ABSTRACT
Colistin is an ultimate line of refuge against multidrug-resistant Gram-negative pathogens. Very recently, the emergence of plasmid-mediated mcr-1 colistin resistance has become a great challenge to global public health, raising the possibility that dissemination of the mcr-1 gene is underestimated and diversified. Here, we report three cases of plasmid-carried MCR-1 colistin resistance in isolates from gut microbiota of diarrhea patients. Structural and functional analyses determined that the colistin resistance is conferred purely by the single mcr-1 gene. Genetic and sequence mapping revealed that mcr-1-harbouring plasmid reservoirs are present in diversity. Together, the data represent the first evidence of diversity in mcr-1-harbouring plasmid reservoirs of human gut microbiota.
IMPORTANCE
The plasmid-mediated mobile colistin resistance gene (mcr-1) challenged greatly the conventional idea mentioned above that colistin is an ultimate line of refuge against lethal infections by multidrug-resistant Gram-negative pathogens. It is a possibility that diversified dissemination of the mcr-1 gene might be greatly underestimated. We report three cases of plasmid-carried MCR-1 colistin resistance in isolates from gut microbiota of diarrhea patients and functionally define the colistin resistance conferred purely by the single mcr-1 gene. Genetic and sequence mapping revealed unexpected diversity among the mcr-1-harbouring plasmid reservoirs of human gut microbiota.
INTRODUCTION
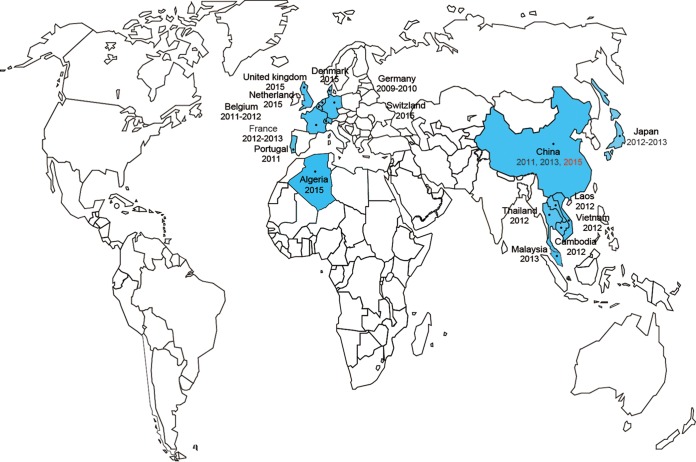
Colistin (polymyxin E), representing a family of cationic polypeptide antibiotics with broad-spectrum antimicrobial activities, is generally regarded as a last line of refuge (drug/therapeutics) against bacterial infections by the multidrug-resistant Gram-negative pathogens (1, 2). The chromosome-encoded mechanism for colistin resistance in certain members of Enterobacteriaceae is associated with two-component systems such as pmrAB (3) and phoPQ (4) and with the regulator mgrB (4), in which the modification of lipid A decreases its affinity to polymyxin (5). Very recently, Liu et al. (5) reported, for the first time, that plasmid-mediated mcr-1 colistin resistance in animal and human isolates of Escherichia coli and Klebsiella pneumoniae has emerged in China (Fig. 1 and 2). They defined an unusual mechanism for colistin resistance in that the mcr-1 gene product belongs to the family of phosphoethanolamine transferase enzymes (Fig. 2b) (5). Our retrospective study showed that the mcr-1 genes have already been detected in no fewer than 16 countries, including 7 countries in Southeast Asia (China [5, 6], Thailand [7], Laos [7], Japan [8], Vietnam [9], Cambodia [10], and Malaysia [6]) (Fig. 1) and 9 European countries (Denmark [11], United Kingdom [England and Wales] [12], the Netherlands [13], France [7, 12], Portugal [6], Switzerland [14], Germany [15], Belgium [16], and Algeria [7]) (Fig. 1). To the best of our knowledge, the plasmid-borne mcr-1 gene has been observed in at least 3 enterobacterial species (E. coli [5, 11, 13], Salmonella enterica [6, 12, 17], and K. pneumoniae [5]) and the host reservoirs included at least three kinds of poultry and livestock (chickens [6, 7], pigs [4, 5], and cattle [6]). Of particular note, animal-to-human transmission of MCR-1 colistin resistance has already been found in China (5), Thailand (7), Laos (11), and Denmark (4), raising serious concern about its possible global dissemination and spread (18). So far, it is very true that plasmid pHNSHP45 from the Chinese swine microbiota (Fig. 2a) is the only one (among hundreds of examples of mcr-1 carriage in animal/human isolates) with the known full genome sequence in China (5).
FIG 1 .
Global distribution of the mcr-1 colistin resistance gene. The countries where the mcr-1 gene was discovered are highlighted in blue.
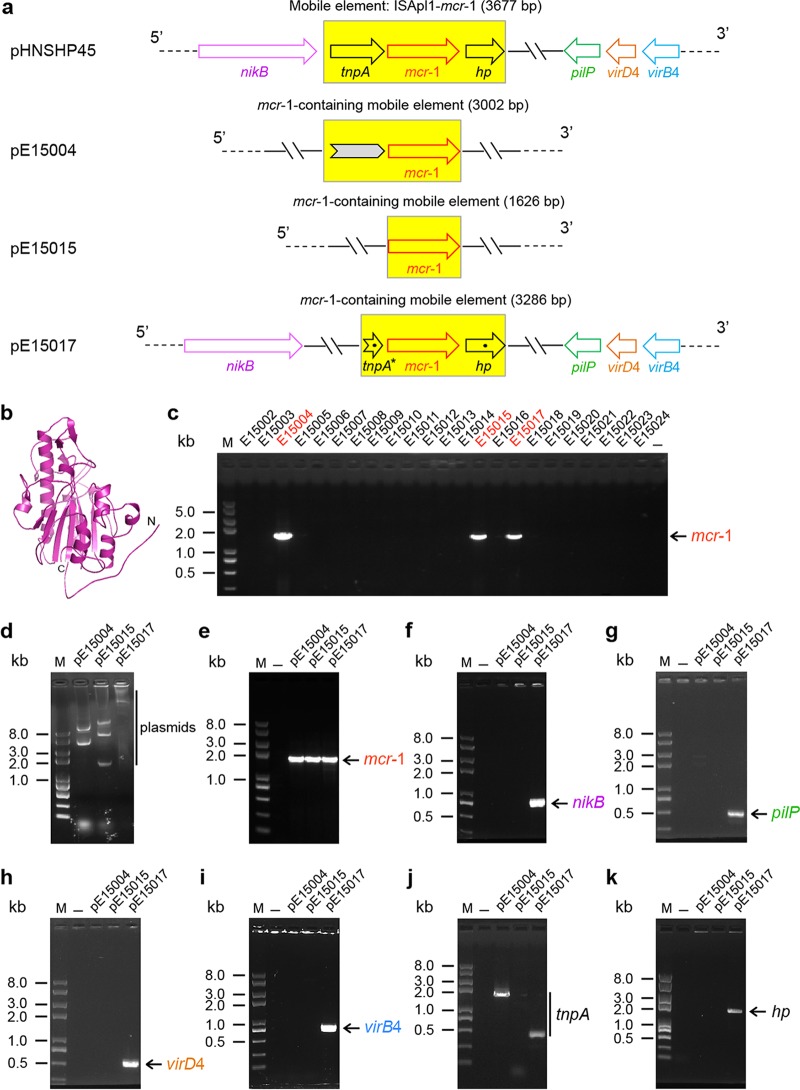
FIG 2 .
Genetic, structural, and molecular characterization of the mcr-1-harbouring plasmids from gut microbiota of diarrhea patients. (a) Scheme for the four mcr-1-harbouring plasmids. pHNSHP45 is a plasmid from the Chinese pig isolate of E. coli with a known genome sequence (5); the other three plasmids (referred to as pE15004, pE15015,and pE15017) were isolated from three clinical E. coli strains (E15004, E15015, and E15017) collected from diarrhea patients admitted to hospitals in Shenzhen City, China, in 2015. Arrows denote the known (and/or putative) genes. A mobile element, ISApl1-mcr-1 (or an mcr-1-containing mobile element), is highlighted with a yellow background. The gray arrow illustrated with plasmid pE15004 (in panel a) represents a DNA fragment of 1,189 bp that shows 100% identity to the equivalent sequence of Salmonella enterica serovar Heidelberg plasmid pSH146-32, whereas the mcr-1 gene of plasmid pE15004 (and/or plasmid pE15015 plus pE15017) is 100% identical to the counterpart gene of E. coli SHP45 strain plasmid pHNSHP45 (5). The mcr-1-harbouring mobile element candidate from the clinical pE15017 plasmid exhibited 99% identity to that of E. coli plasmid pHNSHP45 (5). The broken arrow (tnpA*) denotes the intergenic sequence that is closely next to the tnpA gene at the 3-terminus determined in our trials, and the dots indicate that point mutations are present in tnpA* and a hypothetical protein-encoding locus (hp gene). PCR-based fine mapping suggested that the pE15017 plasmid carries all four genes (nikB, pilP, virD4, and virB4), similarly to E. coli plasmid pHNSHP45, but that such is not the case for the other two clinical mcr-1-harbouring plasmids (pE15004 andpE15015) that we have reported here. The results validated the idea that the diversified plasmid background is linked to a mcr-1 colistin resistance gene. (b) Modeled structure of the enzymatic domain of MCR-1 protein. Structural modeling was performed using the program of Swiss model and Neisseria lipooligosaccharide phosphoethanolamine transferase A (LptA) as the structural template (PDB: 4KAV), and the resultant output (ribbon photograph) was given using PyMol software. (c) mcr-1-based screening of clinical isolates from gut microbiota of diarrhea patients. The expected amplicon of the full-length (~1.6-kb) mcr-1 gene is indicated with a red arrow, and the clinical mcr-1-positive isolates (E15004, E15015, and E15017) are highlighted in red. (d) Profile for plasmids extracted from the clinical mcr-1-positive isolates (E15004, E15015, and E15017) determined on the basis of 0.7% agarose gel electrophoresis. (e to i) Molecular detection of the acquired three plasmids using five pairs of specific primers that separately target the mcr-1 colistin resistance gene (e), nikB (f), pilP (g), and type IV secretion system-encoding genes virD4 (h) and virB4 (i). (j and k) PCR screening for the two neighboring genes of mcr-1, the transposase-encoding tnpA gene (j) and the hypothetical protein (hp)-encoding gene (k). M denotes a Trans2K Plus II DNA ladder (TransGen Biotech, Beijing, China), and minus refers to the negative control.
RESULTS
Discovery of three mcr-1-positive clinical isolates from human gut microbiota.
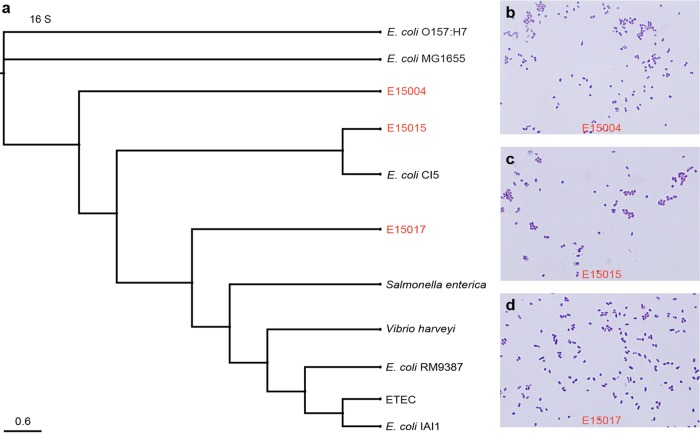
Given the fact that current knowledge on the genetic evolution of both the mcr-1 gene and the mcr-1-harbouring vectors and plasmids is extremely limited (and has even lagged), we anticipated that genetic diversity of mcr-1-carrying plasmid backbones/reservoirs is probably present in gut microbiota of animals as well as human beings. Here we report that this is the case. We analyzed fecal samples collected from diarrhea patients admitted to hospitals in Shenzhen City, China, in 2015. The patients involved in this study comprised neonates (3 months old), adults (18 to 55 years old), and older persons (83 years old) (not shown). Following routine examination procedures such as 16S sequencing, we investigated 48 isolates of Escherichia coli and 27 isolates of K. pneumoniae (not shown). PCR screening for the mcr-1 gene was carried out using a pair of specific primers covering the full length of the mcr-1 coding sequence (1,626 bp). As a result, 3 of 48 clinical E. coli isolates were shown to be mcr-1 positive in our PCR assays (Fig. 2c), whereas none of the human K. pneumoniae isolates were found to be PCR positive for mcr-1 (not shown). To further confirm the identity of the acquired 3 isolates, 16S-based phylogenetic analyses (Fig. 3a), as well as Gram staining assays, were conducted (Fig. 3b to d). In light of the fact that Liu and coworkers successfully acquired a big plasmid (pHNSHP45; 64,105 bp in length) from a pig E. coli isolate (5), we attempted to subject the three mcr-1-positive strains (designated E15004, E15015, and E15017 in Fig. 2c) to plasmid isolation by employing manual extraction with the alkaline lysis method. It seems likely that different plasmids are present in those clinical human strains in that their profiles are completely different, at least in 0.7% agarose gel electrophoresis (Fig. 2d). As expected, all three of the acquired plasmids, namely, pE15004, pE15015, and pE15017, contained the mcr-1 gene, evidenced by our PCR-based determination with these plasmids as the templates (Fig. 2e). Direct DNA sequencing of the three mcr-1 genes (1,626 bp) that we obtained from human clinical isolates showed that they were 100% identical to all the mcr-1 genes of diversified origins with known sequences (such as those carried by pHNSHP45 [5] and even the human gut microbiota contig from N009A [6]), indicating that the mcr-1 colistin resistance gene itself is highly conserved and seems to be under low selective pressure right now.
FIG 3 .
16S-based identification and Gram staining analyses of the three mcr-1-harbouring isolates. (a) 16S-based phylogenetic tree of the three mcr-1-containing isolates. (b) Gram staining analyses for the E15004 isolate. (c) Gram staining analyses for the E15015 isolate. (d) Gram staining analyses for the E15017 isolate.
In vivo evidence for MCR-1-mediated colistin resistance.
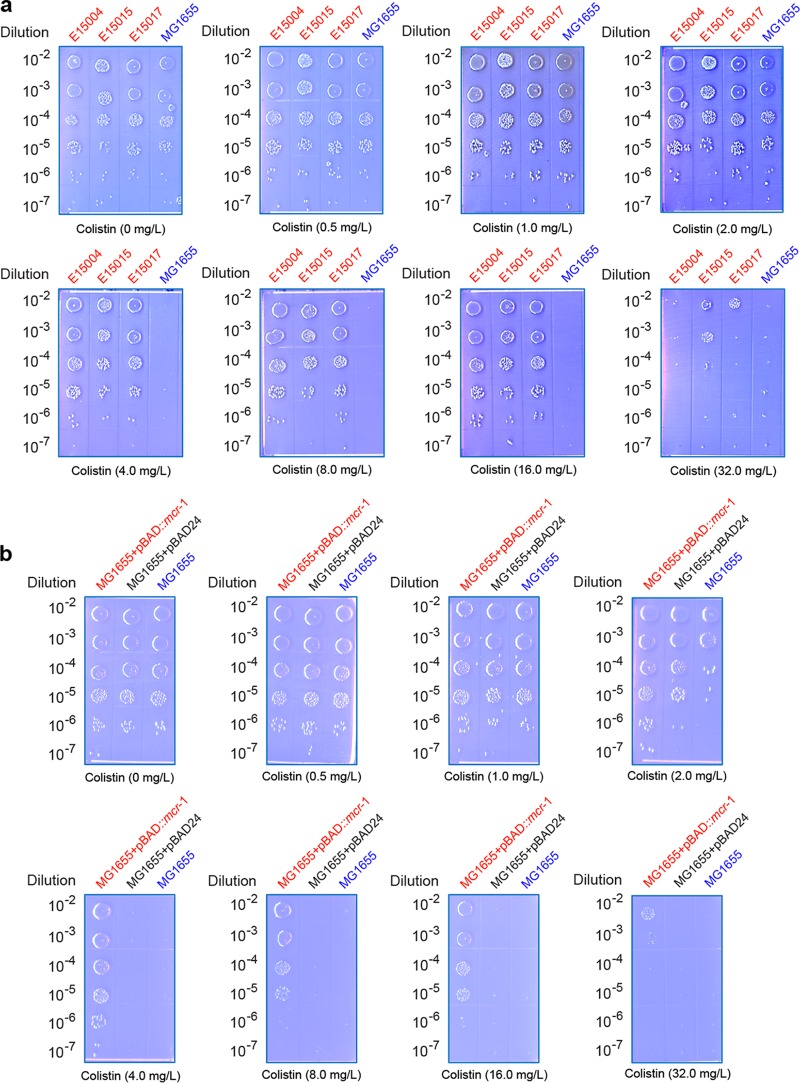
Structural modeling of MCR-1 showed that its architecture is similar to that of Neisseria lipo-oligosaccharide phosphoethanolamine transferase A (Fig. 2c), implying similar enzymatic mechanisms by which bacterial lipid A modification proceeds. To address the function of mcr-1 in vivo, we employed two approaches, one of which was testing the colistin tolerance of the three clinical human Escherichia isolates with a natural mcr-1-positive plasmid and the other of which was visualizing the effect of colistin-susceptible E. coli strain MG1655 by regulated expression of the pure mcr-1 gene. As we expected, the prototypical strain of E. coli MG1655 as the negative control grew on the Luria-Bertani agar (LBA) plates supplemented with no more than 2.0 mg/liter of colistin (Fig. 4a) (note that the resistance of 2.0 mg/liter refers to a diagnostic cutoff/breakpoint [13]). In contrast, the clinical human enteric strains (E15004, E15015, and E15017) consistently exhibited appreciable growth on LBA plates with up to 16 mg/liter of colistin, and the MIC was 32 mg/liter for colistin (Fig. 4a), which is much higher than the 8 mg/liter seen with pHNSHP45 (5). This discrepancy might be in part due to different evaluation methods. When we engineered the mcr-1 gene into an arabinose-inducible expression vector, pBAD24, for E. coli, similar scenarios were observed in our experiments. Although basal expression of mcr-1 (without the addition of any colistin into growth media) confers resistance of the recipient MG1655 strain to 4 mg/liter colistin on LBA plates, induced expression of mcr-1 in the presence of 0.2% arabinose increased its colistin tolerance to 16 mg/liter (Fig. 4b). The results showed that all of the mcr-1 genes in these clinical isolates from gut microbiota of diarrhea patients are functional in colistin resistance.
FIG 4 .
Determination of colistin resistance conferred by the plasmid-borne mcr-1 gene. (a) Determination of colistin MIC of the clinical E. coli isolates from diarrhea patients. (b) Expression of mcr-1 augments resistance of colistin-susceptible strain E. coli MG1655 to the colistin antibiotics. The three clinical mcr-1-positive E. coli strains (E15004, E15015, and E15017) are highlighted in red, whereas the prototypical wild-type strain (also mcr-1-negative strain) of E. coli, MG1655, is referred to the negative control (in blue). To determine the MIC of colistin, the mid-log-phase cultures (optical density at 600 nm [OD600] = 0.7) in serial dilution were spotted on LBA plates supplemented with colistin at various levels (0, 0.5, 1.0, 2.0, 4.0, 8.0, 16.0, and 32.0 mg/liter) and maintained overnight at 37°C. Expression vector pBAD24 is used for functional cloning of the mcr-1 gene in E. coli. Expression of mcr-1 is induced by the addition of 0.2% arabinose to LBA media.
Unexpected diversity in mcr-1-harbouring plasmid reservoirs.
To further figure out whether or not our plasmids harbour key genetic elements similar to those of pHNSHP45, the only mcr-1-harbouring plasmid with a sequenced genome, we designed six more pairs of primers (Table 1) targeting the following six genes (Fig. 2a): first, nikB (Fig. 2f) and pilP (Fig. 2g) separately encode a relaxase for transposal function and a type IV pilus biogenesis protein; second, virD4 (Fig. 2h) and virB4 (Fig. 2i) are two genes encoding two components (VirD4 and VirB4 with an ATPase activity) of a type IV secretion system; third, the two neighboring DNA fragments adjacent to the mcr-1 gene correspond to the tnpA transposase-encoding gene (Fig. 2j) and to a gene encoding a hypothetical protein (hp) (Fig. 2k), respectively. To our surprise, PCR assays showed that only the pE15017 plasmid is positive for the first four genes of those six genes (nikB [Fig. 2f], pilP [Fig. 2g], virD4 [Fig. 2h], and virB4 [Fig. 2i]), suggesting that pE15017 probably has high homology to pHNSHP45, whereas the remaining two plasmids (pE15004 and pE15015 [Fig. 2c]) exhibit a different plasmid backbone in that at least those four genes are lacking (Fig. 2f to i). A similar scenario was also noted in the PCR amplification of the hp gene adjacent to 3′ end of the mcr-1 gene (i.e., the pE15017 plasmid is the only hp gene-positive one, as shown in Fig. 2k). This speculation was also supported by subsequent direct DNA sequencing of the four genes (nikB, pilP, virD4, and virB4) amplified from the pE15017 plasmid in that all of them are 100% identical to the counterparts of pHNSHP45 (Fig. 2a). In contrast, the DNA region covering tnpA, another gene that neighbors the mcr-1 gene, exhibited a differential profile in that the PCR amplicon (~1.5 kb) is present in the pE15004 plasmid, a shortened version of around 0.5 kb (tnpA*) appears in the pE15017 plasmid, but no such band exists in the pE15015 plasmid (Fig. 2j). Further sequence assembly of the DNA fragments overlapping the mcr-1 gene revealed that unexpected diversity is present the mcr-1-harbouring plasmid backbones/reservoirs (Fig. 2a). First, as in the case of the mcr-1-containing mobile element (note that only a 3,002-bp sequence is available right now), use of the Basic Local Alignment Search Tool (BLAST) delineated that the sequence upstream of the mcr-1 gene (1,189 bp) in the pE15004 plasmid is 100% identical to that in S. enterica serovar Heidelberg plasmid pSH146-32, whereas the remaining mcr-1 part completely matches the counterpart of pHNSHP45 (5) (Fig. 2a). Second, the adjacent sequences of mcr-1 similar to those of pHNSHP45 are absent in the pE15004 plasmid (Fig. 2a). Third, the mcr-1-barboring mobile sequence (note that only a 3,286-bp sequence is acquired) in pE15004 showed 99% identity to the equivalent part of pHNSHP45 (5) in that point mutations are present in the shortened version of tnpA (namely, tnpA*) and in the hp gene (Fig. 2a). Our findings provided direct molecular evidence that the mcr-1-carrying plasmid backbones/reservoirs present in the gut microbiota of diarrhea patients are of diversified/hybrid origins, which is somewhat consistent with an in silico speculation by Tse and Yuen (17) in a study of Salmonella and validates a similar hypothesis by Arcilla et al. (13).
TABLE 1 .
Primers used in this studya
| Primer | Primer sequence | Target gene (bp) |
|---|---|---|
| 16S-F | 5′ AAATTGAAGAGTTTGATCATGG 3′ | 16S rRNA gene (1,554) |
| 16S-R | 5′ GCTTCTTTAAGGTAAGGAGGT 3′ | |
| mcr-1-F | 5′ ATGATGCAGCATACTTCTGTG 3′ | mcr-1 (1,626) |
| mcr-1-R | 5′ TCAGCGGATGAATGCGGTG 3′ | |
| nikB-F | 5′ GATGAACTTGATCATCGTGTTGT 3′ | nikB (705) |
| nikB-R | 5′ GTAATTCTGACGAAAAAGAGGA 3′ | |
| pilP-F | 5′ TTAAAGAATAAGCTGGCGTTTC 3′ | pilP (495) |
| pilP-R | 5′ ATGTTAAAAATAATTAAACCAACG 3′ | |
| virD4-F | 5′ AATGTCAACATGATTGTTAC 3′ | virD4 (552) |
| virD4-R | 5′ GAACATAACCCGGACCTGAAAT 3′ | |
| virB4-F | 5′ AACTCTTTTTCAGTAAGCCCAAT 3′ | virB4 (780) |
| virB4-R | 5′ TTAATGTTTGTTGTGGATTACAACC 3′ | |
| tnpA-F | 5′ GGT TTT CGG GCT TTT TAA GAG 3′ | tnpA (1,504) |
| tnpA-R | 5′ TAG CAC ATA GCG ATA CGA TG 3′ | |
| hp-F | 5′ GAT AAG CAA ACT GGC ATC ACG 3′ | hp gene (1,646) |
| hp-R | 5′ GAA CCC TGT ATA TAG CCT GTC 3′ |
All of the primers listed originated in this study.
DISCUSSION
In summary, it is reasonable to surmise that diversified transfer of plasmid-mediated mcr-1 colistin resistance might be present, and confirmation requires further epidemiological investigations. Given that (i) colistin as a veterinary medicine is extensively applied in agricultural (poultry and livestock) production worldwide, (ii) widespread distribution of the mcr-1 colistin resistance gene by transposal elements/plasmids occurs in animals, meat/food samples, and human gut microbiota, (iii) the relatively widespread occurrence of mcr-1 in isolates from no fewer than 10 countries is known right now, and (iv) colistin resistance represents a major breach in our last line of defense against multidrug-resistant bacterial pathogens, it might greatly necessary to clinically monitor the diversified mcr-1-harbouring plasmids/reservoirs, reevaluate the efficacy (safety) of colistin in veterinary use, and formulate a comprehensive strategy to fight against plasmid-mediated mcr-1 colistin resistance, especially in pan-drug-resistant Gram-negative bacterial strains (19).
MATERIALS AND METHODS
Bacterial isolations and identification.
Fecal samples were routinely collected from diarrhea patients admitted to hospitals in Shenzhen City, China, in 2015. Luria-Bertani (LB) liquid media (and/or solid agar plates) were applied to isolate the enterobacterial species. To initially determine the bacterial identity, the acquired bacteria were routinely subjected to biochemical tests as well as to colony PCR assays with 16S-specific primers. Gram staining assays were routinely performed. As a result, either Escherichia coli or Klebsiella pneumoniae was predominantly assigned to the gut microbiota of the diarrhea patients.
DNA manipulations.
To probe the presence of the mcr-1 gene, all of the bacterial isolates harvested (75 in total [48 for E. coli and 27 for K. pneumoniae]) were screened via PCR-based diagnostics with mcr-1-specific primers (Table 1). The mcr-1-positive bacteria were further subjected to plasmid isolations with the manual alkaline lysis method. Then, the plasmid-harbouring isolates were dissected finely using multiplex PCR with six pairs of specific primers targeting six different loci (such as virD4 and virB4, encoding a type IV secretion system) (Table 1). All of the PCR-amplified DNA fragments were verified by direct DNA sequencing and analyzed by ClustalW-aided multiple-sequence alignments. The mcr-1 gene was cloned into the arabinose-inducible expression vector pBAD24 in E. coli (20), giving the recombinant plasmid pBAD24::mcr-1.
Determination of colistin MIC.
Mid-log-phase bacterial cultures in a dilution series were spotted on LBA plates supplemented with various levels of colistin (0, 0.5, 1.0, 2.0, 4.0, 8.0, 16.0, and 32.0 mg/liter) and kept at 37°C overnight. In the assays of inhibitory colistin concentrations, the findings for all three of the mcr-1-containing strains (E15004, E15015, and E15017) were confirmed, whereas the negative control corresponded to MG1655, the prototypical strain of E. coli. To further examine the function of the single mcr-1 gene in vivo, we monitored the resistance of the colistin-susceptible MG1655 strain conferred by expression of arabinose-inducible, pBAD24::mcr-1-borne mcr-1.
Bioinformatics analyses.
The acquired 16S sequences in full length were subjected to phylogenetic analyses using the webserver of Clustal Omega. The coding sequences of the mcr-1 gene and the other target genes such as virD4 and virB4 were aligned with their counterparts with known sequences using BLAST.
ACKNOWLEDGMENTS
We would like to thank the startup package from Zhejiang University (Y.F.). Y.F. is a recipient of the “Young 1000 Talents” Award.
Y.F. designed this project; Y.L., H.Y., Z.L., R.G., and H.Z. performed experiments and analyzed the data; G.F.G, Q.H., and R.W. contributed reagents and tools; Y.F. wrote the manuscript.
Funding Statement
This work was funded by National Natural Science Foundation of China (NSFC) (31570027) and Zhejiang Provincial Natural Science Foundation for Distinguished Young Scholars (LR15H190001).
Footnotes
Citation Ye H, Li Y, Li Z, Gao R, Zhang H, Wen R, Gao GF, Hu Q, Feng Y. 2016. Diversified mcr-1-harbouring plasmid reservoirs confer resistance to colistin in human gut microbiota. mBio 7(2):e00177-16. doi:10.1128/mBio.00177-16.
REFERENCES
- 1.Nation RL, Li J, Cars O, Couet W, Dudley MN, Kaye KS, Mouton JW, Paterson DL, Tam VH, Theuretzbacher U, Tsuji BT, Turnidge JD. 2015. Framework for optimisation of the clinical use of colistin and polymyxin B: the Prato polymyxin consensus. Lancet Infect Dis 15:225–234. doi: 10.1016/S1473-3099(14)70850-3. [DOI] [PubMed] [Google Scholar]
- 2.Paterson DL, Harris PNA. 2016. Colistin resistance: a major breach in our last line of defence. Lancet Infect Dis 16:132–133. doi: 10.1016/S1473-3099(15)00463-6. [DOI] [PubMed] [Google Scholar]
- 3.Gunn JS. 2008. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol 16:284–290. [DOI] [PubMed] [Google Scholar]
- 4.Cannatelli A, D’Andrea MM, Giani T, Di Pilato V, Arena F, Ambretti S, Gaibani P, Rossolini GM. 2013. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother 57:5521–5526. doi: 10.1128/AAC.01480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu LF, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu JH, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 6.Hu Y, Liu F, Lin IY, Gao GF, Zhu B. 18 December 2015. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis doi: 10.1016/S1473-3099(15)00533-2. [DOI] [PubMed] [Google Scholar]
- 7.Olaitan AO, Chabou S, Okdah L, Morand S, Rolain JM. 18 December 2015. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis doi: 10.1016/S1473-3099(15)00540-X. [DOI] [PubMed] [Google Scholar]
- 8.Suzuki S, Ohnishi M, Kawanishi M, Akiba M, Kuroda M 7 January 2016. Investigation of a plasmid genome database for colistin-resistance gene mcr-1. Lancet Infect Dis doi: 10.1016/S1473-3099(16)00008-6. [DOI] [PubMed] [Google Scholar]
- 9.Malhotra-Kumar S, Xavier BB, Das AJ, Lammens C, Hoang HT, Pham NT, Goossens H 7 January 2016. Colistin-resistant Escherichia coli harbouring mcr-1 isolated from food animals in Hanoi, Vietnam. Lancet Infect Dis doi: 10.1016/S1473-3099(16)00014-1. [DOI] [PubMed] [Google Scholar]
- 10.Stoesser M, Mathers AJ, Moore CE, Day NP, Crook DW 7 January 2016. Colistin resistance gene mcr-1 and pHNSHP45 plasmid in human isolates of Escherichia coli and Klebsiella pneumoniae. Lancet Infect Dis doi: 10.1016/S1473-3099(16)00010-4. [DOI] [PubMed] [Google Scholar]
- 11.Hasman H, Hammerum AM, Hansen F, Hendriksen RS, Olesen B, Agersø Y, Zankari E, Leekitcharoenphon P, Stegger M, Kaas RS, Cavaco LM, Hansen DS, Aarestrup FM, Skov RL. 2015. Detection of mcr-1 encoding plasmid-mediated colistin-resistant Escherichia coli isolates from human bloodstream infection and imported chicken meat, Denmark 2015. Euro Surveill 20:pii=30085. doi: 10.2807/1560-7917.ES.2015.20.49.30085. [DOI] [PubMed] [Google Scholar]
- 12.Webb HE, Granier SA, Marault M, Millemann Y, den Bakker HC, Nightingale KK, Bugarel M, Ison SA, Scott HM, Loneragan GH. 18 December 2015. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis doi: 10.1016/S1473-3099(15)00538-1. [DOI] [PubMed] [Google Scholar]
- 13.Arcilla MS, van Hattem JM, Matamoros S, Melles DC, Penders J, de Jong MD, Schultsz SC; COMBAT Consortium . 18 December 2015. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis doi: 10.1016/S1473-3099(15)00541-1. [DOI] [PubMed] [Google Scholar]
- 14.Haenni M, Poirel L, Kieffer N, Châtre P, Saras E, Métayer V, Dumoulin R, Nordmann P, Madec JY 7 January 2016. Co-occurrence of extended spectrum β-lactamase and MCR-1 encoding genes on plasmids. Lancet Infect Dis doi: 10.1016/S1473-3099(16)00007-4. [DOI] [PubMed] [Google Scholar]
- 15.Falgenhauer L, Waezsada SE, Yao Y, Imirzalioglu C, Käsbohrer A, Roesler U, Michael GB, Schwarz S, Werner G, Kreienbrock L, Charkaborty T; RESET Consortium 7 January 2016. Colistin resistance gene mcr-1 in extended-spectrum β-lactamase-producing and carbapenemase-producing gram-negative bacteria in Germany. Lancet Infect Dis doi: 10.1016/S1473-3099(16)00009-8. [DOI] [PubMed] [Google Scholar]
- 16.Malhotra-Kumar S, Xavier BB, Das AJ, Lammens C, Butaye P, Goossens H 7 January 2016. Colistin resistance gene mcr-1 harboured on a multidrug resistant plasmid. Lancet Infect Dis doi: 10.1016/S1473-3099(16)00012-8. [DOI] [PubMed] [Google Scholar]
- 17.Tse H, Yuen KY 18 December 2015. Dissemination of the mcr-1 colistin resistance gene. Lancet Infect Dis doi: 10.1016/S1473-3099(15)00532-0. [DOI] [PubMed] [Google Scholar]
- 18.Reardon S. 21 December 2015. Spread of antibiotic-resistance gene does not spell bacterial apocalypse—yet. Nature doi: 10.1038/nature.2015.19037. [DOI] [Google Scholar]
- 19.Yang C. 2015. Emergence and spread of a plasmid-mediated polymyxin resistance mechanism, MCR-1: are bacteria winning? Infect Dis Trans Med 1:56. [Google Scholar]
- 20.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J Bacteriol 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]