Abstract
A variety of microbially mediated metabolic pathways impact biogeochemical cycling in terrestrial subsurface environments. However, the role that viruses have in influencing microbial mortality and microbial community structure is poorly understood. Here we investigated the production of viruses and change in microbial community structure within shallow alluvial aquifer sediment slurries amended with 13C-labeled acetate and nitrate. Biostimulation resulted in production of viruses concurrent with acetate oxidation, 13CO2 production and nitrate reduction. Interestingly, change in viral abundance was positively correlated to acetate consumption (r2=0.6252, P<0.05) and 13CO2 production (r2=0.6572, P<0.05); whereas change in cell abundance was not correlated to acetate consumption or 13CO2 production. Viral-mediated cell lysis has implications for microbial community structure. Betaproteobacteria predominated microbial community composition (62% of paired-end reads) upon inoculation but decreased in relative abundance and was negatively correlated to changes in viral abundance (r2=0.5036, P<0.05). As members of the Betaproteobacteria decreased, Gammaproteobacteria, specifically Pseudomonas spp., increased in relative abundance (82% of paired-end reads) and was positively correlated with the change in viral abundance (r2=0.5368, P<0.05). A nitrate-reducing bacterium, Pseudomonas sp. strain Alda10, was isolated from these sediments and produced viral-like particles with a filamentous morphology that did not result in cell lysis. Together, these results indicate that viruses are linked to carbon biogeochemistry and community structure in terrestrial subsurface sediments. The subsequent cell lysis has the potential to alter available carbon pools in subsurface environments, additionally controlling microbial community structure from the bottom-up.
Keywords: carbon cycle, nitrate, subsurface microbial ecology, viruses
Introduction
Microbial life in the terrestrial subsurface has been estimated to represent one-third of all microorganisms on Earth, bearing an estimated 22–215 Pg of C (Whitman et al., 1998). Although such a significant proportion of life on Earth survives in these subsurface environments, we know very little about the interactions between indigenous microorganisms, specifically between bacteria and viruses. Recent observations of viruses in shallow and deep subsurface environments have reported viral abundance ranging from 105 to 107 ml−1, often exceeding cell abundance, with virus-to-cell ratios (VCRs) ranging from 0.4 to 18 (Kyle et al., 2008; Eydal et al., 2009; Roudnew et al., 2012). These results are consistent with VCRs found in surface environments (Srinivasiah et al., 2008). In addition to viral enumeration studies in the subsurface, bacteriophage have also been isolated (Eydal et al., 2009) as well as identified in metagenomic data sets collected from subsurface environments (Anderson et al., 2011; Wrighton et al., 2012) and in completed genome sequences of subsurface isolates (Coombs, 2009). Together, these results suggest active viral infection of subsurface microbial communities that could further influence subsurface biogeochemical cycling.
Nutrient and energy limitations in terrestrial subsurface environments result in respiration rates that are orders of magnitude that are slower relative to surface soil (Chapelle and Lovley, 1990; Phelps et al., 1994; Onstott et al., 1999; Price and Sowers, 2004). The availability of photosynthetically derived organic carbon in subsurface systems hydrologically connected to the surface results in bottom-up control of community structure (Gibert and Deharveng, 2002; Foulquier et al., 2010,2011). Top-down controls of microbial community structure by viral predation (Bouvier and Del Giorgio 2007; Šimek et al., 2007; Zhang et al., 2007; Boras et al., 2009) remain poorly understood in subsurface environments relative to surface aquatic systems. Viral predation in surface pelagic ecosystems has been demonstrated to control food webs as well as biogeochemical cycles (carbon, nitrogen and phosphorus cycling) by liberation of cell-associated carbon and nutrients (Noble and Fuhrman, 1999; Middelboe et al., 2003; Lennon and Martiny, 2008). Viral-mediated bacterial mortality may also increase microbial diversity by ‘killing the winner' as well as increasing genetic diversity through horizontal gene transfer (Thingstad and Lignell, 1997; Fuhrman, 1999; Wommack and Colwell, 2000; Weinbauer, 2004; Suttle, 2007; Winter et al., 2010). Although the relationship between viruses and microbial communities in surface pelagic systems has been firmly established, a similar relationship between viruses and bacterial hosts in subsurface soils and sedimentary environments remains poorly understood (Kimura et al., 2008). Enhanced cell lysis mediated by top-down viral predation further facilitates the liberation of organic carbon and nutrients stored in cellular biomass, cellular debris or metabolites such as acetate or amino acids (Middelboe and Jørgensen, 2006) that can subsequently influence community structure from the bottom-up. Increases in labile organic carbon can thus further promote microbial activity/carbon turnover especially in organic carbon limited environments such as the subsurface. The extent to which this occurs in terrestrial subsurface systems is not yet understood. These interactions are not only important in natural systems, but viral-mediated cell lysis could also alter biostimulated microbial communities, altering the efficacy of bioremediation technologies.
Given the majority of biomass on Earth is contained within terrestrial soil and sedimentary environments, it is important to understand the role of viruses. In an effort to begin to assess the significance of viral infection in terrestrial subsurface environments, we investigated the production of viruses in response to biostimulation of nitrate contaminated alluvial aquifer sediment with 13C-labeled acetate and nitrate. Here we report the change in bacterial community composition and viral abundance concurrent with acetate and nitrate consumption and 13CO2 production. In addition, we isolated a predominant member of the nitrate-reducing microbial community and revealed an infection with a filamentous bacteriophage. Here we demonstrate a link between the production of viruses, members of the bacterial community and mineralization of organic matter (CO2 production) in terrestrial subsurface sediments.
Materials and methods
Source of sediment
Subsurface sediment was collected using direct push Geoprobe coring (Geoprobe Systems, Salinas, KS, USA) from the saturated zone of a nitrate contaminated (0.48–0.61 mM or 30–38 p.p.m.) sand and gravel alluvial aquifer (6.1–19.5 m below surface) in central Nebraska, USA in the Platte River floodplain (40°52′40.28′′N 98°28′32.31′′W). Core liners were sterilized by rinsing with 80% ethanol and placed under UV in a sterile biological cabinet and sealed before use. Sediment cores were capped, sealed with paraffin and placed into gastight sheaths under 100% argon immediately after collection to maintain anoxic conditions (Snow, 1996). Anoxic conditions within the borehole were verified (−22 to −27 mV) immediately after groundwater collection by a handheld pH/ORP probe (pH100, YSI Environmental, Yellow Springs, OH, USA). Gastight sheaths containing sealed sediment cores were transported back to the laboratory in a cooler and placed in a 4 °C cold room until processing. Sediment cores were sectioned under anoxic conditions in a Coy anaerobic chamber (N2:CO2:H2 atmosphere; 76:19:5). Each section was homogenized, sealed under vacuum in sterile, gastight glass jars and stored in the dark at 4 °C. Sediment sections harboring the most abundant nitrate-reducing microbial community (17.9–18.6 m; Supplementary Figure S1) were selected for the stimulation experiments described below.
Stimulation of viral production in nitrate-reducing sediment slurries
Anoxic (100% argon headspace) artificial groundwater medium (0.235 g l−1 KH2PO4, 0.09 g l−1 K2HPO4, 0.1 g l−1 NH4Cl, 0.0248 g l−1 MgCl2, 0.141 g l−1 CaCl2 and 5 ml l−1 vitamin and mineral mix (Lovley and Phillips, 1988)) amended with acetate (final concentration 10 mM) and nitrate (final concentration 10 mM) was inoculated (25% w/v) with subsurface sediment (17.9–18.6 m below surface). Dissolved organic carbon in the form of acetate and an electron acceptor nitrate was added in excess of natural concentrations in order to stimulate a nitrate-reducing microbial community in an effort to observe the production of viruses. Inputs of electron donor and acceptor in excess of natural concentrations would occur in environmental systems experiencing fluxes either naturally or via bioremediation or bioaugmentation. Uniformly labeled 13C-acetate (99%) was used to distinguish amended acetate from the endogenous organic carbon. Treatments with nitrate and/or 13C-acetate omitted served as controls. Sediment slurries were incubated statically in the dark at room temperature. Samples were periodically collected for enumeration of cells and viruses, quantification of CO2, CH4, acetate, NO3−, NO2−, NH4+ and SO42−, and microbial community analyses.
Mitomycin C treatment
Anoxic (headspace 100% N2) artificial groundwater medium (10% w/v) amended with acetate and nitrate (final concentrations 6.25 and 10 mM, respectively) was inoculated with subsurface sediment. Following a 2-day pre-incubation, mitomycin C, a DNA cross-linker commonly used to induce lysogens (Driggers and Schmidt, 1970), was added to a final concentration of 1.0 μg ml−1 to triplicate slurries. Treatments with acetate and nitrate and treatments with mitomycin C omitted served as controls. The slurries were incubated statically in the dark at room temperature for 24 h. Samples for cell and viral enumeration were collected and enumerated as described above.
A nitrate-reducing bacterium, Pseudomonas sp. Alda10 (Supplementary Figure S2), isolated from the subsurface aquifer sediments in this study, was treated with 1.0 μg ml−1 mitomycin C immediately after transferring (1:10 inoculum) a late log-phase culture into fresh artificial groundwater medium amended with 6.25 mM acetate and 10 mM nitrate. Cultures not treated with mitomycin C served as a negative control. All treatments were performed in triplicate. Cultures were incubated statically in the dark at room temperature for 24 h before collection. Samples for cell and viral enumeration were collected and enumerated as described below.
Enumeration of cell and viral abundance
Immediately after collection, sediment slurry samples were treated to separate cells and viruses from sediments. Sodium pyrophosphate (final concentration 10 mM) and sodium ethylene diaminetetraacetic acid (EDTA; pH 8, final concentration 5 mM) were added to the sample (Hewson and Fuhrman, 2003) and incubated at room temperature for 15 min (Wommack et al., 2009). Each sample was sonicated (Branson 450D, 30% power, 0.3 s duty cycle) 1 min on ice three times and manually shaken between each round for 30 s (Wommack et al., 2009). Samples were then filtered through a 0.45-μm PVDF filter for bacterial cells. Following sonication, samples for viral enumeration were treated with DNase I (10 U ml−1 final concentration) to remove extracellular unencapsidated DNA (Corinaldesi et al., 2010) and filtered through 0.2 μm PVDF filters to remove bacteria. This technique would also result in the removal of viruses >0.2 μm. Samples were fixed with electron microscopy grade glutaraldehyde (0.5% final concentration) for 15–30 min at 4 °C and flash frozen in liquid N2 before storage at −80 °C (Brussaard, 2009).
Viruses are operationally defined here as DNase-resistant SYBR Green I fluorescent particles <0.2 μm. SYBR Green I binds dsDNA efficiently, whereas ssDNA and RNA binds dsDNA less efficiently. DNase I treatment was found to influence virus enumeration in this study and was thus necessary for virus enumeration and removal of free DNA or DNA associated with particle surfaces via adsorption using the methods used. Viruses were enumerated via flow cytometry, which is reliable for enumerating most dsDNA viruses. However, viruses that lie near the limit of detection (such as RNA or ssDNA viruses) may be underestimated (Tomaru and Nagasaki, 2007) using this technique. Frozen samples were diluted in Tris-EDTA (10 mM Tris, 1 mM EDTA, pH 8.0) and stained with SYBR Green I (1:20 000 dilution of the commercial stock). Samples were then heated to 80 °C for 10 min in the dark, followed by a period of cooling at room temperature for 5 min. Samples were enumerated on a BD FACSCantoII flow cytometer (488 nm, 20-mW solid-state laser; Franklin Lakes, NJ, USA) with trigger set to green fluorescence, 1 min acquisition time maintaining an event rate of 200–1000 events per second (Brussaard, 2009) with a detection limit ranging between 103 and 104 viruses per ml. Fluorescence from SYBR Green I-stained Tris-EDTA was subtracted from the sample counts. Samples below the detection limit are counted as 0. All samples were enumerated within 1 month of collection. Although the technique used here was consistent with phage titers, this operational definition does not prevent defective viruses, gene transfer agents, RNA and other unknown particles from inclusion in total counts. The majority of particles that stain positive with SYBR Green I are likely infective viruses (Suttle and Fuhrman, 2010).
Bacterial cells (>0.2 μm) were enumerated by epifluorescence microscopy (Hobbie et al., 1977; Noble and Fuhrman, 1998). Frozen samples were thawed and diluted in Tris-EDTA buffer (10 mM Tris, 1 mM EDTA, pH 8.0) and filtered onto 0.2 μm black polycarbonate filters. The filters were stained with SYBR Green I (1:80 dilution) (Suttle and Fuhrman, 2010). Ten to 20 fields were counted with a minimum of 200 counts per filter.
Analyses of stimulated microbial community
Pelleted samples (6000 g, 20 min) were collected daily from acetate and nitrate stimulated treatments and stored at −20 °C until DNA extraction. To study how the microbial community changes with fluxes of organic carbon and nitrate, we specifically followed the bacterial community in the amended sample over the experimental time series. DNA was extracted using the PowerSoil DNA Extraction kit (MoBio, Carlsbad, CA, USA). The V5 hypervariable region of the 16S rRNA gene was amplified and sequenced via Illumina HiSeq2000 analyzer (Illumina Inc., San Diego, CA, USA) (forward primer: 5′-GGMTTAGATACCC-3′, reverse primer: 5′-CCGYCAATTYMTTTRAGTTT-3′ (Lazarevic et al., 2009) (Eureka Genomics, Hercules, CA, USA). Paired-end sequences (2 × 100 bp) were aligned to form 120 bp contigs. Alignments that did not match perfectly or had ambiguous base calls were discarded (Bartram et al., 2011). Downstream analyses were performed on the QIIME 1.4.0 pipeline (Caporaso et al., 2010). Sequences were grouped into operational taxonomic units (OTUs) based on 97% sequence similarity using Uclust v.1.2.22 (Edgar, 2010; Caporaso et al., 2011) and assigned representative sequences. Representative sequences were aligned using the GreenGenes NAST server (DeSantis et al., 2006b), and assigned taxonomies by GreenGenes (DeSantis et al., 2006a; Caporaso et al., 2011). Maximum likelihood with Jukes Cantor correction (FastTree) was used to build a phylogenetic tree of representative sequences from the filtered sequence alignment (Price et al., 2010). Rarefaction curves (10 sampling repetitions without replacement) of observed OTUs were plotted, and richness was estimated (Chao1 estimator) using QIIME (Caporaso et al., 2010). Beta diversity was estimated by weighted UniFrac distance between samples. Samples were clustered based on between-sample distance using unweighted pair group method with arithmetic mean hierarchical clustering. Jackknifing was performed by resampling with replacement at a depth of 100 sequences per sample (Lozupone and Knight, 2005).
Taxonomic identity of the dominant members of the microbial community after enrichment was verified by construction of a small clone library. Thirty-eight clones were prepared on a randomly chosen replicate from day 4 of incubation. The 16S rRNA gene sequence was amplified using Bacterial Universal primers 27F and 1492R (Lane, 1991). Amplified sequences were then ligated into pCR4 vectors and cloned into TOP10 chemically competent Escherichia coli cells (Life Technologies, Carlsbad, CA, USA). Ligated sequences were amplified from transformed cells via colony PCR and sequenced using the M13 forward and reverse primers, resulting in 700 bp partial 16S rRNA gene sequences. Forward sequences were trimmed for quality before being assigned taxonomies through GreenGenes (DeSantis et al., 2006a).
The 16S rRNA gene sequence of Pseudomonas sp. Alda10 and clone library sequences are available on Genbank (KF286228 and KF286229-KF286266, respectively). Illumina sequences were deposited to the Sequence Read Archive: accession numbers SAMN02212409–SAMN02212423.
Viral production lags bacterial growth. As such, changes in predominant bacterial taxa were compared with viral abundance collected on the next sampling period. Lagged correlation between viral abundance change and change in the relative abundance of taxa was calculated using Pearson's product moment correlation method.
Chemical analyses
Acetate, NO2−, NO3− and SO42− were quantified via ion chromatography on a Dionex ICS-3000 (Thermo Scientific, Sunnyvale, CA, USA) with a IonPac AS9-HC column (4 × 250 mm) (Weber et al., 2006). NH4+ was measured colorimetrically (Wetzel and Likens, 1991). Gas samples, CO2 and CH4, were collected via gastight syringe and analyzed on a Varian 430-GC outfitted with a TCD detector and Porapak N 8/10 column (12′ × 1/8′′ × 0.085′′ SS) at a temperature of 40 °C running a carrier gas of UHP argon. Production of 13CO2 and 12CO2 in headspace was measured separately on an Agilent (Santa Clara, CA, USA) 5972 GC-MSD with GS-GASPRO PLOT column (0.32 μm × 30 m) at a temperature of 40 °C using UHP grade helium as the carrier. Gastight syringe samples of headspace were manually injected, and injection/MS transfer line temperatures were set at 200 °C.
Transmission electron microscopy
Viral-like particles (VLPs) were collected from cultures of Pseudomonsas sp. Alda10 following an incubation on 10 mM nitrate and 6.25 mM acetate with a mitomycin C treatment (final concentration of 1 μg ml−1) at mid log phase. Samples were filtered through a 0.2-μm PVDF filter to remove bacterial cells and spotted onto a copper transmission electron microscopy grid containing a Formvar carbon film, stained with uranyl acetate (final concentration 2%) and imaged by transmission electron microscopy (Hitachi H7500, Tokyo, Japan) as previously described (Ackermann, 2009).
Statistics
Linear regression and Pearson's product moment correlations were performed in GraphPad Prism (v5.02, GraphPad Software, Inc., La Jolla, CA, USA). All correlation coefficients are reported as Pearson's product moment correlations. Significance level was set at a P-value of <0.05. Beta diversity statistics were conducted in QIIME 1.4.0 (Caporaso et al., 2010).
Results
13C-Acetate stimulated production of viruses in subsurface sediments under nitrate-reducing conditions
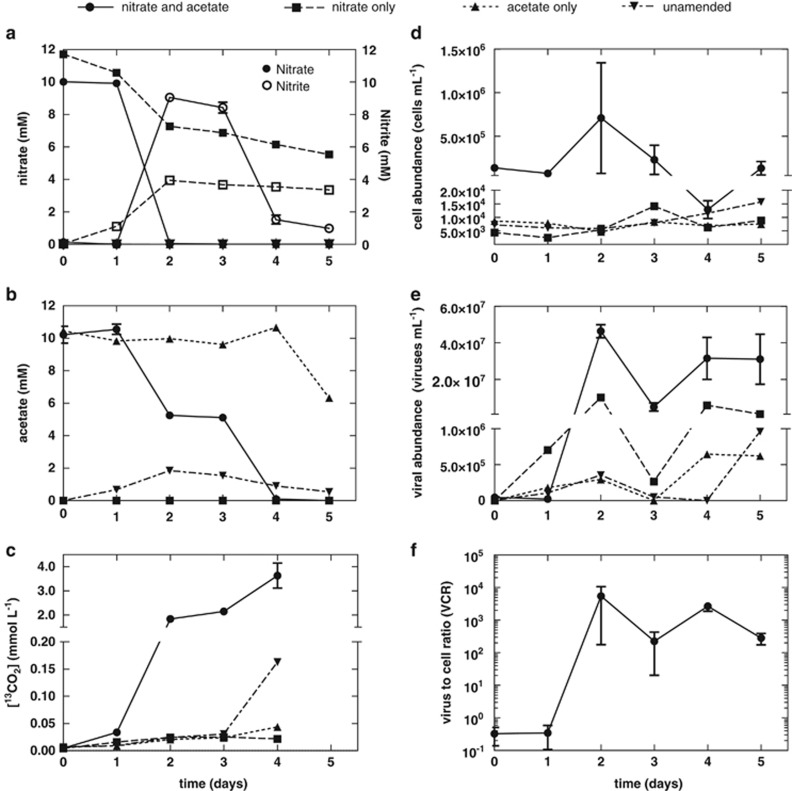
The production of viruses was concomitant with nitrate reduction and acetate oxidation in triplicate subsurface sediment slurries (Figure 1). Although the mean bacterial cell abundance increased to 5.7 × 105 cells per ml sediment slurry within 2 days of incubation in response to acetate oxidation and nitrate reduction to nitrite (Figure 1d), this was a result of the behavior of one replicate that led to a high mean and standard error measurement. No significant changes in bacterial abundance were observed given the high-standard error measurement. However, viral abundance increased 4.6 × 107 viruses per ml (Figure 1e) in response to acetate oxidation and nitrate reduction to nitrite (Figures 1a and b). Fluctuations of the VCR (Figure 1f) were observed over the course of the study. The mean VCR increased from 0.33 to 5400 during this period (Figure 1f), but variability in the lag phase and response between sediment slurry replicates (Supplementary Figure S3) led to a high-standard error measurement (±5200 (s.e.)).
Figure 1.
The production of viruses following inoculation (25% w/v) of subsurface sediment during nitrate reduction. Triplicate slurries were amended with 10 mM 13C-acetate and 10 mM nitrate and compared with singular control treatments amended with 10 mM nitrate only, 10 mM 13C-acetate only, or without any electron donor or acceptor. Over time changes in (a) nitrate and nitrite concentration, (b) acetate concentration, (c) 13CO2 production, (d) cell abundance, (e) viral abundance, and (f) VCR are observed. 13CO2 datum was not collected on day 5 because of destructive sampling. The y axis of panels c, d and e have been split. Symbols denote mean values. Error bars represent s.e.m.
Viral abundance represented the activity within the experimental treatment, whereas cell abundance did not. During the 24-h period between nitrate consumption and nitrite reduction/acetate oxidation, viral abundance rapidly decreased (Figure 1e). Interestingly, acetate oxidation and nitrite reduction did not result in an increase in bacterial abundance but did result in viral production (4.6 × 107 viruses per ml sediment slurry) (Figures 1a, b, d and e). Although N2 and N2O were not measured in this study, N2 or N2O are assumed end products, as nitrite was reduced and no significant ammonium production was observed (Supplementary Figure S4). Sulfate concentrations remained constant, indicating that sulfate reduction did not occur. Methane was not detected in the headspace.
Slurries in which acetate or both acetate and nitrate were omitted did not result in significant increases in viral abundance or increases in bacterial cell abundance (Figures 1d and e). However, nitrate reduction to nitrite was observed (Figure 1a) in controls in which acetate was omitted, which could be a result of the oxidation of naturally occurring organic carbon. Oxidation of organic carbon is supported by the production of 12CO2 (Supplementary Figure S5). This resulted in a slight increase in cell (9.7 × 103 cells per ml sediment slurry) and viral abundance (9.4 × 106 viruses per ml sediment slurry) (Figures 1d and e). Viral production was not significant in acetate-amended/nitrate-omitted cultures because of the unavailability of an electron acceptor.
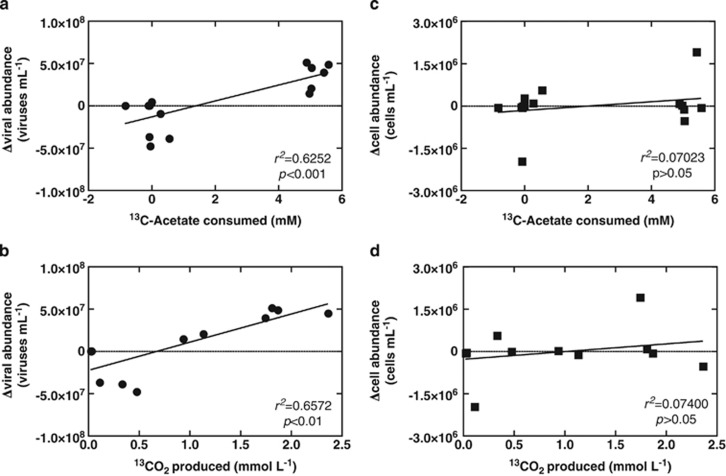
Analysis of the production of viruses was positively correlated with acetate consumption (r2=0.6252, P=0.0004) and 13CO2 production (r2=0.6572, P=0.0014) (Figures 2a and b) in each replicate slurry amended with acetate and nitrate. Interestingly, changes in bacterial abundance were not correlated to acetate consumption (r2=0.07023, P =0.3398) nor 13CO2 production (r2=0.07400, P=0.3924) (Figures 2c and d). No significant 13CO2 production was observed when unlabeled acetate was added or acetate was omitted from sediment slurries, further supporting uptake of the 13C-acetate or respiration of cell debris rather than endogenous sedimentary 13C-organic carbon. These data reflect that periods of viral production during the incubation are associated with periods of high carbon mineralization within the sediment slurry, whereas periods of low carbon transformation are associated with decreases in viral abundance. The same observation is not reflected in cell abundance because of the likely consequence of viral-induced cell lysis.
Figure 2.
Linear regression analyses of changes in viral and cell abundance relative to 13C-acetate and 13CO2. Positive correlations are observed for linear regressions of (a) changes in viral abundance on 13C-acetate consumed and (b) changes in viral production on 13CO2 produced, but no correlation was observed for linear regressions of (c) changes in cell abundance on 13C-acetate consumed and (d) changes in cell abundance on 13CO2 produced.
Changes in bacterial community composition
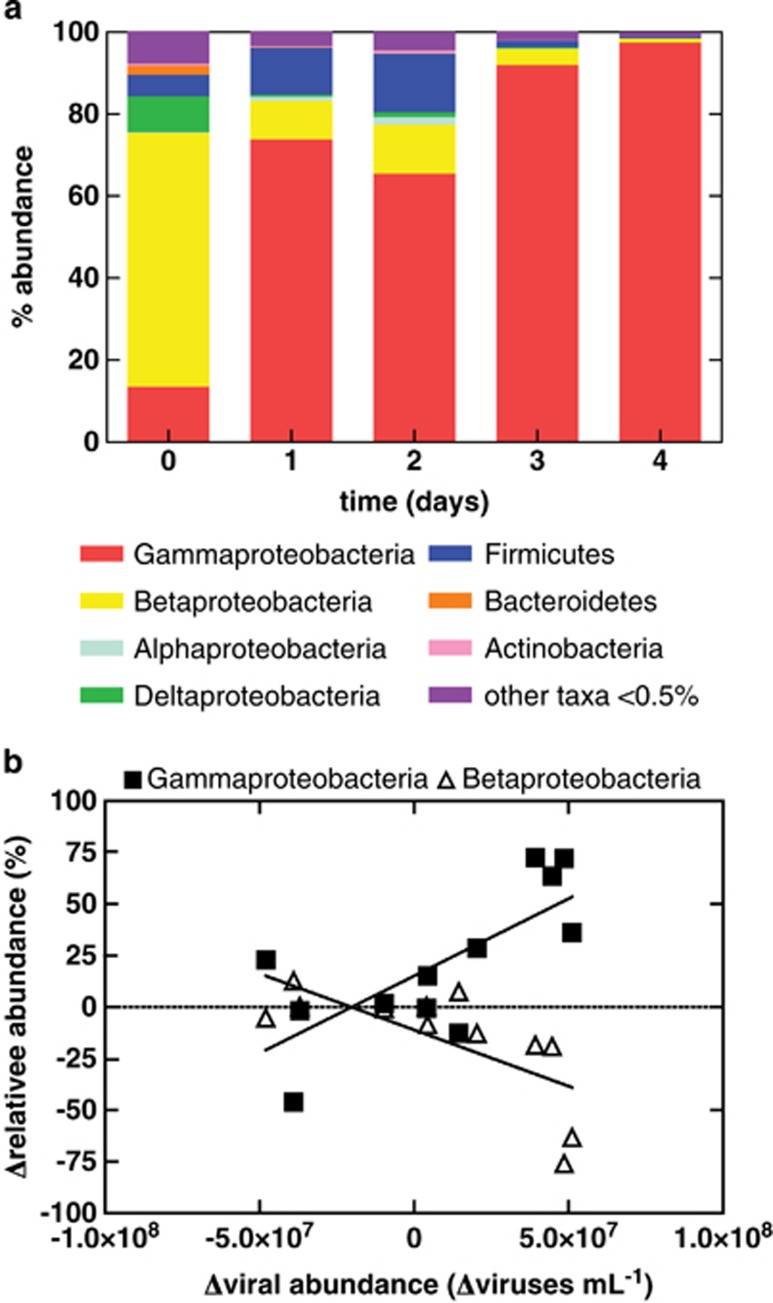
Replicate samples of the indigenous shallow subsurface sediment bacterial community was dominated by the Betaproteobacteria (62% of paired-end reads, Figure 3a), specifically unclassified members of the Oxalobacteraceae (57%) and Commonadacae, Curvibacter (2%) (Table 1). After 1 day of incubation, the slurry bacterial community composition shifted to a community dominated by Gammaproteobacteria (74% of paired-end reads, Figure 3a), specifically Pseudomonas spp., (69%, Table 1) that represented 11% (Table 1) of the microbial community at the beginning of the experiment. A transient increase in members of the Clostridia (Desulfotomaculum sp., 9% Acetobacterium sp., 2% and Desulfosporosinus, 1% Table 1) was also observed. Members of the Betaproteobacteria (Curvibacter, 5% Dechloromonas, 3% and Azoarcus, 1%) also transiently increased. However, after this initial increase after 3 days of incubation, these members decrease in relative abundance, whereas members of the Gammaproteobacteria continue to increase in relative abundance (Figure 3a). By day 3, Pseudomonas spp. accounted for over 82% of all sequences and continued to increase, representing 88% of the microbial community by the end of the experiment (Table 1). The relative abundance of the Betaproteobacteria decreased, representing only 1% of the microbial community by the end of the experiment. The most abundant organisms were cloned and sequenced confirming Pseudomonas spp. as the dominant taxa. Of the 38 clones sequenced, only two non-Pseudomonas spp. clones were identified and classified as Dechloromonas sp.
Figure 3.
(a) Taxonomic composition of bacteria within the sediment enrichment over 4 days of incubation. Relative abundance was averaged across all three replicates. Error bars denote s.e.m. ‘Other taxa' represent OTUs with a relative abundance <0.5%. Specific taxa identified are detailed in Table 1. (b) Correlation analyses between changes in the relative abundance of Gammaproteobacteria vs viral abundances (r2=0.5368 and 0.5036) and changes in the relative abundance of Betaproteobacteria vs viral abundances (P=0.0067 and 0.0097).
Table 1. Average relative abundance of taxa that exceeded 0.5% abundance over the course of the incubation period within nitrate and acetate treatments.
| Day 0 (%) | Day 1 (%) | Day 2 (%) | Day 3 (%) | Day 4 (%) | |
|---|---|---|---|---|---|
| Proteobacteria | |||||
| Gammaproteobacteria | |||||
| Pseudomonadales | |||||
| Pseudomonadaceae | |||||
| Pseudomonas | 10.99 (4.28) | 69.09 (27.85) | 54.64 (19.45) | 82.47 (8.40) | 87.59 (1.62) |
| Unclassified | 0.40 (0.34) | 4.38 (1.88) | 10.29 (7.23) | 9.19 (0.85) | 9.80 (1.35) |
| Xanthomonadales | |||||
| Xanthomonadaceae | |||||
| Lysobacter | 2.25 (0.96) | 0.00096 (0.00096) | 0.00064 (0.00064) | 0 | 0 |
| Sinobacteraceae | |||||
| Unclassified | 0.45 (0.22) | 0 | 0 | 0 | 0 |
| Aeromonadales | |||||
| Aeromonadaceae | |||||
| Aeromonas | 0 | 0.29 (0.088) | 0.52 (0.24) | 0.31 (0.19) | 0.070 (0.011) |
| Betaproteobacteria | |||||
| Burkholderiales | |||||
| Oxalobacteraceae | |||||
| Unclassified Oxalobacteraceae | 57.07 (20.90) | 2.76 (1.51) | 3.56 (1.70) | 0.80 (0.41) | 0.15 (0.022) |
| Comamonadaceae | |||||
| Curvibacter | 1.71 (1.35) | 4.92 (3.18) | 4.79 (2.64) | 1.16 (0.76) | 0.31 (0.043) |
| Rhodocyclales | |||||
| Rhodocyclaceae | |||||
| Dechloromonas | 0.067 (0.057) | 1.34 (0.63) | 2.90 (1.63) | 1.71 (0.96) | 0.41 (0.059) |
| Azoarcus | 0.0045 (0.0045) | 0.44 (0.17) | 0.80 (0.34) | 0.26 (0.14) | 0.11 (0.0098) |
| Propionivibrio | 2.50 (2.50) | 0.0033 (0.0033) | 0.0026 (0.0026) | 0.0030 (0.0016) | 0.00057 (0.00057) |
| Hydrogenophilales | |||||
| Hydrogenophilaceae | |||||
| Thiobacillus | 0.22 (0.22) | 0.0029 (0.0029) | 0.0064 (0.0064) | 0 | 0 |
| Deltaproteobacteria | |||||
| Desulfuromonadales | |||||
| Desulfuromonadaceae | |||||
| Unclassified Desulfuromonadaceae | 8.79 (8.31) | 0.55 (0.23) | 1.17 (0.47) | 0.35 (0.22) | 0.057 (0.0093) |
| Alphaproteobacteria | |||||
| Rhizobiales | |||||
| Bradyrhizobiaceae | |||||
| Bradyrhizobium | 0.10 (0.023) | 0.78 (0.72) | 1.40 (1.30) | 0.033 (0.022) | 0.0060 (0.0049) |
| Phyllobacteriaceae | |||||
| Phyllobacterium | 0.032 (0.016) | 0.19 (0.18) | 0.36 (0.34) | 0.014 (0.0069) | 0.0014 (0.00077) |
| Bacteroidetes | |||||
| Bacteroidetes incertae sedis | |||||
| Prolixibacter | 2.27 (1.91) | 0.0087 (0.0024) | 0.012 (0.0043) | 0.0072 (0.0044) | 0 |
| Firmicutes | |||||
| Clostridia | |||||
| Clostridiales | |||||
| Peptococcaceae | |||||
| Peptococcaceae 2 | |||||
| Desulfotomaculum | 0.0015 (0.0015) | 8.65 (5.44) | 10.82 (6.67) | 1.03 (0.62) | 0.21 (0.016) |
| Peptococcaceae 1 | |||||
| Desulfosporosinus | 5.24 (4.76) | 1.01 (0.50) | 1.93 (0.70) | 0.43 (0.23) | 0.10 (0.017) |
| Eubacteriaceae | |||||
| Acetobacterium | 0 | 1.85 (1.49) | 1.52 (0.80) | 0.23 (0.19) | 0.045 (0.017) |
| Actinobacteria | |||||
| Actinobacteria | |||||
| Actinobacteridae | |||||
| Actinomycetales | |||||
| Micrococcineae | |||||
| Micrococcaceae | |||||
| Arthrobacter | 0.48 (0.23) | 0.13 (0.11) | 0.32 (0.28) | 0.0059 (0.0023) | 0.00076 (0.00075) |
| Unclassified Micrococcineae | 0 | 0.10 (0.065) | 0.33 (0.24) | 0.037 (0.023) | 0.0041 (0.0015) |
| Unclassified bacteria | 0.47 (0.24) | 0.00096 (0.00096) | 0.0013 (0.0013) | 0 | 0 |
| Taxa that did not exceed 0.5% of community in any replicate | 6.74 (0.17) | 3.50 (1.44) | 4.62 (2.11) | 1.94 (0.70) | 1.14 (0.061) |
Standard error of triplicate treatments is listed in parentheses.
0 denotes no detection.
Rarefaction curves of bacterial communities at the beginning of the incubation experiment did not reach an asymptote, indicating that not all taxa were identified (Supplementary Figure S6a). Samples were sequenced at an average depth of 50 000 reads per sample. As few as 33 OTUs were sufficient to describe over 90% of the microbial community for all samples. OTUs that composed fewer than 0.5% of the bacterial community (181–189 taxa) represented only 6.7% of the microbial community. The inoculum bacterial communities had a higher richness than the final enriched bacterial community (Supplementary Figures S6a and b).
Changes in viral abundance are correlated to changes in the relative abundance of Gammaproteobacteria (r2=0.5368, P=0.0067) (Figure 3b), whereas changes in viral abundance are negatively correlated to changes in Betaproteobacteria (r2=0.5036, P=0.0097) (Figure 3b). No statistically significant correlation to viral abundance was observed for other groups. The OTU identified as Pseudomonas was the only Gammaproteobacteria to be positively correlated to changes in viral abundance (r2=0.3551, P=0.041). Initially, the most abundant OTU (∼3–66% of all sequences) was classified as an unclassified Oxalobacteraceae. This OTU was negatively correlated to changes in viral abundance (r2=0.3664, P=0.037). Two other Betaproteobacteria OTUs, both also classified as unclassified Oxalobacteraceae, were also negatively correlated to changes in viral abundance (r2=0.4201, P=0.023 and r2=0.3341, P=0.049).
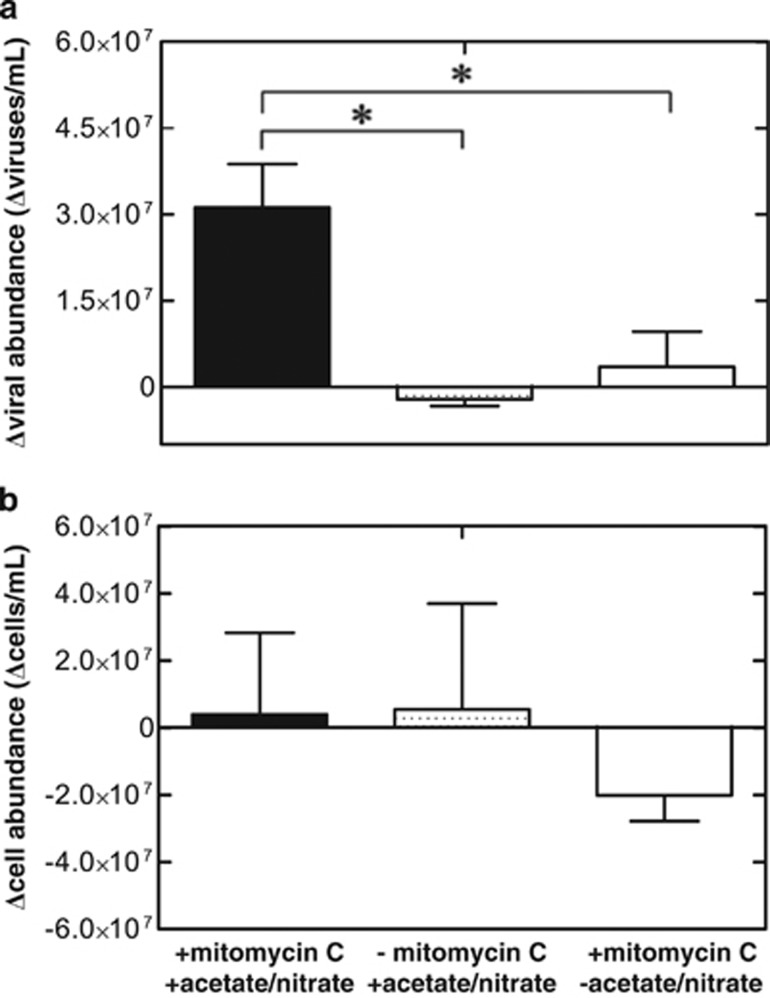
Viral production in mitomycin C-treated sediments and pure culture isolate
Production of viruses (3.1 × 107 viruses per ml sediment slurry) resulted from mitomycin C additions after sediment slurry pre-incubation with nitrate (final concentration 10 mM) and acetate (6.25 mM) for 48 h (Figure 4a). Cell abundances following mitomycin C treatment did not change relative to an unamended control (Figure 4b). When mitomycin C was omitted, triplicate pre-incubated sediment slurries did not result in a change in viral abundance, indicating that indigenous nitrate-reducing cells harbor a bacteriophage. Sediment slurries containing acetate and nitrate, but not pre-incubated for 48 h before mitomycin C treatment, were not significantly different from an unamended control (Supplementary Figure S7). Thus, demonstrating that mitomycin C alone was not sufficient to induce viral production and pre-incubation with acetate and nitrate was necessary to observe significant viral production.
Figure 4.
Mitomycin C treatment with 1.0 μg ml−1 to a sediment slurry (1:10 inoculum w/v) pre-incubated for 2 days with 10 mM nitrate and 6.25 mM acetate resulted in an (a) increase in viral abundance compared with control replicates omitting mitomycin C and control replicates with mitomycin C omitting nitrate and acetate. (b) Changes in cell abundances are not significantly different from controls. ‘*' indicates statistical significance (Bonferonni's multiple comparison test, P<0.05). Bars represent average values from triplicates. Error bars denote s.e.m.
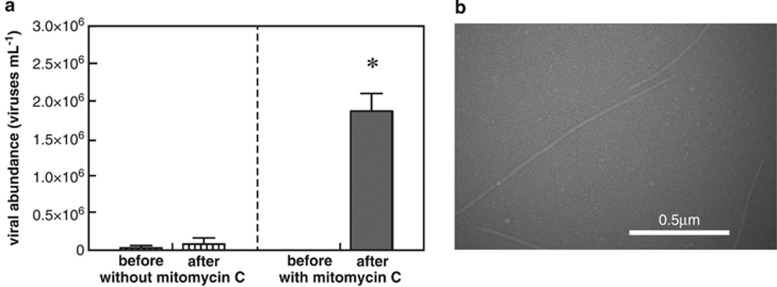
A culture dependent approach was also undertaken to determine lysogenic viral production from a nitrate-reducing bacterium isolated from these sediments. The near complete 16S rRNA gene sequence of the bacterium, strain Alda10, isolated from nitrate-reducing most probable number enrichments was classified as a Pseudomonas species (Supplementary Figure S2). Pseudomonas sp. Alda10 had a 98% 16S rRNA gene sequence identity with the Pseudomonas spp. clones identified in this study. Production of viruses (an increase of 1.9 × 106 viruses per ml) were observed following mitomycin C treatment (1.0 μg ml−1) of Pseudomonas sp. strain Alda10 (Figure 5a). Cultures in which mitomycin C was omitted produced substantially lower numbers of viruses (Figure 5a). Chloroform lysis of cells grown under similar conditions without mitomycin C resulted in viral production greater than cultures amended with mitomycin C (Supplementary Figure S8); thus indicating that mitomycin C may not have necessarily been inducing a lysogen but rather increasing the release of viruses from the cell. Upon examination by transmission electron microscopy, mitomycin C-treated cell-free cultures contained putative VLPs (Figure 5b) that were 17 nm wide and had a variable length up to 1500 nm. Variability in the length of observed VLPs is likely related to breakage during sample preparation. These VLPs are morphologically similar to other filamentous bacteriophage. Similarly sized filamentous phage have been identified in pure culture as well as environmental samples: KSF-1Φ 1200 nm long 7 nm wide (Faruque et al., 2005), Pf1 2000 nm wide (Lorieau et al., 2008), VFJΦ 1400 nm long 7 nm wide (Wang et al 2013), B5 620 nm long 12 nm wide (Chopin et al., 2002); alpine lake filamentous viruses 450–730 nm long 30–40 nm wide (Hofer and Sommaruga, 2001). Together, these results indicate that Pseudomonas sp. Alda10 harbors a bacteriophage infection.
Figure 5.
Mitomycin C amendment (1.0 μg ml−1) of isolate Pseudomonas sp. Alda10 resulted in (a) enhanced viral production compared with control cultures without mitomycin C. ‘*' indicates statistical significance (two-tailed unpaired t-test, P<0.05). Columns represent average of triplicates and error bars denote s.e.m. (b) Transmission electron micrograph of Pseudomonas sp. Alda10 supernatant revealing VLPs with a filamentous morphology.
Discussion
Here we have demonstrated that viruses are significantly linked to carbon biogeochemistry and changes in bacterial community structure within terrestrial subsurface sedimentary systems. The production of viruses within an indigenous shallow subsurface microbial community was stimulated with the addition of nitrate and 13C-labeled acetate. Interestingly, this stimulation led to viral production positively correlated with oxidation of dissolved organic carbon (13C-labeled acetate) and 13CO2 production (Figure 2); whereas changes in bacterial abundance were not correlated with acetate consumption nor 13CO2 production (Figure 2). These results indicate that viruses may be contributing to microbial cell lysis. Lysis in the subsurface not only can influence community structure from the top-down but also has implications for bottom-up control of populations by releasing organic carbon substrates that can be utilized by other members of the microbial community (Noble and Fuhrman, 1999; Middelboe et al., 2003; Lennon and Martiny, 2008). In contrast to surface environments, subsurface microbial communities frequently experience energy limitations that directly impact cell survival and maintenance. Carbon in subsurface environments may be highly recalcitrant (Stevens, 1997), thus viral-induced microbial cell lysis could release labile dissolved organic carbon and particulate organic carbon (Middelboe and Jørgensen, 2006). The release of labile carbon from infected cells may relieve carbon limitation and stimulate metabolism. This is especially important within subsurface environments where microbial communities are carbon and energy limited. In deep-sea benthic sediments, release of 0.37–0.63 Gt C per year from lysed cells is estimated to occur, contributing to 35% of total benthic prokaryotic metabolism (Danovaro et al., 2008). Based on the observation of viral production following stimulation of the host community by addition of electron donor and acceptor, it would not be unreasonable to infer that viral-induced cell mortality would have a similar role in terrestrial subsurface sediments that experience fluxes of carbon and/or an electron acceptor.
The production of viruses in response to carbon and nitrate addition is consistent with previous studies, demonstrating that amendments of organic carbon and/or inorganic nutrients stimulated viral production in pelagic systems (Tuomi et al., 1995; Noble and Fuhrman, 1999; Hewson et al., 2001; Weinbauer et al., 2003; Motegi and Nagata, 2007). However, in our studies, organic carbon alone did not result in significant viral production and required the addition of a terminal electron acceptor (nitrate). This would indicate that nutrients alone were not limiting viral production, but rather the combination of substrates that cells can convert into energy via respiratory processes. These results are consistent with in situ observations reporting higher viral abundance in productive pelagic environments, whereas oligotrophic environments report lower viral abundance (Weinbauer et al., 1993; Hewson et al., 2001; De Corte et al., 2012). Within deep-sea sediments, viral abundance was the lowest where low bacterial growth rates were observed, leading Danovaro et al. (2002) to hypothesize that growth may have a role in limiting virus development. Bacterial growth as a factor controlling virus development is supported by experiments within model bacteriophage. When E. coli host cells are grown in a nutrient-rich complex medium relative to cells grown in a nutrient-limited defined medium, production of virions is greater (Clark et al., 1986). This result is dependent on levels of cAMP, an indicator of host cell energy conditions (Rolfe et al., 1973), demonstrating that viruses may be highly sensitive to the energy available to the host.
Viral production was correlated with the decrease of specific microbial taxa, (Betaproteobacteria), inferring that cell lysis could be controlling taxa from the top-down as influenced by predator–prey dynamics occurring between viruses and microbial community members. However, microbial community structure may also have shifted in response to changes in organic matter pools as a result of viral-mediated cell lysis, in effect altering microbial community structure from the bottom-up. This study did not attempt to distinguish between top-down and bottom-up controls. Associations between total viral abundance and certain OTUs have been observed in microbial association network studies in natural, unmanipulated marine systems using local similarity analysis (Ruan et al., 2006; Fuhrman and Steele, 2008; Steele et al., 2011), but viral abundances were relatively invariant. By manipulating our experimental system with acetate and nitrate, we induced viral production and thus discovered significant correlations with changes in microbial community structure over time relative to the initial inoculum. This effect would be smaller in unmanipulated natural conditions closer to steady-state conditions. Although our use of 16S Illumina tags are not likely to completely capture the strain and subspecies level dynamics predicted by the ‘kill the winner' hypothesis, they are sufficient to observe general associations. In a natural environment with more evenness in the microbial community, high-resolution methods delineating species or subspecies will be necessary to observe associations with viral production.
Interestingly, positive correlation between change in the relative abundance of Gammaproteobacteria and change in viral abundance is likely a result of infection by a filamentous virus that do not typically lyse hosts but are rather extruded from the cell without significant cell lysis (Bradley, 1973; Stanisich, 1974). The nitrate-reducing Pseudomonas sp. Alda10 isolated in this study is infected with a filamentous bacteriophage, and the predominance of Pseudomonas spp. at the conclusion (88%) may be a result of infection with the filamentous phage. Thus, infection with a filamentous phage may confer a competitive advantage to the host as cell lysis would not necessarily occur (Cheng et al., 1999; Kimsey and Waldor, 2004; Berngruber et al., 2010).
Conclusion
Together, these results indicate a link between viral abundance and carbon biogeochemistry in the terrestrial subsurface. Viral productivity was significantly correlated with acetate consumption and CO2 production, whereas cell abundance was not, which was likely because of viral-mediated cell lysis. As environmental conditions alter host metabolic activity and growth, those changes are reflected in the VCR (Figure 1f). Furthermore, the consequences of viral lysis on cell abundance and specific microbial populations may also explain why cell abundance in a meta-analysis of different biogeochemical processes failed to predict microbially mediated biogeochemical fluxes (Röling, 2007). Thus, these rapid changes in VCR can potentially serve as indicators of microbial activity in environmental systems when cell abundance fails. In addition to long-term effects on population structure, viral-mediated cell lysis can also influence bacterial community structure within a short period of time (∼24 h). Thus, viral lysis may cause misleading interpretations of community dynamics monitored in the laboratory or field when sampling is infrequent. Viral productivity may be especially important in environments where there are fluctuating conditions such as redox boundary zones or in environments where bioremediation by biostimulation or bioaugmentation with additions of electron donor or acceptors are performed. However, the extent to which viral-mediated cell lysis alters organic matter pools subsequently influencing microbial community structure and function from the bottom-up remains a critical question in subsurface microbial ecology and carbon biogeochemical dynamics.
Acknowledgments
We would like to thank members of the Weber laboratory and Soil Conservation Survey for assistance with sample collection and MPN enumeration. We also thank Drs Charles Kuszynski and Davide Quaranta for their assistance with flow cytometry; Dr Han Chen for assistance with TEM; and Drs Jim Van Etten and Paul Blum for their experimental discussions. This project was supported by The US Department of Energy Subsurface Biogeochemistry Program to KAW (DE-SC0004113). Sample collection was supported by the US Geological Survey WRRI 104b Program (2009NE183B) to KAW and DDS. Additional support for DP and RW and DW was provided by the National Science Foundation IGERT Fellowship (0903469) and UNL UCARE Program, respectively.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Ackermann HW. (2009). Basic Phage Electron Microscopy. In: Clokie MJ, Kropinski A, (eds). Bacteriophages. Humana Press: New York, NY, USA, pp 113–126. [DOI] [PubMed] [Google Scholar]
- Anderson RE, Brazelton WJ, Baross JA. (2011). Is the genetic landscape of the deep subsurface biosphere affected by viruses? Front Microbiol 2: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartram AK, Lynch MDJ, Stearns JC, Moreno-Hagelsieb G, Neufeld JD. (2011). Generation of multimillion-sequence 16S rRNA gene libraries from complex microbial communities by assembling paired-end Illumina reads. Appl Environ Microbiol 77: 3846–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berngruber TW, Weissing FJ, Gandon S. (2010). Inhibition of superinfection and the evolution of viral latency. J Virol 84: 10200–10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boras JA, Sala MM, Vázquez-Domínguez E, Weinbauer MG, Vaqué D. (2009). Annual changes of bacterial mortality due to viruses and protists in an oligotrophic coastal environment (NW Mediterranean). Environ Microbiol 11: 1181–1193. [DOI] [PubMed] [Google Scholar]
- Bouvier T, Del Giorgio P. (2007). Key role of selective viral-induced mortality in determining marine bacterial community composition. Environ Microbiol 9: 287–297. [DOI] [PubMed] [Google Scholar]
- Bradley DE. (1973). The adsorption of the Pseudomonas aeruginosa filamentous bacteriophage Pf to its host. Can J Microbiol 19: 623–631. [DOI] [PubMed] [Google Scholar]
- Brussaard CPD. (2009). Enumeration of bacteriophages using flow cytometry. Methods Mol Biol 501: 97–111. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Paszkiewicz K, Field D, Knight R, Gilbert JA. (2011). The Western English Channel contains a persistent microbial seed bank. ISME J 6: 1089–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapelle FH, Lovley DR. (1990). Rates of microbial metabolism in deep coastal plain aquifers. Appl EnvironMicrobiol 56: 1865–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CM, Wang HJ, Bau HJ, Kuo TT. (1999). The primary immunity determinant in modulating the lysogenic immunity of the filamentous bacteriophage cf. J Mol Biol 287: 867–876. [DOI] [PubMed] [Google Scholar]
- Chopin MC, Roualt A, Ehrlich SD, Gautier M. (2002). Filamentous phage active on the gram-positive bacterium Propionibacterium freudenreichii. J Bacteriol 184: 2030–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark DW, Meyer H-P, Leist C, Fiechter A. (1986). Effects of growth medium on phage production and induction in Escherichia coli K-12 lambda lysogens. J Biotechnol 3: 271–280. [Google Scholar]
- Coombs JM. (2009). Potential for horizontal gene transfer in microbial communities of the terrestrial subsurface. Horizontal Gene Transfer: Genomes in Flux. In: Gogarten MB, Gogarten JP, Olendzenski LC (eds). Humana Press: New York, NY, USA, pp 413–433. [DOI] [PubMed] [Google Scholar]
- Corinaldesi C, Dell'Anno A, Magagnini M, Danovaro R. (2010). Viral decay and viral production rates in continental-shelf and deep-sea sediments of the Mediterranean Sea. FEMS Microbiol Ecol 72: 208–218. [DOI] [PubMed] [Google Scholar]
- Danovaro R, Manini E, Dell'Anno A. (2002). Higher abundance of bacteria than of viruses in deep Mediterranean sediments. Appl Environ Microbiol 68: 1468–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danovaro R, Dell'Anno A, Corinaldesi C, Magagnini M, Noble R, Tamburini C et al. (2008). Major viral impact on the functioning of benthic deep-sea ecosystems. Nature 454: 1084–1087. [DOI] [PubMed] [Google Scholar]
- De Corte D, Sintes E, Yokokawa T, Reinthaler T, Herndl GJ. (2012). Links between viruses and prokaryotes throughout the water column along a North Atlantic latitudinal transect. ISME J 6: 1566–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006. a). Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl EnvironMicrobiol 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis Jr.TZ, Hugenholtz P, Keller K, Brodie EL, Larsen N, Piceno YM et al. (2006. b). NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 34: W394–W399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driggers LJ, Schmidt JM. (1970). Induction of defective and temperate bacteriophages in Caulobacter. J Gen Virol 6: 421–427. [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Eydal HSC, Jagevall S, Hermansson M, Pedersen K. (2009). Bacteriophage lytic to Desulfovibrio aespoeensis isolated from deep groundwater. ISME J 3: 1139–1147. [DOI] [PubMed] [Google Scholar]
- Faruque SM, Naser IB, Fujihara K, Diraphat P, Chowdhury N, Kamruzzaman M et al. (2005). Genomic sequence and receptor for the Vibrio cholerae phage KSF-1Φ: Evolutionary divergence among filamentous vibriophages mediating lateral gene transfer. J Bacteriol 187: 4095–4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulquier A, Simon L, Gilbert F, Fourel F, Malard F, Mermillod-Blondin F. (2010). Relative influences of DOC flux and subterranean fauna on microbial abundance and activity in aquifer sediments: new insights from 13C-tracer experiments. Freshwater Biol 55: 1560–1576. [Google Scholar]
- Foulquier A, Malard F, Mermillod-Blondin F, Montuelle B, Dolédec S, Volat B et al. (2011). Surface water linkages regulate trophic interactions in a groundwater food web. Ecosystems 14: 1339–1353. [Google Scholar]
- Fuhrman JA. (1999). Marine viruses and their biogeochemical and ecological effects. Nature 399: 541–548. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Steele JA. (2008). Community structure of marine bacterioplankton: patterns, networks, and relationships to function. Aquat Microbial Ecol 53: 69–81. [Google Scholar]
- Gibert J, Deharveng L. (2002). Subterranean ecosystems: a truncated functional biodiversity. Bioscience 52: 473–481. [Google Scholar]
- Hewson I, O'Neil JM, Fuhrman JA, Dennison WC. (2001). Virus-like particle distribution and abundance in sediments and overlying waters along eutrophication gradients in two subtropical estuaries. Limnol Oceanography 46: 1734–1746. [Google Scholar]
- Hewson I, Fuhrman JA. (2003). Viriobenthos production and virioplankton sorptive scavenging by suspended sediment particles in coastal and pelagic waters. Microbial Ecol 46: 337–347. [DOI] [PubMed] [Google Scholar]
- Hobbie JE, Daley RJ, Jasper S. (1977). Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microbiol 33: 1225–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer J, Sommaruga R. (2001). Seasonal dynamics of viruses in an alpine lake: importance of filamentous forms. Aquat Microb Ecol 26: 1–11. [Google Scholar]
- Kimsey HH, Waldor MK. (2004). The CTXφ repressor RstR binds DNA cooperatively to form tetrameric repressor-operator complexes. J Biol Chem 279: 2640–2647. [DOI] [PubMed] [Google Scholar]
- Kimura M, Jia Z, Nakayama N, Asakawa S. (2008). Ecology of viruses in soils: Past, present and future perspectives. Soil Sci Plant Nutr 54: 1–32. [Google Scholar]
- Kyle JE, Eydal HSC, Ferris FG, Pedersen K. (2008). Viruses in granitic groundwater from 69 to 450 m depth of the Äspö Hard Rock Laboratory, Sweden. ISME J 2: 571–574. [DOI] [PubMed] [Google Scholar]
- Lane DJ. (1991). 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (ed) Nucleic Acid Techniques in Bacterial Systematics. John Wiley and Sons: New York, NY, USA, pp 115–175. [Google Scholar]
- Lazarevic V, Whiteson K, Huse S, Hernandez D, Farinelli L, Østerås M et al. (2009). Metagenomic study of the oral microbiota by Illumina high-throughput sequencing. J Microbiol Meth 79: 266–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon JT, Martiny JB. (2008). Rapid evolution buffers ecosystem impacts of viruses in a microbial food web. Ecol Lett 11: 1178–1188. [DOI] [PubMed] [Google Scholar]
- Lorieau JL, Day LA, McDermott AE. (2008). Conformational dynamics of an intact virus: Order parameters for the coat protein of Pf1 bacteriophage. Proc Natl Acad Sci USA 105: 10366–10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley DR, Phillips EJ. (1988). Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl Environ Microbiol 54: 1472–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71: 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middelboe M, Riemann L, Steward GF, Hansen V, Nybroe O. (2003). Virus-induced transfer of organic carbon between marine bacteria in a model community. Aquat Microb Ecol 33: 1–10. [Google Scholar]
- Middelboe M, Jørgensen NO. (2006). Viral lysis of bacteria: an important source of dissolved amino acids and cell wall compounds. J Mar Biol Assoc UK 86: 605–612. [Google Scholar]
- Motegi C, Nagata T. (2007). Enhancement of viral production by addition of nitrogen or nitrogen plus carbon in subtropical surface waters of the South Pacific. Aquat Microb Ecol 48: 27–34. [Google Scholar]
- Noble RT, Fuhrman JA. (1998). Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat Microb Ecol 14: 113–118. [Google Scholar]
- Noble RT, Fuhrman JA. (1999). Breakdown and microbial uptake of marine viruses and other lysis products. Aquat Microb Ecol 20: 1–11. [Google Scholar]
- Onstott T, Phelps T, Kieft T, Colwell F, Balkwill D, Fredrickson J et al. (1999). A global perspective on the microbial abundance and activity in the deep subsurface. Enigmatic Microorganisms and life in Extreme Environments. Kluwer Academic Publishers: Dordrecht, The Netherlands, pp 487–500. [Google Scholar]
- Phelps T, Murphy E, Pfiffner S, White D. (1994). Comparison between geochemical and biological estimates of subsurface microbial activities. Microb Ecol 28: 335–349. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. (2010). FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price PB, Sowers T. (2004). Temperature dependence of metabolic rates for microbial growth, maintenance, and survival. Proc Natl Acad Sci USA 101: 4631–4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe B, Schell J, Becker A, Heip J, Onodera K, Schell-Frederick E. (1973). A colicin-tolerant mutant of Escherichia coli with reduced levels of cyclic AMP and a strong bias towards λ lysogeny. Mol Gen Genet 120: 1–16. [DOI] [PubMed] [Google Scholar]
- Röling WF. (2007). Do microbial numbers count? Quantifying the regulation of biogeochemical fluxes by population size and cellular activity. FEMS Microbiol Ecol 62: 202–210. [DOI] [PubMed] [Google Scholar]
- Roudnew B, Seymour JR, Jeffries TC, Lavery TJ, Smith RJ, Mitchell JG. (2012). Bacterial and virus-like particle abundances in purged and unpurged groundwater depth profiles. Ground Water Monit Remediat 32: 72–77. [Google Scholar]
- Ruan Q, Dutta D, Schwalbach MS, Steele JA, Fuhrman JA, Sun F. (2006). Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors. Bioinformatics 22: 2532–2538. [DOI] [PubMed] [Google Scholar]
- Šimek K, Weinbauer MG, Hornák K, Jezbera J, Nedoma J, Dolan JR. (2007). Grazer and virus-induced mortality of bacterioplankton accelerates development of Flectobacillus populations in a freshwater community. Environ Microbiol 9: 789–800. [DOI] [PubMed] [Google Scholar]
- Snow DD. (1996). Geochemistry, hydrology, and environmental applications of uranium-series nuclides in the Platte River drainage basin. ETD Collection for University of Nebraska—Lincoln Paper AAI9703790, p 126. Available at http://digitalcommons.unl.edu/dissertations/AAI9703790.
- Srinivasiah S, Bhavsar J, Thapar K, Liles M, Schoenfeld T, Wommack KE. (2008). Phages across the biosphere: contrasts of viruses in soil and aquatic environments. Res Microbiol 159: 349–357. [DOI] [PubMed] [Google Scholar]
- Stanisich VA. (1974). The properties and host range of male-specific bacteriophages of Pseudomonas aeruginosa. J Gen Microbiol 84: 332–342. [DOI] [PubMed] [Google Scholar]
- Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY et al. (2011). Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J 5: 1414–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens T. (1997). Lithoautotrophy in the subsurface. FEMS Microbiol Reviews 20: 327–337. [Google Scholar]
- Suttle CA. (2007). Marine viruses—major players in the global ecosystem. Nat Rev Microbiol 5: 801–812. [DOI] [PubMed] [Google Scholar]
- Suttle CA, Fuhrman JA. (2010). Enumeration of virus particles in aquatic or sediment samples by epifluorescence microscopy. In Wilhelm S, Weinbauer M, Suttle C (eds) Manual of Aquatic Viral Ecology. American Society of Limnology and Oceanography: Waco, TX, USA, pp 145–153. [Google Scholar]
- Thingstad TF, Lignell R. (1997). Theoretical models for the control of bacterial growth rate, abundance, diversity and carbon demand. Aquat Microb Ecol 13: 19–27. [Google Scholar]
- Tomaru Y, Nagasaki K. (2007). Flow cytometric detection and enumeration of DNA and RNA viruses infecting marine Eukaryotic microalgae. J Oceanogr 63: 215–221. [Google Scholar]
- Tuomi P, Fagerbakke KM, Bratbak G, Heldal M. (1995). Nutritional enrichment of a microbial community: The effects on activity, elemental composition, community structure and virus production. FEMS Microbiol Ecol 16: 123–134. [Google Scholar]
- Wang Q, Kan B, Wang R. (2013). Isolation and characterization of the new mosaic filamentous phage VFJ Φ of Vibrio cholerae. PLoS One 8: e70934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber KA, Pollock J, Cole KA, O'Connor SM, Achenbach LA, Coates JD. (2006). Anaerobic nitrate-dependent iron (II) bio-oxidation by a novel lithoautotrophic betaproteobacterium, strain 2002. Appl Environ Microbiol 72: 686–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinbauer MG. (2004). Ecology of prokaryotic viruses. FEMS Microbiol Rev 28: 127–181. [DOI] [PubMed] [Google Scholar]
- Weinbauer MG, Christaki U, Nedoma J, Simek K. (2003). Comparing the effects of resource enrichment and grazing on viral production in a meso-eutrophic reservoir. Aquat Microb Ecol 31: 137–144. [Google Scholar]
- Weinbauer MG, Fuks D, Peduzzi P. (1993). Distribution of viruses and dissolved DNA along a coastal trophic gradient in the northern Adriatic Sea. Appl Environ Microbiol 59: 4074–4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel RG, Likens GE. (1991) Limnological Analyses. Springer-Verlag: New York, USA. [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. (1998). Prokaryotes: The unseen majority. Proc Natl Acad Sci USA 95: 6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter C, Bouvier T, Weinbauer MG, Thingstad TF. (2010). Trade-offs between competition and defense specialists among unicellular planktonic organisms: the "killing the winner" hypothesis revisited. Microbiol Mol Biol Rev 74: 42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack KE, Colwell RR. (2000). Virioplankton: viruses in aquatic ecosystems. Microbiol Mol Biol Rev 64: 69–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wommack KE, Williamson KE, Helton RR, Bench SR, Winget DM. (2009). Methods for the isolation of viruses from environmental samples. Methods Mol Biol 501: 3–14. [DOI] [PubMed] [Google Scholar]
- Wrighton KC, Thomas BC, Sharon I, Miller CS, Castelle CJ, VerBerkmoes NC et al. (2012). Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337: 1661–1665. [DOI] [PubMed] [Google Scholar]
- Zhang R, Weinbauer MG, Qian PY. (2007). Viruses and flagellates sustain apparent richness and reduce biomass accumulation of bacterioplankton in coastal marine waters. Environ Microbiol 9: 3008–3018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.