Abstract
Interactions among microbes and stratification across depths are both believed to be important drivers of microbial communities, though little is known about how microbial associations differ between and across depths. We have monitored the free-living microbial community at the San Pedro Ocean Time-series station, monthly, for a decade, at five different depths: 5 m, the deep chlorophyll maximum layer, 150 m, 500 m and 890 m (just above the sea floor). Here, we introduce microbial association networks that combine data from multiple ocean depths to investigate both within- and between-depth relationships, sometimes time-lagged, among microbes and environmental parameters. The euphotic zone, deep chlorophyll maximum and 890 m depth each contain two negatively correlated ‘modules' (groups of many inter-correlated bacteria and environmental conditions) suggesting regular transitions between two contrasting environmental states. Two-thirds of pairwise correlations of bacterial taxa between depths lagged such that changes in the abundance of deeper organisms followed changes in shallower organisms. Taken in conjunction with previous observations of seasonality at 890 m, these trends suggest that planktonic microbial communities throughout the water column are linked to environmental conditions and/or microbial communities in overlying waters. Poorly understood groups including Marine Group A, Nitrospina and AEGEAN-169 clades contained taxa that showed diverse association patterns, suggesting these groups contain multiple ecological species, each shaped by different factors, which we have started to delineate. These observations build upon previous work at this location, lending further credence to the hypothesis that sinking particles and vertically migrating animals transport materials that significantly shape the time-varying patterns of microbial community composition.
Introduction
Microbial communities throughout the water column show long-term, seasonal and short-term dynamics that relate to various biological, chemical and physical environmental parameters (Gilbert et al., 2012; Giovannoni and Vergin, 2012; Chow et al., 2013; Hatosy et al., 2013; Needham et al., 2013; Cram et al., 2014a; Fuhrman et al., 2015). Microbial interactions have been observed experimentally (Jurgens et al., 1999; Miller and Bassler, 2001; Jürgens and Matz, 2002; Bonilla-Findji et al., 2009) through physical attachment (Malfatti and Azam, 2009; Malfatti et al., 2009) and inferred through genomic and physiological information (for example, Bothe et al., 2010). Analysis of statistical association networks enables exploration of correlated dynamics of taxa that either directly interact or respond similarly to environmental variation (Steele et al., 2011; Chow et al., 2014). Interactions (such as predator–prey, mutualisms, parasitism and competition) appear to be major drivers of the ecological dynamics of many marine bacterioplankton (see Strom, 2008).
Pairwise association analysis techniques (reviewed in Faust and Raes, 2012; Cram et al., 2014b) have proven to be valuable tools in looking at trends in the many statistical associations in microbial communities in a variety of environments including lake systems (Eiler et al., 2012; Kara et al., 2012), soil (Zhou et al., 2010; Barberán et al., 2011), the human microbiome (Arumugam et al., 2011; Faust et al., 2012) and globally through meta-analysis across diverse sampling sites (Chaffron et al., 2010; Freilich et al., 2010). In marine surface waters, studies using pairwise association analysis over time have suggested that the abundance of particular bacteria tend to be best predicted by the abundance of other microorganisms, rather than variability of measured parameters (Fuhrman and Steele, 2008; Steele et al., 2011; Chow et al., 2013, 2014). Network analysis has identified many associations between marine microorganisms that are driven by seasonal variability, especially at locations where seasonality is strong (Gilbert et al., 2012), and this seasonal pattern has been de-convoluted in lake environments to show different inter-organismal associations independent of seasonality (Kara et al., 2012). Seasonality has also been seen just above the sea floor and suggests surface influences on deeper depths by way of sinking particle flux and/or migrating zooplankton (see Cram et al., 2014a).
Previously, Chow et al. (2013) examined similarities between association networks in the surface and deep chlorophyll maximum (DCM) at the San Pedro Ocean Time-series (SPOT) and observed some associations that were consistent between depths, specifically associations between operational taxonomic units (OTUs) from the SAR11 clade and other OTUs. Other associations differed between depths such as the relationships between temperature, salinity and nutrient concentrations to the abundances of many OTUs. Time scale appears to be an important determinant of organismal associations. We previously reported that members of the SAR11 cluster were correlated on a daily time scale but not monthly (Fuhrman and Steele, 2008; Steele et al., 2011; Needham et al., 2013; Fuhrman et al., 2015), reflecting patterns of temporal variability in network structure seen in other systems (Alarcón et al., 2008). A common feature of networks is the presence of modules, that is, highly interconnected groups of nodes (Newman, 2006; Olesen et al., 2007; Ings et al., 2009) and microbial networks are no exception (Steele et al., 2011; Chow et al., 2013). Modules in other studies have been shown to represent groups of organisms that have similar seasonal patterns (Gilbert et al., 2012) that sometimes occur independently of environmental parameters and may represent organisms that are associated with each other through symbiosis, allelopathy, common niches or through other means (Steele et al., 2011; Chow et al., 2013). Here, we extend these prior network analyses of euphotic zone depths to the rest of the water column. The dynamics of microbial communities throughout the entire deep (900 m) water column at SPOT were recently described, and one of the most notable results was the discovery of seasonality at the bottom of the water column (Cram et al., 2014a). We aim to elucidate how organisms interact with each other and are shaped by the flux of particles in different water column depths by investigating associations between organisms at different water column depths. Particles take time to sink; at SPOT, they have been reported to sink at an average rate of about 83 m per day or faster (Collins et al., 2011), which suggests that it should take an average of about 11 days to travel from the surface to the bottom of the water column. As this is an average for all particles, it is likely that smaller and less dense particles sink more slowly, while denser and larger particles sink more quickly. Particles take time to decompose or be consumed by microorganisms, and it has been suggested that particles take longer to decompose in deeper waters than in the mixed layer (Kiørboe, 2001). Furthermore, as microbes metabolize sinking particles, it takes time for their abundance to respond in a detectable way. Slow sinking times and delays in particle decomposition mean that particle flux, if it links surface and bottom environments, likely does so with a temporal delay.
Given the complexity of marine microbial networks and our need to quantitatively compare multiple networks, particularly those describing associations at different depths, we leverage various network statistics that are common in social, biological (Albert and Barabási, 2002) and ecological networks (Dunne et al., 2002; Montoya et al., 2006; Olesen et al., 2007; Suweis and D'Odorico, 2014), including microbial ecological ones (Chaffron et al., 2010; Zhou et al., 2010; Kara et al., 2012), but less common in marine microbial ecology. Network topological statistics are used to describe ‘global' network properties, and we utilize three related properties to comparatively and globally describe networks: network density, average clustering coefficients and average path length. The definitions and implications of these quantities are given in the methods section or in the Supplementary Materials.
This paper aims to identify patterns of associations between microbes within and between depths, especially time-lagged associations, to identify links between ecosystems at different depths. It also aims to elucidate the ecology of less studied groups of marine organisms, especially those that are abundant in the deep water column.
Methods
Data source
This project applies a novel perspective to data analyzed by Cram et al. (2014a). These data are publicly available at BCO-DMO <http://www.bco-dmo.org/dataset/537137> (physical and chemical data) and <http://www.bco-dmo.org/dataset/535915> (biological data).
Briefly, the samples analyzed here were collected monthly from the San Pedro Ocean Time-series station between August 2003 and January 2011 from five depths: 5 m, the Deep Chlorophyll Maximum Layer (between 5 m and 40 m), 150 m, 500 m and 890 m. DNA collection, processing and analysis procedures were described previously (Cram et al., 2014a) (see also Brown et al., 2005; Chow et al., 2013). Relative abundance of bacterial OTUs were identified by automated ribosomal intergenic spacer analysis (ARISA) and identities of ARISA peaks were ascertained using clone libraries, again as described previously (Cram et al., 2014a). These clones are from several depths throughout the water column, but OTUs are identified only once in the data set using the previously published priority scheme (Cram et al., 2014a). Environmental parameters (from Beman et al., 2008; Cram et al., 2014a) are summarized in Supplementary Table S1.
Network analysis
Networks were generated using the extended local similarity analysis (eLSA) program (Ruan et al., 2006; Xia et al., 2013) which implements the local similarity analysis with latest improvements for high-throughput data. We used eLSA to identify global, time-lagged, Spearman correlations between bacterial and environmental nodes at all depths. Previous analysis at SPOT have used ‘local similarity analysis', which identifies time-lagged associations as well as ‘local' associations, which are those that only occur over a portion of the data set. In contrast, in this study, we were most interested in relationships that were consistent throughout the study period, so we identified possibly shifted ‘global' Spearman correlations that identify correlations that are present throughout the entire time span. eLSA returned a P-value (calculated permutation test value) for each correlation, which is the probability that a correlation between two OTUs is at least as high as the observed value if they are not associated. From the P-values, false discovery rates or Q-values were calculated using the qvalue package (Dabney and Storey, 2004) in R (Team RDC, 2011). The Q-value measures the proportion of false positives incurred for a given P-value threshold. The eLSA metric that we used allowed 1 month time lags; meaning that correlations were considered between samples taken approximately 1 month apart in time (additional details in Supplementary Information; Network analysis).
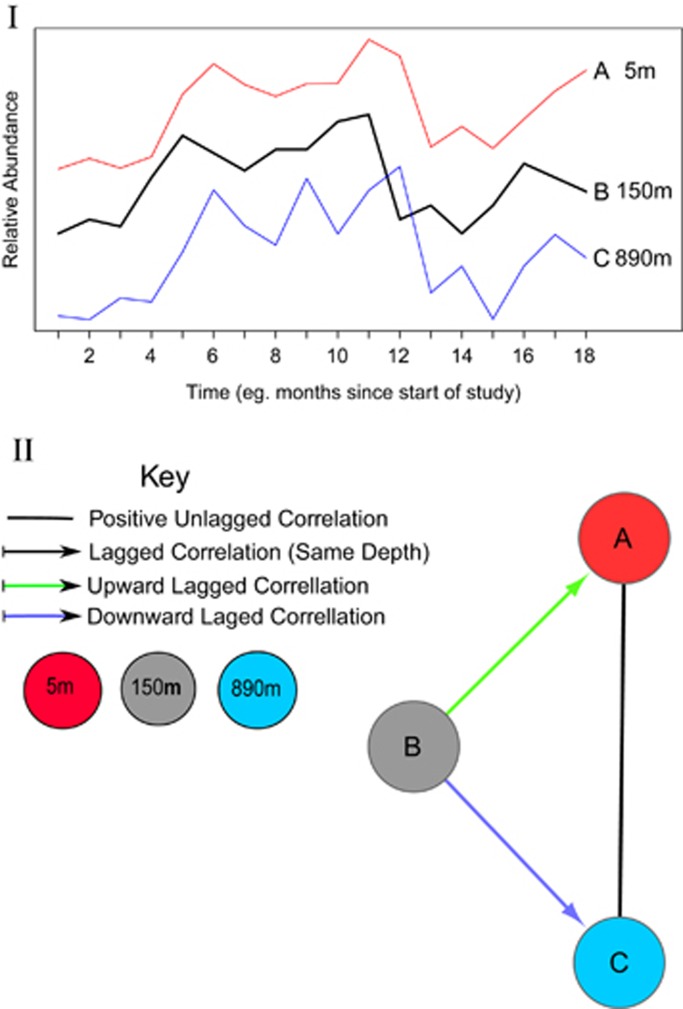
We imported our eLSA networks as well as metadata about variables including variable names, and for bacterial nodes, mean abundance and taxonomic data into Cytoscape 2.8.3 (Smoot et al., 2011). Subnetworks containing nodes from individual depths, or bacterial nodes only, were generated by filtering one master network (Supplementary Information; Organization of networks). Different associations were visualized as edges (lines) connecting nodes (shapes) that represented the parameters (Figure 1).
Figure 1.
An example of a time-lagged correlation between OTUs at different depths. I. Three hypothetical organisms that change in abundance over time. Note that (B), located at 150 m, leads both OTUs A (at 5 m) and C (at 890 m). The correlation between A and B is ‘upward' because the deeper node, leads the shallower node. The correlation between B and C is ‘downward' because the shallower node leads the deeper node. The correlations between A and C is ‘unlagged' because both changes happen at the same time. II. A network showing correlations between all pairs of parameters.
Network statistics
Directionality
'Directionality', defined for the first time in this paper, quantifies whether changes in shallower depths generally happened before associated changes in deeper depths. We investigated the ‘bacterial all-vs-all', ‘positive bacterial all-vs-all' (containing only positive associations), the ‘bacterial between depth networks' and the ‘positive bacterial between depth networks'. In all cases, we examined the edges that connected nodes from different depths and asked what fraction of those edges were time-lagged. Of the time-lagged edges, we asked what fraction of those edges were time-lagged such that the changes in the node that was at the shallower depth in the water column preceded correlated changes in the node that was deeper in the water column. These edges were referred to as ‘downward' lagged. Conversely, ‘upward' lagged edges are those in which changes in the deeper node lead changes in the shallower node.
Density
Network density quantifies how highly connected a network is, given its size (Coleman and Moré, 1983) (Supplementary Information; Density). It is essentially the probability that two OTUs or environmental factors are statistically associated either through direct interactions or some intermediates. The density metric was applied to investigate edges both within depths and between depths (Supplement Information; Application of density measurement).
Intra-depth density is defined as density within a network between nodes from the same depth. Inter-depth density is defined as density of edges connecting nodes from different depths within a network. The ‘density ratio' for each depth is intra-depth density of nodes from that depth, divided by the inter-depth density of edges connecting nodes from that depth to nodes at other depths. Density ratio for each pair of depths is the density of the edges connecting nodes within each depth divided by the density of edges connecting nodes from one depth to nodes at the other depth. This value quantifies how much more common connections are within a depth vs between depths.
Clustering coefficient and path length
Average clustering coefficient (Cl) describes whether the network associates into clumps of highly interconnected organisms, where high values suggest greater modularity. Highly clustered networks are those that contain groups of statistically associated organisms, which may result from physical association such as symbiosis or shared habitats among particular groups of organisms. Average path length (L) is the average shortest path (fewest number of intermediate connections) between each pair of nodes. Low path length indicates that most organisms can be connected through a few intermediates. Clustering coefficient, path length and the associated statistical significance (P-values) were calculated (Supplementary Information; Calculation of clustering and path length). Because both clustering coefficient and path length are dependent partially on network size, we normalized these values to the clustering coefficient and path length of random networks. To do this, we determined Cl/ClR and L/LR, the ratios of observed to random network clustering coefficient and path length for each of the thousand random networks, respectively, and determined the median value. A network is referred to as highly clustered if the Cl of an observed network is at least as high as the 95% percentile of ClR for the 1000 randomized networks. Similarly, a network is referred to as highly connected if the path length L of an observed network is at most the lower 5% of the path length LR for the 1000 randomized networks.
Modularity
Modularity was determined qualitatively, by visually inspecting networks. We identified groups of nodes that were connected by enriched positive correlations. Although we identified negative associations connecting different modules, these negative correlations were not considered in defining modules, as we were looking for groups of positively associated organisms.
Hub and spoke networks
We created hub and spoke networks (as seen in Fuhrman and Steele, 2008; Steele et al., 2011; Chow et al., 2014) around nodes from particular taxonomic groups. To generate these networks from the ‘all-vs-all' network, we first filtered this network to reduce complexity and highlight the strongest correlations. Specifically, we included only bacterial nodes that occurred at least 36 times in the dataset and had a mean relative abundance of at least 1%. We kept only edges with Spearman's absolute ρ values greater than 0.57. These filter cutoff values were chosen to allow us to focus our networks on the strongest correlations and most commonly occurring OTUs. We then selected OTUs of particular interest, specifically from the SAR11, Aegean-169, Deltaproteobacteria, Flavobacteria and Marine Group A (MGA) taxa, which were previously shown to be abundant and ecologically important in the SPOT water column (Cram et al., 2014a), and created sub-networks from those nodes and their nearest neighbors along with immediate adjoining edges.
Results
Association networks identified a prevalence of ‘downward' time-lagged interactions between nodes and parameters at different depths (Figures 1 and 2). By downward we mean that changes in shallower environments preceded changes in deeper environments. Furthermore, we observed modular structures in networks in the euphotic zone and the bottom of the water column (Figure 3, Supplementary Figures S1 and S2). Patterns for positive correlations (Tables 1 and 2) reflect patterns for both positive and negative correlations together (Supplementary Tables S2 and S3), meaning both positive and negative associations propagate in a similar downward manner.
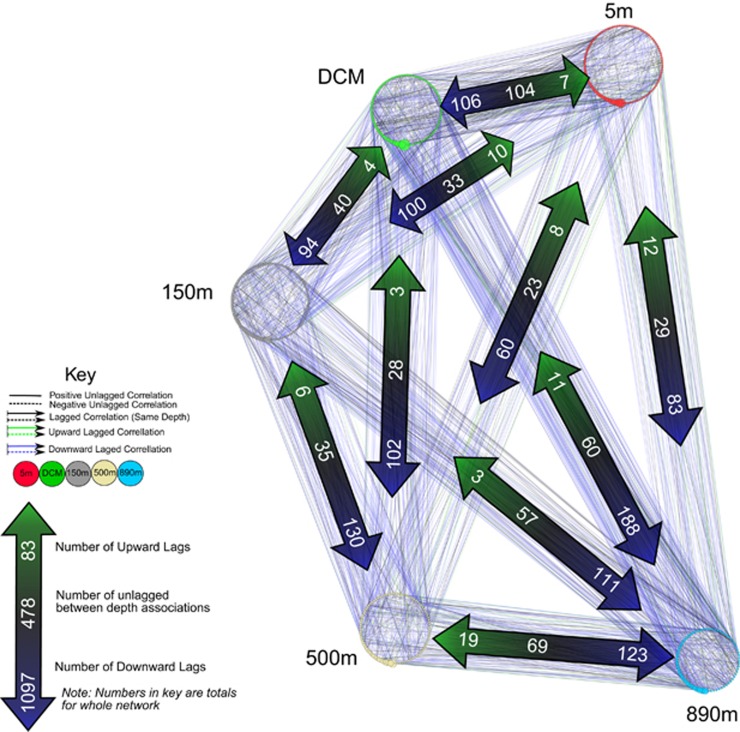
Figure 2.
Association network showing significant, time-lagged and non-time-lagged correlations between bacterial nodes both within and between depths. Nodes (circles) represent bacterial OTUs at each depth. Edges (lines) represent correlations, sometimes time-lagged, between the bacterial OTUs. Shown are bacteria that occur at least 25 times and edges that have lagged Spearman correlations such that |ρ|>0.5, P<0.01, Q<0.05. Edges are color-coded to indicate whether they represent unlagged correlations, ‘downward' time-lagged correlations in which changes in the shallower node precede changes in the deeper node, or ‘upward' lagged correlations in which changes in the deeper node precede changes in the shallower node. Large arrows show total numbers of edges of each type connecting depths.
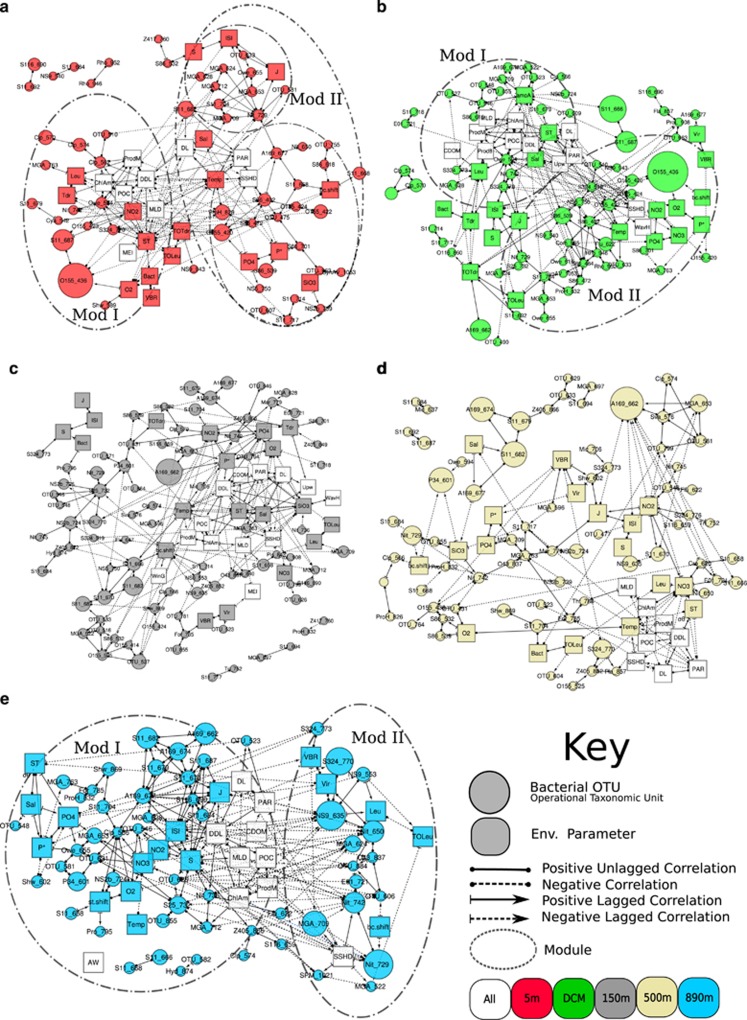
Figure 3.
Association network showing statistically significant, time-lagged and non-time-lagged correlations between bacterial and environmental nodes at each depth (a, 5 m; b, DCM; c, 150 m; d, 500 m; e, 890 m). Nodes represent bacterial OTUs (circles) and environmental parameters (squares) at each depth. Shown are bacteria that occur at least 25 times and associations that have lagged Spearman correlations such that |ρ|>0.55, P<0.01, Q<0.05. Node identities are indicated in Supplementary Table S1. Modules, clusters of highly connected nodes, are circled. In all cases, Mod I corresponds to the module with nodes connected to high surface chlorophyll_a and increasing daylight (spring bloom) and Mod II corresponds to nodes that are correlated positively to each other and negatively to nodes in Mod I.
Table 1. Summary statistics for networks that examine cross correlations between bacteria at different depths.
| Variable/Formula | AllvAll | 5 m-DCM | 5 m-150 m | 5 m-500 m | 5 m-890 m | DCM-150 m | DCM-500 m | DCM-890 m | 150 m-500 m | 150 m-890 m | 500 m-890 m | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nodes | N | 463 | 179 | 179 | 173 | 157 | 182 | 179 | 162 | 182 | 163 | 159 |
| Edges | E | 2301 | 441 | 396 | 327 | 390 | 391 | 396 | 525 | 436 | 466 | 489 |
| Within depth nodes | Ni | 372 | 150 | 156 | 155 | 130 | 160 | 159 | 134 | 165 | 140 | 139 |
| Within depth edges | Ei | 643 | 224 | 253 | 236 | 266 | 253 | 236 | 266 | 265 | 295 | 278 |
| Cross depth edges | Eo | 1658 | 217 | 143 | 91 | 124 | 138 | 160 | 259 | 171 | 171 | 211 |
| Intra-depth depth density | Ei/Ei_Max | 4.6% | 4.0% | 4.2% | 4.0% | 6.3% | 4.0% | 3.8% | 5.9% | 3.9% | 5.9% | 5.7% |
| Inter-depth density | Eo/Eo_max | 1.9% | 2.2% | 1.4% | 1.0% | 1.8% | 1.4% | 1.8% | 3.9% | 1.8% | 2.4% | 3.3% |
| Density ratio | (Ei/EiM)/(Eo/EoM) | 2.39 | 1.82 | 3.05 | 4.09 | 3.54 | 2.89 | 2.13 | 1.52 | 2.21 | 2.45 | 1.72 |
| Upward | U | 83 (5.0%) | 7 (3.2%) | 10 (7.0%) | 8 (8.8%) | 12 (9.7%) | 4 (2.9%) | 3 (2.3%) | 11 (4.2%) | 6 (3.5%) | 3 (1.8%) | 19 (9.0%) |
| Unlagged | F | 478 (28.8%) | 104 (47.9%) | 33 (23.1%) | 23 (25.3%) | 29 (23.4%) | 40 (29.0%) | 28 (21.1%) | 60 (23.2%) | 35 (20.5%) | 57 (33.3%) | 69 (32.7%) |
| Downward | D | 1097 (66.2%) | 106 (48.8%) | 100 (69.9%) | 60 (65.9%) | 83 (66.9%) | 94 (68.1%) | 102 (76.7%) | 188 (72.6%) | 130 (76.0%) | 111 (64.9%) | 123 (58.3%) |
Abbreviation: DCM, deep chlorophyll maximum. The other columns reflect the nodes within and connecting pairs of depths. Nodes (N) are the number of bacterial OTUs at a specific depth that occur at least 25 times in the data set connected by at least one edge (correlation) in the specified network. Edges (E) are the total number of Spearman correlations with |R|>0.5 in that network. Within depth nodes (Ni) are the nodes in these networks that are connected by at least one edge to another node at that same depth. Within depth edges (Ei) are those edges that connect pairs of nodes from the same depth. Cross depth edges (Eo) are those edges that connect two nodes from different depths. Within depth density reflects the probability that a pair of nodes from the same depth are connected by an edge; it is the quotient of the number of edges found connecting nodes that are from the same depth, divided by the maximum number of edges that could possibly connect nodes of the same depth in that network. Cross depth density (Eo/Eo_max) reflects the probability that two nodes from different depths are connected by an edge; it is the total number of edges connecting nodes from different depths, compared with the number of possible edges that could connect nodes from those depths. The density ratio (ei/eim)/(eo/eom) reflects how much likelier two nodes are to be connected if they are from the same depth, than if they are from different depths; it is the quotient of the within depth density and the cross depth density of a network. Upward (U) is the number of time-lagged edges that connect pairs of nodes from different depths such that changes in the node at the deeper depth precede changes in the done at the shallower depth. Unlagged (F) is the number of non-time-lagged edges that connect pairs of nodes from different depths. Downward is the number of time-lagged edges that connect pairs of odes from different depths such that the changes in the shallower node precede correlated changes in the deeper node.
The AllvAll column reflects the network shown in Figure 2.
Table 2. Topological statistics for networks of bacteria at each depth (Figure 1), and for a network of OTUs at all depths (Figure 2).
| 5 m | DCM | 150 m | 500 m | 890 m | AllvAll | |
|---|---|---|---|---|---|---|
| Eligible nodes | 110 | 102 | 113 | 106 | 106 | 537 |
| Nodes (N) | 73 | 77 | 83 | 82 | 57 | 463 |
| Edges (E) | 112 | 112 | 141 | 124 | 154 | 2301 |
| Cl | 0.34 | 0.33 | 0.23 | 0.21 | 0.34 | 0.17 |
| L | 2.37 | 2.02 | 2.33 | 2.37 | 2.14 | 3.46 |
| ClR | 0.04 | 0.04 | 0.04 | 0.04 | 0.09 | 0.02 |
| LR | 3.73 | 3.92 | 3.61 | 3.88 | 2.54 | 2.91 |
| Cl/ClR | 8.33 | 8.78 | 5.75 | 6.02 | 3.59 | 8.10 |
| L/LR | 0.63 | 0.51 | 0.65 | 0.61 | 0.84 | 1.19 |
| Intra-depth density | 4.3% | 3.8% | 4.1% | 3.7% | 9.6% | 2.2% |
| Inter-depth density | 1.6% | 2.1% | 1.7% | 1.8% | 2.8% | NA |
| Density ratio | 2.71 | 1.82 | 2.47 | 2.12 | 3.43 | NA |
Abbreviation: DCM, deep chlorophyll maximum. This table complements Supplementary Figure S1 which visually depicts the network described here. These networks include only nodes for bacteria that are present at >0.01% abundance greater than 25 times (eligible nodes) and edges that have a possibly time-lagged, global, absolute Spearman ρ value of greater than 0.5 or less than −0.5. Nodes are the bacterial OTUs that are connected by at least one edge to another node. Edges are the number of correlations between bacterial OTUs. Density is the number of edges (E) divided by the number of possible edges {N*(N-1)/2} such that {Density=E/(N*(N-1)/2)}. Cl is the clustering coefficient for the network. L is the mean path length for the network. ClR and LR are the median clustering coefficients and path lengths of 1000 equivalently sized (same number of nodes and edges) randomly distributed networks. Cl/ClR is the ratio of the clustering coefficient to the median random network's clustering coefficient. Permutation tests suggest that clustering and path length coefficients are statistically significantly different than those for random (P<0.001). It is apparent that 890 m has higher density than the other depths. Clustering coefficient relative to random networks (Cl/ClR) is highest in the DCM. Like the DCM, 890 m has a high level of absolute clustering, which reflects as several highly connected groups in Supplementary Figure S1E.
Patterns between OTUs at different depths
Analysis of statistical connections between nodes from different depths revealed that many pairs of bacterial OTUs, including ones from different depths, were correlated in a time-lagged manner. Figure 1 provides a theoretical example of time-lagged relationships between bacterial OTUs at different depths.
The ‘all-vs-all bacterial network' (Figure 2) shows the grand summary of pairwise associations between bacteria at each depth. In this network, more than half of the correlations between bacteria at different depths were time-lagged by 1 month such that changes in the OTU in the shallower depth led changes in the abundance of the OTU in the deeper depth by 1 month (Figure 2, Table 1). Some pairs of depths have particularly high fractions of downward lagged edges (for example, DCM and 890 and 150 m and 890 m both had 76% of their edges downward lagged) whereas the surface and nearby DCM had a high fraction of unlagged edges. Significantly, the overwhelming majority (1097 out of 1180 edges, see the key of Figure 2 and Table 1) of the lagged connections between depths were ‘downward'.
Patterns within individual depths
Networks examining correlations (Spearman's |ρ|>0.5, Q<0.01) between bacterial OTUs at each depth (Supplementary Figures S1 and S2) as well as networks showing correlations among bacterial OTUs and environmental parameters (Figure 3) showed different overall association patterns between depths. When we looked only at connections between bacterial OTUs, several patterns were immediately apparent. At 890 m, when we examined only positive associations between bacteria, the network formed two ‘modules' or highly interconnected groups of organisms (Figure 3e, Supplementary Figure S1E). If we also examine negative associations, it is clear that there are also many negative correlations between these modules (Figure 1e, Supplementary Figure S2E). The networks at 5 m and the DCM (Figures 3a and b, Supplementary Figure S1A and B, S2A and B) show two large modules, but neither of these is as highly interconnected as the network at 890 m (Table 2). In all cases, one module had many positively associated nodes while the other had a few nodes that were positively associated in a more diffuse pattern (most nodes only connected to one or two other nodes). Both of these networks had pairs of organisms that associated only with each other but not with other OTUs. As at 890 m, there were negative correlations between nodes in different modules. Nodes at 150 m and 500 m (Figures 3c and d, Supplementary Figures S1C and D, S2C and D), in contrast, appeared to lack a discernable structure, both when negative correlations were included (Figures 3c and d, Supplementary Figure S1C and D) and when they were not (Supplementary Figure S2C and D).
Topological analysis of the bacterial networks showed that 890 m has the highest edge density, the 5 m, the DCM and 890 m have higher clustering than intermediate depths, and that the path lengths of all depths are similar to those of random networks (Table 2, Supplementary Table S3).
Density
The network density at 890 m, relative to the other depths (Table 2), indicated that, for the number of nodes present in this depth, there was a higher probability of any two nodes being correlated than at other depths. There were just as many bacterial OTUs at 890 m that occurred at least 25 times as there were at other depths. While more than half of the OTU nodes at 890 m were associated with several other nodes, just under half of the nodes did not correlate in abundance with any other nodes at 890 m. The networks at the other depths all had similar densities indicating intra-depth density is roughly even across depths. Network density was lowest at 500 m. When only positive associations between nodes were considered (Supplementary Table S3), similar patterns emerged.
At all depths, nodes were about twice as likely to correlate with another node at that same depth (intra-depth density) as they were to connect to a node at another depth (inter-depth density), relative to the number of possible edges that could occur within or across depths (Table 2, Supplementary Table S3). Compared with other depths, nodes at 890 m had the highest density of connections both to other nodes at 890 m as well as to nodes at other depths. Meanwhile, both 5 m and 890 m depths had high intra-depth edge density relative to inter-depth edge density, while the DCM, 150 m and especially 500 m depths had the lower edge densities relative to intra-depth edge density.
Clustering and path length
At all depths, networks were more clustered than the same size Erdos-Renyi random networks (P<0.01). DCM, 5 m and 890 m have higher clustering coefficients than other depths (Table 2). However, only 5 m and DCM but not the 890 m depth has a high clustering coefficient ratio (clustering coefficient divided by the coefficient of a similarly sized random network; Cl/ClR). This is because a same-sized random network as the 890 m network would also be more clustered than other networks. When only positive associations were considered, similar clustering patterns were seen suggesting that the positive and negative associations show similar patterns (Supplementary Table S3). Mean path lengths between nodes at all depths were shorter than in randomly generated networks, when depths were considered individually (Table 2). However, for networks that considered at all depths together at once, path lengths were longer than for random networks, indicating that nodes were more connected within depths than between depths.
Environmental parameters in within depth networks
Networks at each depth that also included environmental parameters (Figure 3) generally showed that most OTUs associated with other OTUs rather than the environmental parameters that we measured. Many environmental parameters, predictably, associated with each other. Usually there were a few OTU nodes among highly interconnected modules that associated with environmental parameters, and we suggest that these associations generally suggest loose relationships between that module and the noted environmental parameters. At 5 m, DCM and 890 m, two modules of positively correlated nodes were evident; in all cases, one of these modules contained several nodes that corresponded to increasing day length (DDL) and high levels of surface chlorophyll, conditions indicating that the community was most abundant in the spring. At each depth, we refer to this module as ‘Mod I'. The other module, which we call ‘Mod II', was comprised of OTUs that associated negatively with the parameters in Mod I.
At 5 m and DCM, Mod II had more nodes than Mod I. In the DCM, several nodes in Mod II associate positively with the abundance of the archaeal ammonia monooxygenase gene (Beman et al., 2011). However, this variable was not measured at the other depths. At 5 m, Mod II appeared to divide into two sub-modules, one of which had a number of OTUs that correlated positively with the bacterial diversity measures Inverse Simpson index and Pielou's evenness index. At 890 m, Mod I had more nodes than Mod II, in contrast to 5 m and the DCM. Many nodes in Mod I were associated with high Inverse Simpson, Richness and Pelou's evenness, and associated with a 1 month lag with the concentration of nitrate and nitrite.
Example networks
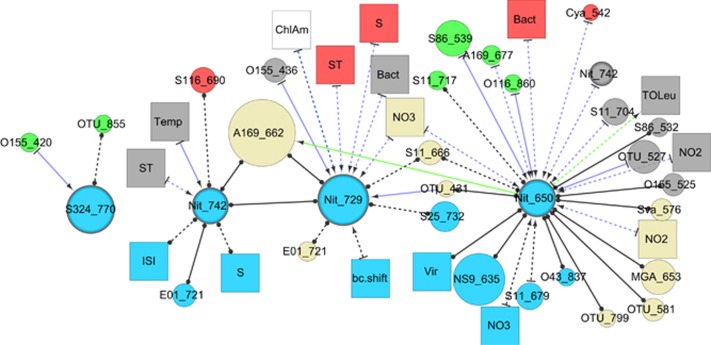
Hub and spoke networks, focusing on particular bacterial taxonomic groups reflected the trends seen in Figure 4, but illustrated that different related OTUs associated with different factors. As an example of how these figures suggest inter-depth associations, Figure 4 shows the four OTUs, from the Deltaproteobacteria class, that were found at 890 m, which were present in the dataset at least 36 times and which associated statistically with other nodes. It is apparent that the Nitrospina OTUs with intergenic spacer length 650 bp associated both positively and negatively, with a time lag of 1 month, with a number of OTUs and parameters from 5 m, the DCM and 890 m. This Nitrospina also correlated without lag to a number of OTUs and parameters at 500 m and 890 m. In contrast, a SAR324 OTU (770 bp) associated only with two OTUs from the DCM and not from other depths. Two other Nitrospina OTUs associated each with a number of parameters, only some of which they shared with the more connected Nitrospina OTU (650 bp). Deltaproteobacterial OTUs from 150 m and 500 m showed different patterns at different depths and followed a general pattern where changes in various parameters in shallower depths preceded changes in deeper depths, cascading through the water column (Supplementary Figure S3).
Figure 4.
Association network of correlations between Deltaproteobacteria OTUs at 890 m and other bacteria and environmental parameters found at every depth throughout the water column. All bacteria shown have a mean abundance of greater than 1% and are found with a relative abundance of greater than 0.01% in more than 36 samples. Edges represent statistically significant, potentially time-lagged correlations (Spearman's |ρ|>0.57, P<0.01, Q<0.05). Abbreviated names are followed by OTU fragment size. Node sizes of bacterial nodes represent the average abundance of OTUs. Abbreviations for the nodes are translated in Supplementary Table S1.
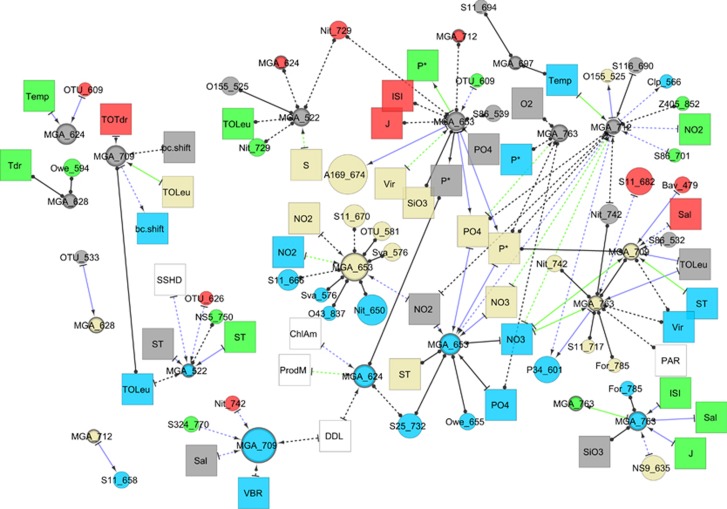
A similar pattern was seen for the Marine Group A OTUs and the parameters and OTUs that associated with them (Figure 5). As with the Deltaproteobacteria, changes at shallower depths led changes at deeper depths. Also as with Deltaproteobacteria, OTUs related to different parameters at different depths. One MGA OTU (653 bp), at 150 m for instance, correlated positively with the abundance of phosphate, and some surface conditions, but not nitrite or nitrate. At 500 m, the same OTU correlated negatively with a 1 month lag to nitrite concentration. At 890 m, it correlated positively, with a 1 month lag to nitrate, nitrite and phosphate. Furthermore, this OTU correlated to the abundances of different sets of bacteria at each depth. Further similar patterns were seen among the different OTUs of the AEGEAN-169 and SAR11 Surface 1 clades throughout the water column (Supplementary Figures S4 and S5).
Figure 5.
Association network of correlations between bacteria from the Marine Group A phylum at 150 m, 500 m and 890 m and other bacteria and environmental parameters found at every depth throughout the water column. Nodes and edges have the same thresholds (ρ, P and Q values for edges, mean abundance and occurrence thresholds for bacterial nodes) as in Figure 4.
Discussion
Time-lagged statistical associations suggest links between depths
The presence of correlations between different bacterial OTUs at different depths implies one or more mechanisms linking these depths. Owing to the absence of mixing between depths, it is most likely that sinking particles (Collins et al., 2011) and/or migrating organisms (Steinberg et al., 2000, 2002; Wilson and Steinberg, 2010; Schnetzer et al., 2011) link microbial communities at different depths. Given that the vast majority of lagged correlations between depths are ‘downward', a likely mechanism would be that sinking particles transport nutrients from the surface, which are in turn utilized by the communities at depth. Note that as our data refer only to the free-living bacteria, the ‘recipient' organisms either use dissolved materials released from the particles or, in some cases, may be shed directly from particles themselves. This is consistent with the long-standing paradigm in biological oceanography that productivity in the euphotic zone drives most of what happens at depth, and, in this case, it is the deep microbial community composition being driven indirectly by the surface environments and communities.
We hypothesized that we would find higher network density within depths than between depths as we would expect OTUs that are in the same location to have more direct associations with each other than we would with OTUs at a remote location, and our data support this. In fact, while OTUs at the same location can be related by way of symbiosis, shared resources, competition or other direct means (Fuhrman and Steele, 2008; Steele et al., 2011), OTUs at different depths can only be related by way of some linking environmental parameter, likely one that was not measured in our dataset such as the particle flux and its decomposition described above.
Negative correlations within and between depths
Just under half of the correlations seen, both between variables from the same depth and between variables from different depths are negative correlations (Tables 1 and 2, Supplementary Table S2 and S3). The time-lagged negative correlations between depths indicate that some change in conditions in the shallower water (that cause an increase of an OTU or other parameter) lead to a decrease in relative abundance of an organism in the deeper water. This could be from sinking particles encouraging the growth of competitors or predators of this organism, or by otherwise changing the deeper environment in a way that is less suitable to that organism (for example, by reduction of processes that had supported growth of that organism in the past). Negative correlations within (or sometimes between) depths may indicate any of a number of interactions between organisms such as competition (Chow et al., 2014) or allelopathy (see Strom, 2008).
Interaction patterns vary between depths
Our results suggest that microbial interactions at each depth show patterns that are common across depths, with key differences between depths. Specifically, all depths show qualitative modular patterns in which there are co-occurring groups of organisms, and that these modules are negatively correlated with other modules reminiscent of alternate states.
We can confirm these qualitative results with quantitative network topography statistics, which show that networks are more highly clustered than random networks, but have path lengths that are short (Table 2). These two conditions together indicate that the networks, by definition, have ‘nonrandom small world' properties (Watts and Strogatz, 1998). The nonrandomness of these networks suggests that the clustering patterns we observed are real and that the short path length is not merely an artifact of the networks being random, as truly random networks also have short path lengths (Watts and Strogatz, 1998). The short path lengths of between two and three mean that most nodes were connected to most other nodes by way of only a few species. This short path length may suggest that although most pairs of OTUs are not directly correlated to each other, they are usually indirectly correlated by way of one or two intermediate OTUs. Thus, changes to OTUs in one part of the network are more likely to affect OTUs throughout the network by way of intermediate variables. This ‘small world' topology is a common feature of microbial association in time series between bacteria, protists and viruses in marine systems (Steele et al., 2011; Chow et al., 2014) lake environments in all seasons (Kara et al., 2012) and many other biological and nonbiological systems (Humphries and Gurney, 2008). The relationship of groups of co-occurring bacteria to one or more environmental parameters suggests that community variability is likely driven by varying environmental conditions, both directly and in a way which may affect the community by cascading inter-microbial interactions. Also apparent are differences in the overall network structures at different depths. DCM, 5 m and especially 890 m appear to be highly modular, perhaps with seasonality in part driving this modularity, as some OTUs in each network are associated with the rate of change of day length. This pattern reflects findings that the surface depths and 890 m are the most seasonal depths (Cram et al., 2014a) and expands these observations to show that only a few species are highly correlated with season itself, while the remaining OTUs are highly correlated with other species, only some of which are particularly seasonal. Thus, while Cram et al. (2014a) observed several species that were statistically significantly seasonal, our findings here expand that observation to suggest that these seasonal organisms may be extending the effect of seasonality to much of the rest of the microbial community.
One interpretation of the defined modules of bacteria at 5 m, DCM and 890 m, which are correlated with biotic and environmental parameters (growth rates and dissolved nutrients at DCM, and temperature and dissolved nutrients at 890 m), is that communities at these depths tend to have two competing states. In this interpretation, microbial communities seem to have a see-saw like dynamic in which the community could be dominated by one module, dominated by the other module or existing in some intermediate state; however, it would be uncommon for subsets of organisms from each module to be particularly abundant at the same time. Some of the OTUs in these modules correlate with environmental parameters, suggesting the state of the environment is likely related to environmental variability and seasonality. However, as most of the connections in these modules are between OTUs, it is likely that ecological interactions between organisms, or else between organisms and unmeasured parameters, play an important role in determining community state. That 150 m and 500 m lack the modular structure identified at other depths suggests these environments have more complicated patterns of interactions, with different OTUs each responding to different environmental or biological stimulus. These findings dovetail with previous findings that there is pronounced seasonality at 5 m, the DCM and 890 m but not at 150 or 500 m, suggesting that seasonal patterns may be the ultimate drivers of the two-module pattern at the euphotic and bottom communities, while more subtle differences shape community structure in the mid-water column depths (Cram et al., 2014a).
Ecological niches of specific organisms
Related organisms, such as different strains of Prochlorococcus marinus (Johnson, 2006), or subtypes of SAR11 (Brown et al., 2012) have been shown to have different ecological niches and spatiotemporal distributions. Less is known about the ecology of many abundant deep water organisms. Our networks provide novel insight into the ecology of a number of little known OTUs. Two of our hub and spoke type networks focus on Gammaproteobacteria, and Marine Group A, both of which have previously been shown to be seasonal in the mid-water column (Cram et al., 2014a) and biogeochemically important (Swan et al., 2011; Allers et al., 2013; Wright et al., 2013). Further networks examine AEGEAN-169 and SAR11, which are particularly abundant throughout the water column (Alonso-Sáez et al., 2007; Carlson et al., 2009; Brown et al., 2012; Cram et al., 2014a). These networks illustrated that previously poorly characterized subgroups such as the species of Nitrospinaand clades of Marine Group A divide into multiple OTUs that associate very differently from each other, suggesting they are each made up of many different ‘ecological species' (Fuhrman, 2009) (Figure 5). That many of these OTUs associate with surface parameters different from other of these OTUs suggests that there must be more than one way that OTUs respond to changes in surface waters. We can envision a number of ways that there could be diverse linkages to surface variability; most of which in some way involve particle flux and migrating organisms.
For instance, rather than many organisms in the deep water column responding only to the magnitude of particle flux, OTUs might respond to the molecular composition of those particles. Some organisms could even be adapted to respond to the flux of particular kinds of particles with particular chemical signatures. In this case, there are a number of hypothetical ways that one could see pairwise correlations between OTUs in different depths; especially time-lagged ones.
For one hypothetical example, suppose that some species of phytoplankton (let us call it phytoplankton X) sometimes varies in its activity or abundance in surface waters. Thus, at some times of year, it both releases more of some kind of dissolved organic matter (through processes reviewed by Thornton, 2014) and forms sinking aggregates (such as in Kiørboe and Hansen, 1993). Surface bacteria that are adapted to break down dissolved organic matter released by plankton X will increase in relative abundance. Meanwhile, bacteria that are adapted to consuming particles produced by this organism (or perhaps material released from the fecal pellets of grazers eating this organism) would increase in abundance deeper in the water column, possibly with a lag related both to the sinking speed of the aggregate and the growth rate of the bacterial species. These bacteria would likely be adapted to deep water environments and would thus be different to the bacteria which respond at the surface. This pattern could happen for many different phytoplankton each of which might produce different suites of dissolved organic matter and promote production of different kinds of aggregates and sinking particles. Together a diverse phytoplankton community, with different plankton OTUs releasing different kinds of dissolved organic matter and producing different particles would produce many pairs of connections between bacteria. Other possibilities that might also produce numerous links by way of sinking particles include the bacteria in the shallower water changing the chemistry of surface-dissolved organic matter and altering the chemical composition of sinking particles. Changes in the concentration of trace nutrients such as vitamins might select for small numbers of surface bacteria and have influences on small subsets of sinking particles.
Beyond these more general trends, more specific patterns are evident within the groups observed and tell us about niches of a number of organisms; both how they relate to surface environments and factors at their own depth.
Marine Group A
The observation that at 890 m one OTU, MGA_653.1, was positively correlated, with a 1 month lag to several nitrogen species and phosphate (Figure 5), suggests it might be adapted to take advantage of relatively nutrient-rich conditions, but slow growing enough that it takes some time to respond to these conditions. MGA_762.8 correlated with a 1 month lag to biodiversity and high salinity in the DCM which suggests it might respond to particles that originate from the DCM during conditions which favor high biodiversity there. Other MGA found from 150 to 500 m lagged the abundance of specific OTUs and environmental conditions in shallower depths likely owing to factors outlined in the previous section. It is generally evident that most MGA found at 150 m and 500 m and below lag changes in parameters shallower in the water column. The exception to this is MGA_712.4 which, at 150 m, is positively correlated to nitrogen species at 500 and 890 m and temperature at 890 m. It is unlikely that this OTU is responding to deep water nitrogen directly, as it is hundreds of meters above the nitrogen with which it is covarying. This OTU's relationship to deep water nitrogen may suggest that its abundance lags some unmeasured factor that contributes to high nitrogen abundance deeper in the water column. Alternatively, as with other upward lags, perhaps it responds to zooplankton that migrate up from those deeper depths, or even buoyant particles that float up from those depths.
Deltaproteobacteria
As for the MGA, various OTUs from the Nitrospina genus at 890 m (Figure 4), have multiple connections to nitrogen species concentration and surface communities, many of them lagged. That two Nitrospina are connected to many species throughout the water column while other species show fewer connections might suggest that those Nitrospina may respond to nutrients released by a kind of particle that also affects many other organisms, whereas the less connected Nitrospina and SAR324 might be adapted to nutrients released by particles to which fewer other OTUs are adapted. Genomic work has provided some recent insight about these organisms potential. Nitrospina, for instance, are thought to be autotrophic nitrite oxidizers (Lücker et al., 2013), and SAR324 are likely particle-associated chemoautotrophs (Swan et al., 2011). However, until now, little had been known about the ecology of these organisms. These data suggest complex niches, beyond what was shown previously.
Alphaproteobacteria
Alphaproteobacterial groups such as SAR11 Surface-1 and AEGEAN-169 like the aforementioned groups show complex ecological interactions with very different patterns shown between different OTUs. The finding that the abundance of SAR11 Surface 1 OTUs from the surface tend to be related to fewer environmental factors than OTUs from deeper in the water column where they are less abundant (Supplementary Figure S4), suggests that although they are able to consistently dominate surface waters, they are more dependent on environmental and community structure variability in deeper waters. Differences in association patterns between different OTUs suggest that even closely related groups of organisms have different ecological niches. This pattern seems to hold for organisms of several different phyla. That many of these association patterns are related to different changes in the surface community suggests impacts of sinking particles are complex and that different surface changes cause different changes to particle flux and manifest in a number of ways in deeper waters.
Pairwise correlations show different patterns than overall seasonality
It is paradoxical that every pair of depths show correlations between them even though only the surface and the bottom show strong seasonal variability (Cram et al., 2014a). One might expect that if all depths are connected, seasonal changes at 5 m should drive seasonality at every other depth and that 890 m, with fewer connections to 5 m than other depths (Figure 2), should be the least seasonal. One possibility is that the seasonality at 5 m and 890 m is driven primarily by dominant organisms, while the connections shown here are often between rarer OTUs whose abundance may not have a strong cumulative effect on overall community structure. Alternatively, non-correlated organisms may change so much in nonseasonal ways that they overwhelm any seasonality from the connections seen here. A final possibility is that the specific correlations are driven by the release of specific nutrients by particles, as described above, whereas the seasonality is driven by a massive flux of diverse, perhaps less labile particles, which are seasonally variable in their magnitude. As productivity in the surface through 150 m is highest in April and productivity at 890 m is highest in August (Cram et al., 2014a), there could be a deposition of particles in the Spring, followed by seasonal succession in response to these particles. This succession would be less dependent on specific characteristics of the dissolved organic matter that are deposited, and lag times might be longer than 1 month; hence, our method would not pick up these correlations.
Conclusions
Our results illustrate that long-term time-series reveal very complex dynamics between bacterial OTUs and their environment; however, high level analysis reveals simple underlying patterns, and more explicit interrogation of particular taxa allows partial description of the niches of basically unknown bacterial types. Our findings are the first, to our knowledge, to show concurrent and lagged changes in community structure between depths in a time series, throughout the ocean water column. In agreement with previous recent findings (Cram et al., 2014a), our data support the inference that community structures of various depths are not isolated from each other but rather are linked. Because vertical profiles suggest that the depth of deepest mixing is ~40 m (Chow et al., 2013), mixing is likely not a viable mechanism for uniting these depths. We hypothesize that sinking particles and/or migrating organisms link the environments by transporting nutrients through the otherwise stratified water column. We furthermore show many associations among bacteria and environmental parameters that help delineate potential interactions and niche characteristics of many previously ecologically undefined, though very abundant, organisms.
Acknowledgments
This study would not have been possible without the support of numerous present and past members of the SPOT team. We especially thank C Chow, J Steele, A Parada, C Roney, D Kim, D Capone and D Caron, and three anonymous reviewers for help and advice. This work was supported by NSF grant numbers 0703159, 1136818 and grant GBMF3779 from the Gordon and Betty Moore Foundation Marine Microbiology Initiative, and the Wrigley Institute for Environmental Studies.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alarcón R, Waser NM, Ollerton J. (2008). Year-to-year variation in the topology of a plant–pollinator interaction network. Oikos 117: 1796–1807. [Google Scholar]
- Albert R, Barabási A-L. (2002). Statistical mechanics of complex networks. Rev Mod Phys 74: 47–97. [Google Scholar]
- Allers E, Wright JJ, Konwar KM, Howes CG, Beneze E, Hallam SJ et al. (2013). Diversity and population structure of Marine Group A bacteria in the Northeast subarctic Pacific Ocean. ISME J 7: 256–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Sáez L, Balagué V, Sà EL, Sánchez O, González JM, Pinhassi J et al. (2007). Seasonality in bacterial diversity in north-west Mediterranean coastal waters: assessment through clone libraries, fingerprinting and FISH. FEMS Microbiol Ecol 60: 98–112. [DOI] [PubMed] [Google Scholar]
- Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR et al. (2011). Enterotypes of the human gut microbiome. Nature 473: 174–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberán A, Bates ST, Casamayor EO, Fierer N. (2011). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6: 343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beman JM, Popp BN, Francis CA. (2008). Molecular and biogeochemical evidence for ammonia oxidation by marine Crenarchaeota in the Gulf of California. ISME J 2: 429–441. [DOI] [PubMed] [Google Scholar]
- Beman JM, Steele JA, Fuhrman JA. (2011). Co-occurrence patterns for abundant marine archaeal and bacterial lineages in the deep chlorophyll maximum of coastal California. ISME J 5: 1077–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonilla-Findji O, Herndl GJ, Gattuso J-P, Weinbauer MG. (2009). Viral and flagellate control of prokaryotic production and community structure in offshore Mediterranean waters. Appl Environ Microbiol 75: 4801–4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe H, Tripp HJ, Zehr JP. (2010). Unicellular cyanobacteria with a new mode of life: the lack of photosynthetic oxygen evolution allows nitrogen fixation to proceed. Arch Microbiol 192: 783–790. [DOI] [PubMed] [Google Scholar]
- Brown MV, Lauro FM, DeMaere MZ, Muir L, Wilkins D, Thomas T et al. (2012). Global biogeography of SAR11 marine bacteria. Mol Syst Biol 8: 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MV, Schwalbach MS, Hewson I, Fuhrman JA. (2005). Coupling 16 S-ITS rDNA clone libraries and automated ribosomal intergenic spacer analysis to show marine microbial diversity: development and application to a time series. Environ Microbiol 7: 1466–1479. [DOI] [PubMed] [Google Scholar]
- Carlson C, Morris R, Parsons R, Treusch A, Giovannoni S, Vergin K. (2009). Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J 3: 283–295. [DOI] [PubMed] [Google Scholar]
- Chaffron S, Rehrauer H, Pernthaler J, von Mering C. (2010). A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res 20: 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C-ET, Kim DY, Sachdeva R, Caron DA, Fuhrman JA. (2014). Top-down controls on bacterial community structure: microbial network analysis of bacteria, T4-like viruses and protists. ISME J 8: 816–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C-ET, Sachdeva R, Cram JA, Steele JA, Needham DM, Patel A et al. (2013). Temporal variability and coherence of euphotic zone bacterial communities over a decade in the Southern California Bight. ISME J 7: 2259–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman T, Moré J. (1983). Estimation of sparse Jacobian matrices and graph coloring blems. SIAM J Numer Anal 20: 187–209. [Google Scholar]
- Collins LE, Berelson W, Hammond DE, Knapp A, Schwartz R, Capone D. (2011). Particle fluxes in San Pedro Basin, California: A four-year record of sedimentation and physical forcing. Deep Sea Res Part Oceanogr Res Pap 58: 898–914. [Google Scholar]
- Cram JA, Chow C-ET, Sachdeva R, Needham DM, Parada AE, Steele JA et al. (2014. a). Seasonal and interannual variability of the marine bacterioplankton community throughout the water column over ten years. ISME J 9: 563–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cram JA, Sun F, Fuhrman JA. (2014. b). Marine bacterial, archaeal and protistan association networks. In:Encyclopedia of Metagenomics, Springer Reference. New York: Springer, 1–10. [Google Scholar]
- Dabney A, Storey JD. (2004). Q-value estimation for false discovery rate control. Medicine 344: 539–548. [Google Scholar]
- Dunne JA, Williams RJ, Martinez ND. (2002). Food-web structure and network theory: The role of connectance and size. Proc Natl Acad Sci USA 99: 12917–12922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiler A, Heinrich F, Bertilsson S. (2012). Coherent dynamics and association networks among lake bacterioplankton taxa. ISME J 6: 330–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust K, Raes J. (2012). Microbial interactions: from networks to models. Nat Rev Microbiol 10: 538–550. [DOI] [PubMed] [Google Scholar]
- Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J et al. (2012). Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol 8: e1002606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freilich S, Kreimer A, Meilijson I, Gophna U, Sharan R, Ruppin E. (2010). The large-scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res 38: 3857–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman JA. (2009). Microbial community structure and its functional implications. Nature 459: 193–199. [DOI] [PubMed] [Google Scholar]
- Fuhrman JA, Cram JA, Needham DM. (2015). Marine microbial community dynamics and their ecological interpretation. Nat Rev Microbiol 13: 133–146. [DOI] [PubMed] [Google Scholar]
- Fuhrman J, Steele J. (2008). Community structure of marine bacterioplankton: patterns, networks, and relationships to function. Aquat Microb Ecol 53: 69–81. [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B et al. (2012). Defining seasonal marine microbial community dynamics. ISME J 6: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Vergin KL. (2012). Seasonality in ocean microbial communities. Science 335: 671–676. [DOI] [PubMed] [Google Scholar]
- Hatosy SM, Martiny JBH, Sachdeva R, Steele J, Fuhrman JA, Martiny AC. (2013). Beta diversity of marine bacteria depends on temporal scale. Ecology 94: 1898–1904. [DOI] [PubMed] [Google Scholar]
- Humphries MD, Gurney K. (2008). Network ‘small-world-ness': a quantitative method for determining canonical network equivalence. PLoS ONE 3: e0002051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ings TC, Montoya JM, Bascompte J, Blüthgen N, Brown L, Dormann CF et al. (2009). Review: Ecological networks – beyond food webs. J Anim Ecol 78: 253–269. [DOI] [PubMed] [Google Scholar]
- Johnson ZI. (2006). Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311: 1737–1740. [DOI] [PubMed] [Google Scholar]
- Jürgens K, Matz C. (2002). Predation as a shaping force for the phenotypic and genotypic composition of planktonic bacteria. Antonie Van Leeuwenhoek 81: 413–434. [DOI] [PubMed] [Google Scholar]
- Jurgens K, Pernthaler J, Schalla S, Amann R. (1999). Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl Environ Microbiol 65: 1241–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kara EL, Hanson PC, Hu YH, Winslow L, McMahon KD. (2012). A decade of seasonal dynamics and co-occurrences within freshwater bacterioplankton communities from eutrophic Lake Mendota, WI, USA. ISME J 7: 680–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiørboe T. (2001). Formation and fate of marine snow: small-scale processes with large-scale implications. Sci Mar 65: 57–71. [Google Scholar]
- Kiørboe T, Hansen JLS. (1993). Phytoplankton aggregate formation: observations of patterns and mechanisms of cell sticking and the significance of exopolymeric material. J Plankton Res 15: 993–1018. [Google Scholar]
- Lücker S, Nowka B, Rattei T, Spieck E, Daims H. (2013). The Genome of Nitrospina gracilis Illuminates the Metabolism and Evolution of the Major Marine Nitrite Oxidizer. Front Microbiol 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfatti F, Azam F. (2009). Atomic force microscopy reveals microscale networks and possible symbioses among pelagic marine bacteria. Aquat Microb Ecol 58: 1–14. [Google Scholar]
- Malfatti F, Samo TJ, Azam F. (2009). High-resolution imaging of pelagic bacteria by Atomic Force Microscopy and implications for carbon cycling. ISME J 4: 427–439. [DOI] [PubMed] [Google Scholar]
- Miller MB, Bassler BL. (2001). Quorum sensing in bacteria. Annu Rev Microbiol 55: 165–199. [DOI] [PubMed] [Google Scholar]
- Montoya JM, Pimm SL, Solé RV. (2006). Ecological networks and their fragility. Nature 442: 259–264. [DOI] [PubMed] [Google Scholar]
- Needham DM, Chow C-ET, Cram JA, Sachdeva R, Parada A, Fuhrman JA. (2013). Short-term observations of marine bacterial and viral communities: patterns, connections and resilience. ISME J 7: 1274–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ. (2006). Modularity and community structure in networks. Proc Natl Acad Sci USA 103: 8577–8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen JM, Bascompte J, Dupont YL, Jordano P. (2007). The modularity of pollination networks. Proc Natl Acad Sci USA 104: 19891–19896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q, Dutta D, Schwalbach MS, Steele JA, Fuhrman JA, Sun F. (2006). Local similarity analysis reveals unique associations among marine bacterioplankton species and environmental factors. Bioinformatics 22: 2532–2538. [DOI] [PubMed] [Google Scholar]
- Schnetzer A, Moorthi SD, Countway PD, Gast RJ, Gilg IC, Caron DA. (2011). Depth matters: Microbial eukaryote diversity and community structure in the eastern North Pacific revealed through environmental gene libraries. Deep Sea Res Part Oceanogr Res Pap 58: 16–26. [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. (2011). Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY et al. (2011). Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J 5: 1414–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg DK, Carlson CA, Bates NR, Goldthwait SA, Madin LP, Michaels AF. (2000). Zooplankton vertical migration and the active transport of dissolved organic and inorganic carbon in the Sargasso Sea. Deep Sea Res Part Oceanogr Res Pap 47: 137–158. [Google Scholar]
- Steinberg DK, Goldthwait SA, Hansell DA. (2002). Zooplankton vertical migration and the active transport of dissolved organic and inorganic nitrogen in the Sargasso Sea. Deep Sea Res Part Oceanogr Res Pap 49: 1445–1461. [Google Scholar]
- Strom SL. (2008). Microbial ecology of ocean biogeochemistry: a community perspective. Science 320: 1043–1045. [DOI] [PubMed] [Google Scholar]
- Suweis S, D'Odorico P. (2014). Early warning signs in social-ecological networks. PLoS ONE 9: e101851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333: 1296–1300. [DOI] [PubMed] [Google Scholar]
- Team RDC. (2011) R: A Language and Environment for Statistical Computing. Vienna: : Austria, http://www.R-project.org/. [Google Scholar]
- Thornton DCO. (2014). Dissolved organic matter (DOM) release by phytoplankton in the contemporary and future ocean. Eur J Phycol 49: 20–46. [Google Scholar]
- Watts DJ, Strogatz SH. (1998). Collective dynamics of ‘small-world' networks. Nature 393: 440–442. [DOI] [PubMed] [Google Scholar]
- Wilson SE, Steinberg DK. (2010). Autotrophic picoplankton in mesozooplankton guts: evidence of aggregate feeding in the mesopelagic zone and export of small phytoplankton. Mar Ecol Prog Ser 412: 11–27. [Google Scholar]
- Wright JJ, Mewis K, Hanson NW, Konwar KM, Maas KR, Hallam SJ. (2013). Genomic properties of Marine Group A bacteria indicate a role in the marine sulfur cycle. ISME J 8: 455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia LC, Ai D, Cram J, Fuhrman JA, Sun F. (2013). Efficient statistical significance approximation for local similarity analysis of high-throughput time series data. Bioinformatics 29: 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Deng Y, Luo F, He Z, Tu Q, Zhi X. (2010). Functional molecular ecological networks. mBio 1: e00169–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.