 srs2::HIS3 | M. Lisby |
| MATα ADE2 leu2-ΔEcoRI LYS2 trp1-1 srs2::HIS3 | ||
| PK95 | MATa ADE2 leu2-ΔBstEII lys2Δ TRP1 srs2-K41R | This study |
| MATα ADE2 leu2-ΔEcoRI LYS2 trp1-1 srs2-K41R | ||
| PK92 | MATa ADE2 leu2-ΔBstEII lys2Δ TRP1 srs2-K41R-SIM* | This study |
| MATα ADE2 leu2-ΔEcoRI LYS2 trp1-1 srs2-K41R-SIM* | ||
| RMY180-5A | MATa ade2-1 ADE2::rDNA | H. Klein |
| PK01-11C | MATa ade2-1 ADE2::rDNA srs2-K1081/K1089R/K1142R | This study |
| PK11-6A | MATa ade2-1 ADE2::rDNA srs2-SIM* | This study |
| PK21-1D | MATa ade2-1 ADE2::rDNA srs2ΔPIM | This study |
| yLK344 | MATa ade2-1 ADE2::rDNA srs2ΔPIM-SIM* | This study |
| IG10-2D | MATa ade2-1 ADE2::rDNA srs2::HIS3 | M. Lisby |
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| Srs2-K1081R | Forward, 5′-gtctaagagaggtgacaaggttagggtggaggaagt-3′ |
| Reverse, 5′-acttcctccaccctaaccttgtcacctctcttagac-3′ | |
| Srs2-K1089R | Forward, 5′-ggaggaagtgatagatttgaggagtgaatttgaggaagatg-3′ |
| Reverse, 5′-catcttcctcaaattcactcctcaaatctatcacttcctcc-3′ | |
| Srs2-K1142R | Forward,5 ′-cagaaattttccaaaaaggtgaggaatgaacctgcatcaagtcaa-3′ |
| Reverse, 5′-ttgacttgatgcaggttcattcctcacctttttggaaaatttctg-3′ | |
| Srs2-I1170A/I1171A/V1172A/I1173A | Forward, 5′-cacgtgcgaaaaaaaagtcaaaattaaacaacggtgaagccgcagccgccgattagtagcactttcatgcctgactacg-3′ |
| Reverse, 5′-cgtagtcaggcatgaaagtgctactaatcggcggctgcggcttcaccgttgtttaattttgacttttttttcgcacgtg-3′ | |
| Srs2-A1170STOP | Forward, 5′-gtcaaaattaaacaacggtgaatgaatagtcatcgattag-3′ |
| Reverse, 5′-ctaatcgatgactattcattcaccgttgtttaattttgac-3′ | |
| Srs2Δ1159–1163 | Forward, 5′-ggatatattttctcagctgtcacgtaaattaaacaacggtgaaatcatag-3′ |
| Reverse, 5′-ctatgatttcaccgttgtttaatttacgtgacagctgagaaaatatatcc-3′ | |
| Srs2-PCR replacement | Forward, 5′-aaccagaaactacatcatcgaattccagctgaccaccatgtgtcagatgatttaatgagacc-3′ |
| Reverse, 5′-cgatcttctacccagaatcacgatccccgggaattgccatgttggcaaatgtctctactgg-3′ | |
| Mre11-I633A/I634A/M635M/V636A | Forward, 5′-aagatgatgttgatattgatgagaatgacgcagctatggccagtactgacgaagaggacgctagttatg-3′ |
| Reverse, 5′-cataactagcgtcctcttcgtcagtactggccatagctgcgtcattctcatcaatatcaacatcatctt-3′ | |
| Rad51-I277A/V278A/V279A | Forward, 5′-gagcgagtctcggttttccttggctgcggccgattctgttatggctctatac-3′ |
| Reverse, 5′-gtatagagccataacagaatcggccgcagccaaggaaaaccgagactcgctc-3′ | |
| Rad51-V321V/V322A/V323A | Forward, 5′-accaatttggtgttgcagccgccgctactaaccaagtggtcgc-3′ |
| Reverse, 5′-gcgaccacttggttagtagcggcggctgcaacaccaaattggt-3′ | |
| Rad52-K43R/K44R | Forward, 5′-ggatatggatgagaggaggcccgttttcggtaacc-3′ |
| Reverse, 5′-ggttaccgaaaacgggcctcctctcatccatatcc-3′ | |
| Rad52-K253R | Forward, 5′-ctcgacgaagaacctggtgcgcatagaaaatacagtaagtcgagg-3′ |
| Reverse, 5′-cctcgacttactgtattttctatgcgcaccaggttcttcgtcgag-3′ | |
| Rad59-K207R | Forward, 5′-gcttgtatggctcaaaaaaaattcgaaatgaagctaacacc-3′ |
| Reverse, 5′-ggtgttagcttcatttcgaattttttttgagccatacaagc-3′ | |
| Rad59-K228R | Forward, 5′-aatagcaagccgacttttatcagactcgaggatgctaaaggcacgc-3′ |
| Reverse, 5′-gcgtgcctttagcatcctcgagtctgataaaagtcggcttgctatt-3′ |
The following expression plasmids have been described previously: (His)9-SRS2::pET11c (20), (His)9-SRS2-K1081/1089/1142R::pET11c (22), AOS1/UBA2::pGEX-4T-1 (52), UBC9::pET21b (53), SMT3::pET-HF (54), SIZ1(1–465)::pET21b (55), MRE11-pPM271 (56), and RAD52-pGEX-3X (57). To create the RAD59-pMAL-TEV plasmid (pLK1086), a PCR fragment of RAD59 was cloned into EcoRI and PstI sites of pMAL-TEV vector.
The yeast two-hybrid plasmids MRE11::pGBT9 (58), POL32::pASΔΔ (59), Rad51::pGBT9, RAD52::pGBT9 (60), RAD59::pGBD-C2 (61), and SMT3::PGAD-C1 (62) have been described. The C-terminal fragment of Srs2 in the construct SRS2(783–1174)::pGADT7 was prepared by PCR and cloned into the EcoRI site of pGADT7. Mutant versions of the aforementioned plasmids were generated by site-directed mutagenesis using specific oligonucleotides (Table 2).
The centromeric plasmids for expression of Srs2 and its mutants in the srs2Δ strain contained the SRS2 gene sequence including 700 bp upstream from the start codon and 180 bp downstream from the stop codon and were cloned into YCplac22 vector (a kind gift from G. Liberi). The SIM mutants were prepared by site-directed mutagenesis using the primers described in Table 2.
Expression and Purification of Recombinant Proteins
The following proteins were expressed and purified as described: Srs2 and Srs2-3KR (63), Aos1/Uba2, His-Ubc9, His-FLAG-Smt3 (64), His-Siz1(1–465) (55), Mre11 (56), GST-Rad52 (57), and Rad59 (65).
In Vitro Sumoylation Assay
The Srs2 sumoylation assay was performed similarly as previously described (42). The 10-μl reaction contained 0.35 μm Aos1/Uba2, 0.1 μm Ubc9, 1.6 μm Smt3, 0.15 μm Siz1(1–465), 0.75 μm Srs2 or Srs2-3KR, 100 μm ATP, and 160 mm KCl in buffer S2 (50 mm HEPES, 10 mm MgCl2). Reactions were incubated for 30 min at 4 °C.
Pulldown Assays
To study Srs2 interactions with Mre11, Rad52, and Rad59, purified His-Srs2 (0.75 μm) was mixed with Mre11 (1 μm), GST-Rad52 (1.5 μm), or MBP-Rad59 (2 μm) and 10 μl of Profinity IMAC nickel-charged resin (Bio-Rad), glutathione-Sepharose 4 Fast Flow (GE Healthcare), or amylose resin high flow (New England BioLabs), respectively. The reaction mixtures were incubated in 50 μl of buffer S2 containing 150 mm KCl for 30 min at 20 °C with mixing at 1000 rpm. The supernatants were then collected and mixed with equal amounts of SDS Laemmli buffer. The beads were washed with 100 μl of buffer S2 containing 150 mm KCl and mixed with 20 μl of SDS Laemmli buffer. The supernatant and the SDS eluate (10 μl each) were analyzed by SDS-PAGE on a 10% gel followed by Coomassie Blue staining.
Electrophoretic Mobility Shift Assay (EMSA) and Helicase Assay
The indicated amounts of Srs2, Srs2-3KR, SUMO-Srs2, or SUMO machinery proteins prepared in corresponding sumoylation reactions were mixed with 3 nm fluorescently labeled 49-mer single- and double-stranded DNA (63) in 10 μl of buffer E (30 mm Tris (pH 7.5), 1 mm DTT, 0.1 mg/ml BSA, 100 mm KCl) and incubated for 15 min at 37 °C. After incubation, 2 μl of loading buffer (60% glycerol, 10 mm Tris (pH 7.5), 60 mm EDTA) were added, and the samples were resolved on 6% native polyacrylamide gel in 1× TAE buffer (40 mm Tris, 20 mm sodium acetate, 2 mm EDTA, pH 7.5) at 4 °C. Gels were scanned using FLA-9000 Starion (Fujifilm) and quantified by MultiGauge software (Fujifilm).
Helicase assay was performed essentially as described previously (63). Briefly, 3′ overhang DNA (3 nm) was incubated with the proteins for 15 min at 30 °C in 10 μl of buffer H (30 mm Tris (pH 7.5), 1 mm DTT, 0.1 mg/ml BSA, 100 mm KCl, 20 mm creatine phosphate, 20 μg/ml creatine kinase, 2.4 mm MgCl2, and 2 mm ATP). Reactions were then treated with 2% SDS and 0.5 mg/ml proteinase K at 37 °C for 5 min. After adding loading buffer, samples were resolved on 10% native polyacrylamide gel in 1× TBE buffer (40 mm Tris-HCl, 20 mm boric acid, 2 mm EDTA (pH 7.5)).
Yeast Two-hybrid Analysis
The assay was performed essentially as described previously (60). Briefly, plasmids containing fusions with GAL4 transcription activation or DNA binding domains were transformed into the haploid S. cerevisiae strain PJ69-4a (MATa) or PJ69-4α strain (MATα), respectively. Diploid strains were grown to A600 ∼ 1 and then 10-fold serially diluted. Activation of the HIS3 reporter gene was analyzed on medium lacking leucine, tryptophan, and histidine. Cells were grown for 3 days at 30 °C before analysis.
DNA Damage Sensitivity Assay
Strains were grown in YPD to A600 ∼ 1 and 10-fold serially diluted. DNA damage sensitivity was assessed on YPD plates without or with the indicated amounts of camptothecin (CPT), methyl methane-sulfonate (MMS), ultraviolet light (UV), hydroxyurea, 4-nitroquinoline 1-oxide, and zeocin (ZEO). Pictures of the plates were taken after 2 days of incubation at 30 °C.
Determination of Mitotic Recombination Rates
Direct-repeat, interchromosomal, and rDNA recombination was determined as previously described (64) with minor modifications. The recombination rates were calculated by the Lea-Coulson median method using FALCOR software (66). To illustrate the variance, median absolute deviation of the individually calculated recombination rates was used. In addition to measuring the rate of ADE2 marker loss at the rDNA by the frequency of half-sectored white versus red colonies, the rate was also determined from the frequency of wholly red colonies similarly to the direct-repeat and interchromosomal recombination. To eliminate the impact of outliers on the statistical evaluation, p values were calculated using the non-parametric Mann-Whitney U test, except that for the ADE2 loss using the half-sectored colonies Yates' χ2 test was used instead.
Results
SUMO Mildly Affects the Biochemical Activities of Srs2
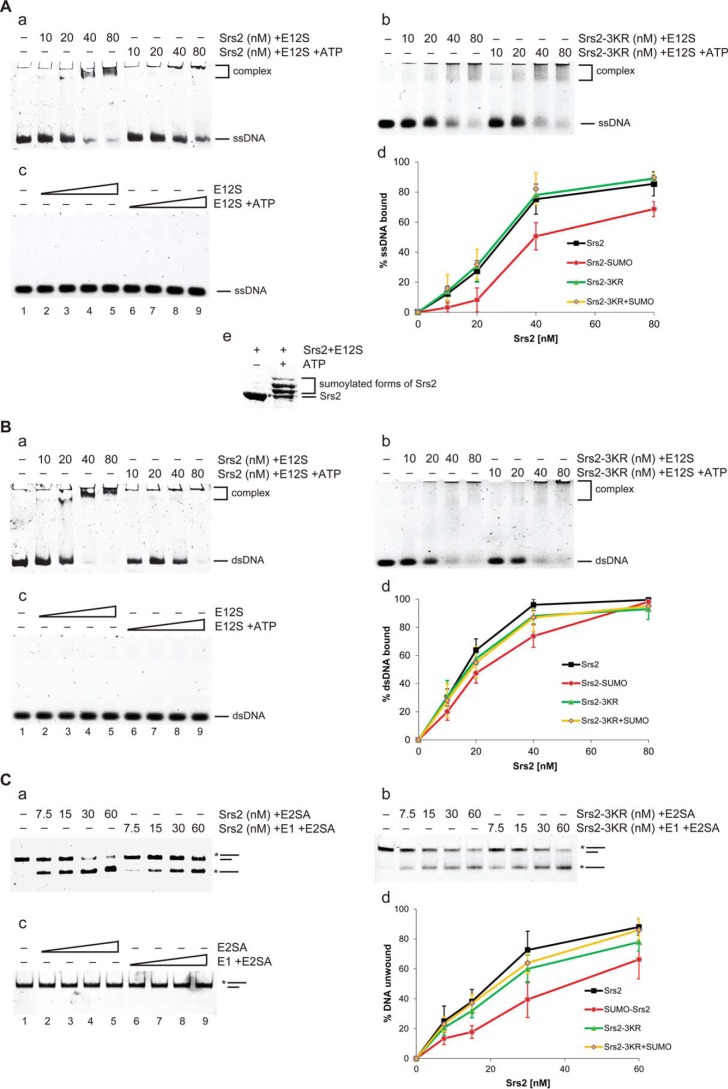
To study the effect of SUMO on Srs2, we first addressed the consequences of Srs2 sumoylation on its biochemical activities in vitro. To precisely compare non-sumoylated and sumoylated forms of Srs2, the sumoylation reaction was performed in the absence or presence of ATP or E1, and the mixtures were analyzed for DNA binding and helicase activities. Although the proportion of sumoylated Srs2 was >90%, only a modest, but significant, decrease in both activities was observed (Fig. 1). The reduction in helicase activity likely resulted from lower affinity of SUMO-Srs2 toward DNA. As the addition of ATP or E1 to the sumoylation reaction did not affect the activities of the non-sumoylatable Srs2-3KR protein and SUMO machinery proteins showed no activity (Fig. 1), sumoylation machinery is not responsible for the observed effects.
FIGURE 1.
DNA binding and helicase activity of sumoylated Srs2 is mildly decreased. A, SUMO-Srs2 shows decreased single-stranded DNA binding activity. a, in vitro sumoylation reaction in the absence or presence of ATP was used to prepare Srs2 or SUMO-Srs2 of the same concentration (in SUMO-Srs2 >90% of the Srs2 protein corresponded to its sumoylated form). The indicated amounts of Srs2 or SUMO-Srs2 were incubated with 3 nm fluorescently labeled 49-mer single-stranded DNA for 15 min at 37 °C. Samples were then applied to electrophoresis on 6% native PAGE, scanned, and quantified by MultiGauge software. In vitro sumoylation reactions containing the non-sumoylatable Srs2-3KR (b) or SUMO machinery proteins (c) in the absence or presence of ATP were used as a control. d, the average values of three independent experiments described in a–c are plotted. Error bars indicate S.D. E12S indicates presence of sumo machinery (E1, E2, and SUMO). e, level of Srs2 in vitro sumoylation used in the studies. B, SUMO-Srs2 shows decreased dsDNA binding activity. Binding of non-/sumoylated Srs2 (a), Srs2-3KR (b), or SUMO machinery proteins (c) to 3 nm 49-bp dsDNA was analyzed as in A. d, the average values of three independent experiments described in a–c are plotted. Error bars indicate S.D. E12S indicates the presence of sumo machinery (E1, E2, and SUMO). C, SUMO-Srs2 shows decreased helicase activity. The indicated amounts of Srs2, SUMO-Srs2 (a), Srs2-3KR (b), or SUMO machinery proteins (c) were mixed with 3 nm 3′ overhang DNA and incubated for 15 min at 30 °C. After the reaction was stopped by the addition of SDS and proteinase K, samples were resolved on 10% native PAGE. d, the average values of three independent experiments described in a–c are plotted. Error bars indicate S.D. E2SA indicates presence of E2, SUMO, and ATP.
SUMO Promotes Srs2 Interactions
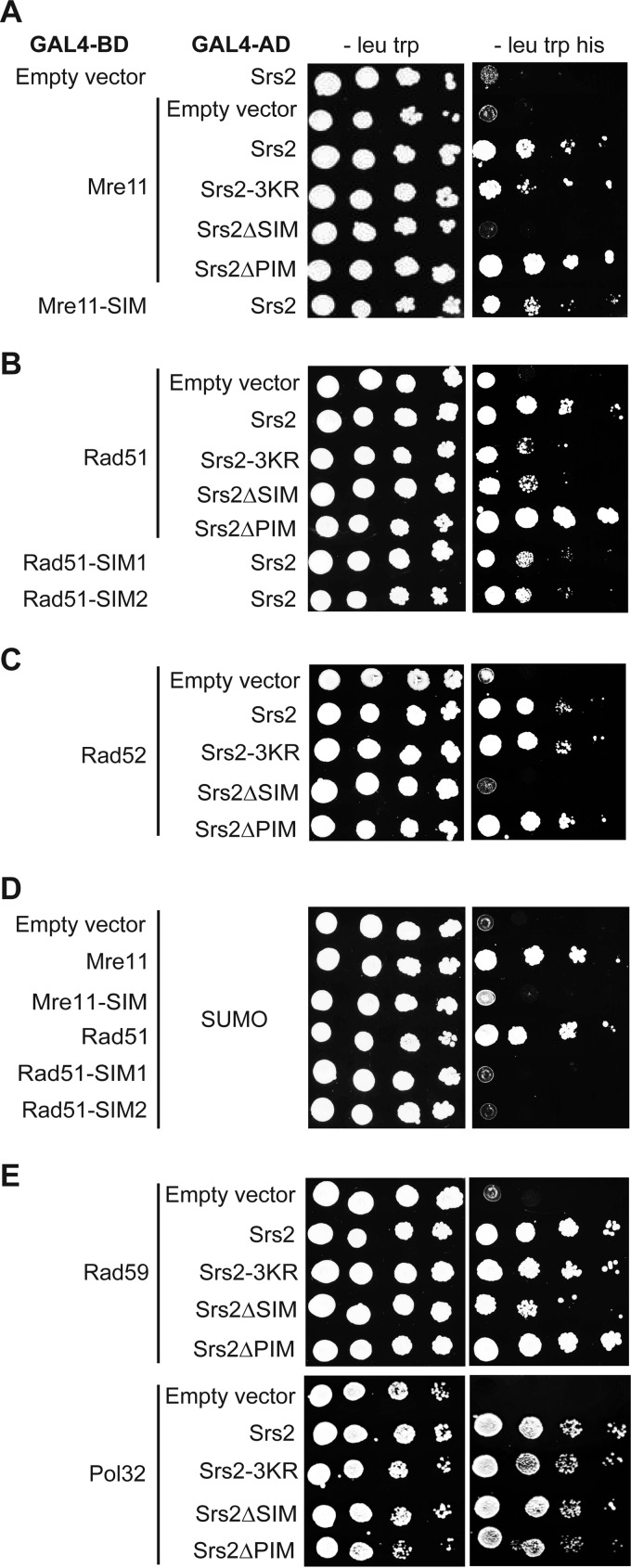
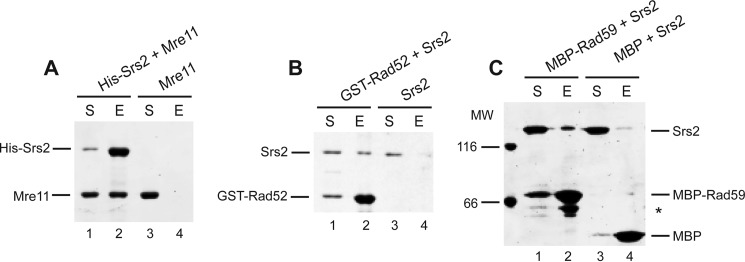
We next asked whether SUMO affects Srs2 protein interactions, as observed in numerous other cases (67, 68), using the yeast two-hybrid system. To evaluate the importance of covalent modification of Srs2 by SUMO, we used the non-sumoylatable Srs2-3KR mutant (42). To study the effect of non-covalent interactions between Srs2 and SUMO, we used the Srs2ΔSIM mutant (missing the last five amino acids), which is devoid of such interaction (42). We previously showed that Srs2 sumoylation is dependent on the SIM of Srs2; therefore, its deletion should also abolish the interactions dependent on Srs2 modification (42). Because the Srs2 SIM motif is known to mediate its interaction with SUMO-PCNA (38, 41, 42, 44), we also sought to differentiate between the PCNA-dependent and -independent roles of the SIM by using the Srs2 mutant lacking the PCNA interaction domain (Srs2ΔPIM). When testing the aforementioned Srs2 mutants for interactions with the known partners, we observed no significant effect in the case of Dun1, Lif1, Mph1, Pol32, and Slx5 (data not shown and Fig. 2E). Nevertheless, SUMO seems to be important to mediate Srs2 interactions with Mre11 and Rad51 (Fig. 2, A and B). Moreover, we identified novel interactions between Srs2 and the recombination mediator Rad52 and its homologue Rad59, which were likewise promoted by SUMO (Fig. 2, C and E). Using in vitro pulldown with purified proteins we confirmed that the observed interactions between Srs2 and Mre11, Rad52, and Rad59 are direct (Fig. 3). In the case of the Mre11 interaction, the Srs2 SIM seems to play a more important role than does Srs2 sumoylation, as Srs2ΔSIM eliminates the interaction (Fig. 2A). Interestingly, Srs2ΔPIM mutant, unlike Srs2ΔSIM, stimulated the interaction. To confirm this SUMO-SIM-dependent interaction, we identified a SUMO-interacting motif within Mre11 and observed a reduction in the interaction of Mre11-SIM mutant (I633A/I634A/M635M/V636A) not only with SUMO (Fig. 2D) but also with Srs2 (Fig. 2A). SUMO also seems to significantly stimulate Srs2 interaction with Rad51, as Srs2-3KR and Srs2ΔSIM displayed decreased interaction (Fig. 2B). Similar to what we observed for Mre11, this effect is independent of Srs2-PCNA interaction, as Srs2ΔPIM has rather a slight stimulatory effect. We also identified two possible SIM motifs within Rad51 (277IVV279 and 321VVV323), and mutation in either motif reduced the interactions with SUMO and Srs2 (Fig. 2, D and B). This suggests that the interaction is mediated by SUMO attached to Srs2 and the Rad51 SIM motifs. Finally, the Srs2ΔSIM mutant completely eliminated the interaction with the Rad52 protein. Srs2 SIM may also play some role in the interaction with Rad59, but this seems to be minor when compared with the cases of Mre11, Rad51, and Rad52 (Fig. 2E). Taken together, these data suggest that SUMO mediates a subset of Srs2 interactions that are independent of interaction with PCNA.
FIGURE 2.
The SUMO-interacting motif of Srs2 or its sumoylation mediate interactions with Mre11, Rad51, and Rad52 in the yeast two-hybrid system. Plasmids containing the GAL4 DNA binding domain fused to the indicated genes were transformed into PJ69-4α strain, and the plasmids containing the GAL4 activation domain were transformed into PJ69-4a. After mating, the diploid strains were spotted as 10-fold serial dilutions on medium lacking leucine and tryptophan or leucine, tryptophan, and histidine. Pictures were taken after 3 days of incubation at 30 °C. Although the Srs2 plasmids contained the C-terminal part of the gene (amino acids 783–1174), other plasmids carried the full-length versions of the corresponding genes. The following mutations were used: Srs2-3KR (K1081R/K1089R/K1142R), Srs2ΔSIM (A1170X), Srs2ΔPIM (Δ1159–1163), Mre11-SIM (I633A/I634A/M635M/V636A), Rad51-SIM1 (I277A/V278A/V279A), and Rad51-SIM2 (V321A/V322A/V323A).
FIGURE 3.
Srs2 directly interacts with Mre11, Rad52, and Rad59. A, Srs2 interacts with Mre11. Recombinant Mre11 (1 μm, lanes 1–4) was incubated with His-tagged Srs2 (0.75 μm, lanes 1 and 2) and nickel-charged resin in buffer S2 containing 150 mm KCl for 30 min at 20 °C. The resin was washed, and the proteins were eluted by SDS Laemmli buffer. The supernatant (S) and the SDS eluate (E) were analyzed by 10% SDS-PAGE followed by staining with Coomassie Blue. B, Srs2 interacts with Rad52. The pulldown assay between purified GST-Rad52 (1.5 μm, lanes 1 and 2) and Srs2 (0.75 μm, lanes 1–4), using glutathione-Sepharose, was performed as in A. C, Srs2 interacts with Rad59. Srs2 (0.75 μm, lanes 1–4) was mixed with either MBP-Rad59 (2 μm, lanes 1 and 2) or MBP alone (2.5 μm, lanes 3 and 4) and amylose resin. The experiment was carried out as in A. The asterisk indicates a degradation product of MBP-Rad59.
Srs2 SIM Promotes Cell Survival Independently of the PCNA Interaction
To determine SUMO-dependent and PCNA-independent regulation of Srs2 in vivo, we constructed yeast strains carrying srs2 mutant alleles similar to the ones used above: srs2-3KR, srs2-SIM* (I1170A/I1171A/V1172A/I1173A), and srs2ΔPIM.
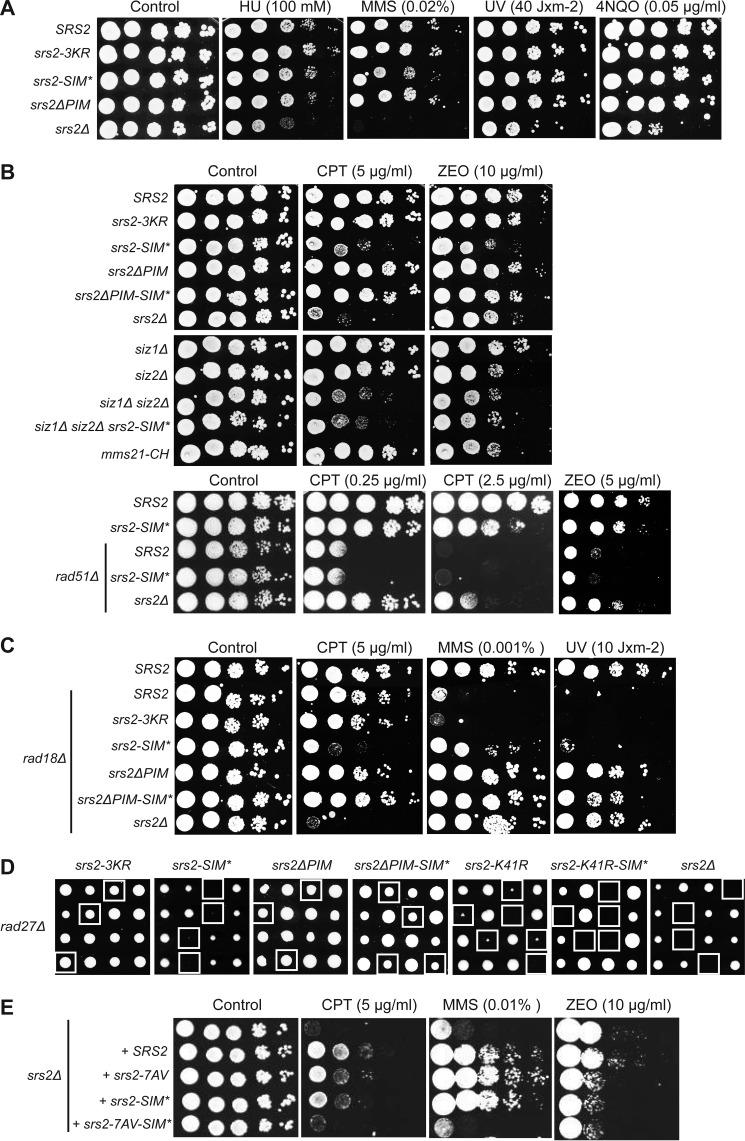
Initially, we evaluated cell survival of these srs2 mutants exposed to various DNA-damaging agents. Although the srs2Δ strain was sensitive to UV, 4-nitroquinoline 1-oxide (4NQO), hydroxyurea (HU), and MMS, the aforementioned mutants exhibited no decrease in cell survival (Fig. 4A). This suggests that SUMO binding and conjugation as well as the interaction with PCNA play no role in the corresponding repair processes. However, srs2-SIM* exhibited severe sensitivity to topoisomerase I inhibitor camptothecin (CPT) and zeocin (ZEO; Fig. 4B), which are drugs causing DNA double-strand breaks (DSBs). The fact that srs2-3KR and srs2ΔPIM were not sensitive to these drugs suggests that non-covalent interaction with SUMO or sumoylated proteins other than PCNA is responsible for reduced survival. To differentiate between these possibilities, we assessed the sensitivity of strains lacking SUMO E3 ligases. The similar phenotype of the double siz1Δ siz2Δ strain and srs2-SIM* and their epistatic relationship suggests that the interactions between SIM of Srs2 and proteins sumoylated by Siz1 together with Siz2 are required for DSB repair. Notably, simultaneous deletion of the PIM and SIM motifs suppressed the sensitivity of srs2-SIM* to CPT and ZEO (Fig. 4B). Because srs2-SIM* shows sensitivity to DSB inducing agents, we next analyzed the interaction between Srs2-SIM* and Rad51 recombinase. An epistatic relationship observed between these two mutants indicates that SIM plays a role in Rad51-dependent HR. On the other hand, deletion of the SRS2 gene led to partial suppression of the rad51 defect, illustrating its multifaceted role in HR.
FIGURE 4.
The SUMO-interacting motif of Srs2 plays PCNA-independent roles. A, DNA damage sensitivity of srs2 mutant strains. The indicated strains were grown in YPD to A600 ∼ 1, 10-fold serially diluted, spotted on YPD plates without or with the indicated amounts of hydroxyurea (HU), methyl methane sulfonate (MMS), ultraviolet light (UV), and 4-nitroquinoline 1-oxide (4NQO), and grown for 2 days at 30 °C. B, Srs2 SIM is important for cell survival after CPT and ZEO treatment. Experiment was performed as in A. C, the srs2-SIM* and srs2ΔPIM mutations suppress the sensitivity of rad18 cells to MMS and UV. The experiment was performed as in A. D, srs2-SIM* is synthetically sick with rad27Δ. Diploid strains heterozygous for the indicated mutations were obtained by crossing the haploid rad27Δ strain to individual srs2 mutant strains. After sporulation, the tetrads were dissected. Four tetrads (positioned in rows) are depicted for each strain. The double mutants are indicated by the white squares. E, Srs2 SIM and its phosphorylation are both important for cell survival after induced DNA damage. srs2Δ cells were transformed with the YCplac22 plasmids containing Srs2 gene or its mutants, grown on media lacking tryptophan, and analyzed as in A.
It has been previously shown that Srs2 sensitizes post-replication repair mutants (rad5, rad6, rad18) due to inhibition of HR in a manner dependent on SUMO-PCNA interaction (6, 11, 42, 44). Therefore, we wanted to determine in more detail the effect of the srs2 mutants on the interaction with SUMO-PCNA by examining their ability to suppress the sensitivity of rad18 cells to various DNA-damaging agents. As rad18 cells were not sensitive to CPT, we observed similar results as in the case of wild-type RAD18 (Fig. 4C). On the other hand, rad18Δ led to a significant decrease in cell survival in response to MMS, UV, hydroxyurea, 4-nitroquinoline 1-oxide, and ZEO, and this was suppressed by srs2-SIM* and srs2ΔPIM but not srs2-3KR (Fig. 4C and data not shown). Moreover, the differences in suppression level of srs2SIM* and srs2ΔPIM indicate that either the SIM has a lower importance for SUMO-PCNA interaction than does its PIM or that the SIM also plays another, PCNA-independent role. To differentiate between these two possibilities, we analyzed the strain lacking both SIM and PIM motifs and observed suppression of the MMS sensitivity of rad18 to the same degree as that of srs2ΔPIM and of UV sensitivity to a slightly lesser extent (Fig. 4C). Therefore, neither possibility can be excluded.
To explore more subtle defects that may be masked due to an existence of parallel repair pathways, we assessed the synthetic effect of the srs2 mutants with deletion of genes known to cause cell death when combined with srs2Δ. Previous studies had shown that the synthetic lethality of srs2 with rad54 and sgs1 is not dependent on the PCNA interactions (41). Accordingly, we observed no genetic interaction between srs2-SIM*, srs2ΔPIM, srs2-3KR, and rad54Δ or sgs1Δ (data not shown), thus indicating that SUMO and PCNA interactions are not important for the role of Srs2 in counteracting toxic recombination intermediates produced in these backgrounds. SRS2 deletion is also lethal with rad27Δ (16, 37) and, interestingly, srs2-SIM* rad27Δ double mutant exhibits severe growth defect similar to that of ATPase dead mutant srs2-K41R (Fig. 4D and Ref. 19). Moreover, this effect is independent of Srs2 sumoylation and PCNA interaction, as srs2-3KR rad27Δ and srs2ΔPIM rad27Δ double mutants are indistinguishable from the rad27Δ strain (Fig. 4D). In agreement with our previous results, deletion of PIM suppressed the growth defect of srs2-SIM*. Combination of the srs2-K41R-SIM* mutations with rad27Δ resulted in synthetic lethality, suggesting that SIM can, albeit only slightly, promote the cell survival of ATPase-deficient Srs2.
Similar to our data for srs2-SIM*, it had previously been observed that the non-phosphorylatable srs2 mutant (srs2–7AV) is more sensitive to zeocin than srs2Δ and is lethal in combination with rad27Δ (46). Therefore, we tested the relationship between SIM and Srs2 phosphorylation by transforming centromeric plasmids containing Srs2 or its mutants into the srs2Δ strain. Although comparison of the CPT and MMS sensitivities revealed an additive effect of srs2-SIM* and srs2–7AV, sensitivity to zeocin was epistatic (Fig. 4E), thus indicating both linked as well as independent roles of SIM and phosphorylation of Srs2 in DNA repair. In summary, these data show that SUMO-interacting motif of Srs2 not only stimulates the Srs2 interaction with SUMO-PCNA, but it also plays a role independent from SUMO-PCNA interaction and Srs2 sumoylation.
Interaction with PCNA, SUMO, and Sumoylation Differently Regulate Srs2 Function in HR
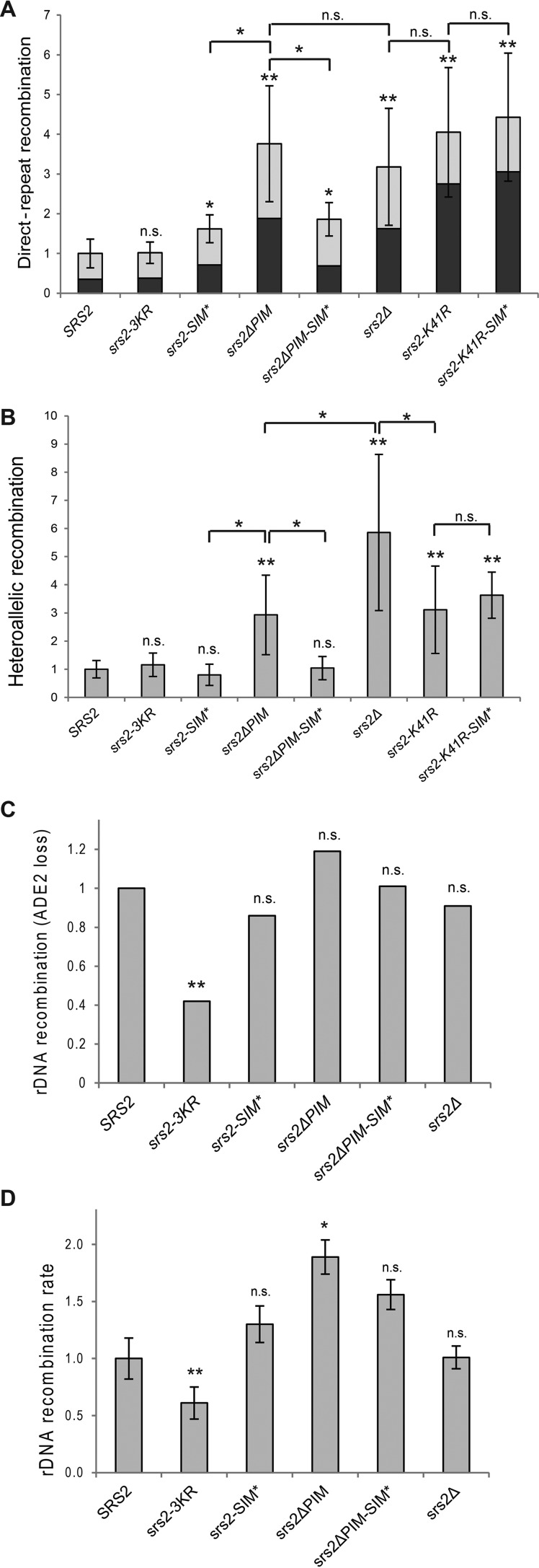
The observations that SIM of Srs2 is important when cells need to deal with DSBs or with the absence of Rad27 is consistent with the idea that it is important to promote HR. To test this possibility, we examined the srs2 mutants in mitotic recombination assays, including direct-repeat (DR), heteroallelic, and rDNA recombination (64, 69, 70). In the DR assay, which measures spontaneous intrachromosomal recombination in haploid cells that is generated mostly by single-strand annealing and gene conversion events, we observed a slightly elevated recombination rate in the case of srs2-SIM* and much greater increase in the case of srs2ΔPIM (Table 3 and Fig. 5A). Moreover, the recombination rate of srs2ΔPIM was statistically indistinguishable from that of the srs2Δ strain, indicating that this domain is important for the anti-recombination function of Srs2 in haploid cells. As the two strains also exhibit the same ratio between deletion and gene conversion events, the Srs2-PCNA interaction seems to be involved in inhibition of both types of events. The lower recombination rate of srs2-SIM* in comparison to srs2ΔPIM may ensue from SIM's minor role in SUMO-PCNA binding (as suggested by Fig. 4C) or from its partly opposing role in regulating recombination. To differentiate between these two possibilities, we tested the srs2ΔPIM-SIM* double mutant, where a PCNA-independent role of SIM could be observed. Indeed, we noticed a nearly 2-fold decrease in recombination level when SIM* was additionally mutated in the ΔPIM strain, thus indicating that SIM promotes recombination and suppresses the inhibitory role of PIM. To analyze whether the ATPase activity of Srs2 is necessary for this pro-recombination role mediated by its SIM, we next tested srs2-K41R and srs2-K41R-SIM* strains. Inasmuch as the two strains showed very similar recombination levels, the pro-recombination role of Srs2 seems to depend on the ATPase activity of Srs2.
TABLE 3.
Effect of srs2 mutants on mitotic direct-repeat and heteroallelic recombination rates
| Allele | Direct-repeat recombination |
Heteroallelic recombination |
||||||
|---|---|---|---|---|---|---|---|---|
| Ratea | -Fold changeb | p valuec | Fraction Ura+d | p valuec | Ratee | Fold changef | p valuec | |
| × 10−5 | × 10−6 | |||||||
| SRS2 | 4.19 ± 1.51 | 1 | NA | 0.35 | NA | 1.94 ± 0.60 | 1 | NA |
| srs2–3KR | 4.29 ± 1.14 | 1.02 | 0.719 | 0.37 | 0.704 | 2.26 ± 0.81 | 1.16 | 0.112 |
| srs2-SIM* | 6.81 ± 1.48 | 1.62 | 0.003 | 0.44 | 0.105 | 1.54 ± 0.73 | 0.8 | 0.88 |
| srs2ΔPIM | 15.78 ± 6.11 | 3.76 | <0.001 | 0.50 | 0.001 | 5.69 ± 2.74 | 2.93 | <0.001 |
| srs2ΔPIM-SIM* | 7.79 ± 1.61 | 1.86 | 0.002 | 0.37 | 0.741 | 2.02 ± 0.80 | 1.04 | 0.58 |
| srs2Δ | 13.34 ± 6.17 | 3.18 | <0.001 | 0.51 | 0.010 | 11.35 ± 5.38 | 5.86 | <0.001 |
| srs2-K41R | 16.97 ± 6.84 | 4.05 | <0.001 | 0.68 | <0.001 | 6.02 ± 3.01 | 3.11 | <0.001 |
| srs2-K41R-SIM* | 18.56 ± 6.73 | 4.43 | <0.001 | 0.69 | <0.001 | 7.02 ± 1.59 | 3.63 | <0.001 |
a Intrachromosomal recombination between direct repeats in a haploid strain was assessed using leu2-ΔEcoRI and leu2-ΔBstEII alleles flanking the URA3 gene. Overall recombination rate (events per cell per generation) calculated from frequency of LEU+ colonies is presented as the median ± mean absolute deviation, as described under “Experimental Procedures.”
b Recruitment by PCNA is important for the Srs2 role in direct-repeat recombination repression. -Fold change is expressed relative to wild type.
c p value for Mann-Whitney U test applied to the direct-repeat recombination rate, Ura+ fractions, and heteroallelic recombination relative to wild type; NA, not applicable.
d Fraction of gene conversion events (LEU+ URA+) from the overall direct-repeat recombination (LEU+).
e Interchromosomal recombination rate between leu2-ΔEcoRI and leu2-ΔBstEII heteroalleles in a diploid strain was calculated as in footnote a.
f PCNA interaction is partly responsible for Srs2 inhibitory effect on heteroallelic recombination, whereas the SIM has no such effect. -Fold change is relative to wild type.
FIGURE 5.
Srs2 interaction with PCNA, its non-covalent interaction with SUMO, and Srs2 sumoylation play different roles in recombination regulation. The recombination rates relative to wild type (Table 3) are plotted. In A, the overall direct-repeat recombination and the part corresponding to gene conversions (dark gray) are depicted. In B, the heteroallelic recombination between homologous chromosomes is shown. In both A and B the median values of 15–19 trials are illustrated. Error bars indicate median absolute deviations. In C, the rDNA recombination rates were measured by the incidence of half-sectored (red-white) colonies from >20,000 total colonies for each strain (Table 4). In D, the rDNA recombination rates were calculated from wholly red colonies (Table 4); median values of 15–19 trials are shown. The statistical analysis was performed using the non-parametric Mann-Whitney U test (A, B, and D), or Yates' χ2 test (C). n.s., no significant difference, *, p < 0.01; **, p < 0.001 versus wild type or between the indicated strains. Error bars show median absolute deviation.
When we monitored heteroallelic recombination between leu2 alleles located on two homologous chromosomes in diploid cells (71), srs2ΔPIM exhibited an increase in recombination rate similar with that observed in the DR assay (Table 3 and Fig. 5B). The rate was significantly more increased in the srs2Δ strain, however, indicating that part of the Srs2 inhibitory effect on the interchromosomal recombination is PCNA-independent. In contrast to srs2ΔPIM, srs2-SIM* led to a slight decrease in recombination rates, and srs2ΔPIM-SIM* suppressed the recombination rate of srs2ΔPIM to a rate undistinguishable from that of wild type. This is consistent with the idea that, despite its PCNA-dependent role, SIM stimulates recombination. Similar to the cases in DR, srs2-K41R and srs2-K41R-SIM* exhibited undistinguishable recombination levels, but those levels were significantly lower than those of the deletion strain. This suggests that ATPase activity of Srs2 is only partially important for repression of interchromosomal recombination but that it is necessary for the pro-recombination function mediated by its SIM.
Although the non-sumoylatable srs2-3KR mutant does not affect DR and heteroallelic recombination rates, the picture changes when rDNA recombination is examined (72). The rate of rDNA-located ADE2 marker loss, which was measured by the incidence of half-sectored (red-white) colonies, was significantly decreased in the srs2-3KR strain (Table 4 and Fig. 5C). Although there was no statistically significant difference between wild-type strain and srs2ΔPIM or srs2-SIM*, their role was better visible when recombination rate was calculated from wholly red colonies, which had lost the ADE2 marker before plating (Table 4 and Fig. 5D). Apart from srs2-3KR, only the difference between SRS2 and srs2ΔPIM was significant. However, Fig. 5D indicates that PCNA interaction inhibits recombination at the rDNA, whereas SIM again probably plays an ambivalent role: stimulation of recombination by mediating Srs2 sumoylation and inhibition by mediating PCNA binding.
TABLE 4.
Srs2 sumoylation promotes rDNA recombination
| Allele | rDNA recombination |
|||||
|---|---|---|---|---|---|---|
| Half-sectored colonies |
Whole-red colonies |
p valuee | ||||
| Ratea | -Fold changeb | p valuec | Rated | -Fold changeb | ||
| × 10−3 | × 10−3 | |||||
| SRS2 | 2.73 | 1 | NA | 4.34 ± 0.80 | 1 | NA |
| srs2-3KR | 1.15 | 0.42 | <0.001 | 2.63 ± 0.59 | 0.61 | <0.001 |
| srs2-SIM* | 2.35 | 0.86 | 0.452 | 5.66 ± 0.69 | 1.30 | 0.100 |
| srs2ΔPIM | 3.63 | 1.19 | 0.327 | 8.19 ± 0.67 | 1.89 | 0.006 |
| srs2ΔPIM-SIM* | 2.76 | 1.01 | 0.946 | 6.79 ± 0.63 | 1.56 | 0.026 |
| srs2Δ | 2.48 | 0.91 | 0.705 | 4.40 ± 0.43 | 1.01 | 0.976 |
a ADE2 marker loss (located in the rDNA locus) in the first generation after plating was assayed by counting half-sectored colonies (80).
b -Fold change relative to wild type.
c p value for Yates' χ2 test applied to the rDNA recombination rate relative to wild type; NA, not applicable.
d ADE2 marker loss was assayed by counting whole-red colonies. Recombination rate (events per cell per generation) was calculated by the Lea-Coulson median method and is presented as the median ± median absolute deviation.
e p value for Mann-Whitney U test applied to rDNA recombination relative to wild-type.
In summary, analysis of the recombination rates suggests that interaction between Srs2 and PCNA is important for Srs2 anti-recombination function, whereas SIM of Srs2 also plays a PCNA-independent role that leads to recombination stimulation. The most visible effect of Srs2 sumoylation is stimulation of the rDNA recombination.
Discussion
SUMO Mediates Several Srs2 Interactions
In this work we aimed to decipher the specific roles of SUMO and PCNA in regulating the multiple Srs2 functions during DNA repair. The previous identification of Srs2 sumoylation sites (Srs2-Lys-1081, -1089, -1142) as well as SUMO (Srs2-SIM) and PCNA (Srs2-PIM) interaction motifs enabled us to study their roles in Srs2 regulation.
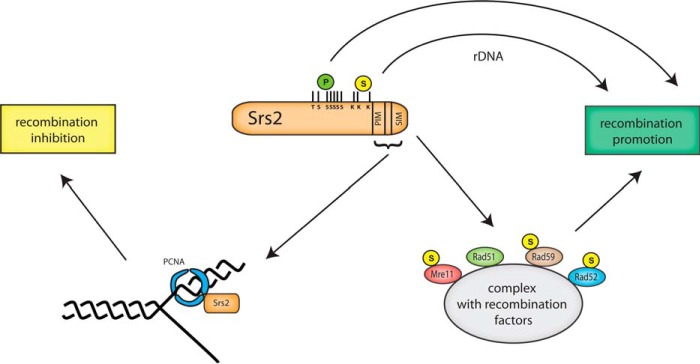
Because we did not observe a major effect of SUMO on Srs2 DNA binding and helicase activities, we proceeded to examine the best documented role of SUMO in stimulation of protein interactions. Because in our experience pulldown experiments have proven largely insensitive in evaluating the effects of SUMO attachment on protein interactions, we proceeded to test the effect of SUMO by analyzing Srs2 mutants using the yeast two-hybrid assay. We noticed that SUMO strengthens two-hybrid Srs2 interaction not only with PCNA but also with other recombination factors, including Mre11, Rad51, Rad52, and to a smaller extent, Rad59 (Fig. 2). Although in the cases of Mre11, Rad52, and Rad59, the SIM motif of Srs2 is important for the interactions, in the case of interaction with Rad51 the Srs2 sumoylation itself is also crucial. Analysis of Srs2ΔPIM shows that stimulation and/or stabilization of Srs2 interactions by SUMO is not promoted by the PCNA interaction. The fact that Srs2ΔPIM shows even increased interactions suggests the existence of two possibly mutually exclusive pools of Srs2, one bound through its SIM with recombination proteins and the second bound through PIM and SIM to PCNA (Fig. 6). The absence of the PCNA binding, then, allows an increase in the pool of free Srs2 and enables more interactions with recombination proteins mediated via SIM. The mutual exclusivity of these two binding pools is also supported by our previous observations showing that sumoylation of Srs2 blocks the interaction with PCNA and that increasing amounts of SUMO and Siz1 can counteract the inhibitory effect of SUMO-PCNA on Srs2 sumoylation (42). Therefore, the clustering of PCNA and SUMO binding motifs with nearby post-translational modification sites in Srs2 may be an intriguing regulatory mechanism required for the multifaceted role of Srs2 during DNA repair.
FIGURE 6.
Model for regulation of Srs2 functions by post-translational modifications. The SIM and PIM motifs of Srs2 both participate in its recruitment by SUMO-PCNA to replication forks, which leads to HR inhibition. Srs2 SIM also stimulates interactions with sumoylated recombination factors, leading to recombination promotion. Recombination is likewise promoted by post-translational modifications of Srs2 itself. Phosphorylation of Srs2 was shown to stimulate SDSA (46), whereas the effect of its sumoylation is particularly exhibited by rDNA recombination promotion.
Srs2 SIM Is Important for HR
To further understand the relationship between the Srs2 interaction and/or modification sites and its biological role, we analyzed the sensitivity of our Srs2 mutant strains under various conditions. In contrast to srs2ΔPIM and srs2-3KR mutants, srs2-SIM* was sensitive to camptothecin and to zeocin and displayed a synthetic growth defect with rad27Δ. This suggests it has a pro-recombination role within Srs2. The HR promotion is likely Rad51-dependent, as indicated by the observation that rad51Δ is epistatic to srs2-SIM*. Based on our interaction data, we propose that SIM of Srs2 stimulates interactions with other sumoylated proteins involved in HR (Fig. 6). This is also supported by the epistatic relationship between srs2-SIM and siz1Δ siz2Δ. The observation that ΔPIM rescues the SIM* mutant is in agreement with the idea that when PCNA does not compete for Srs2 binding, the SIM of Srs2 is no longer essential for efficient formation of recombination complexes. This further supports the notion of mutual exclusivity of the two Srs2 pools.
A previous study of Srs2 regulation by post-translational modifications had shown that, similarly to our srs2-SIM* mutant, the non-phosphorylatable SRS2 mutant (srs2–7AV) is required for recombination, as it is lethal in combination with rad27Δ and more sensitive to zeocin compared with srs2Δ (46). Moreover, phosphorylation of Srs2 is needed to reorganize protein complexes involving Mre11, Srs2, and Sgs1 after DNA damage (47). Our analysis of the srs2–7AV-SIM* mutant showed clear synergistic relationships of the two mutants with respect to CPT and MMS, thus suggesting that the Srs2 SIM and its phosphorylation work independently in promoting recombination (Figs. 4E and 6). We hypothesize that the DNA damage-induced assembly of DNA-repair complexes is jointly promoted by their phosphorylation and sumoylation, and phosphosites and SIM of Srs2 play an important part in the multiple undergoing phosphate- and SUMO-dependent interactions.
Srs2 SIM Promotes Recombination
Our data indicate that Srs2 SIM also plays a PCNA-independent role in promoting recombination, and therefore, we analyzed the effects of corresponding Srs2 mutants in various mitotic recombination assays (Fig. 5). In the direct-repeat recombination assay, the srs2-SIM* showed only a small increase in recombination, whereas recombination in the case of srs2ΔPIM was increased significantly and indistinguishably from that of the srs2Δ strain. The importance of the PCNA interaction for the inhibitory effect of Srs2 on DR suggests it occurs at the replication forks, where Srs2 first needs to be recruited by PCNA. Because SIM of Srs2 also stimulates interaction with SUMO-PCNA and thus participates in HR inhibition, the recombination promotion of SIM was only visible in the ΔPIM background, where the PCNA interaction is disrupted. Furthermore, the SIM is likely responsible for promoting gene conversions, as particularly the gene conversion part of recombination was decreased when SIM* was combined with ΔPIM.
When we measured recombination between heteroalleles located on homologous chromosomes in diploid cells, the rates behaved similarly to those from the DR assay. Loss of Srs2 SIM-mediated protein interactions with HR factors suppressed the increased recombination rates of srs2ΔPIM. We nevertheless observed two considerable differences, and these are likely linked. First, the recombination in srs2-SIM* was identical to that in the wild type, suggesting that the pro-recombination role of Srs2 plays a greater part in this type of recombination. Second, the srs2Δ strain showed a significantly greater increase in recombination than did srs2ΔPIM, indicating that the PCNA interaction plays a less important part in Srs2's role in suppressing recombination between homologous chromosomes. This is probably due to the different requirements of the two types of recombination. The recombination between sister chromatids is dependent on DNA replication during S phase, when the PCNA sumoylation also appears to recruit Srs2 and inhibit recombination (38–40). In the case of recombination between homologous chromosomes, DNA replication is not necessary, and it is, therefore, less affected by PCNA-mediated recruitment of Srs2. This lesser role of SUMO-PCNA in Srs2 inhibition of heteroallelic recombination is also manifested by the pronounced stimulatory role of SIM.
Our recombination results are in agreement with those of Pfander et al. (38), whose interchromosomal and direct-repeat recombination results with srs2ΔC136 and srs2ΔC6 strains resemble those of our srs2ΔPIM-SIM* and srs2-SIM* strains, pointing to an important role of Srs2-PCNA interaction for inhibition of sister chromatid recombination. The more recent results of Miura et al. (34) are in various ways both similar to and dissimilar from our study's findings. In contrast to our data, they observed that SUMO-PCNA interaction is necessary for Srs2's role in SDSA promotion. This may be caused by the differences between our chromosome-based and their plasmid-based recombination assays, which demands homology search at two different chromosomal sites. On the other hand, their double Holliday junction-mediated assay shows similarities with ours, with srs2ΔPIM leading to a significant increase in recombination, whereas srs2-SIM* barely shows any effect. Moreover, both studies suggest that the pro-recombination role of Srs2 requires its ATPase activity (Fig. 5 and Ref. 34).
Interestingly, we observed that the sumoylation of Srs2 itself is particularly important in promoting recombination at the rDNA (Fig. 5, C and D). This finding is not unexpected, as the importance of SUMO in regulation of rDNA recombination has already been described by several studies (73).
The Pro- and Anti-recombination Roles of Srs2 Need to Be Balanced
Overall, our results indicate that SUMO plays opposing roles in Srs2 regulation. It not only takes part in Srs2 recruitment by SUMO-PCNA, thereby leading to HR inhibition at replication forks, but it also plays a PCNA-independent role in stimulating the formation of pro-recombination complexes by stabilizing interactions of Srs2 with recombination proteins (Fig. 6). The efficient formation of Srs2 complexes with PCNA or recombination proteins is dependent on two sets of transient interactions, protein- and SUMO-specific. Neither of these interaction types seems to be sufficient by itself. The protein-specific interactions need to be stabilized by SUMO-SIM interactions for efficient complex formation in the wild-type cells, a mechanism that enables their rapid assembly and disassembly according to the actual cellular needs.
The importance of Srs2 SIM for both the PCNA-dependent anti-recombination and PCNA-independent pro-recombination complexes leads to competition for the binding of Srs2's SIM. The observation that srs2-SIM* causes cell death under conditions necessitating repair by HR suggests its particular importance for the pro-recombination complex formation. Although these complexes are not formed efficiently in the srs2-SIM* strain, it seems there is still a residual PCNA binding. When this residual binding is abolished by the simultaneous PIM deletion, the phenotype of cells lacking SIM is rescued. This suggests the absence of PCNA competition for Srs2 binding enables efficient pro-recombination complex formation, even in the absence of its SIM. This indicates that PCNA-PIM interaction is stronger than the specific interactions between Srs2 and its recombination partners, as we indeed observed in the pulldown experiments. Our results also show that rather than SIM alone, the proper balance between the pro- and anti-recombination pools of Srs2 is crucial, as the srs2ΔPIM-SIM* mutants behave similarly to wild type in both survival and recombination assays.
The underlying molecular mechanism of the Srs2 pro-recombination function is still unclear. Although the possible mechanisms of Srs2 function in SDSA promotion have been outlined, the exact function of Srs2 in this process remains elusive. Even though Srs2 is able to unwind synthetic D-loop structures in vitro (32), it is not able to do so when the Rad51-mediated strand invasion and DNA synthesis are reconstituted (33). We hypothesize that rather than disrupting the D-loop intermediate, Srs2 prevents reloading of Rad51 on the displaced extended strand to promote annealing to the complementary part at the other end of the DSB, similarly to human RECQ5 helicase (74). That would be in line with the observed interaction with Rad52 and Rad59 proteins, implicated to take part in this process (61, 75, 76). Alternatively, Srs2 can be involved in the resolution of the recombination intermediates (77).
The facts that Srs2 plays a multifaceted role in HR regulation and interacts with a plethora of recombination proteins indicate a complex and robust DNA damage response mechanism. Such a mechanism requires regulation and fine-tuning of proper repair scenarios. We show that sumoylation plays an important part in this process, and formation of SUMO-SIM interactions facilitates recombination complex assembly necessary for efficient DNA repair. As human protein PARI (PCNA-associated recombination inhibitor) functionally and structurally resembles Srs2 (78, 79), a similar SIM-dependent process may exist in human cells. The described mechanism is likely to serve for other repair proteins whose functions need to be closely regulated and coordinated.
Author Contributions
P. K. and L. K. designed the experiments and wrote the paper. P. K. and V. A. performed the experiments. S. S. prepared some reagents. P. K., V. A., M. L., and L. K. analyzed the data.
Acknowledgments
We thank X. Zhao, H. Klein, and G. Liberi for providing yeast strains and plasmids and S. Gangloff for critical reading of the manuscript.
This work was supported by the Czech Science Foundation (GACR 13-26629S and P207/12/2323; to L. K.), European Regional Development Fund- Project FNUSA-ICRC (CZ.1.05/1.1.00/02.0123; to L. K.) and Research Support Programme (GAMU-MUNI/M/1894/2014; to L. K), “Employment of Newly Graduated Doctors of Science for Scientific Excellence” co-financed from the European Social Fund (CZ.1.07/2.3.00/30.0009 (to V. A.) and CZ.1.07/2.3.00/30.0037 (to P. K.), funds from the Faculty of Medicine, Masaryk University to junior researcher (to V. A.), the Danish Agency for Science, Technology and Innovation, the Villum Kann Rasmussen Foundation, the Lundbeck Foundation, European Research Council (ERC) (to M. L.), and Fundação para a Ciência e Tecnologia (FCT; to S. S.). The authors declare that they have no conflicts of interest with the contents of this article.
- HR
- homologous recombination
- SUMO
- small ubiquitin-like modifier
- SIM
- SUMO-interacting motif
- PCNA
- proliferating cell nuclear antigen
- PIM
- PCNA-interacting motif
- CPT
- camptothecin
- MMS
- methyl methane-sulfonate
- ZEO
- zeocin
- DSB
- double-strand break
- DR
- direct-repeat
- SDSA
- synthesis-dependent strand annealing.
References
- 1. Krejci L., Altmannova V., Spirek M., and Zhao X. (2012) Homologous recombination and its regulation. Nucleic Acids Res. 40, 5795–5818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. San Filippo J., Sung P., and Klein H. (2008) Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 3. Rong L., and Klein H. L. (1993) Purification and characterization of the SRS2 DNA helicase of the yeast Saccharomyces cerevisiae. J. Biol. Chem. 268, 1252–1259 [PubMed] [Google Scholar]
- 4. Marini V., and Krejci L. (2010) Srs2: the “Odd-Job Man” in DNA repair. DNA Repair 9, 268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lawrence C. W., and Christensen R. B. (1979) Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J. Bacteriol. 139, 866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aboussekhra A., Chanet R., Zgaga Z., Cassier-Chauvat C., Heude M., and Fabre F. (1989) RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair: characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 17, 7211–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schiestl R. H., Prakash S., and Prakash L. (1990) The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics 124, 817–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ulrich H. D. (2001) The srs2 suppressor of UV sensitivity acts specifically on the RAD5- and MMS2-dependent branch of the RAD6 pathway. Nucleic Acids Res. 29, 3487–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rong L., Palladino F., Aguilera A., and Klein H. L. (1991) The hyper-gene conversion hpr5–1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics 127, 75–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Aguilera A., and Klein H. L. (1988) Genetic control of intrachromosomal recombination in Saccharomyces cerevisiae: I. Isolation and genetic characterization of hyper-recombination mutations. Genetics 119, 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palladino F., and Klein H. L. (1992) Analysis of mitotic and meiotic defects in Saccharomyces cerevisiae SRS2 DNA helicase mutants. Genetics 132, 23–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Aboussekhra A., Chanet R., Adjiri A., and Fabre F. (1992) Semidominant suppressors of Srs2 helicase mutations of Saccharomyces cerevisiae map in the RAD51 gene, whose sequence predicts a protein with similarities to procaryotic RecA proteins. Mol. Cell. Biol. 12, 3224–3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chanet R., Heude M., Adjiri A., Maloisel L., and Fabre F. (1996) Semidominant mutations in the yeast Rad51 protein and their relationships with the Srs2 helicase. Mol. Cell. Biol. 16, 4782–4789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lee S. K., Johnson R. E., Yu S. L., Prakash L., and Prakash S. (1999) Requirement of yeast SGS1 and SRS2 genes for replication and transcription. Science 286, 2339–2342 [DOI] [PubMed] [Google Scholar]
- 15. Gangloff S., Soustelle C., and Fabre F. (2000) Homologous recombination is responsible for cell death in the absence of the Sgs1 and Srs2 helicases. Nat. Genet. 25, 192–194 [DOI] [PubMed] [Google Scholar]
- 16. Klein H. L. (2001) Mutations in recombinational repair and in checkpoint control genes suppress the lethal combination of srs2Delta with other DNA repair genes in Saccharomyces cerevisiae. Genetics 157, 557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Krejci L., Van Komen S., Li Y., Villemain J., Reddy M. S., Klein H., Ellenberger T., and Sung P. (2003) DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423, 305–309 [DOI] [PubMed] [Google Scholar]
- 18. Veaute X., Jeusset J., Soustelle C., Kowalczykowski S. C., Le Cam E., and Fabre F. (2003) The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423, 309–312 [DOI] [PubMed] [Google Scholar]
- 19. Krejci L., Macris M., Li Y., Van Komen S., Villemain J., Ellenberger T., Klein H., and Sung P. (2004) Role of ATP hydrolysis in the antirecombinase function of Saccharomyces cerevisiae Srs2 protein. J. Biol. Chem. 279, 23193–23199 [DOI] [PubMed] [Google Scholar]
- 20. Colavito S., Macris-Kiss M., Seong C., Gleeson O., Greene E. C., Klein H. L., Krejci L., and Sung P. (2009) Functional significance of the Rad51-Srs2 complex in Rad51 presynaptic filament disruption. Nucleic Acids Res. 37, 6754–6764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Antony E., Tomko E. J., Xiao Q., Krejci L., Lohman T. M., and Ellenberger T. (2009) Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol. Cell 35, 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Burkovics P., Sebesta M., Sisakova A., Plault N., Szukacsov V., Robert T., Pinter L., Marini V., Kolesar P., Haracska L., Gangloff S., and Krejci L. (2013) Srs2 mediates PCNA-SUMO-dependent inhibition of DNA repair synthesis. EMBO J. 32, 742–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pâques F., and Haber J. E. (1997) Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol. Cell. Biol. 17, 6765–6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sugawara N., Ira G., and Haber J. E. (2000) DNA length dependence of the single-strand annealing pathway and the role of Saccharomyces cerevisiae RAD59 in double-strand break repair. Mol. Cell. Biol. 20, 5300–5309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hegde V., and Klein H. (2000) Requirement for the SRS2 DNA helicase gene in non-homologous end joining in yeast. Nucleic Acids Res. 28, 2779–2783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wilson T. E. (2002) A genomics-based screen for yeast mutants with an altered recombination/end-joining repair ratio. Genetics 162, 677–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ira G., and Haber J. E. (2002) Characterization of RAD51-independent break-induced replication that acts preferentially with short homologous sequences. Mol. Cell. Biol. 22, 6384–6392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ira G., Malkova A., Liberi G., Foiani M., and Haber J. E. (2003) Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115, 401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carter S. D., Vigasová D., Chen J., Chovanec M., and Aström S. U. (2009) Nej1 recruits the Srs2 helicase to DNA double-strand breaks and supports repair by a single-strand annealing-like mechanism. Proc. Natl. Acad. Sci. U.S.A. 106, 12037–12042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vaze M. B., Pellicioli A., Lee S. E., Ira G., Liberi G., Arbel-Eden A., Foiani M., and Haber J. E. (2002) Recovery from checkpoint-mediated arrest after repair of a double-strand break requires Srs2 helicase. Mol. Cell 10, 373–385 [DOI] [PubMed] [Google Scholar]
- 31. Aylon Y., Liefshitz B., Bitan-Banin G., and Kupiec M. (2003) Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 23, 1403–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dupaigne P., Le Breton C., Fabre F., Gangloff S., Le Cam E., and Veaute X. (2008) The Srs2 helicase activity is stimulated by Rad51 filaments on dsDNA: implications for crossover incidence during mitotic recombination. Mol. Cell 29, 243–254 [DOI] [PubMed] [Google Scholar]
- 33. Sebesta M., Burkovics P., Haracska L., and Krejci L. (2011) Reconstitution of DNA repair synthesis in vitro and the role of polymerase and helicase activities. DNA Repair 10, 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miura T., Shibata T., and Kusano K. (2013) Putative antirecombinase Srs2 DNA helicase promotes noncrossover homologous recombination avoiding loss of heterozygosity. Proc. Natl. Acad. Sci. U.S.A. 110, 16067–16072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Friedl A. A., Liefshitz B., Steinlauf R., and Kupiec M. (2001) Deletion of the SRS2 gene suppresses elevated recombination and DNA damage sensitivity in rad5 and rad18 mutants of Saccharomyces cerevisiae. Mutat. Res. 486, 137–146 [DOI] [PubMed] [Google Scholar]
- 36. Ruiz J. F., Gómez-González B., and Aguilera A. (2009) Chromosomal translocations caused by either pol32-dependent or pol32-independent triparental break-induced replication. Mol. Cell. Biol. 29, 5441–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Debrauwère H., Loeillet S., Lin W., Lopes J., and Nicolas A. (2001) Links between replication and recombination in Saccharomyces cerevisiae: a hypersensitive requirement for homologous recombination in the absence of Rad27 activity. Proc. Natl. Acad. Sci. U.S.A. 98, 8263–8269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pfander B., Moldovan G.-L., Sacher M., Hoege C., and Jentsch S. (2005) SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436, 428–433 [DOI] [PubMed] [Google Scholar]
- 39. Papouli E., Chen S., Davies A. A., Huttner D., Krejci L., Sung P., and Ulrich H. D. (2005) Crosstalk between SUMO and ubiquitin on PCNA is mediated by recruitment of the helicase Srs2p. Mol. Cell 19, 123–133 [DOI] [PubMed] [Google Scholar]
- 40. Hoege C., Pfander B., Moldovan G.-L., Pyrowolakis G., and Jentsch S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 41. Le Breton C., Dupaigne P., Robert T., Le Cam E., Gangloff S., Fabre F., and Veaute X. (2008) Srs2 removes deadly recombination intermediates independently of its interaction with SUMO-modified PCNA. Nucleic Acids Res. 36, 4964–4974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kolesar P., Sarangi P., Altmannova V., Zhao X., and Krejci L. (2012) Dual roles of the SUMO-interacting motif in the regulation of Srs2 sumoylation. Nucleic Acids Res. 40, 7831–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burgess R. C., Lisby M., Altmannova V., Krejci L., Sung P., and Rothstein R. (2009) Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J. Cell Biol. 185, 969–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Armstrong A. A., Mohideen F., and Lima C. D. (2012) Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature 483, 59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim S. O., Yoon H., Park S. O., Lee M., Shin J.-S., Ryu K.-S., Lee J.-O., Seo Y.-S., Jung H. S., and Choi B.-S. (2012) Srs2 possesses a non-canonical PIP box in front of its SBM for precise recognition of SUMOylated PCNA. J. Mol. Cell Biol. 4, 258–261 [DOI] [PubMed] [Google Scholar]
- 46. Saponaro M., Callahan D., Zheng X., Krejci L., Haber J. E., Klein H. L., and Liberi G. (2010) Cdk1 targets Srs2 to complete synthesis-dependent strand annealing and to promote recombinational repair. PLoS Genet. 6, e1000858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chiolo I., Carotenuto W., Maffioletti G., Petrini J. H., Foiani M., and Liberi G. (2005) Srs2 and Sgs1 DNA helicases associate with Mre11 in different subcomplexes following checkpoint activation and CDK1-mediated Srs2 phosphorylation. Mol. Cell. Biol. 25, 5738–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Thomas B. J., and Rothstein R. (1989) Elevated recombination rates in transcriptionally active DNA. Cell 56, 619–630 [DOI] [PubMed] [Google Scholar]
- 49. Zhao X., Muller E. G., and Rothstein R. (1998) A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol. Cell 2, 329–340 [DOI] [PubMed] [Google Scholar]
- 50. Sherman F., Fink G. R., and Hicks J. B. (1989) Laboratory Course Manual for Methods in Yeast Genetics 186 S., 10 Abb., 2 Tab. Cold Spring Harbor 1986. Cold Spring Harbor Laboratory; ISBN: 0–87969-197–2 [Google Scholar]
- 51. Erdeniz N., Mortensen U. H., and Rothstein R. (1997) Cloning-free PCR-based allele replacement methods. Genome Res. 7, 1174–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bencsath K. P., Podgorski M. S., Pagala V. R., Slaughter C. A., and Schulman B. A. (2002) Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J. Biol. Chem. 277, 47938–47945 [DOI] [PubMed] [Google Scholar]
- 53. Johnson E. S., and Blobel G. (1997) Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272, 26799–26802 [DOI] [PubMed] [Google Scholar]
- 54. Johnson E. S., Schwienhorst I., Dohmen R. J., and Blobel G. (1997) The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 16, 5509–5519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takahashi Y., Toh-E A., and Kikuchi Y. (2003) Comparative analysis of yeast PIAS-type SUMO ligases in vivo and in vitro. J. Biochem. 133, 415–422 [DOI] [PubMed] [Google Scholar]
- 56. Trujillo K. M., and Sung P. (2001) DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J. Biol. Chem. 276, 35458–35464 [DOI] [PubMed] [Google Scholar]
- 57. Krejci L., Song B., Bussen W., Rothstein R., Mortensen U. H., and Sung P. (2002) Interaction with Rad51 is indispensable for recombination mediator function of Rad52. J. Biol. Chem. 277, 40132–40141 [DOI] [PubMed] [Google Scholar]
- 58. Chamankhah M., and Xiao W. (1999) Formation of the yeast Mre11-Rad50-Xrs2 complex is correlated with DNA repair and telomere maintenance. Nucleic Acids Res. 27, 2072–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Huang M. E., de Calignon A., Nicolas A., and Galibert F. (2000) POL32, a subunit of the Saccharomyces cerevisiae DNA polymerase delta, defines a link between DNA replication and the mutagenic bypass repair pathway. Curr. Genet. 38, 178–187 [DOI] [PubMed] [Google Scholar]
- 60. Krejci L., Damborsky J., Thomsen B., Duno M., and Bendixen C. (2001) Molecular dissection of interactions between Rad51 and members of the recombination-repair group. Mol. Cell. Biol. 21, 966–976 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61. Davis A. P., and Symington L. S. (2001) The yeast recombinational repair protein Rad59 interacts with Rad52 and stimulates single-strand annealing. Genetics 159, 515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Takahashi Y., Kahyo T., Toh-E A., Yasuda H., and Kikuchi Y. (2001) Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for septin components and functions as an adaptor between conjugating enzyme and substrates. J. Biol. Chem. 276, 48973–48977 [DOI] [PubMed] [Google Scholar]
- 63. Marini V., and Krejci L. (2012) Unwinding of synthetic replication and recombination substrates by Srs2. DNA Repair 11, 789–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Altmannova V., Eckert-Boulet N., Arneric M., Kolesar P., Chaloupkova R., Damborsky J., Sung P., Zhao X., Lisby M., and Krejci L. (2010) Rad52 SUMOylation affects the efficiency of the DNA repair. Nucleic Acids Res. 38, 4708–4721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Shi I., Hallwyl S. C., Seong C., Mortensen U., Rothstein R., and Sung P. (2009) Role of the Rad52 amino-terminal DNA binding activity in DNA strand capture in homologous recombination. J. Biol. Chem. 284, 33275–33284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hall B. M., Ma C.-X., Liang P., and Singh K. K. (2009) Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics. 25, 1564–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kerscher O. (2007) SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO Rep. 8, 550–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Altmannová V., Kolesár P., and Krejčí L. (2012) SUMO Wrestles with Recombination. Biomolecules. 2, 350–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Smith J., and Rothstein R. (1999) An allele of RFA1 suppresses RAD52-dependent double-strand break repair in Saccharomyces cerevisiae. Genetics 151, 447–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Mortensen U. H., Erdeniz N., Feng Q., and Rothstein R. (2002) A molecular genetic dissection of the evolutionarily conserved N terminus of yeast Rad52. Genetics 161, 549–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Smith J., and Rothstein R. (1995) A mutation in the gene encoding the Saccharomyces cerevisiae single-stranded DNA-binding protein Rfa1 stimulates a RAD52-independent pathway for direct-repeat recombination. Mol. Cell. Biol. 15, 1632–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kaeberlein M., McVey M., and Guarente L. (1999) The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13, 2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Eckert-Boulet N., and Lisby M. (2009) Regulation of rDNA stability by sumoylation. DNA Repair 8, 507–516 [DOI] [PubMed] [Google Scholar]
- 74. Paliwal S., Kanagaraj R., Sturzenegger A., Burdova K., and Janscak P. (2014) Human RECQ5 helicase promotes repair of DNA double-strand breaks by synthesis-dependent strand annealing. Nucleic Acids Res. 42, 2380–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lao J. P., Oh S. D., Shinohara M., Shinohara A., and Hunter N. (2008) Rad52 promotes postinvasion steps of meiotic double-strand-break repair. Mol. Cell 29, 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sugiyama T., Kantake N., Wu Y., and Kowalczykowski S. C. (2006) Rad52-mediated DNA annealing after Rad51-mediated DNA strand exchange promotes second ssDNA capture. EMBO J. 25, 5539–5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Chavdarova M., Marini V., Sisakova A., Sedlackova H., Vigasova D., Brill S. J., Lisby M., and Krejci L. (2015) Srs2 promotes Mus81-Mms4-mediated resolution of recombination intermediates. Nucleic Acids Res. 43, 3626–3642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Moldovan G.-L., Dejsuphong D., Petalcorin M. I., Hofmann K., Takeda S., Boulton S. J., and D'Andrea A. D. (2012) Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell 45, 75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Burkovics P, Dome L, Juhasz S, Altmannova V, Sebesta M, Pacesa M, Fugger K, Sorensen CS, Lee MY, Haracska L, Krejci L (2016) The PCNA-associated protein PARI negatively regulates homologous recombination via the inhibition of DNA repair synthesis. Nucleic Acids Res. 10.1093/nar/gkw024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Merker R. J., and Klein H. L. (2002) hpr1Delta affects ribosomal DNA recombination and cell life span in Saccharomyces cerevisiae. Mol. Cell. Biol. 22, 421–429 [DOI] [PMC free article] [PubMed] [Google Scholar]