Abstract
The role of bacterioplankton in the cycling of marine dissolved organic matter (DOM) is central to the carbon and energy balance in the ocean, yet there are few model organisms available to investigate the genes, metabolic pathways, and biochemical mechanisms involved in the degradation of this globally important carbon pool. To obtain microbial isolates capable of degrading semi-labile DOM for growth, we conducted dilution to extinction cultivation experiments using seawater enriched with high molecular weight (HMW) DOM. In total, 93 isolates were obtained. Amendments using HMW DOM to increase the dissolved organic carbon concentration 4x (280 μM) or 10x (700 μM) the ocean surface water concentrations yielded positive growth in 4–6% of replicate dilutions, whereas <1% scored positive for growth in non-DOM-amended controls. The majority (71%) of isolates displayed a distinct increase in cell yields when grown in increasing concentrations of HMW DOM. Whole-genome sequencing was used to screen the culture collection for purity and to determine the phylogenetic identity of the isolates. Eleven percent of the isolates belonged to the gammaproteobacteria including Alteromonadales (the SAR92 clade) and Vibrio. Surprisingly, 85% of isolates belonged to the methylotrophic OM43 clade of betaproteobacteria, bacteria thought to metabolically specialize in degrading C1 compounds. Growth of these isolates on methanol confirmed their methylotrophic phenotype. Our results indicate that dilution to extinction cultivation enriched with natural sources of organic substrates has a potential to reveal the previously unsuspected relationships between naturally occurring organic nutrients and the microorganisms that consume them.
Introduction
Marine dissolved organic matter (DOM) supports a considerable fraction of the carbon, energy and nutrient requirements of bacterioplankton in the ocean, yet the biological processes that control DOM turnover are poorly understood. This is due in part to the chemical complexity of naturally occurring DOM and the current level of molecular characterization, along with a lack of information on the variety of bacteria and metabolic strategies involved in DOM degradation.
Marine DOM characterization has been largely limited to defining its bulk chemical properties. Marine DOM can be operationally separated into two different fractions based on size: low molecular weight (<1 kDa) and high molecular weight (HMW: ⩾1 kDa). The HMW DOM fraction is isolated from seawater by ultrafiltration and typically accounts for 25–40% of the total dissolved organic carbon (DOC) in marine waters (Benner, 2002). The elemental composition of HMW DOM (carbon:nitrogen:phosphorus=298:18:1; Benner, 2002) contrasts with marine plankton (Redfield et al., 1963) in that HMW DOM is depleted in nitrogen and phosphorus relative to carbon. HMW DOM from open ocean-surface waters contains 40–60 nmol nitrogen per μmol carbon primarily in the form of amides (McCarthy et al., 1997; Aluwihare et al., 2005) and only 3–4 nmol phosphorus per μmol carbon as part of phosphate esters and phosphonate molecules (Clark et al., 1998; Kolowith et al., 2001). The 13C NMR spectrum of HMW DOM from marine surface waters indicates it is largely composed of carbohydrates (50%), humic substances (25%) and a lesser amount of lipids and proteins (Benner et al., 1992; Aluwihare et al., 1997; Benner, 2002).
Radiocarbon analysis offers insight of the fate and reactivity of DOM. HMW DOM from the North Pacific has a Δ14C of 10‰ and its constituting neutral monosaccharides derived from acid hydrolysis have Δ14C values of 47–67‰. This is similar to the Δ14C of dissolved inorganic carbon in surface waters (72±7‰ Repeta and Aluwihare, 2006) and indicates recent production. In contrast, total DOC is radiocarbon depleted (Δ14C of −146‰ and −540‰ in surface and deep North Pacific waters, respectively) likely owing to the presence of refractory carbon (Williams and Druffel, 1987). This data suggests that subcomponents of HMW DOM from surface waters, including polysaccharides, cycle faster (1–25 years; Repeta and Aluwihare, 2006) than the total pool of DOC (6000 years; Williams and Druffel, 1987). These chemical characteristics, along with field observations of semi-labile DOM utilization in natural seawater (Carlson et al., 2004; McCarren et al., 2010), indicate HMW DOM can fuel microbial metabolism on ecologically relevant timescales. Difficulties in identifying the specific chemical moieties supporting such growth, however, limits current understanding of DOM transformation, remineralization and ultimate impact on the flux of carbon and nutrients through the marine food web.
Bacteria and Archaea are the dominant degraders of DOM (Carlson, 2002), but the specific taxa, genes and molecular mechanisms most responsible for DOM metabolism remain relatively obscure. Recently, several culture-independent studies have begun to identify the microbial community members that respond to DOM enrichment, though the majority of these studies have focused on DOM isolated from phytoplankton cultures (Poretsky et al., 2010; Nelson and Carlson, 2012; Sarmento and Gasol, 2012; Landa et al., 2013; Sharma and Becker et al., 2013; Beier et al., 2014), which is presumed to be more labile than HMW DOM collected in oligotrophic environments. This is likely because of the presence of labile proteins and amino acids (Sarmento et al., 2013) and homopolysaccharides (Meon and Kirchman, 2001) in phytoplankton DOM. Aluwihare et al. (1997) and Aluwihare and Repeta (1999) observed rapid drawdown of phytoplankton DOM and evolution of polysaccharides from homopolysaccharides (energy storage products) to heteropolysaccharides with a composition like HMW DOM. In contrast, there have been fewer culture-independent studies focusing on the cycling of HMW DOM (McCarren et al., 2010; Sharma and Becker et al., 2013). In all of these studies, DOM additions resulted in the enrichment of select members of the microbial community and in several cases a temporal succession of DOM-degrading bacteria (McCarren et al., 2010; Sharma and Becker et al., 2013). Although these “omic”-based studies have generated hypothesis as to the functional and metabolic roles of DOM-degrading bacteria, appropriate model systems upon which to test such hypothesis and dissect the genes, metabolic pathways and chemical transformation reactions driving DOM degradation are still lacking.
The goal of this study was to isolate HMW DOM metabolizing bacteria from marine environments to develop model systems for investigating the biochemical and metabolic pathways driving DOM transformations in the ocean. Dilution to extinction cultivation has greatly expanded the collection of marine bacterial isolates (Button et al., 1993; Connon and Giovannoni, 2002), and has proved instrumental in isolating organisms resistant to traditional cultivation techniques, including members of the SAR11 and SAR116 clades (Rappé et al., 2002; Stingl et al., 2007a, 2008), oligotrophic marine gammaproteobacteria (Cho and Giovannoni, 2004), members of the SUP05 clade of gammaproteobacterial sulfur oxidizers (Marshall and Morris, 2013) and pyschropiezophilic alphaproteobacteria (Eloe et al., 2011). Field studies, genome analysis and pure cultures experiments enabled by the cultivation of bacterial clades like SAR11 have expanded our understanding of the ecology and physiology of marine bacteria and their role in the ocean carbon cycle (Malmstrom et al., 2004; Schwalbach et al., 2010; Sun et al., 2011; Grote et al., 2012). In this study we expanded on the technique by enriching dilution to extinction cultures with HMW DOM collected directly from ocean surface waters, leveraging the high-throughput nature of dilution to extinction cultivation to screen a microbial assemblage for HMW DOM-utilizing organisms.
Results
Molecular characterization of HMW DOM
DOM was characterized by 1H nuclear magnetic resonance spectroscopy to determine major constituents. The spectrum looked similar to previously published 1H NMR spectra from samples collected at the Natural Energy Laboratory study site (Repeta and Aluwihare, 2006) with strong resonances at 5.5 p.p.m. from carbohydrate anomeric (O-CH-O), 4.0 p.p.m. from carbohydrate (HCOH), 2.7 p.p.m. from N-acetyl N-methyl aminosugar (H3C-N-C(O)CH3), 2.0 p.p.m. from N-acetyl aminosugars N-C(O)CH3) and 1.3 p.p.m. from 6-deoxysugar protons. The spectral region between 0.9–3.0 p.p.m. also included a broad unresolved baseline attributed to co-occurring humic substances. Monosaccharide analysis of hydrolysable sugars showed the presence of similar amounts of glucose, galactose, mannose, rhamnose, fucose and xylose+arabinose. This distribution is typical of HMW DOM hydrolysable sugars in open ocean surface seawater (Aluwihare et al., 1997). The sum of hydrolysable neutral sugars represent 10% of total HMW DOC.
Experimental overview
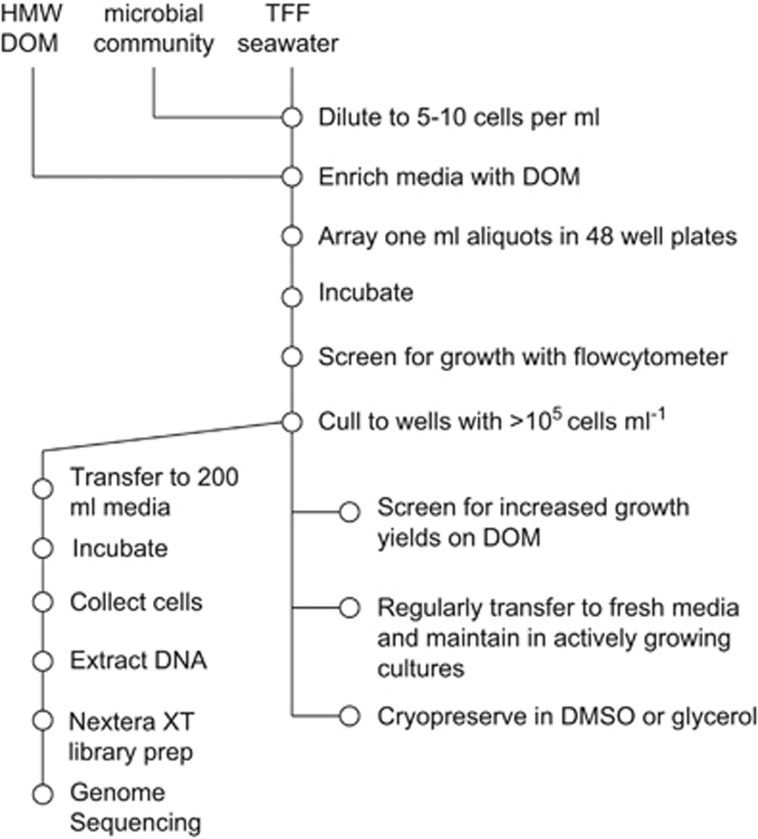
The culturing method applied here is a modification of the 'high-throughput' culturing technique described by Connon and Giovannoni (2002) to isolate marine bacteria in seawater media using dilution to extinction. Our main modification consisted of supplementing filter-sterilized natural seawater with HMW DOM (>1 kDa) isolated by ultrafiltration from oligotrophic ocean surface waters (Figure 1). To limit the amount of background DOM in the natural seawater and to increase the probability of obtaining isolates responding to the HMW DOM additions we used relatively nutrient deplete, Sargasso Sea seawater sterilized by tangential-flow filtration (TFF) to prepare dilution to extinction samples.
Figure 1.
Procedure for the setup, incubation, screening and downstream analysis of dilution to extinction cultures enriched with high molecular weight dissolved organic matter (HMW DOM), including purity screening and phylogenetic identification via whole-genome sequencing. TFF, tangential-flow filtered.
The inoculum was collected from Nahant Bay in the coastal North Atlantic and had an initial cell concentration of 1.5 × 106 (±2.2 × 105 s.d.) cells per ml. The inoculum was diluted into TFF seawater to a final concentration of 3 cells per ml and then divided into three treatments: a no DOM addition (70 μM DOC background) and two HMW DOM additions (4x DOC (280 μM) and 10x DOC (700 μM)). One mililiter aliquots were placed in a 48-well cultivation plates and incubated at 24 °C in the dark. After 4 weeks of incubation, the cultures were diluted 10 000-fold into fresh TFF media containing the appropriate DOM concentrations to ensure viability before the screening.
Growth screen
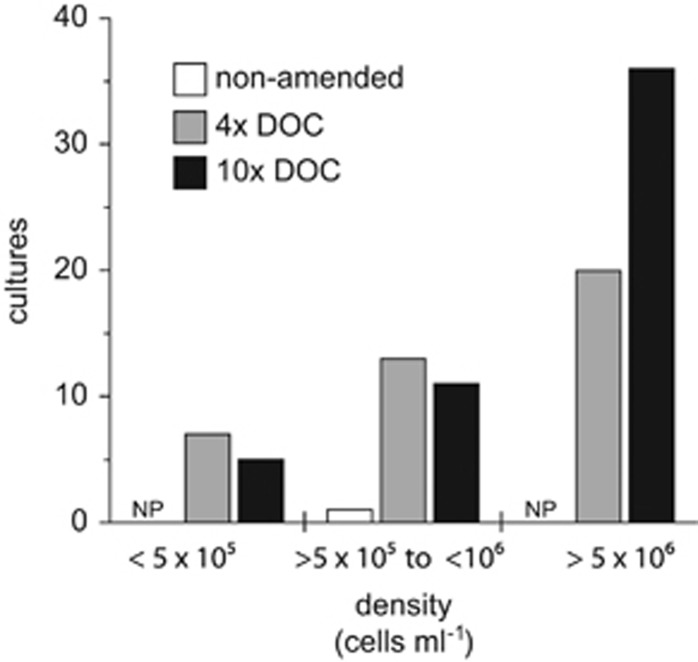
After 6–7 weeks of incubation, the cultures were screened for growth by flow cytometry and any well with >105 cells per ml was scored growth positive. Out of 2520 potential extinction cultures, 93 (3.7%) scored positive. Only 1 of the 840 non-DOM-amended inoculated wells (0.1%) scored growth positive (Table 1). By contrast, the HMW DOM amendments had significantly more wells scoring growth positive (Table 1; P<0.05, bootstrap comparison with 106 iterations), increasing the percent recovery to 4.7% and 6.0% for the 4x and 10x DOC treatments, respectively. Cell concentrations also tended to increase with higher DOM dose (Figure 2), with upper concentrations reaching >106 cells per ml (max 2.4 × 106), the majority of which occurred in the 10x DOC treatment. No growth was detected in any of the non-inoculated control wells (Table 1). The single culture obtained from the non-DOM-amended treatment showed little sustained growth, and could not be maintained in successive transfers of the culture collection grown on 4x DOC.
Table 1. HMW DOM-enriched dilution to extinction cultivation experimental design and growth screen results. Forty-eight well plates were filled with TFF oligotrophic seawater amended with HMW DOM at 4x (280 μM) DOC, 10x (700 μM) DOC or non-amended and then inoculated with the diluted coastal bacterioplankton community.
| non-DOM | 4 × DOM | 10 × DOM | Total | |
|---|---|---|---|---|
| Wells | ||||
| Innoculated | 840 | 840 | 840 | 2520 |
| Controls | 54 | 54 | 54 | 162 |
| Cultures detected | ||||
| Innoculated | 1 | 40 | 52 | 93 |
| Controls | 0 | 0 | 0 | 0 |
Abbreviation: DOM, dissolved organic matter.
The 'controls' consisted of six wells on each treatment plate, as well as an entire microtiter plate, that contained the same media and DOM enrichment, but were not inoculated. Positive growth ('cultures detected') was defined as a well having a density of 105 or more cells per ml after 6–7 weeks of incubation and one round of redilution.
Figure 2.
Effect of HMW DOM enrichment on dilution to extinction cultures' final cell density. Only wells that scored positive for growth (⩾105 cells per ml) are included. NP, no positive cultures detected; 4x DOC, ~280 μM DOC; 10 × DOC, ~700 μM DOC.
HMW DOM dose response
To verify that the isolates had a discernible growth response to the HMW DOM, the 93 cultures were tested for increased cell yields with DOM additions. The TFF seawater was first UV-oxidized to reduce background DOC concentrations from 70 μM DOC to 25 μM DOC. HMW DOM was then added at 4x (280 μM), 20x (1400 μM) or 100x (7000 μM) open ocean DOC concentrations. Sixty six of the cultures showed an increase in cell yields with DOM addition, reaching >5 × 105 cells per ml in the DOM-amended treatments and having no growth in the non-DOM-amended controls (Supplementary Figure S1). Furthermore, 36 cultures showed a direct proportional increase in cell yields with increasing HMW DOM concentrations (100x>20x>4x).
Identification and purity screen
In an initial phylogenetic screen of the isolates, 88 small subunit (SSU) rRNA gene sequences were determined via PCR amplification and subsequent Sanger sequencing. Ten of these sequences clustered within the gammaproteobacteria Alteromonadaceae (7 cultures including 3 members of the SAR92 clade and 1 of the OM60 clade), Pseudoalteromonadaceae (1 culture) and Vibrio (2 cultures). The remaining 78 sequences all clustered within the OM43 clade of Methylophilaceae betaproteobacteria. There was no significant difference in the relative recovery of any specific taxonomic group in the 4x versus 10x DOC treatments.
The quality of the SSU rRNA Sanger sequencing provided an initial assessment of culture purity, with positive PCR amplifications yielding poor quality Sanger sequences being potentially indicative of mixed cultures. We applied an additional approach to assess the purity of cultures and identify their phylogenetic origins using 'tagmentation' sequencing library preparation and Illumina paired-end sequencing for whole-genome shotgun (WGS) sequencing (Figure 1 and see Materials and methods section). Sixty-eight cultures were successfully sequenced, with an average of 120 000 paired reads (200 × 200 bases) per sample.
The purity of the cultures was assessed using the WGS data in three ways. In the first approach, the reads were de novo assembled into contigs which were then searched via BLASTn against the SILVA rRNA database to identify SSU rRNA and large subunit rRNA sequences. Samples having multiple rRNA genes binning to different organisms were considered mixed. Ninety percent of the 68 cultures had a single copy of small and large subunit rRNA gene binning to one organism with high identity (Table 2), either an OM43 clade betaproteobacterium (82%) or miscellaneous gammaproteobacteria (9%). Five samples contained multiple rRNA genes hitting two different organisms. These were primarily an OM43 clade betaproteobacteria with a SAR92 clade gammaproteobacteria, although one culture (NB0016) contained an OM43 clade betaproteobacteria and a flavobacterium.
Table 2. Phylogenetic identification of HMW DOM-enriched dilution cultures via assembled SSU and LSU rRNA genes from whole-genome sequencing.
| Genes found | Best matching described taxa via 16 S rRNA | Best matching described taxa via 23 S rRNA | ||||||
|---|---|---|---|---|---|---|---|---|
| IDa | 16 S | 23 S | Best match | Percentage IDb | Lengthc | Best match | Percentage IDb | Lengthc |
| Md | 1 | 1 | Methylophilaceae OM43 clade uncultured bacterium | 99.4 | 1534 | OM43 clade betaproteobacterium KB13 | 98.9 | 2874 |
| 11 | 1 | 1 | Simiduia agarivorans SA1 DSM 21679 | 93.9 | 1538 | Teredinibacter turnerae T7901 | 92.3 | 2624 |
| 15 | 1 | 1 | SAR92 clade uncultured bacterium | 98.5 | 1533 | Congribacter; gamma proteobacterium HTCC2207 | 97.9 | 2909 |
| 30 | 1 | 0 | Algicola bacteriolytica | 99.1 | 1497 | ND | ||
| 59 | 1 | 1 | Vibrio splendidus LGP32 | 97.9 | 1548 | Vibrionales bacterium SWAT-3 | 99.9 | 2902 |
| 70 | 1 | 1 | Methylophilaceae OM43 clade uncultured bac. 580 | 99.9 | 1532 | Methylophilaceae OM43 clade uncultured bac. 580 | 99.7 | 2883 |
| 72 | 1 | 1 | Vibrio splendidus LGP32 | 99.8 | 1548 | Vibrio splendidus LGP32 | 99.9 | 2879 |
| 77 | 1 | 1 | Simiduia agarivorans SA1 DSM 21679 | 93.9 | 1538 | Teredinibacter turnerae T7901 | 92.5 | 2871 |
| 80 | 1 | 1 | Vibrio splendidus LGP32 | 99.6 | 1548 | Vibrio splendidus LGP32 | 99.4 | 2905 |
| 93 | 1 | 1 | Methylophilaceae OM43 clade uncultured bac. 580 | 99.9 | 1532 | Methylophilaceae OM43 clade uncultured bac. 580 | 99.7 | 2883 |
| 94 | 1 | 1 | Alteromonadaceae uncultured bacterium | 98.3 | 1490 | Glaciecola sp. HTCC2999 | 91.9 | 2882 |
| 16 | 2 | 2 | Methylophilaceae OM43 clade uncultured bacterium | 99.9 | 1534 | Methylophilaceae OM43 clade uncultured bac. 580 | 99.4 | 2874 |
| Polaribacter; uncultured Flavobacteriaceae | 99.8 | 976 | Polaribacter irgensii 23-P | 95.1 | 2402 | |||
| 48 | 2 | 2 | Methylophilaceae OM43 clade uncultured bac. 580 | 99.8 | 1533 | Methylophilaceae OM43 clade uncultured bac. 580 | 99.7 | 2883 |
| SAR92 clade uncultured bacterium | 98.2 | 1507 | Congribacter; gamma proteobacterium HTCC2207 | 97.9 | 2903 | |||
| 54 | 3 | 2 | Methylophilaceae OM43 clade uncultured bacterium | 99.3 | 1535 | OM43 clade betaproteobacterium KB13 | 98.8 | 2874 |
| SAR92 gamma proteobacterium HTCC230 | 99.7 | 758 | Congribacter; gamma proteobacterium HTCC2207 | 98.2 | 1348 | |||
| Microbulbifer variabilis Ni-2088 | 92.7 | 562 | ND | — | — | |||
| 71 | 2 | 2 | Methylophilaceae OM43 clade uncultured bacterium | 99.7 | 1546 | OM43 clade betaproteobacterium KB13 | 99.7 | 2893 |
| Microbulbifer variabilis Ni-2088 | 99.6 | 1097 | Congribacter; gamma proteobacterium HTCC2207 | 97.9 | 2283 | |||
| 76 | 2 | 2 | Methylophilaceae OM43 clade uncultured bacterium | 99.7 | 1534 | OM43 clade betaproteobacterium KB13 | 99.7 | 2874 |
| SAR92 clade uncultured bacterium | 99.6 | 1492 | Congribacter; gamma proteobacterium HTCC2207 | 97.9 | 1587 | |||
Abbreviations: ND, not detected; bac., bacterium. The number of rRNA genes detected in a given sample was used as an indication of culture purity. The rRNA gene phylogeny was identified by a BLASTn similarity search against the SILVA SSU and LSU database.
Culture identification number (NB00##).
Percent matching identities.
Gene length.
The 53 cultures of 68 cultures screened with a single 16 S and 23 S rRNA gene and which had the top BLASTn match for both genes to the OM43 clade of Methylophilaceae. The mean (±s.d.) perecent identites for these 53 cultures was 99.4 (±0.4) for the 16 S rRNA and 98.9 (±0.05) for the 23 S rRNA.
To verify that rRNAs did not escape detection in the course of sequence assembly we searched for SSU rRNA sequences in the unassembled reads. The quality controlled (trimmed and paired-end joined) unassembled reads were searched against each sample's assembled SSU rRNA sequence. Samples having a large portion of reads matching at low identity were taken as an indication of a mixed culture. Four of the five same samples that yielded multiple, divergent rRNA reads after assembly (Table 2) had high levels of low identity rRNA matching reads (5–17% Supplementary Figures S2 and S3).
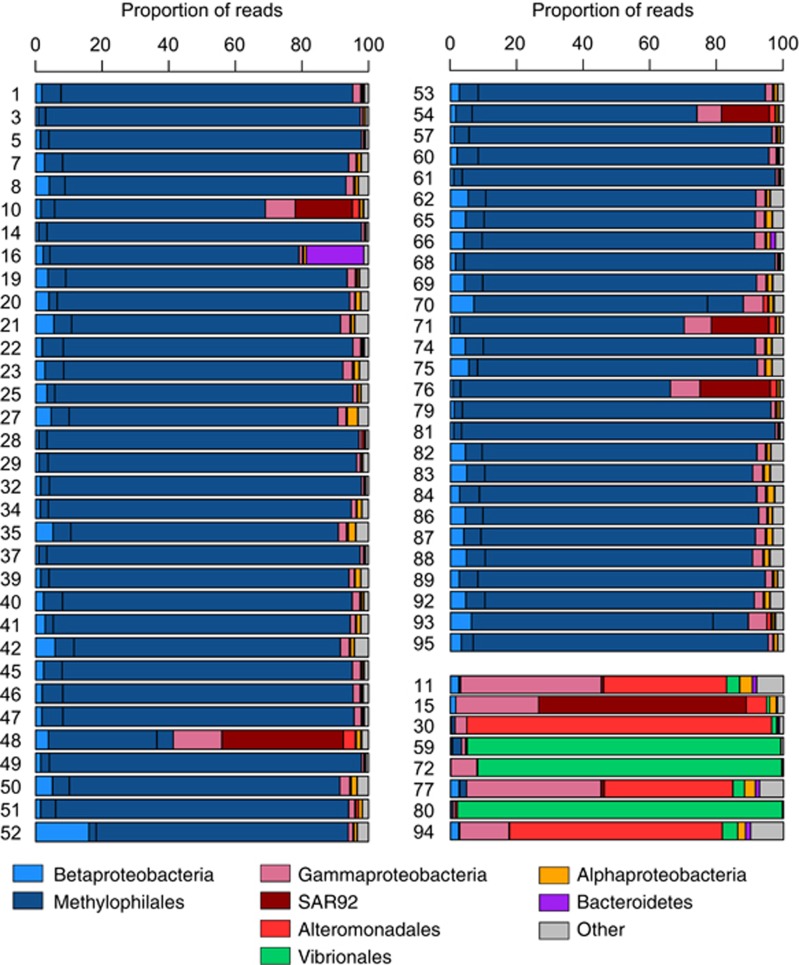
Finally, a metagenomic approach was taken, taxonomically binning the unassembled reads for each sample based on the top hit from a LAST sequence similarity search against NCBI's RefSeq database. For 54 of the 60 cultures with betaprotebacterial SSU rRNA sequences the majority of reads binned to betaproteobacterial reference genomes (Figure 3). The remaining six cultures had a substantial enrichment in reads binning to non-betaprotebacterial reference genomes (17–53% of total reads). Five of these cultures were also identified as mixed using the SSU rRNA analyses above but the sixth (NB0010) was only confidently detected with the metagenomic approach because the unassembled SSU rRNA analysis revealed only a marginal proportion of low identity SSU rRNA reads (1.4%).
Figure 3.
Taxonomic binning of unassembled reads based on the top hit of a LAST sequence similarity search against NCBI's REFSeq database. The culture identification number (NB00##) is located on the left of the sample bar.
In summary, these three screening approaches produced congruent assessments of culture identity and purity, with only 6 out of 68 of WGS-sequenced cultures indicating the presence of mixed populations.
Isolate phylogeny
The WGS-assembled SSU rRNA sequences matched excellently with the Sanger reads and provided phylogenetic placement of the culture collection. Of the 58 WGS SSU rRNA sequences (length=1542 nucleotides) that had overlapping regions with quality Sanger sequences (489–1444 bp, quality>97%), only five sequences had mismatches between the Sanger and WGS data (NB0034 (10 mismatches), NB0054 (11), NB0057 (32), NB0068 (12) and NB0072 (194)), with the remaining 53 cultures matching at 100% identity between the two sequencing methods.
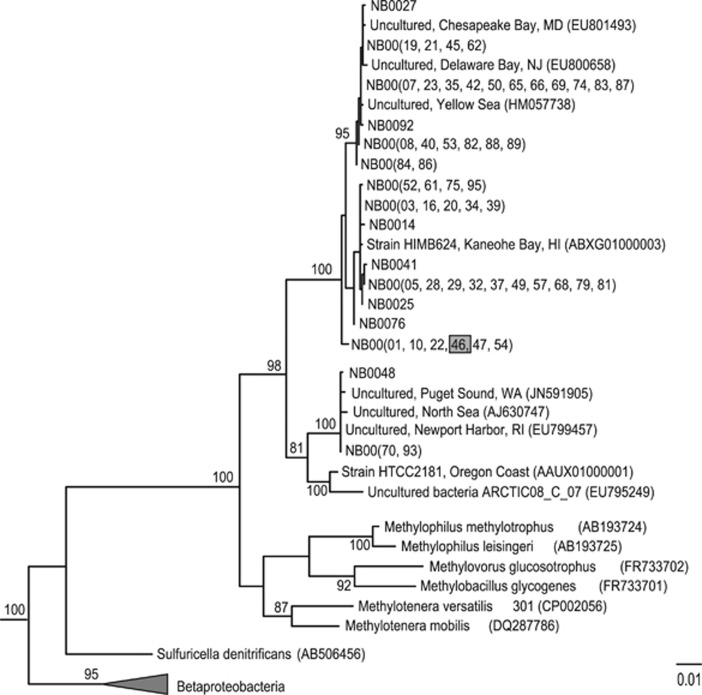
The full-length WGS-assembled rRNA sequences were used to determine the phylogenetic placement of cultures. The OM43 clade cultures consisted of 16 unique groups of sequences that clustered with sequences obtained from coastal metagenomes and clone libraries (Figure 4). The majority of methylotroph isolates were >99% identical to one another at the SSU rRNA level with the greatest sequence divergence occurring between isolate NB0027 and NB0070 (96.6% SSU rRNA identities, Figure 4). The gammaproteobacteria sequences were more phylogenetically diverse, spanning several Alteromonadales clades and Vibrio (Supplementary Figure S4). The SSU rRNA sequence of NB0015 clustered with the SAR92 clade isolate HTCC2207 (SSU 97% identity). Four additional SAR92 clade SSU rRNA sequences were identified as mixed with OM43 clade cultures and were 98–100% similar to the NB0015 sequence. The remaining Alteromonadales sequences clustered with Teredinibacter turnerae (NB0011 and NB0077), Alteromonas macleodii (NB0094) and Psychrosphaera saromensis (NB0030). Vibrio sequences (NB0059, NB0072 and NB0080) were closely related to marine strains Vibrio chagasii, Vibrio lentus and Vibrio pomeroyi (Supplementary Figure S4).
Figure 4.
Phylogenetic relationships based on the SSU rRNA gene extracted from the whole-genome sequences of Nahant Bay isolates (NB00##) belonging to the OM43 clade of Betaproteobacteria. The grey box highlights strain NB0046 used in additional growth experiments (Figure 5). The tree was inferred from 1350 alignment positions from sequences curated in the ARB software (reference) using the RAxML (maximum likelihood) method. RAxML bootstrap values (1000 replicates) are shown (>80%) on nodes. Neighbor-joining inference method also produced bootstrap values >80% for these nodes. The scale bar indicates substitutions per site. Gammaproteobacteria and Alphaproteobacteria sequences (not shown) were used as outgroups.
Growth responses to differing media and carbon substrates
Two isolates representative of the most commonly identified bacterial groups, OM43 clade betaproteobacterium NB0046 and SAR92 clade gammaproteobacterium NB0015, were selected for further characterization.
Growth was undetectable for the OM43 betaproteobacterium NB0046 on rich media (Marine Broth 2216), methanol amended agar plates (Janvier et al. 1985) or TFF seawater amended with glucose and succinate (100 μM each). In non-carbon amended TFF oligotrophic seawater, NB0046 will typically reach cell concentrations of 1 × 105 cells per ml, although we have found this to be variable, with some cultures showing zero growth in the no carbon amendments, whereas others can reach almost 106 cells per ml. We also tested cultures in media prepared with UV-oxidized seawater. UV oxidation lowers the total amount of naturally occurring DOM in the seawater, but the oxidation process can also produce a broad suite of low molecular weight, highly oxidized organic compounds that could serve as substrates or inhibitors to our isolates (Moran and Zepp, 1997). However, in our experiments, UV oxidation of the seawater medium consistently reduced growth yields in non-carbon amended cultures to much lower cell concentrations (⩽103 cells per ml).
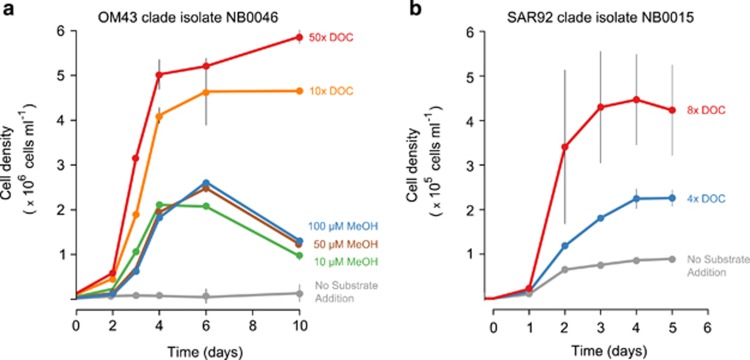
The addition of methanol to seawater media significantly increased OM43 isolate cell yields (Figure 5), with maximum cell concentrations achieved between 10–100 μM methanol; higher methanol concentrations (>1 mM) tended to inhibit, rather than increase cell yields, consistent with previous observations of OM43 clade cultures (Halsey et al., 2012). The highest cell yields were achieved with the HMW DOM additions, where isolate NB0046 reached >5 × 106 cells per ml in HMW DOM-containing media at 50x ambient DOC concentrations (~4 mM DOC, Figure 5). However, normalizing the observed cell yields to carbon added to the media indicates that methanol supports higher growth yields (2.5 × 107 to 2.1 × 108 cells per μmol of carbon) than HMW DOC treatments (1.6 × 106 to 6.4 × 106 cells per μmol of carbon).
Figure 5.
Growth profiles of select cultures amended with HMW DOM. Cells from either (a) OM43 clade betaproteobacterium isolate NB0046 or (b) SAR92 clade gammaprotebacterium isolate NB0015 were inoculated into TFF oligotrophic seawater amended with HMW DOM and inorganic nutrients (OM43 culture: 400 μM NH4, 30 μM PO4; SAR92 culture: 200 μM NH4, 200 μM NO3, 25 μM PO4). Additional methanol (MeOH) treatments were included for the OM43 clade isolate. Points are the mean cell concentrations of four (OM43) or three replicates (SAR92), with error bars representing the s.d. (where not visible, error bars are smaller than the symbols).
SAR92 clade gammaproteobacterium NB0015 grew in rich media (Marine Broth 2216), although only at dilute concentrations (1:10 and 1:100 strength). No growth was detected on full strength media, marine agar, seawater amended with methanol, or seawater amended with glucose and succinate (100 μM each). In non-carbon amended TFF oligotrophic seawater, NB0015 reached 1x105 cell per ml. Supplementing TFF oligotrophic seawater with HMW DOM (4x and 8x DOC) increased NB0015 growth to 2–5 × 105 cells per ml (Figure 5). The cell yield normalized to carbon added as HMW DOC was 7.1 × 105 cells per μmol of carbon, an order of magnitude lower than for the OM43 clade isolate NB0046. In contrast, cultures of NB0015 reached on average a maximum cell concentration of 1.5 × 106 cells per ml when grown in an artificial seawater medium (Kester et al., 1967) supplemented with a mix of simple carbon compounds (D-glucose (55 μM), succinate (85 μM), pyruvate (114 μM), glycerol (109 μM), N-acetyl D-glucosamine (45 μM) and ethanol (434 μM); Supplementary Figure S5), consistent with the previous isolation of SAR92 clade strains (Cho and Giovannoni, 2004).
Discussion
Dilution to extinction cultivation has proven a useful approach for obtaining highly abundant but difficult to cultivate bacteria. It enables separation of slower growing cells from faster, more easily cultured microorganisms, and it also allows the growth of cells in media that more closely mimics the native environment. Although several studies have supplemented natural seawater media with defined carbon substrates in efforts to improve the cell yields (Cho and Giovannoni, 2004) we applied the alternative approach of enriching seawater media with HMW DOM collected directly from the environment. We postulated that the native, complex mixture of marine HMW DOM could increase the probability of obtaining relevant DOM-degrading microorganisms. Such isolates could then be used as tools to help define the chemical characteristics of marine HMW DOM and the biological processes that degrade it. A similar tactic was successfully applied by Hutalle-Schmelzer et al. (2010) who enriched bacterioplankton dilution cultures with the humic fraction of lake-derived DOM.
In our study, dilution to extinction cultures enriched with HMW DOM increased culturing efficiency and yielded over 90 organisms with DOM-degrading capabilities. Our non-DOM-amended cultures had lower culturing efficiency (0.1%) than previous dilution culturing studies, which typically range from 3–25% (Connon and Giovannoni, 2002; Cho and Giovannoni, 2004; Song et al., 2009). The reduced culturing efficiency in our experiments may be explained by the deliberate use of oligotrophic Sargasso Sea surface water to prepare the media, which likely has low levels of vitamins (for example, <0.1 pM vitamin B12 (Menzel and Spaeth, 1962) and 20–25 pM thiamin (Carini et al., 2014)), inorganic nutrients (often below detection limits, 0.05 μmol per kg for nitrate+nitrite and 0.03 μmol per kg for phosphate; http://batsftp.bios.edu/BATS/bottle) and DOC concentrations (70 μM; as measured for this study) in the basal media than are typical of coastal seawater. Although this oligotrophic medium may have prohibited the enrichment of some coastal microorganisms, its low DOC and nutrient background (inorganic nitrogen and phosphorus were not added) provided greater sensitivity for detecting DOM addition effects without requiring extensive manipulation of the natural media. The fact that culturing efficiencies significantly increased in DOM-amended treatments suggested that DOM addition provides, at least partially, growth factor(s) or organic sources of nitrogen and phosphorus that were missing in oligotrophic seawater.
Establishing the purity of the culture collection is essential for linking the metabolism of DOM to specific taxa, genes and biochemical mechanisms. Determining culture purity, however, can be challenging, especially when a culture contains a secondary isolate at very low abundance (Shrestha et al., 2013). While SSU rRNA gene sequencing or screens such as restriction fragment length polymorphisms (RFLP) are most often used to identify cultures, they suffer from primer design and preferential amplification biases which can decrease sensitivity for detecting mixed cultures. The WGS screen used in this study does not suffer from oligonucleotide primer mismatch and excludes most other PCR biases thereby providing a more sensitive and universal method for determining culture purity. Although PCR-based SSU rRNA gene sequencing revealed no mixed cultures in our study, WGS sequencing showed that 6 of the 68 screened cultures were mixed as congruently determined by three different bioinformatic analyses: (i) number of contigs containing SSU rRNA genes, (ii) unassembled read's sequence similarity to assembled SSU rRNA sequences and (iii) unassembled read taxon binning based on sequence similarity search against NCBI's RefSeq database.
The ability of WGS sequencing to identify mixed cultures is directly dependent on the relative abundance of the secondary culture in relation to the sequencing depth, as well as on the phylogenetic relatedness of the co-isolates. In terms of resolving mixed cultures of closely related isolates, we found cultures with no other indication of being mixed had the majority of unassembled SSU rRNA reads at ⩾98% identity to the assembled SSU rRNA sequence (Supplementary Figure S2), suggesting that two different populations with <2% SSU rRNA divergence would be difficult to detect. We note that this would include many of the betaproteobacterial isolates obtained here. The metagenomic approach of identifying mixed cultures via taxonomic binning will be less sensitive than the SSU rRNA bioinformatic approaches to resolving closely related populations owing to database limitations. The breadth of genes examined using the metagenomic approach, however, should provide greater sensitivity for detecting low abundance but phylogenetically diverse secondary populations. For the betaproteobacterial genomes examined here, 5–10% of unassembled reads had significant matches to non-betaproteobacterial genomes likely because of factors such as short reads lengths, horizontal gene transfer events and limited reference genomes in the database. This suggests a lower limit for detecting co-isolates at around 10% total abundance, though we note differences in genome size will also affect this sensitivity. Alternative approaches to increase power for detecting closely related co-isolates at low abundance would likely require more sensitive bioinformatic analysis, such as the statistical frequency of genome single-nucleotide polymorphisms (Shrestha et al., 2013) focusing on less conserved genes such as those typically used in multi-locus sequence typing.
De novo assembly of the WGS data produced contigs containing full-length SSU and LSU sequences that were as accurate, and often had 100% identities, with overlapping Sanger reads. Phylogenetic placement of these WGS SSU rRNA sequences revealed the culture collection was dominated by members of the OM43 clade of betaproteobacteria.
The OM43 clade, initially described by Rappé et al. (1997), was first isolated from the Oregon coast by Connon and Giovannoni (2002). Their distribution is generally limited to the coastal zone, where they can compose 1–3% of the bacterioplankton community (Suzuki et al., 2004), though their abundance has been shown to significantly increase during phytoplankton blooms (Morris et al., 2006; Rich et al., 2011). Metatranscriptomic and metaproteomic data suggest they are active components of coastal bacterioplankton communities (Sowell et al., 2010; Gifford et al., 2014). The OM43 isolates obtained in this study were closely related to those previously characterized coastal strains and sequences (Figure 4). The majority of our isolates clustered most closely with the Hawaiian strain HIMB624 (Huggett et al., 2012) and related SSU rRNA phylotypes identified in diverse coastal locations. Three of our isolates formed an outgroup with the Oregon coastal isolate HTCC2181, as well as sequences obtained primarily from higher latitudes. Though closely related, the presence of SSU rRNA microdiversity within our isolate collection suggests the prevalence of OM43 clade members in the dilution to extinction cultures was not because of the enrichment of a single clonal population.
Genome sequences of OM43 betaproteobacteria are missing the E1 subunit of the α-ketoglutarate dehydrogenase complex of the TCA cycle, a trait thought to be indicative of an obligatory methylotrophic lifestyle (Halsey et al., 2012; Huggett et al., 2012). A preliminary genome analysis of NB0046 shows that it is also missing the E1 α-ketoglutarate dehydrogenase subunit. Furthermore, NB0046 shares similar characteristics with other OM43 lineage genomes (Giovannoni et al., 2008; Huggett et al., 2012), including a small genome size (<1400 genes), the presence of an alternative methanol dehydrogenase (XoxF) instead of the canonical Mxa or Mdh methanol dehydrogenase (Chistoserdova, 2011), genes for formaldehyde oxidation via tetrahydrofolate but not via tetrahydromethanopterin, and carbon assimilation through the RUMP cycle instead of the serine cycle. These observations suggest the methylotrophs isolated on HMW DOM have a similar 'obligate' methylotrophic lifestyle as other OM43 clade members, although a more detailed and thorough analysis of the 60 OM43 genomes sequenced here will be required to confirm this.
Given their reliance on single carbon substrates, the predominance of C1 compound specialists in our cultures amended with HMW DOM was unexpected. Our results, however, are predicated in a previous study by McCarren et al. (2010), who observed that HMW DOM added to oligotrophic ocean bacterioplankton microcosms increased the relative abundance and transcriptional activity of methylotrophs, particularly Methylophaga species belonging to the gammaproteobacteria. McCarren et al. (2010) postulated that HMW DOM polysaccharides containing a large fraction of methylated sugars (Quan and Repeta, 2007) were the source of single carbon compounds, including methanol, which provided the methylotrophs their growth substrate. We further propose that the prevalence of pure cultures of methylotrophs in our HMW DOM-enriched dilution to extinction samples is owing to their ability to cleave C1 groups from the methyl sugars and uronic acid methyl esters in the DOM polysaccharide. In addition, low levels of abiotic oxidation of the HMW DOM during collection and storage may possibly degrade the methylated sugars, releasing low molecular weight compounds that can be directly consumed by methylotrophs. In either case, the data indicated that HMW DOM can serve as a sole source of carbon and energy for these methylotrophs.
The order of magnitude difference in growth yields (cell yield normalized to carbon added) between methanol (6.8 × 107 to 5.3x108 cells per μmol of carbon) and HMW DOM (1.6 × 106 to 6.4 × 106 cells per μmol of carbon) suggests that growth on HMW DOM is less efficient than on methanol. Similar low growth yields (7.1 × 105 cells per μmol of carbon) were observed for SAR92 clade strain NB0015 in cultures amended with HMW DOM. Alternatively, the low growth yields observed may indicate that only a small fraction of HMW DOM may be available to a single organism with limited metabolic potential, like isolates NB0046 and NB0015, despite the total amount of DOC added to cultures. The efficiency at which DOC is converted into biomass, typically <20% in the open ocean (Carlson and Ducklow, 1996) has implications on the magnitude of carbon that cycles through bacterioplankton. Measurements of DOC drawdown and respiration, though not obtained in HMW DOM dose-response experiments, will be necessary to determine bacterial growth efficiencies on HMW DOC and to estimate the bacterial carbon demand that this organic carbon pool can support.
The second major taxonomic group found in the HMW DOM-enriched cultures was the gammaproteobacteria. This included isolates belonging to the Alteromonadales and Vibrionales. The Vibrio (NB0059, NB0072 and NB0080), Alteromonad (NB0094) and Pseudoalteromonad (NB0030) isolates did not exhibit a proportional increase in cell yields in response to added HMW DOM in dose-response experiments even though these isolates were initially obtained from the HMW DOM-enriched dilution to extinction cultures. Among the gammaproteobacteria only strains NB0011 and NB0077, and the SAR92 clade isolates exhibited increased growth in the presence of HMW DOM. The SAR92 clade isolate NB0015 exhibited the strongest growth response to HMW DOM although cell yields were low (2–4 × 105 cells per ml) compared with the OM43 clade isolates (>5 × 106 cells per ml). However, NB0015 growth yields reached >1x106 cells per ml when cultured in defined media with a mixture of simple carbon substrates indicating that this isolate may require a variety of carbon compounds and nutrients not available in HMW DOM to supplement its metabolism. The SAR92 clade was the most frequently recovered gammaproteobacteria group in the dilution to extinction cultures including four SAR92 clade-related SSU rRNA sequences found mixed with cultures of OM43 clade strains, more than any other taxa. Including these additional sequences, the SAR92 clade was the second most common group identified in our dilution to extinction cultivation experiment.
The growth of SAR92 clade strains in our HMW DOM-amended samples may partially be explained by its numerical abundance in coastal bacterioplankton assemblages (Stingl et al., 2007b) and their ease of recovery by dilution to extinction cultivation (Cho and Giovannoni, 2004). These factors alone, however, do not solely account for the prevalence of SAR92 in our cultures because SAR92 was only identified in the HMW DOM-enriched dilution to extinction cultures and not in the non-DOM-amended cultures. We postulate that the carbohydrate-rich component of HMW DOM in particular may have stimulated the growth of SAR92 bacteria. Some of the closest relatives of the SAR92 clade (90–93% SSU rRNA identity) include cultured isolates with carbohydrate degrading capabilities. For example, Microbulbifer hydrolyticus (Gonzalez et al., 1997), Saccharophagus degradans 2–40 (Andrykovitch and Marx, 1988; Weiner et al., 2008) and Simiduia agarivorans SA1 (Shieh et al., 2008) can breakdown several recalcitrant polysaccharides, including agar, alginate, cellulose or chitin. Another relative, Teredinibacter turnerae T7902, a bacterium associated with wood-boring bivalves, is capable of digesting cellulose (Distel et al., 2002). It is plausible that SAR92 clade strains may directly degrade HMW DOM polysaccharides in contrast to the OM43 clade methylotrophs which may utilize C1 compounds that decorate the polysaccharides. The availability of model laboratory organisms able to grow on HMW DOM now allows us to test these hypotheses. Future work chemically characterizing the HMW DOM before and after microbial degradation, as well as examining the transcriptional and proteomic responses of the isolates during DOM metabolism will help to determine the bonded nature of the carbon sustaining the cultures.
In summary, dilution to extinction cultures enriched with HMW DOM extended the power of the dilution cultivation technique by enriching for cells in media closely mimicking their native environment, and also by stimulating growth using naturally derived organic substrates. This approach is useful for obtaining model DOM-degrading isolates as both the carbon substrates and the organisms acting upon them are unknown. Using this technique we found organisms ranging from obligate methylotrophs to less fastidious heterotrophs that were able to grow using oligotrophic ocean HMW DOM as a substrate, suggesting there are multiple metabolic strategies involved in the degradation of HMW DOM. We note, however, that only a fraction of the total DOM added to our cultures was remineralized, suggesting that there were other growth-limiting factors, or potential requirement for syntrophic microbial partners to further degrade the DOM polymers (McCarren et al., 2010). Co-culture experiments will likely be essential to further elaborate the biological, physiological and biochemical details of consortial DOM degradation processes in the sea.
Materials and methods
Concentration of HMW DOM from seawater
DOM collection was conducted using the method described by Repeta and Aluwihare (2006). Briefly, the HMW fraction of DOM (>1 kDa) was concentrated from 24 000 l of surface seawater (15 m) collected 2 km offshore of the Island of Hawaii, Hawaii at the National Energy Laboratory Hawaii Authority (NELHA) in February 2006. The HMW DOM-concentrated seawater was then filtered through a 30 kDa ultrafiltration membrane to remove cell debris and viral particles, diafiltered to remove salts and freeze-dried. A total of 9.5 g of freeze-dried HMW DOM was obtained that was 32% carbon, 2.7% nitrogen with a C/N ratio of 13.9 representing 18% of the DOC in the original raw seawater.
Molecular characterization of HMW DOM
DOM was characterized by 1H nuclear magnetic resonance spectroscopy to determine major constituents. Spectra were recorded on a Bruker Avance DPX 400 MHz spectrometer fitted with an inverse broadband 5 mm probe. Approximately 1–2 mg of sample was dissolved in 100% D2O. Chemical shifts were referenced to residual HOD at δ=4.80 p.p.m. Acid hydrolysis (2.8 M trifluoroacetic acid, heated at 120°C for 4 hours under nitrogen) of HMW DOM yields a suite of neutral sugars (arabinose, fucose, galactose, glucose, mannose, rhamnose, xylose) that were separated by reverse-phase high-pressure liquid chromatography (Ascentis C-18 column, Sigma-Aldrich, St Louis, MO, USA; 150x1 mm, 3 μm, eluted at 120 μl min−1 with 10/90 (v/v) acetonitrile/water) and quantified at 307 nm as the their aminobenzoate ethyl ester derivatives (Baik and Cheong, 2007). Our chromatographic analysis does not separate xylose and arabinose which are reported as the sum xylose+arabinose. Analyses of unhyrdolyzed HMW DOM does not yield any aminobenzoate ethyl ester products that interfere with our monosaccharide analyses.
DOC measurements
Samples for DOC concentration analysis were transferred into combusted glass vials and acidified with 25% phosphoric acid solution before sealing with acid-washed Teflon septa. Sample concentrations were determined using the high-temperature combustion method on a Shimadzu TOC-VCSH with platinized alumina catalyst alongside potassium hydrogen phthalate standards and consensus reference materials provided by the DOC-CRM program.
Preparation of seawater media
The water for extinction culturing and subsequent cultivation media was collected off Bermuda (33.2497°N 65.7103°W) during the KN207-01 cruise, aboard the R/V Knorr on 03 May 2012 via the ship's flow-through system with a 0.2 μm filter. After collection and storage, the Sargasso seawater was sterilized by TFF as described by Becker et al. (2007), with minor modifications. The seawater was pre-filtered through a 142-mm diameter, 0.2-μm pore-size Supor-membrane (Pall, Ann Arbor, MI, USA) and a 0.1-μm pore-size Supor capsule filter (Pall) and collected in an acid-cleaned polycarbonate carboy. Pre-filtered water circulated through a Pellicon 2 Mini tangential-flow ultrafiltration system (Millipore, Billerica, MA, USA) consisting of a 30 kDa cassette of regenerated cellulose (Millipore). The tangential-flow filtrate was collected in autoclaved, acid-cleaned polycarbonate carboys and stored at 4°C. Seawater remained sterile at room temperature after the addition of nutrients and sterile rich media. The final TFF seawater medium had a total organic carbon concentration of 70 μM.
Extinction culturing with HMW DOM additions
The inoculum was collected in Nahant Bay, MA (42°25.195 N, 70°54.463 W) on 23 August 2012. Bacterioplankton cell densities were obtained by direct-cell counts of DNA-stained cells using SYBR green I (Molecular Probes, Life Technologies, Carlsbad, CA, USA) ~1 hour after sample collection. The inoculum was diluted into sterile seawater to an estimated cell density of 3 cells per ml in a final volume of 5 l. The dilute inoculum was split into three 1-l aliquots, one left untreated (70 μM DOC) and two were supplemented with HMW DOM to 210 and 630 μM HMW DOC to obtain media with a final 280 (4x) and 700 (10x) μM DOC, respectively. One-ml aliquots from each treatment were then distributed into non-tissue culture treated polystyrene 48-well plates (BD Biosciences, Franklin Lakes, NJ, USA). Each experimental treatment consisted of 840 potential extinction cultures in twenty 48-well plates. Every plate included six negative controls consisting of wells filled with the corresponding treatment media made up with sterile seawater. Plates were covered in foil to reduce evaporation and incubated at 24 °C in the dark for 4–5 weeks. In order to ensure that potential extinction cultures remained viable and capable of growth in the corresponding cultivation media, after the 4–5-week incubation the samples were re-diluted 10 000-fold into new plates containing fresh TFF seawater and the appropriate DOM concentrations. These were then incubated for 6–7 weeks under the same conditions as before.
Detection of culture growth
Extinction cultures were detected using a guava easyCyte 8HT flow cytometry system (Millipore) by transferring 50 μl from each cultivation plate well to a 96-well microtitter plate containing 10 μl of 500-fold diluted SYBR green I and 140 μl of sterile seawater. Samples were stained for at least 30 min before flow cytometry. Each well was analyzed for 15 s at a flow rate of 0.59 μl s−1 with a blue laser (488 nm excitation) to detect green fluorescence. Wells were scored positive for cell growth when the concentration was ⩾105 cells per ml.
HMW DOM dose response screen of the culture collection
Cultures scoring positive for growth were examined for increased cell yields under different HMW DOM concentrations. To limit cell growth to consumption of HMW DOM added, background DOC in the TFF seawater was photo-oxidized via exposure to high-intensity UV light for 4 h, reducing DOC concentrations by 30–40%. During this process, approximately an eighth of the volume was lost owing to evaporation, and this was replenished with ultrapure water. The UV-oxidized TFF seawater (25 μM DOC) was then supplemented with 210, 1330 and 6930 μM DOC using HMW DOM to obtain ~4x, ~20x or ~100x DOC media, respectively, in similitude to media prepared for extinction culturing. Non DOM-amended UV-oxidized seawater served as control. No inorganic nutrients or vitamins were added to the media. Each treatment media was distributed into 1-ml aliquots in 48-well cultivation plates. Wells were inoculated with 10-μl sub-samples of the dilution to extinction cultures and incubated at 24°C in the dark. Subsamples were taken every 5–7 days for cell-density enumeration using the SYBR green I flow cytometry assay described above.
SSU rRNA sequencing
A total of 400–800 μl from the 93 cultures scoring positive for growth were transferred to a 0.2 μm Supor-membrane 96-well filter plate (Pall) and vacuum filtered. The cells were resuspended from the filter with two separate aliquots of 125 μl of lysis buffer (40 mM EDTA, 50 mM Tris at pH 8.3, 0.73 M sucrose 1.15 mg ml−1 lysozyme (Sigma-Aldrich), 200 μg ml−1 RNase (Qiagen, Hilden, Germany) and transferred to a 96 deep-well plate, which was then placed at −80°C. After freezing, the plate was thawed, Proteinase K and SDS were added to final concentrations of 0.65 mg ml−1 and 10% SDS, respectively, and allowed to incubate for 2 h at 55°C. The lysate was then transferred to new 2-ml deep-well plate and DNA extracted using a DNeasy 96-well Blood and Tissue kit (Qiagen).
The SSU rRNA gene was PCR amplified using universal Eubacterial primers pA Escherichia coli 8-28 F (5′-AGA GTT TGA TCC TGG CTC AG-3′) and E. coli 1510-1492R (5′- GGT TAC CTT GTT ACG ACT T -3') in 50-μl reactions consisting of 1 μM each forward and reverse primer, 1 μl of FailSafe Enzyme Mix (Epicentre, Madison, WI, USA), 25 μl of 2x FailSafe PCR PreMix E (Epicentre) water and 20-μl DNA template. After amplification, the PCR amplicons were run on 1% agarose gel and bands running at ~1500–1600 nt were excised and PCR-purified with a 96-well gel extraction kit (Qiagen) followed by an additional purification using the QIAquick 96-well PCR purification kit (Qiagen) and eluted in 80 μl elution buffer. Sanger sequencing was conducted following the BigDye v3.1 sequencing protocol (Applied Biosystems, Foster City, CA, USA) on a ABI 3730 DNA analyzer (Applied Biosystems) using the SSU rRNA universal primers described above (pA E. coli 8-28, E. coli 1510-1492) and with bacterial primers 519F 5′-CAGCMGCCGCGGTAATWC-3′ and 800R 5′-TACCAGGGTATCTAATCC-3′.
Whole-genome sequencing
Seventy cultures were selected for whole-genome sequencing based on their growth yields and response to HMW DOM substrate addition. To obtain sufficient amounts of DNA for the NexterXT library preparation, 10-μl of culture was inoculated into acid-cleaned polycarbonate bottles containing 200 ml TFF seawater amended with HMW DOM to 3x DOC. After incubating at 22°C in the dark for 4–5 weeks, the cells were collected on 0.1 μm pore-size, 25 mm diameter Durapore membranes (Millipore) using Swinnex filter-holders (Millipore) and peristaltic pumping. Filters were stored frozen (−80°C) in 600 μl of tissue lysis buffer (Qiagen). DNA was extracted from thawed filters using a DNeasy Blood and Tissue Kit (Qiagen) following the manufacturer's instructions. DNA samples were prepared for sequencing and barcoded using the Nextera XT DNA 96-sample preparation kit (Illumina, San Diego, CA, USA), and sequenced with one 250 × 250 nt paired-end MiSeq run (Illumina). Sequences are deposited in the NCBI sequence read archive under study SRP045600.
Bioinformatic and Phylogenetic analysis
FastQ files from the MiSeq run were imported into the CLC Genomics Workbench (CLC bio, Aarhus, Denmark). Paired reads were joined and then assembled into contigs using CLC's de novo assembler with automatic word and bubble sizes, a minimum contig length of 200, insertion and deletion costs set to 3, mismatch cost set to 2, length fraction set to 0.5 and the similarity fraction set to 0.8. Only contigs ⩾1000 nt in length were further examined. Contigs containing small and large subunit rRNA sequences were identified and annotated by a BLASTn sequence similarity search against the SILVA database (http://www.arb-silva.de, version 111).
To ensure that low abundance reads with a divergent SSU rRNA were not missed during the assembly process the unprocessed miSeq reads were trimmed and paired-end joined using trimomatic (http://www.usadellab.org/cms/?page=trimmomatic) and PandaSeq (https://github.com/neufeld/pandaseq/wiki/PANDAseq-Assembler), respectively. The trimmed and joined reads were compared with the assembled SSU rRNA for each sample via a BLASTn sequence similarity search, with hits having an e-value<0.001, a read length>50 nt and 45% identity over the BLAST alignment considered significant. The unassembled, but trimmed and joined reads, were also taxonomically binned based on a LAST (Frith et al., 2010; Kielbasa et al., 2011) sequence similarity search against NCBI's REfSeq database (version 64) with a score penalty for frameshift of 500 and an initial match multiplicity of 10. Top hits with a score ⩾40 and alignment length ⩾75 nt were considered significant and used for the taxonomic annotation.
Partial SSU rRNA gene sequences obtained via Sanger sequencing were quality-trimmed, manually inspected and assembled with Sequencher version 5.1 (GeneCodes, Ann Arbor, MI, USA). SSU rRNA gene sequences from Sanger or WGS were aligned using the SILVA incremental aligner online tool (Pruesse et al., 2012) and curated in ARB (Ludwig et al., 2004) using the All-Species Living Tree project release 115 (Munoz and Yarza et al., 2011). Full-length SSU rRNA gene sequences obtained from WGS were used to build phylogenetic trees and were deposited in NCBI GenBank under accession numbers KP770034 through KP770106. For the betaproteobacteria OM43 clade strains a tree was inferred from 1350 alignment positions using the RAxML maximum likelihood method (Stamatakis, 2014) and neighbor joining (Saitou and Nei, 1987). For gammaproteobacteria isolates, a tree was constructed using 1293 unambiguous alignment positions using RAxML. RAxML 8.0.24 was implemented on Cipres Science Gateway (Miller et al., 2010) and neighbor joining was implemented on MEGA5 (Tamura et al., 2004, 2011). RAxML trees were curated in EMBL's interactive Tree of Life tool (Letunic and Bork, 2006, 2011).
Acknowledgments
We thank Jamie Becker for DOC concentration analyses, technical assistance and providing advice and feedback. We are also indebted to Tsultrim Palden for assistance in DNA extraction and sequencing. This research was funded by the Gordon and Betty Moore Foundation through Grant GBMF3298 to DJR and EFD, GBMF #3777 (to EFD) and NSF Science and Technology Center grant EF0424599 (to EFD), and the Simons Foundation (to EFD and DR). This work is a contribution of the Center for Microbial Oceanography: Research and Education (C-MORE) and the Simons Collaboration on Ocean Processes and Ecology (SCOPE).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Aluwihare LI, Repeta DJ, Chen RF. (1997). A major biopolymeric comoponent of dissolved organic carbon in surface sea water. Nature 387: 166–169. [Google Scholar]
- Aluwihare LI, Repeta DJ. (1999). A comparison of the chemical characteristics of oceanic DOM and extracellular DOM produced by marine algae. Mar Ecol Prog Ser 186: 105–117. [Google Scholar]
- Aluwihare LI, Repeta DJ, Pantoja S, Johnson CG. (2005). Two chemically distinct pools of organic nitrogen accumulate in the ocean. Science 308: 1007–1010. [DOI] [PubMed] [Google Scholar]
- Andrykovitch G, Marx I. (1988). Isolation of a new polysaccharide-digesting bacterium from a salt marsh. Appl Environ Microbiol 54: 1061–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baik YS, Cheong WJ. (2007). Determination of molecular weight distribution and average molecular weights of oligosaccharides by HPLC with a common C18 phase and a mobile phase with high water content. Bull Korean Chem Soc 28: 847–850. [Google Scholar]
- Becker JW, Brandon ML, Rappé MS, Hurst CJ, Crawford RL, Garland JL et al. (2007). Cultivating microorganisms from dilute aquatic environments: melding traditional methodology with new cultivation techniques and molecular methods. Manual of Environmental Microbiology 3: 399–406. [Google Scholar]
- Beier S, Rivers AR, Moran MA, Obernosterer I. (2014). The transcriptional response of prokaryotes to phytoplankton-derived dissolved organic matter in seawater. Environ microbiole doi:10.1111/1462-2920.12434. [DOI] [PubMed]
- Benner R, Pakulski JD, McCarth M, Hedges JI, Hatcher PG. (1992). Bulk chemical characteristics of dissolved organic matter in the ocean. Science 255: 1561–1564. [DOI] [PubMed] [Google Scholar]
- Benner R. (2002). Chemical composition and reactivity. In: Hansel DA, Carlson CA (eds), Biogeochemistry of marine dissolved organic matter. Academic Press: San Diego, CA, USA, pp 59–90. [Google Scholar]
- Button DK, Schut F, Quang P, Martin R, Robertson BR. (1993). Viability and isolation of marine bacteria by dilution culture: theory, procedures, and initial results. Appl Environl Microbiol 59: 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carini P, Campbell EO, Morré J, Sañudo-Wilhelmy SA, Thrash JC, Bennet SE et al. (2014). Discovery of a SAR11 growth requirement for thiamin's pyrimidine precursor and its distribution in the Sargasso Sea. ISME J 8: 1727–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CA. (2002) Production and Removal Processes. In: Hansel DA, Carlson CA (eds), Biogeochemistry of marine dissolved organic matter. Academic Press: : San Diego, USA, pp 91–151. [Google Scholar]
- Carlson CA, Ducklow HW. (1996). Growth of bacterioplankton and consumption of dissolved organic carbon in the Sargasso Sea. Aquat Microb Ecol 10: 69–85. [Google Scholar]
- Carlson CA, Giovannoni SJ, Hansell DA, Goldberg SJ, Parsons R, Vergin K. (2004). Interactions among dissolved organic carbon, microbial processes, and community structure in the mesopelagic zone of the northwestern Sargasso Sea. Limnol Oceanogr 49: 1073–1083. [Google Scholar]
- Chistoserdova L. (2011). Modularity of methylotrophy, revisited. Environ Microbiol 13: 2603–2622. [DOI] [PubMed] [Google Scholar]
- Cho J, Giovannoni SJ. (2004). Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl Environ Microbiol 70: 432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LL, Ingall ED, Benner R. (1998). Marine phosphorus is selectively remineralized. Nature 393: 426. [Google Scholar]
- Connon S, Giovannoni SJ. (2002). High-throughput methods for culturing microorganisms in very-low-nutrient media yield diverse new marine isolates. Appl Environ Microbiol 68: 3878–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Distel DL, Morrill W, MacLaren-Toussaint N, Franks D, Waterbury J. (2002). Teredinibacter turnerae gen. nov., sp. nov., a dinitrogen-fixing, cellulolytic, endosymbiotic gamma-proteobacterium isolated from the gills of wood-boring molluscs (Bivalvia: Teredinidae). Int J Syst Evol Microbiol 52: 2261–2269. [DOI] [PubMed] [Google Scholar]
- Eloe EA, Malfatti F, Gutierrez J, Hardy K, Schmidt WE, Pogliano K et al. (2011). Isolation and characterization of a psychropiezophilic alphaproteobacterium. Appl Environ Microbiol 77: 8145–8153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Wan R, Horton P. (2010). Incorporating sequence quality data into alignment improves DNA read mapping. Nucleic Acids Res 38: e100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford SM, Sharma S, Moran MA. (2014). Linking activity and function to ecosystem dynamics in a coastal bacterioplankton community. Front Microbiol 5: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Hayakawa DH, Tripp HJ, Stingl U, Givan SA, Cho JC et al. (2008). The small genome of an abundant coastal ocean methylotroph. Env Microb 10: 1771–1782. [DOI] [PubMed] [Google Scholar]
- Gonzalez JM, Mayer F, Moran MA, Hodson RE, Whitman WB. (1997). Microbulbifer hydrolyticus gen. nov., sp., nov., and Marinobacterium georgiense gen. nov., sp. nov., two marine bacteria from a lignin-rich pulp mill waste enrichment community. Int J Syst Bacteriol 47: 369–376. [DOI] [PubMed] [Google Scholar]
- Grote J, Thrash C, Huggett MJ, Landry Z, Carini P, Giovannoni S et al. (2012). Streamlining and core genome conservation among highly divergent members of the SAR11 clade. mBio 3: e00252–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halsey KH, Carter AE, Giovannoni SJ. (2012). Synergistic metabolism of a broad range of C1 compounds in the marine methylotrophic bacterium HTCC2181. Environ Microbiol 14: 630–640. [DOI] [PubMed] [Google Scholar]
- Huggett MJ, Hayakawa DH, Rappé MS. (2012). Genome sequence of strain HIMB624, a cultured representative from the OM43 clade of marine Betaproteobacteria. Stand Genomic Sci 6: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutalle-Schmelzer KML, Zwirnmann E, Krüger A, Grossart HP. (2010). Enrichment and cultivation of pelagic bacteria from a humic lake using phenol and humic matter additions. FEMS Microbiol Ecol 72: 58–73. [DOI] [PubMed] [Google Scholar]
- Janvier M, Frehel C, Grimont F, Gasser F. (1985). Methylophaga marina gen. nov., sp. nov. and Methylophaga thalassica sp. nov., marine methylotrophs. Int J Syst Bacteriol 35: 131–139. [Google Scholar]
- Kester DR, Duedall IW, Connors DN, Pytkowicz RM. (1967). Preparation of artificial seawater. Limnol Oceanogr 12: 176–179. [Google Scholar]
- Kielbasa SM, Wan R, Sato K, Horton P, Frith MC. (2011). Adaptive seeds tame genomic sequence comparison. Genome Res 21: 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolowith LC, Ingall ED, Benner R. (2001). Composition and cycling of marine organic phosphorus. Limnol Oceanogr 46: 309–320. [Google Scholar]
- Landa M, Cottrell MT, Kirchman DL, Kaiser K, Medeiros PM, Tremblay L et al. (2013). Phylogenetic and structural response of heterotrophic bacteria to dissolved organic matter of different chemical composition in a continuous culture study. Environ Microbiol 16: 1668–1681. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. (2006). Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23: 127–128. [DOI] [PubMed] [Google Scholar]
- Letunic I, Bork P. (2011). Interactive Tree of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39: W475–W478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Ritcher L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malmstrom RR, Kiene RP, Cottrell MT, Kirchman DL. (2004). Contribution of SAR11 bacteria to dissolved dimethylsulfoniopropionate and amino acid uptake in the North Atlantic ocean. Appl Environ Micorbiol 70: 23–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall T, Morris RM. (2013). Isolation of an aerobic sulfur oxidizer from the SUP05/Arctic96BD-19 clade. ISME J 7: 452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarren J, Becker JW, Repeta DJ, Shi Y, Young CR, Malmstrom RR et al. (2010). Microbial community transcriptomes reveal microbes and metabolic pathways associated with dissolved organic matter turnover in the sea. Proc Natl Acad Sci USA 107: 16420–16427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy M, Pratum T, Hedges J, Benner R. (1997). Chemical composition of dissolved organic nitrogen in the ocean. Nature 390: 150–154. [Google Scholar]
- Menzel DW, Spaeth JP. (1962). Occurrence of vitamin B12 in the Sargasso Sea. Limnol Oceongr 7: 151–158. [Google Scholar]
- Meon B, Kirchman DL. (2001). Dynamics and molecular composition of dissolved organic material during experimental phytoplankton blooms. Mar Chem 75: 185–199. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November 2010, New Orleans, LA, USA, pp 1–8.
- Moran MA, Zepp RG. (1997). Invited Review Role of photoreactions in the formation of biologically labile compounds from dissolved organic matter. Limonol Oceanogr 42: 1307–1316. [Google Scholar]
- Morris RM, Longnecker K, Giovannoni SJ. (2006). Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ Microbiol 8: 1361–1370. [DOI] [PubMed] [Google Scholar]
- Munoz R, Yarza P, Ludwig W, Euzéby J, Amann R, Schleifer KH et al. (2011). Release LTPs104 of the all-species living tree. Syst Appl Microbiol 34: 169–170. [DOI] [PubMed] [Google Scholar]
- Nelson CE, Carlson CA. (2012). Tracking differential incorporation of dissolved organic carbon types among diverse lineages of Sargasso Sea bacterioplankton. Environ Microbiol 14: 1500–1516. [DOI] [PubMed] [Google Scholar]
- Poretsky RS, Sun S, Mou X, Moran MA. (2010). Transporter genes expressed by coastal bacterioplankton in response to dissolved organic carbon. Environ Microbiol 12: 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. (2012) SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. [DOI] [PMC free article] [PubMed]
- Quan TM, Repeta DJ. (2007). Periodate oxidation of marine high molecular weight dissolved organic matter: evidence for a major contribution from 6-deoxy-and methyl sugars. Mar Chem 105: 183–193. [Google Scholar]
- Rappé M, Kemp PF, Giovannoni SJ. (1997). Phylogenetic diversity of marine coastal picoplankton 16S rRNA genes cloned from the continental shelf off Cape Hatteras, North Carolina. Limnol Oceanogr 42: 811–826. [Google Scholar]
- Rappé MS, Connon SA, Vergin KL, Giovannoni SJ. (2002). Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature 418: 630–633. [DOI] [PubMed] [Google Scholar]
- Redfield AC, Ketchum BH, Richards FA. (1963) The influence of organisms on the composition of seawater. In: Hill MN (ed), Comparative and descriptive oceanography. Wiley: : New York, NY, USA, pp 26–77. [Google Scholar]
- Repeta DJ, Aluwihare LI. (2006). Radiocarbon analysis of neutral sugars in high-molecular-weight dissolved organic carbon: Implications for organic carbon cycling. Limnol Oceanogr 51: 1045–1053. [Google Scholar]
- Rich VI, Pham VD, Eppley J, Shi Y, DeLong EF. (2011). Time-series analyses of Monterey Bay coastal microbial picoplankton using a ‘genome proxy'microarray. Environ Microbiol 13: 116–134. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Sarmento H, Gasol JM. (2012). Use of phytoplankton-derived dissolved organic carbon by different types of bacterioplankton. Environ Microbiol 14: 2348–2360. [DOI] [PubMed] [Google Scholar]
- Sarmento H, Romera-Castillo C, Lindh M, Pinhassi J, Sala MM, Gasol JM et al. (2013). Phytoplankton species-specific release of dissolved free amino acids and their selective consumption by bacteria. Limnol Oceanogr 58: 1123–1135. [Google Scholar]
- Schwalback MS, Tripp HJ, Steindler L, Smith DP, Giovannoni SJ. (2010). The presence of glycolysis operon in SAR11 genomes is positively correlated with ocean productivity. Environ Microbiol 12: 490–500. [DOI] [PubMed] [Google Scholar]
- Sharma AK, Becker JW, Ottesen EA, Bryant JA, Duhamel S, Karl DM et al. (2013). Distinct dissolved organic matter sources induce rapid transcriptional responses in coexisting populations of Prochlorococcus, Pelagibacter and the OM60 clade. Environ Microbiol 16: 2815–2830. [DOI] [PubMed] [Google Scholar]
- Shieh WY, Liu TY, Lin SY, Jean WD, Chen JS. (2008). Simiduia agarivorans gen. nov., sp. nov., a marine, agarolytic bacterium isolated from shallow coastal water from Keelung, Taiwan. Int J Syst Evol Microbiol 58: 895–900. [DOI] [PubMed] [Google Scholar]
- Shrestha PM, Nevin KP, Shrestha M, Lovley DR. (2013). When is a microbial culture “pure”? Persistent cryptic contaminant escapes detection even with deep genome sequencing. mBio 4: e00591–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Oh HM, Cho JC. (2009). Improved culturability of SAR11 strains in dilution-to-extinction culturing from the East Sea, West Pacific Ocean. FEMS Microbial Lett 295: 141–147. [DOI] [PubMed] [Google Scholar]
- Sowell SM, Abraham PE, Shah M, Verberkmoes NC, Smith DP, Barofsky DF et al. (2010). Environmental proteomics of microbial plankton in a highly productive coastal upwelling system. ISME J 5: 856–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. (2014). RAxML Version 8: A tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl U, Cho JC, Foo W, Vergin KL, Lanoil B, Giovannoni SJ. (2008). Dilution-to-extinction culturing of psychrotolerant planktonic bacteria from permanently ice-covered lakes in the McMurdo Dry Valleys, Antarctica. Microb Ecol 55: 395–405. [DOI] [PubMed] [Google Scholar]
- Stingl U, Desiderio RA, Cho JC, Vergin KL, Giovannoni SJ. (2007. a). The SAR92 clade: an abundant coastal clade of culturable marine bacteria possessing proteorhodopsin. Appl Environ Microbiol 73: 2290–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingl U, Tripp HJ, Giovannoni SJ. (2007. b). Improvements of high-throughput culturing yielded novel SAR11 strains and other abundant marine bacteria from the Oregon coast and the Bermuda Atlantic Time Series study site. ISME J 1: 361–371. [DOI] [PubMed] [Google Scholar]
- Sun J, Steindler L, Thrash JC, Halsey KH, Smith DP, Carter AE et al. (2011). One carbon metabolism in SAR11 pelagic marine bacteria. Plos One 6: e23973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki MT, Preston CM, Béjà O, De La Torre JR, Steward GF, DeLong EF. (2004). Phylogenetic screening of ribosomal RNA gene-containing clones in bacterial artificial chromosome (BAC) libraries from different depths in Monterey Bay. Microb Ecol 48: 473–488. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M, Kumar S. (2004). Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA 101: 11030–11035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner RM, Taylor LE II, Henrissat B, Hauser L, Land M, Coutinho PM et al. (2008). Complete genome sequence of the complex carbohydrate-degrading marine bacterium, Saccharophagus degradans strain 2-40T. PLoS Genet 4: e1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams PM, Druffel ERM. (1987). Radiocarbon in dissolved organic matter in the central North Pacific Ocean. Nature 330: 246–248. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.