Abstract
Background and Aims The origin of limes and lemons has been a source of conflicting taxonomic opinions. Biochemical studies, numerical taxonomy and recent molecular studies suggested that cultivated Citrus species result from interspecific hybridization between four basic taxa (C. reticulata, C. maxima, C. medica and C. micrantha). However, the origin of most lemons and limes remains controversial or unknown. The aim of this study was to perform extended analyses of the diversity, genetic structure and origin of limes and lemons.
Methods The study was based on 133 Citrus accessions. It combined maternal phylogeny studies based on mitochondrial and chloroplastic markers, and nuclear structure analysis based on the evaluation of ploidy level and the use of 123 markers, including 73 basic taxa diagnostic single nucleotide polymorphism (SNP) and indel markers.
Key Results The lime and lemon horticultural group appears to be highly polymorphic, with diploid, triploid and tetraploid varieties, and to result from many independent reticulation events which defined the sub-groups. Maternal phylogeny involves four cytoplasmic types out of the six encountered in the Citrus genus. All lime and lemon accessions were highly heterozygous, with interspecific admixture of two, three and even the four ancestral taxa genomes. Molecular polymorphism between varieties of the same sub-group was very low.
Conclusions Citrus medica contributed to all limes and lemons and was the direct male parent for the main sub-groups in combination with C. micrantha or close papeda species (for C. aurata, C. excelsa, C. macrophylla and C. aurantifolia – ‘Mexican’ lime types of Tanaka’s taxa), C. reticulata (for C. limonia, C. karna and C. jambhiri varieties of Tanaka’s taxa, including popular citrus rootstocks such as ‘Rangpur’ lime, ‘Volkamer’ and ‘Rough’ lemons), C. aurantium (for C. limetta and C. limon – yellow lemon types – varieties of Tanaka’s taxa) or the C. maxima × C. reticulata hybrid (for C. limettioides – ‘Palestine sweet’ lime types – and C. meyeri). Among triploid limes, C. latifolia accessions (‘Tahiti’ and ‘Persian’ lime types) result from the fertilization of a haploid ovule of C. limon by a diploid gamete of C. aurantifolia, while C. aurantifolia triploid accessions (‘Tanepao’ lime types and ‘Madagascar’ lemon) probably result from an interspecific backcross (a diploid ovule of C. aurantifolia fertilized by C. medica). As limes and lemons were vegetatively propagated (apomixis, horticultural practices) the intra-sub-group phenotypic diversity results from asexual variations.
Keywords: Citrus, phylogenetics, SNP, SSR, indel, lime and lemon origin.
INTRODUCTION
Lemons and limes are cultivated under all Mediterranean, sub-tropical and inter-tropical climates worldwide. With 15 Mt produced, limes and lemons represent the third largest citrus horticultural group (FAO, 2014). Nearly 1·6 Mt of lemons and limes are traded, ranking them 11th worldwide. Lemons and limes are sold both as fresh fruit and as processed products. After expanding up to 2007, the lemon market is currently stagnating and is in direct competition with the market for limes, whose consumption has increased dramatically since the 1980s (Duportal et al., 2013). Only a few cultivars are cultivated worldwide for the production of fresh fruit and essential oils or for use as rootstock, even though significant diversity exists in this horticultural group.
Citrus fruits and their relatives originated in South-east Asia, New Caledonia and Australia (Swingle and Reece, 1967). The genus Citrus L., which includes limes and lemons, has been cultivated in tropical and temperate parts of the world for >2000 years. In the classification of Swingle and Reece (1967) lemons and limes are classified as two separate species, Citrus limon (L.) Burm. F. and C. aurantifolia (Christm.) Swing. In the classification of Tanaka (1954), limes and lemons are classified as 37 species. These conflicting classifications result from the total sexual compatibility between Citrus species and the frequent occurrence of apomixis due to nucellar polyembryony (Scora, 1975; Barrett and Rhodes, 1976), which led many taxonomists to consider interspecific hybrids fixed by apomixis or vegetative propagation (cuttings or grafting) as new species.
Thus, the origin of the lemon has been a source of conflicting opinions by both historians and taxonomists. Bonavia (1888) concluded that the lemon reached India relatively late, whereas Tolkowsky (1938) believed lemons are mentioned in early Sanskrit texts dated around 800 BC. Webber et al. (1967) suggested Southern China or possibly upper Burma as the native area of the lemon. Early taxonomists hypothesized that C. limon was a derivative or hybrid of C. medica L., the citron. Gallesio (1811) and Tolkowsky (1938) reported that, in the Middle Ages, the lemon was considered to be a variety of citron, and de Candolle (1886) considered lemon to be a close relative of citron. Lemon, lime and citron were included in the species C. medica by Linnaeus (1753) and other early taxonomists. More recently, authors including Risso (1813), Swingle (1914, 1943), Tanaka (1954) and Bhattacharya and Dutta (1956) gave separate specific names to citron and lemon. Swingle (1914) named citron C. medica, lime C. aurantifolia and lemon C. limonia Osbeck, but in 1943 he reclassified lemon as C. limon. Swingle also considered it likely that lemon is a relative of citron and suggested that it originated from hybridization between citron and lime. Hodgson (1955) grouped citron, lemon and lime together. Early taxonomic works thus recognized that citrons, limes and lemons are related, but failed to propose clear definitive conclusions on the origin of the species. Biochemical studies (Malik et al., 1974; Scora, 1975), numerical taxonomy (Barrett and Rhodes, 1976) and, more recently, molecular studies (Federici et al., 1998; Nicolosi et al., 2000; Barkley et al., 2006; Garcia-Lor et al., 2012, 2013b; Ollitrault et al., 2012; Curk et al., 2014, 2015b) suggested that all cultivated Citrus species result from interspecific hybridization between four basic taxa [mandarin, C. reticulata Blanco; pummelo, C. maxima (Burm.) Merr.; citron, C. medica L.; and a papeda, C. micrantha Wester]. It is now generally accepted that limes and lemons are related to citron (C. medica L.) (Nicolosi et al., 2000; Barkley et al., 2006; Bayer et al., 2009), but opinions differ on the origin of specific varieties. In agreement with Swingle and Reece (1967), Barrett and Rhodes (1976) and Federici et al. (1998) argued that the classical ‘Eureka’- or ‘Lisbon’-type yellow lemons resulted from direct hybridization between citron and lime, whereas, more recently, Nicolosi et al. (2000) proposed that the ‘Lisbon’-type yellow lemon was the product of direct hybridization between sour orange (C. aurantium L.) and citron. Sour orange is itself presumed to be a hybrid between C. maxima and C. reticulata (Swingle and Reece, 1967; Scora, 1975, 1988; Green et al., 1986; Yamamoto et al., 1993; Nicolosi et al., 2000; Wu et al., 2014; Curk et al., 2015b). ‘Mexican’ lime was proposed to be a direct hybrid between citron and C. micrantha (Scora, 1975; Nicolosi et al., 2000). These hypotheses concerning ‘Lisbon’-type yellow lemons and ‘Mexican’-type limes were recently confirmed by relatively large single nucleotide polymorphism (SNP) analysis (Ollitrault et al., 2012; Garcia-Lor et al., 2013b; Curk et al., 2015b). However, the origin of the other lemons and limes remains controversial or is unknown.
The aim of the present work was to perform an extended analysis of the diversity, genetic structure and origin of lime and lemon germplasm. It was based on 93 lime and lemon accessions, 33 representatives of the four basic taxa and seven accessions of the other secondary species, all available in the germplasm depositories of the Citrus Biological Resource Centre (CRB Citrus, INRA-CIRAD, San Giuliano, France) and of the IVIA (Valencia, Spain). It combined maternal phylogeny studies based on mitochondrial indels (Froelicher et al., 2011) and chloroplastic simple sequence repeats (SSRs) (Bryan et al., 1999; Weising and Gardner, 1999), and analysis of nuclear genome diversity. Concerning nuclear genome diversity, Barkley et al. (2009) showed that homoplasy may limit the usefulness of SSR markers in identifying the phylogenetic origin of DNA fragments in citrus, and Garcia Lor et al. (2012), Ollitrault et al. (2015) and Curk et al. (2015b) showed that insertion/deletion polymorphisms (indels) and SNPs were more useful to select efficient specific diagnostic markers. However, most selected markers for specific alleles of the ancestral taxa fail to identify intraspecific variation (Garcia-Lor et al., 2013b; Curk et al., 2015b; Ollitrault et al., 2015). Therefore, we combined eight indels and 96 SNP markers to reveal the interspecific structure of the different accessions, and 19 SSR markers to obtain information on intraspecific polymorphism. A ploidy analysis of all accessions by flow cytometry completed our characterization of the nuclear genome.
MATERIALS AND METHODS
Plant material
Leaves from 133 accessions of the Citrus genus were collected from the IVIA Citrus Germplasm Bank of pathogen-free plants (Valencia, Spain; accessions with an IVIA identification number) and from the Citrus Biological Resource Centre (CRB Citrus, INRA-CIRAD, NFS96-900) in San Giuliano (Corsica, France; accessions with an SRA identification number) (Supplementary Data Table S1). Tanaka (1961) and Swingle and Reece (1967) botanical classifications are given when the accessions are already classified. According to Swingle and Reece’s classification, 24 accessions are representative of C. aurantifolia (Christm.) Swing, and 54 of C. limon (L.) Burm. In addition, we included 15 unclassified accessions phenotypically related to the lemon and lime groups, 33 accessions representative of the four basic taxa [12 mandarins (nine C. reticulata and three C. tachibana), 11 pummelos (C. maxima), eight citrons (C. medica) and two papedas (C. micrantha)], and seven other secondary species [two sour oranges (C. aurantium), two grapefruits (C. paradisi), one clementine (C. clementine) and two sweet oranges (C. sinensis)].
Evaluation of ploidy level
Ploidy level was determined by flow cytometry. Each sample consisted of a small piece of leaf (0·5 mm2) collected from each accession together with a similar piece of leaf taken from a tetraploid control plant [doubled-diploid ‘Shamouti’ sweet orange (Aleza et al., 2011)]. The leaf samples were chopped together with a razor blade in a nuclei isolation solution (High Resolution DNA Kit Type P, solution A; Partec, Munster, Germany). Nuclei were filtered through a 30 μm nylon filter and stained with a 4',6-diamino-2-phenylindole (DAPI) solution (High Resolution DNA Kit Type P, solution B; Partec). After a 5 min incubation period, the stained samples were run in a Ploidy Analyzer (Partec PA) flow cytometer equipped with a HBO 100 W high-pressure mercury bulb and both KG1 and BG38 filter sets. Histograms were analysed using dpac v2.0 software (Partec), which determines peak position, the coefficient of variation (CV) and the relative ploidy index of the samples. When only one peak was observed, an additional analysis was performed with an internal diploid control.
DNA extraction
High molecular weight genomic DNA was extracted from the leaf samples using the DNeasy Plant Mini Kit (Qiagen S.A., Madrid, Spain) according to the manufacturer’s instructions.
SSR and indel marker genotyping
All accessions were genotyped with 19 SSRs (Kijas et al., 1997; Froelicher et al., 2008; Luro et al., 2008; Cuenca et al., 2011), eight indel nuclear markers (Garcia-Lor et al., 2012), five chloroplastic SSRs (Bryan et al., 1999; Weising and Gardner, 1999) and three mitochondrial indels (Froelicher et al., 2011) (Supplementary Data Table S2; Fig. S1). PCR amplifications were performed using a thermocycler Ep gradient S (Eppendorf®) in a final volume of 10 μL containing 0·8 U of Taq DNA polymerase (Fermentas®), 2 ng mL–1 citrus DNA, 0·2 mm wellRED (Sigma®) dye-labelled forward primer, 0·2 mm non-dye-labelled reverse primer, 0·2 mm of each dNTP, 10× PCR buffer (Fermentas) and 1·5 mm MgCl2. The PCR protocol was as follows: denaturation at 94 °C for 5 min followed by 40 repeats of 30 s at 94 °C, 1 min at 50 or 55 °C (depending on the melting temperature of the primers), 45 s at 72 °C; and a final elongation step for 4 min at 72 °C. Capillary electrophoresis was carried out using a CEQ™ 8000 Genetic Analysis System (Beckman Coulter Inc., Fullerton, CA, USA). Data collection and analysis were carried out with GenomeLab GeXP (Beckman Coulter Inc.) version 10.0 software. Allele dosage was calculated using the MAC-PR (microsatellite DNA allele counting-peak ratio) method (Esselink et al., 2004), validated in citrus by Cuenca et al. (2011).
KASPar genotyping
Ninety-six SNP markers (Ollitrault et al., 2012; Garcia-Lor et al., 2013a; Curk et al., 2015b) (Supplementary Data Table S2) were used for genotyping with KASPar technology by KBioscience® (http://www.kbioscience.co.uk/). The KASPar™ Genotyping System is a competitive, allele-specific dual Förster resonance energy transfer (FRET)-based assay. Primers were designed by LGC Genomics®, based on the SNP locus-flanking sequence. Detailed information on all SNP markers is given in Table S2 and their distribution over the nine scaffolds is schematized in Fig. S1. Further details about this genotyping method can be found in Cuppen (2007). The fluorescence signals of the PCR products were measured with Fluostar Omega (BMG), and genotype calling was performed with KlusterCaller software (LGC Genomics). For triploid and tetraploid limes, the allele dose was estimated from their respective allele signal according to the method described by Cuenca et al. (2013).
Genetic analyses
Expected heterozygosity (He), observed heterozygosity (Ho), and F Stat parameters [FW (Wright, 1978) and FST] were calculated using GENETIX v. 4.03 software (Belkhir et al., 1996–2004). In population genetics, Wright’s F-statistics or FW (also known as fixation indices) describe the deviation to the expected level of heterozygosity in a population according to Hardy–Weinberg expectation. One of the main factors that induce such deviation is the sub-division of the global population into sub-populations, with limited gene flow between sub-populations. The index (FST) is a measure of differentiation between a priori defined sub-populations. A convergence between FST and FW indicates that the differentiation between the a priori defined sub-populations is the main component of the global population structure. The potential of each marker to distinguish one of the four basic taxa from the three others was estimated based on the GST parameter (Nei, 1973) considering two sub-populations: (1) the taxa concerned and (2) a theoretical population of the three other basic taxa. GST estimations were computed with GENETIX v. 4.03 software (Belkhir et al., 1996–2004).
Genetic relationships were identified by Neighbor–Joining analysis with DARwin software (Perrier and Jacquemoud-Collet, 2006), using the simple matching dissimilarity index. Cluster robustness was tested with 1000 bootstraps.
Population structure was inferred with STRUCTURE version 2.3.4 (Pritchard Lab, 2014), which implements a model-based clustering method using genotype data (Pritchard et al., 2000; Falush et al., 2003). No prior population structure was defined. The option we used was the linkage model, with correlated allele frequencies and the computed probability of the data for estimating K. Analyses were performed with the K value (number of sub-populations) varying between 1 and 10. The statistics used to select the correct K value were those used by Evanno et al. (2005): mean likelihood, L(K); mean difference between successive likelihood values of K, L'(K); the absolute value of this difference, |L''(K)|; and ΔK, which is the mean of the absolute values of L''(K) divided by the standard deviation of L(K). The likelihood distribution L(K) and ΔK were the main values used to choose the optimal K value of the population. STRUCTURE was run ten times with 50 000 burn-in steps followed by 50 000 Monte Carlo Markov Chain (MCMC) repetitions. For the best K value, the output clusters of ten independent STRUCTURE runs were permuted and aligned, and the average frequency and standard error of the contribution of each basic population were estimated.
RESULTS
Evaluation of ploidy level
Five triploid (‘Ambilobe’, ‘Coppenrath’, ‘Madagascar’, ‘Mohtasseb’ and ‘Tanepao’) and one tetraploid (‘Giant Key’) limes were identified among the 12 accessions classified as C. aurantifolia by Tanaka (1961). The five accessions of C. latifolia (‘Bears’, ‘El Kseur’, ‘IAC-5', ‘Persian’ and ‘Tahiti’) were triploid. All the other accessions analysed were found to be diploid (Suppelementary Data Table S1).
Cytoplasmic diversity
The three indel mitochondrial markers revealed five mitotypes within Citrus (Supplementary Data Fig. S2; Table S3), in agreement with Froelicher et al. (2011). Mitotype 1 was characteristic of true citrons and was not found in any other accessions. Mitotype 2 included the two C. micrantha, the 12 C. aurantifolia, C. aurata, the two C. excelsa, and C. macrophylla accessions. Mitotype 3 included the eight edible mandarins and the clementine, but no limes or lemons. Mitotype 4 corresponded to the mandarins called acid mandarins by Froelicher et al. (2011). It included four non-edible or wild mandarins, C. jambhiri, two C. karna, three C. limonia and six Citrus sp. accessions. Mitotype 5 previously identified as a C. maxima mitotype (Froelicher et al., 2011) included all C. maxima accessions, two C. aurantium, C. bergamia, five C. latifolia, three C. limetta, 39 C. limon, three C. lumia, three C. limettioides, two C. paradisi, two C. sinensis, the C. meyeri and the C. pyriformis accessions, as well as nine Citrus sp. accessions.
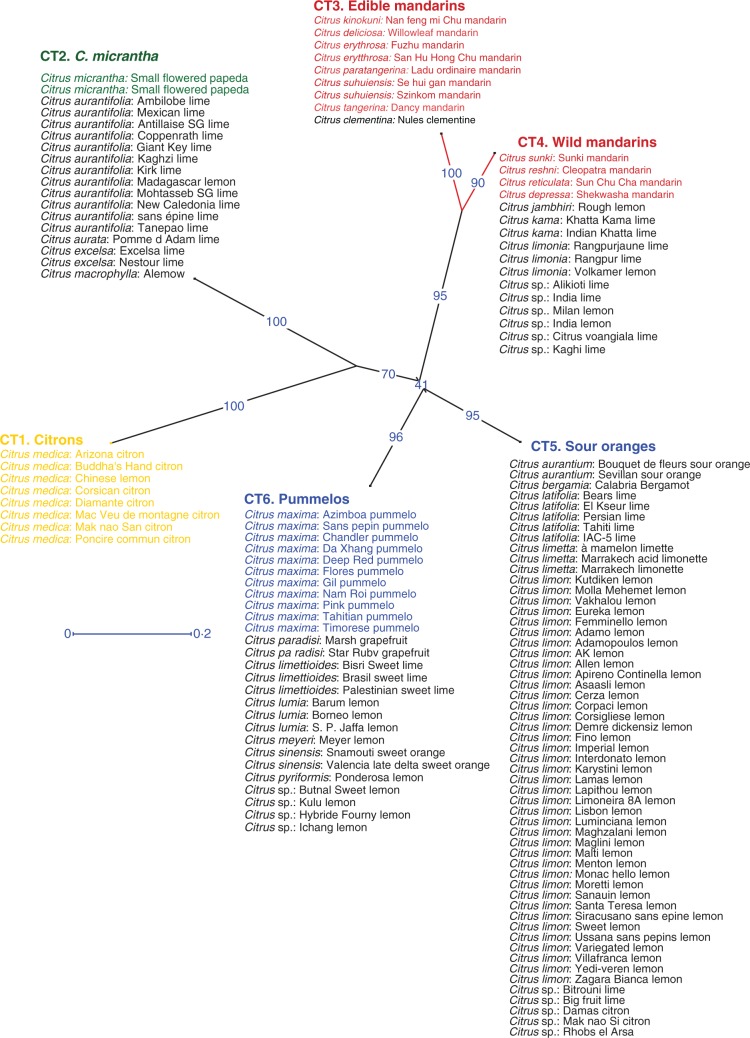
Chloroplastic markers revealed six chlorotypes, mostly in agreement with the mitotypes (Supplementary Fig. S2; Table S3), but with one difference: mitotype 5 was divided into two sub-groups. The first sub-group (chlorotype 5) shared the sour orange chlorotype with C. bergamia, five C. latifolia, three C. limetta, 39 C. limon and four C. Citrus sp. The second sub-group (chlorotype 6) was found in the 11 C. maxima, three C. lumia, three C. limettioides, two C. paradisi, two C. sinensis, the C. meyeri and the C. pyriformis accessions, as well as four Citrus sp. Thus six cytoplasmic types (CTs) corresponding to the six chlorotypes were identified (Fig. 1; Fig. S2). Lime and lemon accessions were found in four of the six CTs: 53, 12, 12 and 16 in sour orange, pummelo, non-edible mandarin and C. micrantha CTs, respectively.
Fig. 1.
Cytoplasmic type of the 133 citrus accessions. Neighbor–Joining tree established from three mitochondrial indels and five chloroplastic simple sequence repeats (SSRs). Blue numbers: bootstrap value given to each edge indicates the frequency of occurrence of this edge in the bootstrapped trees. Blue line: scale of edge lengths.
Nuclear genome diversity at the diploid level
Potential of the different kind of markers for differentiation between ancestral taxa.
Simple sequence repeats were the most polymorphic markers, with an average of 8·11 alleles/locus across the 33 accessions representative of the four basic taxa, while the selected indels and SNPs had 2·75 and two alleles/locus, respectively (Table 1). The same higher SSR polymorphism was observed within accessions (average observed heterozygosity Ho = 0·42 ± 0·02 compared with 0·10 ± 0·07 for indels and 0·07 ± 0·02 for SNPs) and within each basic taxon [expected heterozygosity (He) ranged from 0·21 to 0·55, 0 to 0·27, and 0·01 to 0·12 for SSRs, indels and SNPs, respectively]. In return, SSRs displayed lower values for the global genetic organization parameters (FW of 0·47 ± 0·09, 0·78 ± 0·14 and 0·78 ± 0·06; FST considering the four basic taxa as sub-populations of 0·51 ± 0·09, 0·85 ± 0·10 and 0·79 ± 0·06 for SSRs, indels and SNPs, respectively (Table 1).
Table 1.
Diversity of the basic taxa
| C. reticulata (n = 12) | C. maxima (n = 11) | C. medica (n = 8) | C. micrantha (n = 2) | Total (n = 33) | ||
|---|---|---|---|---|---|---|
| 19 SSRs | NA | 4·47 | 3·26 | 1·89 | 1·44 | 8·11 |
| Ho | 0·61 ± 0·11 | 0·45 ± 0·15 | 0·10 ± 0·06 | 0·41 ± 0·24 | 0·42 ± 0·018 | |
| He | 0·55 ± 0·10 | 0·47 ± 0·11 | 0·21 ± 0·11 | 0·21 ± 0·12 | 0·77 ± 0·04 | |
| FW | –0·11 ± 0·07 | 0·05 ± 0·22 | 0·46 ± 0·26 | −0·91 ± 0·19 | 0·47 ± 0·09 | |
| FST | 0·513 ± 0·085 | |||||
| MLG | 12 | 10 | 8 | 2 | 32 | |
| 8 indels | NA | 1·63 | 1·36 | 1 | 1·13 | 2·75 |
| Ho | 0·21 ± 0·17 | 0·06 ± 0·07 | 0 | 0·13 ± 0·24 | 0·10 ± 0·07 | |
| He | 0·27 ± 0·09 | 0·05 ± 0·06 | 0 | 0·06 ± 0·12 | 0·44 ± 0·05 | |
| FW | –0·10 ± 0·18 | –0·08 ± 0·04 | N | –1 | 0·78 ± 0·14 | |
| FST | 0·85 ± 0·10 | |||||
| MLG | 10 | 3 | 1 | 1 | 15 | |
| 96 SNPs | NA | 1·39 | 1·14 | 1·14 | 1·02 | 2 |
| Ho | 0·13 ± 0·04 | 0·04 ± 0·02 | 0·04 ± 0·02 | 0·01 ± 0·02 | 0·07 ± 0·02 | |
| He | 0·12 ± 0·04 | 0·04 ± 0·03 | 0·04 ± 0·02 | 0·01 ± 0·01 | 0·33 ± 0·03 | |
| FW | –0·06 ± 0·08 | 0·11 ± 0·21 | –0·07 ± 0·17 | –1 | 0·78 ± 0·06 | |
| FST | 0·79 ± 0·06 | |||||
| MLG | 10 | 10 | 7 | 1 | 28 | |
| 123 markers | NA | 1·89 | 1·48 | 1·24 | 1·05 | 2·99 |
| Ho | 0·21 ± 0·05 | 0·10 ± 0·04 | 0·05 ± 0·02 | 0·07 ± 0·04 | 0·13 ± 0·03 | |
| He | 0·19 ± 0·04 | 0·11 ± 0·04 | 0·06 ± 0·03 | 0·04 ± 0·02 | 0·40 ± 0·04 | |
| FW | –0·1 ± 0·06 | 0·06 ± 0·14 | 0·16 ± 0·18 | –0·92 ± 0·15 | 0·73 ± 0·05 | |
| FST | 0·75 ± 0·06 | |||||
| MLG | 12 | 11 | 8 | 2 | 33 |
n, number of accessions analysed; NA, mean number of alleles/locus; Ho, observed heterozygosity; He, expected heterozygosity; FW, Wright’s fixation index; FST: intertaxa structuration parameter; MLG. number of different multilocus genotypes.
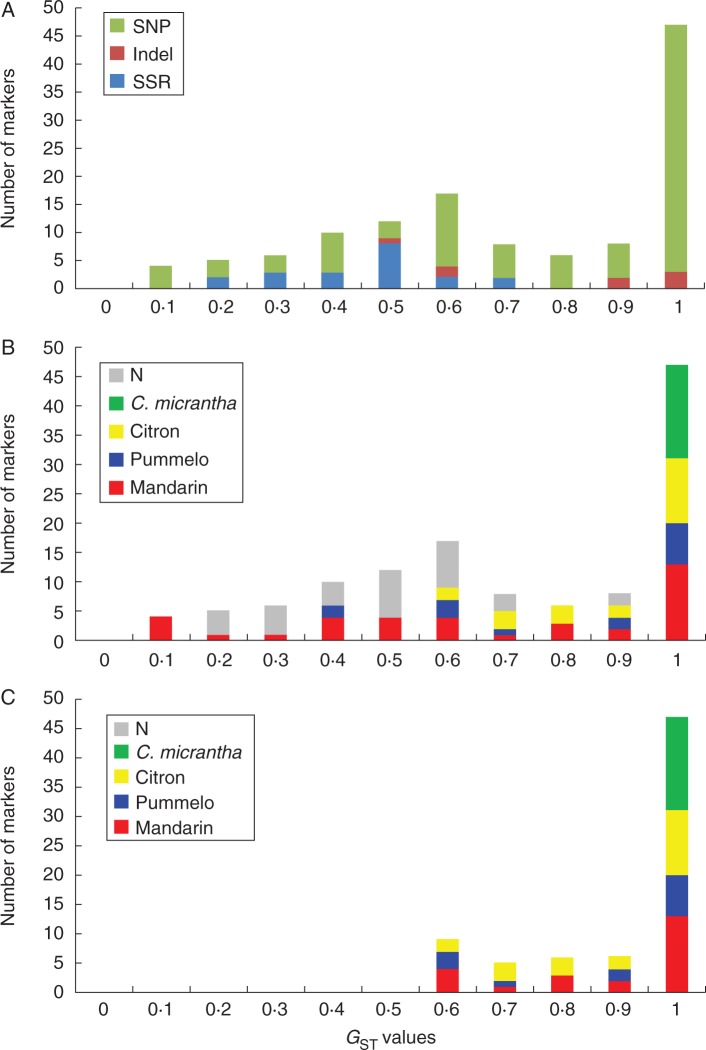
The analysis of species GST confirmed the relatively low efficiency of SSRs for specific differentiation compared with selected indels and SNPs (Fig. 2A). Among the low GST values (<0·5), some SNP markers developed from clementine BAC (bacterial artificial chromosome) end-sequencing data (Ollitrault et al., 2012) revealed intraspecific variability of C. reticulata and C. maxima (Fig. 2B). To analyse the interspecific admixture of limes and lemons, we selected the 73 markers with a GST value >0·5 and unambiguously identified specific alleles (Fig. 2C). For markers with GST <1, an allele was considered to be specific when it was only observed in one of the basic taxa.
Fig. 2.
Distribution of the highest specific GST values. (A) Comparative distribution of the three kinds of markers [simple sequence repeats (SSRs), indels and single nucleotide polymorphisms (SNPs)]. (B) Diagnostic value of the 123 markers for the ancestral taxa. (C) Diagnostic value of the 73 selected markers for STRUCTURE and specific allele homozygosity/heterozygosity (Hom/Het) analysis of the ancestral taxa. N, non-diagnostic markers.
Nuclear structure of limes and lemons.
Dissimilarities (simple matching index) were calculated between each pair of diploid accessions from the 123 markers. Ninety multilocus genotypes (MLGs) were identified. Twenty-seven yellow lemons (C. limon) displayed the same MLG (Table 2; Supplemetnary Data Table S1). Identical MLGs were also observed in two C. lumia (‘Barum’ and ‘Borneo’ lemons), two C. limettioides (‘Bisri’ and ‘Brazil’ sweet limes), two C. limetta (‘A mamelon limette’ and ‘Marrakech acid limonette’), two C. sinensis, two C. paradisi and two C. aurantium. Genetic parameters were evaluated from the MLG matrix.
Table 2.
Diversity of Swingle and Reece’s and Tanaka’s lime and lemon species based on the MLG matrix and 123 molecular markers
| Swingle and Reece | Tanaka | n | MLGs | Ho | He | FW |
|---|---|---|---|---|---|---|
| C. aurantifolia (Christm.) Swing. | C. aurantifolia | 6 | 6 | 0·45 ± 0·07 | ||
| C. bergamia | 1 | 1 | 0·39 ± 0·09 | |||
| C. excelsa | 2 | 2 | 0·46 ± 0·09 | |||
| C. limettioides | 3 | 2 | 0·53 ± 0·09 | |||
| C. macrophylla | 1 | 1 | 0·44 ± 0·09 | |||
| Total C. aurantifolia | 13 | 12 | 0·47 ± 0·06 | 0·34 ± 0·04 | –0·30 ± 0·07 | |
| C. limon (L.) Burm. | C. aurata | 1 | 1 | 0·45 ± 0·09 | ||
| C. jambhiri | 1 | 1 | 0·59 ± 0·09 | |||
| C. karna | 2 | 2 | 0·53 ± 0·09 | |||
| C. limetta | 3 | 2 | 0·56 ± 0·09 | |||
| C. limon | 39 | 13 | 0·51 ± 0·08 | |||
| C. limonia | 3 | 3 | 0·58 ± 0·08 | |||
| C. lumia | 3 | 2 | 0·31 ± 0·07 | |||
| C. meyeri | 1 | 1 | 0·55 ± 0·09 | |||
| C. pyriformis | 1 | 1 | 0·25 ± 0·08 | |||
| Total C. limon | 54 | 26 | 0·50 ± 0·06 | 0·34 ± 0·04 | –0·34 ± 0·06 | |
| Citrus species | 15 | 15 | 0·47 ± 0·05 | 0·36 ± 0·04 | –0·26 ± 0·05 | |
| All limes and lemons | 82 | 53 | 0·49 ± 0·05 | 0·37 ± 0·03 | –0·25 ± 0·05 | |
n, number of accessions; MLG, multilocus genotype; Ho, observed heterozygosity; He, expected heterozygosity; FW, Wright’s fixation index.
With an average of 0·49 ± 0·05, all Tanaka’s (1961) and Swingle and Reece’s (1967) lime and lemon species displayed high heterozygosity (Ho; Table 2) compared with the four basic taxa (Table 1). Similar average values (0·47 ± 0·05) were observed for the unclassified lime- and lemon-like accessions. Citrus pyriformis had the lowest Ho (0·25), while the values of all other Tanaka species ranged from 0·31 for C. lumia and 0·59 for C. jambhiri. Both Swingle and Reece’s C. limon and C. aurantifolia species showed similar intraspecific polymorphisms (He = 0·34 ± 0·04) and an excess of heterozygosity (negative FW values) explained by the asexual diversifications (mutations, transposition and epigenetic variations) of many actual cultivars issued from the ancestral hybrid by vegetative propagation.
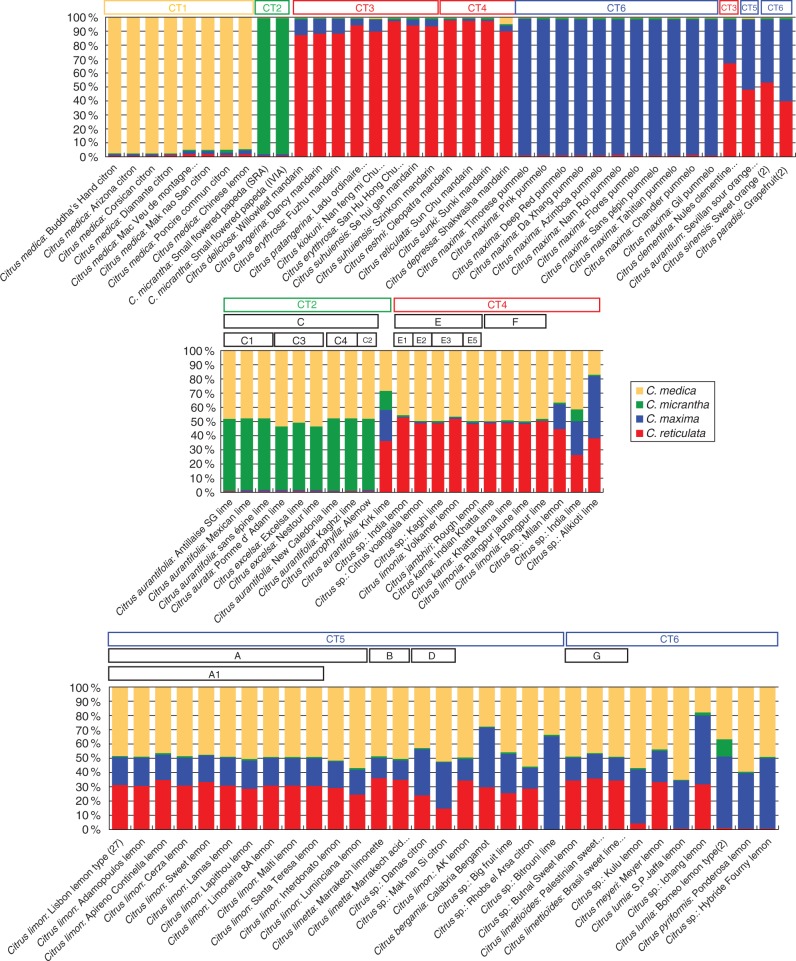
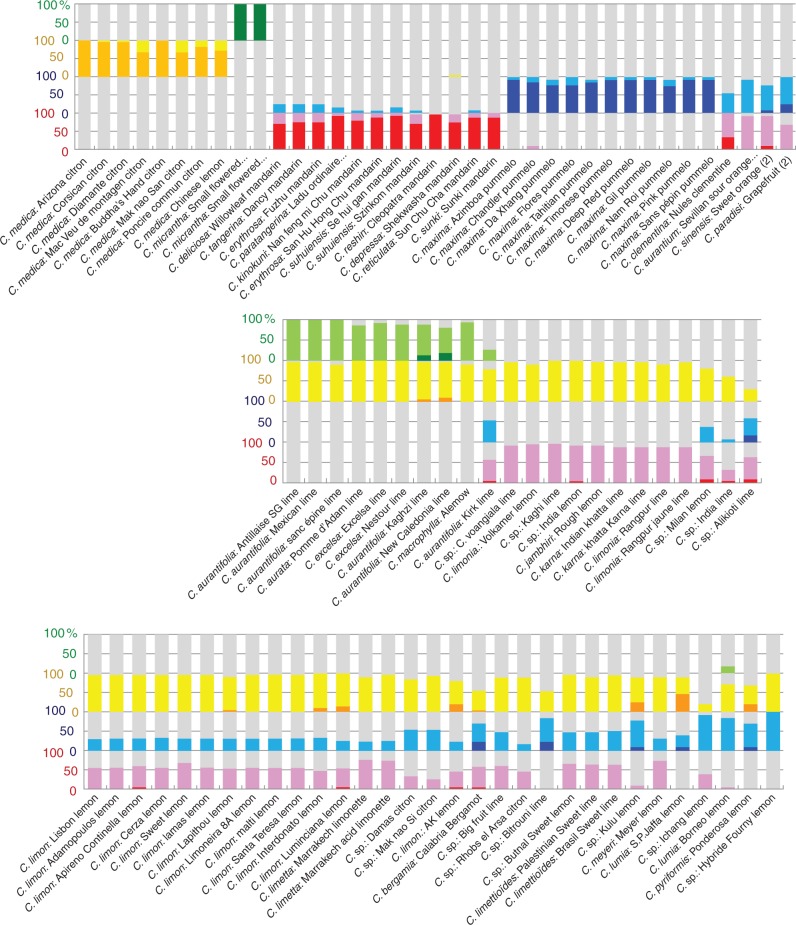
Neighbor–Joining analysis of the 90 MLGs (53 limes and lemons, 33 basic taxa and four other secondary species) was performed using the 123 molecular markers, and revealed several clusters combining lime and lemon accessions with high bootstrap values (>80 %; Supplementary Data Fig. S3). Accessions in the same cluster systematically shared the same CT. For that reason, the nuclear results [clusters on the Neighbor–Joining tree, STRUCTURE analysis (Fig. 3) and study of the frequencies of homozygous and heterozygous diagnostic alleles in the four ancestral taxa (Fig. 4)] are organized according to the CTs. STRUCTURE software analysis and homozygosity/heterozygosity (Hom/Het) for the four species-diagnostic marker sets were based on the 73 markers with a specific GST >0·5 and unambiguous specific alleles. STRUCTURE analysis with the linkage option was performed without prior definition of a population, and ten replicate runs were performed for K values ranging from 1 to 10. Analysis of ΔK showed that optimal results were obtained with K = 4 (Supplementary Data Table S4). Low variability of estimated frequencies was observed among the ten runs with K = 4 (Table S5). The average values of the ten runs are shown in Fig. 3.
Fig. 3.
Percentage contribution of the four ancestral taxa to the 90 MLGs. STRUCTURE analyses using 73 indels and SNP markers (average values for ten runs with K = 4) (blue, Citrus maxima; red, C. reticulata; green, C. micrantha; yellow, C. medica). The boxes at the top of each panel show the distribution of lemons and limes in the different nuclear clusters and sub-clusters (A–G) and the different types of cytoplasm (CT1–CT6)
Fig. 4.
Contribution of the four ancestral taxa to the 90 MLGs. Analyses of the frequency of homozygosity and heterozygosity (%) using the four specific sets of diagnostic markers (total of 73 specific markers). Orange, C. medica homozygosity; yellow, C. medica heterozygosity; red, C. reticulata homozygosity; pink, C. reticulata heterozygosity; deep blue, C. maxima homozygosity; light blue, C. maxima heterozygosity; dark green, C. micrantha homozygosity; light green, C. micrantha heterozygosity).
Nine of the ten MLGs sharing CT2 (C. micrantha CT) formed the main cluster (C), which was sub-divided into three sub-clusters. Sub-cluster C1 included three C. aurantifolia accessions (‘Antillaise’, ‘Mexican’ and ‘Sans épine’ limes). Sub-cluster C3 included the C. aurata accession (‘Pomme d’Adam’) and the two C. excelsa accessions (‘Nestour’ and ‘Excelsa’ limes). Two C. aurantifolia accessions were included in C4 (‘Kagzhi’ and ‘New Caledonia’ limes). The C. macrophylla accession was also part of cluster C (C2). These nine MLGs displayed very similar structural patterns (Fig. 3) in which C. medica and C. micrantha each contributed half. The sub-clusters C1 and C3 and C. macrophylla also displayed very similar Hom/Het patterns, with most of the C. medica- and C. micrantha-specific alleles in heterozygosity and no other taxon contribution (Fig. 4). All these genotypes presumably result from direct hybridization between C. medica and C. micrantha (or closely related) gene pools. Several SSR markers pointed to heterozygous genotypes in sub-clusters C1, C3 and C. macrophylla, each with different alleles. They thus probably resulted from three independent reticulation events. Analysis of the proportion of markers out of the 123 for which a C. micrantha × C. medica hybridization (based on our limited sample) presumably produced the genotypes of these secondary species (Supplementary Data Table S6) revealed higher congruence for ‘Mexican’ lime (95·1 %) than for ‘Excelsa’ lime (88 %) and ‘Alemow’ lime (89·3 %). In addition to the heterozygous patterns observed in C1, C3 and C. macrophylla, the two accessions in the C4 cluster displayed small proportions of C. medica- and C. micrantha-specific alleles in homozygosity (Fig. 4). Their origin is therefore more complex and presumably results from a backcross (BC) or from F2 hybridization events, or possibly from the loss of chromosome fragments. Among the 123 markers, the direct C. micrantha × C. medica model showed only 86 % congruence. The best sexual hybridization model tested was the backcross model (‘Mexican’ lime × citron) with 93·4 % congruence; however, this does not explain the homozygous C. micrantha alleles. Variability within a sub-cluster was very low in all the C sub-clusters and mostly corresponded to heterozygous/homozygous variations (14 Het/Hom polymorphisms, only five Het/Het and 0 Hom/Hom were observed). This variability within a sub-cluster may correspond to sporadic mutations [or possibly to genotyping errors due to PCR drift (i.e. allele competition) for the Het/Hom variations].
The ‘Kirk’ lime, which shared the CT2, differed completely from the other accessions at the nuclear level. It combined significant contributions from all four ancestral taxa (Fig. 3) in heterozygosity (Fig. 4). It displayed close to 50 % C. reticulata-, 50 % C. maxima-, 75 % C. medica- and 25 % C. micrantha-specific alleles in heterozygosity. ‘Kirk’ lime therefore probably results from the hybridization of two complementary direct interspecific hybrids. For example, considering its C. micrantha CT (C. micrantha × C. medica) × (C. maxima × C. reticulata), hybridization is a possibility. We tested the congruence of the ‘Mexican’ lime × sour orange and ‘Mexican’ lime × sweet orange models across the 123 markers (Supplementary Data Table S6), but their congruence was relatively low (82·8 and 89·7 %, respectively). However, considering the diversity of C. maxima and C. reticulata, the ‘Mexican’ lime × (C. maxima/C. reticulata) model remains possible (93·1 % congruence).
Cytoplasmic type 4 (wild mandarin cytoplasm type) showed two main clusters (Figs 1 and 3; Supplementary Data Fig. S3). The first cluster (E) included one sub-cluster (E3) and three additional MLGs (E1, E2 and E4). The E cluster joined the F cluster with a lower bootstrap value (Fig. S3). The E and F clusters included all C. limonia (‘Volkamer’ lemon in cluster E, and ‘Rangpur’ and ‘Yellow Rangpur’ limes in cluster F) and C. karna accessions (‘Indian Khatta’ and ‘Khatta Karna’ limes in cluster F) and C. jambhiri (‘Rough’ lemon in cluster E) and three Citrus sp. (‘Citrus voangiala’ lime, ‘Kaghi’ lime and ‘SP India’ lemon). The patterns revealed by STRUCTURE analysis of all these accessions (Fig. 3) and the Hom/Het diagnostic allele (Fig. 4) were very similar, showing that C. medica and C. reticulata (Fig. 3) each contributed close to 50 % of heterozygosity (Fig. 4). The differentiation between the isolated accessions and clusters E and F (heterozygous markers with different alleles) suggests that each group resulted from different reticulation events of C. reticulata × C. medica hybridizations. This hypothesis was validated for ‘Rangpur’ lime, ‘Volkamer’ and ‘Rough’ lemons with around 97 % of congruence for the C. reticulata × C. medica model (Table S6). Among the different mandarins, ‘Sun Shu Cha’ shared the wild mandarin cytoplasm type and displayed the best nuclear congruencies (89·3 % for ‘Rangpur’ lime, 94·2 % for ‘Volkamer’ lemon and 95·1 % for ‘Rough’ lemon). The three other accessions which shared the CT4 displayed a more complex genomic structure. Two accessions (‘Milan’ lemon and ‘Alikioti’ lime) combined C. reticulata, C. maxima and C. medica contributions in variable proportions. With a close to 50 % contribution from C. reticulata, ‘Milan’ lemon presumably results from a hybridization between C. reticulata × (C. maxima × C. medica). ‘Alikioti’ lime had homozygous C. maxima and C. reticulata alleles and only a slightly more than 10 % contribution from C. medica, pointing to a complex origin with, for example, one parent with C. maxima/C. reticulata heterozygosity and the other of tri-specific origin (C. reticulata, C. maxima and C. medica). In the case of the ‘India’ lime, there was a discrepancy between the STRUCTURE analysis, which suggests a contribution from C. micrantha, and the Hom/Het pattern of diagnostic SNPs. Consequently no hypothesis can be proposed concerning its origin.
Cytoplasmic type 5 (sour orange cytoplasm type) showed three main nuclear clusters (Figs 1 and 3; Supplemtary Data Fig. S3). The first cluster (A) is only composed of C. limon MLGs. It includes a sub-clade (A1) of very close genotypes, in particular an MLG representing 27 lemons (MLGLem). The second cluster (B) branched with cluster A but with a low bootstrap value. This cluster included the three C. limetta accessions (‘Marrakesh acid’ and ‘à mamelon’ limes represented by the same MLG, and ‘Marrakesh’ sweet lime). The third cluster (D) included two citron hybrids (‘Mak Nao Si’ and ‘Damas’). All CT5 accessions, except the ‘Bitrouni’ lime, had the same three ancestors (C. reticulata, C. maxima and C. medica). The STRUCTURE and Hom/Het patterns of all the accessions in cluster A1 were very similar, with 49, 31 and 20 % contributions from C. medica, C. reticulata and C. maxima, respectively (values for the MLGLem) and heterozygosity of specific alleles. These results are in full agreement with a (C. maxima × C. reticulata) × C. medica hybridization. This hypothesis was then validated across the 123 markers with 99·2 % congruence (Table S6). All genotypes in sub-cluster A1 were very close and, considering the highly complex heterozygous interspecific structure resulting from three-way hybridization, they could not be the result of different hybridization events or by further hybridizations after the creation of the yellow lemon prototype. These varieties are therefore mutants or somaclonal variants that appeared during clonal propagation. Two additional accessions in cluster A (‘Interdonato’ and ‘Luminciana’ lemons) displayed slightly higher contributions from citron (54 and 57 %, respectively) with specific homozygous alleles. Interestingly, the differentiation of these two lemons from the MLGLem results from homozygosity, mostly of C. medica alleles with apparent non-random distribution. In particular, ‘Luminciana’ lemon displayed homozygosity for C. medica alleles with five consecutive heterozygous markers at the end of chromosome 9, and two consecutive heterozygous markers on chromosomes 5 and 8. Given that the complex structure resulting from the three-species combinations is conserved in all other regions of the genome, thus excluding the possibility of sexual recombination, this observation suggests significant deletion events in these two genotypes. The profiles of the three C. limetta accessions resembled those of the A cluster but with a higher proportion of C. reticulata. In addition, they displayed heterozygosity for different alleles from those in cluster A. The three C. limetta accessions very probably result from the same kind of evolutionary sequence as C. limon in cluster A, (C. maxima × C. reticulata) × C. medica, but from different segregations in the C. maxima × C. reticulata gamete. Like yellow lemons, congruence with the C. aurantium × C. medica model was very high across the 123 markers (98·3 %; Supplementary Data Table S6). The ‘Rhobs el Arsa’ citron hybrid had a very similar structure and probably has the same origin as C. limetta. The two accessions in cluster D (‘Mak Nao Si’ and ‘Damas’ citron hybrids) probably also resulted from the same evolution but with a higher proportion of C. maxima transmitted by the gamete of the C. maxima × C. reticulata parent. The ‘AK’ lemon displayed homozygosity for the C. medica allele and therefore comes from a more complex origin. Citrus bergamia had a smaller proportion of C. medica (30 %) and close to 50 % C. maxima alleles, as well as the specific homozygous alleles of C. maxima, C. reticulata and C. medica. Citrus bergamia therefore probably does not result from direct interspecific hybridization. Among the three model tested with the 123 markers (Table S6), the C. limon (‘Lisbon’) × C. aurantium (‘Sevilla’) model was the best (96·7 % congruence). With 67 % contribution from C. maxima and 31 % from C. medica and homozygous C. maxima alleles, the ‘Bitrouni’ lime probably results from a C. maxima × (C. maxima × C. medica) backcross.
Cytoplasmic type 6 displayed one main nuclear cluster (G) (Figs 1 and 3; Supplementary Data Fig. S3), joining the three C. limettioides accessions (‘Palestinian’ and ‘Brazil’ sweet limes, and ‘Bisri’ lime; the last two accessions having the same MLG), and ‘Butnal’ sweet lemon. These accessions had a three-species nuclear structure (C. reticulata, C. maxima and C. medica in heterozygosity) similar to that observed in yellow lemons. With almost 50 % contribution from C. medica, they are probably (C. maxima × C. reticulata) × C. medica hybrids but with C. maxima × C. reticulata parents different from C. limon or C. limetta, as testified by the CT. The STRUCTURE and Hom/Het pattern of ‘Meyer’ lemon resembled that of C. limettioides and probably has a similar origin (C. maxima × C. reticulata) × C. medica. This origin was confirmed across the 123 markers with 98·3 and 95·8 % congruence, respectively (Table S6). However, none of the accessions with a C. reticulata/C. maxima constitution [sweet orange, ‘Ichang’ lemon tested (Table S6)] and with CT6 provided conclusive results. Most of the other accessions sharing this CT displayed complex structures with contributions from three or four ancestral taxa. However ‘Jaffa’ lemon, C. pyriformis (‘Ponderosa’) and ‘Hybride Fourny’ lemon showed contributions only from C. maxima and C. medica. ‘Hybride Fourny’ lemon is probably a direct hybrid (96·7 % congruence across 123 markers; Table S6), while ‘Ponderosa’ and ‘Jaffa’ lemons displayed C. maxima and C. medica homozygous alleles, suggesting a more complex origin (possibly an F2-like origin). The accessions classified as C. lumia appeared to have very different origins: ‘Jaffa’ lemon showed a C. medica/C. maxima constitution while ‘Borneo’ and ‘Barum’ lemons showed a C. maxima/C. medica/C. micrantha constitution.
The global genetic structure was rather disconnected from the two main taxonomic classifications of limes and lemons, as confirmed by the F Stat analysis (Table 3). Swingle and Reece’s and Tanak’s FST values were respectively very low (0·094) and low (0·232), while the high negative FIS values (–0·227 and –0·475, respectively) probably testify to fixed within-species heterozygosity due to vegetative propagation. Considering the main genetic clusters in our study (Supplementary Data Fig. S3) as sub-populations, the F parameters revealed improved intergroup organization compared with classical taxonomic classifications (FST = 0·315 ± 0·036) and FIS = –0·813 ± 0·059).
Table 3.
Genetic structuration of lime and lemon germplasm according to Tanaka (1961), Swingle and Reece (1967) and our cluster analysis
| FST | FIS | FIT | |
|---|---|---|---|
| Tanaka sub-division | 0·232 ± 0·027 | –0·475 ± 0·072 | –0·133 ± 0·068 |
| Swingle sub-division | 0·094 ± 0·022 | –0·227 ± 0·068 | –0·112 ± 0·069 |
| Genetic clusters | 0·315 ± 0·036 | –0·813 ± 0·059 | –0·267 ± 0·071 |
Nuclear diversity at the triploid and tetraploid levels
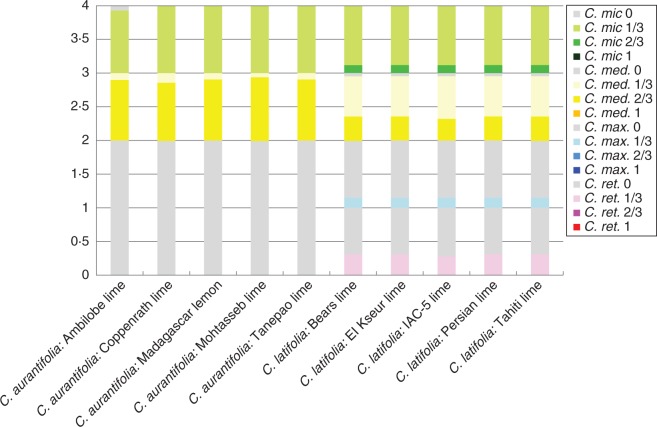
Ten triploid limes and one tetraploid lime belonging to C. aurantifolia and C. latifolia species were analysed with molecular markers by inferring allele dosage. Figure 5 gives an example for the 9P25060404 SNP marker (AG) displaying a citron-specific allele (G). Among the four triploid limes analysed on the plate, an AGG genotype was inferred for ‘Tanepao’ and ‘Coppenrath’ limes, and AAG for ‘Tahiti’ and ‘Persian’ limes. When it was not possible to infer allele doses for heterozygous genotypes, the data were considered missing. Missing data ranged from 4 to 30 %, with an average of 11 % for SSRs and indels and between 0 and 6 % for SNPs, with an average of 2 % (Table 4).
Fig. 5.
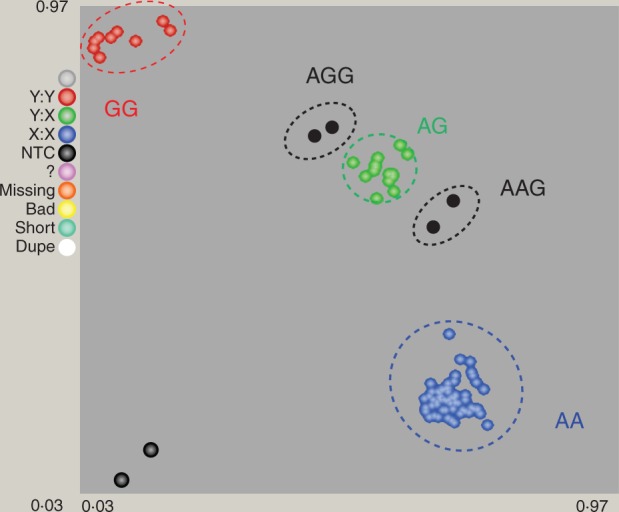
Pattern of relative allele fluorescence for the 9P25060404 SNP marker and inference of allele dosage for four triploid limes (AGG, ‘Tanepao’ and ‘Coppenrath’ limes; AAG, ‘Tahiti’ and ‘Persian’ limes). Horizontal axis, dose of allele A; vertical axis, dose of allele G; red cluster (Y:Y), GG homozygote accessions; green cluster (Y:X), AG heterozygote accessions; blue cluster (X:X), AA homozygote accessions; black cluster (NTC) AGG, triploid accessions with two doses of allele G and one dose of allele A; black cluster (NTC) AAG, triploid accessions with two doses of allele A and one dose of allele G.
Table 4.
Heterozygosity and multiallelism of polyploid limes and lemons
| Names according toTanaka’s classification | Ploidy | n | SSR and indels |
SNPs |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Common name | Hom | Di | Tri | n | Hom | Het | ||||
| C. aurantifolia | Madagascar lemon | 3 | 0·15 | 0·26 | 0·57 | 0·17 | 0·00 | 0·57 | 0·43 | |
| C. aurantifolia | Ambilobe lime | 3 | 0·15 | 0·30 | 0·48 | 0·22 | 0·01 | 0·59 | 0·41 | |
| C. aurantifolia | Coppenrath lime | 3 | 0·07 | 0·28 | 0·52 | 0·20 | 0·02 | 0·56 | 0·44 | |
| C. aurantifolia | Mohtasseb lime | 3 | 0·30 | 0·32 | 0·42 | 0·26 | 0·02 | 0·60 | 0·40 | |
| C. aurantifolia | Tanepao lime | 3 | 0·11 | 0·29 | 0·46 | 0·25 | 0·02 | 0·56 | 0·44 | |
| C. latifolia | Bears lime | 3 | 0·07 | 0·20 | 0·36 | 0·44 | 0·02 | 0·46 | 0·54 | |
| C. latifolia | Persian lime | 3 | 0·07 | 0·20 | 0·36 | 0·44 | 0·00 | 0·46 | 0·54 | |
| C. latifolia | El Kseur lime | 3 | 0·11 | 0·21 | 0·38 | 0·42 | 0·06 | 0·46 | 0·54 | |
| C. latifolia | IAC-5 lime | 3 | 0·11 | 0·21 | 0·38 | 0·42 | 0·02 | 0·46 | 0·54 | |
| C. latifolia | Tahiti lime | 3 | 0·07 | 0·20 | 0·36 | 0·44 | 0·03 | 0·46 | 0·54 | |
| C. aurantifolia | Giant Key lime | 4 | 0·04 | 0·19 | 0·81 | 0·00 | 0·02 | 0·59 | 0·41 | |
n, frequency of missing data; Hom, frequency of homozygous loci; Di, frequency of heterozygous loci with two alleles; Tri, frequency of heterozygous loci with three alleles.
Among the SSRs and indels, the C. latifolia and C. aurantifolia accessions displayed, respectively, 29 and 20 % homozygous markers, 49 and 37 % heterozygous markers with two alleles, and 22 and 43 % heterozygous markers with three alleles. Citrus latifolia accessions were also more heterozygous for diallelic SNP markers, with an average of 54 % heterozygous markers compared with 42 % in the triploid C. aurantifolia. In the tetraploid ‘Giant Key’ lime, no markers displayed three alleles and the average heterozygosity was 41 %. Concerning SSRs and indels, the relative peak ratios of ‘Giant Key’ lime were equivalent to those of ‘Mexican’ lime and the respective allele fluorescence for SNPs was also similar to that of the ‘Mexican’ lime. We therefore assumed that the allele doses at heterozygous loci were 2/2. With this inferred dosage, ‘Giant Key’ is probably a doubled diploid of a ‘Mexican’ type of C. aurantifolia.
The hierarchical classification of the ten triploid limes revealed two strong clusters corresponding to Tanaka’s C. latifolia and C. aurantifolia species (Fig. 6). Diversity within each cluster was very low.
Fig. 6.
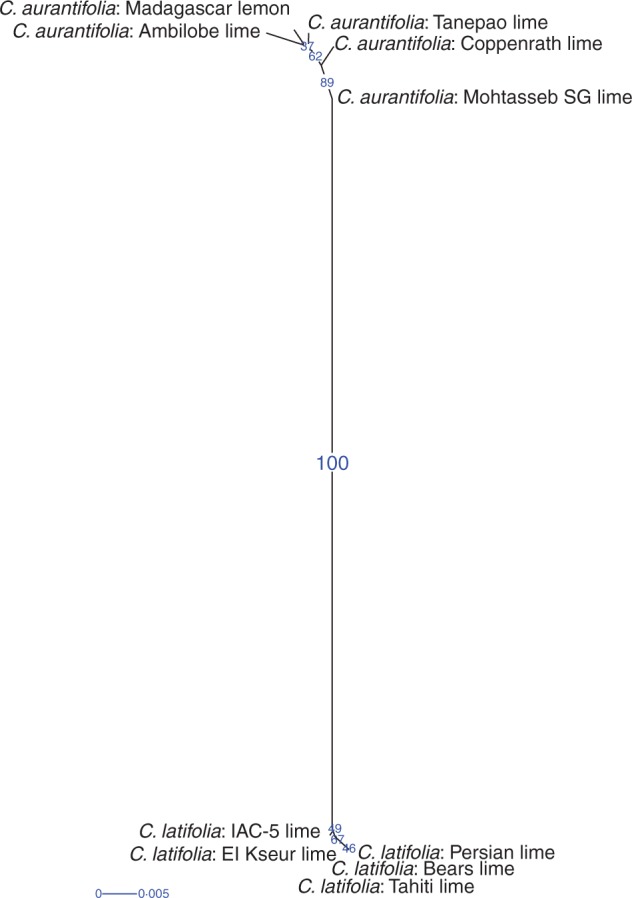
Hierarchical classification of triploid limes using 123 genetic markers. Blue numbers, the bootstrap value given to each edge indicates the frequency of occurrence of this edge in the bootstrapped trees. Blue line, scale of edge lengths.
We then evaluated the frequency of homozygosity, 2/3, 1/3 and 0 doses for the diagnostic alleles of the four specific marker sets (total of 73 markers) (Fig. 7). None of the specific alleles was found in homozygosity. The five triploid C. aurantifolia displayed very similar patterns, with most of the C. medica diagnostic alleles found in double doses and C. micrantha alleles in single doses (Fig. 7). This pattern and the C. micrantha-like cytoplasm suggest that these triploid limes result from an interspecific backcross (C. micrantha × C. medica) × C. medica with a double contribution (2 × gamete) of the interspecific hybrid. The hypothesis of an F2 origin was invalidated by the presence of triallelic SSR and indel markers. We tested the hypothesis of the interspecific BC model by estimating the proportion of the 123 markers able to generate the triploid C. aurantifolia genotypes from a 2× ‘Mexican’ lime gamete and a 1× citron gamete (from the citron population). The test showed that from 96·3 to 98·8 % of the markers fitted the model (Supplementary Data Table S6).
Fig. 7.
Contribution of the four ancestral taxa to the ten triploid limes. Analysis of the frequency of homozygosity and heterozygosity doses 2/3, 1/3 for the four specific sets of diagnostic markers (total of 73 markers). C., Citrus; mic, micrantha; med., medica; max., maxima; ret., reticulata.
The C. latifolia accessions displayed a much more complex genomic structure, with a contribution from the four basic taxa. Specific C. medica and C. micrantha alleles were found in single (around 60 and 87 %, respectively) or double doses (35 and 13 %, respectively), and some C. reticulata- (32 %) and C. maxima- (15 %) specific alleles were observed in single doses. Considering that C. latifolia accessions share the sour orange/yellow lemon CT, a (C. maxima × C. reticulata) × C. medica) × C. (micrantha × C. medica) model with a 2× male gamete probably explains the genomic structure of the C. latifolia accessions. We tested this hypothesis using a C. limon (‘Lisbon’ lemon) × C. aurantifolia (‘Mexican’ lime) model [C. limon ‘Lisbon’ being a potential (C. maxima × C. reticulata) × C. medica) hybrid and C. aurantifolia ‘Mexican’ lime a C. micrantha × C. medica hybrid]. Ninety-nine percent of the 123 markers fitted the model (Table S6). Moreover, all specific C. maxima and C. reticulata alleles observed in the C. latifolia accessions were also present in C. limon ‘Lisbon’.
DISCUSSION
The lime and lemon citrus horticultural group is genetically highly complex, involving four ancestral species with diploid, triploid and tetraploid compartments.
While the other main citrus horticultural groups (sweet oranges, sour oranges, grapefruits and mandarins) result only from diploid C. reticulata and C. maxima gene pools (Nicolosi et al., 2000; Barkley et al., 2006; Ollitrault et al., 2012; Garcia-Lor et al., 2013b), the genomic structure and the origin of limes and lemons appear to be much more complex.
Lime and lemon accessions shared four of the six types of cytoplasm identified within the Citrus genus, testifying to four maternal phylogenetic origins. Most C. limon, all C. limetta and C. latifolia as well as C. bergamia and several unclassified accessions had an identical CT to C. aurantium. Earlier molecular marker studies also concluded that C. aurantium and ‘yellow lemons’ had the same CT (Nicolosi et al., 2000; Bayer et al., 2009; Froelicher et al., 2011; Carbonell-Caballero et al., 2015; Curk et al., 2015a). Froelicher et al. (2011) also showed that C. aurantium, ‘yellow lemons’ and triploid limes (‘Bears’ and ‘Tahiti’) shared the same mitotype. All C. limettioides, all C. lumia, C. meyeri, C. pyriformis and several unclassified accessions shared a C. maxima CT also found in sweet orange and grapefruit. Chloroplast DNA (Nicolosi et al., 2000) and mitochondrial analysis (Froelicher et al., 2011) already showed that C. limettioides and C. meyeri had a C. maxima maternal phylogeny. All C. limonia, C. jambhiri and C. karna, as well as six unclassified accessions shared the same non-edible mandarin CT. Differentiation between two main mandarin CTs as well as the association of ‘Rangpur’ lime, ‘Rough’ and ‘Volkamer’ lemons and the ‘Cleopatra’ mandarin CT was previously described by Froelicher et al. (2011) at the mitochondrial level. The C. micrantha CT was found in all C. aurantifolia and all C. excelsa, C. aurata and C. macrophylla accessions. The extreme similarity between the ‘Mexican’ lime and ‘Alemow’ CT and the CT of C. micrantha was already proposed based on chloroplastic and mitochondrial marker studies (Nicolosi et al., 2000; Bayer et al., 2009; Froelicher et al., 2011; Penjor et al., 2013; Curk et al., 2015a).
The natural variation in ploidy levels among accessions observed in limes and lemons is rather rare in citrus. Spontaneous tetraploid plants (doubled diploids) have been reported in seedlings of diploid polyembryonic genotypes (Aleza et al., 2011), and 2n gametes are quite frequent (Esen and Soost, 1971; Ollitrault et al., 2008), but the resulting natural 3× genotypes have never previously been observed in the mandarin/pummelo groups outside breeding programmes. The triploidy of C. latifolia cultivars has already been described (Bacchi, 1940), and ‘Giant Key’ lime was known to be a spontaneous tetraploid selected in a seedling of the diploid ‘Key’ lime (‘Mexican’ lime type) in Florida in 1973 by H.C. Barrett (US Horticultural Research Laboratory, Orlando; USDA, 2015). The triploidy of the ‘Tanepao’-like accessions was unknown and unsuspected because these triploid accessions produce seedy fruits. while triploidy is mainly associated with sterility or highly reduced fertility and seedlessness in citrus (Ollitrault et al., 2008).
Based on 123 co-dominant markers (SSRs, indels and SNPs), nuclear analysis revealed generalized high heterozygosity of limes and lemons MLGs (Ho = 0·49 ± 0·05 on average) when compared with that of the basic taxa (0·13 ± 0·03). This confirms their probable interspecific origin, as already suggested by several molecular studies (Herrero et al., 1996; Federici et al., 1998; Gulsen and Roose, 2001a; Barkley et al., 2006). All individuals in the same nuclear cluster shared the same CT. The Neighbor–Joining tree revealed seven main clusters (with bootstrap values of >90 %) of lime and lemon MLGs. Two of them (C and E) were sub-divided into three and four sub-clusters , respectively, with high (between 99 and 100 %) bootstrap values. Inter-sub-cluster variability revealed several heterozygous loci in the different sub-clusters but with different allelic constitutions, suggesting that they arose from different hybridization events. Cluster A displayed a sub-cluster (A1) of closely related genotypes (including an MLG representative of 27 C. limon accessions), and two slightly more differentiated C. limon accessions. Considering that differentiation was only due to the loss of heterozygosity in these two accessions compared with the highly redundant MLG in sub-cluster A1 (Lisbon lemon type) and the complex tri-specific origin of cluster A (C. medica/C. reticulata, C. maxima), we consider it probable that all MLGs in cluster A derived from the same hybrid ancestor without an additional sexual event. Nineteen MLGs remained alone, i.e. with strong clustering with other lime or lemon genotypes, and may have originated from independent hybridization events. We therefore hypothesize that the limes and lemons we analysed arose from 29 independent reticulation events [clusters A, B, D, F and G, sub-clusters C1, C3, C4 and E3, one MLG in cluster C (C2), three MLGs in cluster E (E1, E2 and E4) and 16 unclustered MLGs]. This hypothesis states that a significant proportion of lime and lemon intra-horticultural group diversity results from different sexual recombination events, while in two other main secondary species and their corresponding horticultural groups (C. sinensis, sweet oranges, C. paradisi and grapefruits) all existing cultivars originated from a single clonal parent via a series of mutations or somatic variations (Bretó et al., 2001; Barkley et al., 2006; Ollitrault et al., 2012; Garcia-Lor et al., 2013b).
The identification of 73 markers out of a total of 123 with high phylogenetic values (allowing differentiation of one basic taxon from the three others), further STRUCTURE analyses and analysis of the frequency of homozygous and heterozygous specific alleles in the diagnostic markers in the four sets of ancestral taxa provided information on the interspecific admixture of each lime and lemon genome. While the C. medica CT was not found in limes or lemons, the contribution of citron at the nuclear level appears to be essential and is further discussed below. Numerous lime and lemon MLGs appeared to originate from direct hybridization between two ancestral taxa. Citrus micrantha × C. medica is probably the model for nine diploid MLGs, and the C. reticulata × C. medica model for nine MLGs. Four MLGs displayed contributions from only C. medica and C. maxima, but only the ‘Hybride de Fourny’ hybrid was found to be a direct hybrid between these two species, as previously hypothesized by Luro et al. (2012). A three-specific origin (C. maxima × C. reticulata) × C. medica is in agreement with 20 MLG patterns. Contributions from these three species – although probably in more complex hybridization schemes – have been reported in several limes and lemons, particularly in C. bergamia. Finally, four MLGs at diploid level and C. latifolia accessions at triploid level displayed a contribution from the four ancestral taxa. The origins of the main lime and lemon groups are further discussed in more detail below.
The geographical distribution of the centres of origin of the different citrus species proposed by Malik et al. (1974) supports the hypothesis that most of the prototypes of limes and lemons originated in Asia where the diversification areas of the presumed parent overlap. Neither the botanical classifications of Swingle and Reece (1967) and Tanaka (1954) nor the usual lime and lemon denomination correctly encompass the organization of genetic diversity, which is logical considering, on the one hand, the oversimplified dichotomy of Swingle and Reece (C. limon/C. aurantifolia) or the usual denomination (lemon/lime), whereas many more different phylogenetic origins are involved, and, on the other hand, the multiplication of taxa proposed by Tanaka, which separates sub-groups of similar origin (i.e. C. karna/C. jambhiri/C. limonia or C. limon/C. limetta).
Citron is the common genetic contributor to limes and lemons but never acted as the female parent
All lime- and lemon-like accessions analysed displayed a contribution from the C. medica genome. Citrons appeared mainly as a direct parent for the 12 identified clusters and independent sub-clusters and six of the unclustered MLGs. However, none of the lime and lemon accessions analysed had a C. medica maternal phylogeny. We therefore conclude that citron was the male parent. Our results extend the hypothesis proposed by several early taxonomists on the relationship between citron, limes and lemons to many more lime and lemon accessions than previous studies. Our results also agree with more recent conclusions based on analyses of cytoplasm and nuclear markers (Nicolosi et al., 2000; Gulsen and Roose, 2001a; Abkenar et al., 2004; Barkley et al., 2006; Bayer et al., 2009; Jena et al., 2009; Froelicher et al., 2011; Luro et al., 2012; Curk et al., 2015a). Many authors consider that C. medica originated in India (Malik et al., 1974; Scora, 1975; Jena et al., 2009). However, chloroplast phylogenetic studies led Beattie et al. (2008) to propose an Australasian origin for C. medica. Indeed their closest relatives are Clymenia polyandra from Papua New Guinea, Oxanthera spp. from New Caledonia, and Microcitrus and Eremocitrus species from Papua New Guinea and Australia. All these authors proposed that 30–35 million years ago, early species dispersed westward – possibly as floating fruit – from north-eastern Australia, probably aided by equatorial currents. Whatever their real geographical origin, India has clearly been a centre for the diversification of citron (Malik et al., 1974). Cytogenetic studies also demonstrated that C. medica is a true parental species of limes and lemons (Carvalho et al., 2005). In a study of citron, limes and lemons, Carvalho et al. (2005) showed that C. medica was the only cytogenetically homozygous accession and that all its chromosome types were clearly represented in limes and lemons, some of them forming heteromorphic pairs. In addition, lemons and limes were heterozygous for all rDNA sites, whereas C. medica was entirely homozygous. Among the citrus groups thought to be true Citrus species, citrons had the lowest observed heterozygosity and diversity, as observed in several molecular studies (Barkley et al., 2006; Garcia-Lor et al., 2012). This low polymorphism may be explained by the cleistogamy of citron flowers. Indeed self-pollination increases homozygosity. Barrett and Rhodes (1976) reported that citrons produce vigorous selfed seedlings and tend to be highly homozygous, which is consistent with our data. Cleistogamy may also explain why none of the limes and lemons which displayed a direct relationship with C. medica have citron as the female parent, while it is a totally monoembryonic species (no apomixes) and is thought to produce hybrid progeny.
Global phylogenomic structure and origin of the main lime and lemon groups
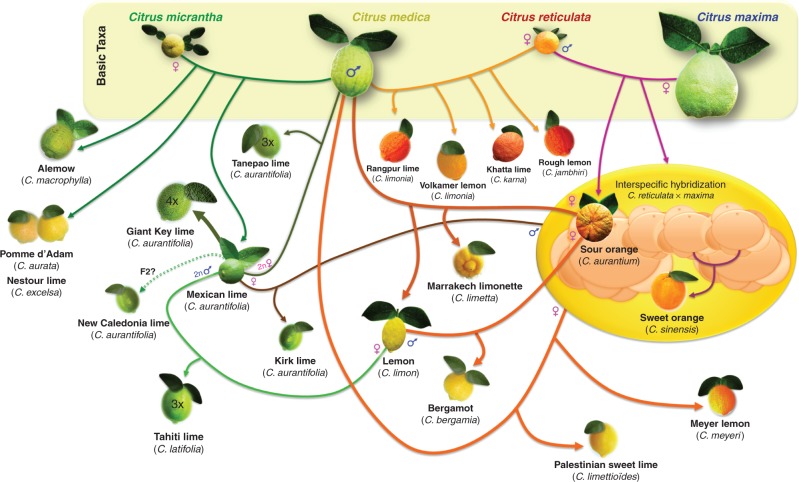
Incomplete congruence was found between Tanaka’s classification and the nuclear interspecific patterns and inferred origins. Our conclusions are schematized in Fig. 8, and those concerning the main groups of lemons and limes previously debated in the citrus literature are discussed below.
Fig. 8.
Origin of the main lime and lemon varietal sub-groups.
C. micrantha × C. medica.
Citrus micrantha × C. medica are presumably the model for C. macrophylla, C. aurata, C. excelsa and diploid C. aurantifolia, including the widely cultivated ‘Mexican’ lime. Concerning ‘Mexican’ lime-like cultivars, our conclusions agree with the hypothesis of a papeda × citron hybridization and, more specifically, C. micrantha × C. medica hybridization proposed by several authors based on biochemical data (Scora, 1975), molecular markers (Nicolosi et al., 2000; Yamamoto et al., 2007; Ollitrault et al., 2012; Garcia-Lor et al., 2013b) and cytogenetic observations (Carvalho et al., 2005). Our results exclude the hypothesis of a tri-hybrid intergeneric cross involving C. medica, C. maxima and a species of Microcitrus, proposed by Barrett and Rhodes (1976), or of a direct hybridization between C. medica and C. maxima, proposed by Liang et al. (2007) from an AFLP (amplified fragment length polymorphism) study. The origin of ‘Alemow’ (C. macrophylla in Tanaka’s classification) is more controversial. Swingle and Reece (1967) considered C. macrophylla to be a hybrid of C. celebica, or some other species of the subgenus Papeda, with a species of the subgenus Citrus, probably C. maxima. Tanaka placed it in the Limonellus section along with C. aurantifolia. Federici et al. (1998) found that C. macrophylla clustered with C. aurantifolia, and the papedas C. hystrix and C. micrantha, and Nicolosi et al. (2000) mentioned that ‘Mexican’ lime and C. macrophylla had similar C. medica/C. micrantha constitutions. A possible C. micrantha × C. medica origin was also proposed by Ollitrault et al. (2012) based on SNP data. Very few or no data are available on the origin of the other Tanaka species which presumably have a similar C. micrantha (or closely related papeda) × C. medica origin. In the case of the ‘New Caledonian’ and ‘Kaghzi’ lime, an F2 (C. micrantha × C. medica) × (C. micrantha × C. medica) model better fits the observed pattern with homozygous C. medica- and C. micrantha- specific alleles than a direct cross between ancestral taxa.
C. reticulata × C. medica.
Based on our data, we propose C. reticulata × C. medica as a model for C. limonia, C. jambhiri and C. karna. The origins of ‘Rough’ lemon (C. jambhiri), ‘Volkamer’ lemon (C. limonia Osbeck) and ‘Rangpur’ lime (C. limonia) have been widely debated. Most authors recognized mandarin as one parent of ‘Rangpur’ lime but combined with different second parents: lime (Webber, 1943; Tatum et al., 1974; Barkley et al., 2006), sour orange (Barrett and Rhodes, 1976), ‘Rough’ lemon (Handa and Oogaki, 1985) and citron, like the conclusion of our study (Federici et al., 1998; Gulsen and Roose, 2001a; Li et al., 2007). Scora (1975) suggested that ‘Rough’ lemon was a natural hybrid between a mandarin and a citron, and several more recent molecular studies agree with this hypothesis (Federici et al., 1998; Nicolosi et al., 2000; Gulsen and Roose, 2001a; Barkley et al., 2006; Ollitrault et al., 2012). The origin of ‘Volkamer’ lemon is more controversial. Barrett and Rhodes (1976) thought mandarin × sour orange was a possible parentage. Nicolosi et al. (2000) considered that citron and sour orange were the ancestors. Based on cytogenetic evidence, Carvalho et al. (2005) considered that ‘Volkamer’ lemon originated from mandarin × citron like ‘Rough’ lemon and ‘Rangpur’ lime. Our results agree with the last hypothesis, and both cytoplasmic and nuclear data point to a very close relationship between ‘Rough’ lemon and ‘Volkamer’ lemon. Cytogenetic studies also provided evidence for mandarin × citron as the origin of ‘Volkamer’ lemon, ‘Rough’ lemon and ‘Rangpur’ lime (Carvalho et al., 2005). These authors also observed that half the ‘Rough’ lemon and ‘Rangpur’ lime karyotypes were identical to the haploid chromosome complement of C. medica, while the other half of the chromosome set perfectly matched the haploid complement of the ‘Cleopatra’ mandarin. This last observation is also in agreement with our cytoplasmic data showing that ‘Rough’ lemon, ‘Volkamer’ lemon and ‘Rangpur’ limes share the CT of acid mandarins (‘Cleopatra’, ‘Sunki’, ‘Shekwasha’ and ‘Sun Chu Cha’).
(C. maxima × C. reticulata)×C. medica.
Three-species (C. maxima × C. reticulata) × C. medica appear to be at the origin of C. limetta, C. limettioides, C. meyeri and C. limon accessions. Cytoplasmic data and analysis of the fit of different concrete parental models suggest that C. aurantium × C. medica are at the origin of C. limetta and C. limon accessions, while other parents derived from C. maxima and C. reticulata gene pools are at the origin of C. limettioides and C. meyeri. Contributions from C. maxima, C. reticulata and C. medica in probably more complex hybridization schemes have been reported in several limes and lemons, particularly in C. bergamia. While Scora (1975) proposed that, by hybridization with lime, citron gave rise to lemon, more recent molecular and cytogenetic studies (Nicolosi et al., 2000; Gulsen and Roose, 2001a; Carvalho et al., 2005; Ollitrault et al., 2012; Garcia-Lor et al., 2013b; Ramadugu et al., 2013) agree that the ‘yellow lemon’ types originated from a C. aurantium × C. medica hybridization, and this was confirmed by the very good fit (>99 %) between our data and this model. Citrus limetta cultivars probably originated in the Mediterranean Basin (Webber et al., 1967) where sour orange and citron had long been present, and our results suggest that they have the same phylogenetic origin as ‘yellow lemons’. Many hypotheses have been proposed concerning the origin of C. limettioides. Citrus aurantifolia has frequently been proposed as one of its parents: C. aurantifolia by C. limetta or citron (Webber, 1943); C. aurantifolia by C. sinensis (Barrett and Rhodes, 1976) or C. aurantifolia × C. medica (Carvalho et al., 2005). Nicolosi et al. (2000) proposed citron and sweet orange as putative male and female parents, respectively. Our cytoplasmic data totally exclude the hypothesis of C. aurantifolia as the female parent of C. limettioides. Moreover, no evidence for a contribution by C. micrantha (one direct parent of C. aurantifolia) was provided by our nuclear analysis. We therefore consider that a contribution by C. aurantifolia to C. limettioides is improbable. The C. sinensis × C. medica model did not fit our data well (85 %), but 98 % of the 123 markers provided coherent patterns for a (C. maxima × C. reticulata) × C. medica model. Scora (1975) and Gulsen and Roose (2001a) proposed that ‘Meyer’ lemon (C. meyeri) is a sweet orange/citron hybrid. Our cytoplasmic results agree with this hypothesis, but the nuclear fit of this model was low (86 %). Therefore, C. meyeri is probably a (C. maxima × C. reticulata) × C. medica hybrid (96 % fit with our data) but whose concrete parents are not yet known. The origin of bergamot (C. bergamia) is also controversial. Gallesio (1811) proposed a sour orange × lemon origin, but several molecular studies disagreed and proposed hybridization between citron and lime (Chen et al., 1991), between sour orange and a sweet lime (Herrero et al., 1996; Federici et al., 2000), or between sour orange and a citron (Nicolosi et al., 2000; Li et al., 2010). We tested these different hypotheses against our data, and the best fit was observed for the C. limon × C. aurantium model (97 %), which is also in agreement with our cytoplasmic data.
Origin of triploid lime
Previous cytoplasmic studies showed that C. latifolia (‘Tahiti’ lime-like accessions) shared the same cytoplasmic pattern as C. limon and C. aurantium (Bayer et al., 2009; Froelicher et al., 2011). However, to our knowledge, this is the first molecular analysis of the nuclear structure which makes it possible to propose a hypothesis concerning the origin of these widely grown triploid limes. The five C. latifolia analysed displayed very similar patterns, with a contribution from the four ancestral taxa (C. maxima, C. medica, C. micrantha and C. reticulata), with all specific C. medica and C. micrantha alleles in single or double doses, while C. maxima and C. reticulata alleles were single dose or absent. Based on this cytoplasmic and nuclear pattern, we propose that ‘Tahiti’ lime-like accessions resulted from the fertilization of a haploid lemon ovule by a diploid gamete of a diploid ‘Mexican’-like lime. Ninety-nine percent of the 123 markers analysed agreed with this model. This diploid gamete probably originated from a natural doubled diploid of ‘Mexican’-like lime, like the ‘Giant Key’ lime selected from seedlings of ‘Key’ lime in 1973 by H.C. Barrett (US Horticultural Research Laboratory, Orlando). It is also possible that the C. aurantifolia diploid gamete was an unreduced gamete from a diploid plant. Based on segregation studies of morphological traits in ‘Tahiti’ lime seedlings, Reece and Childs (1962) proposed that this cultivar resulted from lime by citron or lemon hybridization but did not recognize the triploid status of ‘Tahiti’ lime. The identity of ‘Persian’, ‘Tahiti’ and ‘Bears’ limes is explained by the diffusion of these cultivars (Morton, 1987). The ‘Tahiti’-like lime is believed to have been introduced into the Mediterranean region via Iran (‘Persian’ lime). Portuguese traders probably transported it to Brazil, from where it was apparently taken to Australia in around 1824. From Tahiti, it reached California between 1850 and 1880 and Florida by 1883. According to Webber (1943), the ‘Bears’ variety originated around 1895 thanks to J.T. Bears, a nurseryman in Porterville, California, presumably as a seedling from a tree grown from seed from a fruit of Tahitian origin.
We also reveal a second group of triploid limes (‘Tanepao’, ‘Coppenrath’, ‘Ambilobe’ and ‘Mohtasseb’ limes and ‘Madagascar’ lemon) of different phylogenetic origin. These accessions, which display a C. micrantha CT and only a C. medica and C. micrantha contribution for the nuclear genome with mostly double doses of C. medica-specific alleles and a single one for C. micrantha, are most probably the result of a (C. micrantha × C. medica) × C. medica hybridization with a diploid gamete of the C. micrantha × C. medica parent. Tested with the ‘Mexican’ lime as genotype for the C. micrantha × C. medica parent, 96·3 % of our data fitted the 123 markers.
Asexual variations are important sources of phenotypic variability in the apomictic lime and lemon groups
All lime and lemon accessions display partial apomixes (polyembryonic seeds with nucellar embryos). Previous molecular (Gulsen and Roose, 2001b; Curk et al., 2015b) and cytogenetic studies (Carvalho et al., 2005) showed that numerous lemon cultivars originated from a single clonal parent via a series of mutations. Similar conclusions were proposed by Snoussi et al. (2012) for several limes and lemons after a survey of Tunisian citrus germplasm. Our results led to the same conclusions concerning the polymorphism observed within the different sub-clusters we identified (Supplementary Data Table S7). Indeed the accessions were highly heterozygous and displayed very limited diversity within sub-clusters, generally in the form of homozygous/heterozygous polymorphisms with a common allele. Such patterns probably do not originate from sexual hybridization of the highly heterozygous prototypes of the sub-groups. Their origin in parallel reticulation events was completely excluded in the sub-group of tri-specific origin. Given the diversity and heterozygosity within these taxa, it also appears unlikely in the sub-group resulting from direct hybridization between two ancestral taxa. Sporadic mutations and the mobility of transposable elements have been proposed as the source of diversity within citrus groups propagated vegetatively by apomictic seeds or by grafting (Bretó et al., 2001; Barkley et al., 2006; Ollitrault et al., 2012; Garcia-Lor et al., 2013b). In our study, the loss of the non-citron allele for five consecutive markers at the end of chromosome 9, and of two alleles on chromosome 5 and two on chromosome 8, while the rest of its tri-specific genomic structure was conserved, suggests that major chromosome deletion events are at the origin of the ‘Luminciana’ lemon. Mutation or epigenetic variations were previously reported to be a major cause of diversification in several apomictic species (Hörandl and Paun, 2007; Nybom, 2007). The contribution of somatic mutations to the evolution of other vegetatively propagated crops, such as grapes (Crespan, 2004), olives (Cipriani et al., 2002), yams (Scarcelli, 2005) and cassava (Sardos et al., 2008), has also been demonstrated. In addition to sexual recombination, human selection of new phenotypes and further clonal propagation are also key factors in the generation of intervarietal phenotypic polymorphism within horticultural groups (Barrett and Rhodes, 1976; Ollitrault et al., 2003).
Conclusions
The lime and lemon horticultural group is genetically highly polymorphic, with diploid, triploid and tetraploid varieties, maternal phylogeny involving four types of cytoplasm out of the six encountered in the Citrus genus, and the nuclear genome contribution of the four basic Citrus taxa (C. medica, C. maxima, C. micrantha and C. reticulata). All lime and lemon accessions we analysed were highly heterozygous and displayed interspecific admixture involving two or three, but also the four ancestral taxa genomes in a few accessions and particularly in the widely cultivated ‘Tahiti’ triploid limes (C. latifolia types). Citrus medica was shown to be the common genomic component of all limes and lemons but never to act as direct female parent due to the cleistogamy of citron accessions. Citrus medica was very probably the direct male parent of the main lime and lemon sub-group in combination with C. micrantha or close papeda species (C. aurata, C. excelsa, C. macrophylla and C. aurantifolia varieties of Tanaka’s taxa), C. reticulata (C. limonia, C. karna and C. jambhiri varieties of Tanaka’s taxa), C. aurantium (C. limetta and C. limon varieties of Tanaka’s taxa) or C. maxima × C. reticulata hybrid (C. limettioides, C. meyeri). Other combinations involving C. medica hybrids were also identified. Two origins were revealed for triploid limes. Citrus latifolia accessions (‘Tahiti’, ‘Bears’, ‘Persian’, ‘El Kseur’ and ‘IAC 5’) probably result from the fertilization of a haploid gamete of C. limon (yellow lemon type) by a diploid gamete of C. aurantifolia (‘Mexican’ lime type; 2n gamete of a diploid parent or a diploid gamete produced by a doubled diploid) while the C. aurantifolia triploid accessions (‘Tanepao’, ‘Coppenrath’, ‘Madagascar’, ‘Ambilobe’ and ‘Mohtasseb’) probably result from an interspecific backcross (a diploid ovule of C. aurantifolia –‘Mexican’ lime type – pollinated by C. medica). The lime and lemon horticultural group therefore results from many independent reticulation events (29 identified in this work) which explains why neither of the botanical classifications [Swingle and Reece’s (1967) with two species or Tanaka’s (1954) with 37 taxa] nor the usual denomination (limes and lemons) correctly reflects the genetic organization. Given the very high interspecific heterozygosity of all the accessions we analysed, we attribute the low intra-sub-group polymorphism to sporadic mutations, transposable elements, epigenetic variations or the deletion of genomic fragments, as clearly revealed at the end of chromosome 9 by our results on the ‘Luminciana’ lemon. Further investigation into interspecific admixture and the inferred phylogenetic origins of the main subgroup of limes and lemons will be essential for a better utilization of citrus biodiversity to create new rootstock and acid citrus cultivars.
SUPPLEMENTARY DATA
Supplementary data are available online at www.aob.oxfordjournals.org and consist of the following. Figure S1: distribution of the markers over the nine scaffolds. Figure S2: Neighbor–Joining analysis of mitochondrial and chloroplastic markers revealed five mytotypes and six chlorotypes. Figure S3: results of Neighbor–Joining analysis of the 90 MLGs with 123 molecular markers. Table S1: list of accessions with Swingle and Tanaka classification names, ploidy level, mytotype, chlorotype, cytoplasmic type and nuclear cluster information. Table S2: detailed marker information. Table S3: amplicon fragment size patterns of the eight cytoplasmic markers for the six identified cytoplasmic types. Table S4: estimation of the best number of sub-populations in structure analysis. Table S5: means and confidence interval of the four basic taxa (from ten permuted and aligned independent STRUCTURE run cluster outputs). Table S6: congruency of molecular data (123 SNP, indel and SSR markers) with several hypotheses of lime and lemon origins. Table S7: genotype data for all markers and all accessions.
LITERATURE CITED
- Abkenar AA, Isshiki S, Tashiro Y. 2004. Phylogenetic relationships in the ‘true citrus fruit trees’ revealed by PCR-RFLP analysis of cpDNA. Scientia Horticulturae 102: 233–242. [Google Scholar]
- Aleza P, Froelicher Y, Schwarz S, et al. 2011. Tetraploidization events by chromosome doubling of nucellar cells are frequent in apomictic citrus and are dependent on genotype and environment. Annals of Botany 108: 37–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacchi O. 1940. Observaçóes citólogicas em citrus. I. Número de cromosômios de algumas espécies e variedades. Jornal de Agronomia, Piracicaba 3: 249–58. [Google Scholar]
- Barkley NA, Roose ML, Krueger RR, Federici CT. 2006. Assessing genetic diversity and population structure in a citrus germplasm collection utilizing simple sequence repeat markers (SSRs). Theoretical and Applied Genetics 112: 1519–1531. [DOI] [PubMed] [Google Scholar]
- Barkley NA, Krueger RR, Federici CT, Roose ML. 2009. What phylogeny and gene genealogy analyses reveal about homoplasy in citrus microsatellite alleles. Plant Systematics and Evolution 282: 71–86. [Google Scholar]
- Barrett HC, Rhodes AM. 1976. A numerical taxonomic study of affinity relationships in cultivated Citrus and its close relatives. Systematic Botany 1: 105–136. [Google Scholar]
- Bayer RJ, Mabberley DJ, Morton C, et al. 2009. A molecular phylogeny of the orange subfamily (Rutaceae: Aurantioideae) using nine cpDNA sequences. American Journal of Botany 96: 668–685. [DOI] [PubMed] [Google Scholar]
- Beattie GAC, Holford P, Mabberley DJ, Haigh AM, Broadbent P. 2008. On the origins of Citrus, Huanglongbing, Diaphorina citri and Trioza erytreae. IRCHLB Proceedings, 23–56. [Google Scholar]
- Belkhir K, Borsa P, Chikhi L, Raufaste N, Bonhomme F. 1996–2004. GENETIX 4.05, logiciel sous Windows™ pour la génétique des populations. Laboratoire Génome, Populations, Interactions, CNRS UMR 5000. [Google Scholar]
- Bhattacharya SC, Dutta S. 1956. Classification of citrus fruits of Assam. Indian Council of Agricultural Research. Scientific Monograph. no. 20. [Google Scholar]
- Bonavia E. 1888. The cultivated oranges and lemons, etc. of India and Ceylon . London: W.H. Allen. [Google Scholar]
- Bretó MP, Ruiz C, Pina JA, Asins MJ. 2001. The diversification of Citrus clementina Hort. ex Tan., a vegetatively propagated crop species. Molecular Phylogenetics and Evolution 21: 285–293. [DOI] [PubMed] [Google Scholar]
- Bryan G, McNicoll J, Ramsay G, Meyer R, De Jong W. 1999. Polymorphic simple sequence repeat markers in chloroplast genomes of Solanaceous plants. Theoretical and Applied Genetics 99: 859–867. [Google Scholar]
- de Candolle A. 1886. Origin of cultivated plants, 2nd edn London: Kegan Paul. [Google Scholar]
- Carbonell-Caballero J, Alonso R, Ibañez V, Terol J, Talon M, Dopazo J. 2015. A phylogenetic analysis of 34 chloroplast genomes elucidates the relationships between wild and domestic species within the genus Citrus. Molecular Biology and Evolution 32: 2015–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho R, Soares Filho WS, Brasileiro-Vidal AC, Guerra M. 2005. The relationships among lemons, limes and citron: a chromosomal comparison. Cytogenetic and Genome Research 109: 276–282. [DOI] [PubMed] [Google Scholar]
- Chen LG, Omura M, Hidaka T. 1991. A study on the taxonomy of citrus with GOT isozymes. Acta Horticulturae Sinica 18: 27–32. [Google Scholar]
- Cipriani G, Marrazzo MT, Marconi R, Cimato A, Testolin R. 2002. Microsatellite markers isolated in olive (Olea europaea L.) are suitable for individual fingerprinting and reveal polymorphism within ancient cultivars. Theoretical and Applied Genetics 104: 223–228. [DOI] [PubMed] [Google Scholar]
- Crespan M. 2004. Evidence on the evolution of polymorphism of microsatellite markers in varieties of Vitis vinifera L. Theoretical and Applied Genetics 108: 231–237. [DOI] [PubMed] [Google Scholar]
- Cuenca J, Froelicher Y, Aleza P, Juarez J, Navarro L, Ollitrault P. 2011. Multilocus half-tetrad analysis and centromere mapping in citrus: evidence of SDR mechanism for 2n megagametophyte production and partial chiasma interference in mandarin cv ‘Fortune’. Heredity 107: 462–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuenca J, Aleza P, Navarro L, Ollitrault P. 2013. Assignment of SNP allelic configuration in polyploids using competitive allele-specific PCR: application to citrus triploid progeny. Annals of Botany 111: 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuppen E. 2007. Genotyping by allele-specific amplification (KASPar). CSH Protocols 2007: pdb.prot4841. [DOI] [PubMed] [Google Scholar]
- Curk F, Ancillo G, Garcia-Lor A, et al. 2014. Next generation haplotyping to decipher nuclear genomic interspecific admixture in Citrus species: analysis of chromosome 2. BMC Genetics 15: 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curk F, Garcia-Lor A, Snoussi H, et al. 2015a. New insights on limes and lemons origin from nuclear and cytoplasmic markers genotyping and targeted nuclear gene sequencing. Acta Horticulturae 1065: 135–146. [Google Scholar]
- Curk F, Ancillo G, Ollitrault F, et al. 2015b. Nuclear species-diagnostic SNP markers mined from 454 amplicon sequencing reveal admixture genomic structure of modern citrus varieties. PLoS One 10: e0125628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duportal M, Jorda E, Sanchez C, Imbert É, Loeillet D, Vannière H. 2013. FruiTrop FOCUS Citron. FruiTrop Cirad Hors Série 2013: 140. [Google Scholar]
- Esen A, Soost RK. 1971. Unexpected triploids in citrus: their origin, identification and possible use. Journal of Heredity 62: 329–333. [Google Scholar]
- Esselink G, Nybom H, Vosman B. 2004. Assignment of allelic configuration in polyploids using the MAC-PR (microsatellite DNA allele counting–peak ratios) method. Theoretical and Applied Genetics 109: 402–408. [DOI] [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. 2005. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FAO. 2014. FAOSTAT. http://faostat3.fao.org/home/E. [Google Scholar]
- Federici CT, Roose ML, Scora RW. 2000. RFLP analysis of the origin of Citrus bergamia, Citrus jambhiri, and Citrus limonia. Acta Horticulturae 535: 55–62. [Google Scholar]
- Federici CT, Fang DQ, Scora RW, Roose ML. 1998. Phylogenetic relationships within the genus Citrus (Rutaceae) and related genera as revealed by RFLP and RAPD analysis. Theoretical and Applied Genetics 96: 812–822. [Google Scholar]
- Froelicher Y, Mouhaya W, Bassene JB, et al. 2011. New universal mitochondrial PCR markers reveal new information on maternal citrus phylogeny. Tree Genetics and Genomes 7: 49–61. [Google Scholar]
- Froelicher Y, Dambier D, Bassene JB, et al. 2008. Characterization of microsatellite markers in mandarin orange (Citrus reticulata Blanco). Molecular Ecology Resources 8: 119–122. [DOI] [PubMed] [Google Scholar]
- Gallesio G. 1811. Traité du citrus, Louis Fantin edn. Paris: Chez Louis Fantin Libraire. [Google Scholar]
- Garcia-Lor A, Luro F, Navarro L, Ollitrault P. 2012. Comparative use of InDel and SSR markers in deciphering the interspecific structure of cultivated citrus genetic diversity: a perspective for genetic association studies. Molecular Genetics and Genomics 287: 77–94. [DOI] [PubMed] [Google Scholar]
- Garcia-Lor A, Ancillo G, Navarro L, Ollitrault P. 2013a. Citrus (Rutaceae) SNP markers based on competitive allele-specific PCR; transferability across the Aurantioideae subfamily. Applications in Plant Sciences 1: 1200406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Lor A, Curk F, Snoussi-Trifa H, et al. 2013b. A nuclear phylogenetic analysis: SNPs, indels and SSRs deliver new insights into the relationships in the ‘true citrus fruit trees’ group (Citrinae, Rutaceae) and the origin of cultivated species. Annals of Botany 111: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RM, Vardi A, Galun E. 1986. The plastome of Citrus. Physical map, variation among Citrus cultivars and species and comparison with related genera. Theoretical and Applied Genetics 72: 170–177. [DOI] [PubMed] [Google Scholar]
- Gulsen O, Roose ML. 2001a. Determination of genetic diversity and phylogenetic relations to citrus ancestors in lemons by DNA markers. Bahce 30: 53–63. [Google Scholar]
- Gulsen O, Roose ML. 2001b. Lemons: diversity and relationships with selected Citrus genotypes as measured with nuclear genome markers. Journal of the American Society for Horticultural Science 126: 309–317. [Google Scholar]
- Handa T, Oogaki C. 1985. Numerical taxonomic study of Citrus L. and Fortunella swingle using morphological characters application of multivariate analysis. Journal of the Japanese Society for Horticultural Science 54: 145–154. [Google Scholar]
- Herrero R, Asins MJ, Pina JA, Carbonell EA, Navarro L. 1996. Genetic diversity in the orange subfamily Aurantioideae. II. Genetic relationships among genera and species. Theoretical and Applied Genetics 93: 1327–1334. [DOI] [PubMed] [Google Scholar]
- Hodgson RW. 1955. Origin of citrus fruits grown in California. California Citrograph 49: 132–136. [Google Scholar]
- Hörandl E, Paun O. 2007. Patterns and sources of genetic diversity in apomictic plants: implications for evolutionary potentials. In: Hörandl E, Grossniklaus U, Van Dijk PJ, Sharbel TF, eds. Apomixis: evolution, mechanisms and perspectives. Koenigstein, Germany: Regnum Vegetabile: 281–298. [Google Scholar]
- Jena SN, Kumar S, Nair NK. 2009. Molecular phylogeny in Indian Citrus L. (Rutaceae) inferred through PCR-RFLP and trnL-trnF sequence data of chloroplast DNA. Scientia Horticulturae 119: 403–416. [Google Scholar]
- Kijas JMH, Thomas MR, Fowler JCS, Roose ML. 1997. Integration of trinucleotide microsatellites into a linkage map of Citrus. Theoretical and Applied Genetics 94: 701–706. [Google Scholar]
- Li H, Futch SH, Syvertsen JP. 2007. Cross-correlation patterns of air and soil temperatures, rainfall and Diaprepes abbreviatus root weevil in citrus. Pest Management Science 63: 1116–1123. [DOI] [PubMed] [Google Scholar]
- Li X, Xie R, Lu Z, Zhou Z. 2010. The origin of cultivated Citrus as inferred from internal transcribed spacer and chloroplast DNA sequence and amplified fragment length polymorphism fingerprints. Journal of the American Society for Horticultural Science 135: 341–350. [Google Scholar]
- Liang G, Xiong G, Guo Q, He Q, Li X. 2007. AFLP analysis and the taxonomy of Citrus. Acta Horticulturae 760: 137–142. [Google Scholar]
- Linnaeus C. 1753. Species Plantarum. Vol. II Holmiae (Stockholm), Impensis Laurentii Salvii [Printed for the Ray Society, London, in 1959.]. [Google Scholar]
- Luro F, Costantino G, Terol J, et al. 2008. Transferability of the EST-SSRs developed on Nules clementine (Citrus clementina Hort ex Tan) to other Citrus species and their effectiveness for genetic mapping. BMC Genomics 9: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luro F, Venturini N, Costantino G, Paolini J, Ollitrault P, Costa J. 2012. Genetic and chemical diversity of citron (Citrus medica L.) based on nuclear and cytoplasmic markers and leaf essential oil composition. Phytochemistry 77: 186–196. [DOI] [PubMed] [Google Scholar]
- Malik MN, Scora RW, Soost RK. 1974. Studies on the origin of the lemon. Hilgardia 42: 361–382. [Google Scholar]
- Morton J. 1987. Tahiti Lime. In: Morton JF, ed. Fruits of warm climates. Miami, FL. [Google Scholar]
- Nei M. 1973. Analysis of gene diversity in subdivided populations. Proceedings of the National Academy of Sciences, USA 70: 3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolosi E, Deng ZN, Gentile A, La Malfa S, Continella G, Tribulato E. 2000. Citrus phylogeny and genetic origin of important species as investigated by molecular markers. Theoretical and Applied Genetics 100: 1155–1166. [Google Scholar]
- Nybom H. 2007. Unique reproduction in dogroses (Ros sect. Canina) maintains successful and highly heterozygous genotypes. In: Hörandl E, Grossniklaus U, Van Dijk PJ, Sharbel TF, eds. Apomixis: evolution, mechanisms and perspectives. Koenigstein, Germany: Regnum Vegetabile, 169–194. [Google Scholar]
- Ollitrault P, Jacquemond C, Dubois C, Luro F. 2003. Citrus. In: Hamon P, Seguin M, Perrier X, Glaszmann J-C, eds. Genetic diversity of cultivated tropical plants. Cirad: Montpellier, 193–217. [Google Scholar]
- Ollitrault P, Dambier D, Luro F, Froelicher Y. 2008. Ploidy manipulation for breeding seedless triploid citrus. Plant Breeding Reviews 30: 323–352. [Google Scholar]
- Ollitrault P, Terol J, Garcia-Lor A, et al. 2012. SNP mining in C. clementina BAC end sequences; transferability in the Citrus genus (Rutaceae), phylogenetic inferences and perspectives for genetic mapping. BMC Genomics 13: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollitrault P, Garcia-Lor A, Terol J, et al. 2015. Comparative values of SSRs, SNPs and InDels for Citrus genetic diversity analysis. Acta Horticulturae 1065: 457–466. [Google Scholar]
- Penjor T, Yamamoto M, Uehara M, et al. 2013. Phylogenetic relationships of Citrus and its relatives based on matK gene sequences. PLoS One 8: e62574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier X, Jacquemoud-Collet JP. 2006. DARwin software. http://darwin.cirad.fr/. [Google Scholar]
- Pritchard Lab. 2014. Structure Software. http://pritchardlab.stanford.edu/structure.html. [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadugu C, Pfeil BE, Keremane ML, Lee RF, Maureira-Butler IJ, Roose ML. 2013. A six nuclear gene phylogeny of Citrus (Rutaceae) taking into account hybridization and lineage sorting. PLoS One 8: e68410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece PC, Childs JFL. 1962. Character differences among seedlings of the Persion lime. Proceedings of the Florida State Horticultural Society 75: 110–116. [Google Scholar]
- Risso JA. 1813. Essai sur l’histoire naturelle des oranges, bigaradiers, limettiers, cédratiers, limoniers ou citroniers, cultivés dans le Départment des Alpes Maritimes. Annales de Musee Paris 20: 169–212. [Google Scholar]
- Sardos J, McKey D, Duval MF, Malapa R, Noyer JL, Lebot V. 2008. Evolution of cassava (Manihot esculenta Crantz) after recent introduction into a South Pacific Island system: the contribution of sex to the diversification of a clonally propagated crop. Genome 51: 912–921. [DOI] [PubMed] [Google Scholar]
- Scarcelli N. 2005. Structure et dynamique de la diversité d’une plante cultivée à multiplication végétative: le cas des ignames au Bénin (Dioscorea sp.). PhD thesis, University of Montpellier II. [Google Scholar]
- Scora RW. 1988. Biochemistry, taxonomy and evolution of modern cultivated citrus. Proceedings of the Sixth International Citrus Congress 277–289. [Google Scholar]
- Scora RW. 1975. On the history and origin of Citrus. Bulletin of the Torrey Botanical Club 102: 369–375. [Google Scholar]
- Snoussi H, Duval MF, Garcia-Lor A, et al. 2012. Assessment of the genetic diversity of the Tunisian citrus rootstock germplasm. BMC Genetics 13: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swingle WT. 1914. Citrus and related genera. In: Bailey LH, ed. Standard cyclopedia of horticulture. New York: The Macmillan Co., 779–785. [Google Scholar]
- Swingle WT. 1943. The botany of citrus and its wild relatives of the orange family. In: Webber HJ, Batchelor LD, eds. The citrus industry. Berkeley, CA: University of California Press, 129–474. [Google Scholar]
- Swingle WT, Reece PC. 1967. The botany of Citrus and its wild relatives. In: Reuther W, Webber HJ, Batchelor LD, eds. The citrus industry, 2nd edn Berkeley, CA: University of California Press, 190–430. [Google Scholar]
- Tanaka T. 1954. Species problem in Citrus (Revisio Aurantiacearum IX). Tokyo, Japan: Japanese Society for Promotion of Science. [Google Scholar]
- Tanaka T. 1961. Citologia: semi-centennial commemoration papers on Citrus studies. Osaka: Citologia Supporting Foundation. [Google Scholar]
- Tatum JH, Berry RE, Hearn CJ. 1974. Characterization of citrus cultivars and separation of nucellar and zygotic seedlings by thin layer chromatography. Proceedings of the Florida State Horticultural Society 87: 75–81. [Google Scholar]
- Tolkowsky S. 1938. Hesperides: a history of the culture and use of citrus fruits. London: J: Bale, Sons & Curnow, Ltd. [Google Scholar]
- USDA 2015, ARS, National Genetic Resources Program. Germplasm Resources Information Network (GRIN). National Germplasm Resources Laboratory, Beltsville, MD. http://www.ars-grin.gov.4/cgi-bin/npgs/acc/display.pl?1705370 (28 October 2015). [Google Scholar]
- Webber HJ. 1943. Cultivated varieties of citrus. In: Webber HJ, Batchelor LD, eds. The citrus industry. Berkeley, CA: University of California Press, 475–668. [Google Scholar]
- Webber HJ, Reuther W, Lawton HW. 1967. History and development of the Citrus industry. In: Reuther W, Webber HJ, Batchelor LD, eds. The citrus industry. Berkeley, CA: University of California Press, 1–39. [Google Scholar]
- Weising K, Gardner RC. 1999. A set of conserved PCR primers for the analysis of simple sequence repeat polymorphisms in chloroplast genomes of dicotyledonous angiosperms. Genome 42: 9–19. [PubMed] [Google Scholar]
- Wright S. 1978. Variability within and among natural populations. Evolution and the genetics of populations. Chicago, IL: University of Chicago Press. [Google Scholar]
- Wu GA, Prochnik S, Jenkins J, et al. 2014. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nature Biotechnology 32: 656–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Kobayashi S, Nakamura Y, Yamada Y. 1993. Phylogenic relationships of citrus revealed by RFLP analysis of mitochondrial and chloroplast DNA. Japanese Journal of Breeding 43: 355–365. [Google Scholar]
- Yamamoto M, Abkenar AA, Matsumoto R, et al. 2007. CMA banding patterns of chromosomes in major Citrus species. Journal of the Japanese Society for Horticultural Science 76: 36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.