Abstract
Broad HIV-1 genetic diversity in Cameroon provides a unique opportunity to monitor HIV-1 evolution and allows the detection of novel strains. We have genetically characterized the HIV-1 subtypes found in 156 samples from 90 drug-naive subjects in Yaoundé, Cameroon collected from 2011 to 2013, using phylogenetic analysis of regions in gag and pol. We identified subtypes CRF02_AG (64.9%), CRF22_01A1 (7.1%), D (4.5%), F2 (3.9%), G (3.2%), CRF18_cpx (3.2%), CRF37_cpx (3.2%), CRF11_cpx (2.6%), CRF13_cpx (1.9%), A1 (1.3%), CRF01_AE (1.3%), CRF09_cpx (1.3%), A2 (0.6%), and H (0.6%). Sequence data for both the gag and pol regions were obtained from 62 subjects; for 59 of these subjects the two regions were identified as the same viral subtype while three subjects were discordant, A1/CRF02_AG (subject MDC006), CRF02_AG/F2 (subject MDC179), and a dual infection with CRF02_AG/F2 (subject MDC131). Longitudinal sequence data were obtained for 28 of these 62 subjects and confirmed the cross-sectional results. These data update subtype information for this area and highlight the necessity of such studies due to the numerous circulating subtypes, the ongoing superinfection, and the risk of emerging novel recombinant viruses.
Cameroon remains an epicenter of the HIV-1 epidemic and has the highest prevalence of HIV infections in West Central Africa.1 Every HIV-1 group, subtype, and many circulating recombinant forms (CRFs) have been identified in Cameroonian infections; furthermore, epidemiological evidence links the origins of groups M, O, N, and P to Cameroon and/or nearby regions.2–7 The recombinant CRF02_AG dominates the epidemic in Cameroon unlike most areas of the world that are dominated by pure subtype infections B and C.2,3 Many other common CRFs, including CRF22_01A1 and CRF01_AE, have also been found to be prevalent in Cameroon.2,3 Second generation recombinants (SGRs), which are strains composed at least in part of one CRF, are increasing in appearance in HIV-1 infections, many containing portions of CFR02_AG.2,3 The increased amount of diversity and recombination in Cameroon provides a unique opportunity to study and monitor the evolution of HIV-1 strains.
Due to the propensity of the HIV-1 genome to recombine, many novel recombinants have been discovered since the viral subtypes were characterized. Novel recombinants and SGRs are indicative of superinfection, which has been found to occur with high frequency in Cameroon.8 Superinfection occurs when a second, subsequent HIV-1 infection takes place following seroconversion after the primary infection.8 It is not clear yet how and why some patients become superinfected and others do not; however, the threat to the epidemic is made clear by the production and circulation of novel recombinants.2,3 It is important therefore to identify and monitor the evolution of HIV-1 diversity particularly in regions of the world, such as Cameroon, where increased amounts of superinfection lead to novel recombinants. Such studies are needed to better inform vaccine design, therapy, diagnosis, and public health intervention approaches for HIV-1 prevention.
We have collected and analyzed cross-sectional and longitudinal patient samples from Cameroon's capital city, Yaoundé, in order to determine the strains that are prevalent and circulating, and to detect the formation of any unreported unique recombinant forms (URFs). Samples from 90 drug-naive patients were randomly selected from the collaborative cohort shared between New York University, New York, and the Medical Diagnostic Center (MDC), Yaoundé, Cameroon. From 74/90 (82.2%) study subjects, we were able to analyze longitudinal samples taken 5–11 months apart from 2011 to 2013, and 16/90 subjects (17.8%) were analyzed only cross-sectionally from 2011 to 2012. Study subjects enrolled were patients referred to the MDC after an HIV-positive result and/or for gratis medical consultation and CD4 count monitoring. The mean CD4+ cell count was 460 cells/mm3 with a range from 92 to 1,306 cells/mm3. Information regarding age, sex, demographics, and concurrent medical conditions was recorded. The median age of the study subjects was 38 years, with a range from 21 to 63 years; 77.8% were women and 22.2% men. The most likely cause of transmission was self-reported as 95.6% heterosexual, 1.1% homosexual, 1.1% blood transfusion, and 2.2% did not self-report any mode of transmission.
Plasma and peripheral blood mononuclear cells (PBMCs) were collected using Ficoll gradient centrifugation from whole blood. Both plasma and PBMCs were stored at −80°C. Viral RNA was extracted from the plasma using the QIAamp viral RNA mini kit according to the manufacturer's protocol (Qiagen Inc., Valencia, CA). Reverse transcription and nested polymerase chain reactions (PCRs) were performed with the SuperScript One-Step RT-PCR system (Life Technologies, Carlsbad, CA) to isolate a portion of gag, p24, HXB2 region 1,577 to 2,040 (463 bp), and a portion of pol, p51 RT, HXB2 region 2,584 to 4,180 (1,596 bp), using primers HIG777, HIP202, HIgag1584, and G17 for the gag region and NYUPOL9, RTPOL1F, RTPOL1R, RTPOL2F, and RTPOL2R for the pol region.3 The resulting reactions were analyzed on a 1% DNA agarose gel by electrophoresis. Reactions with a positive band at the appropriate size were submitted for sequencing. For regions of pol in which overlapping sequences were obtained using forward and reverse PCR products, assembled sequences were produced using Staden Package 1.6.0. The sequences were aligned with all known subtyped HIV-1 group M reference strains using CLUSTAL Omega and phylogenetic trees were generated with MEGA software using the Kimura two-parameter method.9–12 Breakpoint analysis was performed for selected samples by creating bootstrap plots using the SimPlot software package, version 3.5.1.21.; bootscanning on neighbor-joining trees used a 180-bp window moving along the alignment with increments of 20 bp, respectively. Each sequence analyzed was queried against all known HIV-1 Group M reference sequences and then queried only with sequences indicated by MEGA and SimPlot analysis to be present.9,10,13
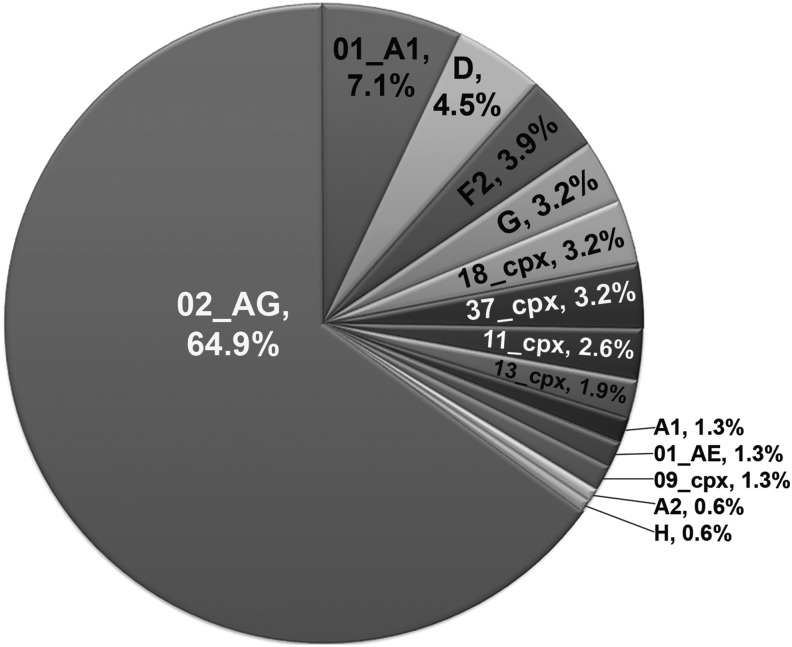
Sequences were successfully amplified in gag, pol, or both regions for the 90 HIV-1-infected subjects studied. CRF02_AG was the predominant strain comprising 64.9% of the gag and/or pol sequences analyzed. This was followed by CRF22_01A1 at 7.1% and D at 4.5%. Numerous other subtypes and CRFs were identified including F2 (3.9%), G (3.2%), CRF18_cpx (3.2%), CRF37_cpx (3.2%), CRF11_cpx (2.6%), CRF13_cpx (1.9%), A1 (1.3%), CRF01_AE (1.3%), CRF09_cpx (1.3%), A2 (0.6%), and H (0.6%) (Fig. 1). These data report on all samples including cross-sectional and longitudinal.
FIG. 1.
Genetic diversity detected in HIV-1-infected subjects in Yaoundé, Cameroon. Combined data of subtype assignments for viral genetic regions gag and pol determined by phylogenetic analysis of viral sequences. The subtypes identified were based on analysis of cross-sectional and longitudinal samples obtained from 90 subjects.
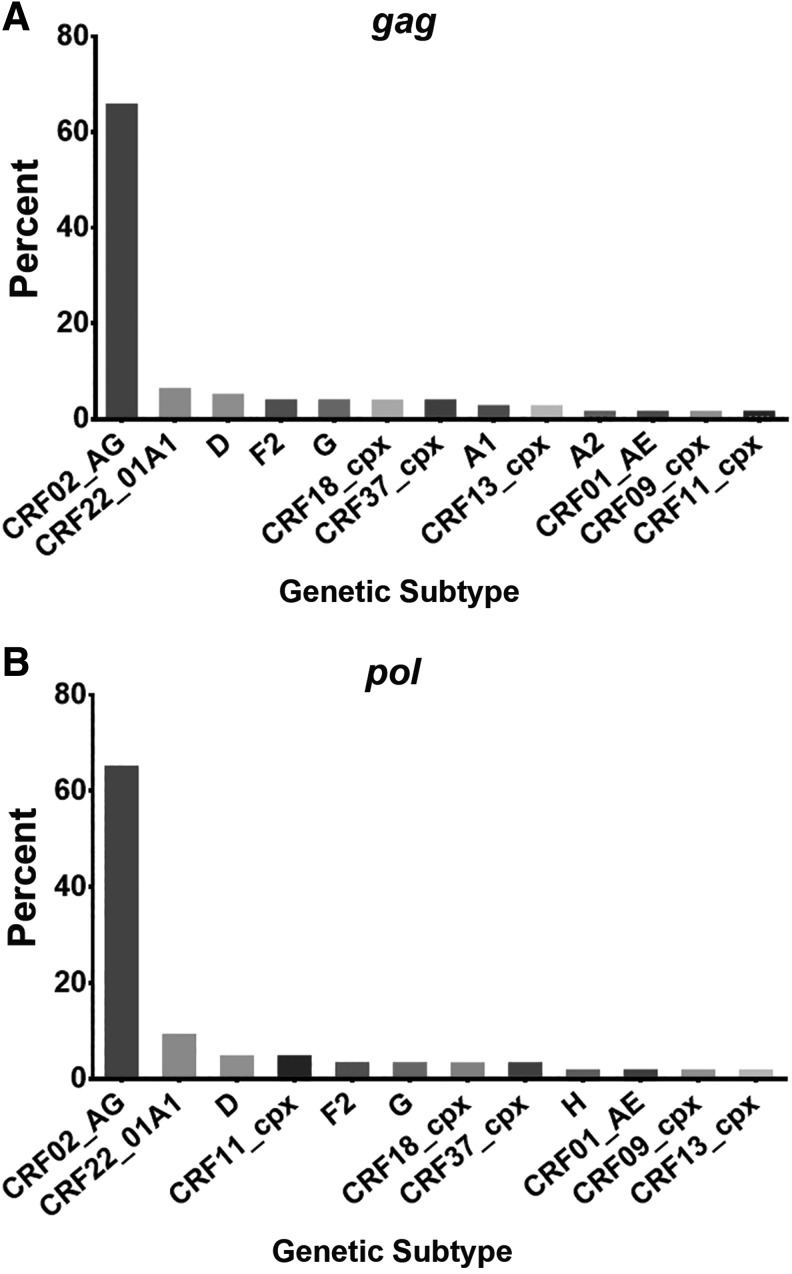
Subtype information was analyzed by region for both gag and pol. Amplification of gag was obtained from 84/90 (93.3%) subjects, including more than one time point for 44/84 (52.4%) subjects (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/aid). The cross-sectional data for the first time point available for each subject are described in Fig. 2A and reveal that CRF02_AG was the predominant subtype, 65.5% (55/84), followed by CRF22_01A1 (6.0%, 5/84), D (4.8%, 4/84), F2 (3.6%, 3/84), G (3.6%, 3/84), CRF18_cpx (3.6%, 3/84), CRF37_cpx (3.6%, 3/84), A1 (2.4%, 2/84), CRF13_cpx (2.4%, 2/84), A2 (1.2%, 1/84), CRF01_AE (1.2%, 1/84), CRF09_cpx (1.2%, 1/84), and CRF11_cpx (1.2%, 1/84). Subtype identities were confirmed for the 44 subjects from whom longitudinal samples were obtained (Supplementary Table S1).
FIG. 2.
Subtype identification by HIV-1 genomic region. Percentage of each HIV-1 subtype and CRF identified from sequences amplified in the (A) gag and (B) pol region of subjects from Yaoundé, Cameroon. All identifications are based on phylogenetic analysis of the first time point available and data represent cross-sectional analysis.
Sequences of the pol region were obtained for 68/90 (75.6%) of the subjects analyzed, including analysis of more than one time point for 30/68 (41.1%) of the study subjects. Described here is the cross-sectional data for the first time point available for each subject (Fig. 2B). Subtypes identified in pol were CRF02_AG (64.7%, 44/68), CRF22_01A1 (8.8%, 6/68), D (4.4%, 3/68), CRF11_cpx (4.4%, 3/68), F2 (2.9%, 2/68), G (2.9%, 2/68), CRF18_cpx (2.9%, 2/68), CRF37_cpx (2.9%, 2/68), H (1.5%, 1/68), CRF01_AE (1.5%, 1/68), CRF09_cpx (1.5%, 1/68), and CRF13_cpx (1.5%, 1/68) (Fig. 2B). Subtype analysis of the longitudinal samples from the 30 subjects from whom sequential samples were obtained also revealed the subtypes as identified in the first time point specimen (Supplementary Table S1).
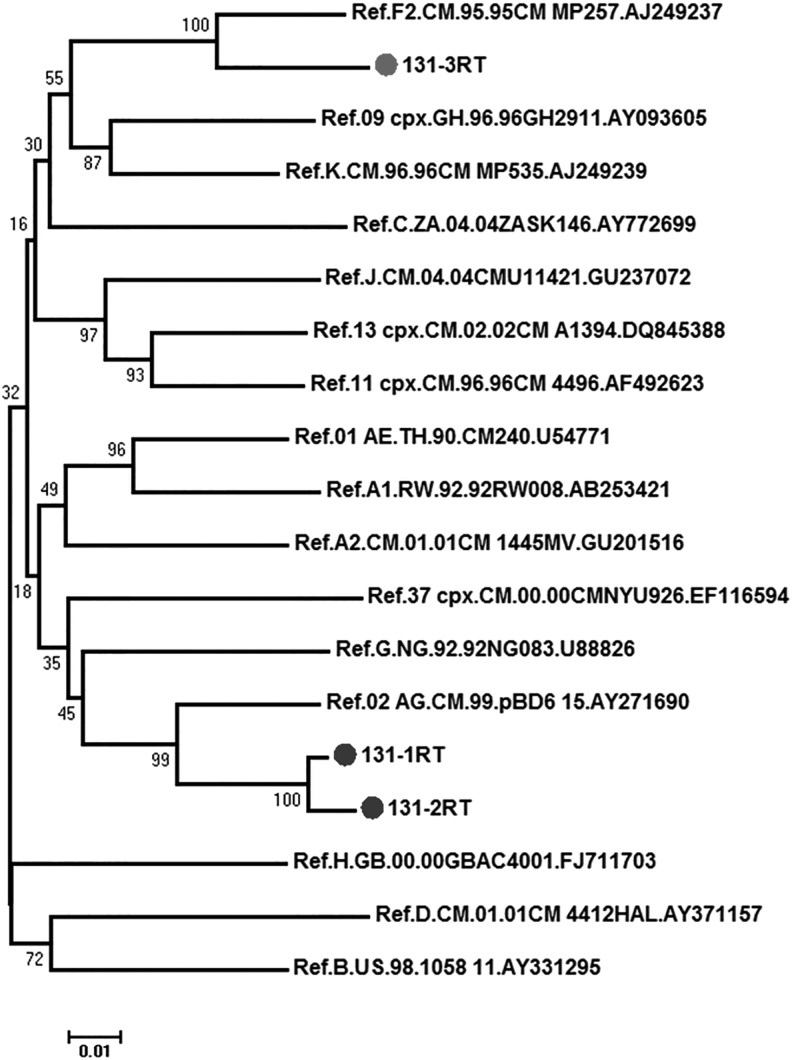
Analysis of the gag and pol regions from the same subject permitted the identification of concordant or discordant subtype infection. A total of 62 study subjects were analyzed successfully for both gag and pol, 21/62 (33.9%) were analyzed using at least two longitudinal samples, and 7/62 (11.3%) were analyzed with three longitudinal samples (Supplementary Table S1). All subjects contained concordant infections with three exceptions: subjects MDC006, MDC179, and MDC131. Sequences from MDC006 including the first and only time point analyzed for both regions were found as A1 in the gag region and CRF02_AG in the pol region. Due to the composition of the CRF02_AG this may not be a true URF but if confirmed would therefore be an SGR with CRF02_AG. Samples from subjects MDC179 and MDC131 were found to have CRF02_AG in the gag region and F2 in the pol region. This was confirmed with two sequential samples taken 6 months apart for MDC179 (Supplementary Table S1). Sequences from subject MDC131 indicated CRF02_AG in both the gag and pol regions in the first two samples spanning from 2011 to 2012. However, analysis of a third time point, taken 5 months after the second sample, revealed the pol region identity to be subtype F2 (Fig. 3). Samples from this subject (MDC131) were analyzed longitudinally for breakpoints using SimPlot and confirmed the presence of CRF02_AG and F2, CRF02_AGgagF2pol. Breakpoints were not detected in these regions and these results together suggest the subject infected with CRF02_AG became superinfected with an F2 virus in the third time point.
FIG. 3.
Phylogenetic tree of the MDC131 pol sequences. MDC131-1 and MDC131-2, 1,526 bp; MDC131-3, 935 bp. Kimura two-parameter, neighbor-joining, bootstrap method. Bootstrap values are indicated at the branch origin.
These data were obtained using bulk PCR and breakpoints were not analyzed between these regions, therefore it is not known if a recombination event led to the CRF02_AGgag and F2pol outcome in the third time point (Fig. 3). Further studies, including cloning and next generation sequencing, are underway to fully characterize the infections in these three subjects and will be described elsewhere.
This study provides an updated analysis of the circulating HIV-1 subtypes in a region of the world that is most likely and continues to be an epicenter of the virus. In particular, Yaoundé, the capital city of Cameroon, is a city that harbors broad human genetic diversity as a result of human migration from other regions of the country allowing for diverse human interaction and HIV-1 cross-transmission. The last report in Cameroon was performed by our group, which analyzed HIV-1 from Limbe samples collected in 2010.3 Our findings agree that CRF02_AG remains the most prevalent subtype, representing 64.9% of the Yaoundé samples tested for gag, pol, or both. This result confirms and updates our findings from 2009 as well and is consistent with our findings for other areas of Cameroon.2–3,8 In addition to CRF02_AG, we also found CRF22_01A1 to be the second most prevalent, 7.1%, and subtype D third, 4.5%. Numerous CRFs were identified in our study, which was the largest proportion of diversity reported for this area. Studies such as these remain essential to provide an assessment of current HIV-1 strains in circulation.
The phylogenetic analyses also allowed us to identify concordant infections by analyzing the two HIV-1 genomic regions. Furthermore, longitudinal samples allowed us to report any evidence of recombination and/or superinfection. We were able to detect the presence of a possible superinfection in one subject, for which we were able to analyze three sequential samples. The subject was infected with CRF02_AG and F2, CRF02_AGgag, and subtype F2pol, indicating a possible novel recombinant. Infection with CRF02_AGgag and subtype F2pol was also found in an additional subject in the first and second time points. Sequences from these subjects when analyzed with any other reported CRF02_AG and F2 recombinant, including those from our previous studies and others accessed in the Los Alamos HIV database, do not cluster with any A1F2, 02F2G, AF2G, or 02F2 strains.9 To date, no CRF has been documented containing these two subtypes, although our group has previously reported multiple subjects infected with these two viruses including CRF02_AG in the protease region and F2 in the envelope region in 2004.14 Superinfection with these two subtypes was found in the gag region in two patients in 2008, one in which CRF02_AG was the superinfecting strain of an F2 infection and another in which F2 was the superinfecting strain of a CRF02_AG infection.8 Taken together with our result the possible recombination between the highly dominant CRF02_AG subtype and the F2 subtype warrants further investigation and may be due in part to an unknown mechanism of favoritism for these events.
Extreme genetic diversity and recombination are monumental challenges for the advancement of HIV-1 vaccination, treatment, and diagnostics. Genetic differences between subtypes are up to 35% in the env region and even 20% between strains of the same subtype.15 Unexplored differences may also lead to an extreme outcome in response to vaccination strategies, which are currently being developed and/or in future studies. Recombination not only adds to viral diversity, it can also quickly pass drug resistance mutations interfering with treatment leading to multidrug-resistant viruses.16 Diagnostic tools that are equipped to detect only known subtypes may miss positive cases due to limited knowledge. It is imperative that we monitor the diversity of viral subtypes, especially in an area known for high proportions of genetic diversity and recombination, in order to ensure that our research efforts in vaccination, treatment, and diagnostics are capable of handling the current status of the pandemic.
Sequence Data
The genomic sequences of the gag and pol regions analyzed in this publication are available in GenBank under accession numbers KT25444–KT255592 and KT758181–KT758289, respectively.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Luzia Mayr, Sonal Soni, Dr. Ralf Duerr, Dr. Aubin Nanfack, and Dr. Viswanath Ragupathy for their support and laboratory expertise. We are grateful to the volunteers who have provided the samples for this study. Funding has been provided by the National Institute of Allergy and Infectious Diseases (NIAID), National Institute of Health (NIH) Grants AI083142 and AI07647, National Cancer Institute (NCI) Grant CA153726, and Fogarty International Center Grant TW009604.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.UNAIDS, Global Report: UNAIDS Report on the Global AIDS Epidemic 2013. Available at www.unaids.org/en/resources/campaigns/globalreport2013/globalreport
- 2.Konings FA, Haman GR, Xue Y, et al. : Genetic analysis of HIV-1 strains in rural Eastern Cameroon indicates the evolution of second-generation recombinants to circulating recombinant forms. J Acquir Immune Defic Syndr 2006;42(3):331–341 [DOI] [PubMed] [Google Scholar]
- 3.Agyingi L, Mayr LM, Kinge T, et al. : The evolution of HIV-1 group M genetic variablity in Southern Cameroon is characterized by several emerging recombinant forms of CRF02_AG and viruses with drug resistance mutations. J Med Virol 2014;86(3):385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan CA, Bodelle P, Coffey R, et al. : The prevalence of diverse HIV-1 strains was stable in Cameroonian blood donors from 1996 to 2004. J Acquir Immune Defic Syndr 2008;49(4):432–439 [DOI] [PubMed] [Google Scholar]
- 5.Simon F, Mauclere P, Roques P, et al. : Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat Med 1998;4(9):1032–1037 [DOI] [PubMed] [Google Scholar]
- 6.Gao F, Bailes E, Robertson DL, et al. : Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 1999;397(6718):436–441 [DOI] [PubMed] [Google Scholar]
- 7.Vallari A, Holzmayer V, Harris B, et al. : Confirmation of putative HIV-1 group P in Cameroon. J Virol 2011;85(3):1403–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Powell RL, Urbanski MM, Burda S, et al. : High frequency of HIV-1 dual infections among HIV-positive individuals in Cameroon, West Central Africa. J Acquir Immune Defic Syndr 2009;50(1):84–89 [DOI] [PubMed] [Google Scholar]
- 9.Los Alamos HIV Database, available at www.hiv.lanl.gov/
- 10.Kumar S, Nei M, Dudley J, and Tamura K: MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 2008;9(4):299–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sievers F, Wilm A, Dineen D, et al. : Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2011;7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura M: A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 1980;16(2):111–120 [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein J: PHYLIP—Phylogeny Inference Package (Version 3.2). Cladistics 1989;5:164–166 [Google Scholar]
- 14.Burda ST, Konings FA, Williams CA, et al. : HIV-1 CRF09_cpx circulates in the north west province of Cameroon where CRF02_AG infections predominate and recombainants strains are common. AIDS Res Hum. Retroviruses 2004;20(12):1358–1363 [DOI] [PubMed] [Google Scholar]
- 15.Gaschen B, Taylor J, Yusim K, et al. : Diversity considerations in HIV-1 vaccine selection. Science 2002;296(5577):2354–2360 [DOI] [PubMed] [Google Scholar]
- 16.Moutouh L, Corbeil J, and Richman DD: Recombination leads to the rapid emergence of HIV-1 dually resistant mutants under selective drug pressure. Proc Natl Acad Sci USA 1996;93(12):6106–6111 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.