Abstract
The Rta (R transactivator) protein plays an essential role in the Epstein–Barr viral (EBV) lytic cascade. Rta activates viral gene expression by several mechanisms including direct and indirect binding to target viral promoters, synergy with EBV ZEBRA protein, and stimulation of cellular signaling pathways. We previously found that Rta proteins with C-terminal truncations of 30 aa were markedly enhanced in their capacity to bind DNA (Chen, L.W., Chang, P.J., Delecluse, H.J., and Miller, G., (2005). Marked variation in response of consensus binding elements for the Rta protein of Epstein–Barr virus. J. Virol. 79(15), 9635–9650.). Here we show that two phenylalanines (F600 and F605) in the C-terminus of Rta play a crucial role in mediating this DNA binding inhibitory function. Amino acids 555 to 605 of Rta constitute a functional DNA binding inhibitory sequence (DBIS) that markedly decreased DNA binding when transferred to a minimal DNA binding domain of Rta (aa 1–350). Alanine substitution mutants, F600A/F605A, abolished activity of the DBIS. F600 and F605 are located in the transcriptional activation domain of Rta. Alanine substitutions, F600A/F605A, decreased transcriptional activation by Rta protein, whereas aromatic substitutions, such as F600Y/F605Y or F600W/F605W, partially restored transcriptional activation. Full-length Rta protein with F600A/F605A mutations were enhanced in DNA binding compared to wild-type, whereas Rta proteins with F600Y/F605Y or F600W/F605W substitutions were, like wild-type Rta, relatively poor DNA binders. GAL4 (1–147)/Rta (416–605) fusion proteins with F600A/F605A mutations were diminished in transcriptional activation, relative to GAL4/Rta chimeras without such mutations. The results suggest that, in the context of a larger DBIS, F600 and F605 play a role in the reciprocal regulation of DNA binding and transcriptional activation by Rta. Regulation of DNA binding by Rta is likely to be important in controlling its different modes of action.
Keywords: EBV Rta, DNA binding
Introduction
Epstein–Barr virus (EBV), the first identified human tumor virus, is involved in the pathogenesis of several malignancies of lymphoid and epithelial cell origin. These include Burkitt’s lymphoma (Komano et al., 1998), nasopharyngeal carcinoma (Klein, 1979), Hodgkin’s lymphoma (Andersson, 2006), gastric cancer (Hoshikawa et al., 2002) as well as lymphoid and non-lymphoid cancers in immunocompromised hosts (Thompson and Kurzrock, 2004). Reactivation of EBV from latency into the lytic cycle is likely to play a role in the pathogenesis of some of these malignancies. For example, patients with Burkitt’s lymphoma and nasopharyngeal carcinoma have markedly elevated serum antibodies to viral lytic products for years prior to diagnosis (Geser et al., 1982; Zeng et al., 1985). The switch from latency to the lytic cycle of EBV is mediated by the products of two EB viral genes, BZLF1 and BRLF1 (Countryman et al., 1987; Countryman and Miller, 1985; Hardwick et al., 1988). These protein products, ZEBRA and Rta, are potent transcriptional activators that drive the lytic cascade of the virus (Carey et al., 1992; Ragoczy et al., 1998; Zalani et al., 1996). Mutational analysis of the EBV genome cloned in bacmids has confirmed that both proteins are required for lytic EBV reactivation and production of mature virions (Feederle et al., 2000). ZEBRA is sufficient to activate the EBV lytic cycle in all cell backgrounds (Countryman and Miller, 1985; Rooney et al., 1988; Takada et al., 1986). The capacity of Rta to induce the lytic cycle appears to be associated with its ability to activate expression of ZEBRA and to stimulate its own synthesis (Ragoczy and Miller, 2001). These activities may vary among cells (Ragoczy et al., 1998; Zalani et al., 1996).
EBV Rta protein does not share obvious homology with cellular proteins. However, homologues of Rta are found among all gamma-herpesviruses (DeWire et al., 2002; Hardwick et al., 1988; Lukac et al., 1998; Sun et al.,1998; Whitehouse et al.,1997). In all these viruses, the Rta-homologous proteins play crucial roles to activate lytic-cycle viral gene expression and to stimulate DNA replication (Ragoczy et al., 1998; Sun et al., 1998; Whitehouse et al., 1997; Wu et al., 2000). EBV Rta protein stimulates expression of early lytic-cycle genes; it can also activate some late lytic-cycle genes, such as gp350 and BLRF2 in the absence of DNA replication (Feederle et al., 2000; Ragoczy and Miller, 1999). The role of Rta in the EBV lytic cycle is not limited to transcriptional activation; Rta also enhances lytic viral DNA replication. Unlike ZEBRA, Rta is not known to bind to the lytic origin of DNA replication (Fixman et al., 1992, 1995). However, both ZEBRA and Rta are recruited to replication compartments at late stages of viral DNA replication (Daikoku et al., 2005; Park et al., 2008).
Rta protein activates lytic genes by at least three distinct mechanisms. Activation of one class of genes, including BMLF1 (a protein involved in mRNA transport and processing) and BLRF2 (a tegument protein), operates through direct binding of Rta to Rta responsive elements (RREs) in their promoters (Chen et al., 2005; Gruffat et al., 1992; Ragoczy and Miller, 1999). Another class of viral genes, activated synergistically by Rta and ZEBRA, include BMRF1 (DNA polymerase accessory protein), BHRF1 (bcl2 homologue) and BHLF1 (function unknown) (Holley-Guthrie et al., 1990; Lieberman et al., 1989). Synergistic activation of the BMRF1 promoter requires that both ZEBRA and Rta bind directly to the promoter DNA (Quinlivan et al., 1993). Direct and synergistic target genes share consensus Rta response elements (RREs) in their upstream promoter sequences (Chen et al., 2005; Gruffat et al., 1990, 1992; Hammerschmidt and Sugden,1988; Quinlivan et al.,1993). Activation of a third class of genes that lack RREs in their promoters involves indirect mechanisms. For example, Rta can activate Zp, the promoter of BZLF1, through activation of PI3 kinase (Darr et al., 2001) and the stress mitogen-activation protein (MAP) kinases (Adamson et al., 2000). Rta can autostimulate its own promoter Rp, through an Sp1/Sp3 binding site (Ragoczy and Miller, 2001). Interaction of Rta with MBD-1 containing chromatin-associated factor (MCAF1) leads to formation of an Sp1–MCAF1–Rta complex and promotes Sp1-mediated transcription (Chang et al., 2005). Rta also can activate the promoter of BALF5 (EBV DNA polymerase) through upstream stimulating factor (USF) and E2F (Liu et al., 1996). The existence of these different modes of action suggest that the DNA binding activity of the protein must be regulated.
Rta is a 605 aa protein. The N-terminus of Rta contains a DNA binding domain (aa 1–280) and a dimerization domain (aa 1–232). The transactivation domain is found in the C-terminal half of the protein (aa 416–605) (Manetet al.,1991). The activation domain of Rta contacts the TATA binding protein and TFIID in vitro (Manet et al., 1993). Rta interacts with CREB binding protein at multiple sites to enhance its transactivation function (Swenson et al., 2001). Rta is post-translationally modified by SUMO-1 at several lysine residues. Modification by SUMO-1 minimally enhances the transactivation function of Rta (Chang et al., 2004, 2008a). Rta is also modified by SUMO2/3 under the influence of the EBV BI’LF4 gene (Calderwood et al., 2008). Rta also binds to retinoblastoma protein (Rb), resulting in displacement of E2F, and stimulation of cells to enter the S phase of the cell cycle (Swenson et al., 1999; Zacny et al., 1998). This interaction may also activate the promoter of BALF5, the viral DNA polymerase (Liu et al., 1996). Interaction of Rta with the transcription factor TSG101 enhances binding of Rta to promoters of late viral genes (Chua et al., 2007).
In previous studies, we demonstrated that deletion of the C-terminal 30 aa of Rta strongly promoted the capacity of Rta protein to bind DNA in vitro. Elimination of this inhibitory region allowed us to develop the first system to study the DNA binding activity of Rta protein that was expressed in human cells and to precisely characterize the Rta response elements (Chen et al., 2005). Here, using smaller deletions, we show that the C-terminal 10 aa of Rta plays an essential role in this DNA binding inhibitory activity. Further point mutational analysis showed that two phenylalanines, F600 and F605, in this C-terminal region were essential for inhibiting the DNA binding function of Rta. We identified a DNA binding inhibitory sequence (DBIS), encompassing the C-terminal 51 aa of Rta, that was able to suppress the minimal DNA binding domain of Rta, located in aa 1–350. F600A/F605A mutations overcame the action of the DBIS. The F600A/ F605A mutations not only enhanced binding by Rta they significantly decreased the transactivation function of Rta in several different biologic assays for activation of EBV lytic gene expression. F600A/ F605A mutations not only weakened the transactivation activity of intact Rta protein, they also decreased the transactivation function of GAL4–Rta fusion proteins. These results suggest that the DNA binding and transactivation function of Rta may be reciprocally regulated by these two amino acids in context of a larger DNA binding inhibitory domain.
Results
Delineation of a DNA binding inhibitory region of Rta using C-terminal deletion mutants
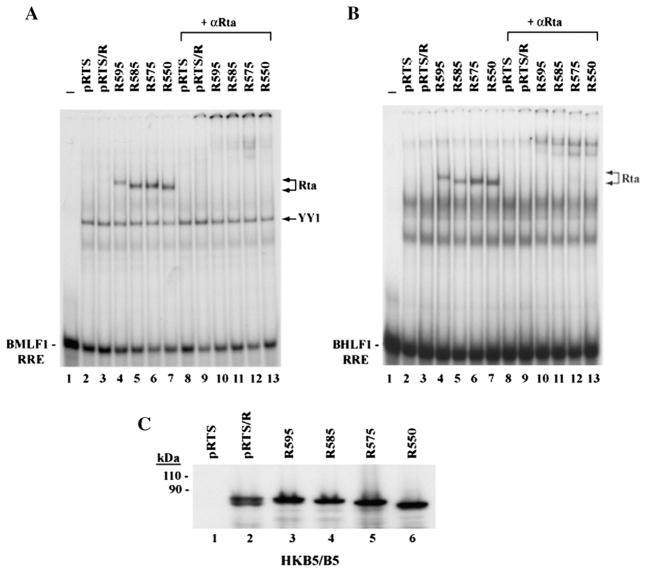
Our previous study of deletion mutants showed that elimination of 30 amino acids (aa 576 to 605) from the C-terminal region of Rta protein markedly enhanced the DNA binding activity of Rta in vitro to the RRE from the BMLF1 promoter (Chen et al., 2005). To further demarcate the region involved in the inhibition of DNA binding, and to learn whether the deletions equally affected binding to the BHLF1 promoter, which also contains a high affinity RRE, we compared the DNA binding activity of wild-type and C-terminal truncated Rta proteins expressed in a human cell line. When extracts of HKB5/B5 cells that had been transfected with a plasmid containing a wild-type BRLF1 gene (pRTS/R) were used in EMSA experiments, the association between full-length Rta protein and RREs from either BMLF1 or BHLF1 promoter was very weak or not detectable (Fig. 1A and Fig. 1B, lane 3). However, four Rta mutants with progressive deletions in the C-terminus displayed stronger DNA binding activity than wild-type Rta protein (Figs. 1A and B, lanes 4 to 7). The full-length and truncated Rta proteins were expressed equally in transfected cells (Fig. 1C); therefore, lack of DNA binding activity by the full-length construct was not due to insufficient levels of protein expression. The specific interaction between the truncated Rta proteins and the RRE DNA was confirmed by supershift with antibody to Rta (aa 1–320) (Figs. 1A and B, lanes 9–13). All the deletion mutants bound more strongly than wild-type to both probes. Even, R595 (aa 1–595), with only a 10 amino acid deletion in the C-terminus, bound DNA more avidly than WT Rta (Figs. 1A and B, lane 4). This data indicated that a component of the DNA binding inhibitory sequence (DBIS) was present in the C-terminal 10 amino acids of Rta, although the entire signal might extend beyond this region.
Fig. 1.
Deletion of the C-terminal 10 amino acids of Rta enhances its capacity to bind to DNA. (A, B) EMSAs. HKB5/B5 cells were transfected with plasmids expressing empty vector (pRTS), full-length Rta protein (pRTS/R) and C-terminal truncated mutants R595 (aa 1–595), R585 (aa 1–585), R575 (aa 1–575) and R550 (aa 1–550). Cells were collected 48 h after transfection. Extracts of transfected cells were incubated with 37 bp 32P-labeled probes containing the BMLF1 RRE (A) or the BHLF1 RRE (B). Purified IgG polyclonal antibody to Rta (aa 1–320) was used to supershift the Rta–DNA complexes. (C) Expression of full-length and C-terminal deletion mutant Rta proteins in HKB5/B5 cells. The transfected cell extracts were analyzed by immunoblotting with polyclonal antibody to Rta.
Rta (F600A/F605A) is enhanced in binding DNA in vitro
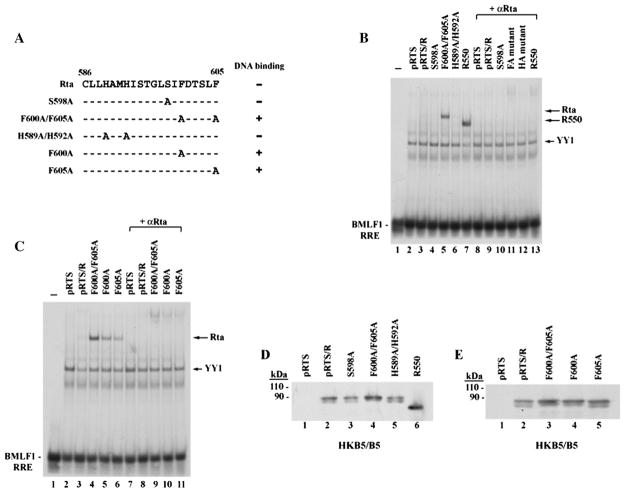
To analyze which amino acids in the C-terminus might contribute to inhibition of DNA binding, we initially explored the hypothesis that DNA binding could be regulated by post-translational modification in a manner similar to cellular transcription factors such as p53 (Jayaraman and Prives, 1995) and viral transcription factors such as ZEBRA (El-Guindy et al., 2007). Therefore, we first mutated S598, a potential phosphorylation site in the DBIS of Rta. However, the S598A mutant protein behaved like wild-type Rta; the interaction between S598A and a duplex oligonucleotide containing the RRE from the BMLF1 promoter was very weak (Fig. 2B, lane 4). The expression level of S598A mutant was similar to full-length Rta (Fig. 2D, lane 3). Next, we examined the role of phenylalanine or histidine residues in the inhibitory region. These amino acids play important roles in certain protein–protein interactions (Ahmed et al., 2001; Clark et al., 1996; Dowd et al., 2002; Ehlers et al., 1996; Lin et al., 2006). The mutant F600A/F605A was markedly enhanced in DNA binding (Fig. 2B, lane 5); however, the mutant H589A/H592A did not bind to BMLF1 RRE (lane 6). R550 (aa 1–550), a C-terminal truncated mutant that eliminated the inhibitory region, served as an example of a form of Rta that was competent to bind DNA (lane 7). All mutant Rta proteins were equally expressed in transfected cells; thus differences in DNA binding activity of these point mutants were not due to their expression levels (Fig. 2D).
Fig. 2.
Phenylalanine to alanine point mutants, Rta F600A and F605A, are enhanced in binding to DNA in vitro. (A) Summary of point mutagenesis in the C-terminal region of Rta. Numbers represent amino acid residues. The effect of three single point mutants (S598A, F600A and F605A) and two double point mutants (F600A/F605A and H589A/ H592A) on DNA binding are shown. (+): DNA binding enhanced. (B) and (C) EMSAs. HKB5/B5 cells transfected with the indicated plasmids were harvested 48 h after transfection, and cell extracts were prepared. EMSAs were conducted with BMLF1 RRE as a probe. Purified IgG antibody to Rta was used for supershift. FA mutant: F600A/ F605A; HA mutant: H589A/H592A (D) and (E) expression of wild-type and mutant Rta proteins in cell extracts used for EMSA was detected by immunoblotting with antibody to Rta. pRTS is an empty vector control.
Two single point mutants, F600A and F605A, each enhanced DNA binding by Rta (Fig. 2C, lanes 5 and 6). The double point mutant F600A/F605A manifested stronger DNA binding activity than either single point mutant, F600A or F605A (Fig. 2C, lane 4). This data suggested that both F600 and F605 contributed to inhibition of DNA binding by Rta. The restoration of DNA binding ability in the mutants F600A/F605A, F600A and F605A was unrelated to their protein expression level; all Rta mutants were expressed similarly in cell extracts used for EMSA (Fig. 2E).
A DNA binding inhibitory function of Rta, encoded in the C-terminal 50 aa, can be transferred to a minimal DNA binding domain of Rta
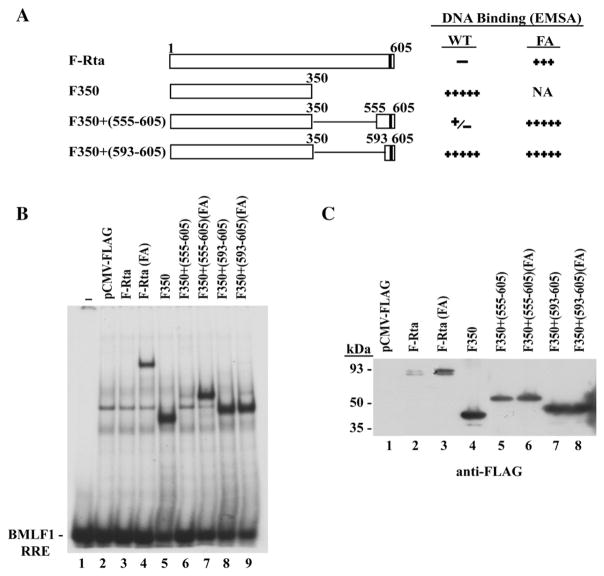
The results shown in Fig. 1 demonstrated that the C-terminal 10 aa were essential for inhibiting binding of Rta protein to DNA. To define a minimal region that was sufficient to inhibit binding of Rta to DNA, two different portions of the C-terminus of Rta protein were fused to a truncated Rta protein, containing only the DNA binding domain of Rta (aa 1–350) (Fig. 3A). A FLAG-tagged version of the DNA binding domain of Rta, F350, manifested strong DNA binding activity in EMSA (Fig. 3B, lane 5). Fusion with the C-terminal 13 aa of Rta (aa 593–605), containing the two essential phenylalanines, failed to inhibit the DNA binding of F350 (Fig. 3B, lane 8). By contrast, fusion of F350 with the C-terminal 51 aa of Rta (aa 555–605) was sufficient to block binding to DNA (Fig. 3B, lane 6). Installing the F600A/F605A mutation in Rta (aa 555–605) restored DNA binding by the F350+ (550–605) Rta deletion mutant protein (Fig. 3B lane 7). Since the expression level of the mutant proteins F350+ (555–605) without or with the FA mutation was similar (Fig. 3C, lanes 5, 6), their capacity to bind DNA was independent of their protein expression. These results indicated that a functional DNA binding inhibitory sequence is located in the C-terminal 51 aa. The C-terminal 13 amino acids, although a component of the DBIS, was not sufficient to inhibit DNA binding.
Fig. 3.
The DNA binding inhibitory activity can be transferred to a C-terminal truncated form of Rta protein (aa 1–350) that is competent to bind DNA. (A) Summary of DNA binding activity of wild-type (WT) and deletion mutant Rta proteins without or with the F600A/F605A (FA) point mutations. F-Rta: CMV-Flag-Rta. F350: CMV-Flag-Rta (aa 1–350). NA: not applicable. (B) An EMSA performed using extracts prepared from HKB5/B5 cells transfected with the indicated plasmids. The BMLF1 RRE was used as a probe. (C) Expression of wild-type and deletion mutant Rta proteins with or without the FA mutation in cell extracts used for EMSA. Proteins were detected by immunoblotting with antibody to FLAG.
The F600A/F605A mutation enhances binding of Rta to DNA in vivo
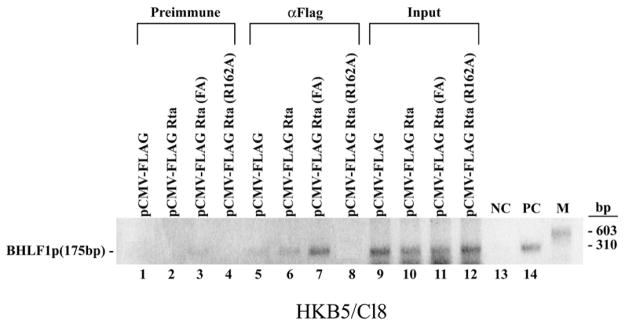
Since EMSA demonstrated enhanced DNA binding activity by Rta (F600A/F605A) in vitro, we carried out chromatin immunoprecipitation (ChIP) experiments to determine whether enhanced DNA binding activity of this mutant was also detected in EBV-infected cells. HKB5/Cl8 cells were transfected with plasmids expressing wild-type FLAG-Rta, FLAG-Rta (F600A/F605A) and FLAG-Rta (R162A), a mutant of Rta that was deficient in binding to DNA by EMSA (data not shown). Chromatin extracts were immunoprecipitated with pre-immune serum or with antibodies to FLAG. The association between proteins and DNA was detected by PCR with primers specific for the promoter region of BHLF1 that is known to contain two high affinity RREs (Gruffat et al., 1990; Manet et al., 1991). As observed in vitro, Rta (F600A/ F605A) showed stronger DNA binding in vivo on the BHLF1 promoter than wild-type Rta (Fig. 4, compare lanes 6 and 7). The R162A mutant did not detectably bind the BHLF1 promoter in this ChIP assay.
Fig. 4.
The mutant Rta (F600A/F605A) is enhanced in binding to DNA in vivo. ChIP assays were performed with extracts of HKB5/Cl8 cells transfected with empty vector pCMV-Flag (lanes 1, 5 and 9), pCMV-Flag-Rta (lanes 2, 6 and 10), pCMV-Flag-Rta (F600A/F605A) (lanes 3, 7 and 11) and pCMV-Flag-Rta (R162A) (lanes 4, 8 and 12). Twenty hours after transfection cell extracts were precipitated with pre-immune antiserum (lanes 1–4) or antibody to FLAG (lanes 5–8). Association between proteins and DNA was detected by PCR using primers specific for the promoter region of BHLF1. Lanes: 9 to 12, input material; 13, negative-control (NC): PCR without added DNA template; 14, positive-control (PC): PCR with plasmid containing BHLF1 promoter as template.
Mutants of Rta that are competent to bind DNA, when expressed in mammalian cells, are markedly diminished in their capacity to synergize with ZEBRA
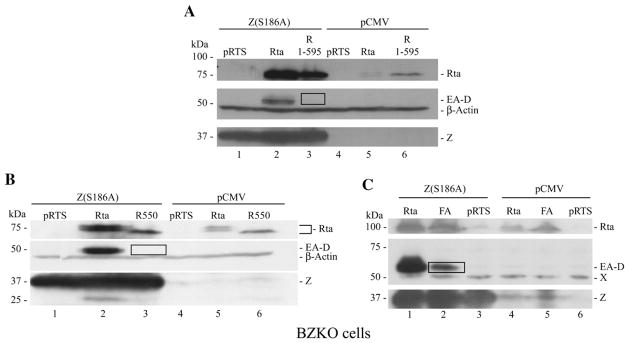
In an initial attempt to characterize the biologic phenotype of Rta mutants with enhanced DNA binding activity two deletion mutants, R (1–595) and R (1–550), and the F600A/F605A double point mutant were evaluated for their capacity to activate expression of EA-D (early antigen-diffuse), the product of the EBV BMRF1 gene, encoding the DNA polymerase processivity factor. EA-D is activated co-operatively by direct binding of EBV Rta and EBV ZEBRA to the BMRF1 promoter (Quinlivan et al., 1993). The ZEBRA mutant Z(S186A) does not by itself activate BMRF1, but can synergize with Rta to activate expression of this gene, both the transcript and the protein (Adamson and Kenney, 1998; Francis et al., 1999). Figs. 5A (lane 3) and 5B (lane 3) show that two deletion mutants of Rta which were enhanced in DNA binding did not detectably activate EA-D protein when they were introduced into BZKO cells together with Z(S186A). Wild-type Rta was always active in this assay. The F600A/F605A mutant had weak activity in the assay of synergy with Z(S186A), approximately 19% of the activity of wild-type Rta protein (Fig. 5C, compare lanes 1 and 2). These experiments demonstrated that those mutants of Rta with enhanced DNA binding were significantly diminished in transcriptional activity in an assay for synergy with ZEBRA.
Fig. 5.
Mutants of Rta that are enhanced in binding to DNA are deficient in synergy with ZEBRA. (A, B). BZKO cells were transfected with Z(S186A) (lanes 1–3) or CMV vector (lanes 4–6) together with pRTS (lanes 1,4), Rta (lane 2,5) or deletion mutants of Rta (lane 3, 6). The deletion mutants were R(1–595) (A) or R(1–550) (B). (C) BZKO cells were transfected with Z(S186A) (lanes 1–3) or CMV vector (lanes 4–6) together with wild-type Rta (lanes 1,4), the FA mutant (F600A/F605A) (lanes 2,5) or pRTS (lanes 3,6). Cell lysates were analyzed for expression of Rta, EA-D, ZEBRA and β actin by immunoblotting with monospecific antisera. In panel C, X denotes a cross-reactive cellular protein that serves as a loading control.
Rta (F600A/F605A) is a weaker transactivator than wild-type Rta
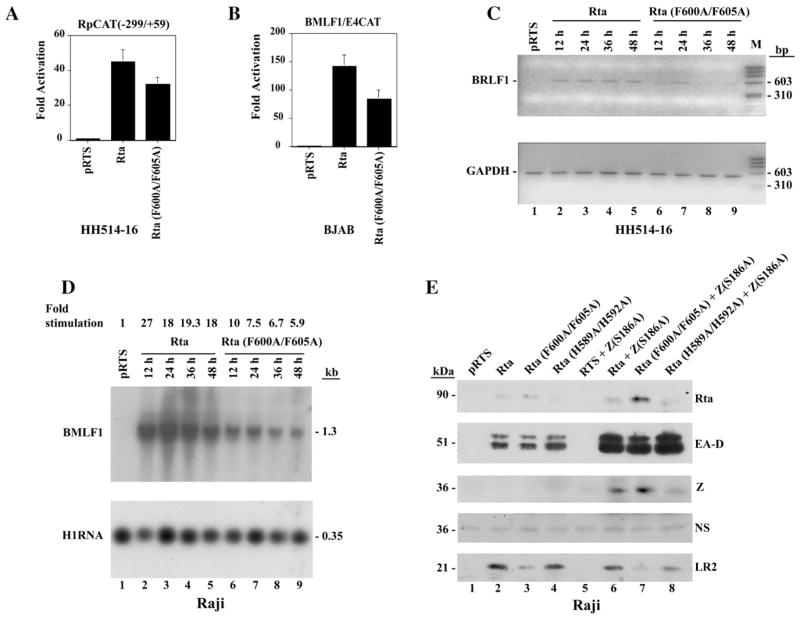
The next series of experiments (Fig. 6) compared transcriptional activation by the Rta (F600A/F605A) (FA) mutant with wild-type Rta in a broader range of biologic assays than synergy with Z (S186A). These included reporter assays for activation of viral promoters and assays for activation of expression of EB viral lytic cycle genes that are direct and indirect targets of Rta action. In reporter assays (Fig. 6A) the FA mutant was approximately 70% as active as wild-type in its capacity to auto-activate Rp, which is an indirect target of Rta action (Ragoczy and Miller, 2001). The BMLF1p, which is a direct target of Rta, by virtue of its high affinity RRE, (Gruffat et al., 1992), was activated by the FA mutant at 60% of wild-type level (Fig. 6B).
Fig. 6.
Compared to wild-type Rta, the Rta (F600A/F605A) mutant is diminished in its capacity to activate target genes. (A) Response of a CAT reporter containing the EBV BRLF1 promoter [RpCAT(−299/+59)] to Rta or Rta (F600A/F605A) proteins in HH514–16 cells. (B) Response of BMLF1/E4CAT promoter to Rta or Rta (F600A/F605A) proteins in BJAB cells. (C) Autostimulation of BRLF1 mRNA by Rta and Rta (F600A/F605A). Total RNA was prepared from HH514–16 cells at 12 h intervals after transfection. Expression of BRLF1 mRNA was analyzed by RT-PCR. GAPDH mRNA served as an internal loading control. (D) Activation of BMLF1 mRNA by Rta and Rta (F600A/F605A). Total RNA was prepared from Raji cells at different times after transfection. Expression of BMLF1 RNA was analyzed by Northern blot hybridization. Hybridization with H1RNA of RNaseP served as a loading control. (E) Activation of EBV lytic cycle proteins in Raji cells by wild-type Rta or F600A/F605A mutant Rta acting alone or together with Z(S186A). Cells were transfected with a total of 10 μg of plasmid DNA. In lanes 2–4, cells received 5 μg of WT or mutant Rta and 5 μg of empty vector (pRTS). In lanes 6–8, cells received 5 μg of WT or mutant Rta and 5 μg of Z(S186A). An immunoblot prepared 48 h after transfection was probed sequentially with antisera to Rta, EA-D, ZEBRA (Z), and BLRF2 (LR2). NS: a non-specific band which controlled for protein loading.
The capacity of the FA mutant to activate expression of the same two target genes from latent EBV genomes was also assessed. EBV Rta auto-stimulates the BRLF1 gene in some EBV-positive cells, such as HH514–16 (Ragoczy et al., 1998; Ragoczy and Miller, 2001). Using RT-PCR, with primers that span a 940 nt intron in the BRLF1 gene (Manet et al., 1989), we found that wild-type Rta activated BRLF1 to approximately the same level at 4 time points 12 to 48 h after transfection into HH514–16 cells. A weak signal of the 550 bp RT-PCR product was detected only once, 24 h after transfection of the FA mutant (Fig. 6C). The level of internal control GAPDH (glyceraldehyde-3-phosphate dehydrogenase) mRNA was the same in cells transfected with wild-type or F600A/F605A (Fig. 6B, compare lanes 2–5 and lanes 6–9).
Activation of BMLF1 expression was analyzed by Northern blotting in Raji cells where this gene is dominantly controlled by Rta (Chen et al., 2005). At 4 time points between 12 and 48 h after transfection, the abundance of the BMLF1 mRNA was approximately 2.8-fold greater after transfection of wild-type Rta than after transfection of the FA mutant (Fig. 6D, compare lanes 2–5 and lanes 6–9).
The effect of the F600A/F605A mutations on the capacity of Rta to activate its target genes and to synergize with Z(S186A) was also examined by immunoblotting in Raji cells (Fig. 6E). In this cell background wild-type Rta weakly activated EA-D (Fig. 6E; lanes 2–4); the addition of the ZEBRA mutant Z(S186A), which by itself manifests no activity (lane 5), leads to strong synergistic activation of EA-D by Rta and ZEBRA (Fig. 6E, lanes 6–8). In Raji cells the Rta (F600A/F605A) mutant was slightly reduced in its ability to activate EA-D (Fig. 6E, lane 3); it could synergize with Z(S186A), but the signal of EA-D in the presence of Z(S186A) was also slightly reduced by comparison to wild-type (Fig. 6E, lanes 3 and 7). The deleterious effects of the FA point mutants on synergy of Rta with Z(S186A) were not as pronounced in Raji cells as in BZKO cells (compare Fig. 5C with Fig. 6E).
Despite the absence of lytic DNA synthesis in Raji cells, Rta activates a late gene, BLRF2, encoding a component of the viral tegument in this cell background (Ragoczy and Miller, 1999). Unlike the synergistic activation of the BMRF1 promoter, the activation of BLRF2 was not enhanced by co-transfection of the Z(S186A) mutant (Fig. 6E, compare lanes 2 and 6) indicating that BLRF2 is a direct target of Rta. The Rta (F600A/F605A) mutant was markedly impaired at activating BLRF2, both in the absence and presence of Z(S186A) (lane 3 and lane 7). The Rta (F600A/F605A) mutant was also markedly defective in activating BLRF2 in BZKO cells (data not shown). The mutant Rta (H589A/H592A) (Fig. 2) behaved like wild-type in activating EA-D, alone and in synergy with Z(S186A), and in activation of BLRF2 (Fig. 6E, lanes 4 and 8). The mutant Rta (F600A/F605A) protein was more abundantly expressed than wild-type Rta or the mutant of Rta (H589A/H592A). Thus, whether assessed by reporter assays or assays for EBV lytic gene expression, at the level of viral transcripts or viral polypeptide expression, Rta (F600A/F605A), which manifests stronger DNA binding activity than wild-type Rta, was consistently defective as a transactivator.
Importance of aromatic amino acids at positions 600 and 605 on transcriptional activity of Rta
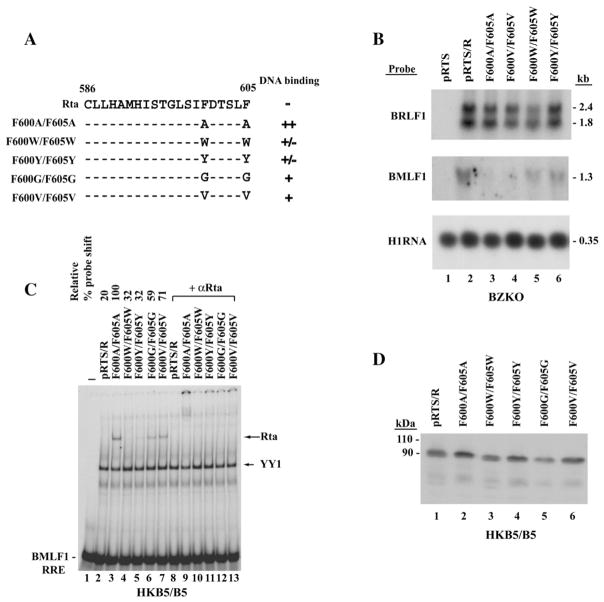
Previous detailed studies of the Herpes Simplex Virus VP16 protein have emphasized the importance of aromatic residues in mediating its transcriptional activation function (Cress and Triezenberg, 1991; Regier et al., 1993). To explore the significance of aromatic residues at positions 600 and 605 in the C-terminal activation domain of Rta, the activity of mutants with substitutions of aromatic residues, tyrosine (Y) and tryptophan (W), was compared with mutants that exchanged alanine (A), glycine (G) and valine (V) at these positions (Fig. 7A). Substitutions of A or V at these positions markedly weakened or eliminated the ability of Rta to activate a direct target, the BMLF1 gene, in BZKO cells, whereas the aromatic substitutions, W and Y, decreased, but did not abolish, this function (Fig. 7B).
Fig. 7.
Effects of different amino acid substitutions of F600 and F605 on the DNA binding and transcriptional activation functions of full-length Rta protein. (A) Summary of effect of amino acid substitutions on DNA binding. Numbers represent amino acid residues. (B) Northern blot analysis of the capacity of Rta and the F600/F605 substitution mutants to activate BMLF1 mRNA in BZKO cells. The indicated plasmids were transfected into BZKO cells. 48 h after transfection total RNA was isolated, electrophoresed and transferred to a nylon membrane. The blots were probed for BRLF1 mRNA, to detect expression from the plasmids, and for an Rta target gene, BMLF1 mRNA. Hybridization with a probe to H1 RNA of RNaseP served as a loading control. (C) Extracts of HKB5/B5 cells harvested 48 h after transfection were used in EMSA with BMLF1 RRE as a probe. Antibody to Rta (aa 1–320) was used for supershift in EMSAs. (D) Expression of Rta and mutant proteins in the same cell lysate prepared for EMSA was detected by polyclonal antibody to Rta (1–320).
Aromatic amino acid substitutions at positions 600 and 605 weaken the DNA binding activity of Rta
The capacity of Rta mutants with aromatic or non-aromatic substitutions at positions F600 and F605 to bind DNA was compared by EMSA with a probe of the Rta response element from the BMLF1 promoter (Fig. 7C). The two mutants with aromatic substitutions, F600W/F605W and F600Y/F605Y, both bound DNA weakly. Substitutions of A, G or V resulted in the acquisition of the ability to bind the probe in vitro. Substitution of alanine conferred more DNA binding activity than substitution of valine or glycine. All the protein mutants were expressed to comparable level (Fig. 7D). Thus, inhibition of DNA binding appears to be mediated by aromatic residues and not by the non-aromatic amino acids that were tested. Furthermore, these experiments showed that a substitution of a non-aromatic residue other than alanine, namely valine, that increased DNA binding also weakened the capacity of Rta to activate a direct target, BMLF1, in BZKO cells.
Effect of fusion of the DNA binding inhibitory sequence (DBIS) of EBV Rta to the DNA binding domain of GAL4
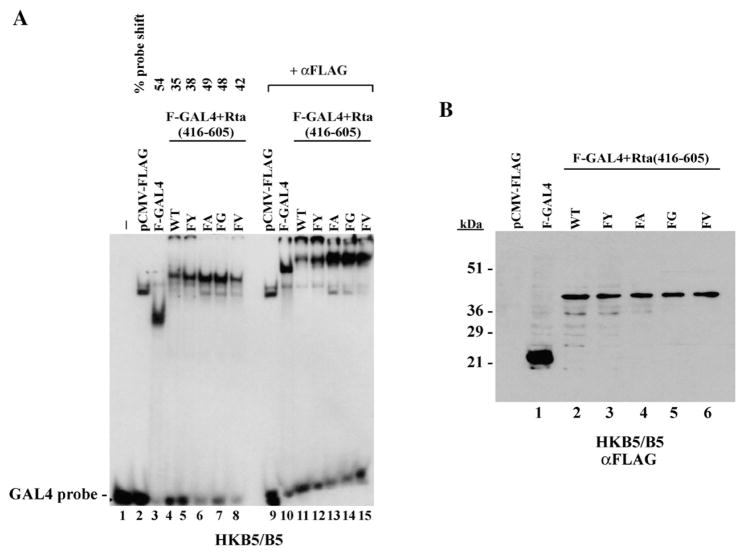
The next experiment asked whether the DBIS of Rta inhibited the function of a heterologous DNA binding domain. For these experiments aa 416–605 of Rta, containing the transcriptional activation domain and DBIS, were fused to GAL4 (1–147). The construct contained an N-terminal FLAG tag to facilitate detection of the proteins. The capacity of the chimeric proteins to bind a GAL4 site in vitro was evaluated by EMSA (Fig. 8A). Although equal amounts of the GAL4/Rta fusion proteins were present in cell extracts used for EMSA, none of the GAL4–Rta fusion proteins was as abundant as GAL4 alone (Fig. 8B, lane1). Addition of Rta (416–605) to GAL4 (1–147) did not eliminate binding to a GAL4 site; GAL4/Rta bound the GAL4 site about 65% as efficiently as GAL4. This could be the result of lower protein levels. Of the five GAL4/Rta fusion constructs tested, GAL4/Rta and GAL4/Rta (FY) bound less avidly than GAL4/Rta with FA or FG substitutions. Similar results were obtained by two of us (L-W.C and V.R) in replicate experiments performed in two different laboratories. In general, the results showed that the DBIS of Rta was much less inhibitory on the GAL4 DNA binding domain than on the Rta–DNA binding domain (Fig. 3). However, relative to wild-type Rta, mutants with amino acid substitutions such as FA and FG that favored DNA binding on an RRE (Fig. 7) also demonstrated stronger DNA binding on a GAL4 site (Fig. 8).
Fig. 8.
DNA binding by GAL4/Rta fusion proteins. FLAG-tagged constructs contained the DNA binding domain of GAL4 (aa 1–147) fused to Rta (aa 416–605) encompassing the transactivation domain. The Rta portion of the constructs were wild-type or contained the indicated amino acid substitutions at positions F600 and F605. (A) EMSA showing binding of GAL4/Rta fusion proteins to a GAL4 probe (lanes 1–8). A supershift was carried out with antibody to FLAG (lanes 9–16). (B) Immunoblot of extracts used for the EMSA probed with antibody to FLAG.
F600A/F605A mutations weaken the transactivation domain of Rta fused to the DNA binding domain of GAL4
The DNA binding inhibitory sequence of Rta is co-linear with a portion of the transactivation domain of Rta protein. Since experiments illustrated in Figs. 5 through 7 demonstrated that the Rta (F600A/F605A) mutant was a relatively deficient activator of EBV lytic genes, we explored the possibility that Rta (F600A/F605A) mutations impinge on the transcriptional activation function of Rta when fused to a heterologous DNA binding domain. GAL4–Rta chimeras containing the GAL4 DNA binding domain (aa 1–147) fused to the Rta transactivation domain (aa 416–605), and similar chimeras containing the Rta (F600A/F605A) mutations, were examined in parallel for their capacity to activate a reporter consisting of five GAL4 binding sites upstream of the adenovirus E1B minimal promoter fused to CAT. In BJAB cells, as has been previously described in other cells, GAL4–Rta was a stronger activator than GAL4 fused to the activation domain of Herpes simplex virus VP16 protein (Hardwick et al.,1992). The activity of the GAL4–Rta (F600A/F605A) mutant was similar to that of GAL4–VP16; it retained only 30% of the activity of GAL4–Rta (Fig. 9A). Since transcriptional activation of the GAL4–Rta fusion proteins was mediated through the DNA binding domain of GAL4 and the transactivation domain of Rta, these experiments suggested that the F600A/F605A mutations directly decreased the transcription activation function of Rta.
Fig. 9.
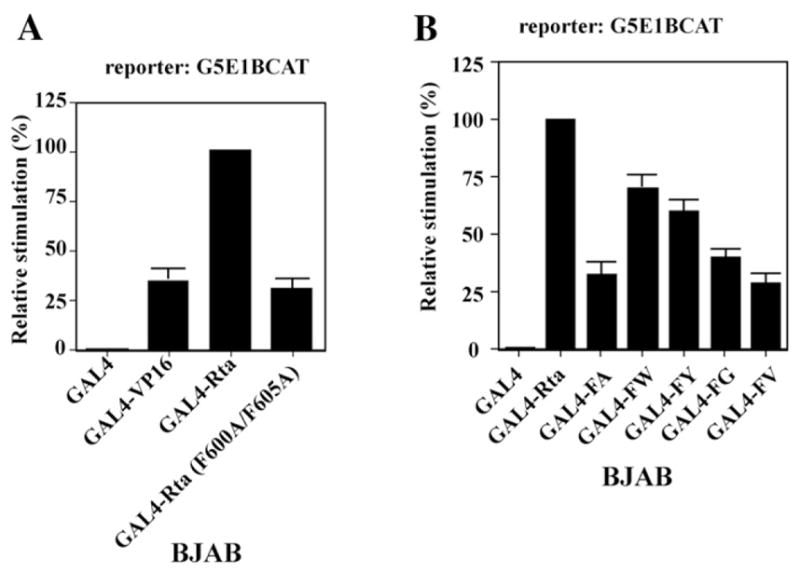
Amino acid substitutions at F600 and F605 diminish the potency of the transactivation domain of Rta (aa 416–605) fused to the GAL4 DNA binding domain. pM plasmids encoding GAL4, GAL4–Rta, GAL4–Rta with amino acid substitutions, or GAL4–VP16 fusion proteins were co-transfected with G5E1BCAT reporter plasmid containing 5 GAL4 binding elements upstream of the adenovirus E1B minimal promoter fused to CAT into BJAB cells. The transcription stimulation data was expressed relative to wild-type GAL4–Rta as 100%. The data in panels (A) and (B) was compiled from three experiments. Error bars represent standard errors of the means.
The transactivation ability of GAL4 (1–147)–Rta (416–605) fusions with aromatic or non-aromatic substitutions at positions F600/F605 was compared in BJAB cells. All substitutions at these positions weakened the activity of GAL4/Rta. However, substitutions with the aromatic amino acids, tyrosine and tryptophan, displayed higher transcriptional activity (60 to 70% wild-type level) than did GAL4–Rta chimeras with substitutions of non-aromatic amino acids, alanine, glycine or valine (25 to 30% of wild-type) (Fig. 9B). These experiments indicated that the two phenylalanines are intrinsic components of the transactivation domain of Rta. Substitution of aromatic amino acids was more effective at maintaining transcriptional activity of GAL4/Rta than was substitution of non-aromatic amino acids.
Discussion
In this paper we report novel findings that may help to explain the strategy used by EBV Rta to activate different classes of target genes. We identify a regulatory sequence, located at the C-terminus of Rta and distinct from the DNA binding domain of Rta, that is likely to play a role in modulating the direct versus the indirect modes of action of Rta protein. An important portion of this DNA binding inhibitory sequence (DBIS) is found in the C-terminal 10 aa of Rta (Fig. 1). The C-terminal 51 aa, encompassing the DBIS, can be transferred to, and functionally inactivate, a minimal DNA binding domain of Rta (Fig. 3). Mutations of two phenylalanines in this sequence, F600 and F605, markedly enhance DNA binding in vitro and in vivo (Figs. 2–4). However, Rta (F600A/F605A) mutants which were enhanced in DNA binding were defective in activating direct targets, such as the BMLF1 promoter (Figs. 6B and D) and BLRF2 promoter (Fig. 6E), indirect downstream target promoters, such as the BRLF1 promoter (Figs. 6A, C) and targets, such as the BMRF1 promoter, that are activated in synergy by Rta and Z(S186A) (Fig. 5C). At amino acid positions 600 and 605 non-aromatic substitutions were found to weaken the transactivation function of Rta and GAL4–Rta (Figs. 7, 9) but enhance the DNA binding activity of Rta. Conversely aromatic substitutions of tyrosine and tryptophan for phenylalanine preserved more transactivation, but, like wild-type, these substitution mutants were impaired in binding DNA, as detected by EMSA. Thus, inhibition of DNA binding appears to be strongly influenced by aromatic amino acids at F600 and F605 in the Rta protein.
Biological function of the DNA binding inhibitory sequence (DBIS)
The finding that deletion of the C-terminal 10 aa of Rta that has been expressed in human cells markedly enhances its capacity to bind DNA (Fig. 1) led to the identification of a transferable DBIS in the C-terminus of EBV Rta protein far removed from the DNA binding domain which is located in aa 1–320 (Manet et al., 1991). In several cellular transcription factors a DBIS that lies outside a functionally defined DNA binding motif can alter the affinity or specificity of DNA binding. Deletion mutants of these transcription factors, including TATA-binding protein (TBP), p53, nuclear factor κB (NF-κB) and Ets-1 show enhanced DNA binding activity relative to that of full-length proteins (Grimm and Baeuerle, 1993; Hupp et al., 1992; Lieberman et al., 1991; Sanchez-Garcia et al., 1993).
Recently, our laboratory identified a DBIS located between aa 520 and 535 in ORF50, the homolog of EBV Rta in Kaposi’s Sarcoma-Associated Herpesvirus (KSHV) (Chang et al., 2008b; Chang and Miller, 2004). The amino acid sequence that is crucial for the DNA binding inhibitory function of ORF50 include basic amino acids, such as K527, K528, R529 and K530. The DNA binding inhibitory sequences of KSHV ORF 50 and EBV Rta proteins differ in at least two respects, namely, the nature of the crucial amino acids and their location within the protein. In KSHV ORF50 protein, basic amino acids are crucial for the inhibitory function (Chang et al., 2008b). As we show here, in EBV Rta aromatic amino acids are important. In KSHV ORF50 the DBIS is internal, whereas in EBV Rta it is located at the C-terminus. It will ultimately be important to understand the location of the two DBIS in the folded proteins.
Point mutants in EBV Rta and KSHV ORF50 protein that are enhanced in DNA binding share the property of being defective in transactivation function. The ORF50 (K527E/K528E) mutant is defective at activating direct targets, such as the promoter of PAN RNA, and indirect targets, such as the promoter of ORF 57 (Chang et al., 2005). Similarly, Rta (F600A/F605A) is defective in activating both direct targets, such as the BMLF1 and BHLF1 promoters, and indirect targets, such as the BRLF1 promoter. The region of KSHV ORF50 that regulates DNA binding overlaps a protein abundance regulatory sequence (PARS). The mutants K527E/K528E enhance DNA binding and promote over-expression of an electrophoretic mobility variant, ORF 50B (Chang and Miller, 2004). The DBIS and PARS signals of ORF50 can be distinguished by point mutagenesis (Chang et al., 2008b). These two regulatory functions are not co-linear in EBV Rta. The (F600A/ F605A) mutations that enhance DNA binding do not invariably increase levels of expression of EBV Rta protein (Figs. 2, 3, 7).
EBV Rta activates many essential genes by an indirect mechanism. Rta may not contact DNA directly on these promoters; instead, it is likely that Rta interacts with cellular proteins which do bind the promoter. For example, Rta activation of indirect targets appears to be mediated by Sp1/Sp3 on the BRLF1 promoter, ATF2 protein on the BZLF1 promoter, USF and E2F on the BALF5 promoter (Adamson et al., 2000; Liu et al., 1996; Ragoczy and Miller, 2001). Nonetheless, Rta still can activate some direct target genes, such as BMLF1 and BLRF2, through a direct DNA binding mechanism (Chen et al., 2005; Gruffat et al., 1992; Ragoczy and Miller, 1999). The switch between an indirect and a direct mechanism is likely to be tightly controlled. It appears that the default conformation of full-length Rta protein in eukaryotic cells is a form that has weak DNA binding activity as detected by EMSA and ChIP (Figs. 1 and 4). Perhaps this form favors BRLF1 auto-stimulation, which operates via an indirect mechanism and is likely to be a very early action of Rta (Ragoczy, Heston, and Miller, 1998).
The DNA binding activity of Rta may be influenced by post-translational modification or protein–protein interaction or both
A surprising finding from our preliminary unpublished experiments is that both wild-type Rta and Rta mutants in which the DBIS has been deleted or inactivated by point mutation that were produced in E. coli bound DNA very weakly or undetectably in an EMSA assay (not shown). This finding suggests that the DNA binding activity of Rta, even in the absence of the DBIS, requires interaction with another protein or modification. Rta is modified by SUMO 1 at three lysine residues, K19, K213, and K517 (Chang et al., 2004). SUMOylation at these sites modestly enhanced the capacity of Rta to transactivate reporter plasmids. Other recent studies demonstrate that the product of an EBV early gene, BI’LF4, a viral tegument component, enhanced SUMOylation of Rta, but repressed the ability of Rta to transactivate via Rta responsive elements (Calderwood et al., 2008). Rta becomes phosphorylated after induction of the EBV lytic cycle in Akata cells treated with anti-IgG (Zacny et al., 1998). Modifications by SUMO or phosphorylation or other uncharacterized modifications may by themselves alter the conformation of Rta allowing it to bind DNA or promote interactions which facilitate DNA binding. It seems likely that both positive and negative regulation of DNA binding by Rta is mediated by modifications and protein–protein interactions.
A number of potentially functionally important interactions between Rta and cellular proteins have been described. Rta interacts with Rb, releasing E2F1, which plays a role in activating BALF5, the viral DNA polymerase gene (Furnari et al., 1992; Zacny et al., 1998). TSG101, a transcriptional co-regulator of nuclear hormone receptors, enhances binding of Rta to the promoters of several late genes encoding capsid and tegument proteins (Chua et al., 2007). Interaction of Rta with MCAF1 promotes transcriptional activation by Rta via Sp1 (Chang et al., 2005). Rta interacts with the histone acetylase, CREB-binding protein (CBP) promoting Rta transcriptional activation (Swenson et al., 2001). Of the protein–protein interactions that have been investigated only EBV BI’LF4 and CBP interact directly with the C-terminus of Rta. All the interactions described could potentially be influenced by modifications or conformational changes resulting from mutation at F600 and F605.
The two C-terminal phenylalanines play an important role in EBV lytic cycle activation
F600 and F605 in the C-terminus of Rta are critical for the capacity of Rta to activate full expression of two direct target EBV lytic cycle genes, BMLF1 and BLRF2, in Raji cells (Fig. 6D, E). The two phenylalanines can only be substituted by two other aromatic amino acids, tyrosine (Y) and tryptophan (W), to preserve partial activation of BMLF1 mRNA by Rta in cells lacking EBV BZLF1 (Fig. 7B). Substitution of two non-aromatic amino acids, alanine(A) and valine (V), abolished the capacity of Rta to activate BMLF1 (Fig. 7B). The Y and W aromatic substitutions also preserve more transcriptional activation function of GAL4/Rta fusions in BJAB cells than do the A and V substitutions, although none of the substitutions is as active as wild-type Rta (Fig. 9).
These results are consistent with other findings about the importance of phenylalanines in transcriptional activation domains. Aromatic residues of the EBV BZLF1 protein (ZEBRA, Zta) are important components of the activation domain and play a role in the interactions with TFIIA/TFIID and CBP (Deng et al., 2001). A phenylalanine residue at position 442 of the Herpes Simplex virion protein VP16 has been found to be critical for its transactivation function (Cress and Triezenberg, 1991). In findings similar to our results, substitution of F442 with other aromatic residues, such as Y and W, decreased but did not abolish transactivation function. Substitutions of VP16 F442 with non-aromatic residues such as alanine were markedly detrimental to activation of transcription (Regier et al., 1993). The activation of transcription by VP16 appears to depend on its association with P-TEFb. A F442A mutant fusion protein did not bind to P-TEFb and was 80% reduced in activity when compared to wild-type VP16 fusion protein (Kurosu and Peterlin, 2004). Therefore, F442 was thought to be essential for protein–protein interaction. Moreover, conserved hydrophobic residues flanking F442 of VP16 also contributed to its transcription activity (Regier et al., 1993). We have also studied the significance of several hydrophobic residues nearby the F600/F605 residues, namely L597, I599 and L604 in the C-terminus of Rta. Three alanine substitution point mutants, L597A, I599A and I604A, did not significantly change the transcriptional activity or DNA binding activity of Rta (data not shown).
Several other studies have documented the important role of phenylalanines in protein–protein interactions. For example, F43 in CD4 makes a significant contribution to the high affinity interaction between CD4 and HIV env (Dowd et al., 2002). F17 of gastrin was shown to be important for binding to the human CCK2 receptor (Ahmed et al., 2001). The aromatic ring of F30 also plays a major role in binding of the homopentameric B subunit of verotoxin (VT1) to the glycosphingolipid receptor globotriaosylceramide (Gb3) (Clark et al., 1996). The role of specific phenylalanines in these proteins may be necessary for maintaining domain structure or may be directly involved in contacts with target proteins.
Why is enhanced DNA binding by Rta (F600A/F605A) associated with a defect in activating viral target genes in vivo?
Mutants of EBV Rta and KSHV ORF50 which are enhanced in DNA binding are reduced in transactivation potency. Because enhanced DNA binding activity of these mutants was detected with EMSA in vitro and with ChIP in vivo, and, in the case of ORF50, with specific DNA oligonucleotide affinity chromatography, it seems unlikely that the phenotypes are artifacts of the conditions of the DNA binding assays. While mutations in the terminal phenylalanines of Rta may affect protein–protein interactions required for indirect mechanisms of gene activation, it was unexpected that the Rta (F600A/F605A) mutant was also defective in activating direct target genes, such as BMLF1 and BRLF2. On direct targets enhanced DNA binding might be expected to facilitate the encounter between the activator and the target DNA. We have considered several hypotheses which might explain the loss of biological activity by mutants which are more proficient in binding DNA. Enhanced DNA binding may decrease the likelihood of a required protein–protein interaction with a co-activator or a component of basal transcriptional machinery. In effect, enhanced DNA binding competes for association with the transcription machinery, independent of any effect the mutation might produce on a specific protein–protein interaction. The gain-of-function mutation may change the on/off rate of Rta protein on the promoter. Enhanced binding to DNA might promote protein degradation (Muratani and Tansey, 2003). Activation of a direct target may require transcription factors with both DNA binding and non-DNA binding mechanisms. Enhanced DNA binding by one transcription factor may decrease the efficiency of activation by reducing the opportunities for interactions with other transcription factors bound on the same promoter.
Reciprocal relationship between DNA binding and transcriptional activity
When the DNA binding activity of an Rta mutant is stronger, as the result of A or G substitutions, the transactivation function of that mutant is weaker. When the transcriptional activation is stronger, as in wild-type, W and Y substitutions, DNA binding activity is weaker (Figs. 7–9). We propose that the DNA binding and transactivation function of EBV Rta must be balanced. Enhancing the DNA binding of Rta will decrease its transactivation function. We believe that this is the first report identifying specific amino acid residues in a regulatory sequence of a transcription factor simultaneously controlling both DNA binding and transactivation function. A key to this reciprocal regulation is the localization of the DBIS in an essential region of the transactivation domain of Rta. Because the two phenylalanines in this regulatory region seem to be critical, we favor the idea that protein–protein interactions are involved in the reciprocal regulation. Identification of the proteins that interact with the DBIS, and whose interaction is influenced by residues at position F600 and F605 will be required to understand the mechanisms for this reciprocal regulation.
In conclusion, we have described an essential feature of the regulation of Rta protein that simultaneously affects its DNA binding and transactivation functions. This regulation may help to advance understanding of the strategy used by Rta to selectively activate expression of different classes of target genes and to participate in EBV lytic DNA replication.
Materials and methods
Cell cultures and transfections
HKB5/B5 is an EBV-negative cell line and HKB5/Cl8 is an EBV-positive cell line formed by fusion of HH514–16 cells with 293T cells (Chang and Miller, 2004). HH514–16 (Cl16) is a clonal derivative of the P3J-HR1 Burkitt’s lymphoma cell line that is efficiently activated into the viral lytic cycle (Heston et al., 1982). BJAB cells are derived from an EBV-negative B cell lymphoma (Menezes et al., 1975). Raji, a human B-cell line derived from a Burkitt’s lymphoma, contains an EBV strain that is defective for DNA replication and late gene expression (Decaussin et al., 1995). BZKO (BZLF1 Knock Out) cells are 293 cells carrying a bacmid containing the EBV genome with an inactivated BZLF1 gene (Feederle et al., 2000). Lymphoid cells were grown in RPM11640 medium plus fetal calf serum; BZKO cells were cultured in DMEM plus fetal calf serum in the presence of 100 μg/ml hygromycin. HKB5/B5, HKB5/Cl8 and BZKO cells were transfected using the DMRIE-C Reagent (Invitrogen) (Chang et al., 2002). HH514–16, BJAB and Raji cells were transfected by electroporation.
Plasmid construction
pRTS, pRTS/Rta, RpCAT (−299/+58), BMLF1(RRE)/CAT and CMV/Z(S186A) have been described (Chen et al., 2005; Ragoczy et al., 1998; Ragoczy and Miller, 2001; Sun et al., 1998). pGAL4 (pM), pG5E1BCAT and pGAL4-VP16 plasmids were obtained from CLON-TECH. To make vectors with C-terminal deletion mutants of the Rta gene that were expressed in mammalian cells, various regions of the Rta gene were amplified by PCR and cloned into pRTS at the XbaI and BglII sites. pCMV-FLAG-Rta contains DNA sequences encoding FLAG peptide upstream of full-length Rta gene. F350 contains DNA sequence encoding FLAG peptide upstream of Rta (aa 1–350) gene. F350+ (555–605) or F350+ (593–605) were constructed by PCR amplification and self-ligation to make internal deletion mutants from aa 35 to aa 554 or from aa 351 to aa 592. Single or double amino acid point mutations were introduced into the Rta gene in pRTS/Rta, pCMV-FLAG-Rta or pGAL4–Rta using the Quickchange site-directed mutagenesis kit. pCMV-FLAG-GAL4–Rta was constructed by PCR amplification of the GAL4–Rta fragment with or without F600/F605 mutations and transferred to the HindIII and XbaI sites of pCMV-FLAG vector. All mutations were confirmed by sequencing.
Cell extracts for EMSA and Western blot analysis
Extracts of total cell proteins for use in DNA binding reactions were prepared by the method of Mosser et al. (1988). Briefly, 107 transfected cells were suspended in 120 μl of lysis buffer (0.42 M NaCl, 20 mM HEPES [pH 7.5], 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 2 μg aprotinin per ml). Lysates were spun at 90,000 rpm at 4 °C for 15 min in a benchtop ultracentrifuge (Beckmann Optima TLX), and the supernatants were aliquoted, flash-frozen, and stored at −70 °C. Protein concentrations were determined by the Bradford method.
EMSAs
Annealed double-stranded oligonucleotides derived from the BMLF1 promoter (nucleotide −390 to −374) or from the BHLF1 promoter (nucleotide −776 to −760) were end-labeled with 32P using T4 polynucleotide kinase (Boehringer Mannheim). The primers for making the GAL4 probe were 5′AGCGGAGTACTGTCCTCCGAG and 5′CTCGGAGGACAGTACTCCGCT. Binding reactions contained 15 μg of cell protein in a solution containing 10 mM HEPES (pH 7.5), 50 mM NaCl, 2 mM MgCl2, 2.5 μM ZnSO4, 0.5 mM EDTA, 1 mM dithiothreitol, 15% glycerol and 0.5 μg poly(dI–dC) in a total volume of 20 μl. After incubation for 5 min at room temperature, 30,000 to 50,000 cpm of labeled oligonucleotide was added in each reaction. For supershift reactions, IgG antibodies were added 10 min following the addition of the probe, and incubation at room temperature continued for 10 min. The reaction mixtures were loaded onto a 4% native polyacrylamide gel in 0.5× Tris–Borate–EDTA buffer and electrophoresed at 200 V. Gels were dried on Whatmann 3MM paper under vacuum and exposed to autoradiography film.
Western blot analysis
Cell extracts were mixed with sodium dodecyl sulfate (SDS) sample buffer and heated to 100 °C for 5 min before separation by electrophoresis in a 10% polyacrylamide SDS gel. Following electrophoresis, the proteins were transferred to nitrocellulose membranes by electroblotting and blocked in 5% nonfat dry milk overnight at 4 °C. The blots were incubated with antiserum at 25 °C for 2 h, washed twice for 20 min in 10 mM Tris–HCl (pH 7.5)–200 mM NaCl–5% Tween 20, incubated with 125I-labeled protein A for 1 h, and washed again. The membranes were exposed overnight with intensifying screens to KODAK XAR-5 film at −70 °C.
Antisera
Anti-Rta is a polyclonal rabbit antiserum raised to an N-terminal fragment of Rta (Ragoczy et al., 1998). Anti-EA-D (R3) is a mouse monoclonal antibody to BMRF1 (Pearson et al., 1983). Anti-ZEBRA and anti-LR2 are polyclonal rabbit antisera raised to TrpE-BZLF1 and TrpE-BLRF2 fusion proteins (Katz et al., 1992; Serio et al., 1996). Antibody to FLAG (M2; Sigma), GAL4 (sc-577; Santa Cruz) and β actin (Sigma A5316) were obtained commercially.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed as described (Chang and Miller, 2004). HKB5/B5 cells (107) were transfected with 4 μg of plasmids expressing Rta protein or Rta mutants by DMRIE-C reagent. Twenty hours after transfection, cultured cells were incubated with 1% formaldehyde for 10 min at 37 °C. The cells were harvested, washed once with phosphate-buffered saline containing 1 mM phenylmethylsulfonyl fluoride, 1 μg of aprotinin per ml and 1 μg of pepstatin A per ml, and resuspended in 1.5 ml of lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris–Cl [pH 8.1]). Cell lysates were immunoprecipitated with 10 μg of anti-FLAG antibody or 50 μl of pre-immune serum and incubated at 4 °C overnight. The immune complexes were then precipitated by 60 μl of 50% protein G-agarose and washed five times with buffers of different stringency (Chang and Miller, 2004). The pelleted protein G complex was eluted twice with 200 μl of elution buffer (1% SDS, 0.1 M NaHCO3) for 15 min at room temperature. The eluted chromatin complexes were treated with 20 μl of 5 M NaCl at 65 °C for 4 h to reverse protein–DNA cross-links. DNA was recovered by phenol-chloroform extraction and ethanol precipitation and used as a template in PCR. The primers for amplification of the BHLF1 promoter region were BH-a (5′ CACCTTTGCACATTTGGTCAGC) and BH-b (5′ TTAGTGTGTCATGGTGAGGC).
CAT assays
BJAB or HH514–16 cells (107) were transfected with 5 μg of reporter DNA plus 5 μg of activator (Rta derivatives) or vector DNA. Chloramphenicol acetyltransferase (CAT) assays were performed 48 h after transfection as described previously (Serio et al., 1996). Activation was calculated as percent acetylation of chloramphenicol in the presence of activator divided by percent acetylation in the presence of vector. Results represent the average of at least two separate transfections.
RT-PCR
Total RNA was isolated from 1.5×107 HH514–16 cells by using RNeasy Mini kit (Qiagen). Each RNA sample was treated with 30 U of DNase I by using the RNase-free DNase set (Qiagen). Total RNA (0.5 μg) was used as a template for reverse transcription (RT)-PCR to amplify a 550 bp fragment from Rta mRNA by using the SuperScript one-step RT-PCR system (Invitrogen). The primer pair (forward, 5′AGGTGT-TCCACTACCTGCTTGCC; reverse, 5′ACAACACCTCACTACACAAACAGA) spans a 940 bp intron upstream of the Rta gene to differentiate the PCR product from amplification of genomic DNA (1.5 kb). PCR products were separated by 2% agarose gel electrophoresis. A multi-intron-spanning primer pair (forward, 5′TGAAGGTCGGAGTCAACG-GATTTGG; reverse, 5′GACTGTGGTCATGAGTCCTTCCACG) was used to amplify a 520 bp fragment from the human glyceraldehyde3-phosphate dehydrogenase (GAPDH) mRNA.
Northern blot analysis
Total cellular RNA, prepared using an RNeasy Mini kit (Qiagen), was electrophoresed through a 1% agarose formaldehyde gel, and transferred to nylon membranes (Hybond-N+; Amersham Pharmacia Biotec). All probes were labeled by the random-primed method (Ragoczy and Miller, 1999). A 1.3-kb EcoRI/BamHI fragment from BamHI-M was used to detect BMLF1 mRNA, and a 600-bp XbaI/XhoI fragment from pRTS/R was used to detect BRLF1 mRNA. A radiolabeled 370-bp NcoI/PstI fragment of H1 component of RNase P was used as a probe to control for RNA loading (Bartkiewicz et al., 1989). Hybridization was carried out in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–5× Denhardt’s–0.5% SDS-100 μg of salmon sperm DNA per ml at 60 °C overnight. Membranes were washed in 2× SSC–0.5% SDS once for 20 min and in 0.1× SSC–0.5% SDS three times for 20 min at 60 °C.
Acknowledgments
Supported by Grants CA16038 and CA12055 from the NIH to G.M. We are grateful to Jill Countryman and Ayman El-Guindy for many helpful discussions.
References
- Adamson AL, Kenney SC. Rescue of the Epstein–Barr virus BZLF1 mutant, Z (S186A), early gene activation defect by the BRLF1 gene product. Virology. 1998;251(1):187–197. doi: 10.1006/viro.1998.9396. [DOI] [PubMed] [Google Scholar]
- Adamson AL, Darr D, Holley-Guthrie E, Johnson RA, Mauser A, Swenson J, Kenney S. Epstein–Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J Virol. 2000;74(3):1224–1233. doi: 10.1128/jvi.74.3.1224-1233.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed SI, Wibowo F, Gembitsky DS, Bozso Z, Murphy RF, Lovas S. Importance of the C-terminal phenylalanine of gastrin for binding to the human CCK(2) receptor. J Pept Res. 2001;58(4):332–337. doi: 10.1034/j.1399-3011.2001.00942.x. [DOI] [PubMed] [Google Scholar]
- Andersson J. Epstein–Barr virus and Hodgkin’s lymphoma. Herpes. 2006;13(1):12–16. [PubMed] [Google Scholar]
- Bartkiewicz M, Gold H, Altman S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989;3(4):488–499. doi: 10.1101/gad.3.4.488. [DOI] [PubMed] [Google Scholar]
- Calderwood MA, Holthaus AM, Johannsen E. The Epstein–Barr virus LF2 protein inhibits viral replication. J Virol. 2008;82(17):8509–8519. doi: 10.1128/JVI.00315-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M, Kolman J, Katz DA, Gradoville L, Barberis L, Miller G. Transcriptional synergy by the Epstein–Barr virus transactivator ZEBRA. J Virol. 1992;66(8):4803–4813. doi: 10.1128/jvi.66.8.4803-4813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PJ, Miller G. Autoregulation of DNA binding and protein stability of Kaposi’s sarcoma-associated herpesvirus ORF50 protein. J Virol. 2004;78(19):10657–10673. doi: 10.1128/JVI.78.19.10657-10673.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang PJ, Shedd D, Gradoville L, Cho MS, Chen LW, Chang J, Miller G. Open reading frame 50 protein of Kaposi’s sarcoma-associated herpesvirus directly activates the viral PAN and K12 genes by binding to related response elements. J Virol. 2002;76(7):3168–3178. doi: 10.1128/JVI.76.7.3168-3178.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang LK, Lee YH, Cheng TS, Hong YR, Lu PJ, Wang JJ, Wang WH, Kuo CW, Li SS, Liu ST. Post-translational modification of Rta of Epstein–Barr virus by SUMO-1. J Biol Chem. 2004;279(37):38803–38812. doi: 10.1074/jbc.M405470200. [DOI] [PubMed] [Google Scholar]
- Chang LK, Chung JY, Hong YR, Ichimura T, Nakao M, Liu ST. Activation of Sp1-mediated transcription by Rta of Epstein–Barr virus via an interaction with MCAF1. Nucleic Acids Res. 2005;33(20):6528–6539. doi: 10.1093/nar/gki956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang LK, Liu ST, Kuo CW, Wang WH, Chuang JY, Bianchi E, Hong YR. Enhancement of transactivation activity of Rta of Epstein–Barr virus by RanBPM. J Mol Biol. 2008a;379(2):231–242. doi: 10.1016/j.jmb.2008.04.011. [DOI] [PubMed] [Google Scholar]
- Chang PJ, Shedd D, Miller G. A mobile functional region of Kaposi’s sarcoma-associated herpesvirus ORF50 protein independently regulates DNA binding and protein abundance. J Virol. 2008b;82:9700–9716. doi: 10.1128/JVI.00862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LW, Chang PJ, Delecluse HJ, Miller G. Marked variation in response of consensus binding elements for the Rta protein of Epstein–Barr virus. J Virol. 2005;79(15):9635–9650. doi: 10.1128/JVI.79.15.9635-9650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua HH, Lee HH, Chang SS, Lu CC, Yeh TH, Hsu TY, Cheng TH, Cheng JT, Chen MR, Tsai CH. Role of the TSG101 gene in Epstein–Barr virus late gene transcription. J Virol. 2007;81(5):2459–2471. doi: 10.1128/JVI.02289-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark C, Bast D, Sharp AM, St Hilaire PM, Agha R, Stein PE, Toone EJ, Read RJ, Brunton JL. Phenylalanine 30 plays an important role in receptor binding of verotoxin-1. Mol Microbiol. 1996;19(4):891–899. doi: 10.1046/j.1365-2958.1996.427962.x. [DOI] [PubMed] [Google Scholar]
- Countryman J, Miller G. Activation of expression of latent Epstein–Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA. 1985;82:4085–4089. doi: 10.1073/pnas.82.12.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Countryman J, Jenson H, Seibl R, Wolf H, Miller G. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein–Barr viruses disrupt latency. J Virol. 1987;61(12):3672–3679. doi: 10.1128/jvi.61.12.3672-3679.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cress WD, Triezenberg SJ. Critical structural elements of the VP16 transcriptional activation domain. Science. 1991;251(4989):87–90. doi: 10.1126/science.1846049. [DOI] [PubMed] [Google Scholar]
- Daikoku T, Kudoh A, Fujita M, Sugaya Y, Isomura H, Shirata N, Tsurumi T. Architecture of replication compartments formed during Epstein–Barr virus lytic replication. J Virol. 2005;79(6):3409–3418. doi: 10.1128/JVI.79.6.3409-3418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darr CD, Mauser A, Kenney S. Epstein–Barr virus immediate-early protein BRLF1 induces the lytic form of viral replication through a mechanism involving phosphatidylinositol-3 kinase activation. J Virol. 2001;75(13):6135–6142. doi: 10.1128/JVI.75.13.6135-6142.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaussin G, Leclerc V, Ooka T. The lytic cycle of Epstein–Barr virus in the nonproducer Raji line can be rescued by the expression of a 135-kilodalton protein encoded by the BALF2 open reading frame. J Virol. 1995;69(11):7309–7314. doi: 10.1128/jvi.69.11.7309-7314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Chen CJ, Zerby D, Delecluse HJ, Lieberman PM. Identification of acidic and aromatic residues in the Zta activation domain essential for Epstein–Barr virus reactivation. J Virol. 2001;75(21):10334–10347. doi: 10.1128/JVI.75.21.10334-10347.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWire SM, McVoy MA, Damania B. Kinetics of expression of rhesus monkey rhadinovirus (RRV) and identification and characterization of a polycistronic transcript encoding the RRV Orf50/Rta, RRV R8, and R8.1 genes. J Virol. 2002;76(19):9819–9831. doi: 10.1128/JVI.76.19.9819-9831.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd CS, Leavitt S, Babcock G, Godillot AP, Van Ryk D, Canziani GA, Sodroski J, Freire E, Chaiken IM. Beta-turn Phe in HIV-1 Env binding site of CD4 and CD4 mimetic miniprotein enhances Env binding affinity but is not required for activation of co-receptor/17b site. Biochemistry. 2002;41(22):7038–7046. doi: 10.1021/bi012168i. [DOI] [PubMed] [Google Scholar]
- Ehlers M, Grotzinger J, Fischer M, Bos HK, Brakenhoff JP, Rose-John S. Identification of single amino acid residues of human IL-6 involved in receptor binding and signal initiation. J Interferon Cytokine Res. 1996;16(8):569–576. doi: 10.1089/jir.1996.16.569. [DOI] [PubMed] [Google Scholar]
- El-Guindy A, Heston L, Delecluse HJ, Miller G. Phosphoacceptor site S173 in the regulatory domain of Epstein–Barr Virus ZEBRA protein is required for lytic DNA replication but not for activation of viral early genes. J Virol. 2007;81(7):3303–3316. doi: 10.1128/JVI.02445-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feederle R, Kost M, Baumann M, Janz A, Drouet E, Hammerschmidt W, Delecluse HJ. The Epstein–Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 2000;19(12):3080–3089. doi: 10.1093/emboj/19.12.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fixman ED, Hayward GS, Hayward SD. trans-acting requirements for replication of Epstein–Barr virus ori-Lyt. J Virol. 1992;66(8):5030–5039. doi: 10.1128/jvi.66.8.5030-5039.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fixman ED, Hayward GS, Hayward SD. Replication of Epstein–Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J Virol. 1995;69(5):2998–3006. doi: 10.1128/jvi.69.5.2998-3006.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis A, Ragoczy T, Gradoville L, El-Guindy A, Miller G. Amino acid substitutions reveal distinct functions of serine 186 of the ZEBRA protein in activation of lytic cycle genes and synergy with the EBV Rta transactivator. J Virol. 1999;73(6):4543–4551. doi: 10.1128/jvi.73.6.4543-4551.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furnari FB, Adams MD, Pagano JS. Regulation of the Epstein–Barr virus DNA polymerase gene. J Virol. 1992;66(5):2837–2845. doi: 10.1128/jvi.66.5.2837-2845.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geser A, de The G, Lenoir G, Day NE, Williams EH. Final case reporting from the Ugandan prospective study of the relationship between EBV and Burkitt’s lymphoma. Int J Cancer. 1982;29(4):397–400. doi: 10.1002/ijc.2910290406. [DOI] [PubMed] [Google Scholar]
- Grimm S, Baeuerle PA. The inducible transcription factor NF-kappa B: structure–function relationship of its protein subunits. Biochem J. 1993;290(Pt 2):297–308. doi: 10.1042/bj2900297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruffat H, Manet E, Rigolet A, Sergeant A. The enhancer factor R of Epstein–Barr virus (EBV) is a sequence-specific DNA binding protein. Nucleic Acids Res. 1990;18(23):6835–6843. doi: 10.1093/nar/18.23.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruffat H, Duran N, Buisson M, Wild F, Buckland R, Sergeant A. Characterization of an R-binding site mediating the R-induced activation of the Epstein–Barr virus BMLF1 promoter. J Virol. 1992;66(1):46–52. doi: 10.1128/jvi.66.1.46-52.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammerschmidt W, Sugden B. Identification and characterization of oriLyt, a lytic origin of DNA replication of Epstein–Barr virus. Cell. 1988;55(3):427–433. doi: 10.1016/0092-8674(88)90028-1. [DOI] [PubMed] [Google Scholar]
- Hardwick JM, Lieberman PM, Hayward SD. A new Epstein–Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J Virol. 1988;62(7):2274–2284. doi: 10.1128/jvi.62.7.2274-2284.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick JM, Tse L, Applegren N, Nicholas J, Veliuona MA. The Epstein–Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J Virol. 1992;66(9):5500–5508. doi: 10.1128/jvi.66.9.5500-5508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heston L, Rabson M, Brown N, Miller G. New Epstein–Barr virus variants from cellular subclones of P3J-HR-1 Burkitt lymphoma. Nature. 1982;295(5845):160–163. doi: 10.1038/295160a0. [DOI] [PubMed] [Google Scholar]
- Holley-Guthrie EA, Quinlivan EB, Mar EC, Kenney S. The Epstein–Barr virus (EBV) BMRF1 promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J Virol. 1990;64(8):3753–3759. doi: 10.1128/jvi.64.8.3753-3759.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshikawa Y, Satoh Y, Murakami M, Maeta M, Kaibara N, Ito H, Kurata T, Sairenji T. Evidence of lytic infection of Epstein–Barr virus (EBV) in EBV-positive gastric carcinoma. J Med Virol. 2002;66(3):351–359. doi: 10.1002/jmv.2152. [DOI] [PubMed] [Google Scholar]
- Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA binding function of p53. Cell. 1992;71(5):875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- Jayaraman J, Prives C. Activation of p53 sequence-specific DNA binding by short single strands of DNA requires the p53 C-terminus. Cell. 1995;81(7):1021–1029. doi: 10.1016/s0092-8674(05)80007-8. [DOI] [PubMed] [Google Scholar]
- Katz DA, Baumann RP, Sun R, Kolman JL, Taylor N, Miller G. Viral proteins associated with the Epstein–Barr virus transactivator, ZEBRA. Proc Natl Acad Sci USA. 1992;89:282–378. doi: 10.1073/pnas.89.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein G. In: The Relationship Of The Virus To Nasopharyngeal Carcinoma. Epstein MA, Achong BG, editors. Springer-Verlag; Berlin: 1979. [Google Scholar]
- Komano J, Sugiura M, Takada K. Epstein–Barr virus contributes to the malignant phenotype and to apoptosis resistance in Burkitt’s lymphoma cell line Akata. J Virol. 1998;72(11):9150–9156. doi: 10.1128/jvi.72.11.9150-9156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu T, Peterlin BM. VP16 and ubiquitin; binding of P-TEFb via its activation domain and ubiquitin facilitates elongation of transcription of target genes. Curr Biol. 2004;14(12):1112–1116. doi: 10.1016/j.cub.2004.06.020. [DOI] [PubMed] [Google Scholar]
- Lieberman PM, Hardwick JM, Hayward SD. Responsiveness of the Epstein–Barr virus NotI repeat promoter to the Z transactivator is mediated in a cell-type-specific manner by two independent signal regions. J Virol. 1989;63(7):3040–3050. doi: 10.1128/jvi.63.7.3040-3050.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman PM, Schmidt MC, Kao CC, Berk AJ. Two distinct domains in the yeast transcription factor IID and evidence for a TATA box-induced conformational change. Mol Cell Biol. 1991;11(1):63–74. doi: 10.1128/mcb.11.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin RJ, Blumenkranz MS, Binkley J, Wu K, Vollrath D. A novel His158Arg mutation in TIMP3 causes a late-onset form of Sorsby fundus dystrophy. Am J Ophthalmol. 2006;142(5):839–848. doi: 10.1016/j.ajo.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Liu C, Sista ND, Pagano JS. Activation of the Epstein–Barr virus DNA polymerase promoter by the BRLF1 immediate-early protein is mediated through USF and E2F. J Virol. 1996;70(4):2545–2555. doi: 10.1128/jvi.70.4.2545-2555.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac DM, Renne R, Kirshner JR, Ganem D. Reactivation of Kaposi’s sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252(2):304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- Manet E, Gruffat H, Trescol-Biemont MC, Moreno N, Chambard P, Giot JF, Sergeant A. Epstein–Barr virus bicistronic mRNAs generated by facultative splicing code for two transcriptional trans-activators. EMBO J. 1989;8(6):1819–1826. doi: 10.1002/j.1460-2075.1989.tb03576.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manet E, Rigolet A, Gruffat H, Giot JF, Sergeant A. Domains of the Epstein–Barr virus (EBV) transcription factor R required for dimerization, DNA binding and activation. Nucleic Acids Res. 1991;19(10):2661–2667. doi: 10.1093/nar/19.10.2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manet E, Allera C, Gruffat H, Mikaelian I, Rigolet A, Sergeant A. The acidic activation domain of the Epstein–Barr virus transcription factor R interacts in vitro with both TBP and TFIIB and is cell-specifically potentiated by a proline-rich region. Gene Expr. 1993;3(1):49–59. [PMC free article] [PubMed] [Google Scholar]
- Menezes J, Leibold W, Klein G, Clements G. Establishment and characterization of an Epstein–Barr virus (EBC)-negative lymphoblastoid B cell line (BJA-B) from an exceptional, EBV-genome-negative African Burkitt’s lymphoma. Biomedicine. 1975;22(4):276–284. [PubMed] [Google Scholar]
- Mosser DD, Theodorakis NG, Morimoto RI. Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells. Mol Cell Biol. 1988;8(11):4736–4744. doi: 10.1128/mcb.8.11.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muratani M, Tansey WP. How the ubiquitin–proteasome system controls transcription. Nat Rev, Mol Cell Biol. 2003;4(3):192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- Park R, Heston L, Shedd D, Delecluse HJ, Miller G. Mutations of amino acids in the DNA-recognition domain of Epstein–Barr virus ZEBRA protein alter its sub-nuclear localization and affect formation of replication compartments. Virology. 2008;382(2):145–162. doi: 10.1016/j.virol.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson GR, Vroman B, Chase B, Sculley T, Hummel M, Kieff E. Identification of polypeptide components of the Epstein–Barr virus early antigen complex with monoclonal antibodies. J Virol. 1983;47(1):193–201. doi: 10.1128/jvi.47.1.193-201.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlivan EB, Holley-Guthrie EA, Norris M, Gutsch D, Bachenheimer SL, Kenney SC. Direct BRLF1 binding is required for cooperative BZLF1/BRLF1 activation of the Epstein–Barr virus early promoter, BMRF1. Nucleic Acids Res. 1993;21(14):1999–2007. doi: 10.1093/nar/21.8.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragoczy T, Miller G. Role of the Epstein–Barr virus Rta protein in activation of distinct classes of viral lytic cycle genes. J Virol. 1999;73(12):9858–9866. doi: 10.1128/jvi.73.12.9858-9866.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragoczy T, Miller G. Autostimulation of the Epstein–Barr virus BRLF1 promoter is mediated through consensus Sp1 and Sp3 binding sites. J Virol. 2001;75(11):5240–5251. doi: 10.1128/JVI.75.11.5240-5251.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragoczy T, Heston L, Miller G. The Epstein–Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J Virol. 1998;72(10):7978–7984. doi: 10.1128/jvi.72.10.7978-7984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regier JL, Shen F, Triezenberg SJ. Pattern of aromatic and hydrophobic amino acids critical for one of two subdomains of the VP16 transcriptional activator. Proc Natl Acad Sci USA. 1993;90(3):883–887. doi: 10.1073/pnas.90.3.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney C, Taylor N, Countryman J, Jenson H, Kolman J, Miller G. Genome rearrangements activate the Epstein–Barr virus gene whose product disrupts latency. Proc Natl Acad Sci USA. 1988;85(24):9801–9805. doi: 10.1073/pnas.85.24.9801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Garcia I, Osada H, Forster A, Rabbitts TH. The cysteine-rich LIM domains inhibit DNA binding by the associated homeodomain in Isl-1. EMBO J. 1993;12(11):4243–4250. doi: 10.1002/j.1460-2075.1993.tb06108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serio TR, Angeloni A, Kolman JL, Gradoville L, Sun R, Katz DA, Van Grunsven W, Middeldorp J, Miller G. Two 21-kilodalton components of the Epstein–Barr virus capsid antigen complex and their relationship to ZEBRA-associated protein p21 (ZAP21) J Virol. 1996;70(11):8047–8054. doi: 10.1128/jvi.70.11.8047-8054.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Lin SF, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi’s sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95(18):10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson JJ, Mauser AE, Kaufmann WK, Kenney SC. The Epstein–Barr virus protein BRLF1 activates S phase entry through E2F1 induction. J Virol. 1999;73(8):6540–6550. doi: 10.1128/jvi.73.8.6540-6550.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson JJ, Holley-Guthrie E, Kenney SC. Epstein–Barr virus immediate-early protein BRLF1 interacts with CBP, promoting enhanced BRLF1 transactivation. J Virol. 2001;75(13):6228–6234. doi: 10.1128/JVI.75.13.6228-6234.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada K, Shimizu N, Sakuma S, Ono Y. trans activation of the latent Epstein–Barr virus (EBV) genome after transfection of the EBV DNA fragment. J Virol. 1986;57(3):1016–1022. doi: 10.1128/jvi.57.3.1016-1022.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MP, Kurzrock R. Epstein–Barr virus and cancer. Clin Cancer Res. 2004;10(3):803–821. doi: 10.1158/1078-0432.ccr-0670-3. [DOI] [PubMed] [Google Scholar]
- Whitehouse A, Carr IM, Griffiths JC, Meredith DM. The herpesvirus saimiri ORF50 gene, encoding a transcriptional activator homologous to the Epstein–Barr virus R protein, is transcribed from two distinct promoters of different temporal phases. J Virol. 1997;71(3):2550–2554. doi: 10.1128/jvi.71.3.2550-2554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TT, Usherwood EJ, Stewart JP, Nash AA, Sun R. Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J Virol. 2000;74(8):3659–3667. doi: 10.1128/jvi.74.8.3659-3667.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacny VL, Wilson J, Pagano JS. The Epstein–Barr virus immediate-early gene product, BRLF1, interacts with the retinoblastoma protein during the viral lytic cycle. J Virol. 1998;72(10):8043–8051. doi: 10.1128/jvi.72.10.8043-8051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalani S, Holley-Guthrie E, Kenney S. Epstein–Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc Natl Acad Sci USA. 1996;93(17):9194–9199. doi: 10.1073/pnas.93.17.9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Zhang LG, Wu YC, Huang YS, Huang NQ, Li JY, Wang YB, Jiang MK, Fang Z, Meng NN. Prospective studies on nasopharyngeal carcinoma in Epstein–Barr virus IgA/VCA antibody-positive persons in Wuzhou City, China. Int J Cancer. 1985;36(5):545–547. doi: 10.1002/ijc.2910360505. [DOI] [PubMed] [Google Scholar]