Abstract
The composition of Bartonella infection was explored in wild Gerbillus andersoni rodents and their Synosternus cleopatrae fleas. Rodent blood samples and fleas were collected in two periods (two different seasons; 4 months apart) from juveniles and adult hosts, and their bartonellae lineages were identified by a 454-pyrosequencing analysis targeting a specific Bartonella citrate synthase gene (gltA) fragment. The rate of Bartonella spp. co-infection was estimated and the assemblage and distribution of bartonellae lineages across the samples with respect to ecological and phylogenetic distance similarities were analyzed. Moreover, environmental factors that could explain potential differences between samples were investigated. Out of the 91 bartonellae-positive samples, 89% were found to be co-infected with more than two phylogenetically distant Bartonella genotypes and additional closely related (but distinguishable) variants. These bartonellae lineages were distributed in a non-random manner, and a negative interaction between lineages was discovered. Interestingly, the overall composition of those infections greatly varied among samples. This variability was partially explained by factors, such as type of sample (blood versus fleas), flea sex and period of collection. This investigation sheds light on the patterns of Bartonella infection and the organization of Bartonella lineages in fleas and rodents in nature.
Keywords: Bartonella, co-infection, fleas, gltA, pyrosequencing, rodents
Introduction
Bartonella organisms are fastidious, facultative intracellular Gram-negative bacteria belonging to the alpha-2-Proteobacteria class (Birtles and Raoult, 1996). Bartonellae establish long-term and often subclinical infections in their mammalian reservoirs, as an outcome of an apparent co-evolution between the bacteria and their hosts (Breitschwerdt and Kordick, 2000; Harms and Dehio, 2012). Additionally, the hemotropic lifestyle of bartonellae has facilitated their transmission from one animal to another by bloodsucking arthropod vectors, promoting high prevalence in host populations worldwide (Kosoy et al., 2012). Bartonella spp. seem to be well-adapted to their vectors, not affecting their feeding and reproductive performance, as demonstrated in Xenopsylla ramesis fleas experimentally infected with Bartonella sp. OE 1-1 (Morick et al., 2013b). Thus, mammals and arthropods are important sources of an immense diversity of bartonellae, including animal-associated species, human-associated species as well as zoonotic species (Harms and Dehio, 2012).
Bartonellae infection is highly prevalent in wild rodents and their associated fleas worldwide (Kosoy, 2010). Rodents are considered to be important reservoirs of bartonellae, as they host a great variety of Bartonella species and strains (Birtles et al., 2001; Kosoy et al., 2012). To date, >17 Bartonella species have been isolated from wild rodents (Gundi et al., 2004, 2009; Kosoy, 2010; Sato et al., 2012). Moreover, several studies have reported a wider diversity of Bartonella genotypes that surpass the current classification of Bartonella species (Pretorius et al., 2004; Inoue et al., 2009; Gundi et al., 2012). For instance, Inoue et al. (2009) described 52 novel Bartonella genotypes from small rodents, closely and distantly related to the recognized Bartonella species, based on a citrate synthase gene (gltA) fragment (Inoue et al., 2009). Noteworthy, co-infection with two or more Bartonella species or variants in a single host have been demonstrated in wild rodents (Kosoy et al., 2004; Telfer et al., 2007; Morick et al., 2011). Fleas are believed to be the main vectors of bartonellae in wild rodent populations (Billeter et al., 2008). Although, only a few flea species were experimentally proven as competent vectors (Bown et al., 2004; Morick et al., 2011, 2013a), several Bartonella organisms have been detected in various flea species, suggesting their role as vectors and possibly as reservoirs (Loftis et al., 2006; Morick et al., 2010; Billeter et al., 2011; Deng et al., 2012). As reported in rodents, it has been documented that some fleas can carry more than one Bartonella species or variant (Abbot et al., 2007; Brinkerhoff et al., 2010). Moreover, some whole-bacterial community analyses of fleas, performed by 16S rRNA gene characterization, have shown that Bartonella is one of the most abundant bacterial members of some flea species (Jones et al., 2008; Hawlena et al., 2013). Despite the current awareness of the widespread of bartonellae in rodent populations and their fleas, together with the increasing reports of Bartonella spp. co-infection, current knowledge on the organization of bartonellae lineages within an individual host or vector and among vectors and hosts is still vague. Thus, further studies are required in order to understand the composition of bartonellae infection in hosts and fleas and how the assemblage of these organisms is influenced by evolutionary, temporal, host and vector-related factors.
The fastidious nature of bartonellae and the potential dominance of certain strains over others in mixed infections (in vivo and in vitro) complicate the identification of distinct members in a single host or vector by isolation methods (Vartoukian et al., 2010). In fact, isolation methods together with modern genetic analyses had given only partial answers on the composition of Bartonella spp. co-infections and on the members hosted by a single infected individual (Abbot et al., 2007; Chan and Kosoy, 2010). However, deeper and more feasible tools for the detection of all co-existing members within a mixed infection are required. Accordingly, the 454-pyrosequencing assay enables the detection of individual sequences obtained from the same source (Dowd et al., 2008), and through an adequate target sequence, it allows the characterization of a specific bacterial group. Thus, the citrate synthase gene (gltA) offers a reliable and specific target for distinguishing between closely and distantly related Bartonella lineages in mixed infections, due to its potent discriminatory power (Birtles and Raoult, 1996; La Scola et al., 2003), its stability (as a housekeeping gene), its extensive GenBank database and its single copy presence in the Bartonella genome (Guy et al., 2013).
This study aimed to identify and compare the bartonellae infection composition in wild rodents and their fleas under natural conditions, using a Bartonella-specific 454-pyrosequencing assay. Factors, such as season, host age, flea sex and host flea load, that could potentially influence the assembly of different bartonellae variants within an individual host or vector or the Bartonella-lineage diversity, were investigated.
Materials and methods
Samples collection
Wild Gerbillus andersoni gerbils were caught through a capture re-capture method from the West Negev, Israel (Yevul, Hevel Shalom, 31° 10′ N), in two different periods. The first capture was held on the rodents' reproduction season (May–June 2011; first period) and the second at the end of the summer, where all individuals are non-reproductive adults (September 2011; second period). Sixteen gerbils were captured and re-captured with live Sherman traps (H.B. Sherman, Tallahasse, FL, USA) distributed on 10 plots (1 hectare size/plot; 40 traps per plot). All captured G. andersoni rodents were locally anesthetized with 0.4% benoxinate hydrochloride (Localin, Fischer Pharmaceutical Labs, Tel Aviv, Israel), blood samples were drawn into EDTA tubes from the retro-orbital sinus and all rodents were tagged with Trovan chips (Electronic Identification Devises LTD, East Yorkshire, UK). Blood samples were transported to the laboratory and stored at −20 °C until further analysis. In addition, all animals were weighed and classified as juveniles (body mass of ⩽18 g) or adults (body mass of >18 g), based on the classification described elsewhere (Hawlena et al., 2006). Flea inspection was performed during both collection periods for each rodent. All fleas found on each animal were counted and collected into 70% ethanol tubes and stored at −20 °C until further taxonomic identification, gender identification and DNA extraction. After the procedure, rodents were released at the exact trapping point. In the second collection period, only recaptured rodents (trapped in the first period) were sampled and included in the study.
The study was approved by the Ben-Gurion University committee for the ethical care and use of animals in experiments (authorization number IL-14-03-2011) and by the Israel Nature and National Parks Protection Authority (approval number 2012/38415).
DNA extraction
DNA was extracted from ETDA blood samples using the BiOstic Bacteremia DNA Isolation kit (MoBio, Carlsbad, CA, USA) according to the manufacturer's instructions. In addition, four fleas (two females and two males, when available) at each period, from each rodent, were selected for DNA extraction and screening. DNA was extracted from each individual flea by the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA, USA), based on the supplementary protocol ‘purification of total DNA from ticks' for detection of Borrelia DNA.
Preliminary molecular screening of Bartonella infection
All DNA samples were screened for Bartonella spp. by conventional PCR targeting a fragment of the Bartonella-gltA. Accordingly, an approximate 360 bp gltA-fragment was amplified using primers Bhcs.781p (5′-GGGGACCAGCTCATGGTGG-3′) and Bhcs.1137n (5′-AATGCAAAAAGAACAGTAAACA-3′) (Norman et al., 1995). The gltA-PCR reactions were performed in 25 μl final volume of PCR-Ready High Specificity ready mix (Syntezza Bioscience Ltd, Jerusalem, Israel) containing 1 μl of 10 μM solution of each primer, 21 μl double-distilled water and 2 μl of each extracted tested DNA. The amplification products were obtained by the following protocol: 2 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 30 sec at 54 °C and 1 min at 72 °C and a final step of 5 min at 72 °C. Additionally, a Bartonella DNA extracted from a cultured Bartonella henselae strain (positive control), a Bartonella-negative DNA and a non-template DNA were used as controls in each PCR reaction. The PCR products were run on 1.5% agarose gel and visualized under ultraviolet light to determine any positive amplification. All negative samples were re-run in the PCR assay twice to confirm their negativity.
454-Pyrosequencing analysis of positive Bartonella samples
In order to detect the majority of Bartonella lineages within each infected rodent and flea, the total genomic-DNA from the samples detected positive by the preliminary Bartonella-gltA screening were selected for pyrosequencing analysis. In addition, one gerbil blood sample negative for Bartonella-gltA DNA was selected for pyrosequencing and used as a negative control. A partial gltA-amplicon pyrosequencing assay using primers Bhcs.781p and Bhcs.1137n was performed by a modified protocol of the bacterial Tag-Encoded-FLX Amplicon Pyrosequencing originally described by Dowd et al. (2008). Briftly, a single-step 30-cycle PCR using HotStarTaq Plus Master Mix Kit (Qiagen) was performed under the following conditions: 94 °C for 3 min, followed by 28 cycles of 94 °C for 30 s; 53 °C for 40 s and 72 °C for 1 min and a final elongation step at 72 °C for 5 min. Following PCR, all amplicon products were diluted in equal concentrations and purified using Agencourt Ampure beads (Agencourt Bioscience Corporation, Beverly, MA, USA). Finally, samples were sequenced utilizing FLX sequencing run on a 70 × 75 GS PicoTiterPlate in 454 FLX titanium instruments (Roche, Nutley, NJ, USA), according to the manufacturer's guidelines.
Bioinformatics analysis
All pyrosequencing gltA sequence reads were cleaned by MOTHUR v1.24 (Schloss et al., 2009). In brief, fasta and quality data were extracted from the raw SFF file. Sequences were grouped according to the barcode and primer, allowing one mismatch to the barcode and two mismatches to the primer. De-noising was achieved by MOTHUR's implementation of the AmpliconNoise algorithm (Quince et al., 2011), which removes both 454 sequencing errors and PCR single base errors. Thereafter, all sequences were trimmed to remove the barcode and primer sequences, sequences with homopolymers (for example, AAAA) >8 bp and sequences of <100-bp long. Then, the sequences were aligned, filtered and overlapped with no overhanging or no-data base pairs. Additionally, to further reduce sequencing errors, samples were pre-clustered using MOTHUR's implementation of the algorithm of Huse et al. (2010). In addition, chimeric reads were removed with MOTHUR's implementation of the UChime method (Edgar et al., 2011). Pairwise distances were calculated between all DNA reads, which were subsequently clustered into operational taxonomic units (OTUs) at the 0.04 level (namely, ⩾96.0% similarity, same OTU group), following recommendations from La Scola et al. (2003) for gltA sequences. From each OTU, a representative sequence was obtained. Each OTU representative sequence was identified using BLASTn against the GenBank database (http://www.ncbi.nlm.nih.gov), taking into account the full length of the sequences (100% coverage). OTUs which their representative sequence did not match with a Bartonella origin or with <87% similarity to any Bartonella spp. were removed from the analyses, following the criterion from La Scola et al. (2003) for gltA sequences. Finally, OTUs were arranged in a data matrix where each data point represented the abundance of the particular OTU in the particular sample, in relation to the sampling effort (that is, the number of 454 gltA reads obtained from that sample). In order to measure the effect of low abundant OTUs, which could represent either rare Bartonella variants or remaining artifacts of the 454-pyrosequencing assay, we performed a sensitivity analysis by eliminating each OTU that represented <0.1, 0.5 and 1% of each sample sequences and generated three new data matrices at each level. All matrices were analyzed for statistical significance as described below.
Data and statistical analyses
Ecological and phylogenetic similarities in the infection composition among samples
The composition of Bartonella infection within the samples were examined and analyzed with respect to their ecological similarities and their phylogenetic distance similarities, following Jones et al. (2010). Ecological similarities in infection composition among samples were based on Bray–Curtis similarities, as follows: First, relative abundances were fourth-root transformed so that highly abundant lineages did not dominate the analyses. Then, the ‘OTU-sample' abundance matrix was converted to a ‘sample–sample' distance matrix. Phylogenetic similarities in the infection composition among samples were based on the relative phylogenetic distance of OTUs present in the samples, using either unweighted Fast UniFrac (based only on phylogenetic distance) or weighted Fast UniFrac (based on phylogenetic and ecological distances) analyses (Hamady et al., 2010). The phylogenetic distance among OTUs was based on a Maximum Likelihood phylogenetic tree built with all the OTUs using the MEGA software (Tamura et al., 2011) following the recommendations of Hall (2013).
Statistical analyses for the three approaches were performed by non-parametric multivariate multiple regression linear models using the DISTLM forward software (McArdle and Anderson, 2001; Anderson, 2003). P-values for the conditional tests (used for the forward-selection procedure) were determined using 9999 permutations of residuals under the reduced model (McArdle and Anderson, 2001). The following independent variables were considered in the analyses: The type of sample (blood versus flea), period of collection (first versus second season), the identity of the rodent host, rodent age (juvenile versus adult), and the flea load per host. To further test the effect of the flea sex, the analyses for the flea samples only were repeated. Then, to decrease the potential host individual variability, data from the individuals that had a complete set of positive samples (that is, blood samples and at least one female and one male flea were found to be Bartonellae positive in both periods) was tested. Significant levels were adjusted for multiple tests, using a sequential Bonferroni's correction (Rice, 1989). The overall contribution of individual OTUs to observed differences was explored by SIMPER, an analysis tool within PRIMER-E (Clarke, 1993). The output of this analysis ranks each OTU based on its percentage of contribution to differences between the groups. Finally, to visualize significant patterns, ordination of multivariate data was generated in 3D principal coordinates analysis plots, using Fast Unifrac online application tool (Hamady et al., 2010).
Lineage diversity and occurrence analyses
Lineage diversity analysis was performed to compare the diversity of OTUs among the different sample types, using General Linear Models with forward stepwise procedure. The dependent variable was Fisher's alpha index, which estimates the true lineage diversity after accounting for sample size, based on the assumption that the abundance of lineages follows the log series distribution (Fisher et al., 1943). The independent variables were the type of sample, period of collection, flea sex, rodent age and the flea load per host.
Co-occurrence patterns of OTUs in the samples were evaluated by EcoSim algorithm (Gotelli and Entsminger, 2009), as follows. First, the observed presence/absence pattern of OTUs within the samples was compared against a distribution of 5000 random assemblages of OTUs. The resulting C-scores measure the tendency for lineages to not occur together. Thus, the larger the C-score obtained, the less the average of co-occurrence among lineage pairs, an indication of competition (Steiner et al., 2008). In a similar way, the number of OTU pairs that never co-occur and the number of unique lineage combinations that co-occur in different samples were evaluated by checkerboard lineage-pairs index and by the number of species combination index, respectively. In a competitively structured community, the observed number of checkerboard lineage-pairs is expected to be greater, and lineage combinations are expected to be lower than expected by chance.
Characterization of OTUs and evaluation of Bartonellaco-infection
In order to assess co-infection rates in a more conservative way (in addition to the above OTU-based analyses), the representative sequence of each OTUs was classified according to three levels of identity percentage to the closest genotype match from the GenBank database (that is, ⩾97% high, 94–96% intermediate and<94% low identity). Then, all OTUs that were classified into a particular genotype within the same identity level group were merged and considered as members of the same Bartonella variant. Finally, infection with a single or multiple genotypes or variants was determined in each sample, and co-infection was defined as the detection of two or more distantly related Bartonella variants (that is, genotypes with different species origin) in the samples.
Results
Animals and ectoparasites
Sixteen G. andersoni gerbils were caught in both collection periods (same individuals caught twice) and included in this study. In the first period, they were classified as 8 juveniles and 8 adults. In the second collection period, all gerbils were classified as adults, and 81% (13/16) of them were collected in the same plots of release. The three remaining rodents were trapped at adjustment plots, with a maximum dispersion of up to 70 m from the original trapping point. All rodents were infested with fleas during the entire study, with an average (±s.d.) of 13.4±8.6 fleas per host in the first collection period and 18.4±7.7 fleas per host in the second period (Supplementary Table S1). All fleas were identified as Synosternus cleopatrae by morphological characteristics.
Bartonella gltA-PCR screening of rodents and fleas
In the first collection period, 12 G. andersoni blood samples (7 juveniles and 5 adults) were positive for the Bartonella-gltA using conventional PCR. In the second period, only six of the gerbils were detected positive by the same screening method. From a total of 126 S. cleopatrae fleas (64 males and 62 females) collected from the animals, 71% (26 males and 18 females) from the first collection period and 58% (18 males and 19 females) from the second period were found positive for the Bartonella-gltA by conventional PCR assay. Noteworthy, all Bartonella-infected and non-infected rodents hosted infected and uninfected fleas.
454-Pyrosequencing data analysis
Bartonella partial-gltA sequences were obtained from the 92% (91/99) of the samples by the 454 gltA-pyrosequencing assay. Hence, 77 flea samples and 14 blood samples were successfully sequenced. Of those, four rodent hosts had a complete set of positive samples (that is, blood samples and at least one female and one male flea were detected Bartonellae-positive in both periods). No Bartonella-gltA amplicons were obtained from the negative control.
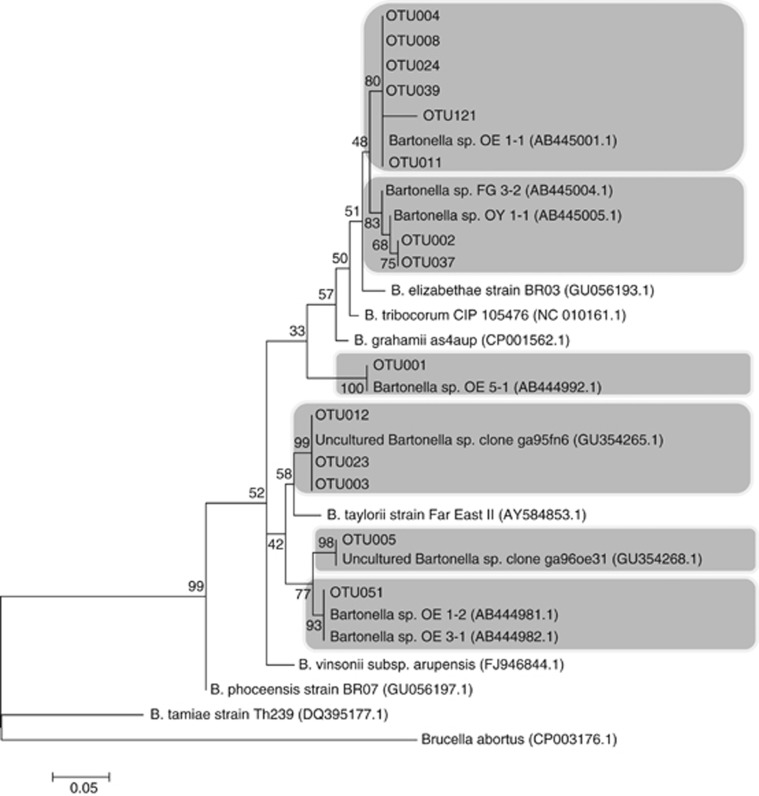
The obtained reads by the pyrosequencing assay had an average length of 250–300 bp. After de-noising, all pyrosequencing reads were successfully overlapped on common fragments of approximately 150 bp (nucleotide position 814–964; based on the gltA gene sequence of Bartonella grahamii as4aup, GenBank accession number NC_012846.1). A total of 69 079 sequences were initially obtained and clustered in 354 OTUs with the 0.04 divergence level. Twenty-three OTUs, representing 92 sequences of them were with a low identity (<87%) to Bartonella and thus were removed from the analysis. Consequently, the posterior analyses were performed on 68 987 Bartonella gltA partial gene sequences, clustered in 331 OTUs. Fourteen OTUs represented the 94.6% (65 237/68 987) of all reads (Figure 1). These 14 OTUs were observed in both fleas and blood samples. Each of the remaining OTUs had an abundance percentage <0.5% of the total reads. Nevertheless, the sensitivity analysis showed that 235 OTUs represented ⩾0.1%, 85 OTUs represented ⩾0.5% and 68 OTUs represented ⩾1% of the relative abundance from each sample.
Figure 1.
Maximun likehood phylogenetic tree of partial (150 bp) gltA-Bartonella sequence, built from the 14 most abundant OTUs found in the study, and the closest similar Bartonella species and genotype sequences from GenBank database, with accession numbers in brackets. Brucella abortus gltA sequence was used as an out-group. Gray boxes highlight the OTUs and their closest associated genotype(s).
Bartonella infection composition
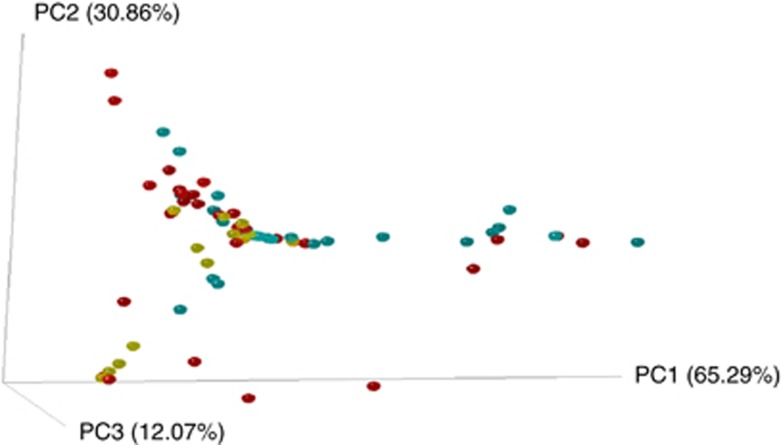
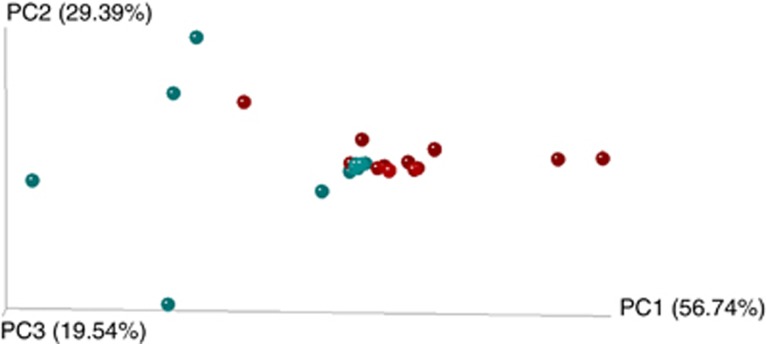
All Bartonella-positive samples included >1 OTU. On average, each blood sample contained 18 different OTUs (ranged from 5 to 35) and fleas contained 17 OTUs per sample (with a range of 2–44 OTUs). Sample type (blood or flea), flea sex and period of collection were found to be significant factors affecting both ecological and phylogenetic similarities in Bartonella infection composition among samples (Table 1, Figures 2 and 3). Six common OTUs, OTU 001, 002, 003, 004, 005 and 008 (Figure 1), were primarily responsible for differences across the groups (blood versus flea, female versus male fleas and period 1 versus period 2 in the case of the four host analysis). Noteworthy, the effect of period observed on the four host analysis (hosts with the complete set of positive samples collected) explained 21% of the variation in phylogenetic similarity (see results of the multivariable regression analyses in Table 1). Through the combined analysis of ecological and phylogenetic similarities, the sample type and period were also found significant factors, explaining 6% and 20% of the variation in the combined similarities, respectively (see results of the multivariable regression analyses in Table 1, Figures 2 and 3).
Table 1. P-values for non-parametric multivariate regression analyses of pairwise blood and flea sample distances based on ecological similarity (similarity in relative abundances of the different OTUs), phylogenetic similarities (similarity in phylogenetic distance of the different OTUs, calculated by unweighted UniFrac) and a combination of the two (calculated by weighted UniFrac).
| Method | Samples included | P-values (cumulative %) |
||||
|---|---|---|---|---|---|---|
| Type of sample | Period | Flea sex | Flea numbers in the host | Age of host | ||
| Ecological similarity | All samples | 0.03 (2) | 0.24 | NA | 0.65 | 0.94 |
| Only flea samples | NA | 0.22 | 0.0003a (4.8) | 0.88 | 0.90 | |
| Four host (fleas+blood) | 0.29 | 0.01a (8.5) | NA | 0.57 | 0.36 | |
| Phylogenetic distance | All samples | 0.008a (4.5) | 0.02a (4.0) | NA | 0.55 | 0.10 |
| Only flea samples | NA | 0.02a (4.2) | 0.02a (4.3) | 0.89 | 0.86 | |
| Four host (fleas+blood) | 0.35 | 0.003a (21) | NA | 0.06 | 0.47 | |
| Phylogenetic+ecological similarity | All samples | 0.005a (6) | 0.11 | NA | 0.80 | 0.38 |
| Only flea samples | NA | 0.14 | 0.14 | 0.37 | 0.89 | |
| Four host (fleas+blood) | 0.49 | 0.0002a (20) | NA | 0.17 | 0.06 | |
Abbreviations: NA, not applicable; OTU, operational taxonomic unit.
Analyses were performed on the whole set of samples, only flea samples (to test the effect of flea sex) or on samples collected from the four host individuals who had a complete set of positive samples. Numbers in parentheses indicate the proportion of explained variance in sample similarity by the variable.
Significant values after adjustment for sequential Bonferroni's correction.
Figure 2.
3D principal coordinates analysis plots generated by Fast Unifrac weighted analysis. Color dots represent the individual samples: yellow dots represent rodent blood samples; blue dots represent male flea samples; and red dots represent female flea samples. Closer dots represent a higher similarity between the bartonellae infection compositions. The full colour version of this figure is available at The ISME Journal online.
Figure 3.
3D principal coordinates analysis plots generated by Fast Unifrac weighted analysis of four hosts (complete set of samples). Color dots represent the individual samples: red dots represent samples from period 1 (May–June); and blue dots represent samples from period 2 (September). Closer dots represent a higher similarity between the bartonellae infection compositions. The full colour version of this figure is available at The ISME Journal online.
The sensitivity analysis showed that the three data matrices, which were generated without low abundant OTUs (those with <0.1, 0.5 or 1% of the sample sequences), had a minimal effect on the results obtained with the whole set of OTUs. For the ecological approach, the three different cutoffs did not affect significantly any of the results. On the other hand, for the phylogenetic distance approaches, most of the significant variables remained significant, with the exception of ‘type of sample', which lost its significance on the analysis of ‘all samples' in the last two cutoff (that is, ⩾0.5% and ⩾1% P=0.12 and P=0.08 unweighted Fast UniFrac, respectively; and P=0.28 and P=0.15 weighted Fast UniFrac, respectively) and flea sex in the case of unweighted Fast UniFrac in the ⩾0.1% cutoff (P=0.12).
Bartonella lineage diversity
None of the tested variables was associated with Bartonella lineage diversity (type of sample: F=0.72, P=0.40; period: F=3.46, P=0.07; host age: F=0.004, P=0.95; number of fleas: F=2.80, P=0.10; flea sex: F=0.57, P=0.45).
Bartonella lineage co-occurrence
The observed presence/absence pattern of OTUs within a sample was non-random. The C-score for the observed matrix was >18 s.ds. greater than the expected C-score (P<0.0001). In addition, the numbers of pairs of OTUs that never co-occurred were found significantly larger (4 s.d. greater) than those expected by chance (P<0.0001). Noteworthy, these results were consistent in both flea and blood samples. However, the number of species combinations (OTU that always co-occurred) was not significantly different from random (P=1.0).
Co-infection at Bartonella-genotype level
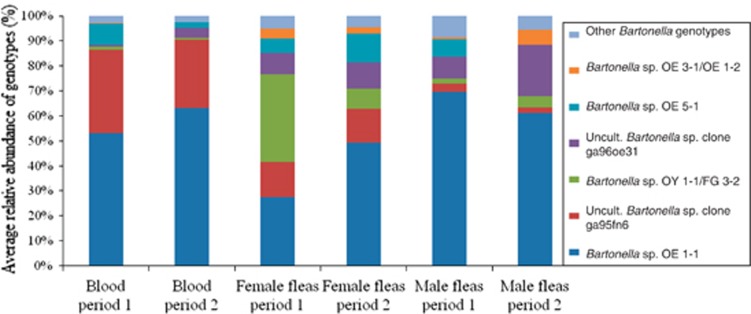
The most common OTUs in the study were closely related to six known Bartonella genotypes (97–100% similarity) (Figures 1 and 4). Less abundant OTUs represented distant variants of those genotypes (94–96% and <94% similarity), and few OTUs matched other Bartonella genotypes (Supplementary Table S2). Co-infection with at least two distantly related Bartonella variants was revealed in 89% (81/91) of all Bartonella-positive samples. This phenomenon was common in both rodent blood and flea samples. The genotype variant with the greater relative abundance within each infection (that is, the dominant genotype) varied according to the sample type (Table 2 and Figure 4). Additionally, the Bartonella genotypes detected in 71% (10/14) of the infected rodents were observed in their associated Bartonella-positive fleas but with a different distribution between the samples (relative abundance). The latter was illustrated for the ‘four host analysis' (Supplementary Figure S1). On the contrary, 86% (12/14) of the infected rodents carried fleas that contained genotypes not detected in the blood.
Figure 4.
Average of the relative abundance of bartonellae genotype diversity in the samples according to their origin (blood, flea sex, collection period). All variants included were 97–100% similar to the indicated genotype of origin.
Table 2. Dominant genotype (that is, the one with the higher relative abundance within the infection) according to the sample type and period of collection.
| Bartonella sp. OE 1-1 | Uncultured Bartonella sp. clone ga95fn6 | Bartonella sp. OY 1-1/FG 3-2 | Uncultured Bartonella sp. clone ga96oe31 | Bartonella sp. OE 5-1 | Bartonella sp. OE 3-1/1-2 | Other bartonellae genotypes | Total of samples | |
|---|---|---|---|---|---|---|---|---|
| Blood | ||||||||
| Period 1 | 7 | 3 | ND | ND | ND | ND | ND | 10 |
| Period 2 | 3 | 1 | ND | ND | ND | ND | ND | 4 |
| Female fleasa | ||||||||
| Period 1 | 5 | 2 | 7 | 1 | 1 | 1 | ND | 17 |
| Period 2 | 9 | 3 | 1 | 2 | 2 | 1 | ND | 18 |
| Male fleas | ||||||||
| Period 1 | 18 | 1 | ND | 2 | 2 | ND | 1 | 24 |
| Period 2 | 11 | ND | 1 | 4 | ND | 1 | ND | 17 |
Abbreviation: ND, no sample was found to be dominated by this genotype.
Numbers (n) indicate the sum of samples where the specific genotype was dominating the infection.
One female flea sample was not included in the table as it had two different genotypes with equal relative abundance (50% each).
Discussion
Co-infection of hosts and vectors with more than one Bartonella species or genotype is a well-known phenomenon (Gurfield et al., 1997; Abbot et al., 2007; Breitschwerdt, 2008; Chan and Kosoy, 2010). However, questions relating to the composition, the potential interchange of bartonellae variants between hosts and vectors and the overall diversity of bartonellae in either environment are not fully clarified. In the present study, we explored the composition of bartonellae infections in wild rodents and their fleas, added supporting evidence for the notion that a single carrier (rodent or flea) in natural communities can harbor multiple closely and distantly phylogenetically related Bartonella lineages and showed that those lineages appear to compete with each other. In addition, we showed that the assemblage of those lineages greatly vary among individual carriers. In contrast to the majority of previous studies, in which bartonellae co-infection represented only a low percentage of the positive samples (Birtles et al., 2001; Kosoy et al., 2004; Telfer et al., 2007), our results indicate that co-infections are more common than single-genotype infections in wild rodents and their fleas. It is likely that previous reports underestimated the occurrence of co-infection due to the challenge that low abundant genotypes represent when most of the detection assay resources are directed towards the predominant genotype. Hence, this latter potential technical obstacle was overcome by the pyrosequencing assay.
Bartonella assemblage composition showed to be more similar among different individuals of the same type of carrier than between different carriers (that is, rodents versus fleas or female versus male fleas; Figure 2 and Table 1). Moreover, a remarkable temporal change in assemblage composition was noticed when samples from the same origin were evaluated (Figure 3). Supporting evidence of shifts in flea-associated bacteria across time was previously indicated through a whole bacterial community analysis (Jones et al., 2010). Our results suggest that selection via ecological processes operates differently in unlike carrier types and produces remarkable genotypic/species diversity over space and time. The co-occurrence analysis highlights an inter-genotype competition as a possible mechanism underlining the great genetic diversity of Bartonella among individuals, as the assemblage of bartonellae lineages was found non-random and pairs of lineages that never co-occurred were found significantly more common than those expected by chance. The fact that the sample type and flea sex explained only a small portion of the variability in Bartonella lineage composition suggests that bartonellae assemblages are affected by additional factors at the individual level not investigated in our study, such as immunological status (Chan and Kosoy, 2010) or co-infection with other pathogens or symbionts (Telfer et al., 2010).
In spite of the noticeable interchange of genotypes between host and vectors, as common genotypes were observed in the blood and flea samples, it is likely that during Bartonella transmission some lineages are lost probably due to a lack of adaptation to a given carrier. First, non-infected rodents hosted infected fleas, and infected rodents carried negative fleas. This lack of correlation between infected and non-infected individuals was previously observed (La Scola et al., 2002; Brinkerhoff et al., 2010; Billeter et al., 2011). Second, when the Bartonella infection at a genotype level was analyzed, a clear trend of some genotypes to dominate a particular niche was revealed. The Bartonella infection compositions in fleas were dominated by any of the six most common genotypes found in the study (Table 2), while blood-associated infections were dominated by only two different genotypes. Therefore, the rodent blood may ‘filter out' or limit the dominance of other Bartonella genotypes introduced by fleas, whereas fleas might serve as a minor selective environment, allowing the proliferation of any bartonellae genotypes. Hence, flea-related traits might be having a positive effect on the diversity of species/variants assembly of the bartonellae infections, as has previously been suggested (Hawlena et al., 2013). Nevertheless, the lineage diversity analysis showed that in a given sample of either flea or rodent blood the diversity of those Bartonella lineages is distributed in a relatively similar manner, is not affected by host and vector characters and is stable over time. Interestingly, no effect in the bartonellae infection composition or lineage diversity with regard to the number of fleas found in each rodent host was detected. This suggests that flea load does not affect the assembly of bartonellae infections and supports the notion that the blood is having a filtering role for bartonellae lineages. Similarly, Telfer et al. (2007) noticed a lack of effect in the flea prevalence/abundance and the flea-transmitted Bartonella species in field voles (Microtus agrestis). Therefore, the dynamics of the transmission of bartonellae lineages need to be further studied in order to understand which factors have a role in the interchange and establishment of bartonellae lineages between hosts and vectors.
The Bartonella genotypes detected in the present study were similar to previously described genotypes or strains from wild rodents of the same geographic area (Inoue et al., 2009, Morick et al., 2010). Interestingly, not only phylogenetic closely related variants but also intermediate and distant variants of those genotypes were detected in the samples. For instance, 36 different OTUs matched with Bartonella sp. clone ga95fn6 (GU354265.1) with a range of 97–99% of identity, other 14 OTUs matched with a 94–96% and 6 OTUs more distantly related to this genotype were also found (Supplementary Table S2). Moreover, in a given sample when a particular genotype was detected, close and distant variants of that genotype were present. These results suggest a potential variation of the bartonellae at individual level, probably by clonal diversification. Unsurprisingly, Bartonella sp. OE 1-1 (AB445001.1) was the most abundant genotype in the study, as was previously detected in high prevalence in S. cleopatrae fleas from G. andersoni rodents (Morick et al., 2010). Nonetheless, other genotypes were similarly distributed in the different sample groups, such is the case of Bartonella sp. clone ga96oe31 in fleas (both sexes). This genotype tended to dominate several flea-associated bartonellae infections (Table 2).
With the development of the molecular techniques, the detection of concurrent multiple infections became more feasible (Abbot et al., 2007). The 454-pyrosequencing assay provided the benefit of receiving the maximal number of amplicons that could be detected in each sample (Dowd et al., 2008). Through this host-vector model, we were able to further characterize the bartonellae infections in rodents and fleas in nature. Nonetheless, we are aware of the limitations of the use of small gltA-fragments (150 bp) and the risk of over-representation of certain variants. For instance, grouping the sequences to OTUs at 0.04 divergence level with short fragment sizes might separate bartonellae lineages that originated from the same Bartonella species. Therefore, to overcome these limitations, we considered OTU groups that had the same Bartonella species origin (for example, B. elizabethae-like organisms) as members of the same species, such as the case of Bartonella OE 1-1 and Bartonella OY 1-1 genotypes (Inoue et al., 2009). Consequently, co-infections were only considered in cases where the bartonellae variants detected had a clear distant phylogenetic origin (Figure 1). Through this conservative approach, cases of co-infection were identified in 89% of the individuals. Moreover, the results of the sensitivity analysis showed that low abundant OTUs, which could represent artifactual OTUs, did not affect our main ecological approach findings. Similar results were obtained for the phylogenetic distance analyses, with a slight loss of significance for only two variables. The latter effect can be explained by the reduced size of the phylogenetic tree, as these analyses are highly dependent on the tree distances. Therefore, we believe that the pyrosequencing technique coupled with the strong discriminatory power of the citrate synthase gene (gltA) gave a first insight into the great complexity of bartonellae variants in mixed infections.
In conclusion, this investigation sheds an insight on the patterns of Bartonella infection compositions in wild rodents and their fleas. Co-infection with multiple genotypes has shown to be a common event as opposed to a single genotype infection. Furthermore, bartonellae lineages were shown to circulate in rodent and flea niches in a non-random manner and under inter-genotype competition, which lead to a great variability. Additionally, it was evident that some lineages tended to dominate or enrich certain niches differently from others, suggesting of a filtering process in the particular niches. Nevertheless, further investigations are required to explore additional factors that may have a role in the determination of bartonellae infection composition across time and space.
Acknowledgments
This research was supported by the ISRAEL SCIENCE FOUNDATION (ISF; Grant No. 30/11 to SH) and by Marie Curie Career Integration Grant (CIG; Grant number FP7-293713 to HH). We thank Dr Zohar Pasternak for his professional assistance and Nadezhda Burdelova (Ben-Gurion University of the Negev, Beer-Sheva, Israel) for the taxonomic identification of fleas.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abbot P, Aviles AE, Eller L, Durden LA. (2007). Mixed infections, cryptic diversity, and vector-borne pathogens: evidence from Polygenis fleas and Bartonella species. Appl Environ Microbiol 73: 6045–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson M. (2003) DISTLM forward 5: a FORTRAN Computer Program to Calculate a Distance-Based Multivariate Analysis for a Linear Model using forward selection. Department of Statistics, University of Auckland: Auckland, New Zealand, https://www.stat.auckland.ac.nz/∼mja/Programs.htm. [Google Scholar]
- Billeter SA, Levy MG, Chomel BB, Breitschwerdt EB. (2008). Vector transmission of Bartonella species with emphasis on the potential for tick transmission. Med Vet Entomol 22: 1–15. [DOI] [PubMed] [Google Scholar]
- Billeter SA, Gundi VA, Rood MP, Kosoy MY. (2011). Molecular detection and identification of Bartonella species in Xenopsylla cheopis fleas (Siphonaptera: Pulicidae) collected from Rattus norvegicus rats in Los Angeles, California. Appl Environ Microbiol 77: 7850–7852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birtles RJ, Raoult D. (1996). Comparison of partial citrate synthase gene (gltA) sequences for phylogenetic analysis of Bartonella species. Int J Syst Bacteriol 46: 891–897. [DOI] [PubMed] [Google Scholar]
- Birtles RJ, Hazel SM, Bennett M, Bown K, Raoult D, Begon M. (2001). Longitudinal monitoring of the dynamics of infections due to Bartonella species in UK woodland rodents. Epidemiol Infect 126: 323–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown KJ, Bennet M, Begon M. (2004). Flea-borne Bartonella grahamii and Bartonella taylorii in bank voles. Emerg Infect Dis 10: 684–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitschwerdt EB, Kordick DL. (2000). Bartonella infection in animals: carriership, reservoir potential, pathogenicity, and zoonotic potential for human infection. Clin Microbiol Rev 13: 428–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitschwerdt EB. (2008). Feline bartonellosis and cat scratch disease. Vet Immunol Immunopathol 123: 167–171. [DOI] [PubMed] [Google Scholar]
- Brinkerhoff RJ, Kabeya H, Inoue K, Bai Y, Maruyama S. (2010). Detection of multiple Bartonella species in digestive and reproductive tissues of fleas collected from sympatric mammals. ISME J 4: 955–958. [DOI] [PubMed] [Google Scholar]
- Chan KS, Kosoy M. (2010). Analysis of multi-strain Bartonella pathogens in natural host population—do they behave as species or minor genetic variants? Epidemics 2: 165–172. [DOI] [PubMed] [Google Scholar]
- Clarke KR. (1993). Nonparametric multivariate analyses of changes in community structure. Aust J Ecol 18: 117–143. [Google Scholar]
- Deng H, Le Rhun D, Buffet JP, Cotte V, Read A, Birtles RJ et al. (2012). Strategies of exploitation of mammalian reservoirs by Bartonella species. Vet Res 43: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Sun Y, Wolcott RD, Domingo A, Carroll JA. (2008). Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathog Dis 5: 459–472. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA, Cobet AS, Williams CB. (1943). The relation between the number of species and the number of individals in a random sample of an animal population. J Anim Ecol 12: 42–58. [Google Scholar]
- Gotelli NJ, Entsminger GL. (2009) EcoSim: Null Models Software for Ecology. Acquired Intelligence Inc & Kesey-Bear: Jericho, VT, USA, http://www.garyentsminger.com/ecosim/index.htm. [Google Scholar]
- Gundi VA, Davoust B, Khamis A, Boni M, Raoult D, La Scola B. (2004). Isolation of Bartonella rattimassiliensis sp. nov. and Bartonella phoceensis sp. nov. from European Rattus norvegicus. J Clin Microbiol 42: 3816–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundi VA, Taylor C, Raoult D, La Scola B. (2009). Bartonella rattaustraliani sp. nov., Bartonella queenslandensis sp. nov. and Bartonella coopersplainsensis sp. nov., identified in Australian rats. Int J Syst Evol Microbiol 59: 2956–2961. [DOI] [PubMed] [Google Scholar]
- Gundi VA, Billeter SA, Rood MP, Kosoy MY. (2012). Bartonella spp. in rats and zoonoses, Los Angeles, California, USA. Emerg Infect Dis 18: 631–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurfield AN, Boulouis HJ, Chomel BB, Heller R, Kasten RW, Yamamoto K et al. (1997). Coinfection with Bartonella clarridgeiae and Bartonella henselae and with different Bartonella henselae strains in domestic cats. J Clin Microbiol 35: 2120–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy L, Nystedt B, Toft C, Zaremba-Niedzwiedzka K, Berglund EC, Granberg F et al. (2013). A gene transfer agent and a dynamic repertoire of secretion systems hold the keys to the explosive radiation of the emerging pathogen Bartonella. PLoS Genet 9: e1003393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG. (2013). Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30: 1229–1235. [DOI] [PubMed] [Google Scholar]
- Hamady M, Lozupone C, Knight R. (2010). Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A, Dehio C. (2012). Intruders below the radar: molecular pathogenesis of Bartonella spp. Clin Microbiol Rev 25: 42–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawlena H, Khokhlova IS, Abramsky Z, Krasnov BR. (2006). Age, intensity of infestation by flea parasites and body mass loss in a rodent host. Parasitology 133: 187–193. [DOI] [PubMed] [Google Scholar]
- Hawlena H, Rynkiewicz E, Toh E, Alfred A, Durden LA, Hastriter MW et al. (2013). The arthropod, but not the vertebrate host or its environment, dictates bacterial community composition of fleas and ticks. ISME J 7: 221–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse SM, Welch DM, Morrison HG, Sogin ML. (2010). Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ Microbiol 12: 1889–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Maruyama S, Kabeya H, Hagiya K, Izumi Y, Une Y et al. (2009). Exotic small mammals as potential reservoirs of zoonotic Bartonella spp. Emerg Infect Dis 15: 526–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RT, McCormick KF, Martin AP. (2008). Bacterial communities of Bartonella-positive fleas: diversity and community assembly patterns. Appl Environ Microbiol 74: 1667–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RT, Knight R, Martin AP. (2010). Bacterial communities of disease vectors sampled across time, space, and species. ISME J 4: 223–231. [DOI] [PubMed] [Google Scholar]
- Kosoy M, Mandel E, Green D, Marston E, Jones D, Childs J. (2004). Prospective studies of Bartonella of rodents. Part II. Diverse infections in a single rodent community. Vector Borne Zoonotic Dis 4: 296–305. [DOI] [PubMed] [Google Scholar]
- Kosoy M, Hayman DT, Chan KS. (2012). Bartonella bacteria in nature: where does population variability end and a species start? Infect Genet Evol 12: 894–904. [DOI] [PubMed] [Google Scholar]
- Kosoy ME. (2010). Ecological associations between bacteria of the genus Bartonella and mammals. Zoologichesky Zhurnal 89: 61–70. [Google Scholar]
- La Scola B, Davoust B, Boni M, Raoult D. (2002). Lack of correlation between Bartonella DNA detection within fleas, serological results, and results of blood culture in a Bartonella-infected stray cat population. Clin Microbiol Infect 8: 345–351. [DOI] [PubMed] [Google Scholar]
- La Scola B, Zeaiter Z, Khamis A, Raoult D. (2003). Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol 11: 318–321. [DOI] [PubMed] [Google Scholar]
- Loftis AD, Reeves WK, Szumlas DE, Abbassy MM, Helmy IM, Moriarity JR et al. (2006). Surveillance of Egyptian fleas for agents of public health significance: Anaplasma, Bartonella, Coxiella, Ehrlichia, Rickettsia, and Yersinia pestis. Am J Trop Med Hyg 75: 41–48. [PubMed] [Google Scholar]
- McArdle BH, Anderson MJ. (2001). Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82: 290–297. [Google Scholar]
- Morick D, Krasnov BR, Khokhlova IS, Shenbrot GI, Kosoy MY, Harrus S. (2010). Bartonella genotypes in fleas (Insecta: Siphonaptera) collected from rodents in the Negev desert, Israel. Appl Environ Microbiol 76: 6864–6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morick D, Krasnov BR, Khokhlova IS, Gottlieb Y, Harrus S. (2011). Investigation of Bartonella acquisition and transmission in Xenopsylla ramesis fleas (Siphonaptera: Pulicidae). Mol Ecol 20: 2864–2870. [DOI] [PubMed] [Google Scholar]
- Morick D, Krasnov BR, Khokhlova IS, Gottlieb Y, Harrus S. (2013. a). Transmission dynamics of Bartonella sp. strain OE 1-1 in Sundevall's jirds (Meriones crassus). Appl Environ Microbiol 79: 1258–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morick D, Krasnov BR, Khokhlova IS, Gutiérrez R, Fielden LJ, Gottlieb Y et al. (2013. b). Effects of Bartonella spp. on flea feeding and reproductive performance. Appl Environ Microbiol 79: 3438–3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman AF, Regnery R, Jameson P, Greene C, Krause DC. (1995). Differentiation of Bartonella-like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol 33: 1797–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretorius AM, Beati L, Birtles RJ. (2004). Diversity of bartonellae associated with small mammals inhabiting Free State province, South Africa. Int J Syst Evol Microbiol 54: 1959–1967. [DOI] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. (2011). Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WE. (1989). Analyzing tables of statistical tests. Evolution 43: 223–225. [DOI] [PubMed] [Google Scholar]
- Sato S, Kabeya H, Miura T, Suzuki K, Bai Y, Kosoy M et al. (2012). Isolation and phylogenetic analysis of Bartonella species from wild carnivores of the suborder Caniformia in Japan. Vet Microbiol 161: 130–136. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner FE, Pinger RR, Vann CN, Grindle N, Civitello D, Clay K et al. (2008). Infection and co-infection rates of Anaplasma phagocytophilum variants, Babesia spp., Borrelia burgdorferi, and the rickettsial endosymbiont in Ixodes scapularis (Acari: Ixodidae) from sites in Indiana, Maine, Pennsylvania, and Wisconsin. J Med Entomol 45: 289–297. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer S, Begon M, Bennett M, Bown KJ, Burthe S, Lambin X et al. (2007). Contrasting dynamics of Bartonella spp. in cyclic field vole populations: the impact of vector and host dynamics. Parasitology 134: 413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer S, Lambin X, Birtles R, Beldomenico P, Burthe S, Paterson S et al. (2010). Species interactions in a parasite community drive infection risk in a wildlife population. Science 330: 243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartoukian SR, Palmer RM, Wade WG. (2010). Strategies for culture of ‘unculturable' bacteria. FEMS Microbiol Lett 309: 1–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.