Abstract
The organophosphorus insecticide chlorpyrifos has been widely used. Its active metabolite chlorpyrifos-oxon (CPO) is a potent anticholinesterase and is detoxified by paraoxonase-1 (PON1). PON1 activity is influenced by numerous factors including a Q192R polymorphism. Using forty human blood samples bearing homozygous genotypes and either high or low activity phenotypes (as determined by high concentration assays of paraoxon and diazoxon hydrolysis) the serum PON1 hydrolysis of high (320 μM) and low (178 nM) CPO concentrations was assessed using direct or indirect spectrophotometric methods, respectively. PON1 activity at high CPO concentration reflected the phenotype and genotype differences; subjects with the high activity phenotype and homozygous for the PON1R192 alloform hydrolyzed significantly more CPO than subjects with the low activity phenotype and/or PON1Q192 alloform (High RR=11023±722, Low RR=9467±798, High QQ=8809±672, Low QQ=6030±1015 μmoles CPO hydrolyzed/min/L serum). However, PON1 hydrolysis of CPO at the lower, more environmentally relevant concentration showed no significant differences between the PON1192 genotypes and/or between high and low activity phenotypes (High RR=231±27, Low RR=219±52, High QQ=193±59, Low QQ=185±43 nmoles CPO/min/L serum). Low CPO concentrations were probably not saturating, so PON1 did not display maximal velocity and the PON1 genotype/phenotype might not influence the extent of metabolism at environmental exposures.
Keywords: Paraoxonase 1, PON1, chlorpyrifos-oxon, polymorphism
1.1 INTRODUCTION
The phosphorothionate insecticide chlorpyrifos is a widely used organophosphorus (OP) insecticide and the question of its safety has been raised. Phosphorothionates are bioactivated by cytochromes P450 to their oxon metabolites (Neal, 1967). Chlorpyrifos is a moderately toxic insecticide (rat oral LD50 155 mg/kg; Gaines, 1969) and can be bioactivated to chlorpyrifos-oxon (CPO), a potent inhibitor of acetylcholinesterase (AChE; E.C. 3.1.1.7) (Aldridge and Davison, 1953) displaying an IC50 (30 min incubation) for rat brain AChE of 4 nM (Chambers et al., 1990). Inhibition of AChE can lead to excess acetylcholine in the peripheral and central nervous systems causing cholinergic overstimulation (Costa, 2008).
Paraoxonase-1 (PON1; E.C. 3.1.8.1) is synthesized in the liver and circulates in the plasma associated with both apolipoprotein A1 and high density lipoproteins (Sorenson et al., 1999). PON1 protects against the acute toxic effects of several OP anticholinesterases through hydrolysis of the parent compounds or their active metabolites (oxons) (Aldridge and Davison, 1953). In fact, paraoxonase takes its name from its ability to hydrolyze paraoxon, the active metabolite of the OP insecticide parathion (Furlong, 2008).
Differences in PON1 activity have been linked to common polymorphisms in both coding and promoter regions (Adkins et al., 1993; Leviev and James, 2000). A common polymorphism in the coding region of PON1 is PON1 Q192R, which influences enzyme activity measured in vitro (Adkins et al.,1993). This polymorphism involves a change from the ancestral arginine (R) to glutamine (Q) at position 192 (www.ncbi.nlm.nih.gov/projects/SNP Reference SNP (refSNP) Cluster Reports 854560 and 662 accessed 2/12/2013).
The involvement of PON1 in the in vivo detoxication of CPO and diazoxon, the active metabolite of the OP insecticide diazinon, has been demonstrated with transgenic animals. Although mice with a deleted Pon1 gene are more susceptible to the toxic effects of CPO and diazoxon than wild mice, surprisingly they are not more susceptible to paraoxon (Cole et al., 2005; Li et al., 2000; Shih et al., 1998). Studies also found that administration of PON1 protein, whether recombinant or purified from plasma, confers additional tolerance of chlorpyrifos and CPO (Costa et al., 1990; Li et al., 1995; Li et al., 2000; Stevens et al., 2008). The effect of the Q192R polymorphism on the catalytic efficiency of PON1 varies based on the substrate involved (Adkins et al., 1993; Davies et al., 1996; Humbert et al., 1993; Li et al., 2000). The PON1R192 alloform provided higher catalytic efficiency of CPO hydrolysis than the PON1Q192 alloform in both in vitro and in vivo testing (Li et al., 2000). Studies using humanized transgenic mice (Pon1 gene replaced with human PON1 genes coding for either PON1Q192 or PON1R192) indicated that mice with PON1R192 were significantly more resistant to toxicity from a high dosage of CPO than mice with PON1Q192, even though the amount of PON1 protein was the same for both (Cole et al., 2003; 2005). Therefore, at high dosages of CPO, the PON1 status, i.e., both the activity level of the PON1 enzyme (phenotype) and the PON1192 polymorphism (genotype), were important determinants of susceptibility. However, these studies did not determine whether PON1 status influenced susceptibility at lower, more environmentally relevant levels of CPO. In routine exposures to chlorpyrifos, CPO would not be expected to be present at high levels because of the kinetics of formation from chlorpyrifos by cytochromes P450. Additionally, because of CPO's high potency as an anticholinesterase, high level environmental or occupational exposures would not be tolerable. The detoxication of CPO by paraoxonase at low (<5 μM) concentrations has been studied (Sogorb et al., 2008; Sogorb and Vilanova, 2010), but this research did not investigate the influence of genotype on the ability of PON1 to degrade CPO.
The present study utilized serum collected previously during a study involving the PON1192 genotyping and PON1 phenotyping (i.e., activity levels for select substrates) of African American and Caucasian male and female subjects (Coombes et al., 2011). The PON1192 genotype was determined using a restriction fragment length polymorphism assay and the PON1 phenotype was assessed with two substrates, paraoxon and diazoxon. A subset of samples and data from forty subjects from the original study sample was chosen who had PON1192QQ or PON1192RR genotypes and either high or low activities towards the two substrates (Figure 1). The in vitro PON1-mediated hydrolysis of high and low CPO concentrations was assessed using either a direct or indirect spectrophotometric method, respectively. The indirect method allows for quantitation of CPO hydrolysis using a lower, more environmentally relevant nanomolar concentration than the high micromolar concentration required by the direct assay for spectrophotometric quantification of the hydrolysis product (Chambers et al., 1994). The indirect assay measures the CPO remaining following PON1 hydrolysis by assessing the inhibition of an exogenous source of AChE by the residual CPO. AChE inhibition can be measured at CPO concentrations well below the detection limits of the direct spectrophotometric method and, therefore, the indirect method can measure CPO concentrations that are more reflective of realistic exposure levels.
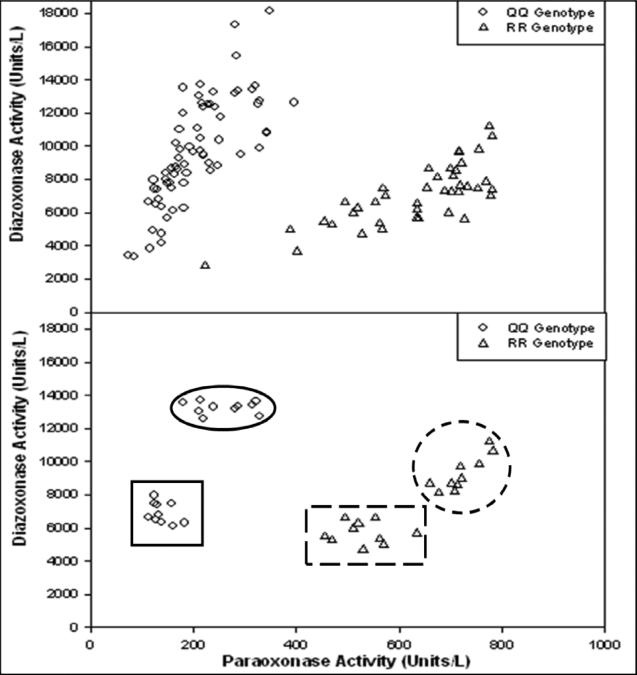
FIGURE 1.
Plot of diazoxonase activities versus paraoxonase activities in the presence of 2M NaCl of human sera showing the separation of homozygous QQ (◇) from homozygous RR (Δ) in the original study sample (top) and the groups selected for the study as high QQ (solid oval outline), low QQ (solid square outline), high RR (dashed circle outline), and low RR (dashed rectangular outline) (bottom). Data derived from Coombes et al. (2011).
1.2 MATERIALS AND METHODS
Subjects of the original study sample were enrolled for blood collection from the clinical practice of Cardiology Associates of North Mississippi, LLC, for the previously described study (Coombes et al., 2011). Serum was stored at −70°C until analysis of PON1 activity with CPO. Written informed consent had been obtained from all participants and the study protocol was approved by both the Institutional Review Board (IRB) of Mississippi State University and the IRB of North Mississippi Medical Center; the latter provides oversight for studies performed at Cardiology Associates.
Chlorpyrifos-oxon was synthesized as previously described and was at least 95% pure (Chambers and Chambers, 1989). All other chemicals were purchased from Sigma Chemical Company (St. Louis, MO). Paraoxon, chlorpyrifos-oxon, and diazoxon are all potent anticholinesterases and were handled with appropriate protection for personnel.
Paraoxonase and diazoxonase activities were determined by the hydrolysis of paraoxon or diazoxon, respectively, and were reported earlier for the entire study sample in Coombes et al. (2011). Paraoxonase and diazoxonase assays conducted in the presence of 2 M NaCl were used to identify four subgroups within the study sample. A subject's PON1 phenotype was designated as “high” or “low” based on the level of diazoxonase and paraoxonase enzyme activities in a plot typically used for assignment of functional genotype (Eckerson et al., 1983). Clusters of ten individuals in each of four subsets whose PON1 phenotypes fell within one of four groups (high QQ, low QQ, high RR, or low RR) were selected from the original study sample (Figure 1). The activities for each of these four clusters of ten individuals each are reported in Table 1.
TABLE 1.
Direct assay measurement of substrate hydrolysis by human serum PON1. Activities are expressed as micromoles of substrate hydrolyzed per minute per liter of serum, means ± SD, n=10. [Data on paraoxon and diazoxon were calculated for this subset of subjects only, but were collected for and methods were described in the Coombes et al. (2011) study.] Values within a substrate with the same superscript letter are not statistically different (p≤0.05).
| Substrate | High RR | Low RR | High QQ | Low QQ |
|---|---|---|---|---|
| Chlorpyrifos-oxon | 11,023 ± 722A | 9,467 ± 798B | 8,809 ± 672B | 6,030 ± 1015C |
| Paraoxon | 720 ± 40A | 530 ± 53B | 259 ± 53C | 139 ± 21D |
| Diazoxon | 9,255 ± 1,040A | 5,699 ± 669B | 13,250 ± 381C | 6,892 ± 620D |
The PON1192 genotype for each individual was originally determined using a restriction fragment length polymorphism assay as reported in Coombes et al. (2011). Subjects selected for this study were homozygous for either the PON1Q192 alloform (QQ) or the PON1R192 alloform (RR). Heterozygous subjects (QR) were excluded.
1.2.1. Direct Assay of Chlorpyrifos-oxonase Activity
PON1 hydrolysis of a high concentration of CPO used the method described in Furlong et al. (1989) with our minor modifications (Pond et al., 1996). Briefly, human serum samples (20 μl serum/ml 0.1 M Tris-HCl buffer at pH 7.4) containing either 1mM EDTA to prevent PON1 activity or 2 mM CaCl2 to stimulate PON1 activity were incubated with shaking at 37°C. CPO in ethanol (320 μM final concentration) was added and the reaction was terminated with 2% sodium dodecyl sulfate (SDS) after 15 min. Absorbance was determined at 315 nm. Differences between the Ca2+-fortified and the EDTA samples were used to calculate 3, 5, 6-trichloropyridinol produced. Data were expressed as micromoles CPO hydrolyzed/min/L serum.
1.2.2 Indirect Assay of Chlorpyrifos-oxonase Activity
The indirect assay of hydrolysis toward a low concentration of CPO was performed as described in Chambers et al. (1994) and Pond et al. (1996) with a few modifications. Human serum samples (10 μl serum/ml 0.1 M Tris-HCl buffer at pH 7.4) containing either 1 mM EDTA or 2 mM CaCl2 were incubated with shaking at 37°C for 15 min. Iso-OMPA in ethanol (tetraisopropylpyrophosphoramide, 10 μM final concentration) was added followed by incubation for 5 min. to inhibit serum butyrylcholinesterase that could stoichiometrically destroy CPO.
CPO in ethanol (final concentration, 178 nM) was added to initiate the reaction. This CPO concentration was selected to result in about 90% AChE inhibition following the PON1 assay with EDTA buffer to account for any non-PON1-mediated CPO hydrolysis. After 15 min, electric eel AChE (0.075 unit/ml final concentration) was added followed by another 15 min incubation. The reaction mixture was diluted with Tris-HCl buffer (0.05 M, pH 7.4) to a final concentration of 0.015 unit electric eel AChE/ml.
AChE activity was measured by our modification (Chambers and Chambers, 1989) of the technique of Ellman et al., (1961). A 15 min incubation was conducted to allow 0.01 M eserine sulfate to inhibit AChE in the blanks. Acetylthiocholine iodide in ethanol (1 mM final concentration) was added followed by a 15 min incubation. The reaction was stopped by addition of 5% SDS/2.6 mM DTNB [5,5′-dithio-bis (2-nitrobenzoic acid)] and the absorbance was measured at 412 nm. A standard curve of CPO concentration versus percent inhibition of electric eel AChE was used to determine the nanomolar concentration of residual CPO. Samples were analyzed in triplicate and data were expressed in nanomoles CPO hydrolyzed per min per liter of serum.
1.2.3 Statistical Analysis
Two-way ANOVA (PROC GLM) with Tukey adjustment of the least square means was performed with SAS for Windows 9.1.3 (SAS Institute Inc., Cary, NC, USA) to determine statistically significant differences of CPO hydrolysis among the four groups. Paraoxonase and diazoxonase data for these selected subjects failed to meet ANOVA assumptions and were analyzed for statistically significant group differences using Kruskal-Wallis. To determine which groups differed, Wilcoxon rank sum testing was used with Bonferroni's correction to adjust for the effect of multiple pairwise comparisons. All statistical comparisons used a p≤0.05 significance level.
1.3 THEORY
This study compared low and high substrate concentrations in assays of human serum PON1-mediated detoxication of chlorpyrifos-oxon to determine whether genotype and phenotype differences were apparent at more environmentally relevant levels of chlorpyrifos-oxon.
1.4 RESULTS
1.4.1 Hydrolysis using Direct Assay
Serum of subjects homozygous for the PON1R192 alloform with a high activity phenotype (high RR) hydrolyzed significantly more CPO than serum of subjects homozygous for the PON1Q192 alloform with a high activity phenotype (high QQ) (p<0.001). Serum of RR homozygous subjects with a low activity phenotype (low RR) hydrolyzed significantly more CPO than serum of homozygous QQ subjects who expressed the low activity phenotype (low QQ) (p<0.001). Within the RR homozygous group, the serum of those with a high activity phenotype (high RR) hydrolyzed significantly more CPO than the serum of those with the low activity phenotype (low RR) (p<0.001). Similarly, serum of the high QQ group hydrolyzed significantly more CPO than subjects who were QQ and expressed a low activity phenotype (low QQ) (p<0.001).
As a point of comparison, the activities of these four subsets in hydrolyzing paraoxon and diazoxon were calculated, using the data from the previous study (Coombes et al., 2011) on only the 40 subjects selected to be in the subset for the present study. Serum of subjects homozygous for the PON1R192 alloform with a high activity phenotype (high RR) hydrolyzed significantly more paraoxon (p<0.001) than the serum of subjects from any of the other groups. Serum of QQ homozygous subjects with a high activity phenotype (high QQ) hydrolyzed significantly more diazoxon (p<0.001) than serum from any other group (Table 1).
1.4.2 Chlorpyrifos-oxon Hydrolysis using the Indirect Assay
There were no significant differences observed in the hydrolysis of CPO between subjects homozygous for the PON1R192 or PON1Q192 alloforms (p=0.075). There were also no significant differences in the hydrolysis of CPO between subjects expressing high or low activity phenotypes (p= 0.742 for QQ homozygotes; p= 0.528 for RR homozygotes). Subjects who were high RR did not hydrolyze CPO more efficiently than high QQ subjects (p=0.077) and low RR subjects did not hydrolyze CPO more efficiently than QQ subjects who expressed a low activity phenotype (p=0.124) (Table 2).
TABLE 2.
Indirect assay measurement of chlorpyrifos-oxon hydrolysis. Activities are expressed as nanomoles of chlorpyrifos-oxon hydrolyzed per minute per liter of serum, means ± SD, n=10. Values with the same superscript letter are not significantly different (p≤0.05) as determined by ANOVA with Tukey adjustment. ^
| High RR | Low RR | High QQ | Low QQ |
|---|---|---|---|
| 231 ± 27A | 219 ± 52A | 193 ± 59A | 185 ± 43A |
1.5 DISCUSSION
In this study using human serum, values for PON1 hydrolysis of a high concentration CPO (320 μM) measured by direct assay agree with previous work, such as the humanized mouse models, that found both serum PON1 phenotype and PON1192 genotype to be important determinants of CPO sensitivity (Cole et al., 2005; Li et al., 2000; Shih et al., 1998). Our data indicate that differences in serum PON1 phenotypes are important for predicting sensitivity towards high concentration CPO since subjects having high PON1 phenotypes hydrolyzed significantly more CPO than subjects with low PON1 phenotypes. Additionally at the high CPO concentration, subjects homozygous for the PON1R192 alloform hydrolyzed significantly more CPO than subjects with the PON1Q192 alloform. These results support previous findings on the importance of the PON1 Q192R polymorphism as a determinant of hydrolytic efficiency in both in vitro and in vivo testing at high concentrations of CPO (Li et al., 2000). Therefore genotype would be expected to influence the degree of toxicity in very high exposures to chlorpyrifos, such as occurs in an accidental poisoning.
Hydrolysis of CPO has been studied at low substrate concentrations, confirming the importance of PON1 in detoxifying CPO (Sogorb et al., 2008; Sogorb and Vilanova, 2010). However this is the first study, to our knowledge, to examine the influence of the PON1 Q192R SNP on human PON1-mediated serum hydrolysis of CPO at a lower, more environmentally relevant concentration (i.e., 178 nM). However, no significant differences were demonstrated between the PON1192 genotypes and/or between high and low serum PON1 phenotypes at this lower CPO concentration, contrasting with our high CPO concentration data discussed above as well as with studies of others using direct assay methods and high concentrations of CPO (Cole et al. 2005; Jansen et al., 2009; Li et al., 2000; Shih et al., 1998). Therefore, at a low concentration more reflective of levels that would occur under realistic exposure scenarios, neither PON1192 genotypes nor serum PON1 phenotypes influence the capacity of PON1 to metabolize CPO. Presumably these low CPO concentrations were unable to saturate the PON1 enzyme, and therefore PON1 was not functioning at maximal velocity, as evidenced by the much lower activities observed with the indirect assay compared to the direct assay. This explanation is supported by work showing that the paraoxonase maximal velocity (Vmax) differs significantly for all the PON1192 genotypes but the Michaelis constant (KM) does not (Poh and Muniandy, 2009). If CPO hydrolysis exhibits the same relationships, at high substrate concentrations enzyme velocity is equal to Vmax, and the genotypic differences would have the dominant effect. At low substrate concentrations, however, KM dominates the kinetic equation so its lack of significant differences among Q192R genotypes would explain the present results showing no genotypic differences at a low CPO concentration.
1.6 CONCLUSIONS
Because of the high potency of CPO as an anticholinesterase (4 nM; Chambers et al., 1990) and the expectation that CPO would be generated from chlorpyrifos relatively slowly by cytochromes P450, at environmentally relevant concentrations it is unlikely that PON1 genotype/phenotype would influence appreciably an individual's ability to metabolize CPO.
Supplementary Material
Highlights.
The influence of the human PON1 Q192R SNP on serum hydrolysis of CPO was examined.
PON1 hydrolysis of high (320 μM) and low (178 nM) CPO concentrations was assessed.
High CPO concentration PON1 activity reflected phenotype and genotype differences.
Low CPO concentration PON1 activity showed no phenotype or genotype differences.
PON1 genotype/phenotype might not affect hydrolysis rate in environmental exposures.
ACKNOWLEDGEMENTS
This research was supported in part by National Institutes of Health R21 ES015107. The funding source had no involvement in the study design; collection, analysis and interpretation of data; writing or the decision to publish. This research was also supported in part by the Center for Environmental Health Sciences, Mississippi State University. This is Center for Environmental Health Sciences publication 130.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adkins S, Gan KN, Mody M, La Du BN. Molecular basis for the polymorphic forms of human serum paraoxonase/arylesterase: glutamine or arginine at position 191, for the respective A or B allozymes. Am. J. Hum. Genet. 1993;52:598–608. [PMC free article] [PubMed] [Google Scholar]
- Aldridge WN, Davison AN. The mechanism of inhibition of cholinesterases by organophosphorus compounds. Biochem. J. 1953;55:763–766. doi: 10.1042/bj0550763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers HW, Brown B, Chambers JE. Non-catalytic detoxication of six organophosphorus compounds by rat liver homogenates. Pestic. Biochem. Physiol. 1990;36:308–315. Doi: 10.1016/0048-3575(90)90040-9. [Google Scholar]
- Chambers JE, Chambers HW. An investigation of acetylcholinesterase inhibition and aging and choline acetyltransferase activity following a high level acute exposure to paraoxon. Pestic. Biochem. Physiol. 1989;33:125–131. Doi; 10.1016/0048-3575(89)90003-5. [Google Scholar]
- Chambers JE, Ma T, Boone JS, Chambers HW. Role of detoxication pathways in acute toxicity levels of phosphorothionate insecticides in the rat. Life Sci. 1994;54:1357–1364. doi: 10.1016/0024-3205(94)00515-x. Doi: 10.1016/0024-3205(94)00515-X. [DOI] [PubMed] [Google Scholar]
- Cole TB, Jampsa RL, Walter BJ, Arndt TL, Richter RJ, Shih DM, Twart A, Lusis AJ, Jack RM, Costa LG, Furlong CE. Expression of human paraoxonase (PON1) during development. Pharmacogenetics. 2003;13:357–364. doi: 10.1097/00008571-200306000-00007. 10.1016/0024-3205(94)00515-X. [DOI] [PubMed] [Google Scholar]
- Cole TB, Walter BJ, Shih DM, Tward AD, Lusis AJ, Timchalk C, Richer RJ, Costa LG, Furlong CE. Toxicity of chlorpyrifos and chlorpyrifos oxon in a transgenic mouse model of the human paraoxonase (PON1) Q192R polymorphism. Pharmacogenet. Genomics. 2005;15:589–598. doi: 10.1097/01.fpc.0000167327.08034.d2. 10.1097/01.fpc.0000167327.08034.d2. [DOI] [PubMed] [Google Scholar]
- Coombes RH, Crow JA, Dail MB, Chambers HW, Wills RW, Bertolet BD, Chambers JE. Relationship of human paraoxonase-1 serum activity and genotype with atherosclerosis in individuals from the Deep South. Pharmacogenet. Genomics. 2011;21:867–875. doi: 10.1097/FPC.0b013e32834cebc6. Doi: 10.1097/FPC.06013e32834cebc6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa LG, McDonald BE, Murphy SD, Omenn GS, Richter RJ, Motulsky AG, Furlong CE. Serum paraoxonase and its influence on paraoxon and chlorpyrifosoxon toxicity in rats. Toxicol. Appl. Pharmacol. 1990;103:66–76. doi: 10.1016/0041-008x(90)90263-t. Doi:10.1016/0041-008X(90)90263-T. [DOI] [PubMed] [Google Scholar]
- Costa LG. Toxic effects of pesticides. In: Klaassen CD, editor. Casarett and Doull's Toxicology: The Basic Science of Poisons. McGraw Hill Medical Inc.; New York: 2008. pp. 883–930. [Google Scholar]
- Davies HG, Richter RJ, Keifer M, Broomfield CA, Sowalla J, Furlong CE. The effect of the human serum paraoxonase polymorphism is reversed with diazoxon, soman and sarin. Nat. Genet. 1996;14:334–336. doi: 10.1038/ng1196-334. 10.1038/ng1196-334. [DOI] [PubMed] [Google Scholar]
- Eckerson HW, Romson J, Wyte C, La Du BN. The human serum paraoxonase polymorphism: Identification of phenotypes by their response to salts. Am. J. Hum. Genet. 1983;35:214–227. [PMC free article] [PubMed] [Google Scholar]
- Ellman GL, Courtney KD, Andres V, Jr., Featherstone RM. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]
- Furlong CE, Richter RJ, Seidel S, Costa LG, Motulsky A. Spectrophotometric assays for the enzymatic hydrolysis of the active metabolites of chlorpyrifos and parathion by plasma paraoxonase/arylesterase. Anal. Biochem. 1989;180:242–247. doi: 10.1016/0003-2697(89)90424-7. Doi: 10.1016/0003-2697(89)90424-7. [DOI] [PubMed] [Google Scholar]
- Furlong CE. Paraoxonases: An historical perspective. In: Mackness B, Mackness M, Aviram M, Paragh G, editors. The Paraoxonases: Their Role in Disease Development and Xenobiotic Metabolism. Springer; The Netherlands: 2008. pp. 3–32. [Google Scholar]
- Gaines TB. Acute toxicity of pesticides. Toxicol. Appl. Pharmacol. 1969;14:515–534. doi: 10.1016/0041-008x(69)90013-1. Doi; 10.1016/0041-008X(69)90013-1. [DOI] [PubMed] [Google Scholar]
- Humbert R, Adler DA, Disteche CM, Hassett C, Omiecinski CJ, Furlong CE. The molecular basis of the human serum paraoxonase activity polymorphism. Nat. Genet. 1993;3:73–76. doi: 10.1038/ng0193-73. Doi: 10.1038/ng0193-73. [DOI] [PubMed] [Google Scholar]
- Jansen KL, Cole TB, Park SS, Furlong CE, Costa LG. Paraoxonase 1 (PON1) modulates the toxicity of mixed organophosphorus compounds. Toxicol. Appl. Pharmacol. 2009;236:142–153. doi: 10.1016/j.taap.2009.02.001. Doi: 10.1016/jtaap.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leviev I, James RW. Promoter polymorphisms of human paraoxonase PON1 gene and serum paraoxonase activities and concentrations. Arterioscler. Thromb. Vasc. Biol. 2000;20:516–521. doi: 10.1161/01.atv.20.2.516. Doi: 10.1161/01.ATV.20.2.516. [DOI] [PubMed] [Google Scholar]
- Li WF, Furlong CE, Costa LG. Paraoxonase protects against chlorpyrifos toxicity in mice. Toxicol. Lett. 1995;76:219–226. doi: 10.1016/0378-4274(95)80006-y. Doi: 10.1016/0378-4274(95)80006-Y. [DOI] [PubMed] [Google Scholar]
- Li WF, Costa LG, Richter RJ, Hagen T, Shih DM, Tward A, Lusis AJ, Furlong CE. Catalytic efficiency determines the in-vivo efficacy of PON1 for detoxifying organophosphorus compounds. Pharmacogenetics. 2000;10:767–779. doi: 10.1097/00008571-200012000-00002. Doi: 10.1097/00008571-200012000-00002. [DOI] [PubMed] [Google Scholar]
- Neal RA. Studies of the enzymatic mechanism of the metabolism of diethyl 4 nitrophenyl phosphorothionate (parathion) by rat liver microsomes. Biochem. J. 1967;105:289–297. doi: 10.1042/bj1050289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poh R, Muniandy S. Evaluation of polymorphism at codon 192 of paraoxonase 1 on its kinetic behavior. Journal of Biological Sciences. 2009;9:541–548. Doi: 103923/jbs.2009.541.548. [Google Scholar]
- Pond AL, Coyne CP, Chambers HW, Chambers JE. Identification and isolation of two rat serum proteins with A-esterase activity toward paraoxon and chlorpyrifos-oxon. Biochemical Pharmacology. 1996;52:363–369. doi: 10.1016/0006-2952(96)00215-8. Doi: 10.1016/0006-2952(96)00215-8. [DOI] [PubMed] [Google Scholar]
- Shih DM, Gu L, Xia YR, Navab M, Li WF, Hama S, Castellani LW, Furlong CE, Costa LG, Fogelman AM, Lusis AJ. Mice lacking serum paraoxonase are susceptible to organophosphate toxicity and atherosclerosis. Nature. 1998;394:284–287. doi: 10.1038/28406. Doi: 10.1038/28406. [DOI] [PubMed] [Google Scholar]
- Sogorb MA, García-Argüelles S, Carrera V, Vilanova E. Serum albumin is as efficient as paraoxonase in the detoxification of paraoxon at toxicologically relevant concentrations. Chem. Res. Toxicol. 2008;21:1524–1529. doi: 10.1021/tx800075x. Doi: 10.1021/tx800075x. [DOI] [PubMed] [Google Scholar]
- Sogorb MA, Vilanova E. Serum albumins and detoxification of anti-cholinesterase agents. Chemico-Biological Interactions. 2010;187:325–329. doi: 10.1016/j.cbi.2010.03.001. Doi: 10.1016/j.cbi.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Sorenson RC, Bisgaier CL, Aviram M, Hsu C, Billecke S, La Du BN. Human serum paraoxonase/arylesterase's retained hydrophobic N-terminal leader sequence associates with HDLs by binding phospholipids: Apolipoprotein A-I stabilizes activity. Arterioscler. Thromb. Vasc. Biol. 1999;19:2214–2225. doi: 10.1161/01.atv.19.9.2214. Doi: 10.1161/01.ATV.19.9.2214. [DOI] [PubMed] [Google Scholar]
- Stevens RC, Suzuki SM, Cole TB, Park SS, Richter RJ, Furlong CE. Engineered recombinant human paraoxonase 1 (rHuPON1) purified from Escherichia coli protects against organophosphate poisoning. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12780–12784. doi: 10.1073/pnas.0805865105. Doi: 10.1073/pnas.0805865105. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB REFERENCES
- [2/12/2013]; www.ncbi.nlm.nih.gov/projects/SNP Reference SNP (refSNP) Cluster Reports 854560 and 662.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.