SUMMARY
The SnRK1 protein kinase balances cellular energy levels in accordance with extracellular conditions and is thereby key for plant stress tolerance. In addition, SnRK1 has been implicated in numerous growth and developmental processes from seed filling and maturation to flowering and senescence. Despite its importance, the mechanisms that regulate SnRK1 activity are poorly understood. Here, we demonstrate that the SnRK1 complex is SUMOylated on multiple subunits and identify SIZ1 as the E3 Small Ubiquitin-like Modifier (SUMO) ligase responsible for this modification. We further show that SnRK1 is ubiquitinated in a SIZ1-dependent manner, causing its degradation through the proteasome. In consequence, SnRK1 degradation is deficient in siz1-2 mutants, leading to its accumulation and hyperactivation of SnRK1 signaling. Finally, SnRK1 degradation is strictly dependent on its activity, as inactive SnRK1 variants are aberrantly stable but recover normal degradation when expressed as SUMO mimetics. Altogether, our data suggest that active SnRK1 triggers its own SUMOylation and degradation, establishing a negative feedback loop that attenuates SnRK1 signaling and prevents detrimental hyperactivation of stress responses.
Keywords: SUMOylation, ubiquitination, SNF1-related protein kinase (SnRK1), energy signaling, stress, SIZ1, Arabidopsis thaliana
INTRODUCTION
The plant Snf1-related Protein Kinase 1 (SnRK1) is a central component of a sophisticated signaling network that translates the plant carbon status into defense, growth and developmental decisions (Lastdrager et al., 2014). SnRK1 downregulates growth-related processes, partly through inhibition of major biosynthetic enzymes of carbon and nitrogen metabolism (Sugden et al., 1999; Polge et al., 2008). In addition, it controls the expression of over 1000 genes involved in metabolism, signaling, transcription, stress tolerance, transport and growth (Baena-Gonzalez et al., 2007; Baena-Gonzalez and Sheen, 2008). The coordinated metabolic and transcriptional regulation by SnRK1 contributes to maintaining cellular homeostasis during stress, thereby promoting tolerance and survival (Hao et al., 2003; Lovas et al., 2003; Schwachtje et al., 2006; Baena-Gonzalez et al., 2007; Lee et al., 2009; Lin et al., 2014). SnRK1 has also been implicated in ABA hormone signaling as well as in numerous developmental processes from seed filling, maturation, and germination to reproduction and senescence (Bhalerao et al., 1999; Thelander et al., 2004; Radchuk et al., 2006, 2010; Schwachtje et al., 2006; Baena-Gonzalez et al., 2007; Lu et al., 2007; Ananieva et al., 2008; Lee et al., 2008, 2009; Jossier et al., 2009; Coello et al., 2012; Tsai and Gazzarrini, 2012; Rodrigues et al., 2013; Lin et al., 2014). Finally, transient systemic silencing of SnRK1 results in growth arrest and premature senescence, highlighting the centrality of the SnRK1 system for normal plant growth and development (Thelander et al., 2004; Baena-Gonzalez et al., 2007).
SnRK1 is the plant ortholog of the budding yeast Snf1 (Sucrose-non-fermenting 1) and mammalian AMPK (AMP-activated protein kinase). All three enzymes function as heterotrimeric complexes composed of an α-catalytic and two β- and γ-regulatory subunits, and in all cases kinase activity requires phosphorylation of a conserved T-loop threonine in the α-subunit (Baena-Gonzalez et al., 2007; Polge and Thomas, 2007; Crozet et al., 2014). However, the intimate connection between T-loop phosphorylation in response to energy deprivation and kinase activation observed in SNF1 and AMPK is not established in plants, where additional regulatory mechanisms may be operating (Baena-Gonzalez et al., 2007; Fragoso et al., 2009; Coello et al., 2012; Rodrigues et al., 2013). Furthermore, SnRK1 kinase activity appears unchanged under conditions that induce SnRK1 signaling as well as in pp2c mutants that display deficient repression of the SnRK1 pathway (Baena-Gonzalez et al., 2007; Rodrigues et al., 2013). Finally, the plant enzyme has incorporated unique regulatory subunits and other distinct features, presumably to respond to plant-specific signals and/or to perform plant-specific functions (Polge and Thomas, 2007; Crozet et al., 2014; Emanuelle et al., 2015). These studies reveal the atypical nature of the plant kinase and underscore our lack of knowledge on the factors that determine the signaling lifetime of such a central component.
SUMO (Small Ubiquitin-like Modifier) is a small protein (about 12 kDa) that is post-translationally conjugated to target proteins in a reversible manner to regulate crucial biological processes. SUMOylation is required for normal growth and development, and consequently, mutants defective in the SUMO pathway are either lethal or display strong phenotypes (Geiss-Friedlander and Melchior, 2007; Saracco et al., 2007). In addition, exposure to environmental or metabolic stresses induces a dramatic accumulation of SUMO conjugates, constituting what is considered to be a cellular protective response in all eukaryotes (Guo and Henley, 2014). Accordingly, many SUMO targets identified in Arabidopsis are stress-related components (Elrouby and Coupland, 2010; Miller et al., 2010) and SUMOylation is important for a wide range of plant stress responses (Castro et al., 2012). SUMOylation has been ascribed very diverse biochemical functions, including changes in stability, activity, and subcellular localization, primarily through the modulation of protein interactions (Jentsch and Psakhye, 2013).
SUMOylation requires the maturation of the SUMO moiety by a SUMO protease, exposing a characteristic C-terminal di-glycine motif (Geiss-Friedlander and Melchior, 2007). Mature SUMO is then activated by the SUMO-activating enzyme E1 enzyme [SAE1/2 in Arabidopsis (Park et al., 2011)], through the formation of a thioester bond, and transferred to the SUMO-conjugating enzyme (E2 enzyme, SCE1 in Arabidopsis) through a transthiolation reaction. Finally, SCE conjugates SUMO to a target lysine, either alone or with the help of a SUMO E3 ligase (SIZ1 and HPY2 in Arabidopsis). Substrates can carry single SUMO moieties or SUMO polymers, and in Arabidopsis, two SUMO E4 ligases were recently implicated in the assembly of such SUMO chains (Tomanov et al., 2014). SUMO chains may have a role on their own (Ulrich, 2008), but can also trigger ubiquitination via SUMO-targeted ubiquitin ligases (STUbLs), hence targeting the protein for proteasomal degradation (Praefcke et al., 2012; Elrouby et al., 2013).
A proteome-wide screen previously identified SnRK1α1 as an interactor of SCE1 (Elrouby and Coupland, 2010). In contrast to ubiquitination, where substrate specificity is provided by the E3 ligase, SUMO substrates can be directly recognized and bound on their target lysine by the E2 conjugation enzyme (Geiss-Friedlander and Melchior, 2007; Park et al., 2011), and hence the SnRK1α1–SCE1 interaction suggests that SnRK1α1 may be a target of SUMOylation. Here, we demonstrate that the SnRK1 complex is SUMOylated by the SIZ1 E3 SUMO ligase, resulting in its ubiquitination and proteasomal degradation. Importantly, SUMO-dependent proteolytic removal targets exclusively active SnRK1, suggesting that SUMOylation acts as a safeguard to avoid sustained activation of stress responses.
RESULTS
The SnRK1 complex is SUMOylated
As a first step to test whether SnRK1 is a target of SUMOylation, we confirmed the reported SnRK1α1–SCE1 interaction in a yeast-two-hybrid assay (Y2H; Figure S1) and found that it occurs mostly through the SnRK1α1 regulatory domain (RD). However, a weaker interaction with the SnRK1α1 kinase domain (KD) could also be detected in less stringent selection media.
To investigate whether the SnRK1α1–SCE1 interaction results into SnRK1 SUMOylation, we initially employed an heterologous system in which the Arabidopsis SUMOylation machinery is reconstituted and co-expressed with individual potential substrates in E. coli (Okada et al., 2009). In the presence of mature SUMO1 or mature SUMO3 (SUMO-GG), SnRK1α1 displayed a clear SUMOylation signal that was absent in the corresponding non-conjugatable isoforms (SUMO-AA), used as negative controls (Figure 1a). We could also observe SUMOylation of SnRK1β1, SnRK1β2 (Figure 1a), and SnRK1γ (Figure S2a), although in the case of the latter the functional connection to SnRK1 remains uncertain (Ramon et al., 2013; Emanuelle et al., 2015). The only tested subunit for which SUMOylation was not detected was SnRK1βγ (Figure 1a), recently proposed to be the only γ-type subunit of the SnRK1 complex (Ramon et al., 2013; Emanuelle et al., 2015). These results show that in this E. coli system SUMOylation occurs specifically on several components of the SnRK1 complex.
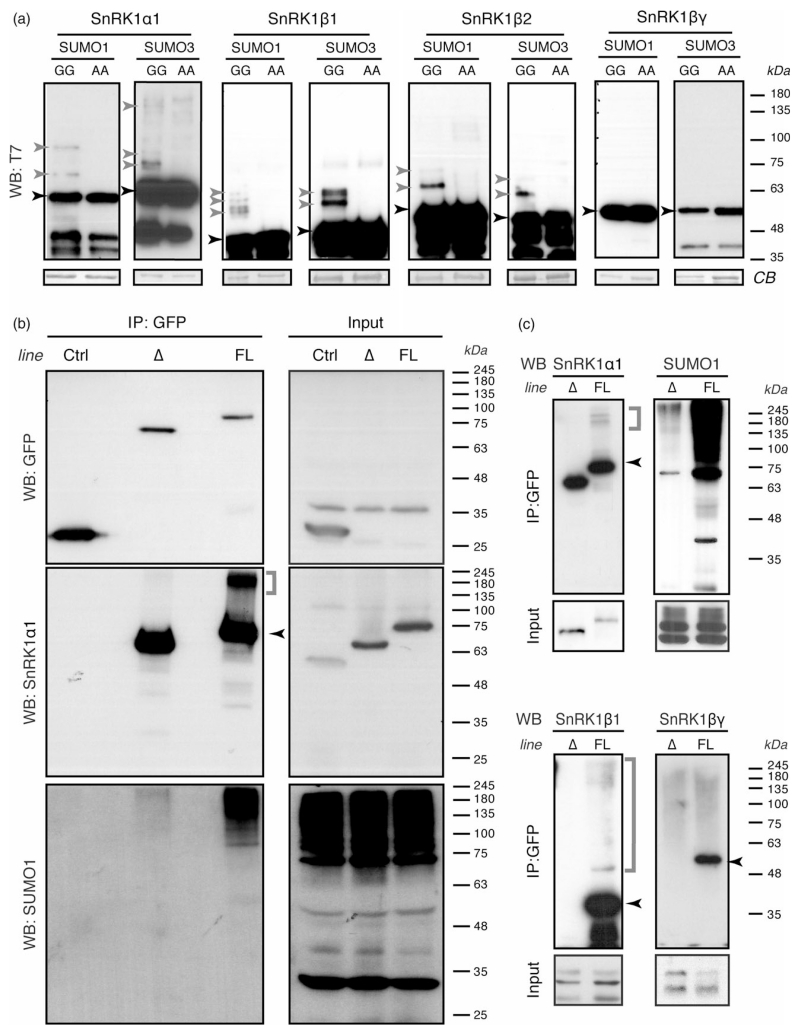
Figure 1.
SnRK1 is SUMOylated
(a) Multiple SnRK1 subunits are SUMOylated in a heterologous E. coli system. SnRK1 subunits harboring 6*His and T7 tags were co-expressed in E. coli with the indicated SUMO isoform together with the SUMO-activating (AtSAE1a/AtSAE2) and SUMO-conjugating (AtSCE1) enzymes. SnRK1 subunits were purified via their His tag by IMAC and immunoblotted against their T7 tag (‘WB: T7’). GG and AA refer to conjugatable and non-conjugatable SUMO variants, respectively. Equal protein loading is shown by Coomassie blue (CB) staining of membranes.
(b, c) The SnRK1 complex is SUMOylated in planta. Leaf crude extracts of plants expressing GFP (Ctrl), SnRK1α1ΔKA1–GFP (Δ) and full-length SnRK1α1–GFP (FL) were used for GFP immunoprecipitation (IP) and immunoprecipitates were analysed by western blot (WB) using antibodies against GFP, SnRK1α1 and SUMO1 (b) or against SnRK1α1, SnRK1β1, SnRK1βγ and SUMO1 (c). The same antibodies were used for assessing the levels of these proteins in the corresponding inputs. Note that in the Ctrl plants GFP is driven by the strong 35S promoter whereas SnRK1α1ΔKA1–GFP and SnRK1α1–GFP are driven by the SnRK1α1 promoter, hence explaining why GFP can only be detected in the input of 35S::GFP plants. Black and grey arrowheads: non-SUMOylated and SUMOylated proteins, respectively. Grey brackets: high molecular weight forms of the indicated proteins.
To determine if SUMOylation of SnRK1 also occurs in planta, we made use of a snrk1α1 knockout mutant complemented with SnRK1α1–GFP driven by its own upstream and downstream regulatory regions (pSnRK1α1:: SnRK1α1–GFP::tSnRK1α1/SnRK1α1-3; hereafter referred as SnRK1α1–GFP; Figure S3). GFP immunoprecipitation followed by Western blot analyses with an anti-SUMO1 antibody revealed a massive accumulation of SUMO1 conjugates in immunoprecipitates from SnRK1α1–GFP plants but not from control plants expressing 35S::GFP (Figure 1b). Interestingly, SUMO1 conjugates associated with SnRK1α1–GFP were not resolved as distinct bands, but rather as a high molecular weight (hMW) ladder, suggesting the formation of (poly)SUMO chains and/or the SUMOylation of multiple residues. This was further supported by the presence of hMW SnRK1α1 forms in the SnRK1α1 immunoblot (Figure 1b, middle panel). Immunodetection with SnRK1β1 and SnRK1βγ antibodies confirmed the association of these subunits with SnRK1α1–GFP (Figure 1c). We could detect hMW forms of SnRK1β1 but not of SnRK1βγ, suggesting that only the former is SUMOylated in planta, in accordance with the results obtained in the E. coli SUMOylation system (Figure 1a). To assess the contribution of the β-subunit(s) to SnRK1 SUMOylation, we generated a transgenic line expressing a truncated SnRK1α1 variant lacking the KA1 domain (Rodrigues et al., 2013), required for the interaction with the β and γ regulatory subunits (Bhalerao et al., 1999; Kleinow et al., 2000) (pSnRK1α1::SnRK1α1ΔKA1–GFP::tSnRK1α1/SnRK1α1-3; hereafter referred as SnRK1α1ΔKA1–GFP, Figure S3). As expected, we could not detect SnRK1β1 or SnRK1βγ associated with SnRK1α1ΔKA1–GFP (Figure 1c). Moreover, in the absence of regulatory subunits the amount of SUMO1 conjugates and hMW SnRK1α1 forms was dramatically reduced (Figure 1b, c), indicating that the regulatory subunits contribute significantly to the overall SUMOylation of the SnRK1 complex. Nevertheless, even if reduced, the presence of SnRK1α1 hMW forms and SUMO1 conjugates in SnRK1α1ΔKA1–GFP immunoprecipitates suggests that the interaction with the regulatory subunits is not strictly necessary for SnRK1α1 SUMOylation.
Collectively, these results indicate that several subunits of the SnRK1 complex are SUMOylated in planta and that this may involve the formation of SUMO chains and/or the modification of multiple residues.
SUMOylation inhibits SnRK1 signaling and is SIZ1-dependent
To investigate whether SUMOylation has an impact on SnRK1 signaling, we first undertook a mutagenesis approach to block SnRK1 SUMOylation. We focused on the major catalytic subunit SnRK1α1, as it accounts for nearly 90% of SnRK1 activity in planta (Jossier et al., 2009). SnRK1 harbors two predicted SUMOylation sites [K144 and K471, Figure S4(a); (Elrouby and Coupland, 2010)], but their mutation to arginine, individually or in combination, did not prevent SUMOylation in the E. coli assay (Figure S4b). To map roughly the site(s) of SUMOylation we used the kinase (KD, 1–293) and the RD (Figure S4a) as substrates in the E. coli assay, and found that SnRK1α1 is SUMOylated on both (Figure S4c). To identify the target lysines, we performed liquid chromatography–tandem mass spectrometry (LC-MS/MS) analyses employing regular mature SUMO3 (SUMO3-GG) and a variant (SUMO3S91R-GG) that facilitates LC-MS/MS analyses by yielding a small tryptic footprint (Okada et al., 2009). We focused these analyses on SUMO3 because it generated similar SUMOylation patterns but with stronger signal intensity than SUMO1, allowing a better yield for LC-MS/MS. We uncovered nine SUMOylated lysines (including K144 but not K471), of which two are located in the KA1 domain and seven in the KD (Figure S4a, d). The structural model of the SnRK1 complex predicts that all of these residues are accessible to solvent (Figure S4(d), residues indicated). A single K390R mutation and a double K34R/K63R mutation were sufficient to abrogate SUMOylation of the RD and KD, respectively (Figure S4e). Furthermore, SUMOylation could no longer be detected in the full-length SnRK1α1K34/63/390R triple mutant (hereafter referred as SnRK1α13K, Figure S4f), suggesting that these three lysines are the genuine targets of SUMOylation in vivo.
We next assessed the functional relevance of SnRK1 SUMOylation in a cell-based reporter gene assay by comparing SnRK1 activity between SnRK1α1, SnRK1α13K, and a SnRK1α1 variant mutated in all SUMOylated lysines (SnRK1α1K20/34/44/56/63/69/390/421R, hereafter referred as SnRK1α18K) except K144, as the K144R mutation abolishes kinase activity [(Cho et al., 2012)]. In this assay, SnRK1 activity is measured as an induction of the pDIN6::LUC reporter and transient SnRK1α1 overexpression is sufficient to trigger strong SnRK1 signaling and reporter activation (Baena-Gonzalez et al., 2007; Rodrigues et al., 2013). As expected, SnRK1α1 overexpression resulted in a 20-fold activation of the pDIN6::LUC reporter, whilst an inactive SnRK1α1T175A kinase [T-loop phosphomutant, (Baena-Gonzalez et al., 2007)] had no effect. However, the two SnRK1α13K and SnRK1α18K variants induced the reporter normally (Figure 2a), suggesting that in planta SnRK1α1 can be SUMOylated on other residues and/or that SUMOylation of other components of the complex (e.g. the β-regulatory subunits) is sufficient to convey the SUMO signal.
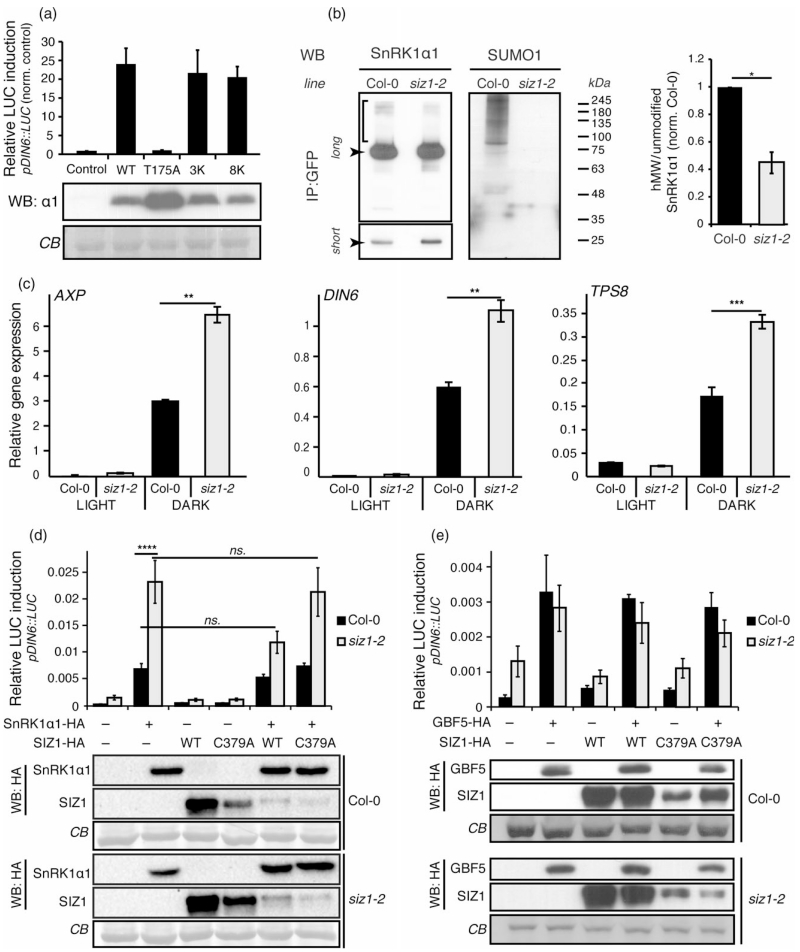
Figure 2.
SIZ1-mediated SUMOylation of SnRK1 represses SnRK1 signaling.
(a) Normal induction of SnRK1 signaling by SnRK1α1 multiple-lysine mutants. Expression of SnRK1α1 in Arabidopsis mesophyll protoplasts triggers SnRK1 signaling, as measured by induction of the pDIN6::LUC reporter. Mutation of lysine residues found to be SUMOylated in the E. coli system does not alter the ability of SnRK1α1 to induce the pDIN6::LUC reporter. SnRK13K, K34R/K63R/K390R; SnRK18K, K20R/K34R/K44R/K56R/K63R/K69R/K390R/K421R. An inactive T-loop phospho-mutant (SnRK1α1T175A) is used as negative control.
(b) SIZ1 is required for SnRK1 SUMOylation. SnRK1α1–GFP was immunoprecipitated (IP) from leaf crude extracts of SnRK1α1–GFP (‘WT’) and SnRK1α1–GFPsiz1-2 (‘siz1-2’) and analysed by Western blot (WB) using antibodies against SnRK1α1 and SUMO1 (as in Figure 1b). The amounts of loaded immunoprecipitates were adjusted to contain approximately similar amounts of SnRK1α1–GFP in WT and siz1-2. The quantification of hMW SnRK1α1 forms in both genotypes is presented on the right and corresponds to the ratio of hMW forms (indicated with a bracket in the longer exposure, ‘long’) per unmodified SnRK1α1 (shorter exposure, ‘short’). The values are normalized to the ratio in control plants (‘WT’).
(c) Overinduction of SnRK1 signaling in the siz1-2 mutant. Relative gene expression (qPCR) of SnRK1 marker genes (AXP, DIN6, TPS8) in Col-0 or siz1-2 mutant plants treated under control (light) or energy stress (dark) conditions.
(d) Overinduction of SnRK1 signaling in siz1-2 is rescued by the catalytic activity of SIZ1. Expression of SnRK1α1 triggers a three-fold higher induction of the pDIN6::LUC reporter in siz1-2 than in Col-0 protoplasts. Normal pDIN6::LUC expression is recovered by co-expression of SIZ1, but not of a catalytically inactive SIZ1C379A variant.
(e) Induction of SnRK1 signaling downstream of SnRK1 is normal in siz1-2. Expression of the GBF5 transcription factor triggers similar induction of the pDIN6::LUC reporter in Col-0 and siz1-2 protoplasts. Expression of all components was confirmed by western blot (WB) with anti-HA antibodies. Equal sample loading was confirmed by Coomassie blue (CB) staining of RubisCO large subunit on membranes. Data are presented as mean ± standard error of the mean (SEM). Statistical significance was determined by ratio pair t-test prior to normalization (b), paired t-test (c), and two-way analysis of variance (anova) (d). n ≥ 3; ns, P > 0.05; *P < 0.05; **P < 0.01; ****P < 0.0001.
As an alternative strategy to globally block SUMOylation of the SnRK1 complex and to assess its functional relevance, we set to identify the E3 ligase(s) responsible for this modification. Even though SCE1 is sufficient for conjugating the SUMO peptide to a substrate in vitro, SUMO E3 ligases are important for substrate specificity and SUMOylation efficiency in vivo (Novatchkova et al., 2012). In Arabidopsis, only two SUMO E3 ligases have been identified, SIZ1 and HPY2 (Miura et al., 2005; Ishida et al., 2009; Miura and Hasegawa, 2010; Novatchkova et al., 2012). Given the strong association of SIZ1 with plant stress responses and the involvement of HPY2/MMS21 mostly in developmental processes (Castro et al., 2012), we hypothesized that SnRK1 SUMOylation is mediated by SIZ1. To test this, we introduced the pSnRK1α1::SnRK1α1–GFP:: tSnRK1α1 construct in siz1-2, a null mutant of SIZ1 [(Miura et al., 2005); hereafter referred as SnRK1α1–GFPsiz1-2] to perform GFP pull-downs and immunoblot analyses as previously described (Figure 1b). As shown in Figure 2(b), western blot analyses of immunoprecipitated SnRK1–GFP revealed a complete absence of SUMO1 conjugates in the siz1-2 mutant, demonstrating that SIZ1 is responsible for SnRK1 SUMOylation by SUMO1 in planta. Importantly, the relative abundance of hMW SnRK1α1 forms in SnRK1α1–GFPsiz1-2 was strongly reduced (more than 50%) when compared to SnRK1α1–GFP plants, indicating that a significant part of these forms corresponds to SUMOylated SnRK1α1 (Figure 2b).
Having established SIZ1 as the E3 ligase responsible for SnRK1 SUMOylation, we next investigated the functional relevance of this modification by comparing SnRK1 signaling in Col-0 (wild-type, WT) and siz1-2 plants. To this end, we treated plants with control (3 h light) or SnRK1 signaling activating conditions (3 h of darkness during the day) and measured the expression of SnRK1 target genes by quantitative RT-PCR (qRT-PCR) as readout of SnRK1 pathway activation (Baena-Gonzalez et al., 2007; Rodrigues et al., 2013). As expected, exposure to darkness triggered a strong induction of SnRK1 target genes (AXP, DIN6 and TPS8) in Col-0 plants (Figure 2c). This induction was two-fold higher in the siz1-2 mutant, showing that SIZ1 is a negative regulator of SnRK1 signaling (Figure 2c). Given that the dwarf growth of siz1-2 is largely caused by elevated salicylic acid (SA) levels (Lee et al., 2007; Miura et al., 2010), we asked if the effect of the siz1-2 mutation on SnRK1 signaling was indirect, and induced by SA. To test this, we treated Col-0 protoplasts transfected with the pDIN6::LUC reporter and SnRK1α1 or control DNA, with SA or ethanol (mock-treated). As shown in Figure S5(a), SA had no effect on SnRK1 signaling during a short (2 h) or extended (15 h) incubation. Furthermore, examination of public microarray datasets with the Genevestigator tool (Hruz et al., 2008) revealed that various SA treatments induce Systemic Acquired Resistance (SAR) marker genes, but not SnRK1 marker genes (DIN6, AXP and TPS8) (Figure S5b), ruling out an indirect effect of the siz1-2 mutation on SnRK1 signaling through SA. To further evaluate potential pleiotropic effects of the siz1-2 mutation on SnRK1 signaling, we next compared reporter gene activation by SnRK1α1 in protoplasts from Col-0 and siz1-2 plants. Expression of SnRK1α1 in Col-0 protoplasts triggered the expected activation of the pDIN6::LUC reporter (Figure 2D). However, this activation was three-fold higher in the siz1-2 mutant, consistent with the hyperactivation of endogenous marker genes in response to dark stress (Figure 2c). Most importantly, co-expression of SIZ1 but not of a catalytically inactive SIZ1 variant [SIZ1C379A; (Cheong et al., 2009)] restored normal activation of the reporter in siz1-2 (Figure 2d), supporting the lack of activity of this SUMO E3 ligase as the cause for hyperactive SnRK1 signaling in the mutant. We next triggered SnRK1 signaling in an alternative manner, by expressing the GBF5 transcription factor that acts downstream of SnRK1 (Baena-Gonzalez et al., 2007). GBF5 induced the expected activation of the pDIN6:: LUC reporter in Col-0 protoplasts and this activation was similar in the siz1-2 mutant (Figure 2e), providing additional evidence that repression of SnRK1 signaling by SIZ1 occurs at the level of the SnRK1 kinase.
Altogether, these results show that SIZ1 inhibits SnRK1 signaling, most probably through SUMOylation of several components of the SnRK1 complex.
SUMOylation triggers SnRK1 degradation
Proteins modified by SUMO acquire novel molecular features that can affect their stability, activity, and/or subcellular localization, primarily through altered interactions with other proteins (Jentsch and Psakhye, 2013). To determine whether the SIZ1-dependent inhibition of SnRK1 involved changes on SnRK1 specific activity, we performed in vitro kinase assays using immunoprecipitated SnRK1α1 and ABF2 as a substrate (Rodrigues et al., 2013). Even though ABF2 phosphorylation was two-fold higher with SnRK1 immunoprecipitated from the siz1-2 mutant than from the Col-0 control, this was fully explained by the two-fold higher levels of SnRK1α1 immunoprecipitated from siz1-2 plants (Figure 3a). This shows that the over-activation of SnRK1 signaling in siz1-2 plants is not caused by changes in SnRK1 specific activity, but rather by an enhanced accumulation of the kinase.
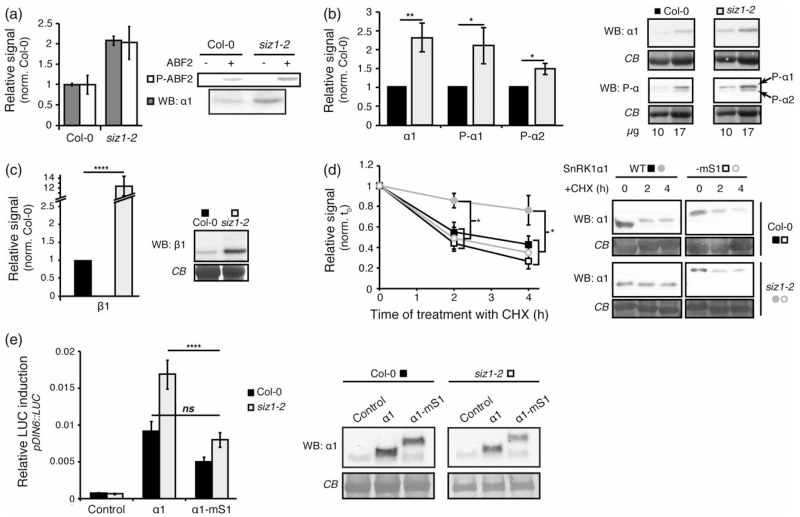
Figure 3.
SnRK1 stability is increased in siz1-2.
(a) SnRK1 specific activity is not altered in the siz1-2 mutant. SnRK1α1 was immunoprecipitated from Col-0 and siz1-2 leaf extracts and incubated in the presence of γ-32P-ATP and a recombinant ABF2 polypeptide as substrate. SnRK1α1 levels and ABF2 phosphorylation were quantified by western blot (WB) and by autoradiography (P-ABF2), respectively.
(b, c) SnRK1 accumulates to higher levels in the siz1-2 mutant. Total leaf protein extracts of Col-0 and siz1-2 plants were analysed by western blot (WB) using antibodies against SnRK1α1 (‘α1’) and the phosphorylated T-loop of SnRK1α1 and SnRK1α2 (‘P-α1/P-α2’) (b, 10 or 17 μg) or against the SnRK1β1 subunit (c, 20 μg). The signals were quantified and normalized to loading. The average quantification in siz1-2 normalized to Col-0 is presented.
(d) Reduced SnRK1α1 degradation in siz1-2 is restored in a SUMO mimetic SnRK1α1 variant. SnRK1α1 fused to mature SUMO1 (–mS1, empty marks) or not (WT, filled marks) was expressed in Col-0 (squares) or siz1-2 (circles) protoplasts. Protoplasts were thereafter treated with cycloheximide (CHX), samples were harvested at the indicated times points, and analysed by western blot using anti-SnRK1α1 antibody (WB: α1). The signal was quantified and normalized to the t = 0 for each kinetics.
(e) Restoring SnRK1 SUMOylation in the siz1-2 mutant by expression of the SUMO mimetic SnRK1α1 variant (α1-mS1) results in normal activation of the pDIN6::LUC reporter. Expression of all SnRK1α1 variants was confirmed by western blot (WB) with anti-SnRK1α1 antibodies. Equal sample loading was confirmed by Coomassie blue (CB) staining of RubisCO large subunit on membranes. Data are presented as mean ± standard error of the mean (SEM). Statistical significance was determined by ratio paired t-test prior to normalization (b, c) or by anova (d, e). n ≥ 3; ns, P > 0.05; *P < 0.05; **P < 0.01; ****P < 0.0001.
To determine more precisely SnRK1 levels, we performed immunoblot analyses using total soluble protein extracts. These analyses confirmed that SnRK1α1 is about 2.5-fold more abundant in the siz1-2 mutant. As a result of increased accumulation, SnRK1α1 T-loop phosphorylation is also 2.5-fold higher in the siz1-2 mutant, consistent with a similar SnRK1 specific activity in Col-0 and siz1-2 plants (Figure 3a). The levels of phosphorylated SnRK1α2 were also higher in siz1-2, suggesting this other catalytic subunit might also be a target of SUMOylation. In accordance with our previous indications that SUMOylation could target the regulatory subunits (Figures 1 and 2a), we also detected an increase in SnRK1β1 protein amounts in siz1-2 (Figure 3c). Similar comparisons were not possible for SnRK1βγ since the available antibodies recognize multiple bands in total protein extracts (Figure S6), despite being adequate for analysing SnRK1 immunoprecipitates (Figure 1c). Interestingly, SnRK1γ displayed a 3.5-fold accumulation in the siz1-2 mutant (Figure S2b), although, in accordance with previous reports (Ramon et al., 2013; Emanuelle et al., 2015), we could not detect this protein in SnRK1α1 immunoprecipitates (Figure S2c).
To investigate the cause of enhanced SnRK1 accumulation in siz1-2, we next performed protein half-life measurements in Col-0 and siz1-2 protoplasts expressing SnRK1α1. Following 6 h of incubation, translation was blocked by the addition of cycloheximide (CHX) and cells were harvested after 2 and 4 h for quantification of SnRK1α1 amounts. As shown in Figure 3(d), SnRK1α1 degradation was mostly abolished in the siz1-2 mutant. To assess whether the lack of SnRK1α1 degradation in siz1-2 is due to the lack of SUMOylation, we generated a translational fusion between SnRK1α1 and a non-conjugatable mature SUMO1-AA (SnRK1α1-SUMO1), thus mimicking SnRK1α1 SUMOylation [‘SUMO mimetic’ (Ulrich, 2009)]. Interestingly, the SnRK1α1 SUMO mimetic was degraded in siz1-2 to a similar extent as SnRK1α1 in Col-0 protoplasts (Figure 3d). Moreover, mimicking SUMOylation with the SnRK1α1-SUMO1 variant bypassed SIZ1 and restored normal pDIN6::LUC reporter activation in the siz1-2 mutant (Figure 3e), altogether supporting the view that SUMOylation of SnRK1α1 promotes its degradation.
SUMOylated SnRK1 is ubiquitinated and degraded by the proteasome
We next asked whether the observed SnRK1 degradation occurred via the ubiquitin proteasome system. To assess this, we determined SnRK1α1 accumulation in the presence of the proteasome inhibitor MG132 or the dimethyl sulphoxide (DMSO) solvent control. In accordance with previous results (Figure 3d), we observed a strong reduction in SnRK1α1 levels 3h after the addition of cycloheximide (Figure 4a). However, this degradation was significantly blocked in cells treated with MG132, showing that SnRK1α1 protein turnover is largely dependent on the proteasome.
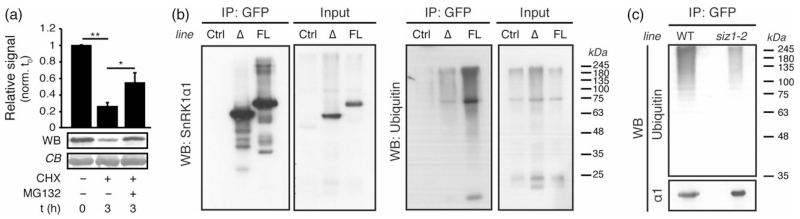
Figure 4.
SUMOylated SnRK1 is ubiquitinated and degraded by the proteasome.
(a) SnRK1α1 is degraded through the proteasome. SnRK1α1 was expressed in Col-0 protoplasts and its levels were assessed by western blot (WB) following a 3 h treatment with cycloheximide (CHX) in the presence or absence of the proteasome inhibitor MG132. Equal sample loading was confirmed by Coomassie blue (CB) staining of RubisCO large subunit on membranes. Quantified levels were normalized to t = 0. Data are presented as mean ± standard error of the mean (SEM). Statistical significance was determined by ratio pair t-test prior to data normalization. n ≥ 3; **P < 0.01; *P < 0.05.
(b) The SnRK1 complex is ubiquitinated in planta. Leaf crude extracts of plants expressing GFP (Ctrl), SnRK1α1ΔKA1–GFP (Δ) and full-length SnRK1α1–GFP (FL) were used for GFP immunoprecipitation (IP) and immunoprecipitates were analysed by western blot (WB) using antibodies against SnRK1α1 and Ubiquitin11. The same antibodies were used for assessing the levels of these proteins in the corresponding inputs.
(c) SnRK1 ubiquitination is largely dependent on SIZ1. SnRK1α1–GFP was immunoprecipitated (IP) from leaf crude extracts of SnRK1α1–GFP (‘WT’) and SnRK1α1–GFPsiz1-2 (‘siz1-2’) and analysed by western blot (WB) using antibodies against SnRK1α1 and Ubiquitin11. The amounts of loaded immunoprecipitates were adjusted to contain approximately similar amounts of SnRK1α1–GFP in WT and siz1-2.
To investigate whether SnRK1 is ubiquitinated for proteasomal degradation in planta, we employed the same system as for assessing SnRK1 SUMOylation (Figure 1b) and performed GFP immunoprecipitation from SnRK1α1–GFP, SnRK1α1ΔKA1–GFP, and 35S::GFP control plants, to check for the presence of ubiquitin conjugates. These analyses revealed a higher accumulation of ubiquitin conjugates in immunoprecipitates from SnRK1α1–GFP than in those from SnRK1α1ΔKA1–GFP plants, while none was detected in the 35S::GFP control (Figure 4b). This pattern was clearly similar to that of SUMO conjugates (Figure 1b), suggesting that SnRK1 ubiquitination and SUMOylation may be interconnected. To test whether SnRK1 SUMOylation is a prerequisite for its ubiquitination, we next immunoprecipitated SnRK1α1–GFP from SnRK1α1–GFPsiz1-2 plants and checked for the presence of ubiquitin. As shown in Figure 4(c), ubiquitin conjugates were markedly reduced in SnRK1α1–GFP immunoprecipitates from SnRK1α1–GFPsiz1-2 compared with SnRK1α1–GFP plants, indicating that SnRK1 SUMOylation is at least partially required for its subsequent ubiquitination.
SnRK1 degradation is strictly dependent on its activity
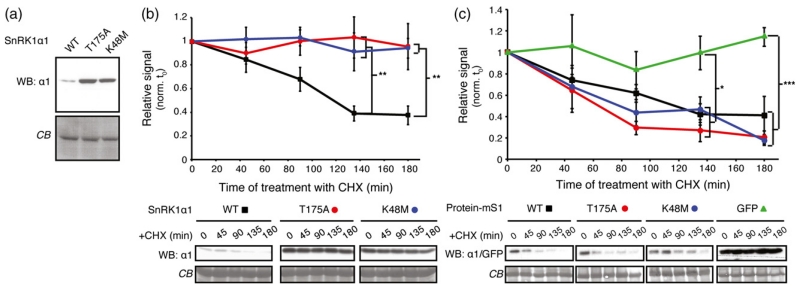
We previously observed that inactive SnRK1α1 variants, such as SnRK1α1T175A and SnRK1α1K48M (impaired in phosphotransfer activity), accumulate to higher levels than active SnRK1α1 (Baena-Gonzalez et al., 2007) (Figure 5a), suggesting a connection between kinase activity and stability. By measuring protein half-life in the presence of cycloheximide in Col-0 cells, we found that the reason for over-accumulation of both inactive variants (SnRK1α1K48M and SnRK1α1T175A) was the lack of protein degradation (Figure 5b).
Figure 5.
Degradation of SnRK1 requires its activity.
(a) Inactive SnRK1α1 variants accumulate to higher levels than the WT protein. Accumulation of WT SnRK1α1 and two inactive SnRK1α1T175A and SnRK1α1K48M mutants transiently expressed in Col-0 protoplasts was analysed by western blot using anti-SnRK1α1 antibodies (WB: α1).
(b) Only active SnRK1α1 undergoes degradation. WT (squares) or inactive (circles) SnRK1α1 variants were expressed in Col-0 protoplasts. Protoplasts were thereafter treated with cycloheximide (CHX), samples were harvested at the indicated time points, and analysed by western blot using anti-SnRK1α1 antibodies (WB: α1). The signal was quantified and normalized to the t = 0 for each kinetics.
(c) Inactive SnRK1α1 variants undergo degradation when expressed as SUMO mimetics. Levels of SnRK1α1-mature SUMO1 (mS1) fusions were followed as in (b). The SnRK1α1 variants used as mS1 fusions are indicated (WT, squares; T175A, red circles, K48M, blue circles). GFP fused to mS1 is provided as a negative control (green triangles). Data are presented as mean ± standard error of the mean (SEM). Statistical significance was determined by two-way anova with Tukey’s multiple comparison. n ≥ 3; *P < 0.05, **P < 0.01; ***P < 0.001.
Given the enhanced stability of SnRK1α1 in the siz1-2 mutant (Figure 3c), we next asked whether the increased stability of the inactive SnRK1α1 variants could also be due to a lack of SUMOylation. To test this, we generated SUMO mimetics of all three SnRK1α1 variants and measured their half-life. Degradation of active SnRK1α1 did not seem to be altered in the corresponding SUMO mimetic (Figures 3d and 5c), suggesting that SUMOylation might not be rate-limiting for degradation of the active kinase. However, the SUMO mimetic forms of the inactive variants were readily degraded, displaying the same degradation profile as active SnRK1α1. In contrast, a SUMO mimetic of a GFP control protein was stable over time, demonstrating a specific effect of SUMO on SnRK1α1 stability.
Collectively, these results confirm the link between SnRK1 activity and turnover and indicate that SUMOylation is an intermediary step in this process.
DISCUSSION
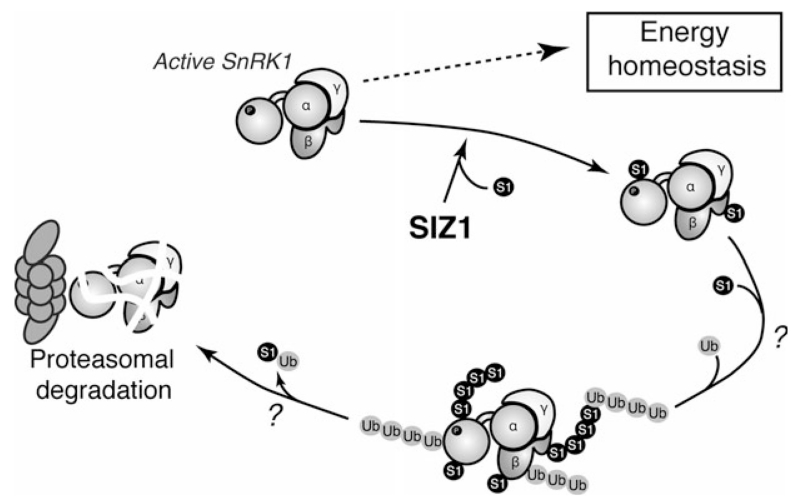
Deregulation of kinase activity often results in cellular or whole-organism dysfunction and even lethality (Lu and Hunter, 2009). Indeed, in yeast, inappropriately high SNF1 activity is deleterious to cell growth, as mutants of the inhibitory phosphatases are only viable if expressing a SNF1 variant with reduced catalytic activity (Ruiz et al., 2011, 2013). Conversely, sugar provision stimulates growth in WT seedlings but not in plants overexpressing SnRK1α1, presumably due to excessive repression of biosynthetic activities when SnRK1 is hyperactive (Baena-Gonzalez et al., 2007). Being a key regulator of the stress response, growth and development, SnRK1 activity must hence be counterbalanced by mechanisms that restrain its action. In this study, we provide strong evidence suggesting that SUMOylation is such a mechanism (Figure 6). SnRK1 is SUMOylated by the SIZ1 E3 SUMO ligase, resulting in its ubiquitination and proteasomal degradation. Mutations that abolish kinase activity prevent SnRK1 degradation, suggesting that SnRK1 activity and SUMOylation are tightly coupled in a negative feedback loop to prevent detrimental pathway hyperactivation.
Figure 6.
Model of SnRK1 regulation by SUMOylation.
Active SnRK1 regulates processes that promote energy homeostasis. As a consequence of its activity SnRK1 is SUMOylated on several subunits in a SIZ1-dependent manner, ubiquitinated, and degraded through the proteasome. The tight coupling between SnRK1 activity and degradation may contribute to establish a balance between stress/defense responses and biosynthetic growth-related processes.
An increasing number of studies show that SUMO E3 ligases often act on preassembled protein complexes, modifying groups of physically interacting components rather than individual proteins (Jentsch and Psakhye, 2013). In agreement with this, our results indicate that SUMOylation occurs at the level of the whole SnRK1 complex (Figures 1, 3, and S2a) and that SIZ1 is responsible for this modification (Figure 2b). The view of collective SnRK1 SUMOylation is further supported by our inability to alter SnRK1 function by mutating the target lysines of the SnRK1α1 subunit (Figures S4f and 2a), as presumably SUMOylation of the remaining subunits compensates for the lack of SUMOylation in SnRK1α1. A ‘SUMOylation wave’ targeting several members of a protein complex has been described, amongst others, for the DNA damage repair complex, in which mutation of single subunits is not sufficient to abrogate SUMOylation of the complex and to cause functional defects (Jentsch and Psakhye, 2013).
Our functional analyses revealed that the SnRK1 pathway is hyperactivated in the siz1-2 mutant, and that the SnRK1 kinase is repressed by SIZ1 (Figure 2c, d). Although we cannot currently rule out the contribution of other proteins to the over-activation of energy signaling in the siz1-2 mutant, the fact that the downstream transcription factor GBF5 induces normal activation of the pathway in siz1-2 cells (Figure 2e) supports the hypothesis that the defect is mainly at the SnRK1 level.
Importantly, normal activation of the SnRK1 pathway in siz1-2 could be restored by transient complementation with SIZ1 but not with a catalytically inactive SIZ1C379A variant (Figure 2d), underscoring the importance of this ligase activity for SnRK1 regulation and ruling out long-term pleiotropic effects of the siz1-2 mutation on SnRK1 function. We could further show that the cause of SnRK1 pathway hyperactivation in the siz1-2 mutant was an increased SnRK1 accumulation (Figures 3a–c and S2c). Interestingly, the increase in protein amounts was not stoichiometric, as SnRK1β1 accumulation in siz1-2 exceeded nearly six-fold that of the SnRK1α1 subunit. This is probably because, in addition to the effect of SIZ1 on SnRK1 protein accumulation, the SnRK1β1 gene is strongly induced by SnRK1 signaling (Baena-Gonzalez et al., 2007), which is higher in siz1-2 (Figure 2)
Protein half-life measurements, using SnRK1α1 as a representative of the SnRK1 complex, revealed that the reason for enhanced SnRK1 accumulation in siz1-2 cells was a defect in protein degradation (Figure 3d). Moreover, mimicking SnRK1α1 SUMOylation was sufficient to rescue its degradation (Figure 3d) and normal pathway activation (Figure 3e) in the siz1-2 mutant, indicating that the cause of impaired SnRK1α1 degradation is the lack of SUMOylation. SnRK1 seems to be degraded by the 26S proteasome, as it is strongly ubiquitinated (Figure 4b) and its degradation is largely blocked by MG132 (Figure 4a). This is consistent with high-throughput analyses in which SnRK1α1 was identified as a target for ubiquitination and was stabilized by MG132 (Maor et al., 2007; Kim et al., 2013). On the other hand, SnRK1 ubiquitination is largely dependent on SUMOylation, as the presence of ubiquitin conjugates in SnRK1α1 immunoprecipitates was greatly reduced in the siz1-2 mutant (Figure 4c). How SUMOylated SnRK1 is targeted to proteasomal degradation is currently unknown but the process is likely to involve the action of the recently discovered E4 SUMO ligases, responsible for (poly)SUMO chain formation (Tomanov et al., 2014), and/or the StUbLs, responsible for the ubiquitination of SUMOylated substrates (Elrouby et al., 2013).
Protein kinases are often targeted by more than one E3 ubiquitin ligases, with some E3 ligases acting specifically on the active kinase (Lu and Hunter, 2009). The fact that in the siz1-2 mutant we could still detect hMW SnRK1α1 forms (Figure 2b) as well as SnRK1α1-associated ubiquitin conjugates (Figure 4c), supports the existence of parallel SUMO-independent ubiquitination pathways. SnRK1α1 interacts with Pleiotropic Regulatory Locus 1 (PRL1) (Bhalerao et al., 1999) and SnRK1α1 degradation is mediated by the DDB1-CUL4-ROC1-PRL1 E3 ubiquitin ligase, in which PRL1 is the putative substrate receptor of the complex (Lee et al., 2008). Proteasomal degradation of SnRK1α1 was also shown to occur in a myoinositol polyphosphate 5-phosphatase 13 (5PTase13)-dependent manner (Ananieva et al., 2008). Whether PRL1, 5PTase13, and SIZ1 act in same pathway or in parallel ones is currently unknown.
We show here that SUMO-mediated degradation of SnRK1 is strictly dependent on SnRK1 kinase activity. Inactive SnRK1α1 variants are not degraded (Figure 5b), accumulating to much higher levels than the active kinase (Baena-Gonzalez et al., 2007) (Figure 5a). The reason for the increased stability of the inactive SnRK1α1 variants appears to be the lack of SUMOylation, as they recover normal turnover when expressed as SUMO mimetics (Figure 5d). Activated kinases can be recognized for ubiquitination and degradation by different mechanisms, often involving the generation of recognition motifs through phosphorylation or the exposure of such motifs through conformational changes (Lu and Hunter, 2009). An intriguing question that remains open is how the SUMO machinery recognizes active SnRK1.
SUMOylation has also been described for AMPK and SNF1, affecting these kinases in various ways. In AMPK, both catalytic (α1/α2) subunits are within a list of ~1600 human proteins found to be SUMOylated using high-resolution MS (Hendriks et al., 2014). AMPKα1 SUMOylation was detected in control samples and was significantly increased in response to SUMO protease inhibition, proteasome inhibition and heat shock treatments, suggesting that, likewise SnRK1 SUMOylation, it might also be involved in signal termination. Interestingly, the lysine identified by Hendriks and colleagues corresponds to K390 in SnRK1α1, also found SUMOylated in our study (Figure S4). Another recent report showed that also AMPKβ2 is SUMOylated (Rubio et al., 2013). However, SUMOylation of AMPKβ2 did not affect its degradation rate but instead enhanced AMPK activity by increasing T-loop phosphorylation. Furthermore, AMPKβ2 SUMOylation and ubiquitination appeared to be antagonistic rather than interdependent processes. In the case of SNF1, the Snf1 catalytic subunit was reported to be SUMOylated in response to glucose and to downregulate SNF1 independently of T-loop phosphorylation in two ways (Simpson-Lavy and Johnston, 2013). Firstly, SUMOylation of Snf1 caused a rapid inhibition of the kinase specific activity, presumably via a conformational switch induced by the interaction between the SUMOylated residue and a SUMO-interacting motif (SIM). It is unlikely that such a mechanism is conserved as the target lysine (K549) implicated in this regulation is not conserved (Figure S7b). However, the possibility that the conserved SIM1 (Figure S7a, T89–V94 in SnRK1α1) plays a role in SnRK1 SUMOylation remains to be established. Furthermore, whilst SNF1 is SUMOylated on a single residue on the catalytic subunit, SnRK1 seems to be SUMOylated on multiple residues/subunits. Secondly, SUMOylation of Snf1 upon glucose feeding induced its degradation, from which the authors concluded that SUMOylation is important to reduce SNF1 levels when cells do no longer need SNF1 activity. Nevertheless, the fact that the Mms21 SUMO (E3) ligase mutant accumulates higher Snf1 levels under low glucose conditions (Simpson-Lavy and Johnston, 2013) suggests that, likewise SnRK1 and AMPK, SNF1 SUMOylation may also prevent pathway hyperactivation when SNF1 is active.
In summary, our work has uncovered a negative feedback loop by which SnRK1 activity triggers its own SUMO-mediated proteasomal degradation. We postulate that this intimate connection between kinase activity and accumulation is evolutionarily conserved and that it may be essential for balancing stress and defense responses with biosynthetic activities, cell proliferation, and growth (Baena-Gonzalez and Sheen, 2008; Huot et al., 2014).
EXPERIMENTAL PROCEDURES
Primers and constructs
A list of all primers, cloning steps, and constructs used in this study is provided in Table S1.
Plant material and growth conditions
Unless otherwise specified, plants were grown in soil under a 12 h light (100μE), 22°C/12h dark, 18°C regime.
All Arabidopsis thaliana plants used in this study are in the Columbia (Col-0) background. The siz1-2 (Miura et al., 2005) and 35S::SnRK1α1 [(Jossier et al., 2009), 35S::SnRK1.1-2] plants have been previously described. The generation of 35S::GFP, SnRK1α1–GFP, SnRK1α1ΔKA1–GFP and SnRK1α1–GFPsiz1-2 lines is fully described in Methods S1.
GFP, SnRK1α1–GFP and SnRK1α1ΔKA1–GFP immunoprecipitation
Proteins from 5-week-old SnRK1α1–GFP, SnRK1α1ΔKA1–GFP, SnRK1α1–GFPsiz1-2 or 35S::GFP plant leaves were extracted with immunoprecipitation (IP) buffer [50 mM Tris–HCl pH 8.0, 50 mm NaCl, 1% (V/V) Igepal CA-630, 0.5% (w/V) sodium deoxycholate, 0.1% (w/V) SDS, 1 mM EDTA pH 8.0, 50 μM MG132, 50 mM N-ethylmaleimide and cOmplete protease inhibitor cocktail (one tablet/10 mL)]. After clearing samples by centrifugation (6785 g, 2°C, 10 min) 800 μL of supernatant were supplemented with fresh MG132 (50 μm) and incubated at 4°C for 1 h with 40 μL of μMACS anti–GFP MicroBeads (μMACS GFP Isolation Kit, Miltenyi, 130-091-125). Samples were thereafter loaded in μColumns (Miltenyi Biotec, Bergisch Gladbach, Germany 130-042-701) pre-equilibrated with 1 mL of IP buffer, and allowed to flow through. Columns were washed three times with 200 μL and once with 600 μL of IP buffer and proteins eluted with 80 μL of elution buffer (Miltenyi, 130-091-125) at 95°C. β-Mercaptoethanol (2%) was added to the eluates prior to boiling for 5 min at 95°C. Proteins were resolved by SDS-PAGE, wet-transferred to a PVDF membrane (30 V, 16 h at 4°C), and analysed by immunoblotting with SnRK1α1, SnRK1β1, SnRK1γ, SnRK1βγ, SUMO1, UBQ11 and GFP antibodies. For each GFP immunoprecipitation experiment, immunodetection with different antibodies was done using equal loading on independent membranes.
SnRK1α1 immunoprecipitation and in vitro kinase assays
For measurements of endogenous SnRK1α1 activity, SnRK1α1 was immunoprecipitated from leaves of 5-week-old Col-0 or siz1-2 plants. Plant material (1 g) was extracted in 2 volumes of buffer C [50 mm HEPES, pH 7.25, 150 mm NaCl, 1 mm EDTA, 0.05% Triton X-100, cOmplete protease inhibitor cocktail (one tablet/50 mL, Roche 11697498001) and 1/500 (v/v) phosphatase inhibitor 2 (Sigma P5726) and 3 (Sigma P0044)]. After two successive centrifugations (20 000 g, 4°C, 10 min), the supernatant was recovered and filtered (0.45 μm) and 1 mg of total protein (quantified using the Bradford protein assay) was incubated with gentle shaking for 3 h at 4°C with 15 μL beads of protein A–antibody complex prepared as follows. For each immunoprecipitation, 15 μL (bed volume) of protein A–agarose (Roche #11719408001) was equilibrated in 1× PBS (Sigma-Aldrich P5493) and incubated with 1.5 μg of anti-SnRK1α1 antibody (anti-AKIN10) in 500 μL of 1× PBS for 1 h at room temperature with gentle shaking. After three washes in buffer C, the beads were used for immunoprecipitation. After incubation for 3 h at 4°C under shaking, the beads were washed five times with buffer C, and one-third (5 μL) was kept for immunoblot analyses with an anti-SnRK1α1 antibody. The remaining 10 μL were used to determine the specific activity of SnRK1 in 30 μL of kinase buffer (50 mm HEPES-NaOH, pH 7.25, 20 mm MgCl2, 0.5 mM DTT and 100 mm ATP) with His-ΔC ABF2 (1 μg) for 1 h at 30°C in the presence of 2 μCi of γ32P-ATP. The reaction products were resolved by 12% SDS-PAGE and detected using a phosphor image system (STORM 860, GE Healthcare, Little Chalfont, United Kingdom).
Antibodies and western-blotting
The SnRK1α1 (1/500, anti-AKIN10, AS10919), SnRK1β1 (1/500, anti-AKINB1, AS09460), SnRK1βγ (1/1000, anti-AKINBG, AS09463, Figure S6) and SnRK1γ (1/2000, anti-AKING1, AS09613) were purchased from Agrisera (Vännäs, Sweden). Phospho-SnRK1α1/2 (T175/176) was detected with an anti–phosphoT172-AMPKα antibody (1/1000 in 5% BSA-TBS-Tween, referred to as P-AMPK; #2535, Cell Signaling Technologies, Danvers, MA, USA). SUMO1 (1/5000, ab5316, Abcam, Cambridge, United Kingdom) and Ubiquitin11 (1/10000, AS08307, Agrisera) antibodies were used to detect the respective protein modifications. Anti-HA (1/1000, Roche, #11867423001), anti-GFP (1/1000, 11814460001, Roche, Basel, Switzerland) and anti-T7 (1/10 000, #69522-3, Novagen, a brand by Merck Biosciences, Darmstadt, Germany) antibodies were used to detect the corresponding tagged proteins.
For immunoblotting all primary antibodies were diluted in 1% non-fat milk in Tris-buffered saline (TBS) (unless otherwise stated) and incubated with the membrane under gentle shaking for 12 h at 4°C. Secondary antibodies (Jackson ImmunoResearch Laboratory, Inc.) were used at 1/10 000 in 1% non-fat milk in TBS for 1 h at RT. In the case of immunoprecipitated samples, secondary antibodies subsequently used in the immunodetection were against the light chain of IgG.
Protoplast transient expression assays
Protoplasts from Col-0 and siz1-2 plants were isolated and transfected as already described (Yoo et al., 2007). All effector constructs (Table S1) were generated by cloning the corresponding coding sequences into a pHBT95 vector harboring a C-terminal HA or GFP tag (Yoo et al., 2007), except for C-terminal fusions with mSUMO1AA, for which the GFP/HA tag was replaced with mSUMO1AA. The indicated mutations were introduced by site-directed mutagenesis and verified by sequencing. SnRK1 signaling was monitored using a pDIN6::LUC reporter, and the pUBQ10::β-glucuronidase reporter as transfection efficiency control (Baena-Gonzalez et al., 2007). For SA treatment, transfected protoplasts were incubated for 4 h to allow protein expression and were thereafter treated with SA (5μm) or ethanol (mock) for 2 h or overnight. For analyses of protein degradation proteins were expressed for 6 h, CHX (100 μm) and/or MG132 (50 μm) were added, and cells were thereafter harvested at the indicated time points. Frozen cell pellets were resuspended in 4× Laemmli solubilization buffer, boiled for 5 min at 95°C and analysed by western blot.
Gene expression analyses
Fully expanded rosettes of 4–5-week-old Col-0 and siz1-2 plants were incubated on sterile MilliQ water in Petri dishes under control (3 h light, 100 μE) or SnRK1 signaling activating conditions (3 h dark). The treatment was always initiated 3 h after the onset of the light period. Total RNA was extracted using TRIzol reagent (Life Technologies, a Thermo Fisher Scientific brand, Waltham, MA, USA), treated with RNase-Free DNase (Promega, Madison, USA), and reverse transcribed (1 μg) using SuperScript III Reverse Transcriptase (Life Technologies), as previously described (Rodrigues et al., 2013). Quantitative real-time RT-PCR (qPCR) analyses were performed using a CFX384TM Real-Time System (Bio-Rad), the iTaq™ Universal SYBR® Green Supermix (Bio-Rad), and the 2−ΔΔCt method for relative quantification (Livak and Schmittgen, 2001). Expression values of SnRK1 target genes were normalized using the CT values obtained for the ACT2 reference gene.
Supplementary Material
SnRK1α1 interacts with the SUMO E2 conjugating enzyme 1 (SCE1) in a yeast two-hybrid assay.
List of primers used in this study.
SnRK1γ is SUMOylated in E. coli and enriched in siz1-2, but is not part of the SnRK1α1 complex in Arabidopsis leaves.
Generation of SnRK1α1–GFP transgenic lines.
SnRK1α1 residues found SUMOylated in the E. coli assay.
Salicylic acid (SA) has no effect on SnRK1 signaling.
Specificity of the anti-SnRK1βγ antibody.
Conservation in SnRK1α1 of the residues implicated in SNF1 SUMOylation.
Supplemental experimental procedures.
ACKNOWLEDGEMENTS
We thank Vera Nunes for excellent plant management and K. Miura, K. Tanaka, and M. Thomas for kindly providing the siz1-2 mutant, the plasmids for the E. coli SUMOylation assay, and the constructs for expression of recombinant SnRK1 subunits, respectively. This work was supported by the Austrian Science Foundation FWF (grant P25488 to AB and P23435 to MT); the EMBO Installation program (to EBG) and grant PTDC/BIA-PLA/3937/2012 (to EBG), UID/Multi/04551/2013_Research unit GREEN-it_’Bioresources for Sustainability’ (to EBG), SFRH/BD/51627/2011 (to LM), SFRH/BPD/79255/2011 (to PC) from Fundação para a Ciência e a Tecnologia.
Footnotes
ACCESSION NUMBERS
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: SnRK1α1, At3g01090; SnRK1α2, At3g29160; SnRK1β1, At5g21170; SnRK1β2, At4g16360; SnRK1γ, At3g48530; SnRK1βγ, At1g09020; GBF5, At2g18160; SIZ1, At5g60410; HPY2, At3g15150; SUMO1, At4g26840; SUMO3, At5g55170; SCE1, At3g57870; DIN6, At3g47340; AXP, At2g33830; TPS8, At1g70290; ACT2, At3g18780; UBQ10, At4g05320.
The authors declare no conflict of interest.
REFERENCES
- Ananieva EA, Gillaspy GE, Ely A, Burnette RN, Erickson FL. Interaction of the WD40 domain of a myoinositol polyphosphate 5-phosphatase with SnRK1 links inositol, sugar, and stress signaling. Plant Physiol. 2008;148:1868–1882. doi: 10.1104/pp.108.130575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Gonzalez E, Sheen J. Convergent energy and stress signaling. Trends Plant Sci. 2008;13:474–482. doi: 10.1016/j.tplants.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J. A central integrator of transcription networks in plant stress and energy signalling. Nature. 2007;448:938–942. doi: 10.1038/nature06069. [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Salchert K, Bako L, Okresz L, Szabados L, Muranaka T, Machida Y, Schell J, Koncz C. Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc Natl Acad Sci U S A. 1999;96:5322–5327. doi: 10.1073/pnas.96.9.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro PH, Tavares RM, Bejarano ER, Azevedo H. SUMO, a heavyweight player in plant abiotic stress responses. Cell. Mol. Life Sci. 2012;69:3269–3283. doi: 10.1007/s00018-012-1094-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong MS, Park HC, Hong MJ, Lee J, Choi W, Jin JB, Bohnert HJ, Lee SY, Bressan RA, Yun DJ. Specific domain structures control abscisic acid-, salicylic acid-, and stress-mediated SIZ1 phenotypes. Plant Physiol. 2009;151:1930–1942. doi: 10.1104/pp.109.143719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YH, Hong JW, Kim EC, Yoo SD. Regulatory functions of SnRK1 in stress-responsive gene expression and in plant growth and development. Plant Physiol. 2012;158:1955–1964. doi: 10.1104/pp.111.189829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coello P, Hirano E, Hey SJ, Muttucumaru N, Martinez-Barajas E, Parry MA, Halford NG. Evidence that abscisic acid promotes degradation of SNF1-related protein kinase (SnRK1) in wheat and activation of a putative calcium-dependent SnRK2. J. Exp. Bot. 2012;63:913–924. doi: 10.1093/jxb/err320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozet P, Margalha L, Confraria A, Rodrigues A, Martinho C, Adamo M, Elias CA, Baena-Gonzalez E. Mechanisms of regulation of SNF1/AMPK/SnRK1 protein kinases. Front Plant Sci. 2014;5:190. doi: 10.3389/fpls.2014.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby N, Coupland G. Proteome-wide screens for small ubiquitin-like modifier (SUMO) substrates identify Arabidopsis proteins implicated in diverse biological processes. Proc Natl Acad Sci U S A. 2010;107:17415–17420. doi: 10.1073/pnas.1005452107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrouby N, Bonequi MV, Porri A, Coupland G. Identification of Arabidopsis SUMO-interacting proteins that regulate chromatin activity and developmental transitions. Proc Natl Acad Sci U S A. 2013;110:19956–19961. doi: 10.1073/pnas.1319985110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelle S, Hossain MI, Moller IE, et al. SnRK1 from Arabidopsis thaliana is an atypical AMPK. Plant J. 2015;82:183–192. doi: 10.1111/tpj.12813. [DOI] [PubMed] [Google Scholar]
- Fragoso S, Espindola L, Paez-Valencia J, Gamboa A, Camacho Y, Martinez-Barajas E, Coello P. SnRK1 isoforms AKIN10 and AKIN11 are differentially regulated in Arabidopsis plants under phosphate starvation. Plant Physiol. 2009;149:1906–1916. doi: 10.1104/pp.108.133298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 2007;8:947–956. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- Guo C, Henley JM. Wrestling with stress: Roles of protein SUMOylation and deSUMOylation in cell stress response. IUBMB Life. 2014;66:71–77. doi: 10.1002/iub.1244. [DOI] [PubMed] [Google Scholar]
- Hao L, Wang H, Sunter G, Bisaro DM. Geminivirus AL2 and L2 proteins interact with and inactivate SNF1 kinase. Plant Cell. 2003;15:1034–1048. doi: 10.1105/tpc.009530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks IA, D’Souza RC, Yang B, Verlaan-de Vries M, Mann M, Vertegaal AC. Uncovering global SUMOylation signaling networks in a site-specific manner. Nat. Struct. Mol. Biol. 2014;21:927–936. doi: 10.1038/nsmb.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY. Growth-defense tradeoffs in plants: a balancing act to optimize fitness. Mol Plant. 2014;7:1267–1287. doi: 10.1093/mp/ssu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T, Fujiwara S, Miura K, Stacey N, Yoshimura M, Schneider K, Adachi S, Minamisawa K, Umeda M, Sugimoto K. SUMO E3 ligase HIGH PLOIDY2 regulates endocycle onset and meristem maintenance in Arabidopsis. Plant Cell. 2009;21:2284–2297. doi: 10.1105/tpc.109.068072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S, Psakhye I. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu. Rev. Genet. 2013;47:167–186. doi: 10.1146/annurev-genet-111212-133453. [DOI] [PubMed] [Google Scholar]
- Jossier M, Bouly JP, Meimoun P, Arjmand A, Lessard P, Hawley S, Grahame Hradie D, Thomas M. SnRK1 (SNF1-related kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J. 2009;59:316–328. doi: 10.1111/j.1365-313X.2009.03871.x. [DOI] [PubMed] [Google Scholar]
- Kim D-Y, Scalf M, Smith LM, Vierstra RD. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. The Plant Cell Online. 2013;25:1523–1540. doi: 10.1105/tpc.112.108613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinow T, Bhalerao R, Breuer F, Umeda M, Salchert K, Koncz C. Functional identification of an Arabidopsis snf4 ortholog by screening for heterologous multicopy suppressors of snf4 deficiency in yeast. Plant J. 2000;23:115–122. doi: 10.1046/j.1365-313x.2000.00809.x. [DOI] [PubMed] [Google Scholar]
- Lastdrager J, Hanson J, Smeekens S. Sugar signals and the control of plant growth and development. J. Exp. Bot. 2014;65:799–807. doi: 10.1093/jxb/ert474. [DOI] [PubMed] [Google Scholar]
- Lee J, Nam J, Park HC, et al. Salicylic acid-mediated innate immunity in Arabidopsis is regulated by SIZ1 SUMO E3 ligase. Plant J. 2007;49:79–90. doi: 10.1111/j.1365-313X.2006.02947.x. [DOI] [PubMed] [Google Scholar]
- Lee JH, Terzaghi W, Gusmaroli G, Charron JB, Yoon HJ, Chen H, He YJ, Xiong Y, Deng XW. Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell. 2008;20:152–167. doi: 10.1105/tpc.107.055418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KW, Chen PW, Lu CA, Chen S, Ho TH, Yu SM. Coordinated responses to oxygen and sugar deficiency allow rice seedlings to tolerate flooding. Sci. Signal. 2009;2:ra61. doi: 10.1126/scisignal.2000333. [DOI] [PubMed] [Google Scholar]
- Lin CR, Lee KW, Chen CY, Hong YF, Chen JL, Lu CA, Chen KT, Ho TH, Yu SM. SnRK1A-interacting negative regulators modulate the nutrient starvation signaling sensor SnRK1 in source-sink communication in cereal seedlings under abiotic stress. Plant Cell. 2014;26:808–827. doi: 10.1105/tpc.113.121939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lovas A, Bimbo A, Szabo L, Banfalvi Z. Antisense repression of StubGAL83 affects root and tuber development in potato. Plant J. 2003;33:139–147. doi: 10.1046/j.1365-313x.2003.016015.x. [DOI] [PubMed] [Google Scholar]
- Lu Z, Hunter T. Degradation of activated protein kinases by ubiquitination. Annu. Rev. Biochem. 2009;78:435–475. doi: 10.1146/annurev.biochem.013008.092711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CA, Lin CC, Lee KW, Chen JL, Huang LF, Ho SL, Liu HJ, Hsing YI, Yu SM. The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell. 2007;19:2484–2499. doi: 10.1105/tpc.105.037887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maor R, Jones A, Nühse TS, Studholme DJ, Peck SC, Shirasu K. Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol. Cell Proteomics. 2007;6:601–610. doi: 10.1074/mcp.M600408-MCP200. [DOI] [PubMed] [Google Scholar]
- Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD. Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci U S A. 2010;107:16512–16517. doi: 10.1073/pnas.1004181107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Hasegawa PM. Sumoylation and other ubiquitin-like post-translational modifications in plants. Trends Cell Biol. 2010;20:223–232. doi: 10.1016/j.tcb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, et al. The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci U S A. 2005;102:7760–7765. doi: 10.1073/pnas.0500778102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Lee J, Miura T, Hasegawa PM. SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol. 2010;51:103–113. doi: 10.1093/pcp/pcp171. [DOI] [PubMed] [Google Scholar]
- Novatchkova M, Tomanov K, Hofmann K, Stuible HP, Bachmair A. Update on sumoylation: defining core components of the plant SUMO conjugation system by phylogenetic comparison. New Phytol. 2012;195:23–31. doi: 10.1111/j.1469-8137.2012.04135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S, Nagabuchi M, Takamura Y, Nakagawa T, Shinmyozu K, Nakayama J, Tanaka K. Reconstitution of Arabidopsis thaliana SUMO pathways in E. coli: functional evaluation of SUMO machinery proteins and mapping of SUMOylation sites by mass spectrometry. Plant Cell Physiol. 2009;50:1049–1061. doi: 10.1093/pcp/pcp056. [DOI] [PubMed] [Google Scholar]
- Park HJ, Kim WY, Park HC, Lee SY, Bohnert HJ, Yun DJ. SUMO and SUMOylation in plants. Mol. Cells. 2011;32:305–316. doi: 10.1007/s10059-011-0122-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polge C, Thomas M. SNF1/AMPK/SnRK1 kinases, global regulators at the heart of energy control? Trends Plant Sci. 2007;12:20–28. doi: 10.1016/j.tplants.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Polge C, Jossier M, Crozet P, Gissot L, Thomas M. Beta-subunits of the SnRK1 complexes share a common ancestral function together with expression and function specificities; physical interaction with nitrate reductase specifically occurs via AKINbeta1-subunit. Plant Physiol. 2008;148:1570–1582. doi: 10.1104/pp.108.123026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke GJ, Hofmann K, Dohmen RJ. SUMO playing tag with ubiquitin. Trends Biochem. Sci. 2012;37:23–31. doi: 10.1016/j.tibs.2011.09.002. [DOI] [PubMed] [Google Scholar]
- Radchuk R, Radchuk V, Weschke W, Borisjuk L, Weber H. Repressing the expression of the SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiol. 2006;140:263–278. doi: 10.1104/pp.105.071167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk R, Emery RJ, Weier D, Vigeolas H, Geigenberger P, Lunn JE, Feil R, Weschke W, Weber H. Sucrose non-fermenting kinase 1 (SnRK1) coordinates metabolic and hormonal signals during pea cotyledon growth and differentiation. Plant J. 2010;61:324–338. doi: 10.1111/j.1365-313X.2009.04057.x. [DOI] [PubMed] [Google Scholar]
- Ramon M, Ruelens P, Li Y, Sheen J, Geuten K, Rolland F. The hybrid Four-CBS-domain KINbetagamma-subunit functions as the canonical gamma~subunit of the plant energy sensor SnRK1. Plant J. 2013;75:11–25. doi: 10.1111/tpj.12192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues A, Adamo M, Crozet P, et al. ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis. Plant Cell. 2013;25:3871–3884. doi: 10.1105/tpc.113.114066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio T, Vernia S, Sanz P. Sumoylation of AMPKb2 subunit enhances AMP-activated protein kinase activity. Mol. Biol. Cell. 2013;24:1801–1811. doi: 10.1091/mbc.E12-11-0806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Xu X, Carlson M. Roles of two protein phosphatases, Reg1-Glc7 and Sit4, and glycogen synthesis in regulation of SNF1 protein kinase. Proc Natl Acad Sci U S A. 2011;108:6349–6354. doi: 10.1073/pnas.1102758108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz A, Xu X, Carlson M. Ptc1 protein phosphatase 2C contributes to glucose regulation of SNF1/AMP-activated protein kinase (AMPK) in Saccharomyces cerevisiae. J. Biol. Chem. 2013;288:31052–31058. doi: 10.1074/jbc.M113.503763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saracco SA, Miller MJ, Kurepa J, Vierstra RD. Genetic analysis of SUMOylation in Arabidopsis: conjugation of SUMO1 and SUMO2 to nuclear proteins is essential. Plant Physiol. 2007;145:119–134. doi: 10.1104/pp.107.102285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwachtje J, Minchin PE, Jahnke S, van Dongen JT, Schittko U, Baldwin IT. SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proc Natl Acad Sci U S A. 2006;103:12935–12940. doi: 10.1073/pnas.0602316103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson-Lavy KJ, Johnston M. SUMOylation regulates the SNF1 protein kinase. Proc Natl Acad Sci U S A. 2013;110:17432–17437. doi: 10.1073/pnas.1304839110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden C, Donaghy PG, Halford NG, Hardie DG. Two SNF1-related protein kinases from spinach leaf phosphorylate and inactivate 3-hydroxy-3-methylglutaryl-coenzyme A reductase, nitrate reductase, and sucrose phosphate synthase in vitro. Plant Physiol. 1999;120:257–274. doi: 10.1104/pp.120.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelander M, Olsson T, Ronne H. Snf1-related protein kinase 1 is needed for growth in a normal day-night light cycle. EMBO J. 2004;23:1900–1910. doi: 10.1038/sj.emboj.7600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomanov K, Zeschmann A, Hermkes R, Eifler K, Ziba I, Grieco M, Novatchkova M, Hofmann K, Hesse H, Bachmair A. Arabidopsis PIAL1 and 2 promote SUMO chain formation as E4-Type SUMO ligases and are involved in stress responses and sulfur metabolism. Plant Cell. 2014;26:4547–4560. doi: 10.1105/tpc.114.131300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AY, Gazzarrini S. AKIN10 and FUSCA3 interact to control lateral organ development and phase transitions in Arabidopsis. Plant J. 2012;69:809–821. doi: 10.1111/j.1365-313X.2011.04832.x. [DOI] [PubMed] [Google Scholar]
- Ulrich HD. The fast-growing business of SUMO chains. Mol. Cell. 2008;32:301–305. doi: 10.1016/j.molcel.2008.10.010. [DOI] [PubMed] [Google Scholar]
- Ulrich HD. SUMO Protocols. Humana Press; New York: 2009. [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SnRK1α1 interacts with the SUMO E2 conjugating enzyme 1 (SCE1) in a yeast two-hybrid assay.
List of primers used in this study.
SnRK1γ is SUMOylated in E. coli and enriched in siz1-2, but is not part of the SnRK1α1 complex in Arabidopsis leaves.
Generation of SnRK1α1–GFP transgenic lines.
SnRK1α1 residues found SUMOylated in the E. coli assay.
Salicylic acid (SA) has no effect on SnRK1 signaling.
Specificity of the anti-SnRK1βγ antibody.
Conservation in SnRK1α1 of the residues implicated in SNF1 SUMOylation.
Supplemental experimental procedures.