Abstract
Background
Y-box binding factor 1 (YB-1) has been associated with prognosis in many tumor types. Reduced YB-1 expression inhibits tumor cell growth, but the mechanism is unclear.
Methods
YB-1 mRNA levels were compared with tumor grade and histology using microarray data from 771 breast cancer patients and with disease-free survival and distant metastasis–free survival using data from 375 of those patients who did not receive adjuvant therapy. Microarrays were further searched for genes that had correlated expression with YB-1 mRNA. Small interfering RNA (siRNA) was used to study the effects of reduced YB-1 expression on growth of three tumor cell lines (MCF-7 breast, HCT116 colon, and A549 lung cancer cells), on tumorigenesis by A549 cells in nude mice, and on global transcription in the three cancer cell lines. Reporter gene assays were used to determine whether YB-1 siRNAs affected the expression of E2F1, and chromatin immunoprecipitation was used to determine whether YB-1 bound to various E2F promoters as well as E2F1-regulated promoters. All P values were from two-sided tests.
Results
YB-1 levels were elevated in more aggressive tumors and were strongly associated with poor disease-free survival and distant metastasis–free survival. YB-1 expression was often associated with the expression of genes with E2F sites in their promoters. Cells expressing YB-1 siRNA grew substantially more slowly than control cells and formed tumors less readily in nude mice. Transcripts that were altered in cancer cell lines with YB-1 siRNA included 32 genes that are components of prognostic gene expression signatures. YB-1 regulated expression of an E2F1 promoter–reporter construct in A549 cells (eg, relative E2F1 promoter activity with control siRNA = 4.04; with YB-1 siRNA = 1.40, difference= −2.64, 95% confidence interval = −3.57 to −1.71, P < .001) and bound to the promoters of several well-defined E2F1 target genes.
Conclusion
YB-1 expression is associated with the activity of E2F transcription factors and may control tumor cell growth by this mechanism.
CONTEXTS AND CAVEATS
Prior knowledge
Elevated levels of Y-box binding factor 1 (YB-1) expression have been associated with higher tumor grade and poor prognosis for several malignancies. The mechanism by which YB-1 expression promotes tumor cell growth was unclear.
Study design
Breast cancer microarray data were used to examine the relationships between YB-1 mRNA levels and tumor grade and histology, patient survival, and transcription patterns of other genes. Three cancer cell lines were transfected with small interfering RNA to examine the effect of YB-1 silencing on their proliferation, tumorigenicity, and global transcription patterns. Because both tumor and in vitro data showed an association of YB-1 with E2F target gene expression, reporter gene assays and chromatin immunoprecipitation were used to determine whether YB-1 regulated E2F-mediated transcription.
Contribution
YB-1 was expressed at higher levels in higher-grade tumors with worse prognosis. Cancer cells in which YB-1 expression was silenced grew more slowly and formed tumors more slowly than control cells. Also, YB-1 small interfering RNA altered the expression of many genes that are components of prognostic signatures in several cancers. Molecular assays showed that YB-1 bound to E2F-regulated promoters and regulated the activity of the E2F1 promoter.
Implication
YB-1 appears to promote tumor growth by modulating the expression of E2Fs and also E2F target genes.
Limitations
Experiments have not been performed to address the exact mechanism whereby YB-1 affects the transcription of E2F target genes.
From the Editors
The Y-box binding protein-1 (YB-1) is a nucleic acid binding protein encoded by the gene YBX1. It was originally identified as a factor that bound to the Y-box (an inverted CCAAT box) in the MHC Class II (HLA-DRA) promoter and repressed transcription (1). Subsequently, YB-1 has been shown to be a multitasking protein with many functions, including the regulation of both transcription and translation of several genes and proteins associated with tumor progression (2), cell survival (3–6), DNA replication (7) and repair (8), drug resistance (9,10), and epithelial–mesenchymal transition (11).
Much work has focused on the association between YB-1 protein levels and patient prognosis. Many immunohistochemical studies have shown elevated YB-1 protein levels in malignant tissues compared with normal tissues, and higher levels of YB-1 have also been associated with higher tumor grade and poorer patient prognosis in a growing list of malignancies, including breast (12), lung (13), colorectal (14), prostate (15), and ovarian cancers (16).
In one compelling study, overexpression of YB-1 from a β-lactoglobulin promoter in two independent strains of transgenic mice induced aberrant proliferation of breast epithelial cells, leading to the development of invasive breast carcinomas of several histological subtypes (17). This finding led to the proposal that YB-1 might be a proto-oncogene. In addition, we previously demonstrated that YB-1 plays an important role in tumor cell growth and survival; sequestration of YB-1 by transfection with an oligonucleotide containing a YB-1 binding site led to apoptosis of many types of tumor cells in vitro (4,5), suggesting that YB-1 might generally control the survival of malignant cells. More recently, RNA interference has been used to reduce the levels of YB-1 in breast (6,18–20), lung (20), and prostate cancer (19); in mesangial (7) and myeloma (21,22) cell lines; and in a breast cancer xenograft model (18). Consistent with our previous work, these studies showed that tumor cell growth was inhibited by the reduction of YB-1 expression.
Historically, YB-1 has been linked to the regulation of cell proliferation because high levels of this mRNA were observed in regenerating liver and because many cell growth–associated genes, for example, thymidine kinase 1 (TK1) and proliferating cell nuclear antigen (PCNA), contain inverted CCAAT boxes in their promoters (23). Subsequently, YB-1 has been demonstrated to activate the transcription of mRNAs for the cyclins CCNA1, CCNB1 (24), and CDC6 (20), as well as for the epidermal growth factor receptor (EGFR) and ERBB2 (6,25,26). YB-1 has also been shown to regulate translation of both the c-MYC and transforming growth factor beta 1 (TGFB1) mRNAs, which also regulate cell proliferation pathways (27,28). Furthermore, reduction of YB-1 protein abundance by RNA interference led to the accumulation of cells in the G1 phase, with a concomitant decrease of cells in S phase (20,29).
The goal of this study was to more fully understand the role that YB-1 plays in tumor biology. We began by analyzing YB-1 mRNA levels in a large breast cancer microarray dataset. This process enabled us to analyze more than 22 268 mRNAs to identify transcripts, molecular pathways, and transcription factors associated with YB-1 expression. To determine whether these patterns were indicative of YB-1 and E2F1 transcriptional activity, we sought to identify the immediate downstream targets of YB-1 by studying three cancer cell lines in vitro. We then used chromatin immunoprecipitation and reporter gene assays to validate the results of our bioinformatic analyses.
Methods and Materials
Bioinformatic Analysis of Breast Cancer Data
To analyze YB-1 mRNA in breast tumors, we obtained Affymetrix HG-U133A and HG-U133PLUS2 microarray data from four published studies [GSE3494, GSE6532, GSE7390, GSE4922, respectively (30–33)] and one unpublished dataset (collected by L. D. Miller), removed duplicate samples, and normalized the data in the R statistical environment using the Robust Multichip Averaging (RMA) algorithm (34) provided by the affy Bioconductor package (35) without background correction. There were 22 268 probe sets represented on both array types, and data from 771 patients’ tumors (HG-U133A and HG-U133PLUS2) were used in subsequent analyses. Intrinsic subtypes were assigned to each tumor using the Single Subtype Predictor algorithm of Hu et al. (36), based on mean-centered data and Spearman correlation with subtype centroids. This cohort included samples from 374 patients who did not receive adjuvant therapy, 301 samples from patients who received endocrine treatment, 61 samples from chemotherapy-treated patients, and 25 samples from patients who were given both endocrine treatment and chemotherapy.
The data had two probe sets that were located at different regions of the YB-1 transcript. The first probe set, 208627_s_at, spanned a region encoding part of the cold shock domain and downstream sequences (nucleotides 463–898: parts of exon 4, 5, and 6). The second probe set, 208628_s_at, hybridized to a region encoding part of the C-terminus and the 3′ untranslated region (nucleotides 985–1260: exons 7 and 8). Northern blot analysis in many tissue types suggests that YB-1 is a single transcript (37). Spearman correlation analysis of these probe sets in tumors from non-adjuvant-treated patients indicated good concordance between these probe sets (Spearman coefficient ρ = .94; data not shown). Therefore, the 208627_s_at probe set was used in all analyses unless stated otherwise. Analysis of variance was performed in R (38).
We performed Kaplan–Meier survival analysis with both log-rank tests (39) and Cox proportional hazards models using the R survival package (40). In these calculations, we excluded samples with data from events that occurred 10 years after diagnosis [78% of such data were from a single cohort (32)). Hazard ratios were estimated from the Cox proportional hazards models. The “cox.zph” function of the survival package in R was used to test the proportionality assumption for the Cox proportional hazards analysis. For both the disease-free survival and distant metastasis–free survival data, plots of Schoenfeld residuals against log(time) showed an approximately zero slope (data not shown), and we could not identify statistically significant nonproportional hazards. Spearman correlation analysis was used to determine which other probe sets detected gene expression patterns that correlated with YBX1 208627_s_at. From these data, 439 individual probe sets corresponding to 379 different RNA transcripts that detected gene expression patterns with the highest positive correlation with YBX1 208627_s_at (98th percentile of ρ) were selected for further analysis. (This cut point was arbitrarily chosen based on the biological coherence of the lists of genes produced.) The 379 genes associated with these probe sets were analyzed for enrichment of E2F1 and E2F consensus binding motifs within their promoters using the GATHER web tool to access the TRANSFAC Pro transcription factor database [http://gather.genome.duke.edu/ (41)]. The GATHER web tool also generated a Bayes Factor to indicate the strength of the model that the 379 genes associated with these probe sets were enriched for E2F1 and/or E2F binding sites within their promoters. We then performed permutation analysis to compare the number of E2F1 sites (V$E2F1_Q3_01 [consensus sequence: TTGGCGCGRAANNGNM] and V$E2F1_Q6 [consensus sequence: TTTSGCGS]) within this list of 379 genes with the number of E2F1 sites within 10 000 randomly selected lists of any 379 genes. The same procedure was repeated to examine whether the 379 YB-1-regulated genes were more likely than other random genes to be part of the Gene Ontology Cell Cycle signature GO:0007049 (42,43).
Principal component analysis was performed as follows. Canonical markers associated with E2F1 activity were identified from the TRANSPATH database (see Supplementary Table 1, available online; http://www.gene-regulation.com/pub/databases.html#transpath). The expression levels of the transcripts encoding these proteins were used to assemble a metagene (the second eigenvalue) representing activity of the E2F1 pathway, which was plotted against increasing YBX1 208627_s_at levels in the non-adjuvant treated breast cancer cohort.
Human Cell Lines
A549 lung adenocarcinoma and MCF-7 and MDA-MB-435S breast cancer cell lines were purchased from the American Type Culture Collection (Manassas, VA), and HCT116 colorectal carcinoma were obtained from B. Vogelstein (Johns Hopkins University, Baltimore, MD) (44). All cell lines were validated for authenticity by CellBank Australia (www.cellbankaustralia.com) and cultured in RPMI-1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% vol/vol fetal bovine serum (FBS; Invitrogen) in humidified air with 5% (vol/vol) CO2 at 37°C.
Small Interfering RNAs (siRNAs) and Transfections
We used Stealth-modified 25-bp duplex siRNAs that were purchased from Invitrogen to reduce YB-1 levels. Guide strand sequences were as follows: for two YB-1-specific siRNAs, siYB-1, 5′-GGUCCUCCACGCAAUUACCAGCAAA-3′; siYB-2, 5′-GACCCUAUGGGCGUCGACCACAGUA-3′; for the control siRNA (siCtrl), 5′-CCACACGAGUCUUACCAAGUUGCUU-3′. The control siRNA has no known human mRNA targets and has been used in previous studies as a control for several Stealth siRNAs (31). siCtrl has a very similar GC-content to siYB-1 and slightly less than siYB-2.
All transfections were performed using a previously optimized method (45). In brief, Stealth siRNAs were reverse transfected at a final concentration of 5 or 10 nM using Lipofectamine RNAiMax (Invitrogen). Both siRNAs and RNAiMax were diluted in medium without serum. After 10 minutes at room temperature, the diluted RNAiMax was added to the siRNAs, and the mixture was incubated for a further 20 minutes. The lipoplexes formed were added to cells for 18 hours. After overnight transfection, the culture medium was replaced with RPMI-1640 supplemented with 10% FBS until the cells were harvested at the indicated times. All transfections were performed either in duplicate (for reverse transcription–quantitative polymerase chain reaction [RT-qPCR] assays) or in triplicate (for growth assays).
Cell Enumeration Assays
Cell enumeration assays were performed to determine the effect of reducing YB-1 levels on tumor cell numbers in vitro. A549, HCT116, MCF-7, and MDA-MB-435S cells were transfected in triplicate, using 1.5 × 103 cells per well with 0.6 pmol of siRNA and 0.1 μL of Lipofectamine RNAiMAX in a 120 μL volume (giving a 5 nM final concentration) in 96-well plates. At the indicated time points (typically 24, 48, 96, and 120 hours) following reverse transfection, the medium was removed, and the plates were frozen at −80°C. When cells at all time points had been harvested, the plates were thawed, and DNA content was determined using a SYBR Green I–based fluorimetric assay, as described previously (45). Briefly, lysis buffer (10 mM Tris–HCl pH 8.0, 2.5 mM EDTA, 1% [vol/vol] Triton X-100) containing SYBR Green 1 (Invitrogen) at 1:4000 (vol/vol) was added to the wells, and the plates were incubated overnight at 4°C and protected from exposure to light. After thorough mixing of the lysates, the plate was directly placed into a Wallac Victor 2 (Turku, Finland) or BioTek Synergy 2 (Winooski, VT) plate reader, and the fluorescence signal indicative of DNA content (as a marker for cell number) was measured for 1 second at an excitation frequency of 485 nm and emission frequency of 535 nm. Growth curves were plotted by normalizing to day 1 fluorescence values.
Real-Time RT-qPCR
To measure the level of YB-1 mRNA after transfection with siRNAs, A549, HCT116, and MCF-7 cells were transfected with 15 pmol of siRNA and 2.5 μL of RNAiMax in 3 mL of media (final concentration of 5nM) using 5 × 105 cells per well in a six-well plate. At 24 hours after transfection, cells were harvested and total RNA extracted using TRI reagent (Sigma-Aldrich; St Louis, MO). Oligo-dT-primed cDNA was synthesized using Moloney Murine Leukemia Virus (M-MLV) reverse transcriptase (Promega, Madison, WI). Quantitative real-time RT-PCR reactions were performed using the Rotor-Gene 6000 system (Qiagen, Venlo, the Netherlands) with QuantiTect SYBR Green PCR Master Mix (Qiagen). Levels of mRNA were normalized to β-Actin mRNA (product of the housekeeping gene, ACTB) and relative change in YB-1 mRNA levels following treatment with YB-1-specific or Ctrl siRNAs was calculated using the ΔΔCT method (46). Primer sequences for RT-qPCR were as follows: YBX1_RT_Forward, 5′-GGAGTTTGATGTTGTTGAAGGA-3′; YBX1_RT_Reverse, 5′-AACTGGAACACCACCAGGAC-3′; ACTB_RT_Forward, 5′-GGATGCAGAAGGAGATCACTG-3′; ACTB_RT_Reverse, 5′-CGATCCACACGGAGTACTTG-3′.
Western Blotting
To determine YB-1 protein levels after siRNA transfections, western blotting was performed. A549, HCT116, MCF-7, or MDA-MB-435S cells were harvested and suspended at a concentration of 2 × 104 cells/μL in ice-cold lysis buffer (10 mM Tris–HCl buffer pH 8, 140 mM NaCl, 1% [vol/vol] NP-40, 0.1% [wt/vol] SDS, 1 mM EDTA, 5 mM DTT, with Complete Protease Inhibitor [Roche; Basel, Switzerland]). The suspension was rotated at 4°C for 30 minutes, and the lysate was clarified by centrifugation at 16 000 × g at 4°C for 20 minutes. For each sample, 2.5 μL of lysate was mixed with 2.5 μL NuPAGE 4× lithium dodecyl sulfate gel loading buffer (Invitrogen), 1 μL NuPAGE 10× sample reducing agent (Invitrogen), and 4 μL H2O. Proteins were denatured by incubation at 70°C for 10 minutes and separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis using pre-cast NuPAGE Bis–Tris 10% mini-gels (Invitrogen) run at 200 V for 1 hour, following the manufacturer’s instructions. Proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (Pall Co., Pensacola, FL) using the X-Cell system (Invitrogen). After protein transfer, the membrane was stained with Ponceau S (Sigma-Aldrich). The membrane was then blocked, probed, and subjected to chemiluminescent detection of YB-1 following instructions for the Western Breeze chemiluminescence substrate (Invitrogen). Blots were probed with a rabbit anti-YB-1 polyclonal antibody that had been raised to a N-terminal peptide of YB-1, as previously described (47), which was used at a dilution of 1:1000 in Western Breeze primary antibody diluent (Invitrogen). To ensure even loading, blots were reprobed with mouse anti-β-Actin (Abcam, Cambridge, UK) at a dilution of 1:10 000.
A549 Xenograft Model
Before we examined the ability of A549 cells containing siRNA to YB-1 to form tumors in nude mice, we used rapid amplification of 5′ complementary ends (5′-RACE) to confirm that siYB-1 reduced endogenous YB-1 mRNA levels by specific cleavage of the YB-1 transcript (data not shown). For mouse experiments, female, homozygous CD-1 nude mice were obtained from Charles River Laboratories (Wilmington, MA) and used when 5–6 weeks old. Then, 1 × 107 A549 cells were reverse transfected with RNAiMAX and 300 pmol of either the siYB-1 or siCtrl siRNA in 30 mL of transfection medium (10 nM final concentration). Untreated cells were included as an additional control. Twenty-four hours after the transfection, the cells were detached from the flasks with trypsin-EDTA, and the enzymatic reaction was stopped by the addition of FBS. After two washes with phosphate-buffered saline (PBS), the cells were resuspended at 8 × 107 cells/mL in PBS. Using a 26-gauge needle, groups of CD-1 nude mice were injected subcutaneously on the flank with 100 μL of the cell suspension, making sure that an equal number of cells were inoculated in each group (five mice were implanted with untransfected cells, six mice with siCtrl-transfected cells, and eight mice with siYB-1-transfected cells). Tumor growth was assessed at the indicated intervals by measuring the tumor size with digital calipers. The greatest longitudinal diameter (length) and the greatest transverse diameter (width) were determined. Tumor volume, based on caliper measurements, was calculated by the formula 0.52 × length × width2. On completion of the experiment, the mice were killed by CO2 asphyxiation, and tumors were excised and weighed. Our mouse experiments were performed with animal care ethics approval by the Genesis Research and Development Animal Ethics Committee, under Code of Ethical Conduct approved by the National Animal Ethics Advisory Committee and Ministry of Agriculture and Forestry, in accordance with the New Zealand Animal Welfare Act of 1999.
Statistical Analysis of Non-Microarray Data
Non-microarray data that received statistical analysis included in vitro growth and RT-qPCR assays of A549, HCT116, MCF-7, and MDA-MB-435S cells following siRNA transfection; A549 xenograft growth and tumor weights; and E2F1 promoter-luciferase reporter gene assays. One-way analysis of variance and post hoc tests (Tukey Honestly Significant Difference test) were performed in the R environment (38).
Microarray Analysis of In Vitro Samples
To identify the immediate downstream targets of YB-1, A549, HCT116, and MCF-7 cells were transfected with 15 pmol of siRNA and 2.5 μL of RNAiMax in 3 mL of media (final concentration of 5 nM) using 5 × 105 cells per well in a six-well plate or left untransfected. After 48 hours, cells were harvested and washed in PBS. Total RNA was extracted using TRI reagent (Sigma-Aldrich), purified, and checked for integrity. RNA was then amplified and labeled using Illumina TotalPrep RNA Amplification Kit (Ambion, Foster City, CA), according to manufacturer’s instructions. Labeled RNA was hybridized to an Illumina BeadChip (Illumina Human Ref 8_V2) and the chips scanned on an Illumina BeadStation bead array scanner.
The data has been submitted to GEO and can be accessed at http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE28433.
Statistical Analysis of In Vitro Microarray Data
Analysis of microarray data generated from A549, HCT116, and MCF-7 cells transfected with YB-1 siRNAs was performed in the R statistical environment (38). The array that had the lowest Euclidean distance from the median of all arrays was chosen as the index array, and the signal intensities from all other arrays were normalized to this array using loess intensity–dependent smoothing. Differential intensity of mRNA signals between the two control samples (untransfected cells and siCtrl) and the two YB-1 siRNA–transfected samples (siYB-1 and siYB-2) were estimated using the limma package for R (48). Although the experimental design (two different control types and only two replicates) prevented a full statistical analysis, linear models analysis (via limma) was used to rank mRNAs according to their mean fold change between the control and siRNA-treated groups and their consistency within the control and siRNA-treated groups. Inspection of hierarchical cluster analyses suggested that P values less than .01, with no correction for multiple testing, provided a reasonable cutoff for this study (Supplementary Figure 1, available online). Lists of transcripts with P values less than .01 were then evaluated using Ingenuity Pathways Analysis (www.ingenuity.com), GATHER [http://gather.genome.duke.edu/ (41)] and scripts developed at our university to identify whether they were enriched for transcripts that encoded proteins with a particular function, proteins that might act as markers of the activity of particular molecular pathways, or proteins that are encoded by genes with particular transcription factor binding motifs in their promoters. The likelihood that the mRNA lists were enriched for any of these mRNA groups due to chance alone was assessed using a permutation analysis: We compared the enrichment identified in the mRNA lists for our experiments with the enrichment seen in each of 10 000 randomly selected mRNA lists the same size as our lists but derived from all genes represented on the Illumina Human Ref 8_V2 microarrays. All statistical tests were two-sided.
E2F1 Reporter Gene Assays
MCF-7 cells were seeded at 1 × 105 cells per well in a six-well plate 18 hours before transfection with 1 μg of an E2F1 promoter–luciferase construct (49) and 10 or 50 ng of E2F1 (50) using Fugene (Roche). To ensure that equivalent quantities of DNA were transfected into the cells, 40 ng pUC19 vector DNA (Invitrogen) was co-transfected with the 10 ng E2F1 transfections. pGL2-Basic (51) was transfected in parallel as a control construct lacking the E2F1 promoter. To test the effect of YB-1 on the activity of the E2F1 promoter, levels of endogenous YB-1 were reduced by transfection of 1 × 105 cells with 15 pmol of siYB-1, or siYB-2, with siCtrl as a negative control, using RNAiMax (Invitrogen). The cells were transfected for 6 hours before the media was replaced. They were harvested after 48 hours, at which time there was little effect on cell numbers from YB-1 depletion (see Figure 4). Cell numbers were determined using ViCell (Beckman Coulter, Brea, CA). Luciferase levels were measured using Luciferase Assay System (Promega) using a TD-20/20 Luminometer (Turner Designs, Sunnyvale, CA). All transfections were performed in triplicate, and for each transfection, the luciferase levels were determined three times. The average reading was normalized to the total cell number to give the relative luciferase activity.
Figure 4.
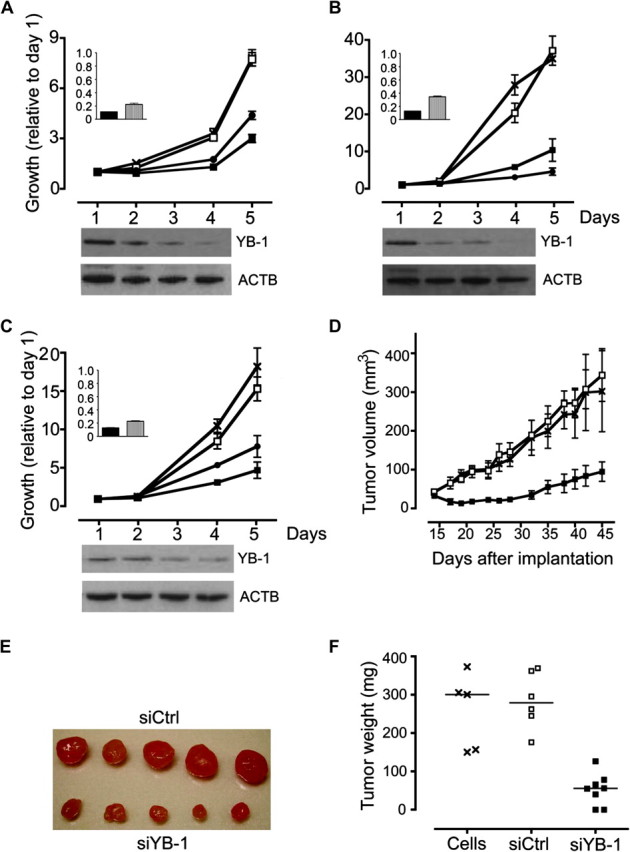
Effect of YB-1 small interfering RNA (siRNA) treatment on growth of cancer cell lines and tumor xenografts. A–C) Effects of YB-1 silencing in three cancer cell lines: (A) MCF-7, (B) HCT116, and (C) A549 cells. Cells were transfected with two different YB-1-specific siRNAs (siYB-1 [■], siYB-2 [●]), a control siRNA (siCtrl [□]), or left untransfected (×). The main graphs show in vitro growth during days 1–5 after transfection, after normalization to the mean fluorescence values on day 1 in triplicate transfections. Error bars show 95% confidence intervals. There were statistically significant differences in cell growth between cells transfected with both YB-1 siRNAs vs siCtrl at day 5 (in MCF-7 cells, P < .001; in HCT116 cells, P < .001; in A549 cells, P < .001 for siYB-1 and P < .002 for siYB-2 [Tukey Honestly Significant Difference (HSD) test]). Insets: levels of YB-1 at 24 hours after transfection as measured by reverse transcription–quantitative polymerase chain reaction in cells transfected with siYB-1 (solid bar) and siYB-2 (hatched bar) normalized to levels in cells treated with siCtrl. Western blots below each graph show the levels of YB-1 protein in cells that were transfected with siYB-1 and harvested on the days corresponding to the coordinates on the x-axes of the graphs above. Blots show levels of YB-1 protein (top) or actin, as a loading control (below) stained with YB-1 antibody or ACTB antibody, respectively. D–F) Effects of YB-1 silencing on tumors in mice with A549 cell xenografts. The mouse model was established by transfection of A549 cells with 10 nM siYB-1 or siCtrl just before implantation into CD1 nude mice. Untransfected cells were transfected in parallel. (D) Tumor volumes at 15–45 days after implantation. Mice were implanted with cells containing siYB-1 (eight mice per group; ■) or siCtrl (six mice per group; □) or untransfected cells (five mice per group; ×). Values represent mean for at least five mice per group. Error bars represent 95% confidence intervals. There was a statistically significant difference in tumor growth between mice implanted with cells containing siYB-1 vs siCtrl (P < .001; Tukey HSD test). (E) Photograph of five tumors excised on day 45 after implantation from mice with siCtrl-treated xenografts (top) and siYB-1-transfected xenografts (bottom). All results show representative data from three or more experiments. (F) Tumor weights of excised A549 cell xenografts. There was a statistically significant difference in siYB-1 vs siCtrl tumor growth (P < .001; Tukey HSD test).
Chromatin Immunoprecipitation (ChIP)
To collect chromatin for immunoprecipitation, MCF-7 cells were grown to near confluence (1 × 107 cells). The cells were harvested, and nuclei were collected by lysing cells for 5 minutes in 1 mL of ice-cold lysis buffer (5 mM HEPES pH 8.0, 85 mM KCl, 0.4% [vol/vol] NP40 with 1% [vol/vol] PMSF, and 2% [vol/vol] complete protease inhibitors [Roche]) and centrifuged at 4°C for 15 seconds at 2300g. To collect chromatin depleted of YB-1 for use as a negative control, nuclei were isolated 72 hours after transfection of 1 × 107 MCF-7 cells with 150 pmol of siYB-1 in 30 mL (5 nM final concentration). The chromatin was fixed within the nuclei for 10 minutes at room temperature in 1 mL of cell lysis buffer that was prepared without NP40 but containing 2.5% (vol/vol) formaldehyde, and then 75 μL of 0.11 mM glycine was added for 5 minutes to quench the fixing reaction. Nuclei were pelleted by centrifugation at 2300g for 15 seconds and then lysed for 15 minutes on ice in 50 μL of 50 mM Tris–HCl, pH 8.1, 1 mM EDTA, 1% (wt/vol) SDS with protease inhibitors. This suspension was diluted in 750 μL of ice-cold Buffer A (20 mM HEPES-KOH pH 7.9, 2 mM MgCl2, 300 mM KCl, 10% [vol/vol] glycerol, 0.1% [vol/vol] Triton X-100, and 1 mM EDTA) before chromatin was sonicated using a Branson 450 Sonicator (Danbury, CT). Sheared chromatin was collected after centrifugation at 16 000g for 10 minutes at 4°C.
For immunoprecipitation of chromatin, 20 μg of YB-1 antibody (47) was incubated with 112 μL sheared chromatin in 225 μL ice-cold immunoprecipitation buffer (20 mM HEPES-KOH pH 7.9, 150 mM KCl, 1.0% [vol/vol] Triton X-100, and 1.5 mM EDTA) with protease inhibitors for 1 hour at 4°C. To this, 40 μL Protein G beads (Roche) were added, and the suspension was then incubated overnight at 4°C on a rocking platform.
For immunodepletion experiments, 20 μg of a rabbit polyclonal E2F1 antibody (Millipore, Billerica, MA) was incubated with 112 μL chromatin for 1 hour at 4°C followed by incubation with 40 μL of Protein G beads as above to remove E2F1-bound chromatin. Then, 20 μg of rabbit polyclonal YB-1 antibody (47) was added to immunoprecipitate any remaining YB-1-bound chromatin, followed by incubation with Protein G beads as above. For controls, normal rabbit IgG–conjugated Protein G agarose beads (Cell Signaling, Danvers, MA) or Protein G beads alone were added to the chromatin in the absence of YB-1 or E2F1 antibodies. After overnight incubation at 4°C, the samples were centrifuged for 30 seconds at 12 000g, and the pellets were washed twice at room temperature with Buffer A and then with TE buffer (10 mM Tris–HCl, pH 8.0, 1 mM EDTA). Before PCR, bound chromatin fragments were eluted in 100 μL Elution buffer (50 mM NaHCO3, 1% [wt/vol] SDS, 30 mg proteinase K) by incubation at 45°C for 3 hours. DNA was purified using a PCR cleanup kit (Qiagen).
For PCR amplification of immunoprecipitated promoters, we used 1 unit of FastStart Taq polymerase (Roche) in 1× PCR buffer with 2 mM MgCl2, 200 μM dNTP, and 0.2 μM forward and reverse primers. The reactions were performed as follows: activation at 95°C for 4 minutes and then 40 cycles of denaturation at 95°C for 30 seconds, annealing for 30 seconds (at 60°C for CDK1, CDC6, CCND1, E2F2, and MCM2, or 59°C for TOP2A or 58°C for E2F5), elongation at 72°C for 1 minute, and then final extension at 72°C for 7 minutes. A number of E2F1 targets were attempted, but the methodology was only optimized for the promoters of CDK1, CDC6, CCND1, E2F2, E2F5, MCM2, and TOP2A genes (52–57). Primer sequences were as follows: CDK1_F, 5′-TGGAGCCTAGGAGTTGGAGA-3′; CDK1_R, 5′-AAGGGCGGCTAGAGAAAAA G-3′; CDC6_F, 5′-TGTTGCTTCCCTCTTTCTAGGC-3′; CDC6_R, 5′-CAAATCCGAATGGCCACAG-3′; CCND1_F, 5′-CGAAGGGGAGAGGGCTTTTT-3′; CCND1_R, 5′-TTAGGGGGTGAGGTGGAGGT-3′; E2F2_F, 5′-TTTGAATGCCAAGGCTCAGA-3′; E2F2_R, 5′-CTGCGACAGTACAGGCTCCA-3′; E2F5_F, 5′-CTTTGGTCAAGAACTTTGGTCA-3′; E2F5_R, 5′-GTGAATGAGGCCCACGAC-3′; MCM2_F, 5′-AAGCGGAGCTATGAAAGCAG-3′; MCM2_R, 5′-GACGGATATTCCCGTGACC-3′; TOP2A_F, 5′-GCTTCCCTACCACTGCTCTG-3′; TOP2A_R, 5′-TCCGACTGGCTACTTTTGCT-3′.
Results
Bioinformatic Analysis of YB-1 Expression in Breast Tumors
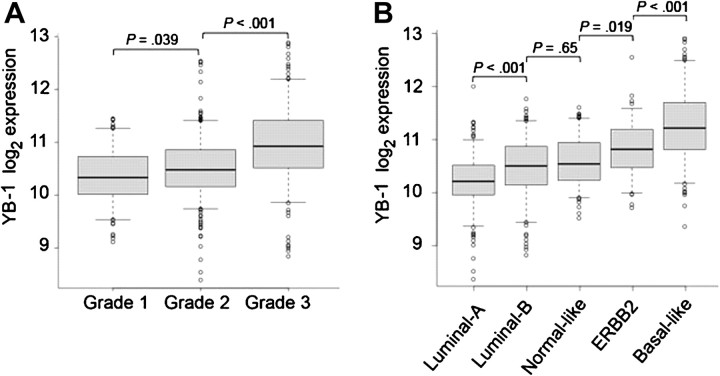
To more fully understand the role played by YB-1 and its associated gene, YBX1, in breast cancer, we performed a bioinformatic analysis of Affymetrix microarray data from a collection of 771 breast tumors derived from four published microarray studies (30–33) and one unpublished study (collected by L. D. Miller). Because histological grade and genetic tumor subtype are important indicators of breast cancer prognosis (58,59), we first compared the abundance of YB-1 mRNA with the histological grade and subtypes of all 771 tumors. YB-1 mRNA levels were found to increase in association with more advanced tumor grade (Figure 1, A). Furthermore, YB-1 mRNA levels were higher in the faster growing estrogen receptor–negative breast cancer subtypes, the ERBB2+ and basal-like subtypes, than in the estrogen receptor–positive luminal A and luminal B subtypes (Figure 1, B), as previously observed for YB-1 protein (6).
Figure 1.
Association of Y-box binding factor 1 (YB-1) mRNA levels with tumor grade and subtype in breast cancers from a cohort of 771 patients. Data from five microarray studies were examined to determine levels of mRNA in the tumors that were complementary to the YBX1 208627_s_at probe set. A) Association of YB-1 mRNA levels with histological grade (one-way analysis of variance [ANOVA], P < 2.2 × 10−16), B) Association of YB-1 mRNA levels with tumor subtype (one-way ANOVA, P < 2.2 × 10−16). In each graph, the solid lines within the boxes represent the median value and the boxes show the 25th to 75th percentile range of YB-1 mRNA levels. Bars represent 95% confidence intervals, with circles representing outliers. All statistical tests were two-sided.
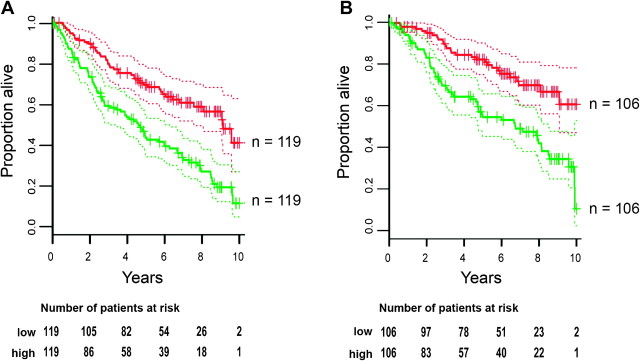
To address the importance of YB-1 in tumor initiation, growth, and metastasis, we focused on the 375 breast cancer patients who did not receive adjuvant therapy. Patients who received treatment were excluded to avoid confounding due to a variety of treatment regimens. We performed survival analysis using Cox proportional hazards models, and follow-up data for 10 years after surgery to determine whether YB-1 mRNA abundance in primary tumors was associated with breast cancer recurrence and occurrence of distant metastasis, measured in terms of disease-free survival and distant metastasis–free survival, respectively. YB-1 mRNA levels were strongly inversely associated with both disease-free survival and distant metastasis–free survival (Cox proportional hazards P values were 1.7 × 10−6 and 5.5 × 10−5, respectively; Figure 2), indicating that presence of higher levels of YB-1 mRNA was associated with poor prognosis. We conclude that patients whose tumors have higher than median levels of YB-1 mRNA are at a greater risk of breast cancer recurrence (hazard ratio = 8.0, 95% confidence interval [CI] = 3.4 to 18.8) and at a greater risk of breast cancer metastasis (hazard ratio = 7.4, 95% CI = 2.8 to 19.4) than patients with lower levels of YB-1 expression.
Figure 2.
Association of Y-box binding factor 1 (YB-1) mRNA expression with prognosis in 375 non-adjuvant treated breast cancer patients. Kaplan–Meier survival plots were prepared based on patient data after placing patients into either of two groups based on whether their tumors had YB-1 mRNA levels (based on microarrays using YBX1 probe set 208627_s_at) that were less than or greater than median levels, as indicated by the red lines and green lines, respectively. Dotted lines indicate 95% confidence intervals (CIs). Number of patients at risk are shown below the plots, with “low” and “high” representing less than or greater than median levels of YB-1 mRNA, respectively. A) Association of YB-1 mRNA levels with disease-free survival (log-rank test, P = 2.9 × 10−6; Cox proportional hazards, P = 1.7 × 10−6; the hazard ratio between the tumors with lowest and highest YB-1 mRNA abundance = 8.0, 95% CI = 3.4 to 18.8). B) Association of YB-1 mRNA levels with distant metastasis–free survival (log-rank test, P = 3.5 × 10−5; Cox proportional hazards, P = 5.5 × 10−5; the hazard ratio between the tumors with lowest and highest YB-1 mRNA abundance = 7.4, 95% CI = 2.8 to 19.4). All statistical tests were two-sided.
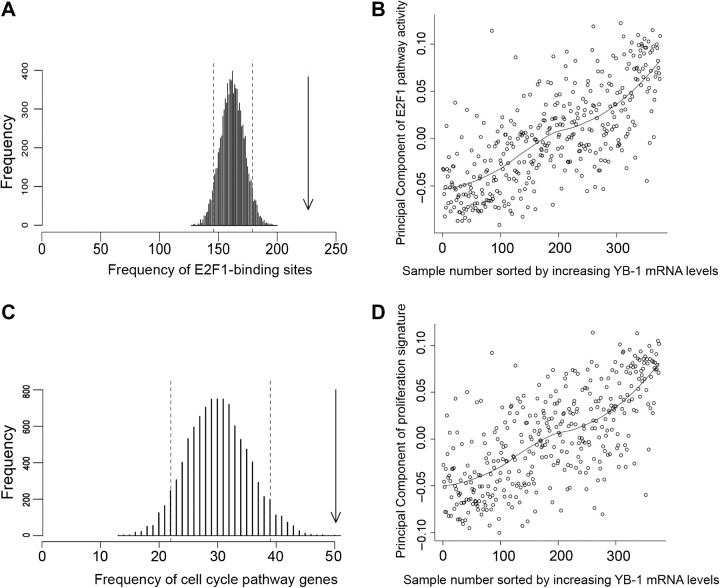
To elucidate how YB-1 mRNA abundance may be associated with poor prognosis in breast cancer, we identified mRNAs with expression profiles that were similar to that of YB-1 mRNA in the 375 tumors from non-adjuvant-treated breast cancer patients. We calculated Spearman correlation coefficients for all of the Affymetrix probe sets compared with the YBX1 probe set 208627_s_at. There were 439 Affymetrix probe sets whose expression was positively correlated with YBX1 208627_s_at expression more highly than the 98th percentile (Spearman ρ > .591; Supplementary Table 2, available online). When we compared the promoters of the 379 genes that corresponded with these 439 probe sets, using the TRANSFAC Pro v8.2 transcription factor binding motif database (60) and the GATHER web tool (41), 334 of the 379 genes had consensus sites within their promoters for a member of the E2F cell cycle–associated transcription factor family (Bayes Factor of 20 for V$E2F1_Q3_01 indicating strong evidence to support this association). A permutation analysis indicated that the probability of randomly selecting a list of 439 probe sets from Affymetrix U133A microarrays with this degree of E2F1 promoter motif enrichment was less than 10−4 (Figure 3, A). We also used Principal Component Analysis to determine whether the activity of the E2F1 pathway was associated with the level of YB-1 mRNA. Affymetrix probe sets that corresponded to 63 canonical E2F1 target genes identified from TRANSPATH (Supplementary Table 1, available online) were used as a statistical summary of E2F1 target gene expression to build a Principal Component of E2F1 pathway activity for each of the tumors. The magnitude of this Principal Component was plotted against tumors sorted by increasing YB-1 mRNA expression (from probe set 208627_s_at) and showed that the activity of the E2F1 pathway correlated with the YBX1 probe set 208627_s_at signal (Spearman ρ = .73; Figure 3, B). The 439 probe sets that detected gene expression patterns that were positively correlated with those detected by YBX1 208627_s_at, as described above, were also compared with the Gene Ontology database (42) using the GATHER web tool. Forty-four of them were associated with the Gene Ontology pathways for the cell cycle, with a Bayes Factor of 13 (empirical P value from permutation analysis < 10−4; Supplementary Table 2, available online; Figure 3, C). In addition, the magnitude of a Principal Component derived from probe sets that correspond to the proliferation gene signature of Whitfield et al. (61) was also correlated with the YBX1 probe set 208627_s signal (Spearman ρ = .69; Figure 3, D). Taken together, these results suggest that 1) YB-1 mRNA expression is correlated with the expression of genes that have E2F1 binding motifs in their promoters, 2) YB-1 mRNA expression is strongly correlated with a linear combination of E2F1 target gene expression, 3) YB-1 mRNA expression is positively correlated with the expression of genes that encode proteins with functions related to cell cycle activity, and 4) YB-1 mRNA expression is correlated with an mRNA signature that has previously been used as a biomarker for cellular proliferation.
Figure 3.
Association of Y-box binding factor 1 (YB-1) mRNA expression with E2F1 and proliferation pathways in tumors from non-adjuvant-treated breast cancer patients. A) Permutation analysis for E2F1 binding sites. Random lists of 439 probe sets were analyzed for the presence of V$E2F1_Q3_01 sites in their promoters, resulting in a Poisson distribution. Dotted lines represent the 5th and 95th percentiles. The arrow shows the abundance of V$E2F1_Q3_01 sites in the promoters of the 379 genes whose transcripts show greater than 98th percentile correlation (Spearman ρ > .591) with the YBX1 208627_s_at probe set (P ≤ 1 × 10−4). B) Principal Component analysis showing the activity of the E2F1 pathway (inferred from a Principal Component of the expression of known E2F1 transcriptional targets) plotted against increasing YB-1 mRNA expression (YBX1 208627_s_at levels). Each individual sample is shown as an open circle. A line of best fit has been drawn to show the trend. C) Permutation analysis showing enrichment of the Gene Ontology cell cycle pathway in 439 probe sets whose expression was positively correlated with YB-1 mRNA (YBX1 208627_s_at) expression. Analysis of the level of enrichment of the Gene Ontology cell cycle pathway in 10 000 random lists of 379 genes resulted in a Poisson distribution. Dotted lines represent 5th and 95th percentiles. The arrow indicates the enrichment of the Gene Ontology cell cycle pathway in the 439 probe sets that had a higher than 98th percentile–positive correlation with YB-1 mRNA (YBX1 208627_s_at) expression (P ≤ 1 × 10−4). D) Principal Component analysis showing a breast cancer proliferation signature (60) plotted against increasing levels of YB-1 mRNA (YBX1 208627_s_at) expression. Each individual tumor sample is shown as an open circle. A line of best fit has been drawn to show the trend. Correlation between YB-1 and the E2F1 pathway, and YB-1 and the breast cancer proliferation signature (Spearman ρ = .73 and.69, respectively) is much greater than correlation between the Principal Components of the E2F1 pathway and the breast cancer proliferation signature (Spearman ρ = .26; data not shown).
Effect of Reduced YB-1 Expression Tumor Cell Growth In Vitro and In Vivo
We next investigated the effect of YB-1 silencing on cell growth in vitro and in xenograft models. Three different cell lines were transfected with two distinct YB-1-targeting siRNAs (siYB-1 and siYB-2); MCF-7 luminal A breast cancer cells, HCT116 colorectal adenocarcinoma cells, and A549 non–small cell lung adenocarcinoma cells. Analysis of YB-1 mRNA levels by RT-qPCR at 1–2 days after transfection confirmed that there were reduced amounts of YB-1 mRNA following transfection of the two YB-1-specific siRNAs, siYB-1 and siYB-2 (Figure 4, A–C inset). Compared with the control siRNA, siYB-1 or siYB-2 reduced YB-1 mRNA by 88% and 78%, respectively, in MCF-7 cells, by 85% and 65%, respectively, in HCT116 cells. and by 87% and 76%, respectively, in A549 cells (Figure 4, A–C inset). Western blotting demonstrated a slight decrease in YB-1 protein levels by 2 days and a substantial reduction by 3 days after transfection (Figure 4, A–C).
In all three cell lines, the silencing of YB-1 expression led to decreased cell numbers with time (Figure 4, A–C). At 2 days after transfection, cells that received YB-1 siRNAs were very slightly reduced in number. After 5 days, A549 cells that received YB-1 siRNAs showed between 49% and 69% reduced cell growth compared with controls (after normalization to numbers on day 1, number of A549 cells with siYB-1 = 4.71, number of A549 cells with siCtrl = 15.18, difference = 10.47, 95% CI = 10.11 to 10.84, P < .001; Figure 4, C). HCT116 cells that received YB-1 siRNAs showed between 72% and 87% growth inhibition (after normalization to numbers on day 1, number of HCT116 cells with siYB-1 = 4.58, number of HCT116 cells with siCtrl = 37.14, difference = 32.56, 95% CI = 32.12 to 32.99, P < .001; Figure 4, B).
To confirm that the ability of YB-1 siRNA to reduce cancer cell growth was not confined to cell lines grown in vitro, we tested the most active YB-1 siRNA in an A549 xenograft model. In these experiments, 240 pmol of siYB-1 or siCtrl were transfected into 8 × 106 A549 cells 24 hours before the cells were subcutaneously injected into the flanks of CD-1 nude mice. Untransfected cells were injected into another group of nude mice in parallel. The average tumor volume was measured during the course of the experiment. Tumors became established in all five mice that were injected with the untransfected cells and all six mice that were injected with siCtrl-transfected cells. By contrast, tumors became established in only six of eight mice that were implanted with cells that had been transfected with siYB-1. The tumors that contained YB-1 siRNA grew more slowly than those in the control groups (on day 45, A549 tumor volume with siYB-1 = 33 mm3, with siCtrl = 174 mm3, difference = 141, 95% CI = 66.36 to 216.40, P < .001; Figure 4, D). On completion of the experiment, the tumors were removed and weighed. The weights of the tumors in mice implanted with siYB-1-transfected A549 cells were considerably lower than those from the control groups (on day 45, A549 tumor weight with siYB-1 = 33 mg, with siCtrl = 285 mg, difference = 232, 95% CI = 134.90 to 329.80, P < .001; Figure 4, E and F).
Effects of Reduced YB-1 Expression in Three Cell Lines
To elucidate the molecular mechanisms by which YB-1 controls cell growth, we examined the effect of YB-1 silencing on global mRNA abundance profiles in vitro. A549, HCT116, and MCF-7 cells were transfected with siYB-1, siYB-2, siCtrl, or left untransfected before analysis using Illumina microarrays. To analyze changes specific to the effects of YB-1 reduction, we focused on the earliest time point at which YB-1 protein levels began to decrease (2 days after transfection; Figure 4). When mRNA expression profiles of cells transfected with the two YB-1 siRNAs were compared with those from the two control samples, YB-1 mRNA was the most dramatically differentially expressed of all mRNAs (reduced by 79%–83% in all cell lines). Linear models analysis [via limma (48)] was used to rank differentially expressed transcripts. Only 25 transcripts were differentially expressed in all three cell lines (Supplementary Table 3, available online).
Thirty-two of the mRNA transcripts that were differentially expressed following YB-1 reduction in each individual cell line are components of prognostic signatures for breast cancer, adenocarcinoma, or colorectal carcinoma (Table 1). In MCF-7 cells, reduction of YB-1 altered the expression of 11 transcripts that are part of the 70-gene signature that is predictive of distant metastasis–free survival in node-negative breast cancer patients (62), as well as the expression of six different transcripts that are part of a 50-gene genetic grade indicator that is strongly associated with tumor proliferation (58) and four others that are part of the 76-gene signature that predicts breast cancer recurrence in lymph node–negative patients (63). Downstream targets of YB-1 in A549 cells included seven transcripts that were identified as part of a 50-gene signature defining low-risk and high-risk adenocarcinoma patients (64) and another four that were molecular components of a 54-gene adenocarcinoma recurrence signature (65).
Table 1.
Association of genes whose expression is modulated by YB-1 levels with prognostic signatures for breast, lung, and colorectal carcinomas*
| Cell line | Prognostic marker genes with differential probe set signal (P < .01) following YB-1 reduction | Study |
| MCF-7 | CCT5, FOXM1, HMGB3, KPNA2, MCM2, PIGV | Breast cancer—Genetic Grade Indicator signature (58) |
| MCF-7 | GBE1, HMGB3, IGFBP5, KIAA1324, NDRG1, PIB5PA, PTDSS1, QDPR, RNB6, STX1A, TGFB3 | Breast cancer—70-gene signature (62) |
| MCF-7 | ACACB, ARHGDIB, DUSP4, KPNA2 | Breast cancer—76-gene signature (63) |
| A549 | CKAP4, HPCAL1, KRT18, MT2A, SLC20A1, SLC2A1, STC1 | Lung adenocarcinoma—50-gene signature (64) |
| A549 | CDC23, GULP1, IL8, PRNP | Lung adenocarcinoma—54-gene signature (65) |
| HCT116 | DNAJB9, PTPLAD1 | Colorectal carcinoma—43-gene signature (66) |
Downstream targets of YB-1 were determined by transfection of MCF-7 breast cancer cells, A549 lung adenocarcinoma cells, or HCT116 colorectal cancer cells with YB-1 siRNAs. Probe sets with differential signals were identified by two-tailed linear model analysis, without false discovery rate, controlled to 1% using the method of Benjamini and Hochberg (48). The prognostic gene signatures examined were: a genetic grade signature for breast cancer that is strongly associated with histological grade (57); the 70-gene signature for breast cancer patients that is predictive of distant metastasis–free survival independent of histological grade, tumor size, and adjuvant treatment in lymph node–negative patients (and used in the Mammaprint assay) (61); the 76-gene signature that predicts recurrence within 5 years in non-adjuvant-treated lymph node–negative breast cancer patients [in which only three genes overlap with the 70-gene signature (63)]; a 50-gene signature for early-stage lung adenocarcinoma that predicts survival independent of stage of disease; a 54-gene signature that predicts disease recurrence in lung adenocarcinoma patients; and a 43-gene signature that could accurately predict 3-year survival in colorectal cancer patients (66). siRNA = small interfering RNA; YB-1 = Y-box binding factor 1.
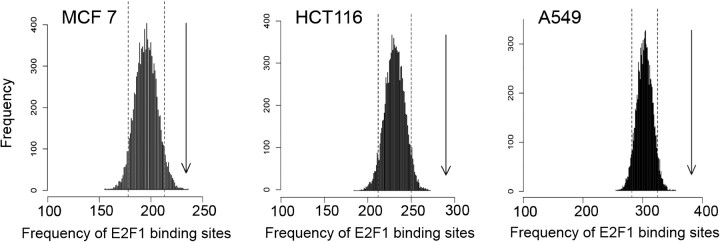
The mRNAs that were differentially expressed following YB-1 reduction (P ≤ .01) were not consistently enriched for any particular function in the Gene Ontology database. However, when the genes encoding these mRNAs were analyzed using the TRANSFAC database and the GATHER web tool (41), the genes that were differentially expressed in each cell line were enriched for E2F1 and/or E2F consensus-binding motifs (V$E2F1_Q3_01 and V$E2F1_Q6) within their promoters. Because E2F and/or E2F1 sites are relatively common in the promoters of genes important for cancer biology (67), permutation analysis was performed to determine whether the transcripts modulated following YB-1 silencing were indeed enriched for E2F1 and/or E2F sites. For all three cell lines, fewer than 5% of 10 000 randomly selected lists of genes had the degree of E2F1 promoter motif enrichment that we had observed (Figure 5). Therefore, although the mRNAs modulated following YB-1 reduction have very little overlap between the three cell lines examined, in all cell lines the promoters of genes encoding the regulated mRNAs were substantially enriched for E2F motifs.
Figure 5.
Analysis of E2F1 consensus binding sites in the promoters of genes whose expression was modulated by YB-1. Permutation analysis was performed to evaluate the number of V$E2F1_Q3_01 sites in the promoters of genes whose expression was changed (P ≤ .01) following treatment of MCF-7, HCT116, or A549 cells with small interfering RNA (siRNA)to YB-1. For each cell line, random sets of the same number of genes were analyzed for the presence of V$E2F1_Q3_01 sites, resulting in a Poisson distribution. The dotted lines represent the 5th and 95th percentiles. The arrows show the abundance of V$E2F1_Q3_01 sites in the promoters of genes whose transcripts were changed following YB-1 reduction (P ≤ .01), indicating that for each cell line, the number of such E2F1 consensus binding sites in the YB-1-modulated mRNA population is greater than expected by chance (P ≤ 1 × 10−4).
Regulation of E2F Family Promoters and YB-1
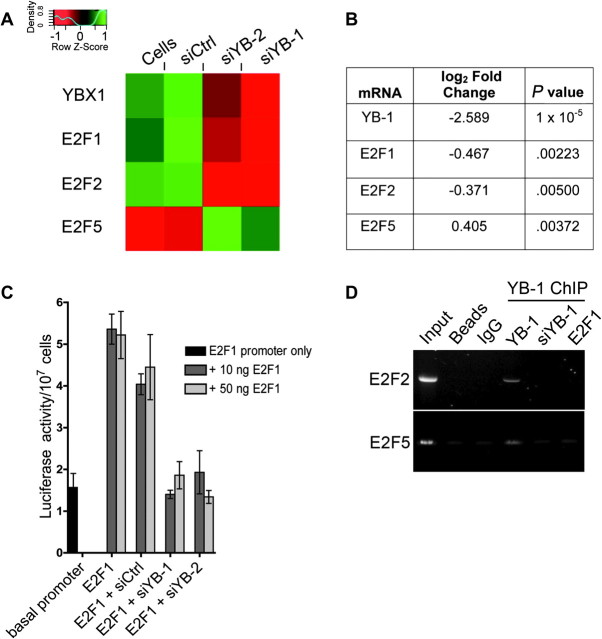
Because of the association between YB-1 mRNA expression and E2F family-mediated transcription that is described above, we analyzed the effect of YB-1 reduction on the expression of individual members of the E2F family. A trend was observed in which YB-1 silencing led to a reduction in the expression of “activator” E2Fs (eg E2F1, E2F2), but an increase in the expression of “repressor” E2Fs [eg, E2F5, E2F7; Figure 6, A and B and Supplementary Figure 2, available online; (68)]. Because E2F1 is the most characterized of all of the E2Fs, and known to regulate cell cycling (68), it was studied further. To determine whether changes in E2F1 mRNA levels were due to transcriptional regulation by YB-1, reporter assays were performed in which a luciferase gene under the control of the E2F1 promoter (49) was transfected into A549 cells. Because E2F1 is autoregulated (49), the E2F1 promoter construct was co-transfected with an E2F1 overexpression vector and either siCtrl or siYB-1 or siYB-2 to reduce YB-1 levels. The in vitro activity of the E2F1 promoter was reduced to basal levels in cells transfected with the YB-1 targeting siRNAs (relative luciferase activity in cells with E2F1 promoter, siCtrl, and 10 ng E2F1 = 4.04; with E2F1 promoter, siYB-1, and 10 ng E2F1 = 1.40; with E2F1 promoter, siYB-2, and 10 ng E2F1 = 1.94; difference, siCtrl vs siYB-1 = −2.64, 95% CI = −3.57 to −1.71, P< .001; difference, siCtrl vs siYB-2 = −2.10, 95% CI = −3.03 to −1.17, P < .001; relative luciferase activity in cells with E2F1 promoter, siCtrl, and 50 ng E2F1 = 4.45; with E2F1 promoter, siYB-1 and 50 ng E2F1 = 1.86; with E2F1 promoter, siYB-2 and 50 ng E2F1 = 1.26; difference, siCtrl vs siYB-1 = −2.59, 95% CI = −3.53 to −1.66, P < .001; difference, siCtrl vs siYB-2 = −3.19, 95% CI = −4.13 to −2.26, P < .001; Figure 6, C). These results suggest that YB-1 is required to transcriptionally activate the E2F1 promoter in A549 cells. A similar result was observed in MCF-7 cells (data not shown).
Figure 6.
Association of YB-1 with transcription of E2F family genes. A) Hierarchical clustering derived from linear models analysis of MCF-7 cells transfected with YB-1 or control siRNAs. The diagram shows changes in the expression levels of particular E2F family transcripts with altered levels of YB-1 expression (red represents decreased expression and green represents increased expression). E2F1 and E2F2 are considered to be “activator” E2Fs, and E2F5 is considered to be a “repressor” E2F. B) Table showing values generated from linear models analysis for YBX1, E2F1, E2F2, and E2F5 probe sets following silencing of YB-1 in MCF-7 cells as described above (“log2 fold change” equals the log2 fold change in expression level of these genes in cells transfected with YB-1 siRNAs compared with control treatments.). C) Effect of YB-1 levels on the activity of the E2F1 promoter in vitro. A549 cells were transfected with either the E2F1 promoter–luciferase reporter construct alone or co-transfected with 10 or 50 ng of E2F1 in the presence of siCtrl, siYB-1, or siYB-2 to determine the effect of decreased YB-1 levels on activity of the E2F1 promoter. Cells were harvested, counted, and assayed for reporter gene activity after 48 hours, and luciferase levels were calculated relative to cell number. Error bars represent 95% confidence intervals. There were statistically significant differences between the siCtrl-transfected cells and those transfected with either of the YB-1 siRNAs (P < .001; Tukey HSD test). Results were generated from triplicate transfections for each treatment and represent typical results generated from at least three independent experiments. D) Chromatin immunoprecipitation of E2F2 and E2F5 promoters in MCF-7 cells using a YB-1 antibody to determine whether YB-1 binds to these promoters in vivo. DNA isolated from cells was cross-linked to permanently couple any proteins to the chromatin. Promoters bound by YB-1 were then immunoprecipitated using a YB-1 antibody. Polymerase chain reaction was performed with primers to the E2F2 and E2F5 promoters to determine whether they were bound by YB-1. As controls, cells transfected with siYB-1 72 hours before immunoprecipitation, or cells pre-incubated with E2F1 antibody (to deplete promoters bound by E2F1) before immunoprecipitation were tested in parallel. Input = DNA before immunoprecipitation; Beads = protein G beads in the absence of DNA; IgG = chromatin immunoprecipitation with a negative control antibody; YB-1 ChIP = E2F2 and E2F5 promoter fragments immunoprecipitated with the YB-1 antibody; YB-1 = YB-1 antibody only; siYB-1 = cells pretreated with siYB-1; E2F1 = cells preincubated with E2F1 antibody. Results shown are typical of experiments performed at least three times.
Taking another approach, ChIP was performed with a YB-1 antibody to determine whether other E2F family promoters were bound by endogenous YB-1 in MCF-7 cells. This experiment showed that the E2F2 and E2F5 promoters were immunoprecipitated by the YB-1 antibody (Figure 6, D), suggesting that YB-1 was bound to these promoters. As a control, when YB-1 levels were reduced by siYB-1 treatment before harvesting the cells, little or no immunoprecipitated product was observed. Furthermore, because E2F family transcription factors can bind to the promoters of other E2F family genes (55,68), we also immunodepleted E2F1-bound chromatin before immunoprecipitation with the YB-1 antibody. In this experiment, again, little or no immunoprecipitated product was detectable. These results suggest that YB-1 binds to several E2F promoters in MCF-7 cells and is a potential transcriptional regulator of these genes.
YB-1 and E2F and/or E2F1 Target Genes
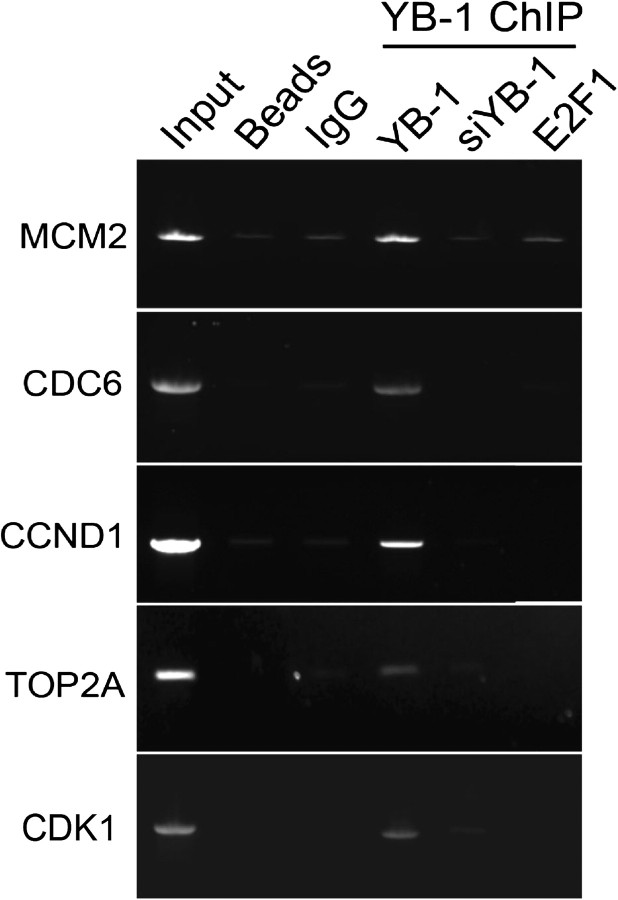
The microarray data suggested that YB-1 might directly or indirectly co-regulate a number of E2F and/or E2F1 target genes. To examine this question, ChIP-PCR was used to determine whether YB-1 bound to some well-defined E2F1-regulated genes in MCF-7 cells. ChIP of promoters from the CDK1, CDC6, CCND1, MCM2, and TOP2A genes was performed as described above. These results showed that endogenous YB-1 binds to the promoters of at least all five of these E2F1-regulated genes in MCF-7 cells (Figure 7) and therefore suggest that YB-1 is a potential transcriptional regulator of these genes.
Figure 7.
Chromatin immunoprecipitation of E2F1 target gene promoters using YB-1 antibody. Chromatin from MCF-7 cells was cross-linked to fix bound proteins to the DNA. Cells were lysed and the chromatin was incubated with a YB-1 antibody to immunoprecipitate promoters bound by YB-1. Polymerase chain reaction was then performed to amplify promoter fragments of known E2F1-regulated genes to determine whether they were bound by YB-1. As controls, MCF-7 cells were transfected with siYB-1 72 hours before they were harvested to deplete YB-1, or chromatin was precleared with an E2F1 antibody to remove E2F1-bound promoters before incubation with YB-1 antibody. Input = MCF-7 DNA before immunoprecipitation; Beads = protein G beads in the absence of DNA; IgG = chromatin immunoprecipitation with the IgG negative control antibody; YB-1 ChIP = promoters immunoprecipitated with the YB-1 antibody; YB-1 = YB-1 antibody only; siYB-1 = cells pretreated with siYB-1; E2F1 = cells preincubated with E2F1 antibody. Figure shows typical results obtained from at least three independent experiments.
Discussion
This study aimed to further our understanding of the role that YB-1 plays in tumor biology. Our initial work focused on the bioinformatic analysis of YB-1 mRNA in breast cancer and showed that YB-1 mRNA was strongly associated with overall survival and recurrence in patients who did not receive adjuvant therapy, as previously observed by immunohistochemical staining of YB-1 protein (69). This analysis also showed a strong association of YB-1 mRNA expression with distant metastasis formation in non-adjuvant-treated patients, which has not been previously reported to our knowledge.
Our study showed that YB-1 mRNA expression was associated with the expression of E2F1 target genes in breast cancer. After performing the same analysis on Gene Expression Omnibus microarray datasets from lung cancer (70) and colorectal cancer (71), we also observed a correlation between YB-1 mRNA levels and the expression of genes that have E2F1-binding motifs within their promoters (Supplementary Figure 3, available online), suggesting that YB-1 may be associated with the expression of E2F1 targets in many tumor types. Potentially one of the most interesting findings was that silencing YB-1 expression in three different tumor cell lines led to changes in the expression of several transcripts that are components of tumor prognostic signatures. These observations suggest that YB-1 may directly regulate the expression of a number of genes that are biomarkers for prognosis, relapse, and metastasis. Consistent with this conclusion, a computational search identified both YB-1 and E2F1 as “upstream Master Regulators” of the 70-gene breast cancer signature, which suggested that these proteins are actually drivers of the tumor phenotype (72). Therefore, YB-1 mRNA levels could act as a surrogate marker for other more complex genomic signatures of recurrence and invasion.
Microarray analysis of in vitro experiments identified E2F1 and/or other E2F target genes as immediately downstream of YB-1. We further demonstrated that endogenous YB-1 could bind to the promoters of a panel of canonical E2F1 transcriptional targets, but our microarray data suggest that YB-1 might actually co-regulate the expression of many E2F1 and/or E2F target genes. Interestingly, analysis of the literature identifies several transcriptional targets of both YB-1 and E2F1, such as CCNA1, CCNB1 (24,73,74), CDC6 (20,53), and TP53 (4,75). Furthermore, analyzing the data from a ChIP-on-chip study that identified over 6000 promoters potentially bound by YB-1 in a basal-like breast cancer cell line, SUM149 (76), we noted that more than 4000 contained E2F1 motifs, supporting our findings. In another basal-like breast cancer cell line, we observed that reduction of YB-1 mRNA levels led to decreased expression of a panel of E2F1 targets, as well as of E2F1 and E2F2 mRNAs (Supplementary Figure 4, available online). We were also interested to note that the E2F1, E2F2, E2F3, E2F4, and E2F7 promoters were bound by YB-1 in a study by Finkbeiner et al. (76), which is consistent with our conclusion that YB-1 might actually regulate both “activator” and “repressor” E2Fs.
Despite our evidence from tumor microarray and cell culture experiments that YB-1 regulates the cell cycle via an E2F1 and/or E2F pathway and our finding that this conclusion is consistent with the literature, caveats still exist. The main limitation is that transcriptional regulation of the E2F pathway by YB-1 is largely inferred from in vitro data (ChIP and reporter assays), and there are no experiments that directly address how YB-1 controls transcription of E2F1-target genes. For example, YB-1 may complex with E2F to aid its binding to promoters, it may interact with the pRb cell cycle check-point regulator to release bound E2F1, or it may affect E2F-mediated transcription by some other mechanism. The establishment of direct correlations between YB-1 levels and the levels of particular E2F-regulated transcripts would add support to the model and begin to address how the E2F family members coordinate to positively and negatively regulate the cell cycle. Some of these questions are currently under investigation in our laboratory.
In summary, here we present several lines of evidence showing that YB-1 is associated with the activity of E2F transcription factors. We propose that YB-1 controls tumor cell growth by regulating transcription of E2Fs and their downstream targets in a cell-type specific manner, which could provide a unifying mechanism for understanding the diverse actions of YB-1 in normal and tumor cells.
Funding
This work was supported by the New Zealand Breast Cancer Research Trust (3621880 to A.L. and C.G.P. and 3607717 supporting C.G.P.); the Health Research Council of New Zealand (07/284 to A.G.W. and A.W.B.); the Cancer Institute NSW in Australia (RLP 01/05 to W.S., R.M., and A.W.B. and 09/RSA/1-38 to J.L.S.); and National Health and Medical Research Council (571193 to J.L.S.).
Supplementary Material
Footnotes
Both C. G. Print and A. W. Braithwaite made equal contributions to this study.
The authors are solely responsible for the design of the study; the collection, analysis, and interpretation of the data; the writing of the article, and the decision to submit the article for publication.
We thank Sheryl Feng, Marika Eszes, Sandra Fitzgerald, Igor Ruza, and Hamish Campbell for technical assistance and Hiromitsu Araki and Daniel Hurley for assistance with R.
References
- 1.Didier DK, Schiffenbauer J, Woulfe SL, Zacheis M, Schwartz BD. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc Natl Acad Sci U S A. 1988;85(19):7322–7326. doi: 10.1073/pnas.85.19.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mertens PR, Harendza S, Pollock AS, Lovett DH. Glomerular mesangial cell-specific transactivation of matrix metalloproteinase 2 transcription is mediated by YB-1. J Biol Chem. 1997;272(36):22905–22912. doi: 10.1074/jbc.272.36.22905. [DOI] [PubMed] [Google Scholar]
- 3.Lasham A, Lindridge E, Rudert F, Onrust R, Watson J. Regulation of the human fas promoter by YB-1, Puralpha and AP-1 transcription factors. Gene. 2000;252(1–2):1–13. doi: 10.1016/s0378-1119(00)00220-1. [DOI] [PubMed] [Google Scholar]
- 4.Lasham A, Moloney S, Hale T, et al. The Y-box-binding protein, YB1, is a potential negative regulator of the p53 tumor suppressor. J Biol Chem. 2003;278(37):35516–35523. doi: 10.1074/jbc.M303920200. [DOI] [PubMed] [Google Scholar]
- 5.Homer C, Knight DA, Hananeia L, et al. Y-box factor YB1 controls p53 apoptotic function. Oncogene. 2005;24(56):8314–8325. doi: 10.1038/sj.onc.1208998. [DOI] [PubMed] [Google Scholar]
- 6.Wu J, Lee C, Yokom D, et al. Disruption of the Y-box binding protein-1 results in suppression of the epidermal growth factor receptor and HER-2. Cancer Res. 2006;66(9):4872–4879. doi: 10.1158/0008-5472.CAN-05-3561. [DOI] [PubMed] [Google Scholar]
- 7.En-Nia A, Yilmaz E, Klinge U, Lovett DH, Stefanidis I, Mertens PR. Transcription factor YB-1 mediates DNA polymerase alpha gene expression. J Biol Chem. 2005;280(9):7702–7711. doi: 10.1074/jbc.M413353200. [DOI] [PubMed] [Google Scholar]
- 8.Das S, Chattopadhyay R, Bhakat KK, et al. Stimulation of NEIL2-mediated oxidized base excision repair via YB-1 interaction during oxidative stress. J Biol Chem. 2007;282(39):28474–28484. doi: 10.1074/jbc.M704672200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldsmith ME, Madden MJ, Morrow CS, Cowan KH. A Y-box consensus sequence is required for basal expression of the human multidrug resistance (mdr1) gene. J Biol Chem. 1993;268(8):5856–5860. [PubMed] [Google Scholar]
- 10.Stein U, Jürchott K, Walther W, Bergmann S, Schlag PM, Royer HD. Hyperthermia-induced nuclear translocation of transcription factor YB-1 leads to enhanced expression of multidrug resistance-related ABC transporters. J Biol Chem. 2001;276(30):28562–28569. doi: 10.1074/jbc.M100311200. [DOI] [PubMed] [Google Scholar]
- 11.Evdokimova V, Tognon C, Ng T, et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 2009;15(5):402–415. doi: 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 12.Bargou RC, Jürchott K, Wagener C, et al. Nuclear localization and increased levels of transcription factor YB-1 in primary human breast cancers are associated with intrinsic MDR1 gene expression. Nat Med. 1997;3(4):447–450. doi: 10.1038/nm0497-447. [DOI] [PubMed] [Google Scholar]
- 13.Shibahara K, Sugio K, Osaki T, et al. Nuclear expression of the Y-box binding protein, YB-1, as a novel marker of disease progression in non-small cell lung cancer. Clin Cancer Res. 2001;7(10):3151–3155. [PubMed] [Google Scholar]
- 14.Shibao K, Takano H, Nakayama Y, et al. Enhanced coexpression of YB-1 and DNA topoisomerase II alpha genes in human colorectal carcinomas. Int J Cancer. 1999;83(6):732–737. doi: 10.1002/(sici)1097-0215(19991210)83:6<732::aid-ijc6>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 15.Giménez-Bonafé P, Fedoruk MN, Whitmore TG, et al. YB-1 is upregulated during prostate cancer tumor progression and increases P-glycoprotein activity. Prostate. 2004;59(3):337–349. doi: 10.1002/pros.20023. [DOI] [PubMed] [Google Scholar]
- 16.Kamura T, Yahata H, Amada S, et al. Is nuclear expression of Y box-binding protein-1 a new prognostic factor in ovarian serous adenocarcinoma? Cancer. 1999;85(11):2450–2454. doi: 10.1002/(sici)1097-0142(19990601)85:11<2450::aid-cncr21>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 17.Bergmann S, Royer-Pokora B, Fietze E, et al. YB-1 provokes breast cancer through the induction of chromosomal instability that emerges from mitotic failure and centrosome amplification. Cancer Res. 2005;65(10):4078–4087. doi: 10.1158/0008-5472.CAN-04-4056. [DOI] [PubMed] [Google Scholar]
- 18.Lee C, Dhillon J, Wang MY, et al. Targeting YB-1 in HER-2 overexpressing breast cancer cells induces apoptosis via the mTOR/STAT3 pathway and suppresses tumor growth in mice. Cancer Res. 2008;68(21):8661–8666. doi: 10.1158/0008-5472.CAN-08-1082. [DOI] [PubMed] [Google Scholar]
- 19.Shiota M, Izumi H, Onitsuka T, et al. Twist promotes tumor cell growth through YB-1 expression. Cancer Res. 2008;68(1):98–105. doi: 10.1158/0008-5472.CAN-07-2981. [DOI] [PubMed] [Google Scholar]
- 20.Basaki Y, Taguchi K, Izumi H, et al. Y-box binding protein-1 (YB-1) promotes cell cycle progression through CDC6-dependent pathway in human cancer cells. Eur J Cancer. 2010;46(5):954–965. doi: 10.1016/j.ejca.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 21.Chatterjee M, Rancso C, Stühmer T, et al. The Y-box binding protein YB-1 is associated with progressive disease and mediates survival and drug resistance in multiple myeloma. Blood. 2008;111(7):3714–3722. doi: 10.1182/blood-2007-05-089151. [DOI] [PubMed] [Google Scholar]
- 22.Cobbold LC, Wilson LA, Sawicka K, et al. Upregulated c-myc expression in multiple myeloma by internal ribosome entry results from increased interactions with and expression of PTB-1 and YB-1. Oncogene. 2010;29(19):2884–2891. doi: 10.1038/onc.2010.31. [DOI] [PubMed] [Google Scholar]
- 23.Ladomery M, Sommerville J. A role for Y-box proteins in cell proliferation. Bioessays. 1995;17(1):9–11. doi: 10.1002/bies.950170104. [DOI] [PubMed] [Google Scholar]
- 24.Jurchott K, Bergmann S, Stein U, et al. YB-1 as a cell cycle-regulated transcription factor facilitating cyclin A and cyclin B1 gene expression. J Biol Chem. 2003;278(30):27988–27896. doi: 10.1074/jbc.M212966200. [DOI] [PubMed] [Google Scholar]
- 25.Sakura H, Maekawa T, Imamoto F, Yasuda K, Ishii S. Two human genes isolated by a novel method encode DNA-binding proteins containing a common region of homology. Gene. 1988;73(2):499–507. doi: 10.1016/0378-1119(88)90514-8. [DOI] [PubMed] [Google Scholar]
- 26.Berquin IM, Pang B, Dziubinski ML, et al. Y-box-binding protein 1 confers EGF independence to human mammary epithelial cells. Oncogene. 2005;24(19):3177–3186. doi: 10.1038/sj.onc.1208504. [DOI] [PubMed] [Google Scholar]
- 27.Cobbold LC, Spriggs KA, Haines SJ, et al. Identification of internal ribosome entry segment (IRES)-trans-acting factors for the Myc family of IRESs. Mol Cell Biol. 2008;28(1):40–49. doi: 10.1128/MCB.01298-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jenkins RH, Bennagi R, Martin J, Phillips AO, Redman JE, Fraser DJ. A conserved stem loop motif in the 5’untranslated region regulates transforming growth factor-β(1) translation. PLoS One. 2010;5(8):e12283. doi: 10.1371/journal.pone.0012283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu YN, Yip GW, Tan PH, et al. Y-box binding protein 1 is up-regulated in proliferative breast cancer and its inhibition deregulates the cell cycle. Int J Oncol. 2010;37(2):483–492. [PubMed] [Google Scholar]
- 30.Miller LD, Smeds J, George J, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci U S A. 2005;102(38):13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loi S, Haibe-Kains B, Desmedt C, et al. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol. 2007;25(10):1239–1246. doi: 10.1200/JCO.2006.07.1522. [DOI] [PubMed] [Google Scholar]
- 32.Desmedt C, Piette F, Loi S, et al. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13(11):3207–3214. doi: 10.1158/1078-0432.CCR-06-2765. [DOI] [PubMed] [Google Scholar]
- 33.Ivshina AV, George J, Senko O, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66(21):10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 34.Irizarry RA, Bolstad BM, Collin F, et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31(4):e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Z, Fan C, Oh DS, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96–108. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohga T, Koike K, Ono M, et al. Role of the human Y box-binding protein YB-1 in cellular sensitivity to the DNA-damaging agents cisplatin, mitomycin C, and ultraviolet light. Cancer Res. 1996;56(18):4224–4228. [PubMed] [Google Scholar]
- 38.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 39.Harrington DP, Fleming TR. A class of rank test procedures for censored survival data. Biometrika. 1982;69(3):553–566. [Google Scholar]
- 40. Therneau T. Survival: survival analysis, including penalized likelihood. R package (version 2.36-5). R foundation for Statistical Computing. 2011.
- 41.Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22(23):2926–2933. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- 42.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25(2):288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bunz F, Dutriaux A, Lengauer C, et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282(5393):1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 45.Reid G, Wallant NC, Patel R, et al. Potent subunit-specific effects on cell growth and drug sensitivity from optimised siRNA-mediated silencing of ribonucleotide reductase. J RNAi Gene Silencing. 2009;5(1):321–330. [PMC free article] [PubMed] [Google Scholar]
- 46.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 47.Cohen SB, Ma W, Valova VA, et al. Genotoxic stress-induced nuclear localization of oncoprotein YB-1 in the absence of proteolytic processing. Oncogene. 2010;29(3):403–410. doi: 10.1038/onc.2009.321. [DOI] [PubMed] [Google Scholar]
- 48.Smyth GK. Bioinformatics and Computational Biology Solutions Using R and Bioconductor. New York, NY: Springer; 2008. Limma: linear models for microarray data; pp. 397–420. [Google Scholar]
- 49.Johnson DG, Ohtani K, Nevins JR. Autoregulatory control of E2F1 expression in response to positive and negative regulators of cell cycle progression. Genes Dev. 1994;8(13):1514–1525. doi: 10.1101/gad.8.13.1514. [DOI] [PubMed] [Google Scholar]
- 50.Hsu SI, Yang CM, Sim KG, Hentschel DM, O’Leary E, Bonventre JV. TRIP-Br: a novel family of PHD zinc finger- and bromodomain-interacting proteins that regulate the transcriptional activity of E2F-1/DP-1. EMBO J. 2001;20(9):2273–2285. doi: 10.1093/emboj/20.9.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stayner CK, Cunliffe HE, Ward TA, Eccles MR. Cloning and characterization of the human PAX2 promoter. J Biol Chem. 1998;273(39):25472–25479. doi: 10.1074/jbc.273.39.25472. [DOI] [PubMed] [Google Scholar]
- 52.Furukawa Y, Terui Y, Sakoe K, Ohta M, Saito M. The role of cellular transcription factor E2F in the regulation of cdc2 mRNA expression and cell cycle control of human hematopoietic cells. J Biol Chem. 1994;269(42):26249–26258. [PubMed] [Google Scholar]
- 53.Yan Z, DeGregori J, Shohet R, et al. Cdc6 is regulated by E2F and is essential for DNA replication in mammalian cells. Proc Natl Acad Sci U S A. 1998;95(7):3603–3608. doi: 10.1073/pnas.95.7.3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Watanabe G, Albanese C, Lee RJ, et al. Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol Cell Biol. 1998;18(6):3212–3222. doi: 10.1128/mcb.18.6.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sears R, Ohtani K, Nevins JR. Identification of positively and negatively acting elements regulating expression of the E2F2 gene in response to cell growth signals. Mol Cell Biol. 1997;17(9):5227–5235. doi: 10.1128/mcb.17.9.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ohtani K, Iwanaga R, Nakamura M, et al. Cell growth-regulated expression of mammalian MCM5 and MCM6 genes mediated by the transcription factor E2F. Oncogene. 1999;18(14):2299–2309. doi: 10.1038/sj.onc.1202544. [DOI] [PubMed] [Google Scholar]
- 57.Kalma Y, Marash L, Lamed Y, Ginsberg D. Expression analysis using DNA microarrays demonstrates that E2F-1 up-regulates expression of DNA replication genes including replication protein A2. Oncogene. 2001;20(11):1379–1387. doi: 10.1038/sj.onc.1204230. [DOI] [PubMed] [Google Scholar]
- 58.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 59.Miller LD, Liu ET. Expression genomics in breast cancer research: microarrays at the crossroads of biology and medicine. Breast Cancer Res. 2007;9(2):206. doi: 10.1186/bcr1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heinemeyer T, Wingender E, Reuter I, et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26(1):362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6(2):99–106. doi: 10.1038/nrc1802. [DOI] [PubMed] [Google Scholar]
- 62.van ‘t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415(6871):530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 63.Wang Y, Klijn JG, Zhang Y, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 64.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8(8):816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 65.Larsen JE, Pavey SJ, Passmore LH, Bowman RV, Hayward NK, Fong KM. Gene expression signature predicts recurrence in lung adenocarcinoma. Clin Cancer Res. 2007;13(10):2946–2954. doi: 10.1158/1078-0432.CCR-06-2525. [DOI] [PubMed] [Google Scholar]
- 66.Eschrich S, Yang I, Bloom G, et al. Molecular staging for survival prediction of colorectal cancer patients. J Clin Oncol. 2005;23(15):3526–3535. doi: 10.1200/JCO.2005.00.695. [DOI] [PubMed] [Google Scholar]
- 67.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Barrette TR, Ghosh D, Chinnaiyan AM. Mining for regulatory programs in the cancer transcriptome. Nat Genet. 2005;37(6):579–583. doi: 10.1038/ng1578. [DOI] [PubMed] [Google Scholar]
- 68.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9(11):785–797. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Habibi G, Leung S, Law JH, et al. Redefining prognostic factors for breast cancer: YB-1 is a stronger predictor of relapse and disease-specific survival than estrogen receptor or HER-2 across all tumor subtypes. Breast Cancer Res. 2008;10(5):R86. doi: 10.1186/bcr2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439(7074):353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 71.Jorissen RN, Gibbs P, Christie M, et al. Metastasis-associated gene expression changes predict poor outcomes in patients with Dukes stage B and C colorectal cancer. Clin Cancer Res. 2009;15(24):7642–7651. doi: 10.1158/1078-0432.CCR-09-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lim WK, Lyashenko E, Califano A. Master regulators used as breast cancer metastasis classifier. Pac Symp Biocomput. 2009;14:504–515. [PMC free article] [PubMed] [Google Scholar]
- 73.Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14(7):804–816. [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu W, Giangrande PH, Nevins JR. E2Fs link the control of G1/S and G2/M transcription. EMBO J. 2004;23(23):4615–4626. doi: 10.1038/sj.emboj.7600459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hallstrom TC, Nevins JR. Specificity in the activation and control of transcription factor E2F-dependent apoptosis. Proc Natl Acad Sci U S A. 2003;100(19):10848–10853. doi: 10.1073/pnas.1831408100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Finkbeiner MR, Astanehe A, To K, et al. Profiling YB-1 target genes uncovers a new mechanism for MET receptor regulation in normal and malignant human mammary cells. Oncogene. 2009;28(11):1421–1431. doi: 10.1038/onc.2008.485. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.