Abstract
Purpose
Human papilloma virus (HPV), HPV-16, is one of the most important prognostic factors for patients with head and neck squamous cell carcinomas (HNSCCs). HPV-positive HNSCCs have a favorable response to radiation therapy (RT) and superior overall survival (OS) compared to HPV-negative HNSCCs. However, an explanation for the mechanisms responsible for the inherent radiosensitivity of HPV-positive HNSCC remains elusive.
Methods and Materials
Records of a cohort of 162 HNSCC patients were reviewed and their outcomes were correlated with their HPV status. Using a panel of HPV-positive and HPV-negative HNSCC cell lines expressing a reporter for cancer stem cells (CSCs) we characterized HPV-positive and HPV-negative lines via flow cytometry, sphereforming capacity assays in vitro, and limiting dilution assays in vivo. Non-CSCs were treated with different doses of radiation and the dedifferentiation of non-CSCs into CSCs was investigated via flow cytometry and qRT-PCR for re-expression of reprogramming factors.
Results
Patients with HPV-positive tumors have superior OS and local-regional control. HPV-positive HNSCC cell lines have lower numbers of CSCs, which inversely correlates with radiosensitivity. HPV-negative HNSCC cell lines lack hierarchy due to enhanced spontaneous dedifferentiation. Non-CSCs from HPV-negative lines show enhanced radiation-induced dedifferentiation compared to HPV-positive lines, and RT induced re-expression of Yamanaka reprogramming factors.
Conclusions
Supporting the favorable prognosis of HPV-positive HNSCCs, we show that HPV-positive HNSCCs have (1) a lower frequency of CSCs, (2) RT can dedifferentiate HNSCC cells into CSCs, and (3) radiation-induced dedifferentiation depends on the HPV status of the tumor.
Keywords: Cancer stem cells, head and neck cancer, proteasome, radiotherapy
Introduction
Approximately 500,000 new cases of head and neck cancer, mostly HNSCC (1), are diagnosed annually and are treated with RT. Despite advances in the treatment of locally advanced HNSCC, survival rates remain largely unchanged over the past 30 years, with a 5-year survival rate of less than 50% (2). Infection with HPV is the cause of some HNSCC, mostly in the oropharynx. HPV-positive HNSCCs have a far better prognosis and an improved response to RT, making HPV infection one of the most important, independent prognostic factors for HNSCC (3, 4)
Previous studies have identified E6/E7-dependent inactivation of p53 and Rb tumor suppressor proteins to be the underlying cause for oncogenic transformation (5, 6). However, studies replicating the clinical radiosensitivity of HPV-positive HNSCC experimentally are sparse and conflicting. Recent reports demonstrated, that HPV-positive HNSCC cell lines are more radiosensitive than HPV-negative cell lines (7, 8) but a clear mechanistic link between overexpression of HPV genes and increased radiosensitivity is still missing, thus indicating a complex role of HPV in HNSCC RT.
The CSC hypothesis postulates that most cancers are organized in a hierarchically, with CSCs at the apex of the hierarchy. First reported in leukemia (9), CSCs are now being identified and characterized in an increasing number of solid cancers (10), including HNSCC (11).
HNSCC cells with low proteasome activity also have CSC traits, and most importantly high numbers of cells with low proteasome subunit expression were inversely correlated with OS (12). Lack of proteasome activity has been shown to identify CSCs in a variety of solid tumors (13–18) and this feature of CSCs can be exploited in an imaging system for CSCs (19). Cells with low proteasome activity can be identified in cell lines expressing the C-terminal degron of murine ODC (cODC) fused to a green fluorescent protein (ZsGreen-cODC-positive). Cells with normal proteasome activity will express, but immediately degrade the fusion protein (ZsGreen-cODC-negative) (14). In HNSCC cells with low proteasome activity comprise a sub-population of ALDH1-positive cells (12).
It has been recently reported that in breast cancer, RT can convert non-CSCs into CSCs (20). Similar results have been reported for lymphoma (21) and hepatocellular carcinoma (22). The underlying mechanisms included radiation-induced re-expression of the transcription factors Oct4, Sox2, Klf4 and Nanog, which are known for reprogramming somatic cells into induced pluripotent stem (iPS) cells (23).
Here, we test the hypothesis that the radiation response of CSCs in HPV-positive and HPV-negative HNSCC differs and is not reflected in the radiation response of the bulk tumor populations, that RT can dedifferentiate non-stem HNSCC cells into CSC, and that radiation-induced dedifferentiation depends on the HPV status.
Materials and methods
Patients
Records of 164 of patients with locally-advanced oropharyngeal SCC treated between 2006–2012 by primary chemoradiation (n=95) or primary surgery followed by adjuvant radiation (n=69) were reviewed. All patients had Karnofsky Performance Status (KPS) greater than 70. 162 patients with available p16 status were stratified based on p16 expression.
Cell culture
Human HPV-negative UM-SCC4, UM-SCC-17B, FaDu and Cal33, and HPV-positive SCC-47 HNSCC cell lines were a gift from the Department of Hematology/Oncology at UCLA and have been previously described (24). SCC-90 was a kind gift from Sanford Ear, Nose, and Throat Clinic, San Diego. ZsGreen-cODC expressing cells were obtained and grown as described previously (25). For culture medium, please see Supplementary Materials and Methods.
Irradiation
Cells grown as monolayer or sphere cultures were irradiated at room temperature using an experimental X-ray irradiator (Gulmay Medical Inc. Atlanta, GA) (5.519 Gy/min; 250kV; 4mm Be, a 3mm Al, and a 1.5mm Cu filter).
Flow Cytometry
ZsGreen-cODC expression was assessed by flow cytometry (MACSquant Analyzer, Miltenyi Biotech, CA; FloJo software package (vers. X.0.7, Tree Star Inc., OR). Cells were defined as "ZsGreen-cODC-positive" if the fluorescence in FL-1H exceeded the fluorescence level of 99.9% of the empty vector-transfected control cells.
Clonogenic Survival Assays
Clonogenic assays were performed immediately after irradiation and survival curves were fitted using a linear-quadratic model. For more experimental details please see Supplementary Materials and Methods.
Animals
Immune-compromised, six-week old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice, originally obtained from The Jackson Laboratories (Bar Harbor, ME) were re-derived, bred and maintained in a pathogen-free environment in the American Association of Laboratory Animal Care-accredited Animal Facilities in accordance to all local and national guidelines for the care of animals.
Tumor xenotransplantation
ZsGreen-cODC-negative and ZsGreen-cODC-positive cells were sorted via high-speed FACS from monolayer cultures of Cal33 and Fadu. For in vivo limiting dilution assays, cells were injected subcutaneously into the thighs and shoulders of 6-week old female NSG mice (106, 105, 104, 103, or 102 cells per inoculum within Matrigel (BD Biosciences)). Tumor growth was assessed weekly.
Statistics
Differences in the prevalence of diabetes mellitus, chronic obstructive pulmonary disease (COPD), anxiety disorder, major depressive disorder, alcohol, tobacco, and marijuana use between the 2 cohorts at cancer diagnosis were compared using either univariate or multivariate analyses. Average pack year smoking history, was analyzed using unpaired, two-tailed t-tests. Kaplan-Meier plots with two-tailed, Log-rank (Mantel- Cox) analysis were used for survival analysis, time to recurrence and distant metastases.
All experimental results are expressed as mean values ± SE of at least 3 biologically independent experiments (see Supplementary Materials and Methods). For the in vivo tumorigenicity experiments, p-values comparing the CSC frequency in the ZsGreen-cODC-positive versus ZsGreen-cDOC-negative populations were calculated using the Extreme Limiting Dilution Analysis (ELDA) software (26). All other statistical analyses were performed in GraphPad Prism 5.0. P-values of ≤ 0.05 were considered to indicate statistically significant differences.
Results
Patients with HPV-positive HNSCC have superior OS and local control
Several independent studies have reported improved OS of HPV-positive HNSCC patients (3, 4). In agreement with these studies, the two-year OS in our patient cohort was significantly higher in patients with HPV-positive tumors compared to HPV–negative (86 vs. 71% p=0.04) (Figure 1A). In search for other factors that could explain the favorable outcome of HPV-positive HNSCC patients we also analyzed the rate of loco-regional recurrence (LRR) and metastasis in these patients. In a multivariate analysis, only the p16 status reached statistical significance (Supplementary Table 1–3). However, in agreement with previous studies (3), we observed a significant higher rate of LRR in the HPV-negative cohort (30 vs. 4%, p<0.0001) (Figure 1B). Patients with HPV-negative tumors showed trends towards increased distant metastases (17.6 vs. 10.8%, p=0.8617) (Supplementary Figure 1), second malignancies, and need for salvage RT (Supplementary Table 3), suggesting that inferior outcome of patients with HPV-negative HNSCCs results from inferior local control after RT.
Figure 1. Patients with HPV-positive HNSCC tumors have superior OS and local control.
Kaplan-Meier estimates for OS (A) and locoregional control (B) for patients with HPV-positive (blue) or HPV-negative (red) HNSCC. 162 patients with locally advanced oropharyngeal HNSCC treated with primary chemoradiation or primary surgery followed by adjuvant radiation were followed to assess OS and time to local-regional recurrence. The patients were grouped based on p-16 positivity or -negativity.
HPV-positive tumors have a lower frequency of CSCs
Since HPV-positive patients have a significantly lower rate of local-regional recurrence (30 vs. 4%, p<0.0001, Figure 1B), we hypothesized that HPV-positive tumors have fewer intrinsic radioresistant CSCs, as well as a lower radiation-induced dedifferentiation rate (20).
It was previously reported that HPV/p16-negative (HPV-negative) HNSCC cell lines contain a subpopulation of cells with low proteasome activity and that the presence of this population of cells is inversely correlated with OS of HNSCC patients (12). Therefore, we sought to determine whether differences in the frequency of CSCs might explain the different clinical outcome (Figure 1A–B). We characterized the frequency of cells with low proteasome activity in four HPV-negative (Cal33, Fadu, SCC4 and SCC17B) and two HPV-positive cell lines (SCC47 and SCC90). In agreement with the literature, HPV-negative HNSCC cell lines contained varying percentages of ZsGreen-cODC-pos cells (Figure 2A–B). We further confirmed that the ZsGreen-cODC-pos subpopulation of cells has higher sphere-forming capacity compared to the ZsGreen-cODC-neg cells (Figure 2C–D, Supplementary Figure 3). ZsGreen-cODC-pos cells from SCC4, SCC17B and Cal33 showed enhanced sphere-forming capacity in primary spheres (Figure 2C), and at least for SCC4, this was maintained in secondary spheres (Figure 2D). However, when the percentage of ZsGreen-cODC-pos cells in two HPV-positive HNSCC lines was compared to the HPV-negative lines, the HPV-positive lines contained a significantly lower (p<0.0001) percentage of ZsGreen-cODC-pos CSCs (Figure 3A). In addition, and in agreement with previous reports (8, 27), HPV-positive cells were significantly more radiosensitive compared to HPV-negative cells (Figure 3B), and had significantly lower plating efficiencies (Figure 3C), which likely reflects the lower numbers of intrinsic CSCs in HPV-positive lines. Consistently, the percentage of ZsGreen-cODC-pos CSCs correlated with the plating efficiency (Figure 3D, top panel) and the surviving fraction at 2Gy (Figure 3D, bottom panel).
Figure 2. Cells with low 26S proteasome activity in HNSCC lines have increased self-renewal.
(A–B) Different cell lines of HNSCC contain a variable percentage of cells with low proteasome activity. Cells with low (ZsGreen-cODC-pos) and high (ZsGreen-cODC-neg) proteasome activity from three different HNSCC lines were sorted and the sphere forming capacity for these two populations was compared via primary (C) and secondary (D) sphere-forming assays. ZsGreen-cODC-pos cells had a significantly higher self-renewal capacity.
Figure 3. HPV-positive HNSCC lines have lower numbers of CSCs.
(A) The frequency of ZsGreen-cODC-positive CSCs in the HPV-positive HNSCC lines is significantly lower compared to the HPV-negative lines (p<0.0001, unpaired t-test with Welch’s correction). (B) HPV-positive lines are significantly more radiosensitive compared to the HPV-negative lines at 2, 4, and 6Gy (statistical test for side panels: two-way ANOVA). (C) HPV-negative lines have an enhanced plating efficiency compared to the HPV-positive lines (p=0.0004, upaired t-test with Welch’s correction). (D) The frequency of intrinsic CSCs in HPV-positive and HPV-negative lines correlated well with their plating efficiency (top panel, r2=0.85), and surviving fraction at 2Gy (bottom panel, r2=0.68)
Spontaneous dedifferentiation accounts for lack of hierarchy in HPV-negative HNSCC lines
Previous reports demonstrated the use of the ZsGreen-cODC protein-reporter in identifying cells with increased in vivo tumorigenicity in breast and glioma cells (14, 28). We investigated whether the ZsGreen-cODC-neg and ZsGreen-cODC-pos cells sorted from two different HPV-negative HNSCC lines (Cal33 and Fadu) differed in their tumorigenicity in an in vivo limiting dilution assay. In the Cal33 cell line the frequency of CSCs was only enriched 3-fold in the ZsGreen-cODC-pos population (1 CSC/8,880; 95% CI: 2,816–28,005) compared to the ZsGreen-cODC-neg population (1 CSC/29,382; 95% CI: 9,254–93,299) (Figure 4A). We did not observe differences in the percentage of tumor formation between ZsGreen-cODC-neg and ZsGreen-cODC-pos Fadu cells (Figure 4B), although tumors derived from ZsGreen-cODC-pos cells were significantly larger (p<0.0001) at 5-weeks post implantation (Figure 4B, green vs. blue symbols). Most strikingly, we obtained 100% tumorigenicity with only 100 Fadu cells, regardless of their source (Figure 4B), thus indicating a very high frequency of CSC in HPV-negative HNSCC, comparable to advanced stage melanoma (29). However, neither our functional in vitro data, nor published literature (11, 30, 31) supports such high CSC frequencies. Therefore, we next tested if loss of hierarchy in HPV-negative HNSCC tumors in vivo resulted from a high rate of dedifferentiation of non-CSCs into CSCs.
Figure 4. HPV-negative HNSCC cell lines lack cellular hierarchy due to enhanced spontaneous dedifferentiation.
ZsGreen-cODC-positive and ZsGreen-cODC-negative cell populations from two different HPV-negative cell lines, Cal33 (A) and Fadu (B) were sorted and an in vivo limiting dilution assay was performed. ZsGreen-cODC-positive cells from Cal33 (A) were enriched ~3-fold in tumorigenic cells, while no enrichment was observed in the Fadu (B) line. The size of the tumors arising from the ZsGreen-cODC population Fadu cells were significantly larger at 5 weeks after innoculation (B, tumor sizes compared via two-way ANOVA). (C) ZsGreen-cODC-negative cells derived from HPV-negative lines spontaneously dedifferentiate into ZsGreen-cODC-positive cells at a significantly higher efficiency (p<0.0001, upaired t-test, with Welch’s correction) and the dedifferentiating rate correlates with the frequency of intrinsic stem cell numbers (D).
We first investigated whether HPV-negative HNSCC cell lines have a high rate of spontaneous dedifferentiation. ZsGreen-cODC-neg cells from all six HNSCC lines were subjected to high-speed FACS and ZsGreen-cODC-pos CSCs were purged. Spontaneous dedifferentiation of non-CSCs (ZsGreen-cODC-neg) into ZsGreen-cODC-pos CSCs was observed in HPV-negative and HPV-positive lines (Figure 4C). However, HPV-negative lines spontaneously dedifferentiated more efficiently than HPV-positive lines (p<0.001, Figure 4C). The percentage of spontaneously dedifferentiated ZsGreen-cODC-pos CSCs correlated with the percentage of intrinsically occurring ZsGreen-cODC-pos CSCs for the HNSCC lines analyzed (Figure 4D). These results suggested that after removal of the ZsGreen-cODC-pos CSCs from HNSCC cultures the CSC pool is restored through spontaneous dedifferentiation and contributing to the lack of hierarchy found in in vivo limiting dilution assays in the HPV-negative lines (Figure 4A–B).
HPV-negative HNSCC cells show enhanced radiation-induced dedifferentiation
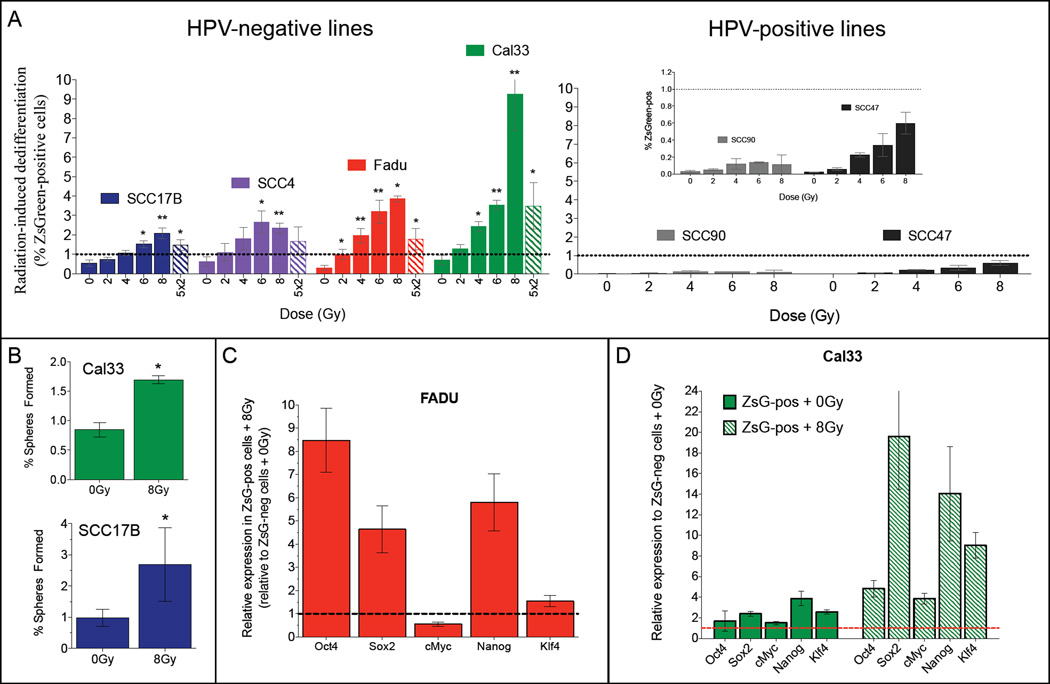
In order to investigate if irradiation can dedifferentiate ZsGreen-cODC-neg HNSCC cells into ZsGreen-cODC-pos CSCs, we sorted ZsGreen-cODC-negative cells from all six HNSCC lines by FACS. After irradiating ZsGreen-cODC-neg cells, we analyzed the percentage of ZsGreen-cODC-pos CSCs after 5 days in culture (Supplementary Figure 2A). Irradiation dose-dependently dedifferentiated ZsGreen-cODC-neg cells from all HPV-negative HNSCC lines into ZsGreen-cODC-pos CSCs cells (Figure 5A and Supplementary Figure 2B). When HPV-negative lines were compared with the HPV-positive lines for their ability to dedifferentiate after RT, HPV-negative lines showed higher radiation-induced dedifferentiation (Figure 5A). Irradiation of ZsGreen-cODC-neg cells with 5 daily doses of 2Gy was less effective in inducing dedifferentiation than a single dose of 8Gy (Figure 5A, stripped bars). In order to confirm that irradiation induced a functional CSC phenotype, we performed sphere-forming capacity assays. A single treatment of 8Gy significantly increased the percentage of cells capable of forming spheres in Cal33 and SCC17B cells (Figure 5B).
Figure 5. Non-CSCs from HPV-negative lines show enhanced radiation-induced dedifferentiation.
ZsGreen-cODC-negative cells were sorted from four different HPV-negative HNSCC lines (A, left panel), and two HPV-positive lines (A, right panel), plated and irradiated. Radiation induced a significant (unpaired two-sided Student’s t-test) dose-dependent dedifferentiation of ZsGreen-cODC-negative cells into ZsGreen-cODC-positive cells in the HPV-negative lines (A, left panel). In contrast, the HPV-positive lines (A, right panel) dedifferentiated much less efficiently. (B) The self-renewal capacity of radiation-induced dedifferentiated cells after a single dose of 8Gy was compared to the non-irradiated cells in two different HPV-negative lines. (C and D) ZsGreen-cODC-neg cells were sorted, plated and irradiated with 0Gy or 8Gy. 5 days later the ZsGreen-cODC-neg and the induced ZsGreen-cODC-pos cells were re-sorted and qRT-PCR was performed for the Yamanaka reprogramming factors in both populations, in two HPV-neg lines, Fadu (C) and Cal33 (D).
Radiation-induced HPV-negative HNSCC stem cells re-express reprogramming factors
It was previously reported that in breast cancer, radiation-induced dedifferentiation is accompanied by the re-expression of Yamanaka reprogramming factors (20), Oct4, Sox2, their downstream target, Nanog, as well as c-Myc and Klf4.
To elucidate the mechanism of irradiation-induced reprogramming of HNSCC non-stem cells into CSCs, we first tested whether irradiation of the bulk population of cells from SCC47, SCC17B and Fadu affects the expression levels of these reprogramming factors. We did not observe any significant differences in expression levels of these factors after radiation with 4Gy (Supplementary Figure 4A–B). We next investigated whether the specific radiation-induced sub-population of CSCs is re-expressing these reprogramming factors after radiation treatment. ZsGreen-cODC-neg cells were first sorted, plated, irradiated with either 0Gy or 8Gy, and 5 days later the ZsGreen-cODC-neg and the induced ZsGreen-cODC-pos populations (from both the 0Gy and 8Gy samples) were re-sorted and analyzed for expression levels of the reprogramming factors. The two HPV-neg lines that reprogram most efficiently, Fadu and Cal33, were chosen in order to obtain a sufficient number of induced ZsGreen-cODC-pos cells to isolate high quality mRNA. 8Gy-induced ZsGreen-cODC-pos cells induced by 8Gy upregulated gene expression of all five reprogramming factors (Figure 5C and D, striped bars), and for some of the genes this radiation-induced re-expression is enhanced in the Cal33 line, in agreement with the more efficient radiation-induced reprogramming observed in this line (Figure 5A). In spontaneously dedifferentiated Cal33 cells these reprogramming factors were also upregulated and irradiation significantly enhanced this re-expression (Figure 5D). These data suggest that radiation-induced dedifferentiation of non-stem HNSCC cells into CSCs correlates with re-expression of reprogramming factors, which have been shown to play an important role in reprogramming of induced pluripotent stem cells (iPS) (23).
Discussion
Previous studies have established clinical and tumorbiological differences between HPV-negative and HPV-positive HNSCCs (32). Adding to this list of inherent differences we show here differences in the frequency and plasticity of CSC.
The search for reliable HNSCC CSC markers is ongoing and CSC enrichment has been reported for CD44-positive (31), ALDH1-positive (33) and CD24+/CD44+ subpopulations. Additionally, HNSCC cells with low proteasome activity also have CSC properties and are a subpopulation of ALDH1-expressing HNSCC cells (12). An imaging system for real-time identification and tracking of cells with low proteasome activity has been validated by a number of groups, different solid cancers (13–18, 34) and it was used to demonstrate that RT can reprogram non-CSCs into CSCs (20).
We have used this system to investigate whether differences exist in the CSC pool, and whether these differences could explain the favorable prognosis of HPV-positive patients. We hypothesized that HPV-positive HNSCC cells that survive RT dedifferentiate into CSCs at low rates, thus explaning the low rate of local recurrences (Figure 1B). We report HPV-dependent, inherent differences in the frequency of intrinsic HNSCC CSCs (Figure 3A), and in the efficiency of RT to induce CSCs (Figure 5A). The significantly lower frequency of CSCs in the HPV-positive lines may explain in part the differences in OS (Figure 1A).
Our findings are in disagreement with a recent report by Zhang et al. showing that HPV-positive HNSCC cells have a greater intrinsic ALDH1+ CSC pool compared to HPV-negative HNSCC (35). While these studies appear contradictory, it is worth noting that ZsGreen-cODC-pos HNSCC CSCs are a sub-population of ALDH1-expressing HNSCC cells (12). The same study showed that ALDH-1 did not predict for OS of either HPV-positive or HPV-negative HNSCC patients (35), while low proteasome subunit expression as a marker for CSCs readily predicted for OS in HNSCC (12).
One of the most surprising findings was the lack of hierarchy in the HPV-negative line, Fadu, and a modest enrichment for CSCs in the ZsGreen-cODC-positive population isolated from the Cal33 line (Figure 4A). Strikingly, we obtained 100% tumor formation from 100 sorted ZsGreen-cODC-pos and ZsGreen-cODC-neg Fadu cells (Figure 4B), suggesting that the observed lack of hierarchy in Cal33 and Fadu was related to high rates of spontaneous dedifferentiation. This was supported by a significant degree of spontaneous dedifferentiation of ZsGreen-cODC-neg cells in HPV-negative lines (Figure 4C), reminiscent of their percentage of intrinsic ZsGreen-cODC-pos cells (Figure 4D). This scenario was re-enforced in experiments investigating radiation-induced dedifferentiation in the HPV-negative and HPV-positive lines. ZsGreen-cODC-neg cells derived from HPV-negative HNSCC lines efficiently dedifferentiate into ZsGreen-cODC-pos cells in a dose dependent manner 5 days after RT. In contrast, the dedifferentiation efficiency of HPV-positive HNSCC cells was significantly lower (Figure 5A). Importantly, we observed that radiation-induced dedifferentiation of HNSCC lines correlated with re-expression of reprogramming factors, Oct4, Sox2, and their downstream target, Nanog. Other reprogramming factors, c-Myc and Klf4 were also upregulated after a single dose of 8Gy.
Conclusions
Our study provides a potential link between the inherent frequency of CSCs in the two main subtypes of HNSCC (HPV-16-negative and HPV-16-positive) and their response to RT. The reasons behind the observed differences in the CSC pool of HPV-negative tumors versus HPV-positive tumors remain elusive and warrant further investigation. Importantly, these studies suggest that HPV-16 status of HNSCC confers a different degree of CSC plasticity, especially in the context of RT. The finding that HPV-negative tumors have a high degree of plasticity and that RT may contribute to tumor recurrence in HPV-negative HNSCC could have major implications for the way these tumors are treated clinically.
Supplementary Material
Summary.
Our studies provide another layer of scientific evidence that offers an alternative explanation for the improved long-term outcome of HPV-positive HNSCC patients: lower frequency of intrinsic CSCs, and an impaired ability to dedifferentiate into HNSCC stem cells after radiation therapy. Thus, preventing spontaneous and radiation-induced dedifferentiation events from occurring could potentially result in significantly improved loco-regional control for these patients.
Acknowledgments
This work was supported by grants from the National Cancer Institute (1RO1CA137110, 1R01CA161294) and the Army Medical Research & Materiel Command’s Breast Cancer Research Program (W81XWH-11-1-0531) to FP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors have declared that no conflict of interest exists.
Author’s contributions
EV and FP conceived of the study, designed the experiments, analyzed the data and wrote the manuscript. AC and CBH collected and analyzed the clinical data. All authors read and approved the final version of the manuscript.
References
- 1.L B. Pathology and genetics of head and neck tumors. Geneva Switzerland: World Health Organization; 2005. [Google Scholar]
- 2.Carvalho AL, Nishimoto IN, Califano JA, Kowalski LP. Trends in incidence and prognosis for head and neck cancer in the United States: a site-specific analysis of the SEER database. Int J Cancer. 2005 May 1;114(5):806–816. doi: 10.1002/ijc.20740. PubMed PMID: 15609302. [DOI] [PubMed] [Google Scholar]
- 3.Lassen P, Eriksen JG, Krogdahl A, Therkildsen MH, Ulhoi BP, Overgaard M, et al. The influence of HPV-associated p16-expression on accelerated fractionated radiotherapy in head and neck cancer: evaluation of the randomised DAHANCA 6&7 trial. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2011 Jul;100(1):49–55. doi: 10.1016/j.radonc.2011.02.010. PubMed PMID: 21429609. [DOI] [PubMed] [Google Scholar]
- 4.Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010 Jul 1;363(1):24–35. doi: 10.1056/NEJMoa0912217. PubMed PMID: 20530316. Pubmed Central PMCID: 2943767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bol V, Gregoire V. Biological basis for increased sensitivity to radiation therapy in HPV-positive head and neck cancers. Biomed Res Int. 2014;2014:696028. doi: 10.1155/2014/696028. PubMed PMID: 24804233. Pubmed Central PMCID: 3996288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kessis TD, Slebos RJ, Nelson WG, Kastan MB, Plunkett BS, Han SM, et al. Human papillomavirus 16 E6 expression disrupts the p53-mediated cellular response to DNA damage. Proceedings of the National Academy of Sciences of the United States of America. 1993 May 1;90(9):3988–3992. doi: 10.1073/pnas.90.9.3988. PubMed PMID: 8387205. Pubmed Central PMCID: 46431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimple RJ, Smith MA, Blitzer GC, Torres AD, Martin JA, Yang RZ, et al. Enhanced radiation sensitivity in HPV-positive head and neck cancer. Cancer Res. 2013 Aug 1;73(15):4791–4800. doi: 10.1158/0008-5472.CAN-13-0587. PubMed PMID: 23749640. Pubmed Central PMCID: 3732540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rieckmann T, Tribius S, Grob TJ, Meyer F, Busch CJ, Petersen C, et al. HNSCC cell lines positive for HPV and p16 possess higher cellular radiosensitivity due to an impaired DSB repair capacity. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2013 May;107(2):242–246. doi: 10.1016/j.radonc.2013.03.013. PubMed PMID: 23602369. [DOI] [PubMed] [Google Scholar]
- 9.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994 Feb 17;367(6464):645–648. doi: 10.1038/367645a0. PubMed PMID: 7509044. [DOI] [PubMed] [Google Scholar]
- 10.Vlashi E, Pajonk F. Cancer stem cells, cancer cell plasticity and radiation therapy. Semin Cancer Biol. 2014 Jul 12; doi: 10.1016/j.semcancer.2014.07.001. PubMed PMID: 25025713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhaijee F, Pepper DJ, Pitman KT, Bell D. Cancer stem cells in head and neck squamous cell carcinoma: a review of current knowledge and future applications. Head & neck. 2012 Jun;34(6):894–899. doi: 10.1002/hed.21801. PubMed PMID: 21850700. [DOI] [PubMed] [Google Scholar]
- 12.Lagadec C, Vlashi E, Bhuta S, Lai C, Mischel P, Werner M, et al. Tumor cells with low proteasome subunit expression predict overall survival in head and neck cancer patients. BMC Cancer. 2014;14:152. doi: 10.1186/1471-2407-14-152. PubMed PMID: 24593279. Pubmed Central PMCID: 3975871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vlashi E, Lagadec C, Chan M, Frohnen P, McDonald AJ, Pajonk F. Targeted elimination of breast cancer cells with low proteasome activity is sufficient for tumor regression. Breast Cancer Res Treat. 2013 Sep;141(2):197–203. doi: 10.1007/s10549-013-2688-6. PubMed PMID: 24013708. Pubmed Central PMCID: 3814133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vlashi E, Kim K, Lagadec C, Donna LD, McDonald JT, Eghbali M, et al. In vivo imaging, tracking, and targeting of cancer stem cells. J Natl Cancer Inst. 2009 Mar 4;101(5):350–359. doi: 10.1093/jnci/djn509. PubMed PMID: 19244169. Pubmed Central PMCID: 2727141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan J, Zhang Q, Wang Y, You M. 26S proteasome activity is down-regulated in lung cancer stem-like cells propagated in vitro. PLoS One. 2010;5(10):e13298. doi: 10.1371/journal.pone.0013298. PubMed PMID: 20949018. Pubmed Central PMCID: 2952619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muramatsu S, Tanaka S, Mogushi K, Adikrisna R, Aihara A, Ban D, et al. Visualization of stem cell features in human hepatocellular carcinoma reveals in vivo significance of tumor-host interaction and clinical course. Hepatology. 2013 Jul;58(1):218–228. doi: 10.1002/hep.26345. PubMed PMID: 23447025. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi K, Tamari K, Ishii H, Konno M, Nishida N, Kawamoto K, et al. Visualization and characterization of cancer stem-like cells in cervical cancer. Int J Oncol. 2014 Sep 24; doi: 10.3892/ijo.2014.2670. PubMed PMID: 25269542. [DOI] [PubMed] [Google Scholar]
- 18.Adikrisna R, Tanaka S, Muramatsu S, Aihara A, Ban D, Ochiai T, et al. Identification of pancreatic cancer stem cells and selective toxicity of chemotherapeutic agents. Gastroenterology. 2012 Jul;143(1):234–245. e7. doi: 10.1053/j.gastro.2012.03.054. PubMed PMID: 22510202. [DOI] [PubMed] [Google Scholar]
- 19.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, et al. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature. 1992 Dec 10;360(6404):597–599. doi: 10.1038/360597a0. PubMed PMID: 1334232. [DOI] [PubMed] [Google Scholar]
- 20.Lagadec C, Vlashi E, Della Donna L, Dekmezian C, Pajonk F. Radiation-induced reprogramming of breast cancer cells. Stem Cells. 2012 May;30(5):833–844. doi: 10.1002/stem.1058. PubMed PMID: 22489015. Pubmed Central PMCID: 3413333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Salmina K, Jankevics E, Huna A, Perminov D, Radovica I, Klymenko T, et al. Up-regulation of the embryonic self-renewal network through reversible polyploidy in irradiated p53-mutant tumour cells. Exp Cell Res. 2010 Aug 1;316(13):2099–2112. doi: 10.1016/j.yexcr.2010.04.030. PubMed PMID: 20457152. Epub 2010/05/12. eng. [DOI] [PubMed] [Google Scholar]
- 22.Ghisolfi L, Keates AC, Hu X, Lee DK, Li CJ. Ionizing radiation induces stemness in cancer cells. PLoS One. 2012;7(8):e43628. doi: 10.1371/journal.pone.0043628. PubMed PMID: 22928007. Pubmed Central PMCID: 3424153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006 Aug 25;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. PubMed PMID: 16904174. [DOI] [PubMed] [Google Scholar]
- 24.Buchhagen DL, Worsham MJ, Dyke DL, Carey TE. Two regions of homozygosity on chromosome 3p in squamous cell carcinoma of the head and neck: comparison with cytogenetic analysis. Head & neck. 1996 Nov-Dec;18(6):529–537. doi: 10.1002/(SICI)1097-0347(199611/12)18:6<529::AID-HED7>3.0.CO;2-4. PubMed PMID: 8902566. [DOI] [PubMed] [Google Scholar]
- 25.Vlashi E, Kim K, Lagadec C, Donna LD, McDonald JT, Eghbali M, et al. In vivo imaging, tracking, and targeting of cancer stem cells. J Natl Cancer Inst. 2009 Mar 4;101(5):350–359. doi: 10.1093/jnci/djn509. PubMed PMID: 19244169. Pubmed Central PMCID: 2727141. Epub 2009/02/27. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. Journal of immunological methods. 2009 Aug 15;347(1–2):70–78. doi: 10.1016/j.jim.2009.06.008. PubMed PMID: 19567251. [DOI] [PubMed] [Google Scholar]
- 27.Sorensen BS, Busk M, Olthof N, Speel EJ, Horsman MR, Alsner J, et al. Radiosensitivity and effect of hypoxia in HPV positive head and neck cancer cells. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2013 Sep;108(3):500–505. doi: 10.1016/j.radonc.2013.06.011. PubMed PMID: 23953409. [DOI] [PubMed] [Google Scholar]
- 28.Lagadec C, Vlashi E, Alhiyari Y, Phillips TM, Dratver MB, Pajonk F. Radiation-Induced Notch Signaling in Breast Cancer Stem Cells. Int J Radiat Oncol Biol Phys. 2013 Aug 27; doi: 10.1016/j.ijrobp.2013.06.2064. PubMed PMID: 23992604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008 Dec 4;456(7222):593–598. doi: 10.1038/nature07567. PubMed PMID: 19052619. Pubmed Central PMCID: 2597380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen YC, Chen YW, Hsu HS, Tseng LM, Huang PI, Lu KH, et al. Aldehyde dehydrogenase 1 is a putative marker for cancer stem cells in head and neck squamous cancer. Biochem Biophys Res Commun. 2009 Jul 31;385(3):307–313. doi: 10.1016/j.bbrc.2009.05.048. PubMed PMID: 19450560. [DOI] [PubMed] [Google Scholar]
- 31.Prince ME, Sivanandan R, Kaczorowski A, Wolf GT, Kaplan MJ, Dalerba P, et al. Identification of a subpopulation of cells with cancer stem cell properties in head and neck squamous cell carcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2007 Jan 16;104(3):973–978. doi: 10.1073/pnas.0610117104. PubMed PMID: 17210912. Pubmed Central PMCID: 1783424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillison ML, D'Souza G, Westra W, Sugar E, Xiao W, Begum S, et al. Distinct risk factor profiles for human papillomavirus type 16-positive and human papillomavirus type 16-negative head and neck cancers. J Natl Cancer Inst. 2008 Mar 19;100(6):407–420. doi: 10.1093/jnci/djn025. PubMed PMID: 18334711. [DOI] [PubMed] [Google Scholar]
- 33.Clay MR, Tabor M, Owen JH, Carey TE, Bradford CR, Wolf GT, et al. Single-marker identification of head and neck squamous cell carcinoma cancer stem cells with aldehyde dehydrogenase. Head & neck. 2010 Sep;32(9):1195–1201. doi: 10.1002/hed.21315. PubMed PMID: 20073073. Pubmed Central PMCID: 2991066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tamari K, Hayashi K, Ishii H, Kano Y, Konno M, Kawamoto K, et al. Identification of chemoradiation-resistant osteosarcoma stem cells using an imaging system for proteasome activity. Int J Oncol. 2014 Sep 24; doi: 10.3892/ijo.2014.2671. PubMed PMID: 25269626. [DOI] [PubMed] [Google Scholar]
- 35.Zhang M, Kumar B, Piao L, Xie X, Schmitt A, Arradaza N, et al. Elevated intrinsic cancer stem cell population in human papillomavirus-associated head and neck squamous cell carcinoma. Cancer. 2014 Apr 1;120(7):992–1001. doi: 10.1002/cncr.28538. PubMed PMID: 24382806. Pubmed Central PMCID: 3961512. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.