Abstract
Recurrent Clostridium difficile infection (CDI) can be effectively treated by infusion of a healthy donor faeces suspension. However, it is unclear what factors determine treatment efficacy. By using a phylogenetic microarray platform, we assessed composition, diversity and dynamics of faecal microbiota before, after and during follow-up of the transplantation from a healthy donor to different patients, to elucidate the mechanism of action of faecal infusion. Global composition and network analysis of the microbiota was performed in faecal samples from nine patients with recurrent CDI. Analyses were performed before and after duodenal donor faeces infusion, and during a follow-up of 10 weeks. The microbiota data were compared with that of the healthy donors. All patients successfully recovered. Their intestinal microbiota changed from a low-diversity diseased state, dominated by Proteobacteria and Bacilli, to a more diverse ecosystem resembling that of healthy donors, dominated by Bacteroidetes and Clostridium groups, including butyrate-producing bacteria. We identified specific multi-species networks and signature microbial groups that were either depleted or restored as a result of the treatment. The changes persisted over time. Comprehensive and deep analyses of the microbiota of patients before and after treatment exposed a therapeutic reset from a diseased state towards a healthy profile. The identification of microbial groups that constitute a niche for C. difficile overgrowth, as well as those driving the reinstallation of a healthy intestinal microbiota, could contribute to the development of biomarkers predicting recurrence and treatment outcome, identifying an optimal microbiota composition that could lead to targeted treatment strategies.
Keywords: faecal microbiota transplantation, C. difficile infection, HITChip, intestinal microbiota
Introduction
During the last decade, the incidence and severity of diarrhoea associated with Clostridium difficile infection (CDI) has markedly increased (Kelly and LaMont, 2008; Khoruts and Sadowsky, 2011). C. difficile is a Gram-positive, anaerobic, toxin-producing bacterium that typically affects the elderly and hospitalized patients following antibiotic use. However, the emergence of more virulent and antibiotic-resistant strains has made CDI an important health issue not only in other populations, such as patients suffering from inflammatory bowel disease (Ananthakrishnan et al., 2009) but also in populations typically considered as healthy (Kelly and LaMont, 2008). Recurrences are a frequent and severe complication of CDI and can affect over 30% of the patients (Bakken et al., 2011; Gough et al., 2011).
Therapies include, among others, antibiotic therapy effective in 70–85% of patients with an initial episode of CDI, but only in ∼30% of patients with subsequent relapses (van Nood et al., 2013). However, currently, the only effective treatment option for longstanding recurrent CDI is faecal microbiota transplantation (FMT) or donor faeces infusion, with a cure rate of 94% in patients with antibiotic refractory disease (van Nood et al., 2013; Vrieze et al., 2013). FMT is defined as the transfer of intestinal microbiota from a healthy donor that leads to the restoration of a stable microbial community in the gut (Borody and Khoruts, 2011; Lo Vecchio and Zacur, 2012). Although FMT has been documented to exist for over 1000 years (Zhang et al., 2012; de Vos, 2013), its application in the medical field was again reported over 50 years ago (Eiseman et al., 1958). Since then, FMT has been used to treat over 500 cases of CDI and other disorders with a disturbed intestinal microbiota (Borody et al., 2003; Landy et al., 2011; Vrieze et al., 2012; Kassam et al., 2013). Although it is generally believed that the efficacy of FMT can be explained by restoration of healthy gut microbiota, there are still few studies addressing the microbial composition and dynamics triggered by FMT (Khoruts et al., 2010; Manichanh et al., 2010; Rea et al., 2011; Petrof et al., 2013).
The intestinal microbiota constitutes an organ by itself, with digestive functions and specific metabolic and signalling networks established between microorganisms and the host (Zoetendal et al., 2008; Aziz et al., 2013). It is shaped by over 1000 species-like phylotypes and dominated by microorganisms, many of them not yet cultured, belonging to five major phyla (that is, Firmicutes, Bacteroidetes, Actinobacteria, Proteobacteria and Verrucomicrobia). The complexity of the intestinal microbial communities requires the application of high-throughput techniques, including metagenomics and 16S rRNA-based approaches. The microbiota composition fluctuates around a stable individual core, and the homoeostasis of this intestinal ecosystem has been shown to be a key factor in the maintenance of health and development of disease (Jalanka-Tuovinen et al., 2011; Rajilic-Stojanovic et al., 2012; Faith et al., 2013). Recently, a metagenomics analysis of European subjects showed that the intestinal microbiota of individuals can be assigned to one of three groups, named enterotypes, based on composition and relative abundance of certain key bacterial groups (Arumugam et al., 2011), which can fluctuate over time (Wu et al., 2011; Rajilic-Stojanovic et al., 2012). Currently, however, this concept is under extensive debate, where researchers rather favour a continuous gradient as opposed to separate enterotypes (Huse et al., 2012; Jeffery et al., 2012; Yong, 2012). Independent of whether these are distinct clusters or gradients, specific patterns of microbiota composition can be assigned not only in the gut but also across the human body (Koren et al., 2013), which could be of relevance in disease (Jeffery et al., 2012; Zupancic et al., 2012).
Patients with CDI harbour an intestinal microbiota with a characteristic reduced diversity owing to antibiotic use (Chang et al., 2008; Shahinas et al., 2012). We hypothesized that patients with recurrent CDI also show a composition favourable for the overgrowth of C. difficile, lacking specific signature microbes that prevent them to return to a sustainable homeostatic status after infection, resulting in recurrences. The goal of the present study was to detail the microbiota of patients with recurrent CDI, and to monitor its dynamics following FMT, by identifying potential signature microbes and key networks present in both healthy donors and patients. These results could expand the microbial knowledge of recurrent CDI, beneficial not only for early diagnostics but also for the development of novel general or individualized treatment strategies. Understanding how microbes behave during FMT and identifying an optimal microbiota composition that prevents recurrences could assist in the selection of optimal microbial features of a transplanted microbiota for a successful outcome.
Materials and methods
Study design and population
Nine patients with recurrent CDI who were treated with FMT in a randomized open label trial (the FECAL trial) (van Nood et al., 2013) were studied. Available isolates of C. difficile were analysed by PCR ribotyping (Paltansing et al., 2007). Five patients were initially randomized to FMT; four patients were initially randomized to vancomycin treatment and received FMT after antibiotic treatment failed. To prepare for FMT, patients received 4 or 5 days vancomycin 500 mg four times a day followed by bowel lavage using a macrogol solution on the last day of antibiotic treatment. The next day, 500 ml of a solution consisting of >75 g of donor faeces mixed with isotonic saline was infused through a nasoduodenal tube at a rate of ∼20 ml min−1. Faeces for FTM were collected from donors on the day of treatment and immediately transported to the hospital in a clean plastic container. The donor faeces solution was infused within 6 h after collection of the faeces by the donor. Faecal samples were collected from donors and patients before FMT (day 0) and from patients at days 14, 21, 35 and 70 after treatment and stored at −80 °C until microbial analysis was performed.
A detailed protocol of donor selection, preparation of the donor faeces solution and follow-up of patients was described previously (van Nood et al., 2013). One of the donors (donor D4) was used in four different patients for FMT, owing to ready access to this donor. The FECAL trial was conducted at the Academic Medical Center Amsterdam, the Netherlands, and the research protocol was approved by its Ethics Committee (Netherlands Trial Register; www.trialregister.nl, number NTR1177). All participants were personally informed, received information and provided written consent before the study. Baseline demographics and clinical information are listed in Supplementary Table 1.
Microbiota composition: networks and dynamics
DNA was isolated from faecal samples as previously described (Salonen et al., 2010) and used for the characterization of the intestinal microbiota composition using the Human Intestinal Tract Chip (HITChip), a phylogenetic microarray containing a duplicated set of over 5000 probes based on 16S rRNA gene sequences of 1140 intestinal bacterial phylotypes (Rajilic-Stojanovic et al., 2009). This microarray provides information regarding both composition and relative quantity of bacteria that make up the human intestinal tract communities. Detailed information on HITChip procedure is provided as Supplementary Material.
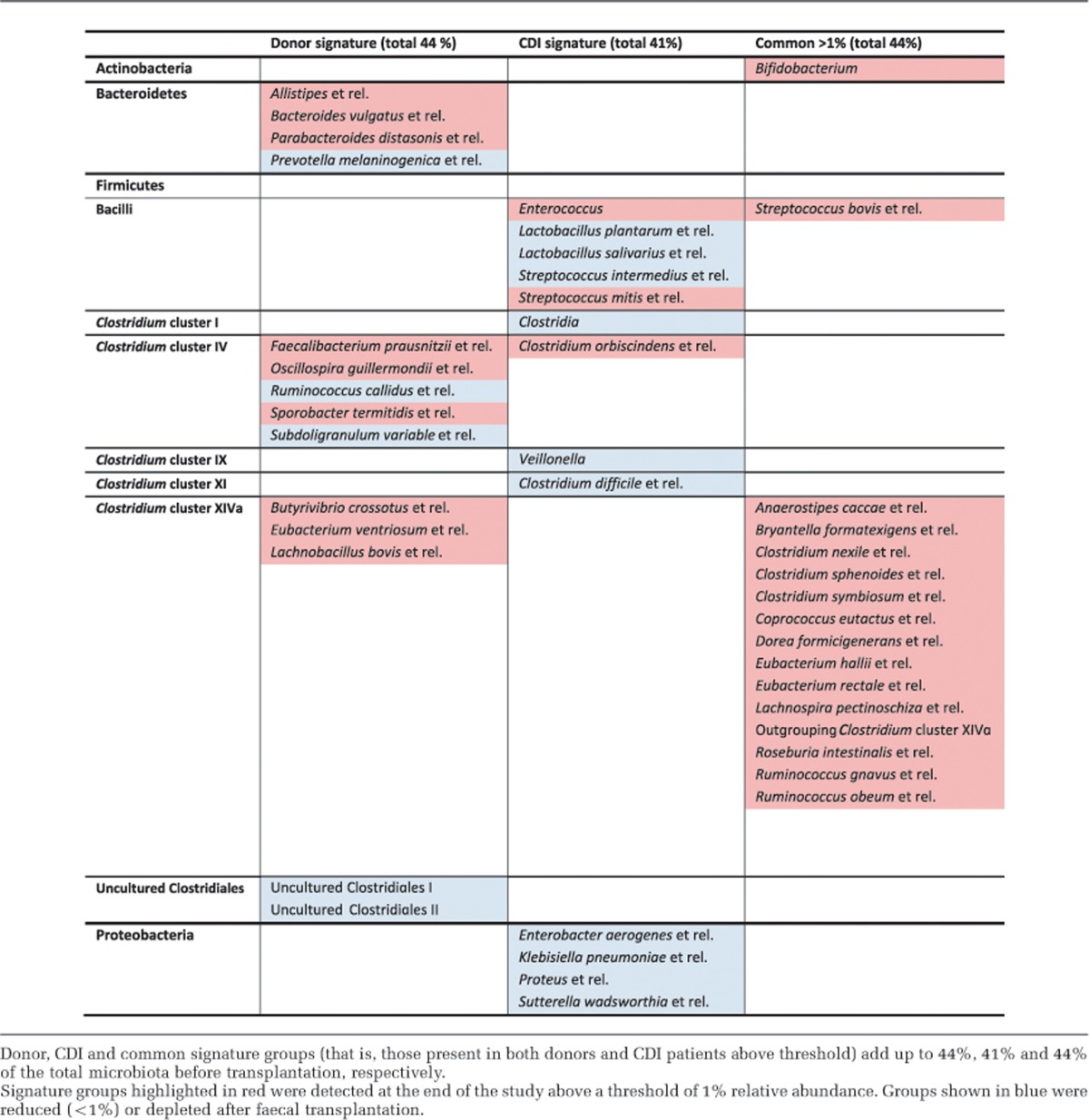
To monitor the microbial dynamics as a result of FMT, a 1% relative abundance threshold was defined and bacterial groups above the threshold were followed for 10 weeks. The cumulative relative abundance of the bacterial groups that passed the threshold (that is, relative abundance above 1%) accounted from 76–90% of the total intestinal microbiota in donors' samples and patients' samples from day 0 to the end of the trial. Phylogenetic groups found above the threshold in donors' samples and absent (or below threshold) in patients' samples at day 0 were called ‘donor signature' groups. Conversely, groups found in patient samples before FMT and not in donor samples were termed ‘CDI signature' groups.
In addition, a series of co-(and anti)occurrence networks were determined across individuals and calculated for donors, and for patients before and for 5 weeks after intervention. These networks were built using Spearman correlations calculated based on the abundance of the 130 bacterial groups (at the approximate genus level) targeted by the HITChip, using thresholds of ρ>|0.8| and relative abundance >0.1%. Networks were visualized using the Gephi network visualization and exploration platform (Bastian et al., 2009).
The stability of the microbial composition established after FMT was evaluated by comparison of the signals for all probes included in the HITChip using the Pearson moving-window correlation of consecutive samples.
Statistical analyses
The bacterial composition of the samples was compared at the phylum level (grouped at the class level for the Firmicutes) and at the approximate genus level (130 phylogenetic groups with >90% 16S rRNA gene sequence similarity; (Rajilic-Stojanovic et al., 2009)) using the Wilcoxon signed-rank test corrected for false discovery rate, in which a corrected P<0.05 was considered significant. Same testing was applied for bacterial diversity (Shannon index; (Magurran, 2004)), richness and evenness (Pielou's evenness index).
Similarity indices were calculated with the Pearson product moment correlation on the log-transformed signal intensities of all probes included in the HITChip microarray. This represents how similar the microbiota profiles are between the different samples.
Multivariate statistical analyses were performed with Canoco 5 (ter Braak and Šmilauer, 2012). Principal component analysis was performed on log-transformed probe signal intensity profiles derived from the HITChip phylogenetic microarray. Principal response curve (PRC) analysis was used to graphically summarize the global differences in the microbiota composition between patients and their respective donors over time (van den Brink and ter Braak, 1999). PRC combines the amounts of taxa, after logarithmic transformation, to a new single variable. The taxa with large deviations weigh high in this combination, whereas taxa that have equal amounts in patients and donors have zero weight. The quality of the PRC graph is expressed by the ratio of the variance explained by the PRC axis and the variance of all deviations across time. For details on interpretation and implementation see Supplementary Material.
Results
Changes in microbiota composition in CDI as a result of FMT
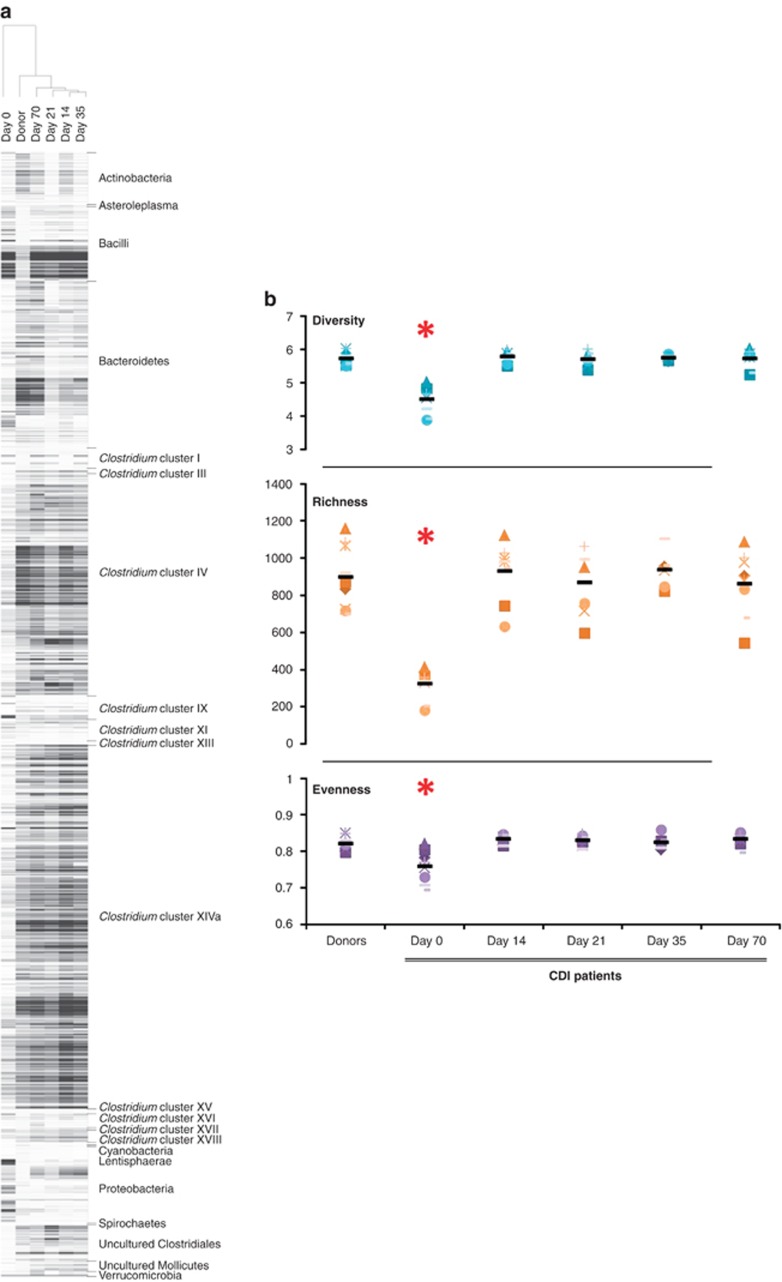
Nine patients with recurrent CDI caused by a variety of C. difficile ribotypes were treated by FMT with microbiota from a total of six healthy donors, one of which (donor D4) donated four times. All patients were cured by FMT without recurrence during follow-up and to date. Faecal samples were collected from donors (at day 0), and from patients before and four times after FMT during a follow-up period of 10 weeks. The microbiota composition of the donors' samples before and after the dilution and filtration showed a high overall similarity (average Pearson correlation coefficient >88±4%, Supplementary Table 3). Hierarchical clustering showed that samples from patients after FMT were notably more similar to donor profiles than to their own profiles at day 0 (Figure 1a; Supplementary Figure 1).
Figure 1.
(a) Hierarchical cluster with a heatmap of samples from patient P4 and its corresponding donor (D4). (b) Diversity, richness and evenness scores for healthy donors and CDI patients before and after faecal transplantation. Symbols correspond to individual patients, and average values are shown with black lines. Day 0 averages for diversity, richness and evenness are significantly different to donors and the rest of the time points (P<0.003).
FMT had a marked effect on diversity, richness and evenness of the microbiota of CDI patients (Figure 1b). Before FMT (day 0), patients showed significantly lower diversity levels compared with healthy donors (4.5±0.4 and 5.7±0.2 respectively, P=0.00006), which were increased and maintained after FMT, with scores at day 70 (5.7±0.3) in the range of those of healthy donors. A similar significant increase was observed for richness and evenness levels, lower in patients with recurrent CDI before FMT and successfully restored and maintained after treatment.
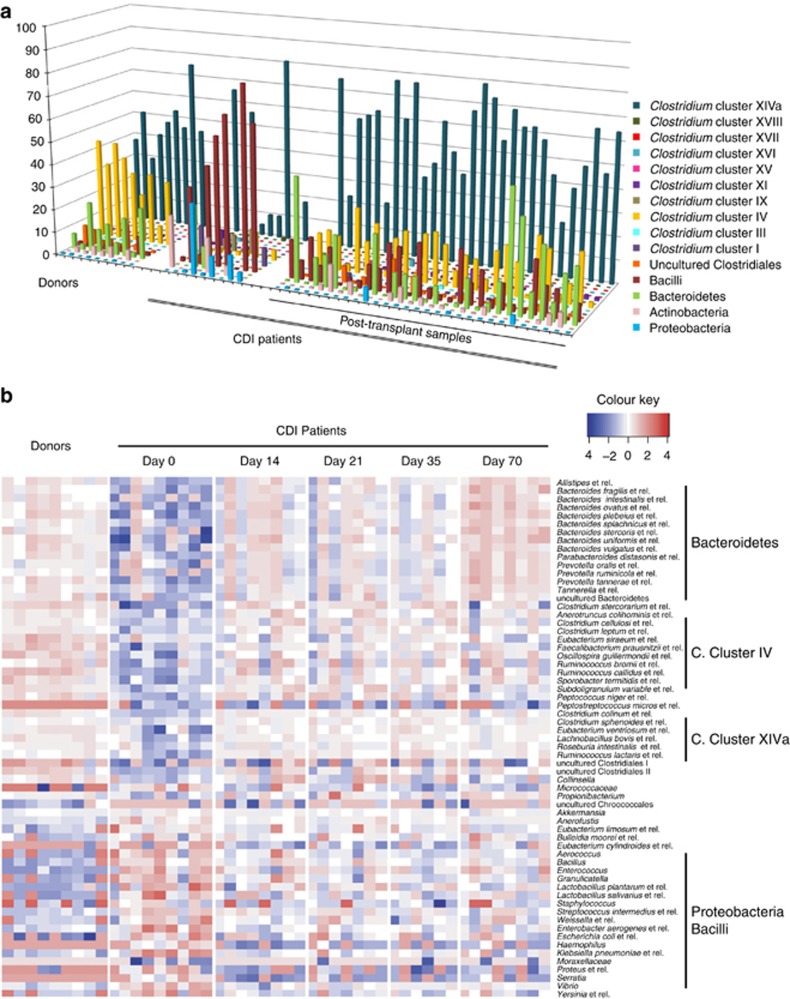
Comparison of the microbiota composition in patients with recurrent CDI revealed an immediate shift after FMT to a distribution more similar to that of healthy donors, both individually per patient and globally in all patients (Figure 2a). Before FMT, CDI patients showed a common characteristic profile dominated by members of the Proteobacteria and Bacilli. All post-transplant samples were enriched with bacteria from the Bacteroidetes and Clostridium clusters IV and XIVa, the latter two groups being recognized to include most of the known intestinal butyrate producers. These Clostridium clusters, normally representative of healthy donor microbiota, also increased in diversity (Supplementary Table 4).
Figure 2.
(a) Faecal microbiota composition at the phylum level (class level for the Firmicutes) of healthy donors and CDI patients before and after faecal transplant (shown are groups with relative abundance >0.5%). (b) Heatmap of relative abundances of bacterial groups (approximate genus level) that differ significantly between donors and CDI patients before faecal transplant and at the end of the study (P<0.05).
Differences in the microbiota were also found when compared at a higher resolution at the approximate genus level (Figure 2b). As expected, a pronounced contrast was found between healthy donors and pre-transplantation samples (day 0). Relative abundances of 41% of the genus-level phylogenetic groups targeted by the HITChip were significantly different between donors and CDI patients before FMT (P<0.05). These differences were largely reduced after FMT, with only 13% of all groups remaining different at day 70 (Supplementary Table 2). The microbiota of the CDI patients were characterized by a high level of signals derived from potentially pathogenic taxa belonging to the Proteobacteria (for example, Enterobacter, Klebsiella, Sutterella or Proteus spp.). After FMT, these groups returned to low levels comparable to those found in the microbiota of healthy individuals.
Effect of the donor microbiota on post-transplant ecosystem
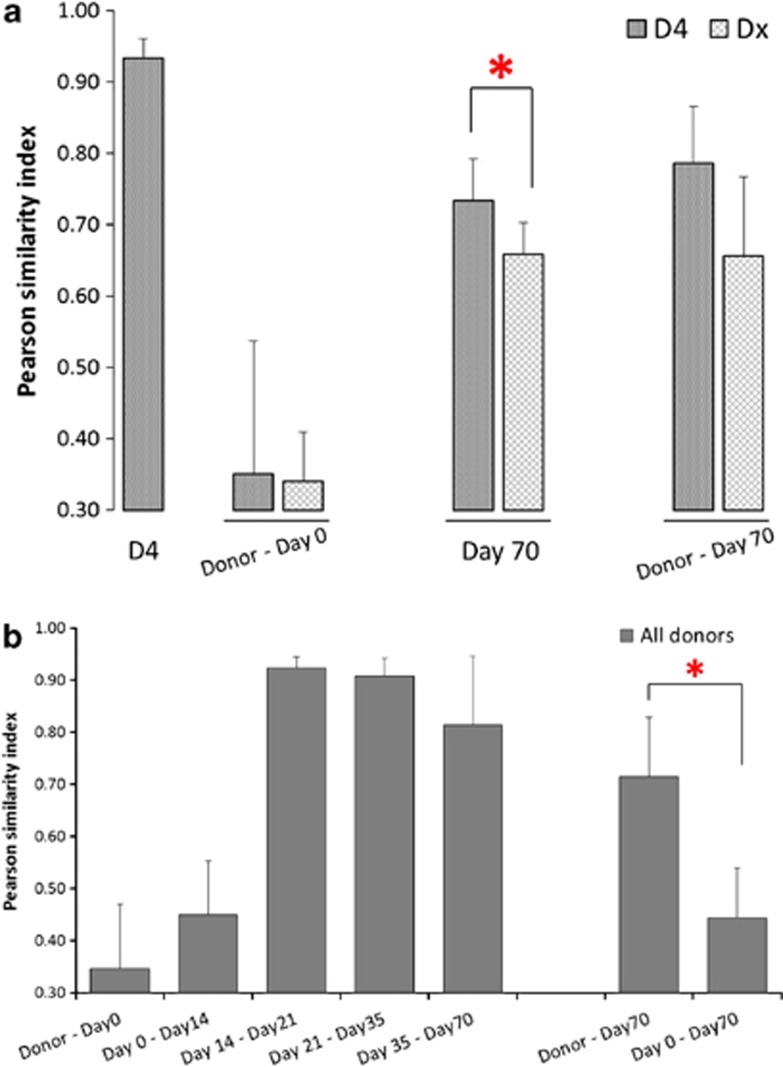
To predict optimal features of a microbiota that leads to a successful FMT, we compared samples from patients that received material from the same donor (donor D4, n=4) and those receiving from different donors (n=5). The microbiota of D4 was stable between the different samples provided, with an average similarity index of 0.93±0.02, a value in line with those previously observed for healthy adults (Rajilic-Stojanovic et al., 2012; Figure 3a).
Figure 3.
Pearson's similarity indices at the probe level of the HITChip microarray. (a) Similarity of microbiota composition of samples of donor D4 (four different FMT samples); donor D4 and other donors and CDI patients at day 0; CDI patients receiving from donor D4 and other donors at the end of the trial (*P=0.01); donor D4 and other donors and CDI patients at day 70. (b) Similarity of microbiota composition of all donors and CDI patients over time; similarity of samples from CDI patients at day 70 compared with their own day 0 and their corresponding donor (*P=0.0001).
When comparing the microbiota composition of patients and donors before FTM, those assigned to D4 and the ones assigned to different donors showed comparable low similarity values with their respective donors (r=0.35±0.19 or r=0.34±0.07, respectively). However, at the end of the trial, samples from patients assigned to D4 were found to be significantly more similar between them (r=0.73±0.05) than when comparing those receiving from different donors (r=0.65±0.04, P=0.01). Interestingly, at the end of the trial the similarity of patients' microbiota with their corresponding donors tended to be higher in patients receiving FMT from D4 (r=0.79±0.08) than in those obtaining from other donors (r=0.66±0.11), although this difference was not significant (P=0.08) (Figure 3a). This suggests that some microbiota compositions may leave a more prevalent and stable imprint on the patients' faecal microbial composition, which is maintained even between different individuals.
A more detailed analysis of the microbiota of D4 revealed a significantly higher abundance of Bacteroides spp. (more specifically those related to B. intestinalis, B. plebeius and B. uniformis) as compared with other donors, with average relative abundances of 3.81±0.31% for D4 and 1±0.12% for other donors (P=0.01).
Reshape and network dynamics of the microbial environment after FMT
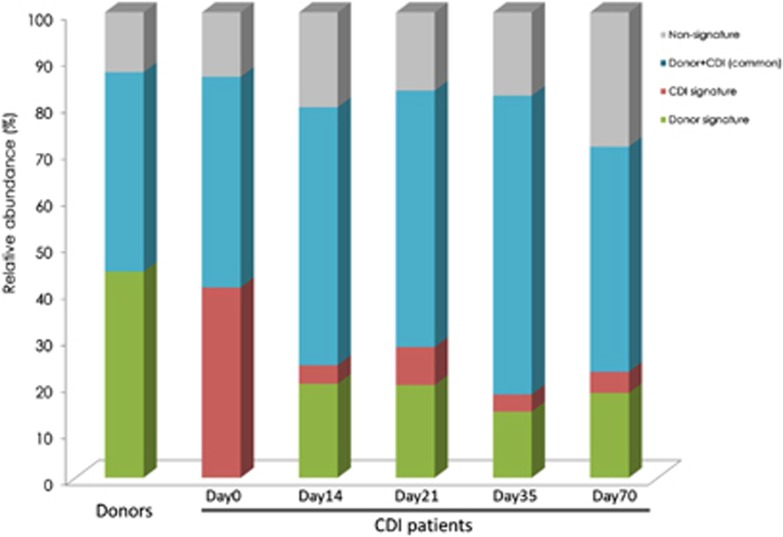
As indicated above, FMT had an immediate repairing effect on the intestinal microbiota of CDI patients. To further investigate how microbes behave during transplant by looking at global dynamics triggered by the treatment, we developed an approach to identify potential signature bacterial characteristic of CDI as well as those responsible for microbiota restoration. These signature bacterial groups were selected based on their relative abundance (over 1%) and monitored throughout the duration of the study (Supplementary Figure 2; Figure 4). ‘CDI signature' groups, initially accounting for almost half of the total microbiota of the patients at day 0 (41±22.1%), were reduced over fivefold just 14 days after FMT (to 6.5±2.8%). Simultaneously, the ‘donor signature' groups absent in CDI patients at day 0 increased to 22.8±8.4%. These changes were maintained for the 10-week duration of the study, as seen by the monitoring of the relative abundance of the different signature groups.
Figure 4.
Reshape of the intestinal microbial environment as a result of faecal transplantation. Bacterial signature groups (relative abundance >1%) were followed up for a period of 10 weeks after FMT. Changes in ‘donor signature', ‘CDI signature', ‘donor+CDI (common)' and ‘non-signature' phylotypes over time.
Among the donor signature bacteria increasing in patients after FMT, members of the Bacteroidetes as well as butyrate producers from Clostridium clusters IV and XIVa were found, including those related to Faecalibacterium prausnitzii and Butyrivibrio crossotus (Table 1). Among the CDI signature groups displaced by the intervention, we found populations of bacteria generally occurring at lower densities in the small intestine such as Enterococcus, Lactobacillus or Veillonella. Remarkably, the levels of potentially pathogenic bacteria were reduced, including C. difficile that dropped immediately from 4±2.1% at day 0 to 0.2±0.2% post transplant, the latter values being similar to those found in healthy donors. Proteobacteria, such as Enterobacter aerogenes and Escherichia coli, changed from 4.6±9.9% at day 0 to 0.1±0.1% 14 days after the intervention. These findings confirm an immediate shift from an ecosystem dominated by potentially pathogenic bacteria to a donor-like microbiota.
Table 1. Signature bacterial groups of donors and CDI patients' microbiota.
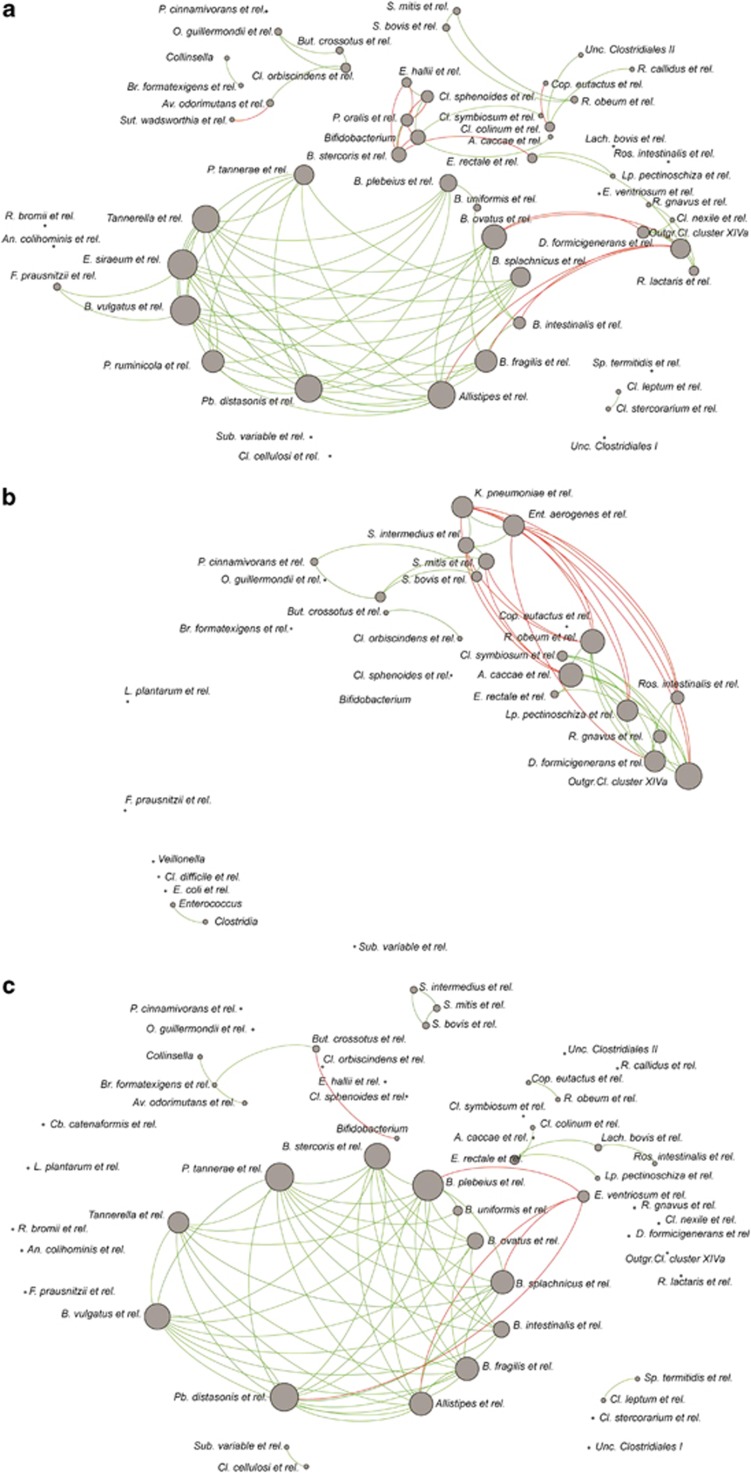
An in-depth analysis of co- and anti-occurrence networks strengthened the concept of the phylum Bacteroidetes as an essential component in this process (Figure 5). A network formed by members of this phylum, central in the microbiota from healthy donors, was absent in networks of CDI patients at day 0, with members of this network being even below the selected threshold. Microbial networks observed in the faecal profiles of CDI patients were characterized by a dominance of potential pathogens such as Klebsiella pneumoniae and E. aerogenes, negatively correlated to known butyrate-producing bacteria such as Eubacterium rectale and Roseburia intestinalis. Network analyses after FMT showed that the Bacteroidetes cluster and its connections were successfully restored after FMT.
Figure 5.
Bacterial networks of (a) donors, (b) patients before FMT and (c) patients for 5 weeks after treatment. Shown groups with Spearman correlations >|0.8| and relative abundance >0.1% in at least 50% of the samples. Green lines and red lines indicate co- and anti-occurrence respectively (that is, positive and negative correlations).
Stability of the intestinal microbiota after FMT
The intestinal microbial ecosystem restored by FMT stabilized quickly over time (Figure 3b). The microbiota of CDI patients was found to be stable from 14 days after FMT, with similarity indices typically found in healthy individuals (r=0.9±0.02 s.d.). This stability was maintained throughout the 10 weeks post-transplant follow-up with no significant changes. As expected, at the end of the trial the microbial composition of patients resembled more their respective donors than each patient's own pre-transplant sample (day 0), with similarity indices of 0.7±0.1 s.d. and 0.4±0.1 s.d., respectively (P=0.0001) (Figure 3b).
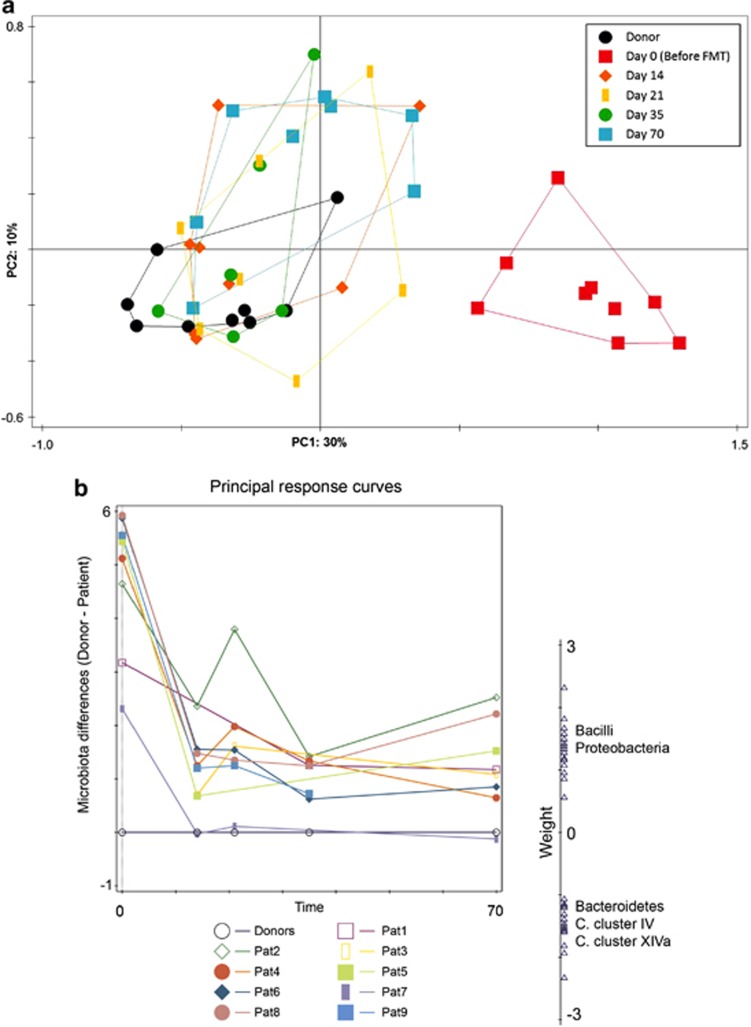
To visualize the global changes of the microbiota of CDI patients after faecal transplant throughout the follow-up period, we performed a multivariate analysis specifically geared towards visualization of trends over time. A principal component analysis (Figure 6a) demonstrated that patients' samples immediately shift and stabilize after FMT towards those from the healthy donor. A principle response curve (PRC, Figure 6b) further confirmed our previous observations that samples at day 0 of patients were highly different from those of their donors and that these differences globally decreased over time, were patients tended to have a microbiota more in line with that of a healthy donor. The main components of the observed changes in composition were Bacilli (mostly small intestinal bacteria) and Proteobacteria (including vancomycin-resistant and potentially pathogenic bacteria). The direction of change of these groups over time was opposite to that of the Bacteroidetes and several members of the Clostridium clusters IV and XIVa, including F. prausnitzii, E. rectale, R. intestinalis and other known butyrate-producing bacteria.
Figure 6.
(a) Principal component analysis of CDI patients over time based on their microbiota composition. First and second ordination axes are plotted, explaining 30 and 10% of the variability in the data set, respectively. (b) Principal response curve analysis summarizing the differences in total microbiota composition between patients and their respective donors over time. The graph shows 35% of all time-dependent differences in microbiota composition between patients and their donors. Bacterial groups shown are the main drivers of the differences between patients and donors: groups that have a positive weight on the response curve follow the observed curves, whereas those with negative weights follow the opposite pattern.
Discussion
In this study we showed that FMT by donor faeces infusion results in a normalization of the disturbed microbiota as seen in patients with longstanding recurrent CDI. In particular diversity, including richness and evenness, significantly reduced in all patients before FMT, recovered to normal values immediately after treatment. We were also able to identify specific signature groups and networks of bacteria that could be involved both in the development of an ecosystem prone to recurrences, as well as in the restoration of a healthy microbiota after donor faeces infusion.
In CDI the use of broad-spectrum antibiotics is believed to create an ecosystem that deviates from the normal predominance of members of the Bacteroidetes and Firmicutes to a one dominated by Bacilli and Proteobacteria (Chang et al., 2008; Mariat et al., 2009). The microbiota of CDI patients is characterized by an absence or reduction of Bacteroidetes spp., as observed both by cultivation (Tvede and Rask-Madsen, 1989) as well as with 16S rRNA gene-targeted molecular approaches (Grehan et al., 2010; Khoruts et al., 2010). This was confirmed in our study, where patients at day 0 showed significant reduction in the relative abundance of several groups belonging to the Bacteroidetes, as well as displaying complete absence of the central co-occurrence networks involving these bacteria. Bacteroides spp. have been shown to have a role in the resistance against C. difficile infection (Tvede and Rask-Madsen, 1989; Hopkins and Macfarlane, 2002, 2003) and are involved in carbohydrate metabolism and regulation of immune functions (Hooper and Gordon, 2001; Wexler, 2007; Vaishnava et al., 2011). In mice, Bacteroides was found to be one of the genera involved in the recovery of disease after bacteriotherapy using a mix of bacteria (Lawley et al., 2012). These results in mice suggest that not a complete microbiota is needed, which may be promising for future therapeutic developments. In a mouse model for obesity, B. uniformis was found to reduce metabolic and immune dysfunction (Gauffin Cano et al., 2012).
In our study, the microbiota of CDI patients before FMT was also characterized by an abnormal overgrowth of Proteobacteria, with considerably higher amounts of low-level pathogens such as E. aerogenes and related species. Network analyses of samples at day 0 were dominated by potential pathogens such as K. pneumoniae. This proteobacterial overgrowth could lead us to hypothesize a constant low-grade intestinal inflammation, that prevents the microbiota to return to a homeostatic status. An increase in Proteobacteria has also been reported in several models of antibiotic treatment and in obesity-induced inflammation (O'Keefe, 2010; Frazier et al., 2011; Verdam et al., 2013).
An enrichment of bacteria generally associated with the small intestine, such as Veillonella, Streptococcus or Lactobacillus species, was previously observed in patients with recurrent CDI before FMT (Khoruts et al., 2010; Zoetendal et al., 2012). Many of these trends can be explained by the history of antibiotic use of these patients (Vrieze, 2013), as most Lactobacillus spp. are intrinsically resistant to vancomycin (Swenson et al., 1990). In our study, bacteria related to Lactobacillus plantarum and Streptococcus intermedius had relative abundances that were between 3 and ⩾50-fold higher in CDI patients than in healthy donors, returning to levels more similar to those observed in donor samples after FMT.
As already mentioned, extended periods of antibiotic treatment have a profound impact on the microbiota composition, which makes it very difficult to separate signature groups owing to the treatment or the disease. In an attempt to uncouple these signatures we compared the microbiota composition of a group of healthy volunteers, receiving a standard vancomycin treatment without developing CDI, with that of the CDI patients at day 0. We were able to assess differences in microbiota composition that could be a trigger for the development of recurrent CDI (Vrieze, 2013). These differences were found predominantly in species within the Bacteroidetes (sixfold higher in non-CDI patients, P=0.009), and the Firmicutes, more specifically members of the Clostridium clusters IV and XIVa (6.5-fold and 11-fold higher, respectively, in non-CDI patients, P<0.002). Species related to the Clostridium cluster IX butyrate producer Megasphaera elsdenii were found 10-fold higher in non-CDI subjects as compared with CDI patients (P=0.01). Butyrate is a short-chain fatty acid shown to have signalling and protective effects on the gut (Wong et al., 2006; Hamer et al., 2008). A deficiency of butyrate in the colon can promote growth and toxin production of C. difficile (O'Keefe, 2010), whereas its delivery to the colon decreases pain sensation (Hamer et al., 2008). This comparison of subjects undergoing antibiotic treatment that leads (or not) to CDI is key for understanding the role of different microbial groups in its pathogenesis, as well as for identifying potential factors for recurrence, and it is currently under research (Antharam et al., 2013).
Faecal transplantation triggered a therapeutic reset in the intestinal community of our patients. By defining a threshold to identify ‘donor signature' and ‘CDI signature' bacterial groups, we were able to monitor the reshaping of the transplanted microbiota over time. As a result of FMT, members of the Bacteroidetes showed the most substantial changes. Networks including these groups, absent in day 0 samples, were immediately re-established after FMT as a central and key component of the co- and anti-occurrence networks. Donor signature bacterial groups were identified including bacteria often associated with health (for example, the potentially anti-inflammatory F. prausnitzii) (Sokol et al., 2008) as well as microbes associated with different steps of the complex carbohydrate fermentation scheme, such as primary complex carbohydrate degradation or secondary fermenters, like butyrate producers. Assessing a resilient and stable colonization of the gastrointestinal tract by a healthy microbiota and its networks is the key to identify an optimal ‘ healthy' microbiota composition, ideal for a successful FMT (Brandt et al., 2011; Vrieze et al., 2013).
A previous study using denaturing gradient gel electrophoresis revealed a durable establishment of donor bacteria up to 24 weeks post transplant, with slight changes in the composition over time (Grehan et al., 2010). In our study, using a more comprehensive microarray-based profiling approach, we confirmed that the microbiota of CDI patients stabilized immediately after FMT, by adopting specific donor signatures that are stably maintained over time.
In addition, our findings suggest that there is a role of the donor microbiota composition in the outcome of FMT. Although all patients successfully recovered, a microbiota imprint could be observed, where some donors can make a larger impact on the microbiota compared with others. Donor D4 seemed to create a more prevalent and stable environment as compared with other donors over time. An in-depth analysis of D4 microbiota revealed a four-time higher relative abundance of Bacteroides spp. (that is, those related to B. intestinalis, B. plebeius and B. uniformis) compared with other donors. In contrast, groups generally considered pathogenic, such as B. fragilis and B. vulgatus, were not found significantly higher in this donor. The higher stability could lead to hypothesize into what makes a better microbiota composition to be used for FMT, and which are the drivers for a durable and stable recovery. Future larger and longer studies are needed to increase our understanding in the effect of donor microbiota on treatment outcome, enabling to look at functions of microbes within a group to select the ‘perfect donor composition' for treatment of CDI.
As demonstrated in this study, the identification of bacterial signatures and central networks as drivers for the restoration of a healthy intestinal microbiota, as well as the dynamics of FMT, could allow us to predict the success of the treatment. Overall, we identified the existence of key groups and networks that represent ecosystem and metabolic drivers, and are likely to be central for the development of recurrent CDI or the maintenance of homeostasis in the microbiota of healthy individuals, and could be therefore used as predictors of recurrence and treatment outcomes. These results could lead not only to early diagnostics of CDI recurrence but also to the development of novel general or individualized targeted treatment strategies. This could ultimately lead to the identification of a defined and essential microbial core as a cure for CDI patients in next-generation therapies (de Vos, 2013), identifying and selecting signature bacterial groups sufficient for treating this potentially fatal disease.
Acknowledgments
We thank the HITChip team for technical assistance in performing experiments and Dr Ed Kuijper for performing the PCR Ribotyping of patient samples in the study.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ananthakrishnan AN, Issa M, Binion DG. (2009). Clostridium difficile and inflammatory bowel disease. Gastroenterol Clin North Am 38: 711–728. [DOI] [PubMed] [Google Scholar]
- Antharam VC, Li EC, Ishmael A, Sharma A, Mai V, Rand KH et al. (2013). Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J Clin Microbiol 51: 2884–2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR et al. (2011). Enterotypes of the human gut microbiome. Nature 473: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz Q, Doré J, Emmanuel A, Guarner F, Quigley EMM. (2013). Gut microbiota and gastrointestinal health: current concepts and future directions. Neurogastroenterol Motil 25: 4–15. [DOI] [PubMed] [Google Scholar]
- Bakken JS, Borody T, Brandt LJ, Brill JV, Demarco DC, Franzos MA et al. (2011). Treating Clostridium difficile infection with fecal microbiota transplantation. Clin Gastroenterol Hepatol 9: 1044–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastian M, Heymann S, Jacomy M. (2009). Gephi: an open source software for exploring and manipulating networks. International AAAI Conference on Weblogs and Social Media; San Jose, CA, USA, 2009.
- Borody TJ, Khoruts A. (2011). Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol 9: 88–96. [DOI] [PubMed] [Google Scholar]
- Borody TJ, Warren EF, Leis S, Surace R, Ashman O. (2003). Treatment of ulcerative colitis using fecal bacteriotherapy. J Clin Gastroenterol 37: 42–47. [DOI] [PubMed] [Google Scholar]
- Brandt LJ, Borody TJ, Campbell J. (2011). Endoscopic fecal microbiota transplantation: ‘first-line' treatment for severe Clostridium difficile infection? J Clin Gastroenterol 45: 655–657. [DOI] [PubMed] [Google Scholar]
- Chang JY, Antonopoulos DA, Kalra A, Tonelli A, Khalife WT, Schmidt TM et al. (2008). Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis 197: 435–438. [DOI] [PubMed] [Google Scholar]
- de Vos WM. (2013). Fame and future of faecal transplantations—developing next-generation therapies with synthetic microbiomes. Microbiol Biotechnol 6: 316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiseman B, Silen W, Bascom GS, Kauvar AJ. (1958). Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery 44: 854–859. [PubMed] [Google Scholar]
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL et al. (2013). The long-term stability of the human gut microbiota. Science 341: 1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazier TH, DiBaise JK, McClain CJ. (2011). Gut microbiota, intestinal permeability, obesity-induced inflammation, and liver injury. JPEN J Parenter Enteral Nutr 35: 14S–20S. [DOI] [PubMed] [Google Scholar]
- Gauffin Cano P, Santacruz A, Moya A, Sanz Y. (2012). Bacteroides uniformis CECT 7771 ameliorates metabolic and immunological dysfunction in mice with high-fat-diet induced obesity. PLoS One 7: e41079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough E, Shaikh H, Manges AR. (2011). Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis 53: 994–1002. [DOI] [PubMed] [Google Scholar]
- Grehan MJ, Borody TJ, Leis SM, Campbell J, Mitchell H, Wettstein A. (2010). Durable alteration of the colonic microbiota by the administration of donor fecal flora. J Clin Gastroenterol 44: 551–561. [DOI] [PubMed] [Google Scholar]
- Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. (2008). Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27: 104–119. [DOI] [PubMed] [Google Scholar]
- Hooper LV, Gordon JI. (2001). Commensal host-bacterial relationships in the gut. Science 292: 1115–1118. [DOI] [PubMed] [Google Scholar]
- Hopkins MJ, Macfarlane GT. (2002). Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J Med Microbiol 51: 448–454. [DOI] [PubMed] [Google Scholar]
- Hopkins MJ, Macfarlane GT. (2003). Nondigestible oligosaccharides enhance bacterial colonization resistance against Clostridium difficile in vitro. Appl Environ Microbiol 69: 1920–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Ye Y, Zhou Y, Fodor AA. (2012). A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS One 7: e34242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalanka-Tuovinen J, Salonen A, Nikkila J, Immonen O, Kekkonen R, Lahti L et al. (2011). Intestinal microbiota in healthy adults: temporal analysis reveals individual and common core and relation to intestinal symptoms. PLoS One 6: e23035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery IB, Claesson MJ, O'Toole PW, Shanahan F. (2012). Categorization of the gut microbiota: enterotypes or gradients? Nat Rev Micro 10: 591–592. [DOI] [PubMed] [Google Scholar]
- Kassam Z, Lee CH, Yuan Y, Hunt RH. (2013). Fecal microbiota transplantation for Clostridium difficile infection: systematic review and meta-analysis. Am J Gastroenterol 108: 500–508. [DOI] [PubMed] [Google Scholar]
- Kelly CP, LaMont JT. (2008). Clostridium difficile—more difficult than ever. N Engl J Med 359: 1932–1940. [DOI] [PubMed] [Google Scholar]
- Khoruts A, Dicksved J, Jansson JK, Sadowsky MJ. (2010). Changes in the composition of the human fecal microbiome after bacteriotherapy for recurrent Clostridium difficile-associated diarrhea. J Clin Gastroenterol 44: 354–360. [DOI] [PubMed] [Google Scholar]
- Khoruts A, Sadowsky MJ. (2011). Therapeutic transplantation of the distal gut microbiota. Mucosal Immunol 4: 4–7. [DOI] [PubMed] [Google Scholar]
- Koren O, Knights D, Gonzalez A, Waldron L, Segata N, Knight R et al. (2013). A guide to enterotypes across the human body: Meta-analysis of microbial community structures in human microbiome datasets. PLoS Comput Biol 9: e1002863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landy J, Al-Hassi HO, McLaughlin SD, Walker AW, Ciclitira PJ, Nicholls RJ et al. (2011). Review article: faecal transplantation therapy for gastrointestinal disease. Aliment Pharmacol Ther 34: 409–415. [DOI] [PubMed] [Google Scholar]
- Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, Raisen C et al. (2012). Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog 8: e1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Vecchio A, Zacur GM. (2012). Clostridium difficile infection: an update on epidemiology, risk factors, and therapeutic options. Curr Opin Gastroenterol 28: 1–9. [DOI] [PubMed] [Google Scholar]
- Magurran AE. (2004) Measuring Biological Diversity. Blackwell: Malden, MA, USA. [Google Scholar]
- Manichanh C, Reeder J, Gibert P, Varela E, Llopis M, Antolin M et al. (2010). Reshaping the gut microbiome with bacterial transplantation and antibiotic intake. Genome Res 20: 1411–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariat D, Firmesse O, Levenez F, Guimaraes V, Sokol H, Dore J et al. (2009). The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol 9: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Keefe SJ. (2010). Tube feeding, the microbiota, and Clostridium difficile infection. World J Gastroenterol 16: 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paltansing S, van den Berg RJ, Guseinova RA, Visser CE, van der Vorm ER, Kuijper EJ. (2007). Characteristics and incidence of Clostridium difficile-associated disease in The Netherlands, 2005. Eur J Clin Microbiol Infect Dis 13: 1058–1064. [DOI] [PubMed] [Google Scholar]
- Petrof EO, Gloor GB, Vanner S, Weese S, Carter DA, Daigneault M et al. (2013). Stool substitute transplant therapy for the eradication of Clostridium difficile infection: ‘RePOOPulating' the gut. Microbiome 1: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M, Heilig HG, Molenaar D, Kajander K, Surakka A, Smidt H et al. (2009). Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol 11: 1736–1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M, Heilig HG, Tims S, Zoetendal EG, de Vos WM. (2012). Long-term monitoring of the human intestinal microbiota composition. Environ Microbiol 15: 1146–1159. [DOI] [PubMed] [Google Scholar]
- Rea MC, Dobson A, O'Sullivan O, Crispie F, Fouhy F, Cotter PD et al. (2011). Effect of broad- and narrow-spectrum antimicrobials on Clostridium difficile and microbial diversity in a model of the distal colon. Proc Natl Acad Sci USA 108(Suppl 1): 4639–4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salonen A, Nikkila J, Jalanka-Tuovinen J, Immonen O, Rajilic-Stojanovic M, Kekkonen RA et al. (2010). Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J Microbiol Methods 81: 127–134. [DOI] [PubMed] [Google Scholar]
- Shahinas D, Silverman M, Sittler T, Chiu C, Kim P, Allen-Vercoe E et al. (2012). Toward an understanding of changes in diversity associated with fecal microbiome transplantation based on 16S rRNA gene deep sequencing. MBio 3: pii e00338–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ et al. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA 105: 16731–16736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson JM, Facklam RR, Thornsberry C. (1990). Antimicrobial susceptibility of vancomycin-resistant Leuconostoc, Pediococcus, and Lactobacillus species. Antimicrob Agents Chemother 34: 543–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Braak CJF, Šmilauer P. (2012) Canoco Reference Manual and User's Guide: Software for Ordination, Version 5.0. Microcomputer Power: Ithaca, NY, USA. [Google Scholar]
- Tvede M, Rask-Madsen J. (1989). Bacteriotherapy for chronic relapsing Clostridium difficile diarrhoea in six patients. Lancet 1: 1156–1160. [DOI] [PubMed] [Google Scholar]
- Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, Koren O et al. (2011). The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334: 255–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink PJ, ter Braak CJF. (1999). Principal response curves: analysis of time-dependent multivariate responses of a biological community to stress. Environ Toxicol Chem 18: 138–148. [Google Scholar]
- van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, de Vos WM et al. (2013). Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 368: 407–415. [DOI] [PubMed] [Google Scholar]
- Verdam FJ, Fuentes S, de Jonge C, Zoetendal EG, Erbil R, Greve JW et al. (2013). Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity (Silver Spring) 21: E607–E615. [DOI] [PubMed] [Google Scholar]
- Vrieze A. (2013) The role of gut microbiota in human metabolism. PhD thesis. Academic Medical Center: Amsterdam. [Google Scholar]
- Vrieze A, de Groot PF, Kootte RS, Knaapen M, van Nood E, Nieuwdorp M. (2013). Fecal transplant: a safe and sustainable clinical therapy for restoring intestinal microbial balance in human disease? Best Pract Res Clin Gastroenterol 27: 127–137. [DOI] [PubMed] [Google Scholar]
- Vrieze A, van Nood E, Holleman F, Salojarvi J, Kootte RS, Bartelsman JF et al. (2012). Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143: 913–916 e7. [DOI] [PubMed] [Google Scholar]
- Wexler HM. (2007). Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev 20: 593–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JM, de Souza R, Kendall CW, Emam A, Jenkins DJ. (2006). Colonic health: fermentation and short chain fatty acids. J Clin Gastroenterol 40: 235–243. [DOI] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen YY, Keilbaugh SA et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334: 105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong E. (2012). Gut microbial ‘enterotypes' become less clear-cut. Nature News & Comment doi:10.1038/nature.2012.10276.
- Zhang F, Luo W, Shi Y, Fan Z, Ji G. (2012). Should we standardize the 1700-year-old fecal microbiota transplantation? Am J Gastroenterol 107: 1755–1755. [DOI] [PubMed] [Google Scholar]
- Zoetendal EG, Raes J, van den Bogert B, Arumugam M, Booijink CC, Troost FJ et al. (2012). The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J 6: 1415–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal EG, Rajilic-Stojanovic M, de Vos WM. (2008). High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57: 1605–1615. [DOI] [PubMed] [Google Scholar]
- Zupancic ML, Cantarel BL, Liu Z, Drabek EF, Ryan KA, Cirimotich S et al. (2012). Analysis of the gut microbiota in the old order amish and its relation to the metabolic syndrome. PLoS One 7: e43052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.