Abstract
Melissococcus plutonius is the causative agent of European foulbrood (EFB), which is a serious brood disease of the European honey bee (Apis mellifera). EFB remains a threat because of a poor understanding of disease epidemiology. We used a recently published multi-locus sequence typing method to characterise 206 M. plutonius isolates recovered from outbreaks in England and Wales over the course of 2 years. We detected 15 different sequence types (STs), which were resolved by eBURST and phylogenetic analysis into three clonal complexes (CCs) 3, 12 and 13. Single and double locus variants within CC3 were the most abundant and widespread genotypes, accounting for 85% of the cases. In contrast, CCs 12 and 13 were rarer and predominantly found in geographical regions of high sampling intensity, consistent with a more recent introduction and localised spread. K-function analysis and interpoint distance tests revealed significant geographical clustering in five common STs, but pointed to different dispersal patterns between STs. We noted that CCs appeared to vary in pathogenicity and that infection caused by the more pathogenic variants is more likely to lead to honey bee colony destruction, as opposed to treatment. The importance of these findings for improving our understanding of disease aetiology and control are discussed.
Keywords: Apis mellifera, European foulbrood, multi-locus sequence typing, insect
Introduction
Recent large-scale colony losses of the honey bee have led to concerns that the sustainability of agriculture and ecosystem service provided by this key pollinator may be threatened (Gallai et al., 2009). The causes of such losses have been reported as being multifactorial and interacting (Vanbergen et al., 2013); however, fungal (Higes et al., 2006), viral (Cox-Foster et al., 2007; Siede et al., 2008; Martin et al., 2012) and bacterial diseases have been implicated in honey bee colony demise. Despite a clear and immediate threat from pathogens, the epidemiological processes underlying honey bee disease remain poorly understood.
European foulbrood (EFB) is a serious disease of honey bee (Apis mellifera) brood caused by the Gram-positive lanceolate coccus M. plutonius, which can lead to the loss of the host colony (White, 1912; Tarr, 1937; Bailey, 1956; Bailey and Collins, 1982; Forsgren et al., 2013). The bacterium is widely distributed (Bailey, 1981) and can also infect and kill the brood of the Eastern honey bee (Apis cerana) and the Himalayan honey bee (Apis laboriosa; Bailey, 1974; Allen et al., 1990). Although early reports suggested a highly genetically homogenous population (Djordjevic et al., 1999), isolates of M. plutonius have been shown to vary in biochemical, serological and in vitro virulence characteristics (Allen and Ball, 1993; Arai et al., 2012; Takamatsu et al., 2013).
Recent decades have seen dramatic increases in the incidence of EFB in both the United Kingdom and Switzerland (Wilkins et al., 2007; Roetschi et al., 2008) and large disease outbreaks have been discovered in areas previously thought to be disease free, such as Norway (Dahle et al., 2011). Regional variations in disease burden have been reported (Forsgren et al., 2005; Budge et al., 2010), and although methods exist to distinguish between systematic patterns of disease and spatial randomness (Besag, 1977; Ripley, 1977), the spatial dependence of EFB has not been formally tested. Confirmation of spatial clustering can lead to important insight into abiotic and biotic risk factors, which may contribute to disease occurrence (for example, see Spósito et al., 2007; Stevens et al., 2009). As yet, little is known about the underlying population structure of M. plutonius that could explain the observed spatial patterns of distribution and many aspects of disease aetiology remain enigmatic (Forsgren et al., 2013).
Multi-locus sequence typing (MLST) is a robust way of distinguishing between genetic variants of a pathogen (Killgore et al., 2008), and concomitantly has been used to unravel the epidemiology of numerous disease systems (for example, Dingle et al., 2002; Ruiz-Garbajosa et al., 2006; Freitas et al., 2009; Lowder et al., 2009). Recently, a modified MLST scheme was developed for M. plutonius, which provided the first insight into the distribution of sequence types (STs) in a limited set of isolates, and disease transmission for some defined case studies (Haynes et al., 2013). However, the distribution of STs at the landscape level remains unknown, and it is unclear whether molecular data reflects underlying phenotypic variation of relevance to disease progression. Here we address this by applying MLST to 206 isolates of M. plutonius sampled from disease cases across England and Wales over the course of 2 years.
Materials and methods
Field sampling and pathogen characterisation
Cases of EFB were identified as part of the government-funded control programme operated by the National Bee Unit and conducted to Good Laboratory Practise. Trained appointed bee inspectors identified colony level symptoms (such as a patchy and erratic brood pattern, sometimes accompanied by an unpleasant odour) and larval symptoms (such as displacement in the cell with a melted appearance). A single symptomatic larva was tested for each honey bee colony with foulbrood symptoms using an EFB Diagnostic Kit following manufacturer's instructions (Vita Europe, Basingstoke, UK). Each kit contained a highly specific monoclonal antibody for M. plutonius incorporated into a Lateral Flow Device (Tomkies et al., 2009). Honey bee colonies with both visual symptoms and a positive result from the Lateral Flow Device were deemed to be positive EFB cases.
Historically, EFB cases are more frequently found in Norfolk, Suffolk and the South of England, when compared with Wales and the North of England (Budge et al., 2010). To investigate these spatial patterns, England and Wales were divided into geographical regions with low (Wales: 0.02% prevalence in 2010) and high (Norfolk and Suffolk: 2.7% in 2010) disease intensity. In these geographical regions, sampling was attempted from the majority of disease cases identified from April 2011 to September 2012, whereas in the remainder of England (1.4% prevalence in 2010) cases were sampled less frequently. For each sampled EFB case, M. plutonius was recovered from a single, diseased larva by streaking a 1-μl loop of larval homogenate directly onto Petri dishes containing M110 agar (Forsgren et al., 2013). Each larva was homogenised using a sterile pellet pestle. Plates were stored for 7–14 days under anaerobic conditions at 35±2 °C and suspect bacterial colonies subcultured before being stored at −80 °C using Protect Select Anaerobe bacterial preservation beads (Technical Service Consultants, Heywood, UK). DNA was extracted from M. plutonius colonies using the Promega Wizard Magnetic DNA Purification System for Food following manufacturer's instructions (Promega, Madison, WI, USA) and stored at −20 °C before use. When culturing failed from a diseased larva, DNA was extracted directly from the larva using the method described above.
DNA preparations were subjected to a species-specific real-time PCR test to confirm the presence of M. plutonius as described previously (Budge et al., 2010). A published four-gene MLST scheme was then used to generate sequence information for galK (galactokinase), argE (acetylornithine deacetylase), gbpB (secreted antigen) and purR (purine operon repressor) for all M. plutonius positive extracts (Haynes et al., 2013). Briefly, 2 μl of DNA sample was added to 12.5 μl of Fermentas 2 × PCR Master Mix (Fermentas, Waltham, MA, USA) with 10 pmol of each primer and made up to a final volume of 25 μl. Each reaction was run on an Applied Biosystems 2720 Thermal Cycler (Applied Biosystems, Grand Island, NY, USA) at 94 °C for 2 min followed by 35 cycles of 94 °C for 30 s, 50 °C for 30 s and 72 °C for 1 min with a final elongation step at 72 °C for 1 min. PCR products were visualised on a 2% agarose gel using Fermentas 6 × DNA Loading Dye, and Bioline HyperLadder 100 bp (Bioline, London, UK). Products of the correct size (579 nt argE; 565 nt galK; 386–632 nt gbpB; 507 nt purR) were purified using the QIAquick PCR purification kit (QIAGEN, Valencia, CA, USA) and sequenced using the Eurofins MWG Operon (Eurofins, Ebersberg, Germany) Sanger sequencing service (Haynes et al., 2013). Alleles and designations were called using the MLST module in Bionumerics version 7.0 (Applied Maths, Sint-Martens-Latem, Belgium).
Population diversity
Potential patterns of evolutionary descent between STs were calculated using the goeBURST algorithm in the PHYLOViZ program (Francisco et al., 2012), a modification of the earlier eBURST algorithm (Feil et al., 2004). The rules of this algorithm, which was designed specifically for exploring MLST data, are fully described by Feil et al. (2004). Briefly, the data set in first subdivided into groups (‘clonal complexes' (CCs)) based on a user-defined threshold level of identity (number of allelic mismatches). In the current study, we used the most conservative approach and defined a CC using goeBURST on the basis that each ST within a CC must share at least three out of four identical alleles with at least one other ST in the group (single locus variants, SLVs). Singletons, defined as those STs differing by at least two alleles from all other STs, were then assigned to CCs on the basis of differing by no more than two alleles with another ST (double locus variants). A ‘founder' is then identified for each CC as the ST which defines the highest number of near neighbours (SLVs), and the founder is then linked to these near neighbours. Remaining STs in the group are then linked as an iterative process to the ST defining the next highest number of SLVs.
A phylogeny was constructed from an alignment of 1914 nucleotide positions representing the concatenated sequences of each ST. The sequences were aligned using ClustalW, as implemented in MEGA 5.2 (Tamura et al., 2011). The phylogenetic analysis was then carried out using the Neighbour-Joining method and the Maximum Composite Likelihood model as implemented in MEGA 5.2. Gaps in the alignment was treated by pairwise deletion, and the confidence of the nodes was gauged using the interior-branch test (500 replications).
Chao1 is a non-parametric estimate of diversity that is particularly suited to microbial studies and allows a comparison of total ST diversity between geographical regions (Hughes et al., 2001). Chao1 estimates of total ST diversity (with corresponding 95% confidence intervals) were calculated for each geographical region using bias-corrected formulae as implemented using the default settings in EstimateS (Chao, 1987; Colwell, 2013).
Population structure
The average geographical distance between all pairs of apiaries sharing a ST was calculated as the interpoint distance (Cuzick and Edwards, 1990). To determine significance, a random sample of the same number of apiaries was taken from the entire data set (that is, those who do not share the same ST), and the interpoint distance was recalculated. This was repeated 500 times and the significance determined by the rank of the test statistic among the random permutations. If ⩾95% of the randomly permuted clusters have an interpoint distance greater than that observed for a given ST, it can be said that apiaries exhibiting that ST are significantly closer together than would be expected from a random sample.
The presence of significant spatial clusters for the most common STs were detected by calculating the K-function 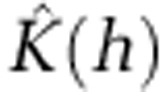 , which is an expectation of the number of events within distance h of an arbitrary event (Ripley, 1977, 1979). To help interpretation of
, which is an expectation of the number of events within distance h of an arbitrary event (Ripley, 1977, 1979). To help interpretation of 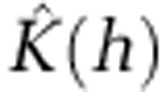 , we subtracted the expectation of the number of events within distance h under the assumption of complete spatial randomness to give
, we subtracted the expectation of the number of events within distance h under the assumption of complete spatial randomness to give 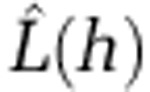 , which when plotted against h has peaks in positive values where clustering occurs (Besag, 1977). Values of zero suggest spatial randomness. Formal assessment of the significance of the observed peaks requires knowledge of the distribution of
, which when plotted against h has peaks in positive values where clustering occurs (Besag, 1977). Values of zero suggest spatial randomness. Formal assessment of the significance of the observed peaks requires knowledge of the distribution of 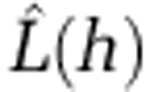 under complete spatial randomness, which is unknown for our data. Instead, upper and lower simulation envelopes were calculated from m independent simulations of n events under the assumption of complete spatial randomness. Observed values of
under complete spatial randomness, which is unknown for our data. Instead, upper and lower simulation envelopes were calculated from m independent simulations of n events under the assumption of complete spatial randomness. Observed values of 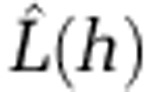 above these simulation envelopes indicate significant clustering (Bailey and Gatrell, 1995).
above these simulation envelopes indicate significant clustering (Bailey and Gatrell, 1995).
Field pathology and honey bee colony fate
EFB is under statutory control in England and Wales and the decision to treat or destroy is primarily based on the level of disease within the colony (Wilkins et al., 2007). If the honey bee colony is heavily diseased (more than 50% of brood frames diseased), then the colony is destroyed; honey bee colonies with less disease can be treated using antibiotics or a husbandry method (Budge et al., 2010). The difference in proportion of honey bee colonies destroyed (as opposed to being treated) was compared between CCs of M. plutonius using a generalised linear model assuming a Bernoulli distribution with a Logit link function (Genstat version 15.1; VSN International Ltd, Hemel Hempstead, UK). Finally, a single brood frame from each sampled EFB case was assessed visually, and the proportion of larvae that appeared to be diseased noted. Differences in the proportion of diseased larvae between CCs of M. plutonius were analysed using Kruskal–Wallis and Mann–Whitney U-tests as appropriate (Genstat version 15.1).
Results
EFB prevalence differed between geographical regions, with six times more disease discovered in Norfolk and Suffolk compared with the region with the lowest prevalence, Wales (Table 1). The number of MLST profiles generated was higher in focal areas with historically high (Norfolk/Suffolk) and low (Wales) disease prevalence compared with the rest of England (Table 1). MLST profiles were obtained for 206 EFB cases across England and Wales from April 2011 to September 2012 (Supplementary Table 1). Allelic profiles demonstrated that EFB in England and Wales between 2011 and 2012 was caused by at least 15 different STs of M. plutonius (Supplementary Table 1). Four new STs were discovered in the current study (STs 21, 22, 23, 24; Supplementary Table 2), compared with those reported previously (Haynes et al., 2013). Some STs were found more frequently (ST2=12; ST3=60; ST5=56; ST13=19; ST23=29) than other rarer STs (ST1=1; ST6=1; ST7=5; ST8=2; ST11=6; ST12=1; ST20=2; ST21=6; ST22=4; ST24=2). After an initial increase, the mean Chao1 estimate for all geographical regions becames relatively level as sample size increased (Supplementary Figure 1), therefore we compared the ST diversity estimates at the highest sample size for each geographical region (Hughes et al., 2001). Chao1 estimates, and corresponding 95% confidence intervals, indicated that ST diversity was significantly lower in Wales (4.00; 95% CIs=4.00, 4.85) than Norfolk/Suffolk (8.48; 95% CIs=7.15, 21.92) or the rest of England (11.48; 95% CIs=10.15, 24.92).
Table 1. EFB prevalence and sampling effort for focal regions of Norfolk and Suffolk, Wales, the remainder of England (England other).
|
Disease prevalence |
Sampling effort |
||||
|---|---|---|---|---|---|
| Region | # Colonies inspected | # EFB cases | Proportion of EFB cases | # EFB cases sampled | Proportion of cases sampled |
| Nor/Suf | 5255 | 208 | 0.040 | 81 | 0.39 |
| Wales | 11 128 | 76 | 0.007 | 40 | 0.53 |
| England other | 56 444 | 1359 | 0.024 | 85 | 0.06 |
Abbreviations: EFB, European foulbrood; MLST, multi-locus sequence typing; Nor/Suf, Norfolk and Suffolk.
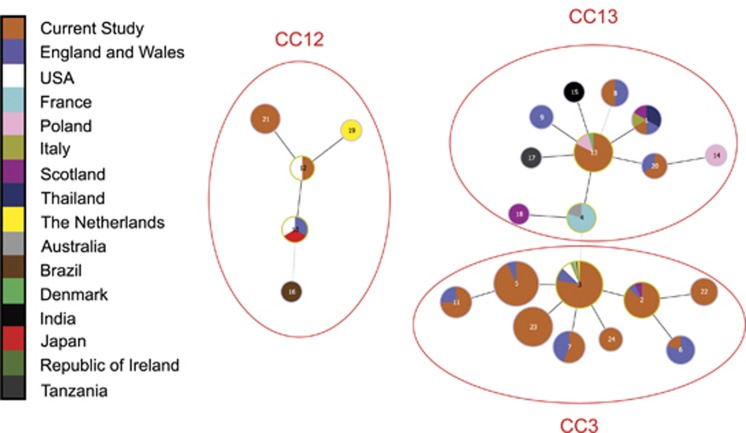
goeBURST supported the presence of three CCs in England and Wales (Figure 1). CC3 and CC13 comprised SLVs and double locus variants from respective founder STs, whereas members of CC12 were characterised by the presence of argE allele 4, which was not noted in any STs belonging to other CCs (Figure 1 and Supplementary Table 2).
Figure 1.
goeBURST diagram using MLST data from the current study (orange) added to those reported previously (Haynes et al., 2013) to show three clonal complexes (CC3, CC12 and CC13) occurring in England and Wales. Each circle represents a different ST, with lines linking closest relatives. Black lines indicate a single allelic change between STs and grey lines indicate differences at two loci. Larger circles indicate more frequent occurrence and those ringed with a yellow outline indicate putative founder genotypes. Colours within circles show the proportion of isolates of a particular type that were found in the countries indicated in the key. Note that all nine CC3 variants, including four newly characterised STs (21, 22, 23, 24), have been discovered in England and Wales.
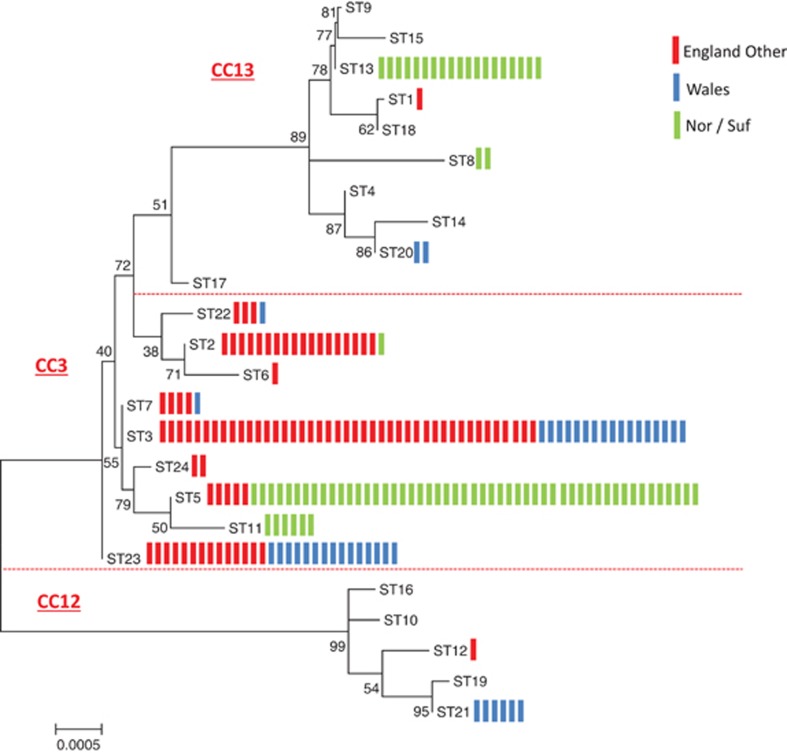
The phylogenetic tree reconstructed from the concatenated allele sequences of all known M. plutonius STs provided groupings that were highly consistent with the goeBURST analysis (Figure 2). The only apparent discrepancy is the positioning of ST17, which is an SLV of ST13 but appears closer to CC3 than to CC13. This is due to the fact that ST17 harbours galK allele 3, which is commonly found in CC3, rather than galK allele 1, which is the common allele in CC13 (Supplementary Table 2). These two alleles differ at three nucleotide positions, hence the presence of galK allele 3 in ST17 is not most parsimoniously explained through identity by descent but by recombination. In other words, this recombination event confounds the phylogenetic signal resulting in the close proximity of ST17 to CC3 rather than CC13.
Figure 2.
Unrooted Neighbour-Joining phylogeny constructed from a 1914-bp concatenated alignment from all four MLST alleles. Bootstrap scores (computed using the internal-branch test) are given on the internal branches. Horizontal red lines demarcate the three clonal complexes (CCs3, CC12 and CC13) as defined by goeBURST (Figure 1). Individual EFB cases characterised in the current study are represented by coloured bars aligned to the corresponding ST; green=Norfolk/Suffolk (Nor/Suf), blue=Wales, red=England (other). STs with no coloured bars represent those reported in the previous study of Haynes et al. (2013) but not in the current study.
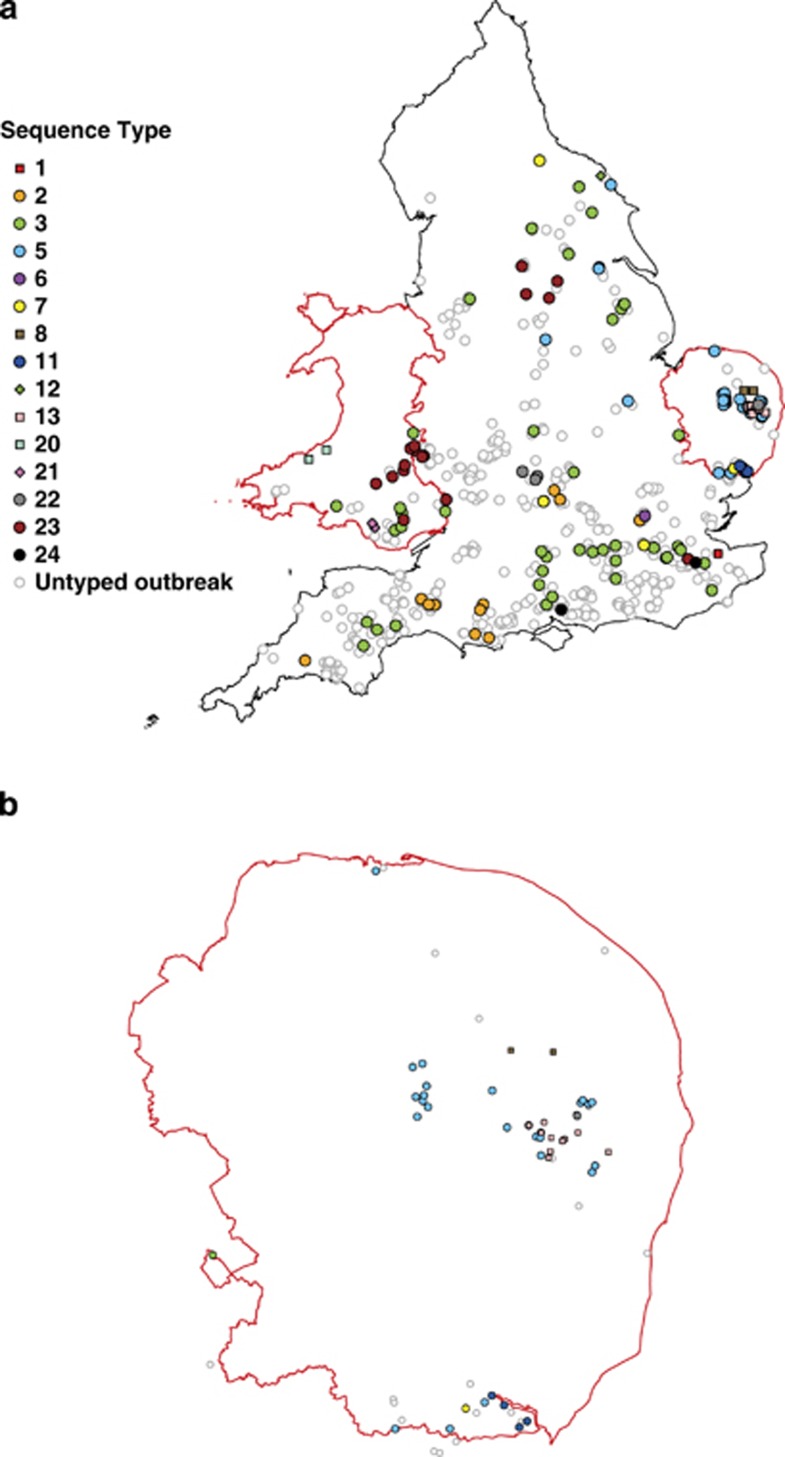
CC3 contains the highest number of STs, and is also most equally distributed between all three geographical regions. In contrast, CC13 was predominantly represented by ST13, which was exclusively found in Norfolk/Suffolk. Similarly, CC12 was predominantly represented by ST21, which was found exclusively in Wales. There is also evidence for crude geographical structuring within CC3. STs 5 and 11 are closely related, and are predominantly found in Norfolk/Suffolk, whereas ST3 and ST23 are common STs in Wales and the rest of England but were absent from Norfolk/Suffolk (Figures 2 and 3).
Figure 3.
Geographic distribution of EFB outbreaks between 2011 and 2012 across (a) England and Wales and (b) Norfolk and Suffolk. Coloured markers indicate the sequence type as derived using a four-gene MLST. Open circles indicate untyped EFB cases. Clonal complexes are depicted using circles (CC3), diamonds (CC12) or squares (CC13). Red borders indicate areas of intensive sampling.
There was insufficient spread in the data to formally test for temporal trends in ST occurrence. However, a summary showing frequency of detection over time indicates no obvious pattern, with common STs being found throughout the beekeeping season (Supplementary Figure 2).
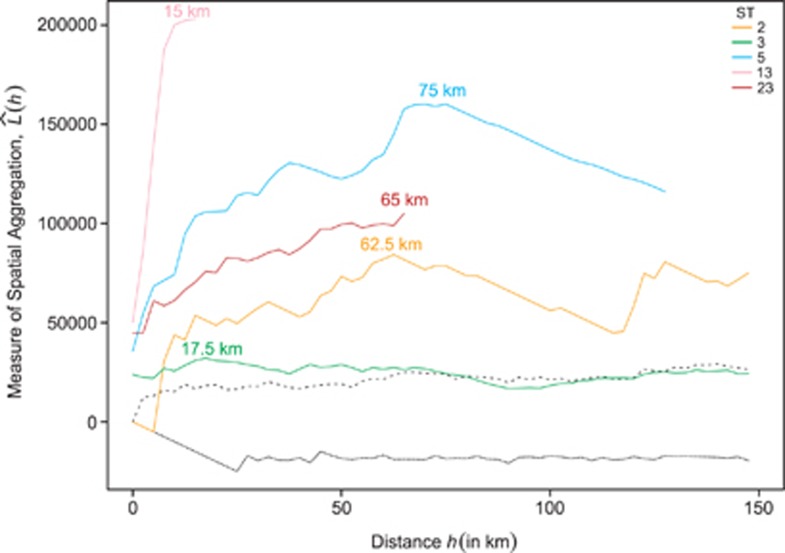
Only those STs found in 10 or more apiaries (that is, ST2, 3, 5, 13, 23) were subjected to further analysis to formally test for spatial clustering. Interpoint distance tests indicated cases were significantly closer together than expected by chance for all STs (ST2: P=0.018, ST3: P=0.038, ST5: P<0.01, ST13: P<0.01 and ST23: P<0.01; Supplementary Figure 3). The average distance between pairs of cases caused by some common STs was over 100 km, suggesting wide-spread dispersal (ST2=135 km; ST3=175 km; ST23=111 km), whereas other STs appeared to have more local distributions (ST5=57 km; ST13=5.5 km; Figure 3 and Supplementary Figure 3).
K-function analysis provided evidence that STs varied in the scale of cluster formation. ST3 and ST13 clustered significantly at much smaller distances (17.5 and 15 km, respectively) than three other common STs (Figure 4). These results suggest that ST3 is widespread across England and Wales, but also forms local clusters, whereas ST13 forms local clusters but is also mostly found in one small region (Figures 3 and 4).
Figure 4.
Estimated number of disease cases within distance h (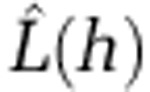 ) for five common STs. The distance (h) at which the maximum amount of clustering is observed for each ST is indicated at the peak value of
) for five common STs. The distance (h) at which the maximum amount of clustering is observed for each ST is indicated at the peak value of 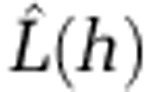 for each ST. Broken lines indicate the simulation envelopes above which significant clustering occurs.
for each ST. Broken lines indicate the simulation envelopes above which significant clustering occurs.
The Bernoulli GLM suggested significant differences in the proportion of honey bee colonies destroyed between M. plutonius CCs (deviation=6.6; df=2; P=0.037). Paired comparisons suggested that honey bee colonies infected with CC3 (0.43; 95% CIs=0.35, 0.50) were more likely to be destroyed, because they contained more diseased brood frames, than those infected with CC13 (0.17; 95% CIs=0.06, 0.37; P=0.021). However, no significant differences were observed in honey bee colony destruction between CC3 and CC12 (0.43; 95% CIs=0.14, 0.77; P=0.62) or between CC12 and CC13 (P=0.46).
The Kruskall–Wallis test suggested significant differences in the proportion of diseased brood between CCs of M. plutonius (H=6.83; df=2; P=0.033), with CC3 providing the highest disease ranking (105.7) followed by CC12 (93.3) and CC13 (73.38). Paired comparisons using Mann–Whitney U-tests suggested a significantly higher proportion of larvae were diseased on brood frames from honey bee colonies infected with CC3 compared with CC13 (P=0.010), but no significant differences were observed between CC3 and CC12 (P=0.62) or between CC12 and CC13 (P=0.46).
Discussion
EFB is a damaging brood disease of a key insect pollinator, which has been under statutory control in the United Kingdome for over 70 years. Despite this, our understanding of population structure and diversity of the causal pathogen has been virtually nonexistent. Here we describe the first large-scale study of M. plutonius diversity on a national scale. We note the presence of 15 distinct STs across England and Wales, which resolve into three CCs based on goeBURST or phylogenetic analysis (CC3, CC12 and CC13), with the exception of the position of ST17 and some internal rearrangement of the relationship of ST4 within CC13. STs from CC3 account for 85% (n=171) of the isolates, with CC12 and CC13 being rare and largely restricted to Wales and Norfolk/Suffolk, respectively (Figures 2 and 3). Interestingly, when all previously characterised STs were considered, including isolates from 14 other countries (Haynes et al., 2013), all nine STs from CC3 were observed in this study. In contrast, the majority of known STs of CC12 (3/5) and CC13 (6/10) were not detected (Figure 1). These patterns suggest that CC3 is an established, endemic complex in England and Wales, whereas CC13 and CC12 may have been introduced more recently and remain geographically restricted. There have been ample opportunities for introductions of new variants of EFB into the United Kingdom through intercontinental trading in honey bees and their hive products (De La Rua et al., 2009). For example, the United Kingdom imports 85% of its honey (USAID, 2012), and honey is known to harbour live M. plutonius (McKee et al., 2003). Assessing the risk posed by the UK honey importation industry by the introduction of new variants of M. plutonius is an interesting question that would require targeted sampling efforts around areas designated as high risk, such as honey packing plants.
Interpoint distance tests and K-function analyses provided strong evidence of highly local clustering of some but not all common STs (Figure 4). The honey bee behaviours of drifting (where adult bees move between honey bee colonies) and robbing (where adult bees steal honey from weaker honey bee colonies) might be important for local transmission of EFB (Forsgren, 2010). Beekeeping practices may also contribute to the sometimes rapid local spread of EFB (Forsgren, 2010). A positive correlation has been described between the proportion of workers infected in symptomless honey bee colonies and the distance to hives with EFB symptoms (Belloy et al., 2007). Indeed, human movements of bees and their diseases alone may account for the regional or national movements observed for ST2, ST3 and ST23, which had average interpoint distances over 100 km, far beyond double the maximum recorded flying distance of a honey bee (13.5 km; Eckert, 1933). Disease movements of a similar scale (between 54 and 84 km) have been characterised recently after honey bees carrying M. plutonius were sold between beekeepers in England (Haynes et al., 2013). Differences in long-range dispersal of STs could reflect different infection behaviours or simply the fact that transmission through commercial activity is a rare event. MLST schemes coupled to clustering tests have successfully been used to confirm transmission routes in other livestock diseases (for example, Olea-popelka et al., 2005). Our results indicate that the epidemiology of EFB is influenced by transmission events that arise at different spatial scales because of combination of host and human behaviours.
Our data provide the first evidence that M. plutonius from different CCs may differ in their virulence at both brood frame and honey bee colony levels, and that severe cases of disease, which ultimately results in colony destruction, might be correlated with particular CCs. MLST schemes have previously provided the additional resolution necessary to observe differences in virulence among genetically distinct STs from within a single species (for example, King et al., 2002). M. plutonius virulence is known to reduce rapidly after culture (Bailey, 1963), and the rapidity of attenuation has been shown to differ between M. plutonius cultures (Arai et al., 2012). Our data represent the first to compare direct observations of virulence from the field, which given the problems of attenuation in culture, are likely the most realistic measure of pathogen behaviour. Intensive regional sampling of the pathogen population revealed that a geographical region with high disease prevalence (Norfolk/Suffolk; 4%) contained different STs and higher pathogen diversity than a region with low disease prevalence (Wales; 0.7% Figures 2, 3 and Supplementary Figure 1). EFB prevalence has been previously shown to vary regionally (Budge et al., 2010) leading to the suggestion that disease development is likely a product of local climate and host susceptibility (Forsgren, 2010). Our data on virulence and regional variation suggest that pathogen variation could make a significant contribution towards explaining regional, national and international variation in disease impact. Some reports suggest the native UK honey bee race (A. m. mellifera) is more susceptible to EFB than commonly imported genotypes such as A. m. ligustica (VanEngelsdorp and Meixner, 2010). Although importation is not regionally focussed, highlighted by frequent A. m. ligustica introgression at different locations (Jensen et al., 2005), host susceptibility provided by more subtle genetic signals could be an important driver of local EFB epidemiology. The majority of observations of CC13 were from Norfolk and Suffolk, so it is important to consider that the observed differences in pathogenicity between CCs could be in part due to regional biotic and abiotic factors mediating disease expression. One such abiotic factor could be rainfall, which tends to be higher in Wales than Norfolk and Suffolk because of greater altitude and exposure to the influence of the Atlantic Ocean. Therefore, although our data indicate that pathogen genetics are a likely driver of local EFB epidemiology, local abiotic factors such as climate, and biotic factors such as host susceptibility were not accounted for here and cannot be ruled out.
Our study offers a greater sampling intensity than any previous study of EFB, resulting in the discovery of four new STs (STs 21, 22, 23 and 24), with one new allele at the galK locus and three at gbpB. In addition, two STs (ST12 and ST13) were found in England and Wales for the first time, having previously been isolated from the United States of America, and Denmark and Poland, respectively (Haynes et al., 2013). An ST notable for its absence in England and Wales during 2011 and 2012 is ST9, which was involved in a movement of disease between beekeepers in Norfolk in 2010 (Haynes et al., 2013). ST9 differed from ST13 (a type circulating locally in Norfolk) by a length variation in the variable number tandem repeat at gbpB. Variable number tandem repeats have been found to show rapid variation in even the most genetically homogenous bacteria (Jackson et al., 1997), and so ST9 may represent a recently evolved type of M. plutonius. During the 2 years following the outbreak, we found no further ST9, either in Norfolk/Suffolk or in the rest of England and Wales. As the honey bee colonies of both beekeepers remained free of EFB, our data suggest that ST9 may have recently evolved and was since eradicated.
Our data provide important context to the species level diversity of M. plutonius. CC12 corresponds to a type previously identified as ‘atypical' M. plutonius (Arai et al., 2012), as it contains the Japanese atypical isolate DAT561 (ST10). Atypical M. plutonius has been shown to have less stringent culturing requirements than the type strain and other ‘typical' M. plutonius isolates (Arai et al., 2012), and has been identified previously in Japan (Arai et al., 2012), the United States of America, the United Kingdom, the Netherlands and Brazil (Haynes et al., 2013). Our results confirm first that the atypical type is more widespread than previously thought, as two STs (ST12 and ST21 both placed within CC12) were found for the first time in England and Wales, and second that atypical M. plutonius remains differentiated from the other, typical M. plutonius despite the increased coverage of species diversity. CC3 and CC13 belong to the typical M. plutonius subtype, with the type strain (ST1) being found within CC13. The interesting exception is ST17, which is a member of CC13 by goeBURST but is phylogenetically close to CC3 because of the presence of a galK allele otherwise found solely in CC3. We confirmed this result twice to avoid sequencing error. If, as seems likely, this is due to recombination between an ST from CC3 and an ST from CC13, this would be the first example of such an event in M. plutonius. If the recombination event did occur in the environment it would mean that, as M. plutonius can only grow in the honey bee larval midgut (Forsgren, 2010), at some point a single bee larva must have been simultaneously infected with two STs. Such an event would result in poor quality sequence when obtaining MLST data directly from a larva. Haynes et al. (2013) reported unambiguous MLST data obtained directly from three larvae, and we report a further seven such results (Supplementary Table 1). Although this does not rule out co-infection of multiple STs within a larva, it appears unlikely that coinfection with multiple STs is a common occurrence in England and Wales. The only representative of ST17 came from a culture collection, after originally being isolated in Tanzania (NCFB 2441). As this isolate has spent some years in culture, it is impossible to say if the recombination event occurred naturally in the environment or subsequently in the laboratory post-collection. The evidence that recombination occurs at all in M. plutonius is a novel and important finding. However, further work is required to resolve the uncertainty surrounding the mechanism of introduction of multiple STs into a single larva and the frequency of events in countries outside the United Kingdom, where higher disease prevalence could increase likelihood of such multiple infections.
We have expanded the known diversity of M. plutonius and provided evidence of multiple CCs operating at different spatial scales across England and Wales. The ability of our data to distinguish between genetically related pathogens within a bacterial species and to formally test for spatial clustering are fundamental to understanding disease epidemiology. The fact that significant structure exists in the pathogen population of a highly mobile pollinator that has been repeatedly exposed to anthropological movements is remarkable. However, significant data gaps remain to understand finer scale, individual and honey bee colony level transmission events. Future developments in whole-genome sequencing could contribute to improved resolution of transmission events and would allow an assessment of the rapidity of evolution in M. plutonius. Our data provide evidence that CCs may differ in their pathogenicity, a noteworthy observation given low levels of genome-wide variation in this peculiar bacterium (Haynes et al., 2013). However, an improved understanding of traits such as latency and infectivity are key to improving the current control regime and to appreciate how current management strategies might influence the distribution and diversity of M. plutonius. The global industry in honey bees and their hive products requires a global context. To this end, we have developed a public MLST database (http://pubmlst.org/) to store and manage international efforts to type M. plutonius and provide insight into the global epidemiology of this important pathogen (Jolley and Maiden, 2010). We offer a national perspective and urge other countries to provide similar data to obtain an international picture of disease movements. Finally, honey bee pathology contains many examples where pathogens, including viruses (for example, see Cox-Foster et al., 2007) fungi (for example, see Higes et al., 2006) and bacteria (for example, see Forsgren, 2010), apparently cause radically different impacts in different territories. Detailed knowledge of pathogen diversity and the biogeography of genetically related sub-types, as demonstrated in the current study, could offer insight to help explain these divergent trans-national observations of pathogen impact.
Acknowledgments
This work was funded jointly by a grant from BBSRC, Defra, NERC, the Scottish Government and the Wellcome Trust, under the Insect Pollinator Initiative (Grants BB/I000801/1; BB/I000518/1; BB/I000836/1). In addition, EGH was funded by a BBSRC CASE studentship in partnership with Bee Disease Insurance and the National Bee Unit. We acknowledge advice on statistics from Dr Stéphane Pietravalle (Fera).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allen MF, Ball BV, Underwood BA. (1990). An isolate of Melissococcus pluton from Apis laboriosa. J Invertebr Pathol 55: 439–440. [Google Scholar]
- Allen MF, Ball BV. (1993). The cultural characteristics and serological relationships of isolates of Melissococcus pluton. J Apic Res 32: 80–88. [Google Scholar]
- Arai R, Tominaga K, Wu M, Okura M, Ito K, Okamura N et al. (2012). Diversity of Melissococcus plutonius from honeybee larvae in japan and experimental reproduction of european foulbrood with cultured atypical isolates. PLoS One 7: e33708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey L. (1956). Ætiology of European foul brood; a disease of the larval honey-bee. Nature 178: 1130. [Google Scholar]
- Bailey L. (1974). An unusual type of Streptococcus pluton from the eastern hive bee. J Invertebr Pathol 23: 246–247. [DOI] [PubMed] [Google Scholar]
- Bailey L. (1981) Honey Bee Pathology 1st edn. Academic Press: London. [Google Scholar]
- Bailey L. (1963). The pathogenicity for honey-bee larvae of microorganisms associated with European foulbrood. J Insect Pathol 5: 198–205. [Google Scholar]
- Bailey L, Collins MD. (1982). Reclassification of ‘Streptococcus pluton' (White) in a new genus Melissococcus, as Melissococcus pluton nom. rev.; comb. nov. J Appl Bacteriol 53: 215–217. [Google Scholar]
- Bailey TC, Gatrell AC. (1995). Introductory methods for point patterns. In:Interactive Spatial Data Analysis. Prentice Hall: Harlow. [Google Scholar]
- Belloy L, Imdorf A, Fries I, Forsgren E, Berthoud H, Kuhn R et al. (2007). Spatial distribution of Melissococcus plutonius in adult honey bees collected from apiaries and colonies with and without symptoms of European foulbrood. Apidologie 38: 136–140. [Google Scholar]
- Besag JE. (1977). Comments on Ripley's paper. J R Stat Soc B 39: 193–195. [Google Scholar]
- Budge GE, Barrett B, Jones B, Pietravalle S, Marris G, Chantawannakul P et al. (2010). The occurrence of Melissococcus plutonius in healthy colonies of Apis mellifera and the efficacy of European foulbrood control measures. J Invertebr Pathol 105: 164–170. [DOI] [PubMed] [Google Scholar]
- Chao A. (1987). Estimating the population size for capture-recapture data with unequal catchability. Biometrics 43: 783–791. [PubMed] [Google Scholar]
- Cox-Foster D, Conlan S, Holmes EC, Palacios G, Evans JD, Moran NA et al. (2007). A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318: 283–287. [DOI] [PubMed] [Google Scholar]
- Colwell RK. (2013) , EstimateS: Statistical estimation of species richness and shared species from samples. Version 9. Persistent URL: http://purl.oclc.org/estimates.
- Cuzick J, Edwards R. (1990). Spatial clustering for inhomogeneous populations. J R Stat Soc B 52: 73–104. [Google Scholar]
- Dahle B, Sørum H, Weidemann J. (2011) European foulbrood in Norway: How to deal with a major outbreak after 30 years absence. In:Proceedings of the COLOSS Workshop: The Future of Brood Disease Research – Guidelines, Methods and Development: Copenhagen, p 8. [Google Scholar]
- Dingle K, Colles F, Ure R, Bolton F, Maiden M. (2002). Molecular characterization of Campylobacter jejuni clones: a basis for epidemiological investigation. Emerg Infect Dis 8: 949–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djordjevic SP, Smith LA, Forbes WA, Hornitzky MA. (1999). Geographically diverse Australian isolates of Melissococcus pluton exhibit minimal genotypic diversity by restriction endonuclease analysis. FEMS Microbiol Lett 173: 311–318. [DOI] [PubMed] [Google Scholar]
- Eckert JE. (1933). The flight range of the honeybee. J Agric Res 47: 257–285. [Google Scholar]
- Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. (2004). eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol 186: 1518–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsgren E. (2010). European foulbrood in honey bees. J Invertebr Pathol 103: S5–S9. [DOI] [PubMed] [Google Scholar]
- Forsgren E, Budge GE, Charrière J-D, Hornitzky MAZ. (2013). Standard methods for European foulbrood research. J Apic Res 52: 1–14. [Google Scholar]
- Forsgren E, Lundhagen AC, Imdorf A, Fries I. (2005). Distribution of Melissococcus plutonius in honeybee colonies with and without symptoms of European foulbrood. Microb Ecol 50: 369–374. [DOI] [PubMed] [Google Scholar]
- Francisco AP, Vaz C, Monteiro PT, Melo-Cristino J, Ramirez M, Carriço JA. (2012). PHYLOViZ: phylogenetic inference and data visualization for sequence based typing methods. BMC Bioinformatics 13: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitas AR, Novais C, Ruiz-Garbajosa P, Coque TM, Peixe L. (2009). Dispersion of multidrug-resistant Enterococcus faecium isolates belonging to major clonal complexes in different portuguese settings. Appl Environ Microbiol 75: 4904–4908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallai N, Salles J-M, Settele J, Vaissière BE. (2009). Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol Econ 68: 810–821. [Google Scholar]
- Haynes E, Helgason T, Young JPW, Thwaites R, Budge GE. (2013). A typing scheme for the honeybee pathogen Melissococcus plutonius allows detection of disease transmission events and a study of the distribution of variants. Environ Microbiol Rep 5: 525–529. [DOI] [PubMed] [Google Scholar]
- Higes M, Martin R, Meana A. (2006). Nosema ceranae, a new microsporidian parasite in honeybees in Europe. J Invertebr Pathol 92: 93–95. [DOI] [PubMed] [Google Scholar]
- Hughes JB, Hellmann JJ, Ricketts TH, Bohannan BKM. (2001). Counting the uncountable: Statistical approaches to estimating microbial diversity. Appl Environ Microbiol 67: 4399–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PJ, Walthers EA, Kalif AS, Richmond KL, Adair DM, Hill KK et al. (1997). Characterization of the variable-number tandem repeats in vrrA from different Bacillus anthracis isolates. Appl Environ Microbiol 63: 1400–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen AB, Palmer KA, Boomsma JJ, Pedersen BV. (2005). Varying degrees of Apis mellifera ligustica introgression in protected populations of the black honeybee, Apis mellifera mellifera, in northwest Europe. Mol Ecol 14: 93–106. [DOI] [PubMed] [Google Scholar]
- Jolley KA, Maiden MC. (2010). BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killgore G, Thompson A, Johnson S, Brazier J, Kuijper E, Pepin J et al. (2008). Comparison of seven techniques for typing international epidemic strains of Clostridium difficile: restriction endonuclease analysis, pulsed-field gel electrophoresis, PCR-ribotyping, multilocus sequence typing, multilocus variable-number tandem-repeat analysis, amplified fragment length polymorphism, and surface layer protein a gene sequence typing. J Clin Microbiol 46: 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S, Leigh J, Heath P. (2002). Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype. J Clin Microbiol 40: 3671–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Rua P, Jaffe R, Dall'Olio R, Munoz I, Serrano J. (2009). Biodiversity, conservation and current threats to European honeybees. Apidologie 40: 263–284. [Google Scholar]
- Lowder BV, Cartwright RA, Guinane CM, Ben Zakour NL, Weinert LA, Conway-Morris A et al. (2009). Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc Natl Acad Sci 106: 19545–19550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Highfield A, Brettell L, Villalobos E, Budge G, Powell M et al. (2012). Global honey bee viral landscape altered by a parasitic mite. Science 336: 1304–1306. [DOI] [PubMed] [Google Scholar]
- McKee B, Djordjevic S, Goodman R, Hornitzky M. (2003). The detection of Melissococcus pluton in honey bees (Apis mellifera) and their products using a hemi-nested PCR. Apidologie 34: 19–27. [Google Scholar]
- Olea-popelka FJ, Flynn O, Costello E, Mcgrath G, Collins JD, O'Keeffe J et al. (2005). Spatial relationship between Mycobacterium bovis strains in cattle and badgers in four areas in Ireland. Prev Vet Med 71: 57–70. [DOI] [PubMed] [Google Scholar]
- Ripley BD. (1977). Modelling spatial patterns. J R Stat Soc B 39: 172–212. [Google Scholar]
- Ripley BD. (1979). Tests of ‘randomness' for spatial point patterns. J R Stat Soc B 41: 368–374. [Google Scholar]
- Roetschi A, Berthoud H, Kuhn R, Imdorf A. (2008). Infection rate based on quantitative real-time PCR of Melissococcus plutonius, the causal agent of European foulbrood, in honeybee colonies before and after apiary sanitation. Apidologie 39: 362–371. [Google Scholar]
- Ruiz-Garbajosa P, Bonten MJM, Robinson DA, Top J, Nallapareddy SR, Torres C et al. (2006). Multilocus sequence typing scheme for Enterococcus faecalis reveals hospital-adapted genetic complexes in a background of high rates of recombination. J Clin Microbiol 44: 2220–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siede R, König M, Büchler R, Failing K, Thiel H. (2008). A real-time PCR based survey on acute bee paralysis virus in German bee colonies. Apidologie 39: 650–661. [Google Scholar]
- Spósito MB, Amorim L, Ribeiro Jr PJ, Bassanezi RB, Krainski ET. (2007). Spatial pattern of trees affected by Black Spot in citrus groves in Brazil. Plant Dis 91: 36–40. [DOI] [PubMed] [Google Scholar]
- Stevens KB, Del Rios Vilas VJ, Guitián J. (2009). Classical sheep scrapie in Great Britain: spatial analysis and identification of environmental and farm-related risk factors. BMC Vet Res 5: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamatsu D, Arai R, Miyoshi-Akiyama T, Okumura K, Okura M, Kirikae T et al. (2013). Identification of mutations involved in the requirement of potassium for growth of typical Melissococcus plutonius strains. Appl Environ Microbiol 79: 3882–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarr HLA. (1937). Studies on European foul brood of bees III. Further experiments on the production of the disease. Ann Appl Biol 24: 614–627. [Google Scholar]
- Tomkies V, Flint J, Johnson G, Waite R, Wilkins S, Danks C et al. (2009). Development and validation of a novel field test kit for European foulbrood. Apidologie 40: 63–72. [Google Scholar]
- USAID (2012) , The world market for honey, Market Survey #1 http://www.fintrac.com/cpanelx_pu/Ethiopia CIAFS/12_06_4949_CIAFS _1 Honey Final Oct 11.pdf.
- Vanbergen A, Adam J,, The Insect Pollinators Initiative (2013). Threats to an ecosystem service: pressures on pollinators. Front Ecol Environ 11: 251–259. [Google Scholar]
- VanEngelsdorp D, Meixner MD. (2010). A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J Invertebr Pathol 103: 80–95. [DOI] [PubMed] [Google Scholar]
- White GF. (1912). The cause of European foul brood. Circ US Dep Agric 157: 1–22. [Google Scholar]
- Wilkins S, Brown MA, Cuthbertson AGS. (2007). The incidence of honey bee pests and diseases in England and Wales. Pest Manag Sci 63: 1062–1068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.