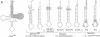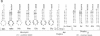Abstract
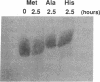
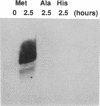
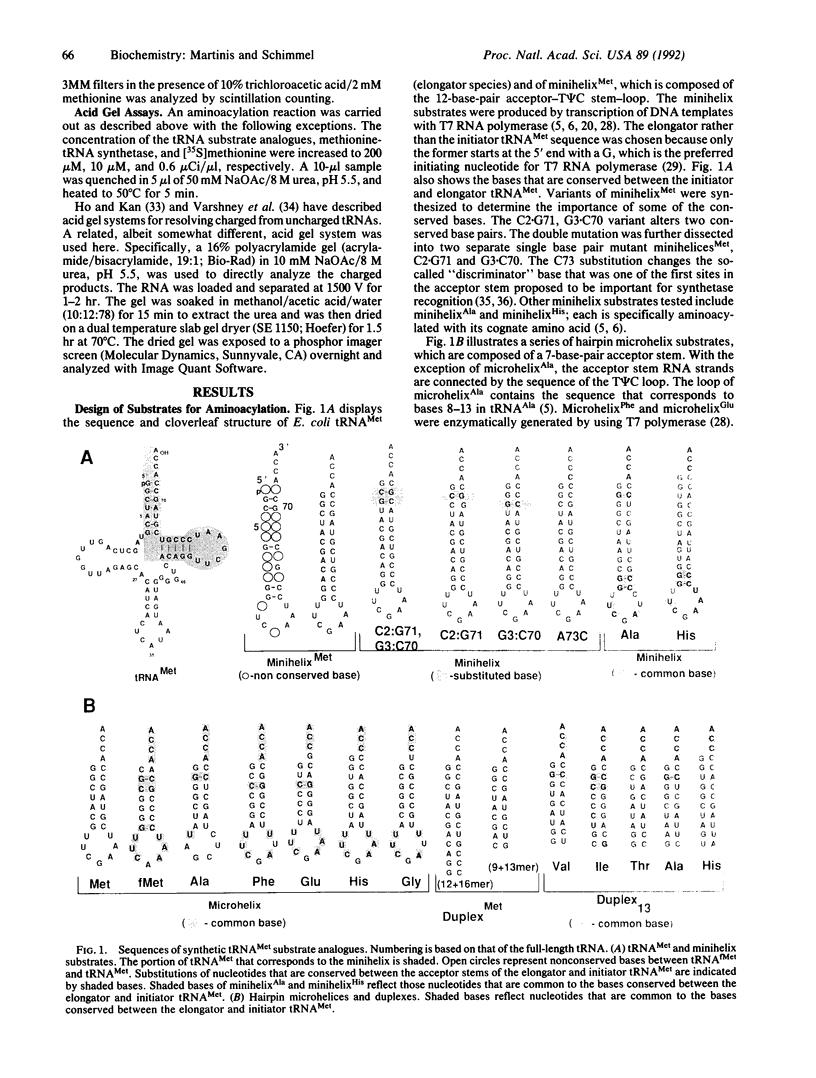
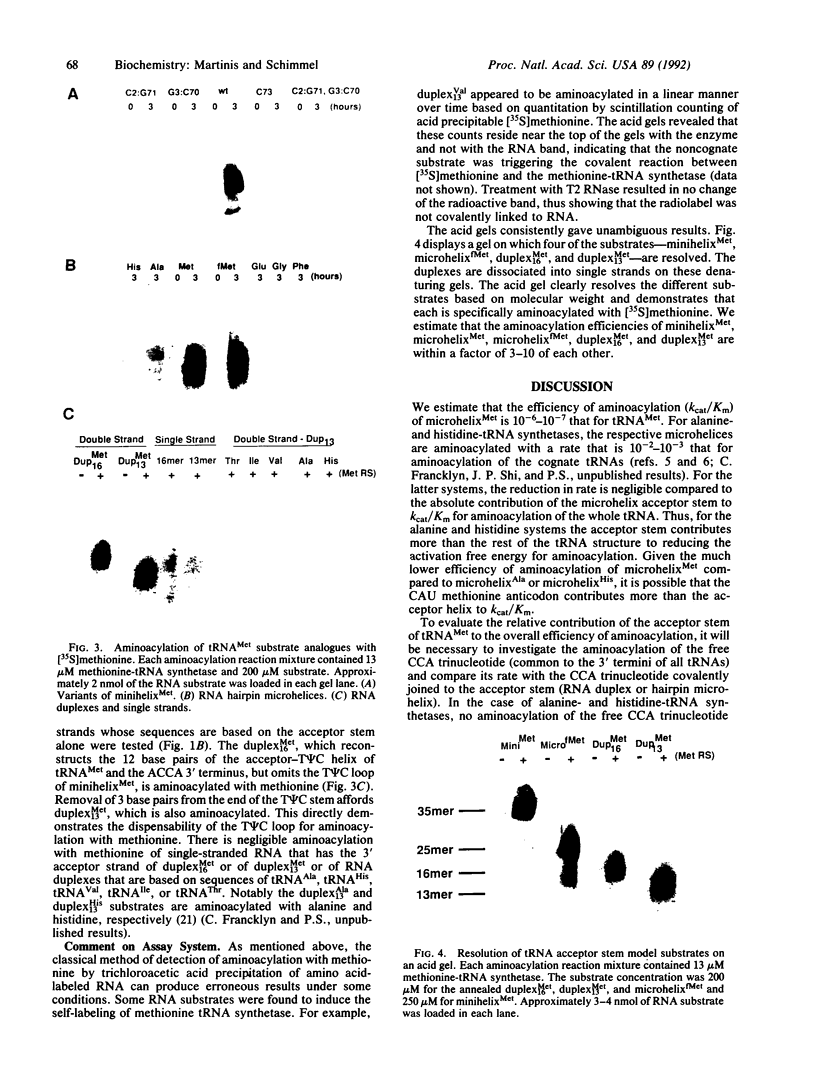
RNA hairpin helices whose sequences are based on the acceptor stems of alanine and histidine tRNAs are specifically aminoacylated with their cognate amino acids. In these examples, major determinants for the identities of the respective tRNAs reside in the acceptor stem; the anticodon and other parts of the tRNA are dispensable for aminoacylation. In contrast, the anticodon is a major determinant for the identity of a methionine tRNA. RNA hairpin helices and hybrid duplexes that reconstruct the acceptor-T psi C stem and the acceptor stem, respectively, of methionine tRNA were investigated here for aminoacylation with methionine. Direct visualization of the aminoacylated RNA product on an acidic polyacrylamide gel by phosphor imaging demonstrated specific aminoacylation with substrates that contained as few as 7 base pairs. No aminoacylation with methionine was detected with several analogous RNA substrates whose sequences were based on noncognate tRNAs. While the efficiency of aminoacylation is reduced by orders of magnitude relative to methionine tRNA, the results establish that specific aminoacylation with methionine of small duplex substrates can be achieved without the anticodon or other domains of the tRNA. The results, combined with earlier studies, suggest a highly specific adaptation of the structures of aminoacyl-tRNA synthetases to the acceptor stems of their cognate tRNAs, resulting in a relationship between the nucleotide sequences/structures of small RNA duplexes and specific amino acids.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brunie S., Zelwer C., Risler J. L. Crystallographic study at 2.5 A resolution of the interaction of methionyl-tRNA synthetase from Escherichia coli with ATP. J Mol Biol. 1990 Nov 20;216(2):411–424. doi: 10.1016/S0022-2836(05)80331-6. [DOI] [PubMed] [Google Scholar]
- Burbaum J. J., Schimmel P. Assembly of a class I tRNA synthetase from products of an artificially split gene. Biochemistry. 1991 Jan 15;30(2):319–324. doi: 10.1021/bi00216a002. [DOI] [PubMed] [Google Scholar]
- Burbaum J. J., Schimmel P. Structural relationships and the classification of aminoacyl-tRNA synthetases. J Biol Chem. 1991 Sep 15;266(26):16965–16968. [PubMed] [Google Scholar]
- Burbaum J. J., Starzyk R. M., Schimmel P. Understanding structural relationships in proteins of unsolved three-dimensional structure. Proteins. 1990;7(2):99–111. doi: 10.1002/prot.340070202. [DOI] [PubMed] [Google Scholar]
- Crothers D. M., Seno T., Söll G. Is there a discriminator site in transfer RNA? Proc Natl Acad Sci U S A. 1972 Oct;69(10):3063–3067. doi: 10.1073/pnas.69.10.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francklyn C., Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989 Feb 2;337(6206):478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- Francklyn C., Schimmel P. Enzymatic aminoacylation of an eight-base-pair microhelix with histidine. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8655–8659. doi: 10.1073/pnas.87.21.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh G., Pelka H., Schulman L. H. Identification of the tRNA anticodon recognition site of Escherichia coli methionyl-tRNA synthetase. Biochemistry. 1990 Mar 6;29(9):2220–2225. doi: 10.1021/bi00461a003. [DOI] [PubMed] [Google Scholar]
- Grodberg J., Dunn J. J. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988 Mar;170(3):1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y. S., Kan Y. W. In vivo aminoacylation of human and Xenopus suppressor tRNAs constructed by site-specific mutagenesis. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2185–2188. doi: 10.1073/pnas.84.8.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988 May 12;333(6169):140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- Jahn M., Rogers M. J., Söll D. Anticodon and acceptor stem nucleotides in tRNA(Gln) are major recognition elements for E. coli glutaminyl-tRNA synthetase. Nature. 1991 Jul 18;352(6332):258–260. doi: 10.1038/352258a0. [DOI] [PubMed] [Google Scholar]
- Kisselev L. L. The role of the anticodon in recognition of tRNA by aminoacyl-tRNA synthetases. Prog Nucleic Acid Res Mol Biol. 1985;32:237–266. doi: 10.1016/s0079-6603(08)60350-5. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Foss K., Jenkins R. A., Schneider J. Nucleotides that determine Escherichia coli tRNA(Arg) and tRNA(Lys) acceptor identities revealed by analyses of mutant opal and amber suppressor tRNAs. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9260–9264. doi: 10.1073/pnas.87.23.9260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T., Mechulam Y., Le Corre D., Panvert M., Blanquet S., Fayat G. Selection of suppressor methionyl-tRNA synthetases: mapping the tRNA anticodon binding site. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):291–295. doi: 10.1073/pnas.88.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu T., Nishikawa K., Nemoto F., Kuchino Y., Nishimura S., Miyazawa T., Yokoyama S. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature. 1988 Nov 10;336(6195):179–181. doi: 10.1038/336179a0. [DOI] [PubMed] [Google Scholar]
- Musier-Forsyth K., Scaringe S., Usman N., Schimmel P. Enzymatic aminoacylation of single-stranded RNA with an RNA cofactor. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):209–213. doi: 10.1073/pnas.88.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musier-Forsyth K., Usman N., Scaringe S., Doudna J., Green R., Schimmel P. Specificity for aminoacylation of an RNA helix: an unpaired, exocyclic amino group in the minor groove. Science. 1991 Aug 16;253(5021):784–786. doi: 10.1126/science.1876835. [DOI] [PubMed] [Google Scholar]
- Nagel G. M., Doolittle R. F. Evolution and relatedness in two aminoacyl-tRNA synthetase families. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8121–8125. doi: 10.1073/pnas.88.18.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J., Abelson J. tRNA identity. Annu Rev Biochem. 1989;58:1029–1049. doi: 10.1146/annurev.bi.58.070189.005121. [DOI] [PubMed] [Google Scholar]
- Pallanck L., Schulman L. H. Anticodon-dependent aminoacylation of a noncognate tRNA with isoleucine, valine, and phenylalanine in vivo. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3872–3876. doi: 10.1073/pnas.88.9.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S. J., Schimmel P. Evidence for interaction of an aminoacyl transfer RNA synthetase with a region important for the identity of its cognate transfer RNA. J Biol Chem. 1988 Nov 15;263(32):16527–16530. [PubMed] [Google Scholar]
- Pütz J., Puglisi J. D., Florentz C., Giegé R. Identity elements for specific aminoacylation of yeast tRNA(Asp) by cognate aspartyl-tRNA synthetase. Science. 1991 Jun 21;252(5013):1696–1699. doi: 10.1126/science.2047878. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Steitz T. A. Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase. Nature. 1991 Jul 18;352(6332):213–218. doi: 10.1038/352213a0. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., DiRenzo A. B., Behlen L. S., Uhlenbeck O. C. Nucleotides in yeast tRNAPhe required for the specific recognition by its cognate synthetase. Science. 1989 Mar 10;243(4896):1363–1366. doi: 10.1126/science.2646717. [DOI] [PubMed] [Google Scholar]
- Scaringe S. A., Francklyn C., Usman N. Chemical synthesis of biologically active oligoribonucleotides using beta-cyanoethyl protected ribonucleoside phosphoramidites. Nucleic Acids Res. 1990 Sep 25;18(18):5433–5441. doi: 10.1093/nar/18.18.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. Parameters for the molecular recognition of transfer RNAs. Biochemistry. 1989 Apr 4;28(7):2747–2759. doi: 10.1021/bi00433a001. [DOI] [PubMed] [Google Scholar]
- Schimmel P. RNA minihelices and the decoding of genetic information. FASEB J. 1991 May;5(8):2180–2187. doi: 10.1096/fasebj.5.8.2022314. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. An anticodon change switches the identity of E. coli tRNA(mMet) from methionine to threonine. Nucleic Acids Res. 1990 Jan 25;18(2):285–289. doi: 10.1093/nar/18.2.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988 Nov 4;242(4879):765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. The anticodon contains a major element of the identity of arginine transfer RNAs. Science. 1989 Dec 22;246(4937):1595–1597. doi: 10.1126/science.2688091. [DOI] [PubMed] [Google Scholar]
- Seong B. L., RajBhandary U. L. Escherichia coli formylmethionine tRNA: mutations in GGGCCC sequence conserved in anticodon stem of initiator tRNAs affect initiation of protein synthesis and conformation of anticodon loop. Proc Natl Acad Sci U S A. 1987 Jan;84(2):334–338. doi: 10.1073/pnas.84.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong B. L., RajBhandary U. L. Mutants of Escherichia coli formylmethionine tRNA: a single base change enables initiator tRNA to act as an elongator in vitro. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8859–8863. doi: 10.1073/pnas.84.24.8859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J. P., Francklyn C., Hill K., Schimmel P. A nucleotide that enhances the charging of RNA minihelix sequence variants with alanine. Biochemistry. 1990 Apr 17;29(15):3621–3626. doi: 10.1021/bi00467a005. [DOI] [PubMed] [Google Scholar]
- Uemura H., Imai M., Ohtsuka E., Ikehara M., Söll D. E. coli initiator tRNA analogs with different nucleotides in the discriminator base position. Nucleic Acids Res. 1982 Oct 25;10(20):6531–6539. doi: 10.1093/nar/10.20.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney U., Lee C. P., RajBhandary U. L. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J Biol Chem. 1991 Dec 25;266(36):24712–24718. [PubMed] [Google Scholar]