Abstract
Lytic polysaccharide monooxygenases (LPMOs) are copper-containing enzymes which oxidatively break down recalcitrant polysaccharides such as cellulose and chitin. Since their discovery LPMOs have become integral factors in the industrial utilization of biomass, especially in the sustainable generation of cellulosic bioethanol. We report here the first structural determination of an LPMO–oligosaccharide complex, yielding detailed insights into the mechanism of action of these enzymes. Using a combination of structure and electron paramagnetic resonance spectroscopy, we reveal the means by which LPMOs interact with saccharide substrates. We further uncover electronic and structural features of the enzyme active site, showing how LPMOs orchestrate the reaction of oxygen with polysaccharide chains.
Introduction
The sustainable production of energy and materials from biomass is of global importance, particularly in the need for carbon-neutral sources as contributors to balanced energy portfolios.1,2 The capacity of biofuels to make a significant contribution, however, depends upon finding a sustainable means of overcoming the recalcitrance of some of the most abundant and widespread polysaccharides, most notably cellulose. Indeed, a viable solution to this problem has been elusive, with recent decades having seen a worldwide research effort directed at providing a means of sustainably deconstructing cellulose or cellulose-related materials. Progress has been slow, but recent advances in the use of cocktails of glycosidic hydrolases identified from the secretomes of saprophytic organisms are promising.3-5 For instance, several newly-opened cellulosic-bioethanol biorefineries which employ enzyme cocktails appear, for the first time, to be close to both commercial and environmental viability. The success of these biorefineries hinges on the efficiency of the enzymatic saccharification step in which lignocellulosic material is decomposed into soluble sugars. The efficacy of this step further rests on advances in the bespoke content of the cocktails, of which perhaps the most conspicuous is the recent inclusion of lytic polysaccharide monooxygenases (LPMOs, also known as PMOs).6
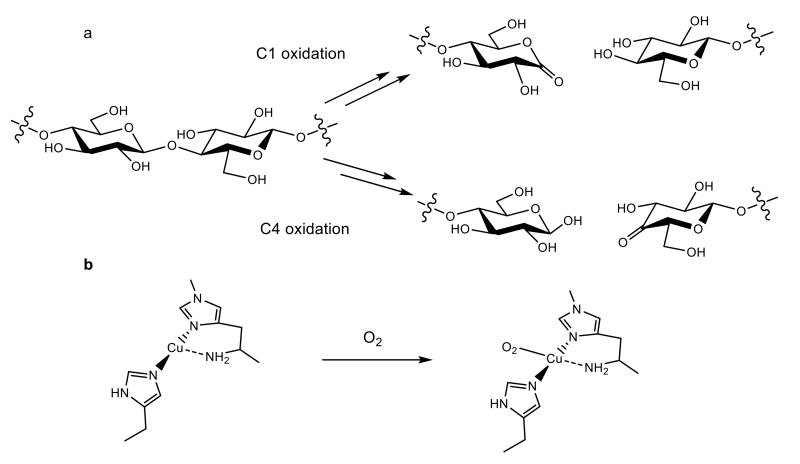
LPMOs are metalloenzymes classified in the Carbohydrate-Active Enzymes database (CAZy) as Auxiliary Activity families AA9–AA11 and AA13.7-10 The ability of LPMOs to boost biomass degradation was reported in the late 2000s11,12 followed in 2010 by the seminal discovery of their oxidative mode of action,13 and then in 2011 by further activity studies14 and identification of copper at the active site.15,16 The unusual activity and active site of LPMOs mean that they are challenging precepts not only in the mechanisms by which biological degradation of biomass occurs, but also in copper bioinorganic chemistry.17,18 LPMOs contribute to the scission of a polysaccharide chain by harnessing the oxidative capacity of molecular oxygen to cleave a glycosidic C–H bond, the strength of which is estimated to be at least 95 kcal mol−1 (Fig. 1a).19-22 To break these bonds LPMOs activate oxygen, in a reducing-agent dependent manner, at a copper-containing active site known as the histidine brace (Fig. 1b).15,17,23,24 In doing so LPMOs boost the actions of canonical glycoside hydrolases by up to two orders of magnitude,12 thereby greatly bringing down the financial and environmental penalties normally associated with using recalcitrant polysaccharides as a feedstock, and moreover, placing LPMOs at the center of considerable research interest.4 This research interest is focused on improving the efficiencies of LPMOs, with particular attention on determining the detailed mechanism of action of the enzymes, an endeavor hindered by the lack of detailed structural information of an LPMO in direct contact with a polysaccharide. It is in this context that we report here the crystal structure of an LPMO–oligosaccharide complex. This much-anticipated structure, in combination with its spectroscopy, reveals the essential molecular features of LPMO interaction with substrates, paving the way forward for improvements in catalytic efficiencies and a greater understanding of the mechanism of action of these enzymes.
Figure 1. LMPO activity and active site.
a, Oxidative cleavage of a polysaccharide chain carried out by LPMOs, showing different routes of oxidation carried out by different sub-groups of LPMOs. b, The histidine brace active site and proposed first step in the reaction with oxygen.19
Results
Isolation and activity
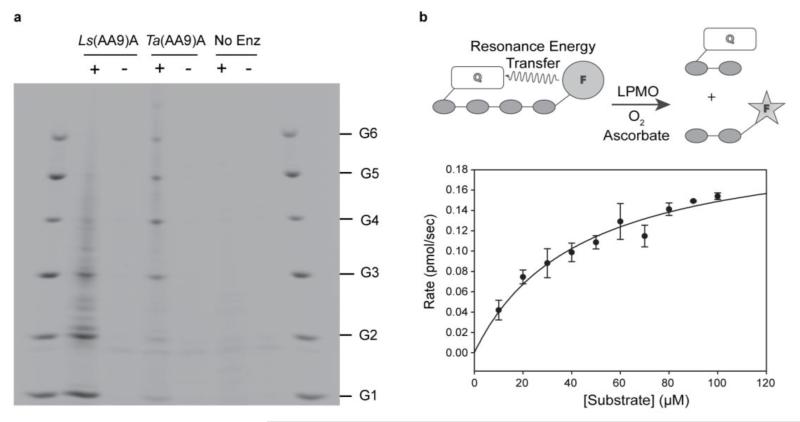
An LPMO (CAZy classification AA9, hereafter Ls(AA9)A) naturally devoid of a carbohydrate-binding-module was selected on the basis of activity-screening experiments of LMPOs from Lentinus similis. Ls(AA9)A was successfully expressed in Aspergillus oryzae, and using PACE gel product analysis, was found to be able to cleave phosphoric acid swollen cellulose (PASC) in the presence of a reducing agent, yielding a range of cello-oligosaccharides dominated by cellobiose and cellotriose (Fig. 2a). This product profile is notably different from that of most other AA9s which yield longer oligosaccharides; for example, Figure 2a shows the action of an AA9 from Thermoascus aurantiacus (Ta(AA9)A).15 Additionally, unlike Ta(AA9)A, Ls(AA9)A also showed cleavage activity on soluble oligosaccharides (cellotriose to cellohexaose, G3–G6, Supplementary Results, Supplementary Fig. 1), a feature reported previously for a handful of other AA9s, and a potentially important factor in the effectiveness of these particular LPMOs to degrade polysaccharides.25-27 The products of Ls(AA9)A action on PASC were analyzed by MALDI-TOF-MS and HPLC, showing that C4 was the sole site of oxidation (Supplementary Fig. 2).24,26,27
Figure 2. Ls(AA9)A and Ta(AA9)A activities on insoluble and soluble cellulose.
a, Comparison of Ls(AA9)A and Ta(AA9)A activities on PASC. Reaction products were analyzed by PACE (performed in triplicate). +, incubation with ascorbate; −, incubation without ascorbate. b, Cleavage of FRET substrate by Ls(AA9)A and subsequent Michaelis-Menten plot, error bars represent random error of triplicate measurements.
Taking advantage of the reactivity of Ls(AA9)A on soluble oligosaccharides, we monitored the kinetics of oxidative cleavage of cellotetraose labelled with fluorescent groups (Fig. 2b, Supplementary Note 1, Supplementary Fig. 3). This Förster-resonance-energy-transfer (FRET) substrate can be used to monitor the action of LPMOs as it fluoresces strongly upon cleavage in dilute solution. Before analysis Ls(AA9)A samples were freshly prepared with careful removal of any excess copper and only initial rates of reaction were used to avoid any complications due to enzyme inactivation.25 The resultant kinetics of cleavage of the cellotetraose by Ls(AA9)A showed no lag period and could be interpreted using classic Michaelis-Menten parameters, affording values of Km = 43±9 μM and kcat = 0.11±0.01 s−1 at pH 7.0 and 37 °C, comparable to previously reported turnover numbers.13,26,28
Structures of Ls(AA9)A and Ls(AA9)A–oligosaccharide
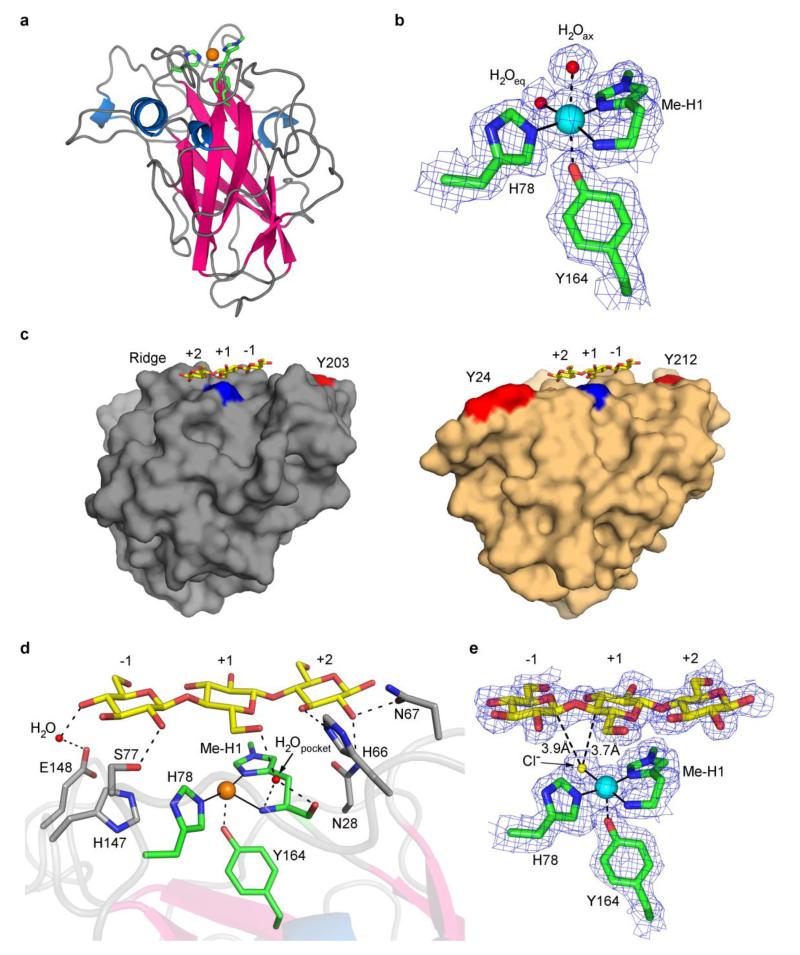
The crystallographic structure of Ls(AA9)A was determined by molecular replacement using PDB deposition 4EIR as a model.24 The resulting structure displayed a known AA9 topology with an immunoglobulin-type fold (Fig. 3a). Initial structures yielded copper coordination geometries consistent with photoreduction of the copper site to Cu(I) by incident X-rays, as seen in previously reported structures of other LPMOs (Fig. 3a, Supplementary Fig. 4a) and confirmed by XAS studies on AA10 LPMOs.23 Accordingly, to mitigate the effects of photoreduction other datasets were also collected at reduced X-ray doses (Fig. 3b, Supplementary Note 2).23,29 For example, in the low-dose X-ray structure of Ls(AA9)A the copper ion had a coordination sphere expected for a Cu(II) oxidation state with three nitrogen atoms of the histidine brace and a water molecule in an approximate square planar geometry (Cu–N(His1) 1.9 Å, Cu–N(His78) 2.1 Å, Cu–NH2 2.2 Å, Cu–OH2 2.2 Å, remote distances Cu…OH2 (axial) 2.8 Å and Cu…OTyr164 2.7 Å) (Fig. 3b).
Figure 3. Structural views of Ls(AA9)A.
with the copper ion depicted as an orange sphere, Cu(I); or a blue sphere, Cu(II)
a, Ribbon view of Ls(AA9)A. b, Electron density of active site obtained with low X-ray dose. c, Comparison of Ls(AA9)A (left) and Ta(AA9)A (right) surfaces. Ls(AA9)A-G3 (lowdose) with Tyr203 in red. Ta(AA9)A (PDB code 3ZUD) with Tyr24 and Tyr212 in red along with superposed structure of the G3 from Ls(AA9)A-G3. The N-terminal methyl-His is shown in blue. d, Principal protein contacts between G3 and the binding surface of Ls(AA9)A in Ls(AA9)A-G3 structure with high dose X-rays. e, Electron density of Ls(AA9)A-G3 with low X-ray dose.
Guided by the activity studies described above, soaking crystals of Ls(AA9)A with substrates cellotriose (G3) or cellohexaose (G6) at pH 5.5 afforded new structures each of which showed unambiguous density associated with the oligosaccharide in contact with the putative binding surface of Ls(AA9)A (Fig. 3c,e, Supplementary Fig. 4b, Supplementary Note 2), confirming long-held proposals that this surface is the site of interaction with substrates.13,15 Using a sugar subsite nomenclature, analogous to that used for glycoside hydrolases, G6 bound to Ls(AA9)A from subsites −4 to +2 (Supplementary Fig. 4b,c), and G3 from −1 to +2 (Fig. 3c-e). The glucosyl units of G6 in subsites −3 and −4 in Ls(AA9)A–G6 were less well structurally defined than other subunits (see Supplementary Note 2), but subunit −3 was sufficiently well resolved to see that it formed close contacts with the aromatic ring of the surface-exposed and highly conserved Tyr203 (C…C distances of 3.7 Å, Supplementary Fig. 4c, Supplementary Fig. 5), presumably via CH…π interactions, again confirming long-held proposals that solvent-exposed aromatic groups are effective at binding poly/oligosaccharides in LPMOs (Supplementary Fig. 4b,c).12,24,30,31 Tyr203 was placed approximately 15 Å away from the copper active site, a general feature which is observed in other AA9 LPMOs, see Figure 3c for example.
At the active site the terminal glucosyl unit at subsite +2 was held within a network of hydrogen bonds with protein residues Asn28, His66, Asn67 anchoring this unit into a fixed position (Fig. 3d, Supplementary Table 2). As illustrated for Ls(AA9)A–G3 in Figures 3d,e and Figure 4, the glucosyl unit at subsite +1 stacked directly on top of His1 via a striking lone-pair…aromatic interaction (ring O5… His1 centroid, 3.4 Å). The glycosidic bond between the units at subsites +1 and −1 arched directly over the putative O2 binding site at the copper ion, bringing the C4–H bond in subsite +1 and the C1–H bond in subsite +1 into close proximity with the exogenous ligand on the copper ion (Fig. 3e). Through a combination of hydrogen bonds (Supplementary Table 2), the lone-pair–aromatic interaction with His1, and (for longer oligosaccharides) the CH…π interaction with Tyr203, the orientation and position of the oligosaccharide was held in a fixed aspect relative to the copper active site. In this regard, it is notable that the array of interactions between G3 substrate and enzyme did not drive any major distortion of torsion or chair conformation angles in the oligosaccharide away from their canonical geometries (Supplementary Table 3).32 This lack of distortion reinforces the prevailing idea that LPMO catalysis is largely mediated by the exceptional chemical environment of the copper histidine brace active site,17,33 rather than aided by the enzyme-mediated distortion of the substrate.
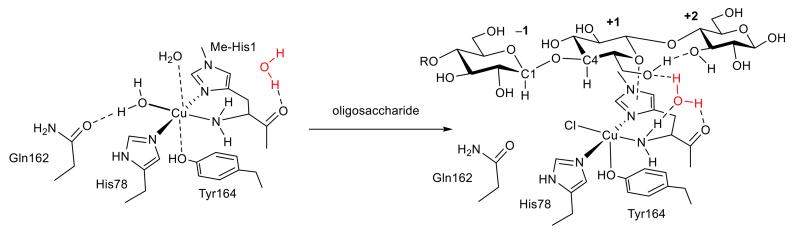
Figure 4. Structure of Ls(AA9)A around active site before and after binding of G3.
‘Pocket’ water molecule is shown in red.
In a comparison of the structures of Ls(AA9)A before and after substrate binding there were clear differences (>0.1 δÅ) in the copper coordination spheres. In the presence of the oligosaccharide the exogenous ligand on the copper was best modeled by a fully occupied chloride ion (Fig. 3e, Cu–Cl, 2.3 Å, Supplementary Note 2), rather than a water molecule at 2.2 Å as in the non-substrate structure (Fig. 3b, chloride was present in both soaking conditions at ca. ~3.5 M, Supplementary Table 4). From space-filling structures (Supplementary Fig. 6) it could be seen that the chloride ion was held within a binding cavity created between the binding surface of Ls(AA9)A and the oligosaccharide substrate. In addition to the change in the equatorial ligand there was a shortening of the Cu…OTyr164 contact from 2.7 to 2.5 Å upon substrate binding. Beyond the immediate copper coordination sphere, the C6 hydroxymethyl group of the unit at subsite +1 was placed close to the copper ion (C…Cu, 3.8 Å) such that the group occluded the axial binding site, displacing the water molecule that was seen in the non-substrate structure (cf. Fig. 3b and Fig. 3e). In the non-substrate structure the main chain carbonyl of His1 formed a hydrogen bond to a water molecule (O…O, 2.8 Å) which was held within a small pocket circumscribed by the side chain of His66 and the main chain peptides of His1 and Ala75 (Fig. 4). Upon substrate binding, the C6 –CH2OH group of the glucosyl unit at subsite +1 formed hydrogen bonds with the C3–OH group of adjacent unit at subsite +2 and with the ‘pocket’ water molecule (O…O, 2.8 Å). The hydrogen bonding pattern around the ‘pocket’ water further included the NH2 terminus (N…O, 2.9 Å), thereby connecting the substrate to the amino terminus and the copper (Fig. 4).
Electron paramagnetic resonance spectroscopy of Ls(AA9)A
To probe the solution phase interaction between Ls(AA9)A and oligosaccharide substrates, continuous wave (cw) X-band EPR spectra of the met-forms of Ls(AA9)A and Ls(AA9)A–G6 were obtained under both high and low chloride conditions (Fig. 5).
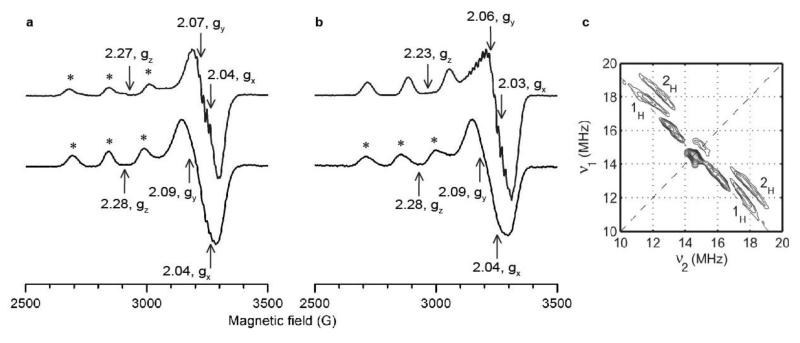
Figure 5. EPR spectra of Ls(AA9)A at 150 K.
* indicates signals from copper site where water was the exogenous ligand, other signals were from species with chloride as exogenous ligand. a, X-band cw EPR spectra in low chloride conditions, Ls(AA9)A-G6 (top) and Ls(AA9)A with no oligosaccharide (bottom). b, X-band cw EPR spectra in high chloride conditions, Ls(AA9)A-G6 (top) and Ls(AA9)A with no oligosaccharide (bottom). c, Contour presentation of the 1H-HYSCORE spectrum of Ls(AA9)A-G6 in the presence of chloride ions (magnetic field 343.6 mT, time between first and second pulse τ = 136 ns, microwave frequency 9.7 GHz). The anti-diagonal cross-peaks are assigned to anisotropic protons, labelled 1H and 2H.
The spectrum of Ls(AA9)A in the absence of substrate is shown in Figure 5a. As with previous EPR spectra of AA9 enzymes, the spin Hamiltonian parameters were diagnostic of a type 2 copper site albeit with some rhombicity evident in the xy direction, gz = 2.28, gy = 2.09, gx = 2.04, |Az| = 458 MHz. The spectrum was sensitive to the presence of chloride in solution and under saturating conditions (~200 mM chloride) the presence of a second, somewhat rhombic minor species (~20%) could be determined gz = 2.25, gy = 2.09, gx= 2.01, |Az| = 455 MHz, (Fig. 5b), although there was some uncertainty in determining the perpendicular g values.
Addition of G6 to Ls(AA9)A under low chloride conditions resulted in a substrate-induced perturbation of the copper active site (Fig. 5a), shown by a small increase in |Az| to 515 MHz, a decrease in gy and gz (gz = 2.27, gy = 2.07, gx = 2.04), and the appearance of well-resolved superhyperfine (SHF) coupling due to an increase in the number of magnetically coupled nitrogen nuclei from two to three (34, 37 MHz in the absence of G6 and 19, 30, 36 MHz with G6, Supplementary Fig. 7a,b,d and Supplementary Table 6). These observations suggested that the addition of substrate drove a larger contribution of the amino terminus to the singly-occupied-molecular-orbital (SOMO) on the Cu(II) ion.34 Under high chloride conditions, additional SHF features appeared due to the presence of a chloride nucleus (Fig. 5b) and there was a significant decrease in gz to 2.23, showing that when chloride was available addition of substrate was accompanied by the coordination of chloride to the Cu(II) site, consistent with crystal structures (Fig. 3e). The synergy of chloride and substrate binding was confirmed by isothermal calorimetry experiments in which the dissociation constant of G6 binding to Ls(AA9)A in the presence of 200 mM chloride was found to be 3.7±0.1 μM, significantly lower than the dissociation constant in solution at the same ionic strength (67 mM Na2SO4) but in the absence of chloride, Kd ~ 1 mM (Supplementary Fig. 8).
In the absence of substrate, both parallel and perpendicular 1H-HYSCORE spectra of Ls(AA9)A exhibited only a central peak due to matrix protons and no cross-peak features arising from magnetically coupled protons (Supplementary Fig. 9a,b), revealing that neither H2O nor the amino terminus were closely coordinated to the Cu(II). After G6 binding however, cross-peaks due to magnetically coupled protons were present at both parallel and perpendicular positions (Supplementary Fig. 9c,d). In particular, at the g⊥ position very well-defined couplings appeared in which two pairs of ridges shifted from the anti-diagonal (1H and 2H in Fig. 5c), arising from two distinct types of protons each with strong dipolar couplings (T = 4.7 and 6.7 MHz, aH ~ 0.5 MHz). Since the X-band cw EPR spectrum demonstrated that chloride occupied the open equatorial coordination position of the Cu(II) site after substrate binding (Fig. 5b), these ridges were unambiguously assignable to the individual protons of the NH2 group, which were then revealed to have distinctly different chemical environments.35 This chemical asymmetry likely arises from the formation of a hydrogen bond between one of the protons and the pocket water molecule upon substrate addition (Fig. 4).
To compare the interactions between the copper site of Ls(AA9)A and oligosaccharide substrates to those with insoluble polysaccharide substrates, the X-band cw EPR spectrum of Ls(AA9)A was collected in the presence of excess PASC (Supplementary Fig. 7c). The spectrum of Ls(AA9)A–PASC was virtually identical to that of Ls(AA9)A with G6, showing that the binding interactions of Ls(AA9)A to polysaccharide substrates like PASC are the same as those observed for oligosaccharide substrates.
Discussion
Exploiting the fact that Ls(AA9)A is able to act on soluble cello-oligosaccharides, we were able to characterize the detailed interaction of an LPMO with a substrate. A comparison of the Ls(AA9)A structures with those of other AA9 LMPOs not able to act on oligosaccharides, exemplified by Ta(AA9)A (Fig. 3c), shows that the latter lack many of the residues that mediate hydrogen bond interactions with the oligosaccharide substrate(s) (Supplementary Fig. 5). Indeed, the topography of the polysaccharide binding surface of Ls(AA9)A is distinctive when compared to the structures of most other AA9s insofar as the active site is found to be located within a contoured binding surface (Fig. 3c). Among the publicly available structures for AA9 LPMOs the structurally most similar to Ls(AA9)A are Nc(AA9)D (PDB code 4EIR), Nc(AA9)C (4D7U) and Nc(AA9)M (4EIS), all with Cα rmsds of 1.4 Å for 202, 199 and 193 aligned residues respectively. The most distant is the structure of Tt(AA9)E (3EJA), with an rmsd of 2.1 Å for 150 aligned residues. As illustrated by the alignment in Supplementary Figure 5 comparison of the active site of Ls(AA9)A-G3 structure to Ta(AA9)A (2YET) shows that most residues able to make hydrogen bonds with oligosaccharides (primarily Asn28, His66 and Asn67, Fig. 3c) are not present in Ta(AA9)A, correlating with the different activities of these two LPMOs on oligosaccharides. Consistent with this notion, Nc(AA9)C, which acts on cellooligosaccharides, has residues which potentially make hydrogen bonds with the substrate, some of which are conserved between Ls(AA9)A and Nc(AA9) (Supplementary Fig. 5).
As part of its contoured surface Ls(AA9)A has a ridge near to the +2/+1 substrate binding subsites (Fig. 3c), a feature which is shared by Nc(AA9)C and Nc(AA9)D, the two other AA9 LPMOs reported to be strict C4 oxidizers.26 This ridge likely prevents binding of Ls(AA9)A to a flat cellulose surface, but could allow binding to more contoured substrates such as oligosaccharides and, for example, the corners of crystalline cellulose fibrils. Other LPMOs such as Ta(AA9)A (Fig. 3c) and Tt(AA9)E lack this ridge, exhibiting much flatter surfaces. From this comparison of structures the general picture which emerges is that different LPMOs have evolved to extend the surface binding area around the active site to different extents towards the + or − subsites. Also NMR studies of the interaction of an AA10 LPMO (CBP21) with chitin show that the proposed binding region of the substrate to the enzyme is similar to that seen in the structure of Ls(AA9)A with oligosaccharides.36
The orientation of substrate binding to Ls(AA9)A brings the C1–H bond of subsite −1 and the C4–H bond of subsite +1 into close proximity to the equatorially-bound ligand at the copper (Fig. 3e, Fig. 4). The exclusive oxidation of the C4–H bond by Ls(AA9)A is commensurate with the closer distance of the exogenous ligand to this bond compared to the C1–H bond (3.7 Å vs 3.9 Å, respectively), consistent with previous suggestions of structural determinants for C1/C4 oxidation,26 although we note that small shifts in substrate positioning could easily alter the balance between C1 and C4 oxidation. The proximity of the C4–H bond to the exogenous ligand atom directly bound to the copper is also consistent with the possibility of a Cu(III)-hydroxide or Cu(II)-oxyl being the reactive intermediate during LPMO catalysis (both of which can be formed by further reduction of a Cu(II)-superoxide species).20,37-39 The potential existence of such copper intermediates in Ls(AA9)A is supported by the formation of a hydrogen-bonding network between the ‘pocket’ water molecule and the NH2 amino terminus upon substrate binding (Fig. 4). Not only does the hydrogen bond network enhance the basicity of the amino terminus it also provides a potential proton transfer channel for deprotonation of the amino terminus, which has been proposed from small model complex studies as a means of stabilizing a highly reactive Cu(III)-hydroxide intermediate.33,40
Further examination of the Ls(AA9)A active site structures shows that oligosaccharide binding leads to the occlusion of the axial binding site on the copper ion by the C6 CH2OH group of subsite +1, displacing an axial water molecule. This displacement moves the copper ion towards Tyr164 (Fig. 3d), thereby linking the copper to the potentially redox-active tyrosine and/or an electron transfer pathway through the enzyme. In addition, the occlusion of the axial binding site prevents the potential loss of a coordinated superoxide anion through the axial site. Such a mechanism of potential deactivation of LPMOs via associative displacement of superoxide was found for Ta(AA9)A in the absence of substrate.19
Our studies also reveal an interaction between the ring oxygen of the glucosyl unit occupying subsite +1 and the face of the imidazole side chain of His1.41 Characterized contacts like this are rare in biological chemistry with only a handful of examples, including a ribose sugar ring-oxygen lone pair interacting with the aromatic face of a guanine group.42 Despite its scarcity in biology however, this type of interaction is prevalent in supramolecular chemistry where it is often labeled as a lone-pair to π* overlap, but is now believed to be principally electrostatic in character.43 The electrostatic nature of the interaction is likely to be enhanced by the presence of alkyl groups on aromatic ring nitrogen atoms, thereby providing some rationale for the previously puzzling δ-N methylation of His1 observed in fungal LPMOs.
Our studies further point to an interesting interdependence of binding of substrate and chloride on the copper ion. As chloride and superoxide have similar size and polarizability, the greatly enhanced affinity for chloride after oligosaccharide binding raises the possibility that binding of dioxygen and the polysaccharide substrate to the LPMOs is similarly synergistic, thereby affording LPMOs a mechanism by which the production of reactive oxygen species such as superoxide is controlled by the presence of the substrate.44 Insight into the origin of this synergistic binding can be found from the crystal structures, where the binding of substrate results in the formation of a cavity at the active site that is of the correct shape to accommodate a chloride or superoxide ion (Supplementary Fig. 6). The change in binding affinity of the exogenous ligand may also arise from changes in the coordinating power of the amino terminus, as described above, which raise the energy of d(x2−y2) orbital and stabilize the Cu(II) oxidation state (Fig. 4).
Finally, the observation that the EPR spectra of Ls(AA9)A binding to PASC and to the oligosaccharide G6 are close to identical indicates that oligosaccharide substrates faithfully mimic the enzyme’s binding to extended polysaccharides. This observation connects our detailed studies of Ls(AA9)A–oligosaccharide complexes with the in vivo activity of LPMOs on extended polysaccharides. Our studies therefore give detailed insight into the means by which LPMOs react with insoluble polysaccharide substrates and help pave the way forward for improving the efficacies of these industrially-important enzymes.
Online Methods
Cloning, expression and purification
The gene encoding Ls(AA9)A was PCR-amplified from genomic DNA of Lentinus similis and cloned in E. coli as described in 45 using the following forward primer F-P247JK: 5′-ACACAACTGGGGATCCACCATGAAGTACTCCATCCTCGGGCT-3′ and reverse primer R-P247JK: 5′-CCCTCTAGATCTCGAGCCTTGTCGAGCGACTCTATCCA-3′, containing insertion sites for the vector pDau109 used for cloning. The sequence was deposited as KT368674 (GenBank). The fragments were then cloned into Bam Hl and Xho I digested pDau109 using an IN-FUSION™ Cloning Kit. Cloning of the genes into Bam Hl-Xho I digested pDau109 resulted in transcription of the Ls(AA9)A gene under the control of a NA2-tpi double promoter. The treated plasmids and inserts were transformed into One Shot TOP10F Chemically Competent E. coli cells (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol, spread onto LB plates supplemented with 0.1 mg of ampicillin per mL and incubated at 37 °C overnight. Colonies of each transformation were cultivated in LB medium supplemented with 0.1 mg of ampicillin per mL and plasmids were isolated using a QIAPREP Spin Miniprep Kit (QIAGEN Inc., Valencia, CA, USA).
Ls(AA9)A was expressed in Aspergillus oryzae MT3568 as also described in 45. A transformant producing the recombinant Ls(AA9)A was inoculated in 2 L of Dap-4C medium and incubated at 30 °C for 4 days. Mycelia were removed by filtration and the broth collected for purification. Ammonium sulfate was added to the sterile filtered broth to a conductivity of 200 mSi/cm and the pH adjusted to 7.5. The broth was applied to a 50/15 Butyl Toyopearl column (Tosoh Biosciences, Stuttgart, Germany) equilibrated with 25 mM Tris, 1.5 M ammonium sulfate, pH 7.5. The column was washed in the same buffer and eluted with a gradient to 25 mM Tris pH 7.5. Fractions containing Ls(AA9)A were combined and washed with milliQ water by ultrafiltration (10 KDa MWCO, PES filter, Sartorius, Goettingen, Germany) to a conductivity of 1.2 mSi/cm. The pH was adjusted to 8.0 and applied to a 50/40 Q Sepharose FF column (GE Healthcare, Pittsburgh, USA) equilibrated with 20 mM Tris pH 8.0. The column was washed in the same buffer and the enzyme eluted with a gradient from 0 to 0.5 M sodium chloride. Fractions containing Ls(AA9)A were combined and concentrated by ultrafiltration using VIVASPIN 20 (10 KDa MWCO) spin concentrators.
Activity measurements by polysaccharide analysis by carbohydrate gel electrophoresis (PACE) and matrix assisted laser-desorption ionisation (MALDI) mass spectrometry
apo-Ls(AA9)A, at 125 μM, was pre-incubated for 1 h at 5 °C in 0.9 stoichiometric Cu(II)NO3 before enzyme reactions. Enzyme reactions on cello-oligosaccharides were in 10 μL containing 5 nmol cello-oligosaccharide, 100 mM ammonium formate pH 6, ± 4 mM ascorbate, ± 5 pmol Ls(AA9)A/Ta(AA9)A, and were incubated at 20 °C for 4 h. Reactions on phosphoric acid-swollen cellulose (PASC) were in 100 μL containing 0.5% (w/v) PASC, 100 mM ammonium formate pH 6, ± 4 mM ascorbate, ± 63 pmol Ls(AA9)A/Ta(AA9)A, and were incubated at 20 °C for 16 h. PASC was prepared beforehand by making a slurry of 1 g of Avicel cellulose (Sigma-Aldrich) with 3 mL H2O, before adding 30 mL ice-cold phosphoric acid and incubating at 0 °C for 1 h. The cellulose was then washed numerous times with water until the flow-through had a neutral pH before use in reactions.
For PACE, reaction products and oligosaccharide standards (Megazyme) were reductively aminated with 8-aminonaphthalene-1,3,6-trisulfonic acid (ANTS; Invitrogen, www.lifetechnologies.com) and separated by acrylamide gel electrophoresis. PACE was performed using a 192 mM glycine, 25 mM tris, pH 8.5 running buffer. The gels contained: resolving gels, 37.5 mL 40% (w/v) acrylamide, 12.5 mL 375 mM Tris-HCl buffer pH 8.8, 100 μL 10% ammonium persulphate, 50 μL tetramethylethylenediamine (TEMED); stacking gels, 2 mL 40% (w/v) acrylamide, 2.5 mL 375 mM Tris-HCl buffer pH 8.8, 5.4 mL water, 100 μL 10% ammonium persulphate, 10 μL TEMED. Electrophoresis was carried out at 100 V for 30 min, 500 V for 30 min and 1000 V for 180–210 min, and were then visualised with a G-box (Syngene) equipped with a short pass detection filter (500–600 nm) and long-wave UV tubes (365 nm emission). Representative gels from replicate experiments are shown.
MALDI mass spectrometry was performed as described previously.7 Trifluoroacetic acid (TFA) hydrolysis was performed by incubating reaction products in 2 M TFA for 1 h at 120 °C before drying in vacuo. High-performance liquid chromatography (HPLC) was performed on a CarboPac PA1 column (high-performance anion-exchange chromatography, HPAEC; Dionex, Camberley, UK) with elution at 0.4 mL min−1 and injections of 20 μL. The elution profile was: 0–3 min, 10 mM NaOH (isocratic); 3–6, 10→1 mM NaOH (linear gradient); 6–19 min, 1 mM NaOH (isocratic); 19–37 min, 45 mM NaOH, 225 mM sodium acetate (isocratic). A pulsed amperometric detector (PAD) with a gold electrode was used. PAD response was calibrated using markers (500 pmol).
Substrate preparation and kinetics measurements
F*-G4-F (Supplementary Note 1) was dissolved in DMSO to a final stock concentration of 10 mM. This was then diluted to 1.33× the working concentration with 20 mM Bis-Tris pH 7 and 133 nM Ls(AA9)A ensuring the DMSO concentration was the same in all samples. 15 μL aliquots were then spread into a 96 well plate and fluorescence was monitored in an Agilent MX3000P QPCR machine using an excitation wavelength of 330 nm and emission wavelength of 492 nm. The increase in fluorescence in triplicate samples was monitored following the addition of 5 μL of ascorbate (prepared in 20 mM Bis-Tris pH 7) giving final concentrations of 100 nM protein and 5 mM ascorbate. Fluorescence was quantified by plotting a standard curve of fluorescence against an equimolar mix of fluorophore (EDANS – Santa Cruz Biosciences) and quencher (DABCYL - Sigma-Aldrich) from 2 to 10 μM concentrations. Inner filter effects were corrected by measuring standard curves in the presence and absence of substrate at each concentration used and calculating correction factors appropriately.46 Data were analysed using SigmaPlot.
Crystallization, Data Collection and Structure determination
All crystals were obtained by vapor diffusion technique set up in sitting drop MRC-plates, with a reservoir volume of 100 μL and at room temperature using an Oryx-8 robot (Douglas Instrument). Crystallization and post-crystallization experimental details are shown in Supplementary Table 4. All data were collected at cryogenic temperatures (100 K) after cryocooling the crystals in liquid nitrogen. Cellotriose (G3) for soaking was purchased from Megazyme, while the cellohexaose (G6) preparation (which contained about 20% cellopentaose as contaminant) was provided by Novozymes/AS.
Inter-grown plates of glycosylated Ls(AA9)A (6.12 mg/mL in 20 mM MES pH 6) were initially obtained in a Morpheus screen. Single plates could be separated and mounted, and a data set extending to a maximum resolution of 2.8 Å was collected at beamline I911-3, Maxlab,Lund, Sweden. The dataset was processed in space group C2221 (this and subsequent data processing carried out with XDS47). A partial structure could be determined by Molecular Replacement with MOLREP 48 using modified coordinates of PDB 4EIR as search model. Despite poor statistics (starting R-factors and R-free around 50%), solutions with 2 mols/asu could be obtained that reproduced well the features seen in a self-rotation function. By alternated manual building in density modified maps in COOT49 and rigid body/restrained refinement in REFMAC550 a partial model (with rather poor geometry and not containing the active site copper) was obtained with an R-factor of 39.2% and R-free of 37.7%, and was used as starting model for further molecular replacement.
For subsequent crystallization trials Ls(AA9)A was deglycosylated in 20 mM MES pH 6.0, 125 mM NaCl by incubation with 0.05–0.06 units/mg Ls(AA9)A of Endoglycosidase H from Roch Diagnostics GmbH (REF: 11 643 053 001), and then buffer exchanged to 20 mM sodium acetate buffer pH5.5. Crystallization trials also included pre-incubation with Cu(II)acetate (see details in Supplementary Table 4).
A relatively large single crystal (maximum dimension 180 μm) was first obtained in an IndexHT screen (Molecular Dimensions) after two weeks, with precipitant containing 0.1 M Citric Acid pH 3.5 and 3.0 M NaCl. The crystals were later found to be reproducible and grew after few days in conditions ranging from 3.0–4.7 M NaCl and pH 3.0–4.0. Crystals could be transferred to buffers at higher pH, to provide conditions closer to the pH of activity and spectroscopy studies, and maintaining high resolution diffraction, in general better than 2.0 Å. The crystals were the basis for determination of a series of structures, a selection of which is presented in this manuscript. For each structure determination a single crystal was used, XDS was used for processing and the resolution cutoff was chosen based on a CC½ above 50%.
The preliminary model obtained for the glycosylated protein was used as a search model for the newly obtained crystal form in space group P4132, for which the first structure could be refined using COOT/REFMAC5. All subsequently determined structures were isomorphous to this. The highest resolution structure obtained (data set denoted Ls(AA9)A_highres) was at 1.28 Å resolution, for which data were collected at λ = 0.965 Å and processed automatically at ID30A-1 of the ESRF using the MASSIF-1 protocol (statistics in Supplementary Table 5). This structure was refined anisotropically for all atoms and had 94.44% of residues in the Ramachandran most favorable regions. Preliminary experiments at ID30A-1 of the ESRF also showed that complexes with cellooligosaccharides could be obtained by soaking. By soaking crystals in pH 5.5 solutions containing cellooligosaccharides G3 (the minimal substrate for Ls(AA9)A) and G6, two complexes were obtained (Ls(AA9)A-G3 and Ls(AA9)A-G6, details in Supplementary Table 4 and Supplementary Table 5) at Maxlab in Lund, Sweden, at beamline I911-2 and I911-3, respectively (with λ = 1.03841 and 1.06883 Å respectively). The final structures had 93.81% and 93.36% of residues in the most favorable regions of the Ramachandran plot, respectively, and the bound ligands were evident in the electron density (Fig. 3e and Supplementary Fig. 4b).
In all the structures described so far, the density suggests that Cu is in a mixture of oxidation states, with significant disorder of the non-protein ligands. In order to obtain optimal structures with Cu(II) for interpretation of the EPR data, data was collected (at λ = 1.0000 Å at I911-3) with short exposure times, beam attenuation (3% transmission) and the minimal number of X-ray images to ensure an acceptable completeness (Ls(AA9)A_lowdose and Ls(AA9)A-G3_lowdose, see Supplementary Tables 4 and 5 for details). The strategy succeeded in obtaining structures where the geometry supports a Cu(II) state (see Supplementary Note 2), although at the cost of somewhat lower quality crystallographic statistics. For the Ls(AA9)A_lowdose and Ls(AA9)A-G3_lowdose structures 92.14% and 92.27% of residues were in the most favorable regions of the Ramachandran plot.
EPR spectroscopy
Continuous wave X-band frozen solution EPR spectra of single sample of 0.2 to 0.6 mM solutions of Cu(II)-Ls(AA9)A and with 2-fold excess of G6 oligosaccharide or 25 mg/mL PASC (in 10% v/v glycerol) at pH 6.0 (20 mM MES buffer, 200 mM NaCl or 20 mM phosphate buffer) and 150 K were acquired on a Bruker EMX spectrometer operating at ~9.30 GHz, with a modulation amplitude of 4 G and microwave power of 10.02 mW.Chloride containing samples were prepared by the addition of an equimolar amount of CuCl2 prepared in MilliQ water to the protein. The sample was then buffer exchanged on a HiLoad 16/600 Superdex 75 column (GE Healthcare) into 20 mM MES pH 6, 200 mM NaCl to remove any excess copper. For chloride-free samples Ls(AA9)A was copper loaded with an equimolar amount of CuSO4 before being buffer exchanged into 20 mM sodium phosphate buffer pH 6 on a HiLoad 16/600 Superdex 75 column (GE Healthcare). Spectral simulations were carried out using EasySpin 5.0.0 integrated into MATLAB R2014a software on a desktop PC. Simulation parameters are given in Supplementary Table 6. gz and |Az| values were determined accurately from the three absorptions at low field. It was assumed that g and A tensors were axially coincident. Accurate determination of gx, gy, |Ax| and |Ay| values was not possible due to the second order nature of the perpendicular region, although it was noted that satisfactory simulation could only be achieved with one particular set of g values. The simulation of Ls(AA9)A-G6 spectrum at low chloride conditions was performed after subtraction of the normalized Ls(AA9)A spectrum recorded in the presence of 200 mM NaCl (~15% of the chloride-bound species was present). Spectra and simulations are shown in Supplementary Figure 7.
1H HYSCORE
Hyperfine sublevel correlation (HYSCORE) spectra were collected on Ls(AA9)A and Ls(AA9)A-G6 samples at 1.2 and 3.2 mM protein concentration, respectively, (the high concentration sample showed a pale blue color) with 2-fold excess G6 oligosaccharide (in 10% v/v glycerol) at pH 6.0 (20 mM MES buffer, 200 mM NaCl) on a Bruker ElexSys E580 spectrometer. Ls(AA9)A was copper loaded as described in the EPR section. Standard dielectric ring Bruker EPR cavities (ER 4118X-MD5 and EN 4118X-MD4) equipped with an Oxford CF 935 helium flow cryostat were used. The 1H HYSCORE spectra were recorded employing the sequence π/2 – τ – π/2 – t1 – π – t2 – π/2 – τ – echo with microwave pulses of length tπ/2 = 16 ns and tπ = 32 ns, τ = 136 ns, starting times t1,2 = 100 ns and time increments Δt1,2 = 20–32 ns at 15 K. The intensity of the inverted echo following the fourth pulse is measured with t1 and t2 varied and constant τ. Unwanted features from the experimental echo envelopes were removed by using a four-step phase cycle.51 256 data points were collected in both dimensions. The relaxation decay was subtracted by baseline corrections (fitting by polynomials of 3–6 degree) in both time domains, subsequently applying apodization (Hamming window) and zero-filling to 1024 data points in both dimensions. After 2D fast Fourier transformation, absolute value spectra were simulated using EasySpin 5.0.0 integrated into MATLAB R2014a software. (Supplementary Note 3 and Supplementary Fig. 9)
Isothermal Titration Calorimetry
Isothermal titration calorimetry was performed in triplicate using an Auto-ITC microcalorimeter (Malvern). Typically protein was present in the cell at 50 μM with a ten-fold more concentrated solution of G6 (Seikagaku Biobusiness) in the syringe. Titrations were performed at 25 °C with an initial 0.5 μL injection which is discarded in the data analysis, followed by 18 2 μL injections with a 2 minute interval between each injection. The chloride containing buffer used was 20 mM MES pH 6.0, 200 mM NaCl. Copper loading of Ls(AA9)A was carried out as for the EPR spectroscopy sample. G6 for the syringe was prepared by dissolving solid G6 in the exact same buffer as used for size exclusion chromatography in the last step of protein preparation for experiments in both the presence and absence of chloride (in the latter case 67 mM Na2SO4 was used to maintain ionic strength). All data were analyzed using the Origin 7 software package (MicroCal). Heats of dilution were subtracted from the data for the titration in the presence of chloride. The analysis typically returned a stoichiometry of between 0.7 and 0.9 for titration in the presence of chloride, probably due to inaccuracies in the measurement of small masses of G6. The G6 concentration was therefore adjusted in the software assuming a 1:1 stoichiometry of G6 to protein (Supplementary Fig. 8)
Supplementary Material
Acknowledgments
This work was supported by the UK Biotechnology and Biological Sciences Research Council (BB/L000423 and BB/L021633/1), Agence Française de l’Environnement et de la Maîtrise de l’Energie (1201C102), Danish Council for Strategic Research (12-134923, 12-134922). L.L.L. acknowledges the experimental assistance of Kathrine Rasmussen and Rasmus M. Borup and thanks MAXLAB, Sweden and the ESRF, France, for provision of synchrotron beamtime and assistance. The Danish Ministry of Higher Education and Science through the Instrument Center DANSCATT and the European Community’s Seventh Framework Programme (FP7/2007-2013) under BioStruct-X (grant agreement N°283570) funded travel to synchrotrons. L.M., S.F., S.C. and H.D. acknowledge support from ICMG FR 2607, LabEx ARCANE (ANR-11-LABX-0003-01), PolyNat Carnot Institute, French Agence Nationale de la Recherche (PNRB2005-11).
Footnotes
Accession codes Atomic coordinates and structure factors for the reported crystal structures were deposited with the Protein Data Bank under accession codes 5ACH (Ls(AA9)A high resolution), 5ACG (Ls(AA9)A low X-ray dose), 5ACI (Ls(AA9)A with G6), 5ACJ (Ls(AA9)A with G3) and 5ACF (Ls(AA9)A with G3 and low X-ray dose). Sequence data for Ls(AA9)A were deposited at GenBank, ref KT368674.
Additional Information The authors declare competing financial interests: M.T. and P.vF. are employees of Novozymes, a producer of enzymes for industrial use.
Supplementary Information is linked to the online version of the paper. Raw data for EPR, ITC and kinetics experiments is available through the University of York Library, DOI: 10.15124/92868bbb-f21a-41e9-a27f-3078b40d144e
References
- 1.Himmel ME, et al. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science. 2007;315:804–807. doi: 10.1126/science.1137016. [DOI] [PubMed] [Google Scholar]
- 2.Sheldon RA. Green and sustainable manufacture of chemicals from biomass: state of the art. Green Chemistry. 2014;16:950–963. [Google Scholar]
- 3.Payne CM, et al. Fungal cellulases. Chemical Reviews. 2015;115:1308–1448. doi: 10.1021/cr500351c. [DOI] [PubMed] [Google Scholar]
- 4.Beeson WT, Vu VV, Span EA, Phillips CM, Marletta MA. Cellulose degradation by polysaccharide monooxygenases. Annu. Rev. of Biochem. 2015;84:923–946. doi: 10.1146/annurev-biochem-060614-034439. [DOI] [PubMed] [Google Scholar]
- 5.Horn SJ, Vaaje-Kolstad G, Westereng B, Eijsink VGH. Novel enzymes for the degradation of cellulose. Biotechnology for Biofuels. 2012;5:45. doi: 10.1186/1754-6834-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Müller G, Várnai A, Johansen KS, Eijsink VGH, Horn SJ. Harnessing the potential of LPMO-containing cellulase cocktails poses new demands on processing conditions. Biotechnology for Biofuels. 2015;8:1–9. doi: 10.1186/s13068-015-0376-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hemsworth GR, Henrissat B, Davies GJ, Walton PH. Discovery of a new family of lytic polysaccharide mono-oxygenases. Nature Chemical Biology. 2014;10:122–126. doi: 10.1038/nchembio.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lo Leggio L, et al. Structure and boosting activity of a starch-degrading lytic polysaccharide monooxygenase. Nature communications. 2015;6:5961. doi: 10.1038/ncomms6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantarel BL, et al. The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic acids research. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Merino S, Cherry J. In: Advances in Biochemical Engineering/Biotechnology. Olsson Lisbeth., editor. Vol. 108. Springer Berlin Heidelberg; 2007. pp. 95–120. Biofuels. Ch. 66. [DOI] [PubMed] [Google Scholar]
- 12.Harris PV, et al. Stimulation of lignocellulosic biomass hydrolysis by proteins of glycoside hydrolase family 61: structure and function of a large, enigmatic family. Biochemistry. 2010;49:3305–3316. doi: 10.1021/bi100009p. [DOI] [PubMed] [Google Scholar]
- 13.Vaaje-Kolstad G, et al. An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science. 2010;330:219–222. doi: 10.1126/science.1192231. [DOI] [PubMed] [Google Scholar]
- 14.Forsberg Z, et al. Cleavage of cellulose by a CBM33 protein. Protein Science. 2011;20:1479–1483. doi: 10.1002/pro.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quinlan RJ, et al. Insights into the oxidative degradation of cellulose by a copper metalloenzyme that exploits biomass components. Proceedings of the National Academy of Sciences. 2011;108:15079–15084. doi: 10.1073/pnas.1105776108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Phillips CM, Beeson WT, Cate JH, Marletta MA. Cellobiose Dehydrogenase and a Copper-Dependent Polysaccharide Monooxygenase Potentiate Cellulose Degradation by Neurospora crassa. ACS Chemical Biology. 2011;6:1399–1406. doi: 10.1021/cb200351y. [DOI] [PubMed] [Google Scholar]
- 17.Hemsworth GR, Davies GJ, Walton PH. Recent insights into copper-containing lytic polysaccharide mono-oxygenases. Curr Opin Struct Biol. 2013;23:660–668. doi: 10.1016/j.sbi.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 18.Aachmann FL, et al. Encyclopedia of Inorganic and Bioinorganic Chemistry. Wiley; 2015. [Google Scholar]
- 19.Kjaergaard CH, et al. Spectroscopic and computational insight into the activation of O2 by the mononuclear Cu center in polysaccharide monooxygenases. Proceedings of the National Academy of Sciences. 2014;111:8797–8802. doi: 10.1073/pnas.1408115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, Ståhlberg J, Sandgren M, Paton RS, Beckham GT. Quantum mechanical calculations suggest that lytic polysaccharide monooxygenases use a copper-oxyl, oxygen-rebound mechanism. Proceedings of the National Academy of Sciences. 2014;111:149–154. doi: 10.1073/pnas.1316609111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao J, Thomas DA, Sohn CH, Beauchamp JL. Biomimetic reagents for the selective free radical and acid–base chemistry of glycans: application to glycan structure determination by mass spectrometry. Journal of the American Chemical Society. 2013;135:10684–10692. doi: 10.1021/ja402810t. [DOI] [PubMed] [Google Scholar]
- 22.Eriksson K-E, Pettersson B, Westermark U. Oxidation: an important enzyme reaction in fungal degradation of cellulose. FEBS Letters. 1974;49:282–285. doi: 10.1016/0014-5793(74)80531-4. [DOI] [PubMed] [Google Scholar]
- 23.Hemsworth GR, et al. The copper active site of CBM33 polysaccharide oxygenases. Journal of the American Chemical Society. 2013;135:6069–6077. doi: 10.1021/ja402106e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Beeson WT, Phillips CM, Marletta MA, Cate JHD. Structural basis for substrate targeting and catalysis by fungal polysaccharide monooxygenases. Structure. 2012;20:1051–1061. doi: 10.1016/j.str.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agger JW, et al. Discovery of LPMO activity on hemicelluloses shows the importance of oxidative processes in plant cell wall degradation. Proceedings of the National Academy of Sciences. 2014;111:6287–6292. doi: 10.1073/pnas.1323629111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borisova AS, et al. Structural and functional characterization of a lytic polysaccharide monooxygenase with broad substrate specificity. Journal of Biological Chemistry. 2015;290:22955–22969. doi: 10.1074/jbc.M115.660183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bennati-Granier C, et al. Substrate specificity and regioselectivity of fungal AA9 lytic polysaccharide monooxygenases secreted by Podospora anserina. Biotechnology for Biofuels. 2015;8:90. doi: 10.1186/s13068-015-0274-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loose JSM, Forsberg Z, Fraaije MW, Eijsink VGH, Vaaje-Kolstad G. A rapid quantitative activity assay shows that the Vibrio cholerae colonization factor GbpA is an active lytic polysaccharide monooxygenase. FEBS Letters. 2014;588:3435–3440. doi: 10.1016/j.febslet.2014.07.036. [DOI] [PubMed] [Google Scholar]
- 29.Gudmundsson M, et al. Structural and electronic snapshots during the transition from a Cu(II) to Cu(I) metal center of a lytic polysaccharide monooxygenase by X-ray photoreduction. Journal of Biological Chemistry. 2014;289:18782–18792. doi: 10.1074/jbc.M114.563494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Asensio JL, Ardá A, Cañada FJ, Jiménez-Barbero J. Carbohydrate–aromatic interactions. Accounts of Chemical Research. 2013;46:946–964. doi: 10.1021/ar300024d. [DOI] [PubMed] [Google Scholar]
- 31.Nishio M. The CH/π hydrogen bond in chemistry. Conformation, supramolecules, optical resolution and interactions involving carbohydrates. Physical Chemistry Chemical Physics. 2011;13:13873–13900. doi: 10.1039/c1cp20404a. [DOI] [PubMed] [Google Scholar]
- 32.Davies GJ, Planas A, Rovira C. Conformational analyses of the reaction coordinate of glycosidases. Accounts of Chemical Research. 2012;45:308–316. doi: 10.1021/ar2001765. [DOI] [PubMed] [Google Scholar]
- 33.Lee JY, Karlin KD. Elaboration of copper-oxygen mediated C-H activation chemistry in consideration of future fuel and feedstock generation. Curr Opin Chem Biol. 2015;25:184–193. doi: 10.1016/j.cbpa.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwaizumi M, Kudo T, Kita S. Correlation between the hyperfine coupling constants of donor nitrogens and the structures of the first coordination sphere in copper complexes as studied by nitrogen-14 ENDOR spectroscopy. Inorganic Chemistry. 1986;25:1546–1550. [Google Scholar]
- 35.Kim D, Kim NH, Kim SH. 34 GHz Pulsed ENDOR characterization of the copper coordination of an amyloid β peptide relevant to Alzheimer’s disease. Angewandte Chemie International Edition. 2013;52:1139–1142. doi: 10.1002/anie.201208108. [DOI] [PubMed] [Google Scholar]
- 36.Aachmann FL, Sørlie M, Skjåk-Bræk G, Eijsink VGH, Vaaje-Kolstad G. NMR structure of a lytic polysaccharide monooxygenase provides insight into copper binding, protein dynamics, and substrate interactions. Proceedings of the National Academy of Sciences. 2012;109:18779–18784. doi: 10.1073/pnas.1208822109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karlin KD, Itoh S. Copper-oxygen chemistry. Wiley; 2011. [Google Scholar]
- 38.Donoghue PJ, et al. Rapid C–H bond activation by a monocopper(III)–hydroxide complex. Journal of the American Chemical Society. 2011;133:17602–17605. doi: 10.1021/ja207882h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu M, et al. Crystal structure and computational characterization of the lytic polysaccharide monooxygenase GH61D from the basidiomycota fungus Phanerochaete chrysosporium. Journal of Biological Chemistry. 2013;288:12828–12839. doi: 10.1074/jbc.M113.459396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dhar D, Tolman WB. Hydrogen atom abstraction from hydrocarbons by a copper(III)-hydroxide complex. Journal of the American Chemical Society. 2015;137:1322–1329. doi: 10.1021/ja512014z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh SK, Das A. The n - π* interaction: a rapidly emerging non-covalent interaction. Physical Chemistry Chemical Physics. 2015;17:9596–9612. doi: 10.1039/c4cp05536e. [DOI] [PubMed] [Google Scholar]
- 42.Egli M, Gessner RV. Stereoelectronic effects of deoxyribose O4′ on DNA conformation. Proceedings of the National Academy of Sciences. 1995;92:180–184. doi: 10.1073/pnas.92.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pavlakos I, et al. Noncovalent lone pair…(No-π!)-heteroarene interactions: the Janus-faced hydroxy group. Angewandte Chemie International Edition. 2015;54:8169–8174. doi: 10.1002/anie.201502103. [DOI] [PubMed] [Google Scholar]
- 44.Roeser D, et al. A general binding mechanism for all human sulfatases by the formylglycine-generating enzyme. Proceedings of the National Academy of Sciences. 2006;103:81–86. doi: 10.1073/pnas.0507592102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Online Methods references
- 45.Spodsberg N, Shaghasi T, Sweeney M, Xu F, Schnorr K. Polypeptides having cellulolytic enhancing activity and polynucleotides encoding same. 2014 Novozymes A/S patent Patent WO2014066141 A2.
- 46.Liu Y, et al. Use of a fluorescence plate reader for measuring kinetic parameters with inner filter effect correction. Analytical biochemistry. 1999;267:331–335. doi: 10.1006/abio.1998.3014. [DOI] [PubMed] [Google Scholar]
- 47.Kabsch W. XDS. Acta Crystallographica Section D. 2010;D66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallographica Section D. 2010;D66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallographica Section D. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 50.Vagin AA, et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallographica Section D. 2004;D60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 51.Gemperle C, Aebli G, Schweiger A, Ernst RR. Phase cycling in pulse EPR. Journal of Magnetic Resonance (1969) 1990;88:241–256. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.