Abstract
Vitamin traffic, the production of organic growth factors by some microbial community members and their use by other taxa, is being scrutinized as a potential explanation for the variation and highly connected behavior observed in ocean plankton by community network analysis. Thiamin (vitamin B1), a cofactor in many essential biochemical reactions that modify carbon–carbon bonds of organic compounds, is distributed in complex patterns at subpicomolar concentrations in the marine surface layer (0–300 m). Sequenced genomes from organisms belonging to the abundant and ubiquitous SAR11 clade of marine chemoheterotrophic bacteria contain genes coding for a complete thiamin biosynthetic pathway, except for thiC, encoding the 4-amino-5-hydroxymethyl-2-methylpyrimidine (HMP) synthase, which is required for de novo synthesis of thiamin's pyrimidine moiety. Here we demonstrate that the SAR11 isolate ‘Candidatus Pelagibacter ubique', strain HTCC1062, is auxotrophic for the thiamin precursor HMP, and cannot use exogenous thiamin for growth. In culture, strain HTCC1062 required 0.7 zeptomoles per cell (ca. 400 HMP molecules per cell). Measurements of dissolved HMP in the Sargasso Sea surface layer showed that HMP ranged from undetectable (detection limit: 2.4 pM) to 35.7 pM, with maximum concentrations coincident with the deep chlorophyll maximum. In culture, some marine cyanobacteria, microalgae and bacteria exuded HMP, and in the Western Sargasso Sea, HMP profiles changed between the morning and evening, suggesting a dynamic biological flux from producers to consumers.
Keywords: vitamins, thiamine, B1, phytoplankton, micronutrient, auxotrophy
Introduction
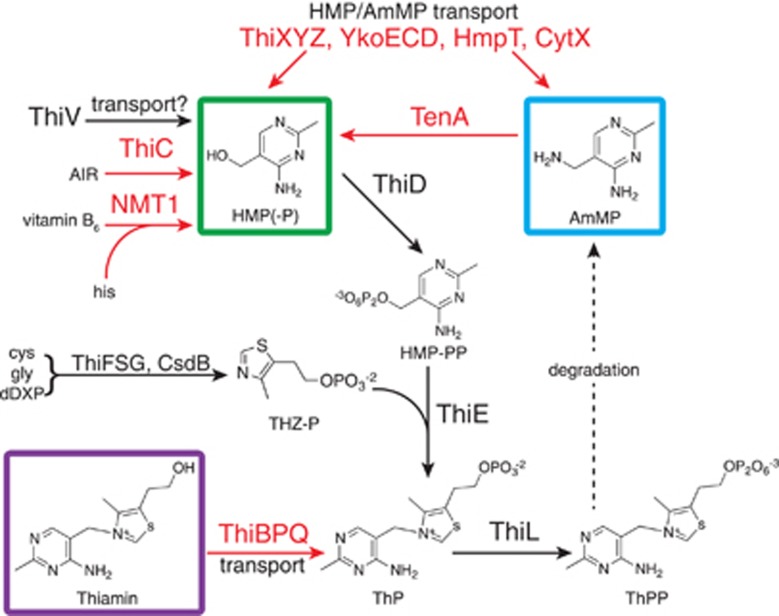
Thiamin (vitamin B1) is an essential coenzyme found in proteins that catalyze crucial transformations of carbon in all living systems. Specifically, thiamin is an essential cofactor for enzymes of the tricarboxylic acid cycle, the non-oxidative portion of the pentose phosphate pathway, the Calvin cycle and for enzymes required for the biosynthesis of branched-chain amino acids and isoprenoids (via the non-mevalonate pathway) (Lengeler et al., 1999). The pathways, enzymes and regulation of de novo thiamin synthesis and salvage have been the topic of extensive research in bacteria, yeasts and some microalgae (Winkler and Breaker, 2005; Croft et al., 2007; Jurgenson et al., 2009). In all organisms capable of de novo thiamin biosynthesis, the formation of thiamin monophosphate (ThP) results from the enzyme-catalyzed linkage of two separately synthesized moieties: 4-amino-5-hydroxymethyl-2-methylpyrimidine diphosphate and 4-methyl-5-(2-phosphoethyl)-thiazole (Figure 1). Phosphorylation of ThP yields the active thiamin coenzyme, thiamin diphosphate (ThPP) (Figure 1) (reviewed in Jurgenson et al., 2009).
Figure 1.
Simplified illustration of thiamin metabolism in Ca. P. ubique. Black colored lines and enzyme abbreviations represent reactions and enzymes encoded by the Ca. P. ubique genome. Red colored lines and enzyme abbreviations represent reactions and enzymes that are absent from the Ca. P. ubique genome. AIR, aminoimidazole ribotide; AmMP, 4-amino-5-aminomethyl-2-methylpyrimidine; cys, cysteine; dDXP, 1-deoxy-D-xylulose 5-phosphate; gly, glycine; his, histidine; HMP(-P), 4-amino-5-hydroxymethyl-2-methylpyrimidine (-phosphate); HMP-PP, 4-amino-5-hydroxymethyl-2-methylpyrimidine diphosphate; ThP, thiamin monophosphate; ThPP, thiamin diphosphate; THZ-P, 4-methyl-5-(2-phosphoethyl)-thiazole.
Renewed interest in vitamin distributions in marine ecosystems has been driven by the development of more sensitive analytical techniques to measure vitamin concentrations (Sañudo-Wilhelmy et al., 2012) and a greater appreciation of the importance of trace compounds to plankton productivity. Whereas the sources, distributions and speciation of trace metals have been extensively researched as they pertain to ocean productivity (reviewed in Morel and Price, 2003), relatively little is known about vitamin biogeochemistry or the affect of vitamins on the structure and composition of planktonic communities. Direct measurements of B-vitamin concentrations in coastal ocean systems found picomolar concentrations and complex patterns in the distributions of several vitamins, including thiamin (Sañudo-Wilhelmy et al., 2012; Barada et al., 2013). In bottle experiments, iron and B-vitamins, particularly vitamin B12, acted synergistically to increase phytoplankton and bacterial productivity, suggesting colimitation (Panzeca et al., 2006; Bertrand et al., 2007). Supporting the view that the exchange of vitamins between species is important, adaptive strategies for coping with low vitamin concentrations have been identified in diatoms (Bertrand et al., 2012). Furthermore, there is evidence that some marine bacteria produce vitamin B12 that is used by phytoplankton (Croft et al., 2005).
Thiamin is a particularly interesting vitamin because the genomes of many environmentally abundant microorganisms do not encode for complete, canonical thiamin biosynthetic pathways (Bertrand and Allen, 2012; Helliwell et al., 2013), suggesting that auxotrophy is common. The distribution of thiamin biosynthetic genes in algal genomes does not correlate well with phylogeny, an indication that thiamin metabolism has evolved and diversified in response to selective pressures that vary with habitat (reviewed in Croft et al., 2006; Helliwell et al., 2013). The evolution of thiamin metabolism in phytoplankton is likely complex, as evidenced by the ability of some strains to use the thiamin moieties 4-methyl-5-thiazolethanol (THZ) or 4-amino-5-aminomethyl-2-methylpyrimidine (AmMP), presumably natural thiamin degradation products, in place of thiamin (Droop, 1958; Lewin, 1962). A specific requirement for the thiamin pyrimidine precursor 4-amino-5-hydroxymethyl-2-methylpyrimidine (HMP) has been described for the protist Plasmodium falciparum (Wrenger et al., 2006) and the bacterium Listeria monocytogenes (Schauer et al., 2009). Moreover, thiamin is exclusively obtained through salvage of thiamin moieties by the bacterium Rhizobium leguminosarum bv. viciae strain 3841 (Karunakaran et al., 2006). Environmental concentrations of these thiamin precursors or degradation products have not been measured, and thiamin metabolism in marine bacteria is a relatively unexplored topic.
This study examines thiamin metabolism in the SAR11 clade of α-proteobacteria (Pelagibacterales). These organisms are the most abundant chemoheterotrophic bacterioplankton in the oceans, often comprising 25–50% of the cells in the euphotic zone (Morris et al., 2002; Carlson et al., 2009). Both in situ studies and those with axenic cultures show that the Pelagibacterales contribute significantly to the cycling of carbon and sulfur in the ocean (reviewed in Tripp, 2013). The first cultivated Pelagibacterales bacterium, ‘Candidatus Pelagibacter ubique' strain HTCC1062 (Ca. P. ubique), contains one of the smallest genomes found in free-living organisms. The small genome of Ca. P. ubique is attributed to streamlining selection (Giovannoni et al., 2005). Gene loss related to streamlining selection has been proposed as an explanation for the unusual combination of amino acids, reduced organosulfur compounds and organic acids required for the growth of Ca. P. ubique (Carini et al., 2013; Tripp, 2013). Although the macronutrient requirements of Ca. P. ubique have been identified, their requirements for vitamins and other trace molecules have not been investigated.
We used comparative genomics to examine the distribution of genes for thiamin metabolism among the Pelagibacterales, and studied the requirement for thiamin or its precursors in Ca. P. ubique. Following up on the surprising finding that Ca. P. ubique requires the thiamin precursor HMP, we applied high-performance liquid chromatography-coupled tandem mass spectrometry (LC-MS) to show that dissolved HMP is present at picomolar concentrations in the oceans. These findings offer important new insights into thiamin cycling, and identify HMP as a growth factor that is likely to have an important role in vitamin-mediated interactions in the ocean.
Materials and methods
Metabolic reconstruction of thiamin biosynthesis in Ca. P. ubique and other Pelagibacterales
To identify putative protein domains involved in thiamin biosynthesis, amino-acid sequences of known Escherichia coli (ThiC, ThiD, ThiE, ThiS, ThiG, ThiL, ThiF, IscS and ThiH), Bacillus subtilis (ThiO) and Saccharomyces cerevisiae (NMT1) thiamin biosynthesis proteins were used as query sequences in an HMMER search against the Pfam database (v.27.0), using the Pfam website (http://pfam.sanger.ac.uk/search) with default settings. Identified Pfam domains were extracted from the Pfam-A database and prepared as an hmmscan (v.3.1b) compliant database. This database was used to search predicted amino-acid sequences of Ca. P. ubique ORFs for putative protein domains involved in thiamin biosynthesis using hmmscan (http://hmmer.janelia.org; v.3.1b) (Supplementary Data set 1). A similar approach was used to identify Ca. P. ubique genes involved in thiamin biosynthesis using the Sifting Families (Sfam) Hidden Markov Model (HMM) database (Sharpton et al., 2012) in place of Pfam (Supplementary Data set 2). When an ORF from Ca. P. ubique was predicted to match a Pfam and/or Sfam identified from a Thi_ query (e-value ⩽1.0 × 10−35), it was assumed that the Ca. P. ubique gene was a homolog of the query. The best hit for E. coli ThiL in the Pfam database (PF00586) is the N-terminal domain of aminoimidazole ribonucleotide synthase-related proteins—a putative ATP-binding domain. Proteins associated with this Pfam model are numerous and functionally diverse. Therefore, ThiL homologs in Ca. P. ubique were assigned based on the strength of their best-hit Sfam model alone.
The Hal pipeline (Robbertse et al., 2011) was used to identify genes encoding Thi biosynthesis homologs, in seven additional Pelagibacterales genomes (HTCC1002, HTCC9565, HTCC7211, HIMB5, HIMB114, HIMB59 and IMCC9063). Orthologous groups were established using the pipeline Hal, as described in Thrash et al. (2014). The Hal pipeline connects the programs BLASTP, MCL, user-specified alignment programs, GBlocks, ProtTest and user-specified phylogenetic programs. Hal uses an all-versus-all BLASTP search and MCL clustering to identify orthologs, as described in detail in Robbertse et al. (2011).
Construction of ThiV phylogenetic trees
RAxML (Stamatakis, 2006) was used for phylogenetic inference, after alignment with MUSCLE (Edgar, 2004), curation with Gblocks (Castresana, 2000) and amino-acid substitution modeling with ProtTest (Abascal et al., 2005). SAR11_0811 was initially identified as a ThiV homolog by searching the amino-acid sequence against others at MicrobesOnline (http://microbesonline.org/). This search identified SAR11_0811 as a member of the COG591 gene family, which had orthologs in the genomes of eight additional organisms: Methylobacillus flagellatus KT, Marinobacter sp. ELB17, Clostridium sp. OhILAs, Haloquadratum walsbyi DSM 16790, Haloarcula marismortui ATCC 43049, Halorhabdus utahensis DSM 12940, Haloferax volcanii DS2 and Halogeometricum borinquense PR3, DSM 11551. Eight SAR11_0811 orthologs in other SAR11 genomes (HTCC1002, HTCC9565, HTCC7211, HIMB5, AAA240-E13, AAA288-G21, HIMB114 and IMCC9063) were identified with the Hal pipeline (Robbertse et al., 2011; Thrash et al., 2014). To provide a fuller phylogenetic context for the trees, additional homologs to ThiV amino-acid sequences from the genomes above were searched against the Sfam HMM database (Sharpton, et al., 2012). Further details are provided in Supplementary Documentation.
Organism source and cultivation details
Ca. P. ubique was revived from 10% glycerol stocks and propagated in AMS1, without added vitamins, amended with oxaloacetate (1 mM), glycine (50 μM), methionine (50 μM) and FeCl3 (1 μM) (Carini et al., 2013). Thiamin or precursors were added as indicated in figure legends and text. All cultures were grown in acid-washed and autoclaved polycarbonate flasks and incubated at 20 °C with shaking at 60 r.p.m. in the dark, unless noted otherwise. Cells for counts were stained with SYBR green I and counted with a Guava Technologies flow cytometer (Millipore, Billerica, MA, USA) at 48–72 h intervals as described elsewhere (Tripp et al., 2008).
Cultures tested for HMP exudation were grown in acid-washed and autoclaved polycarbonate flasks, incubated at 20 °C with shaking at 60 r.p.m. on a 14-h/10-h light (140–180 μmol photons m−2 s−1)/dark cycle and monitored by flow cytometry as described for Ca. P. ubique. For HMP exudation assays, axenic batch cultures of Synechococcus sp. WH8102 and Prochlorococcus sp. MED4 (CCMP2389) were grown in PCRS-11 Red Sea medium (Rippka et al., 2000). Dunaliella tertiolecta (CCMP1320) was grown in AMS1 medium without vitamins (Carini et al., 2013). The OM43 clade isolate, sp. HTCC2181, was grown in natural seawater with no added vitamins as described elsewhere (Giovannoni et al., 2008).
All AMS1 constituents, reagents and vitamins were of the highest available quality (labeled ‘ultrapure' when possible). To minimize unintended traces of vitamins from glassware, all nutrient and vitamin stocks were prepared in combusted glassware (450 °C for 4 h) with nanopure water, 0.1 μm filter sterilized and frozen in amber tubes immediately after preparation. HMP was synthesized as described in Reddick et al. (2001). AmMP was synthesized as described in Zhao et al. (2012). HMP was purified by chromatography and then recrystallized. It was characterized by 1H and 13C nuclear magnetic resonance spectroscopy and by mass spectrometry. AMP was purified by crystallization and was characterized by 1H and 13C nuclear magnetic resonance spectroscopy spectroscopy. No impurities were detected.
HMP and thiamin concentrations in seawater
Seawater for vitamin analysis was collected from Hydrostation S (32°10′N, 64°30′W) from casts at 2000 hours (local time) on 19 September 2012, and 0800 hours (local time) on 20 September 2012. At the time of collection, samples were filtered through nanopure water-rinsed 0.2 μm pore-size supor filters into acid-washed amber polypropylene bottles and frozen immediately.
HMP and thiamin were extracted from 300 ml seawater to a reverse-phase C18 silica bead solid phase (Agilent HF-Bondesil, Agilent Technologies, Santa Clara, CA, USA) as described in Sañudo-Wilhelmy et al. (2012). For quantification purposes, standard curves were constructed from aged seawater (collected from Hydrostation S in July of 2009) spiked with known amounts of HMP and thiamin (ranging from 0 to 100 pM). These standard curves (Supplementary Figures S1 and S2) were extracted alongside samples using identical procedures.
Extracts were reconstituted in 125 μl high-performance liquid chromatography-grade water. Samples were centrifuged to pellet insoluble matter and the supernatant was transferred to sampling vials. HMP was quantified using an Applied Biosystems MDS Sciex 4000 Q TRAP (Foster City, CA, USA) mass spectrometer coupled to a Shimadzu high-performance liquid chromatography system. An Agilent Zorbax SB-Aq (Agilent Technologies) (2.1 × 100 mm2, 3.5 μm) high-performance liquid chromatography column was used for separation over a 10-min gradient flow with mobile phases of pH 4 (formic acid) methanol (MeOH) and pH 4 (formic acid) 5 mM ammonium formate (AmF). The flow rate was 0.4 ml min−1 and a gradient starting at 98% AmF:2% MeOH for 1 min changing to 75% AmF:25% MeOH over 3 min, 50% AmF:50% MeOH over 0.2 min, and finally to 10% AmF:90% MeOH over 0.8 min. The retention time of HMP was approx. 1.8 min.
For HMP quantification, the mass spectrometer was run in ‘Multiple Reaction Monitoring' mode. The HMP parent ion m/z was 140.2, and ion transitions of 81.1 and 54.1 were used for quantification and qualification, respectively. Peaks were analyzed using the Analyst software package v.1.5.2 (AB SCIEX, Concord, ON, Canada). Measured HMP values are the average of technical LC-MS replicates. The greatest standard deviation of replicate measurements was 3.5 pM (coefficient of variation=10%) in the 120 m 0800 hour sample, and the lowest was 0.22 pM (coefficient of variation=3.5%) in the 200 m 20:00 hour sample. Thiamin was detected and quantified as described in Sañudo-Wilhelmy et al. (2012) The limit of detection is defined as three times the standard deviation of the procedural controls and the limit of quantification as 10 times the standard deviation of the procedural controls. The limit of detection for HMP was 2.4 pM (limit of quantification: 8.0 pM) and for thiamin it was 0.81 pM (limit of quantification: 2.7 pM; from Sañudo-Wilhelmy et al. (2012)).
Cell harvesting of marine microbes for HMP exudation assays and detection of HMP background in AMS1
During mid-logarithmic growth (approx 1.0 × 107 cells ml−1), 100 ml of culture was gently filtered (to prevent cell lysis) through 0.1 or 0.2 μm pore-size supor filters to remove cells. The filtrate was collected in an acid-washed amber polypropylene bottle and frozen immediately. Uninoculated media (negative control) for each media type (AMS1, PCRS-11 Red Sea medium and natural seawater medium for HTCC2181) was extracted alongside spent medium treatments for comparison. HMP extraction and detection by LC-MS were performed as described for natural seawater samples.
Results
Thiamin biosynthetic pathways were incomplete in all eight Pelagibacterales genomes we studied (Table 1). Despite the apparent inability to synthesize thiamin de novo, multiple genes encoding ThPP-dependent enzymes were identified in Ca. P. ubique, indicating thiamin is necessary for normal metabolism (Supplementary Figure S3). Four Pelagibacterales strains contained the same thiamin biosynthesis and transport genes as Ca. P. ubique (Table 1). Two additional Pelagibacterales strains, IMCC9063 and HIMB114, have complements of thiamin biosynthesis and transport genes similar to Ca. P. ubique, except both are missing thiL (Table 1). Additionally, IMCC9063 encodes the AmMP salvage enzyme, tenA (Table 1). In Pelagibacterales str. HIMB59, thiC, thiD, thiG, thiE and thiE2 and thiS are absent. However, a gene encoding a thiamin-specific periplasmic binding protein (thiB) (Webb et al., 1998) was identified in HIMB59 (Table 1).
Table 1. Comparative genomics of thiamin biosynthesis in the Pelagibacterales.
| Strain | thiC | thiD | thiE_0583a | thiE_0360a | thiF | thiS | thiG | csdBb | thiL | thiB | tenA |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Ca. P. ubique | Absent | 637671479 | 637671458 | 637671224 | 637671266 | 637671603 | 637671604 | 637671616 | 637671913 | Absent | Absent |
| HTCC1002 | Absent | 639129819 | 639129840 | 639130075 | 639130033 | 639129702 | 639129701 | 639130662, 639129689 | 639130810 | Absent | Absent |
| HTCC7211 | Absent | 2503353714 | 2503353735 | 2503352435 | 2503352394 | 2503352877 | 2503352878 | 2503352890 | 2503353193 | Absent | Absent |
| HTCC9565 | Absent | 2503364149 | 2503364170 | 2503364413 | 2503364372 | 2503364883 | 2503364884 | 2503364896 | 2503365124 | Absent | Absent |
| HIMB5 | Absent | 2504109247 | 2504109269 | 2504109551 | 2504109508 | 2504108506 | 2504108507 | 2504108519, 2504109389 | 2504108893 | Absent | Absent |
| HIMB114 | Absent | 2503356000 | 2503356022 | 2503356319 | 2503356274 | 2503355884 | 2503355883 | 2503355872 | Absent | Absent | Absent |
| IMCC9063 | Absent | 2505688345 | 2505688367 | 2505687345 | 2505687250 | 2505687878 | 2505687879 | 2505688259 | Absent | Absent | 2505687352 |
| HIMB59 | Absent | Absent | Absent | Absent | 2504110146 | Absent | Absent | 2504110964 | 2504110802 | 2504111022 | Absent |
Gene numbers are IMG/ER Gene IDs (https://img.jgi.doe.gov/er).
There are two copies of thiE in Ca. P. ubique: SAR11_0583 and SAR11_0360.
csdB is predicted to encode the cysteine desulfurase activity necessary for thiazole biosynthesis (see Supplementary Methods).
Genes encoding the HMP synthase (thiC) are absent from all Pelagibacterales genomes (Table 1). ThiC catalyzes the molecular rearrangement of the purine nucleotide biosynthetic intermediate 5-aminoimidazole ribotide to form HMP (Figure 1) (Martinez-Gomez and Downs, 2008) and is essential for de novo thiamin biosynthesis in bacteria, archaea and plants. Genes that encode alternate HMP synthesis or salvage proteins were not identified in the Ca. P. ubique genome. For example, Ca. P. ubique lacks genes encoding for NMT1, which synthesizes HMP from vitamin B6 and histidine in Saccharomyces cerevisiae (Figure 1) (Wightman and Meacock, 2003). Genes encoding TenA homologs, which catalyze the hydrolysis of AmMP to yield HMP (Jenkins et al., 2007), were also not present in Ca. P. ubique (Figure 1). Some organisms can transport thiamin intact with the thiamin-specific ABC transporter encoded by thiBPQ. No homologs of the thiamin-specific binding protein, ThiB, were identified in Ca. P. ubique genomes (Figure 1). Further, Ca. P. ubique does not encode homologs of the predicted bacterial HMP/AmMP ABC transport complexes ThiXYZ (Jenkins et al., 2007) and YkoEDC, or for the putative HMP/AmMP permeases HmpT and CytX (Rodionov et al., 2002, 2008).
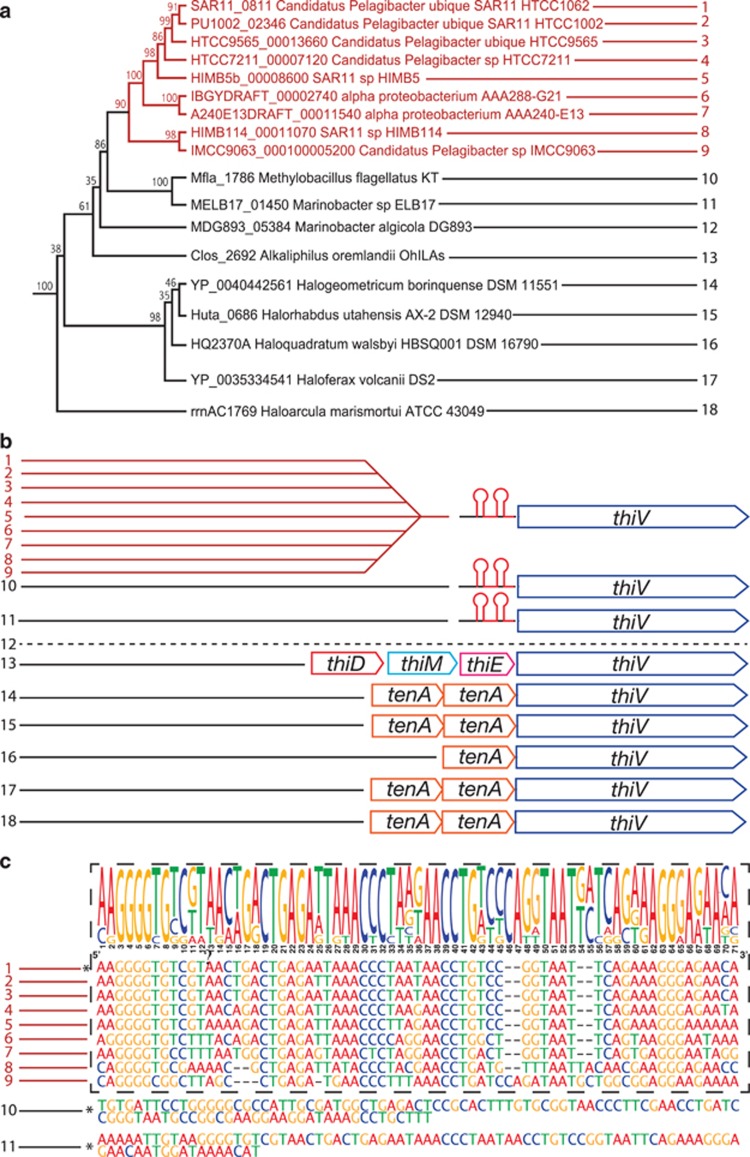
A single predicted ThPP-activated RNA riboswitch was identified in the Ca. P. ubique genome (Meyer et al., 2009) in an unusual configuration upstream of a coding sequence annotated as a sodium:solute symporter family protein (encoded by Ca. P. ubique ORF SAR11_0811). A similarly configured riboswitch was previously identified in the genome of Methylobacillus flagellatus, upstream of a coding sequence for an uncharacterized putative transporter named thiV (Rodionov et al., 2002). Maximum-likelihood phylogenetic analysis of the Pelagibacterales thiV homologs showed that they form a monophyletic group with the thiV sequences from M. flagellatus and a diverse group of microbes, including, Haloarchaea, Gram-positive bacteria and β- and γ-proteobacteria (Figure 2a and Supplementary Figure S4). Genes orthologous to thiV in all organisms (except for Marinobacter algicola) are either (i) in an operon with genes encoding enzymes that enable the salvage of HMP and THZ moieties for thiamin synthesis (thiD, thiM and thiE; Figure 2b); (ii) in an operon with one or two copies of the tenA gene (encoding an AmMP salvage enzyme; Figure 2b); or (iii) are preceded by a ThPP-riboswitch motif (Figures 2b and c).
Figure 2.
Gene phylogeny, synteny and conservation of riboswitch structure for the Pelagibacterales ThiV family sodium:solute symporter. Pelagibacterales genome elements are highlighted in red. (a) Maximum-likelihood phylogenetic tree showing a subset of amino-acid sequences extracted from a complete tree (Supplementary Figure S4). (b) For the same taxa shown in a, the chromosomal colocalization of thiV genes with putative ThPP-binding riboswitches (red stem-loop structure) and genes encoding thiamin salvage enzymes (thiDME or tenA). Dashed line indicates no ThPP-binding riboswitch or associated salvage genes were identified. (c) Nucleotide sequences of predicted ThPP-binding riboswitches depicted in b. Dashed box encapsulates the riboswitch sequences from nine Pelagibacterales genomes and their consensus sequence (illustrated at the top). Sequences that are marked with (*) were predicted to contain ThPP-binding motifs using the rfam (http://rfam.sanger.ac.uk) sequence search tool.
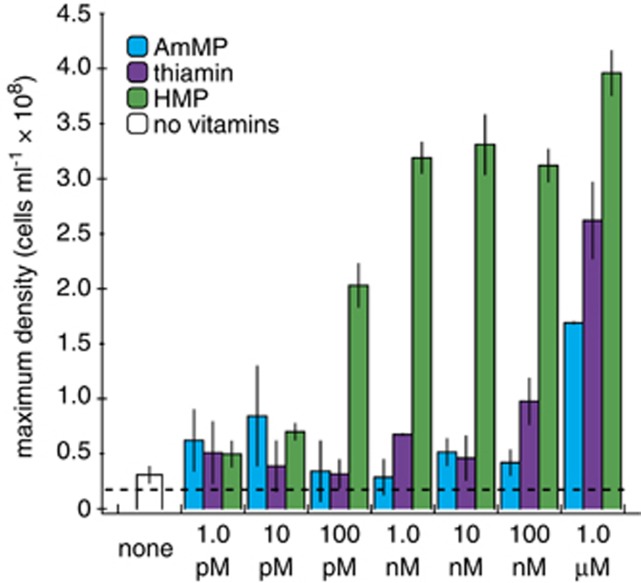
We hypothesized that Ca. P. ubique is auxotrophic for HMP because genes coding for known HMP synthesis pathways (thiC and NMT1) and AmMP salvage mechanisms (tenA) were absent (Figure 1 and Table 1). To test this hypothesis, the growth responses of Ca. P. ubique to HMP, AmMP and thiamin were investigated across seven orders of magnitude (Figure 3). Cultures grown in medium containing no added HMP, without additional thiamin or precursors, attained maximum cell densities of 3.09±0.75 × 107 cells ml−1 (mean±s.d., n=3) (Figure 3). Cell yields responded linearly to HMP additions between 1 and 100 pM (Supplementary Figure S5) and reached maximal cell yields (ca. 3.5 × 108 cells ml−1) at HMP concentrations ⩾1 nM (Figure 3). The cellular HMP requirement was calculated to be 0.66 zeptomoles (396 molecules) per cell from the slope of the linear regression between 1 and 100 pM (Supplementary Figure S5). Thiamin and AmMP were ineffective at restoring thiamin-limited growth at pico- or nanomolar concentrations; these compounds restored growth only when supplied at 1.0 μM (Figure 3). The average growth rate of Ca. P. ubique was 0.29±0.03 per day (mean±s.d., n=123) and did not vary with vitamin or precursor treatments (for example, see Supplementary Figure S6).
Figure 3.
Maximum cell yields of Ca. P. ubique batch cultures in response to AmMP, thiamin and HMP additions. Cells were grown in AMS1 amended with thiamin, HMP or AmMP as indicated. Bar heights are the average densities of biological replicates±s.d. (n=3). The dashed line represents the calculated maximum density expected (∼1.8 × 107 cells ml−1) from the ‘background' level of HMP (see text for details). We attribute the growth with 1 μM thiamin or AmMP to ‘contaminating' HMP (see text for details).
To rule out NMT1 activity as a potential source of HMP, thiamin was replaced with histidine and vitamin B6 (NMT1's substrates; Ishida et al., 2008). Consistent with the prediction that Ca. P. ubique lacks the ability to synthesize HMP through NMT1 activity, histidine+vitamin B6 did not alleviate thiamin-limited growth (Supplementary Figure S7). Thiamin-limited growth was not relieved by pantothenate or THZ addition (Supplementary Figure S7) as has been reported previously for other organisms (Droop, 1958; Downs, 1992).
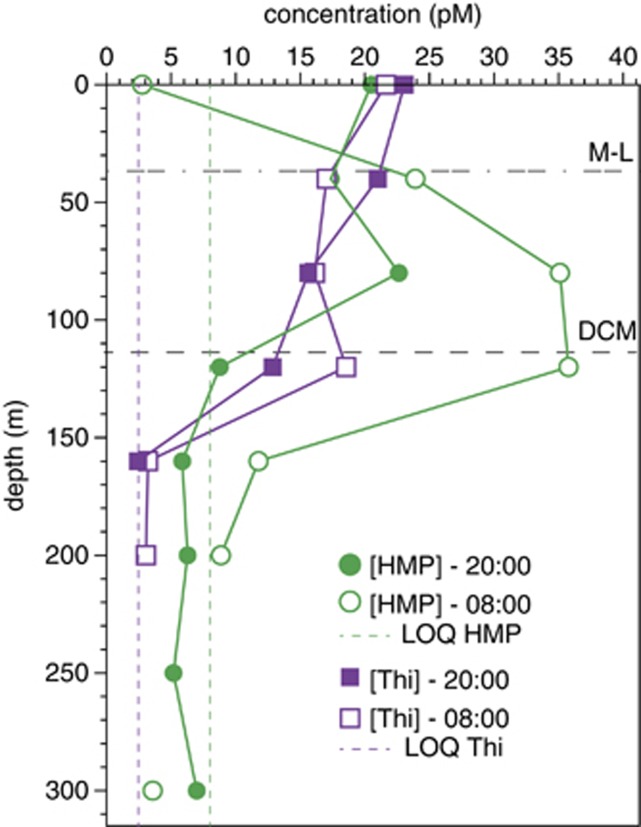
To date, measurements of HMP or AmMP concentrations in the environment have not been reported. To determine if HMP is present in an environment where Pelagibacterales bacteria are also found, thiamin and HMP were extracted from Sargasso Sea seawater collected at two different times of the day (2000 and 0800 hours local time, approximately 1 h after sunset and sunrise, respectively) and quantitatively measured by LC-MS. HMP ranged from undetectable (detection limit: 2.4 pM) to 35.7 pM (Figure 4). The maximum concentration of HMP was observed in samples collected at 0800 hours near the deep chlorophyll maximum (Figure 4). HMP concentrations at 2000 hours were substantially higher at 0 m depth, but lower at depths of 40, 80, 120, 160 and 200 m, compared with samples collected at 0800 hours (Figure 4). HMP was not detected in the 250 m sample collected at 0800 hours. Thiamin was measured in the same samples and ranged from undetectable (detection limit: 0.81 pM; Sañudo-Wilhelmy et al., 2012) to 23 pM, and was present in samples from 0 to 160 m, but not detected in samples from 250 to 300 m (Figure 4).
Figure 4.
Depth distribution of dissolved HMP and thiamin in the Sargasso Sea. Times of collection are presented in local time. HMP values are the average of technical replicate analyses for each sample. There was no technical replication for the thiamin measurements owing to insufficient sample. HMP was not detected in the 250 m sample collected at 0800 hours. Thiamin was not detected in samples collected from 200 m at 2000 hours or at 250 and 300 m at either time. DCM, deep chlorophyll maximum; LOQ, limit of quantification; M-L, mixed layer.
To determine whether marine microbes exude HMP, we measured HMP concentrations in growth media before and after cell growth in strains known to have a complete complement of thiamin biosynthetic genes (Table 2). The marine cyanobacterium Synechococcus sp. WH8102 and the marine chlorophyte D. tertiolecta exuded nanomolar amounts of HMP during growth (Table 2). Moderate amounts of excess HMP were also detected in spent media from cyanobacterium Prochlorococcus MED4 and the OM43 clade of marine β-proteobacteria isolate, strain HTCC2181 (Giovannoni et al., 2008). Two Pelagibacterales cultures were also tested: Ca. P. ubique and Pelagibacterales sp. strain HTCC7211. In both cases, HMP was not detected after cell growth (Table 2).
Table 2. HMP concentrations in uninoculated and partially spent media.
| Organism | HMP (pM) | |
|---|---|---|
| Uninoculated | Partially spent | |
| Synechococcus sp. WH8102 | N/D | 2909.6 |
| Dunaliella tertiolecta | 11.6 | 1584.3 |
| Prochlorococcus sp. MED4 | N/D | 32.8 |
| OM43 isolate HTCC2181 | 12.9 | 33.0 |
| Ca. P. ubique strain HTCC1062 | 11.6 | N/D |
| Pelagibacterales sp. strain HTCC7211 | 11.6 | N/D |
Abbreviations: HMP, 4-amino-5-hydroxymethyl-2-methylpyrimidine; N/D, not detected.
Limit of detection=2.4 pM.
Discussion
Thiamin has long been recognized as an important vitamin for microalgal growth (reviewed in Croft et al., 2006). The physiological requirement for thiamin led to the hypothesis that environmental concentrations of thiamin may exert control over some phytoplankton populations (Natarajan, 1968; Panzeca, et al., 2006). Environmental distributions of thiamin, as determined by bioassay, were variable, and in some cases, coupled to productivity (Natarajan and Dugdale, 1966; Natarajan, 1968, 1970). Studies of thiamin auxotrophy in the laboratory showed that thiamin moieties or degradation products were able to satisfy the thiamin requirement of some microalgae (Droop, 1958; Lewin, 1962). However, research pursuing the ecological importance of these findings tapered off. The experimental results presented here reintroduce the idea that thiamin pyrimidines are important growth determinants in marine ecosystems. We show that the thiamin pyrimidine precursor, HMP, is required for growth of the marine chemoheterotrophic bacterium Ca. P. ubique (Figure 3), a representative isolate of one of the most abundant groups of organisms on the planet. Surprisingly, neither thiamin itself nor AmMP satisfied this requirement (Figure 3). Comparative genomics extended the significance of this requirement to multiple members of the Pelagibacterales clade (Table 1). The importance of these findings were further supported by the detection of dissolved HMP in the Sargasso Sea (Figure 4), one of the most oligotrophic ocean systems on Earth, at concentrations often exceeding those of thiamin. This discovery shows that fundamental information needed to understand thiamin biogeochemistry in marine ecosystems is incomplete—specifically that environmental measurements of thiamin alone may only partially explain interactions related to the thiamin requirements of planktonic cells.
The inability of Ca. P. ubique to use thiamin or its degradation product AmMP was surprising given that many algal species are able to use these compounds (Droop, 1958; Lewin, 1962). The Ca. P. ubique genome encodes no thiamin transporter (Figure 1 and Table 1), consistent with the observation that exogenous thiamin does not support growth (Figure 3). Likewise, we propose that the absence of the tenA gene (Figure 1 and Table 1), necessary for the conversion of AmMP to HMP, explains why AmMP does not substitute for HMP in thiamin biosynthesis. However, genome analysis of Pelagibacterales strain HIMB59 shows that this strain lacks genes required for de novo synthesis of thiamin (thiC, thiD, thiG, thiE and thiS), as well as the AmMP salvage enzyme (tenA; Table 1) and thiV; therefore, we postulate that this strain requires exogenous thiamin. Supporting this idea, thiB, encoding the periplasmic subunit of a thiamin-specific thiamin ABC transporter, was identified in HIMB59 (Table 1).
The new data reported here indicate that thiamin cycling in the oceans may follow complex patterns and involve multiple processes and intermediates. Whereas we show that marine microbes can release HMP into the surrounding environment (Table 2), some phytoplankton exude thiamin (Carlucci and Bowes, 1970a, 1970b). Although thiamin is labile in seawater (Gold, 1968; Gold et al., 1966), its decomposition products in seawater have not been fully characterized and the effect of various environmental factors on degradation are poorly understood. For example, thiamin is a light-sensitive molecule that is readily cleaved by UV-B radiation to AmMP and other products (Okumura, 1961; Machlin, 1984). Although no measurements of AmMP concentrations in the environment have been reported, the physiological responses of phytoplankton to AmMP (Droop, 1958; Lewin, 1962) and the presence of tenA genes in some bacterial genomes that lack the thiC gene (Supplementary Table S1), including Pelagibacterales sp. strain IMCC9063 (Table 1), suggest that environmental AmMP is present, and might also be an important growth determinant in marine ecosystems.
Light-mediated decay of thiamin may be an important factor in thiamin geochemistry and influence HMP production patterns in marine surface waters. The depth profiles showing that the dissolved HMP maximum coincides with the deep chlorophyll maximum (Figure 4) suggest that marine phytoplankton may be important HMP producers. Intriguingly, previous studies reported diel periodicity in the transcription and translation of thiC (the HMP synthase) in laboratory cultures of Prochlorococcus MED4 (Waldbauer et al., 2012). Similarly, the abundance of environmental transcripts mapping to thiC of Synechococcus sp. followed a diel pattern (Ottesen et al., 2013). In both reports, maximum thiC transcript levels were observed in the mid-afternoon, shortly after the periods of highest light intensity. We speculate that the large differences in dissolved HMP concentrations from profiles collected at different times (Figure 4) may be an indication that HMP exudation by thiC-containing cyanobacteria also follows a diel pattern. Although measurements of dissolved vitamins (and precursors) reflect equilibrium concentrations, not fluxes, reports of rapid rates of 3H-thiamin uptake by plankton communities (Koch et al., 2012) suggest that rapid water column vitamin depletion due to biological scavenging is feasible. The notable production of HMP by Synechococcus sp. WH8102 and modest exudation by Prochlorococcus MED4 batch cultures (Table 2) is consistent with the idea that cyanobacteria are important HMP producers; however, diel patterns of HMP production were not tested in our experiments.
The absence of thiC, and thus the requirement for exogenous thiamin pyrimidines, is not unique to the Pelagibacterales, but is broadly and unevenly distributed among diverse microbial taxa inhabiting marine waters. Incomplete thiamin biosynthetic gene complements were previously reported in the genomes of the uncultivated SAR86 clade of marine γ-proteobacteria (Dupont et al., 2012) and in some phytoplankton (reviewed in Bertrand and Allen, 2012; Helliwell et al., 2013). Genes for ThiC are also absent from the genomes of many other ecologically important marine bacteria and archaea (Supplementary Table S1). The observation that canonical thiamin biosynthetic pathways are incomplete in sequenced organisms was further mirrored in metagenomic data sets. Comparisons of the abundances of thiC, thiD and thiG across a metagenomic depth profile from the Sargasso Sea found that thiC genes were depleted relative to thiD and thiG genes at 0, 40 and 80 m, but near the deep chlorophyll maximum, copies of thiC exceeded those of thiD (Supplementary Figure S8). The relative deficiency of thiC to other essential thiamin biosynthesis genes in shallow waters is consistent with the idea that HMP salvage is important for thiamin synthesis at those depths.
We postulate that ThiV sodium:solute symporters constitute a new family of thiamin pyrimidine transport proteins. Previously Worden et al. hypothesized that ThiV and its homologs (identified as ‘SSSF-P') might transport thiamin precursors in some eukaryotic phytoplankton and SAR11 that lacked canonical thiamin biosynthesis genes (Worden et al., 2009). Noting conservation of the relationship between TPP and ThiV across taxa, they concluded ThiV and its homologs might represent ‘ancient thiamine-pathway components', but their function remained uncertain. A phylogeny of bacterial and archaeal ThiV orthologs supports this interpretation by showing that thiV genes colocalize with genes encoding for thiamin pyrimidine salvage enzymes (tenA in archaeal genomes and with thiD, thiM and thiE in the Alkaliphilus oremlandii genome) (Figure 2), implying that ThiV orthologs transport thiamin pyrimidines (HMP or AmMP). We speculate that Ca. P. ubique regulates the acquisition of HMP from the environment by controlling the expression of ThiV with a ThPP-binding riboswitch, in a manner akin to the ThPP-binding riboswitch regulation of de novo HMP synthesis (via ThiC) in other organisms (Winkler et al., 2002). When thiamin is bound to ThPP riboswitches, transcription and translation of the downstream coding sequence is repressed, and thus the detection of ThiV and other ThPP-regulated gene products in metaproteomes may be useful indicators of thiamin deprivation in the environment. For example, peptides mapping to Pelagibacter ThiV orthologs were detected in environmental metaproteomes from the Sargasso Sea (Sowell et al., 2009), but not the Southern Ocean (Williams et al., 2012), perhaps indicating differences in the thiamin status of the two biomes.
The dependence of Ca. P. ubique, and likely other Pelagibacterales, on HMP implies that these cells gain an advantage by outsourcing HMP production to other plankton, in essence relying on HMP as a publically available commodity. This perspective is consistent with genome streamlining theory, and previous reports of unusual nutrient requirements associated with genome reduction in Pelagibacterales (Tripp et al., 2008; Carini et al., 2013). Streamlining theory predicts that atypical nutrient requirements can arise in microorganisms that have large effective population sizes in response to selection favoring small cell size and the efficient use of limiting nutrient resources (Giovannoni et al., 2005). The ‘Black Queen Hypothesis' explored the coevolutionary implications of genome streamlining theory, examining the broader context of adaptive gene loss in a framework that considered competition for public goods (Morris et al., 2012). In this context, because the Pelagibacterales depend on environmental HMP, there is potential for Pelagibacter growth limitation by HMP, intimately tying the success of these organisms to HMP producers.
Because Ca. P. ubique cells are among the smallest known, and replicate efficiently at very low nutrient concentrations, elucidating the trace nutrient requirements of these cells is technically challenging. Even in a defined minimal medium, when precautions were taken to minimize trace vitamin background, Ca. P. ubique reached 2–3 × 107 cells ml−1 in the absence of added vitamins or precursors (Figure 3 and Supplementary Figures S6 and S7). These yields are within a factor of two of theoretical yields (1.8 × 107 cells ml−1) based on the cellular HMP requirement (Supplementary Figure S5) and the amount of ‘background' HMP measured in the medium (12 pM). This ‘background' HMP disappeared in the presence of Ca. P. ubique, implying consumption of the nutrient (Table 2). Previously, background levels of vitamins in heterotrophic growth medium were proposed to underlie scant growth of vitamin auxotrophs in the absence of added vitamins (Norman et al., 1981; Wu et al., 2005), and the difficulty associated with thiamin removal from growth medium has been noted (Button, 1968). The number of HMP molecules required per Ca. P. ubique cell is on the order of 400 molecules per cell (Supplementary Figure S5). Assuming each HMP molecule is used to make one thiamin molecule, and an estimate of 6 fg carbon per cell (unpublished data), the thiamin/carbon ratio of Ca. P. ubique was calculated to be 25 ng thiamin per mg carbon—similar to the values measured for marine phytoplankton (5–100 ng thiamin per mg carbon; Carlucci and Bowes, 1972; Brown et al., 1999). Thus, the cell titers we observed in the absence of added HMP are consistent with the explanation that even pure reagents (e.g., 98–99%) and water from reverse osmosis purifiers can contain very low concentrations of vitamins and vitamin precursors—enough to support the growth of cells that require miniscule amounts of vitamins.
Contaminating HMP was detected in the thiamin stock solution that was added to thiamin-amended treatments. The level of HMP ‘contamination' in the concentrated thiamin stock was measured (via LC-MS) to be ∼2.6 nM HMP per 1 μM thiamin (=0.0012 g HMP per g thiamin) (Supplementary Figure S9). The unintended addition of approximately 2.6 nM HMP as a contaminant of the thiamin stock is the probable explanation for the growth restoration by thiamin at culture concentrations of 1 μM (Figure 3). The source of contaminating HMP appears to be the result of the commercial thiamin manufacturing process. Contaminating amounts of HMP in the AmMP stock could not be determined because HMP and AmMP have similar liquid chromatography retention times, and thus the application of large amounts of AmMP to the chromatography column obscured the detection of possible traces of HMP. We propose that HMP contamination in the AmMP preparation is also a plausible explanation for the slightly elevated yields at high AmMP concentrations.
This investigation illustrates the value of combining metabolic reconstruction from genomes with experimentation in the laboratory and field measurements of specific compounds to explore biogeochemical cycles. The demonstration that HMP exclusively satisfies the thiamin requirement of a highly abundant marine organism (Figure 3), is found in the ocean (Figure 4), and is exuded by some marine organisms (Table 2), identifies this compound as an important, previously unknown growth factor in marine systems. It is particularly surprising that thiamin and AmMP were not used by Ca. P. ubique, implying that HMP-producing organisms potentially could exert control over Pelagibacterales populations. Extending these findings outside of the Pelagibacterales, multiple genomes of cosmopolitan marine bacteria display incomplete thiamin synthesis pathways (Supplementary Table S1), suggesting that thiamin moiety scavenging may be a common strategy in marine waters. The specific mechanism of HMP exudation by marine phytoplankton is unknown. It is possible that in high light environments, intracellular thiamin is relatively unstable, preventing repression of the ThPP-regulated HMP synthase gene (thiC), and resulting in HMP overproduction. However, HMP might also partition to the membrane and from there to the extracellular environment because it is relatively hydrophobic, or its exudation could be driven by coevolutionary interactions. As yet, there is no evidence that favors one of these alternatives over another. A more complete understanding of HMP production patterns, as they pertain to vitamin cycling, will likely be important for understanding turnover and connectedness in plankton communities (Fuhrman et al., 2006; Steele et al., 2011).
Acknowledgments
This work was supported by the Gordon and Betty Moore Foundation's Marine Microbiology Initiative and National Science Foundation grant OCE-0802004. We thank Diana Downs, Woongye Chung, Alyson Santoro, William Orsi and Jeff H Chang for useful dialogue pertinent to experimental design and manuscript preparation, Kimberly Halsey for phytoplankton cultures and manuscript revisions, Brateen Shome for synthesizing and characterizing HMP and AmMP and the crew of the R/V Atlantic Explorer for assistance during seawater collection. Mass spectrometry was performed at the Oregon State University Mass Spectrometry Facility.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abascal F, Zardoya R, Posada D. (2005). ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21: 2104–2105. [DOI] [PubMed] [Google Scholar]
- Barada LP, Cutter L, Montoya JP, Webb EA, Capone DG, Sañudo-Wilhelmy SA. (2013). The distribution of thiamin and pyridoxine in the western tropical North Atlantic Amazon River plume. Front Microbiol 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand EM, Allen AE. (2012). Influence of vitamin B auxotrophy on nitrogen metabolism in eukaryotic phytoplankton. Front Microbiol 3: 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand EM, Allen AE, Dupont CL, Norden-Krichmar TM, Bai J, Valas RE et al. (2012). Influence of cobalamin scarcity on diatom molecular physiology and identification of a cobalamin acquisition protein. Proc Natl Acad Sci USA 109: E1762–E1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand EM, Saito MA, Rose JM, Riesselman CR, Lohan MC, Noble AE et al. (2007). Vitamin B12 and iron colimitation of phytoplankton growth in the Ross Sea. Limnol Oceanogr 52: 1079–1093. [Google Scholar]
- Brown MR, Mular M, Miller I, Farmer C, Trenerry C. (1999). The vitamin content of microalgae used in aquaculture. J Appl Phycol 11: 247–255. [Google Scholar]
- Button D. (1968). Selective thiamine removal from culture media by ultraviolet irradiation. Appl Microbiol 16: 530–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carini P, Steindler L, Beszteri S, Giovannoni SJ. (2013). Nutrient requirements for growth of the extreme oligotroph ‘Candidatus Pelagibacter ubique' HTCC1062 on a defined medium. ISME J 7: 592–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K. (2009). Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J 3: 283–295. [DOI] [PubMed] [Google Scholar]
- Carlucci A, Bowes PM. (1970. a). Production of vitamin B12, thiamine, and biotin by phytoplankton. J Phycol 6: 351–357. [Google Scholar]
- Carlucci A, Bowes PM. (1972). Vitamin B12, thiamine, and biotin contents of marine phytoplankton. J Phycol 8: 133–137. [Google Scholar]
- Carlucci A, Bowes PM. (1970. b). Vitamin production and utilization by phytoplankton in mixed culture. J Phycol 6: 393–400. [Google Scholar]
- Castresana J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17: 540–552. [DOI] [PubMed] [Google Scholar]
- Croft M, Lawrence AD, Raux-Deery E, Warren MJ, Smith AG. (2005). Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nature 438: 90–93. [DOI] [PubMed] [Google Scholar]
- Croft M, Warren M, Smith A. (2006). Algae need their vitamins. Eukaryotic Cell 5: 1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft MT, Moulin M, Webb ME, Smith AG. (2007). Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci USA 104: 20770–20775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs DM. (1992). Evidence for a new, oxygen-regulated biosynthetic pathway for the pyrimidine moiety of thiamine in Salmonella typhimurium. J Bacteriol 174: 1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droop MR. (1958). Requirement for thiamine among some marine and supra-littoral protista. Mar Biol Assoc UK 37: 323–329. [Google Scholar]
- Dupont CL, Rusch DB, Yooseph S, Lombardo M-J, Richter RA, Valas R et al. (2012). Genomic insights to SAR86, an abundant and uncultivated marine bacterial lineage. ISME J 6: 1186–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S. (2006). Annually reoccurring bacterial communities are predictable from ocean conditions. Proc Natl Acad Sci USA 103: 13104–13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Hayakawa DH, Tripp HJ, Stingl U, Givan SA, Cho J-C et al. (2008). The small genome of an abundant coastal ocean methylotroph. Environ Microbiol 10: 1771–1782. [DOI] [PubMed] [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, Podar M, Vergin KL, Baptista D et al. (2005). Genome streamlining in a cosmopolitan oceanic bacterium. Science (New York, NY) 309: 1242–1245. [DOI] [PubMed] [Google Scholar]
- Gold K. (1968). Some factors affecting the stability of thiamine. Limnol Oceanogr 13: 185–188. [Google Scholar]
- Gold K, Roels OA, Bank H. (1966). Temperature dependent destruction of thiamine in seawater. Limnol Oceanogr 11: 410–413. [Google Scholar]
- Helliwell KE, Wheeler GL, Smith AG. (2013). Widespread decay of vitamin-related pathways: coincidence or consequence? Trends Genet 29: 469–478. [DOI] [PubMed] [Google Scholar]
- Ishida S, Tazuya-Murayama K, Kijima Y, Yamada K. (2008). The direct precursor of the pyrimidine moiety of thiamin is not urocanic acid but histidine in Saccharomyces cerevisiae. J Nutr Sci Vitaminol (Tokyo) 54: 7–10. [DOI] [PubMed] [Google Scholar]
- Jenkins AH, Schyns G, Potot S, Sun G, Begley TP. (2007). A new thiamin salvage pathway. Nat Chem Biol 3: 492–497. [DOI] [PubMed] [Google Scholar]
- Jurgenson CT, Begley TP, Ealick SE. (2009). The structural and biochemical foundations of thiamin biosynthesis. Annu. Rev. Biochem. 78: 569–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunakaran R, Ebert K, Harvey S, Leonard ME, Ramachandran V, Poole PS. (2006). Thiamine is synthesized by a salvage pathway in Rhizobium leguminosarum bv. viciae strain 3841. J Bacteriol 188: 6661–6668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch F, Hattenrath-Lehmann TK, Goleski JA, Sanudo-Wilhelmy S, Fisher NS, Gobler CJ. (2012). Vitamin B1 and B12 uptake and cycling by plankton communities in coastal ecosystems. Front Microbiol 3: 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengeler JW, Drews G, Schlegel HG. (1999) Biology of the Prokaryotes. Wiley-Blackwell: Malden, MA, USA. [Google Scholar]
- Lewin RA (ed.). (1962). Organic micronutrients. In: Physiology and Biochemistry of Algae. Academic Press: New York, NY, USA, pp 141–156. [Google Scholar]
- Machlin LJ (ed.). (1984) Handbook of Vitamins: Nutritional, Biochemical, and Clinical Aspects. Marcel Dekker, Inc.: New York, NY, USA. [Google Scholar]
- Martinez-Gomez N, Downs D. (2008). ThiC is an [Fe-S] cluster protein that requires AdoMet to generate the 4-amino-5-hydroxymethyl-2-methylpyrimidine moiety in thiamin synthesis. Biochemistry 47: 9054–9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Ames T, DP S, Weinberg Z, Schwalbach M, Giovannoni S et al. (2009). Identification of candidate structured RNAs in the marine organism ‘Candidatus Pelagibacter ubique'. BMC Genom 10: 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel FMM, Price NM. (2003). The biogeochemical cycles of trace metals in the oceans. Science (New York, NY) 300: 944–947. [DOI] [PubMed] [Google Scholar]
- Morris JJ, Lenski RE, Zinser ER. (2012). The Black Queen Hypothesis: evolution of dependencies through adaptive gene loss. mBio 3: e00036–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris R, Rappé MS, Connon SA, Vergin KL, Siebold WA, Carlson CA et al. (2002). SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420: 806–810. [DOI] [PubMed] [Google Scholar]
- Natarajan K. (1970). Distribution and significance of vitamin B12 and thiamine in the subarctic Pacific Ocean. Limnol Oceanogr 15: 655–659. [Google Scholar]
- Natarajan K, Dugdale R. (1966). Bioassay and distribution of thiamine in the sea. Limnol Oceanogr 11: 621–629. [Google Scholar]
- Natarajan KV. (1968). Distribution of thiamine, biotin, and niacin in the sea. Appl Microbiol 16: 366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman SM, Maier VP, Echols LC. (1981). Development of a defined medium for growth of Cercospora-rosicola Passerini. Appl Environ Microbiol 41: 334–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura K. (1961). Decomposition of thiamine and its derivatives by ultraviolet radiation. J Vitaminol (Kyoto) 24: 158–163. [Google Scholar]
- Ottesen EA, Young CR, Eppley JM, Ryan JP, Chavez FP, Scholin CA et al. (2013). Pattern and synchrony of gene expression among sympatric marine microbial populations. Proceedings of the National Academy of Sciences 110: E488–E497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzeca C, Tovar-Sanchez A, Agusti S, Reche I, Duarte C, Taylor G et al. (2006). B vitamins as regulators of phytoplankton dynamics. EOS Trans 87: 593–596. [Google Scholar]
- Reddick JJ, Nicewonger R, Begley TP. (2001). Mechanistic studies on thiamin phosphate synthase: evidence for a dissociative mechanism. Biochemistry 40: 10095–10102. [DOI] [PubMed] [Google Scholar]
- Rippka R, Coursin T, Hess W, Lichtlé C, Scanlan DJ, Palinska KA et al. (2000). Prochlorococcus marinus Chisholm et al. 1992 subsp. pastoris subsp. nov. strain PCC 9511, the first axenic chlorophyll a2/b2-containing cyanobacterium (Oxyphotobacteria). Int J Syst Evol Microbiol 50(Part 5): 1833–1847. [DOI] [PubMed] [Google Scholar]
- Robbertse B, Yoder RJ, Boyd A, Reeves J, Spatafora JW. (2011). Hal: an automated pipeline for phylogenetic analyses of genomic data. PLoS Curr 3: RRN1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Hebbeln P, Eudes A, Beek ter J, Rodionova IA, Erkens GB et al. (2008). A novel class of modular transporters for vitamins in prokaryotes. J Bacteriol 191: 42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. (2002). Comparative genomics of thiamin biosynthesis in procaryotes. J Biol Chem 277: 48949–48959. [DOI] [PubMed] [Google Scholar]
- Sañudo-Wilhelmy SA, Cutter LS, Durazo R, Smail EA, Gomez- Consarnau L, Webb EA et al. (2012). Multiple B-vitamin depletion in large areas of the coastal ocean. Proc Natl Acad Sci USA 109: 14041–14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer K, Stolz J, Scherer S, Fuchs TM. (2009). Both thiamine uptake and biosynthesis of thiamine precursors are required for intracellular replication of Listeria monocytogenes. J Bacteriol 191: 2218–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpton TJ, Jospin G, Wu D, Langille MG, Pollard KS, Eisen JA. (2012). Sifting through genomes with iterative-sequence clustering produces a large, phylogenetically diverse protein-family resource. BMC bioinformatics 13: 264–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowell SM, Wilhelm LJ, Norbeck AD, Lipton MS, Nicora CD, Barofsky DF et al. (2009). Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J 3: 93–105. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY et al. (2011). Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J 5: 1414–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrash JC, Temperton B, Swan BK, Landry ZC, Woyke T, Delong EF et al. (2014). Single-cell enabled comparative genomics of a deep ocean SAR11 bathytype. ISME J e-pub ahead of print 23 January 2014; doi:10.1038/ismej.2013.243. [DOI] [PMC free article] [PubMed]
- Tripp HJ. (2013). The unique metabolism of SAR11 aquatic bacteria. J Microbiol 51: 147–153. [DOI] [PubMed] [Google Scholar]
- Tripp HJ, Kitner JB, Schwalbach MS, Dacey JWH, Wilhelm LJ, Giovannoni SJ. (2008). SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature 452: 741–744. [DOI] [PubMed] [Google Scholar]
- Waldbauer JR, Rodrigue S, Coleman ML, Chisholm SW. (2012). Transcriptome and proteome dynamics of a light-dark synchronized bacterial cell cycle. PLoS One 7: e43432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb E, Claas K, Downs D. (1998). thiBPQ encodes an ABC transporter required for transport of thiamine and thiamine pyrophosphate in Salmonella typhimurium. J Biol Chem 273: 8946–8950. [DOI] [PubMed] [Google Scholar]
- Wightman R, Meacock PA. (2003). The THI5 gene family of Saccharomyces cerevisiae: distribution of homologues among the hemiascomycetes and functional redundancy in the aerobic biosynthesis of thiamin from pyridoxine. Microbiology 149: 1447–1460. [DOI] [PubMed] [Google Scholar]
- Williams TJT, Long EE, Evans FF, Demaere MZM, Lauro FMF, Raftery MJM et al. (2012). A metaproteomic assessment of winter and summer bacterioplankton from Antarctic peninsula coastal surface waters. ISME J 6: 1883–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR. (2002). Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419: 952–956. [DOI] [PubMed] [Google Scholar]
- Winkler WC, Breaker RR. (2005). Regulation of bacterial gene expression by riboswitches. Annu Rev Microbiol 59: 487–517. [DOI] [PubMed] [Google Scholar]
- Worden AZ, Lee J-H, Mock T, Rouze P, Simmons MP, Aerts AL et al. (2009). Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science (New York, NY) 324: 268–272. [DOI] [PubMed] [Google Scholar]
- Wrenger C, Eschbach M-L, Müller IB, Laun NP, Begley TP, Walter RD. (2006). Vitamin B1 de novo synthesis in the human malaria parasite Plasmodium falciparum depends on external provision of 4-amino-5-hydroxymethyl-2-methylpyrimidine. Biol Chem 387: 41–51. [DOI] [PubMed] [Google Scholar]
- Wu H, Ito K, Shimoi H. (2005). Identification and characterization of a novel biotin biosynthesis gene in Saccharomyces cerevisiae. Appl Environ Microbiol 71: 6845–6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Ma X-D, Chen F-E. (2012). Development of two scalable syntheses of 4-amino-5-aminomethyl-2-methylpyrimidine: key intermediate for vitamin B1. Org Process Res Dev 16: 57–60. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.