Abstract
Hypersaline meromictic lakes are extreme environments in which water stratification is associated with powerful physicochemical gradients and high salt concentrations. Furthermore, their physical stability coupled with vertical water column partitioning makes them important research model systems in microbial niche differentiation and biogeochemical cycling. Here, we compare the prokaryotic assemblages from Ursu and Fara Fund hypersaline meromictic lakes (Transylvanian Basin, Romania) in relation to their limnological factors and infer their role in elemental cycling by matching taxa to known taxon-specific biogeochemical functions. To assess the composition and structure of prokaryotic communities and the environmental factors that structure them, deep-coverage small subunit (SSU) ribosomal RNA (rDNA) amplicon sequencing, community domain-specific quantitative PCR and physicochemical analyses were performed on samples collected along depth profiles. The analyses showed that the lakes harbored multiple and diverse prokaryotic communities whose distribution mirrored the water stratification patterns. Ursu Lake was found to be dominated by Bacteria and to have a greater prokaryotic diversity than Fara Fund Lake that harbored an increased cell density and was populated mostly by Archaea within oxic strata. In spite of their contrasting diversity, the microbial populations indigenous to each lake pointed to similar physiological functions within carbon degradation and sulfate reduction. Furthermore, the taxonomy results coupled with methane detection and its stable C isotope composition indicated the presence of a yet-undescribed methanogenic group in the lakes' hypersaline monimolimnion. In addition, ultrasmall uncultivated archaeal lineages were detected in the chemocline of Fara Fund Lake, where the recently proposed Nanohaloarchaeota phylum was found to thrive.
Introduction
Meromictic (permanently stratified) lakes are distinct hydrological environments characterized by a persistent physicochemical stratification that circumvents deep water recirculation. As a consequence, they commonly comprise an upper stratum where mixing occurs (mixolimnion), a stagnant lower one (monimolimnion) and an interposing boundary layer (chemocline) (Boehrer and Schultze, 2009). These lakes are important model systems for aquatic biology research, because their constant vertical stratification, steep chemical gradients and the presence of oxygen-deprived zones favor niche partitioning among prokaryotic populations, enabling the study of vertical zonations in microbially-mediated biogeochemical cycling (Lauro et al., 2011; Lopes et al., 2011; La Cono et al., 2013).
Ursu and Fara Fund Lakes are 2 of the 41 permanent saline water bodies occurring in the Transylvanian Basin (Romania) that were formed between the eighteenth and nineteenth centuries (Alexe, 2010) on the salt deposits produced by the Paratethys Sea evaporation (during the Badenian salinity crisis, ca. 14 Myr ago). These two temperate lakes are unique among the other Romanian salt lakes because of their heliothermy (that is, trapping of solar radiation by a ‘saline lens'), size (that is, Ursu Lake is one of the largest heliothermal salt lakes in Europe), conservation status (that is, Fara Fund Lake is a protected area) and lake processes (that is, Ursu Lake is a natural lake whereas Fara Fund Lake is an artificial one) (Alexe, 2010; Máthé et al., 2014). In spite of their different genesis, they belong to the same type of meromictic lakes, where the water column overturn is hampered by strong salinity gradients. Although studies investigating the microbiota and substance turnover in meromictic lakes are available (Biderre-Petit et al., 2011; Habicht et al., 2011; Hamilton et al., 2014), most of the ones concerning the hypersaline water bodies were performed on Arctic (Pouliot et al., 2009; Comeau et al., 2012) or Antarctic (Bowman et al., 2000; Lauro et al., 2011) lakes. Although information on temperate neutral hypersaline meromictic lakes microbiota are scarce, previous investigations on Ursu Lake prokaryotes (using community-level physiological profiling, culture-dependent and PCR–denaturing gradient gel electrophoresis methods) have shown the presence of active and diverse communities (Cristea et al., 2014; Máthé et al., 2014); yet, a detailed molecular diversity study is not available to date. The microbial diversity of Fara Fund Lake has not been previously investigated, and hence the lake could constitute a valuable site for monitoring long-term dynamics of microbial communities that are not under the impact of human activities.
The purpose of this study was to compare the prokaryotic diversity and link it with limnological data in two distinct meromictic hypersaline lakes that differ in their hydric regimen, physicochemical characteristics, genesis and conservation status. This would enable novel inferences regarding the microbial ecology and geochemical cycles in bioenergetically constrained stratified environments. To achieve this aim, the vertical distribution and community structure of the prokaryotes thriving in the two lakes was investigated by combining deep-coverage small subunit (SSU) ribosomal RNA (rDNA) amplicon sequencing, community domain-specific quantitative PCR, chemical analyses and bioinformatics. Concentration and stable C isotope composition of dissolved methane and heavier alkanes were also analyzed for a preliminary assessment of microbial or other sources of hydrocarbons.
Materials and methods
Site description and sampling
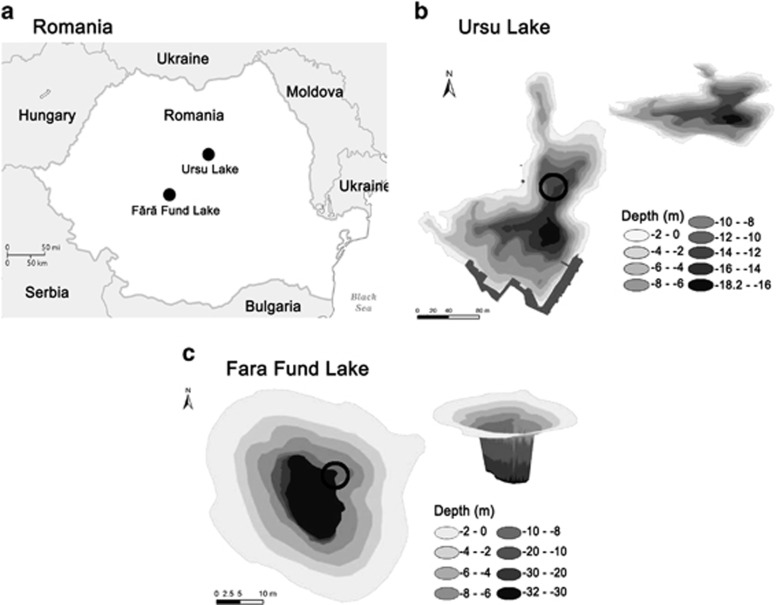
Ursu Lake (46°36'15 N; 25°05'09 E) is a natural meromictic lake located at 505 m elevation in the eastern part of the Transylvanian Basin. Having a surface of 41 270 m2 and a maximum depth of 18.2 m (Figure 1b), it was formed in a depression caused by an intense process of salt dissolution between 1875 and 1880 (Alexe, 2010; Máthé et al., 2014). Currently, it is exploited for recreational purposes during late-May to mid-August. Fara Fund (45°52'34 N; 24°04'03 E) is an artificial meromictic pit lake situated at 383 m elevation in the southern area of the Transylvanian Basin (Figure 1a). Having a relatively small surface (∼1700 m2) and a maximum depth of 32 m (Figure 1b), this athalassic (sensu, Hammer, 1986) lake was formed in 1775 on the area of a late medieval bell-shaped salt mine that was shut down at the end of the seventeenth century. Presently, it is declared as a protected area, as a consequence of its strong heliothermy (Alexe, 2010).
Figure 1.
Ursu Lake and Fara Fund Lake sampling sites. (a) Map showing the location of the lakes within Transylvanian Basin (Romania). The symbol size is not proportional with the sizes of the lakes. (b) Topo-bathymetric profile of Ursu and (c) Fara Fund lakes' basins. The circles indicate the sampling points.
The vertical sampling was carried out in October 2013 using an inflatable boat positioned near the center of the lakes (Figures 1b and c). In situ measurements (for example, temperature, pH, dissolved oxygen, conductivity and redox potential) were performed using a portable water multiparameter system HI 9828 (Hanna Instruments, Woonsocket, RI, USA). The sampling points were chosen in correspondence with the stratification of physicochemical parameters. From each lake, five water samples (0.5, 2.5, 3.5, 9 and 11 m depth in the Ursu Lake; 0.5, 2, 3, 11 and 13 m depth in the Fara Fund Lake) were collected using a submersible 12 V electric pump, with a flow rate of 166 cm3 s−1 and stored in sterile 2-l polypropylene bottles. The samples were transported to the laboratory, where they were immediately processed. The bottom water samples (9 and 11 m in Ursu Lake; 11 and 13 m in Fara Fund Lake) were used for gas analyses. A fraction of the collected water was vacuum filtered using 0.22 μm pore size sterile polycarbonate filters (47 mm diameter) that were further stored at −20 °C until DNA extraction.
Analytical techniques
Analyses of water chemical composition were performed at the Research Institute for Analytical Instrumentation, Cluj-Napoca, Romania. Ammonium, nitrate, and nitrite ions were quantified by colorimetry using a Lambda 25 spectrophotometer (Perkin Elmer, Beaconsfield, UK). Sulfate (SO42−) ions were measured by ion chromatography on ICS-1500 (Dionex, Sunnyvale, CA, USA). The concentration of sulfides was determined by methylene blue method after fixation of samples with 2% (v/v) Zn-acetate (Trüper and Schlegel, 1964). Chloride (Cl−), carbonate (as CaCO3) and bicarbonate (HCO3−) anions were measured by titrimetric methods. The contents of Mg2+, Ca2+, Na+ and K+ were determined by inductively coupled plasma atomic emission spectrometry using OPTIMA 5300 DV spectrometer (Perkin Elmer, Norwalk, CT, USA). The contents of total nitrogen as bound nitrogen (including free ammonia, ammonium, nitrite, nitrate and organic nitrogen, but not dissolved nitrogen gas) was assessed by combustion followed by oxidation to nitric dioxide and subsequent chemoluminescence detection. Organic nitrogen in the form of dissolved organic nitrogen was calculated by subtracting ammonium nitrogen from the total nitrogen quantified by Kjeldahl digestion method (Clesceri et al., 1999). Total organic carbon was determined by sample acidification followed by combustion and infrared detection of CO2 released. Both parameters (total nitrogen and total organic carbon) were measured according to EN 12260 and EN 1484, respectively, using the multi N/C 2100S Analyzer (Analytik Jena, Jena, Germany). Relative expanded measurement uncertainty was calculated according to International Organization for Standardization—Guide to the expression of Uncertainty Measurement using a coverage factor (k) of 2 (k=2), equivalent to a confidence of ∼95%. Uc ranged from 4.5% to 13% depending on the compound as follows: 4.5% for Mg2+; 8% for HCO3−, particulate Fe and Ca2+; 9.5% for PO43−; 9.8% for Na+; 10% for total nitrogen, total organic carbon, CO32−, SO42−, HS− and K+; 11% for NO3− and ON; 13% for NO2−, NH4+ and Cl−.
Gas analyses were performed on water samples collected within the monimolimnion of the two lakes (9 and 11 m samples from Ursu Lake; 11 and 13 m samples from Fara Fund Lake). After static headspace extraction at room temperature, a wide range of gaseous hydrocarbons were analyzed at the Instituto Nazionale di Geofisica e Vulcanologia in Rome (Italy). CH4 was determined by tunable diode laser absorption spectroscopy (West Systems, Pontedera, Italy). Heavier hydrocarbons (C2H6, C2H4, C3H8, n-C4H10, i-C4H10, n-C5H12, i-C5H12 and C6H6) were analyzed by a Fourier transform infrared spectrometer (Gasmet DX-4030, Gasmet, Finland; lower detection limit of all hydrocarbons: 3 p.p.m.v.) with a standard spectra library. The stable carbon isotope composition of CH4 (δ13CCH4) was analyzed by cavity ring-down spectroscopy with a Picarro analyzer G2112-I (Picarro Inc., Santa Clara, CA, USA).
Cell counts, chlorophyll a and total carotenoids analyses
Water samples were fixed in 1% glutaraldehyde and filtered through black, gridded cellulose ester membrane filters (0.45 μm, d=47 mm). Subsequently, they were stained directly using 5 μg ml−1 of DAPI (4',6-diamidino-2-phenylindole, dihydrochloride) solution and examined by epifluorescence (BX60, Olympus Optical, Tokyo, Japan). Chlorophyll a and total carotenoids concentrations were determined as described by Wetzel and Likens (1991).
DNA extraction and quantitative real-time PCR (qPCR)
The filters were cut in small pieces (~3–5 mm) using sterile scissors, and processed for DNA extraction using the ZR Soil Microbe DNA MiniPrep kit (Zymo Research, Irvine, CA, USA) and the PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, CA, USA), following the manufacturer's instructions. The DNA was extracted in duplicate from each sample and stored at −20 °C until further use. The qPCR was used to evaluate the relative abundances of Archaea and Bacteria along the water columns of the two lakes, by targeting the SSU rDNA, and was performed as described elsewhere (Baricz et al., 2014). Both archaeal and bacterial reactions were carried out in triplicate using the Archaea 931F/M1100R (Einen et al., 2008) and Bacteria 338F/518R (Lane, 1991; Muyzer et al., 1993) primer sets.
Amplicon sequencing and analysis
Sequencing of bacterial and archaeal SSU rRNA gene (V4 region) amplicons was performed on the Illumina MiSeq platform (San Diego, CA, USA) using the protocol of Lundberg et al. (2013). Sequence data were processed and quality controlled through a combination of the UPARSE and QIIME pipelines (Caporaso et al., 2010a; Edgar, 2013). Cutadapt (Martin, 2011) was used in ‘paired-end mode' to trim sequencing primers from the forward and reverse reads while simultaneously discarding those read pairs in which both the forward and reverse primers were not detected (allowed for 10% mismatches for primer search). Paired-ends were merged using the -fastq_mergepairs option of usearch (v.7.0.1001; Edgar, 2013). An in-house python script was used to remove unused barcodes of paired-end sequences that did not survive merging. The QIIME (v1.7) script, split_libraries_fastq.py, was used to demultiplex the sequence data with the quality filter set to zero. Quality control processing and singleton removal was carried out via the UPARSE pipeline (for example, usearch -fastq_maxee 0.5, usearch -sortbysize -minsize 2) including de novo and reference-based chimera detection (Edgar, 2013). The resulting operational taxonomic unit (OTU) table was converted to Biological Observation Matrix (BIOM) format (McDonald et al., 2012a). Taxonomy was assigned using the Ribosomal Database Project (RDP) classifier (Wang et al., 2007) against the updated May 2013 ‘13_5/13_8' Greengenes database (Werner et al., 2011; McDonald et al., 2012b) via the parallel_assign_taxonomy_rdp.py script in QIIME. A phylogeny was constructed using FastTree (Price et al., 2010) from a masked PyNAST (Caporaso et al., 2010b) alignment. The resulting phylogeny was manually rooted to Archaea via Dendroscope (v3; Huson and Scornavacca, 2012). Finally, various diversity metrics were calculated via QIIME. Bacterial and archaeal data were separated into their own OTU tables for independent analysis of each group. Representative sequences of OTUs belonging to Nanohaloarchaeota and Parvarchaeota candidate phyla (from the sample collected at 3 m depth in Fara Fund Lake) were further investigated by phylogenetic placement against known relatives within the SILVA SSU Ref database (v115; Quast et al., 2013). The representative OTU sequences were aligned with SINA (Pruesse et al., 2012) and then imported into the ARB software package (Ludwig et al., 2004). The sequences were inserted into the SILVA SSU Ref database with domain-specific position variable filters via parsimony insertion.
Nucleotide sequence accession number
All sequence data are available through the National Center for Biotechnology Information (NCBI) via Sequence Read Archive (SRA) database, under accession numbers: SRR1569016, SRR1560515, SRR1560527, SRR1569024, SRR1560549, SRR1569028, SRR1560574 and SRR1560593.
Data not shown in the manuscript were deposited at the Dryad Digital Repository (www.datadryad.org) and are referenced in the text using the following doi: 10.5061/dryad.2gm06.
Results
Limnological characterization of the meromictic lakes
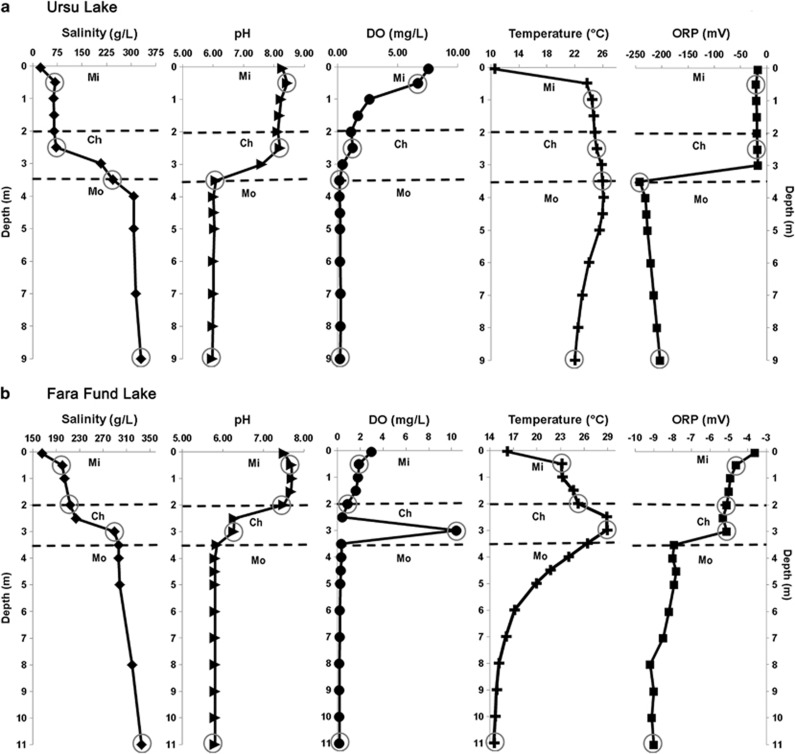
Physicochemical measurements were performed along water columns, and the samples were collected in agreement with the lakes' stratification patterns (Figure 2). The chemical and biological characteristics of the water collected from Ursu and Fara Fund lakes are shown in Table 1. Among the several hydrocarbons analyzed, only methane was detected in the suboxic monimolimnia, with concentrations and stable C isotope composition of ∼0.2 mg l−1 and δ13CCH4: −59.7 to −63.1‰ in the Ursu Lake, and 0.9 mg l−1 and δ13CCH4: −62.1 to −63.0‰ in the Fara Fund Lake, respectively (Table 2). All alkanes from C2 to C6 were below the detection limit (3 p.p.m.v. in the headspace in equilibrium with water).
Figure 2.
Vertical distribution of physicochemical parameters (salinity, dissolved oxygen, pH, temperature and reduction potential) measured in situ along the water columns of Ursu (a) and Fara Fund (b) lakes during October 2013. The three water layers resulted from physicochemical partitioning were separated by dashed lines. Sampling points for DNA and chemical analyses are indicated by circles. Ch, chemocline; DO, dissolved oxygen; Mi, mixolimnion; Mo, monimolimnion; ORP, oxidoreduction potential.
Table 1. Biological and chemical characteristics of the water samples from Ursu and Fara Fund lakes.
| Parameter | Ursu Lake | Fara Fund Lake | ||||||
|---|---|---|---|---|---|---|---|---|
|
Depth (m) |
Depth (m) |
|||||||
| 0.5 | 2.5 | 3.5 | 9 | 0.5 | 2 | 3 | 11 | |
| TCh (μg l−1) | 14.91 | 394.80 | 67.16 | 18.78 | 1.83 | 1.43 | 4.01 | 2.82 |
| TCa (μg l−1) | 668.13 | 1624.35 | 975.62 | 587.08 | 290.82 | 299.19 | 165.67 | 150.50 |
| NH4+-N (mg l−1) | 0.3 | 11.2 | 33.2 | 30.2 | 6.3 | 7.4 | 22.7 | 33.3 |
| NO3−-N (mg l−1) | 0.30 | 0.45 | 0.55 | 0.24 | 0.56 | 1.14 | 0.83 | 0.95 |
| NO2−-N (mg l−1) | 0.36 | 0.55 | 0.27 | 0.14 | <0.05 | <0.05 | <0.05 | <0.05 |
| ON (mg l−1) | 2.0 | 2.2 | 1.4 | 0.6 | 3.1 | 2.5 | 1.3 | 1.0 |
| TN (mg l−1) | 3.34 | 16.88 | 34.80 | 33.63 | 10.28 | 11.18 | 25.23 | 29.40 |
| TOC (mg l−1) | 41.4 | 48.4 | 66.1 | 45.9 | 58.6 | 54.5 | 38.2 | 49.1 |
| aCO3− (mg l−1) | 200 | 245 | 1170 | 968 | 100 | 163 | 652 | 682 |
| HCO3− (mg l−1) | 366 | 427 | 1830 | 1647 | 153 | 183 | 915 | 1220 |
| Cl− (mg l−1) | 41 000 | 44 000 | 165 000 | 205 000 | 128 000 | 131 000 | 180 000 | 217 000 |
| SO42− (mg l−1) | 231 | 299 | 724 | 1363 | 1269 | 1247 | 1792 | 1534 |
| HS− (mg l−1) | <0.04 | <0.04 | 151 | 114 | <0.04 | <0.04 | <0.04 | 36 |
| PO43− (mg l−1) | 0.62 | 6.14 | 8.81 | 3.75 | 0.80 | <0.34 | 0.47 | 0.34 |
| K+ (mg l−1) | 31.8 | 53.6 | 57.0 | 68.4 | 119 | 128 | 110 | 105 |
| Fe (mg l−1) | 0.177 | 0.413 | 0.632 | 0.642 | 0.132 | 0.069 | 0.225 | 11.1 |
| Ca2+ (mg l−1) | 112 | 246 | 738 | 967 | 422 | 568 | 1057 | 1183 |
| Mg2+ (mg l−1) | 20.1 | 27.6 | 35.4 | 101 | 96 | 122 | 143 | 145 |
| Na+ (mg l−1) | 26 300 | 26 500 | 109 000 | 136 000 | 78 000 | 85 800 | 117 500 | 137 300 |
Abbreviations: NNH4+, ammonium nitrogen; ON, organic nitrogen; TCa, total carotenoids; TCh, total chlorophyll a; TN, total nitrogen; TOC, total organic carbon.
Carbonate anions were determined as CaCO3.
Table 2. Dissolved methane concentration and its stable C isotope composition.
| Site | Depth (m) | Dissolved CH4 (mg l−1) | δ13CCH4‰ (VPDB) |
|---|---|---|---|
| Ursu Lake | 9 | 0.26 | −59.7 |
| 11 | 0.24 | −63.1 | |
| Fara Fund Lake | 11 | 0.92 | −62.1 |
| 13 | 0.91 | −63.0 |
Abbreviation: VPDB, Vienna Pee Dee Belemnite standard.
C2 to C6 hydrocarbons, not shown in the table, were below the detection limit (3 p.p.m.v. in the headspace in equilibrium with water).
The measured transparency values (Secchi depths) for the two lakes were 1.9 m for Ursu and 1.5 m for Fara Fund. These measurements were further used to estimate the euphotic depth (Zeu) as described by Wetzel and Likens (1995), and the results indicated that the euphotic zone extended to the superior part of the monimolimnion in Ursu Lake (Zeu: 4.6 m) and to the lower limit of the chemocline in Fara Fund Lake (Zeu: 3.6 m). Chlorophyll a concentrations (expected to be a substitute for phytoplankton-derived autochthonous organic carbon input) varied between lakes and depths and ranged from 1.43 to 394.8 μg l−1. Noteworthy, the chlorophyll a values peaked at the chemocline in both Ursu and Fara Fund lakes.
The vertical profile of temperature (Figure 2) revealed that the water strata were in a transitional state between direct and inverse thermal stratification, with the highest temperatures registered in the chemocline region for both lakes. The monimolimnia were suboxic, had a lower pH, and a higher redox potential and salinity than the overlaying water masses, corroborating with our previous measurements (doi: 10.5061/dryad.2gm06). Notably, the redox potential of the Ursu Lake was more reduced and the vertical salinity profiles fluctuated over a wider range than the water in Fara Fund Lake (Figure 2).
The levels of total organic carbon were similar within and between the lakes and varied from 38.2 to 66.1 mg l−1. As the level of the total nitrogen increased with depth, mostly because of rise in ammonium concentration, the organic nitrogen showed an inverse pattern, decreasing in the deeper strata of both lakes after the chemocline. The water ionic composition analyses showed that the monovalent cations (Na+ and K+) were dominant over the divalent ones (Mg2+ and Ca2+). The main anions were Cl−, SO42−, HCO3− and PO43−, revealing a composition similar to that of sea water.
Sequencing statistics
Paired-end sequencing of 16S rDNA hypervariable region V4 amplicons generated 697 529 high-quality reads (347 773 for Bacteria and 349 756 for Archaea) with an average length of 254 nt. The number of sequences per sample ranged from 64 650 (3.5 m depth sample from Ursu Lake) to 141 896 (2 m depth sample from Fara Fund Lake) (Supplementary Table S1). In order to test the reproducibility of the sequencing method, one sample (9 m depth from Ursu Lake) was analyzed in duplicate, and the results showed that the obtained phylogenetic profile was almost identical between replicates (weighted normalized UniFrac distance=0.029). In addition, Pearson's correlation coefficients between the qPCR results and the number of archaeal and bacterial sequences obtained showed positive significant correlations for both Ursu (r=0.735, P<0.05) and Fara Fund (r=0.728, P<0.05) lakes.
Abundance and vertical distribution of prokaryotic communities
DAPI epifluorescence microscopy showed that in both lakes the mixolimnion was largely populated by small-sized (0.5–1 μm) coccus-shaped organisms, and that the monimolimnion was characterized by an increased abundance of medium-sized (2–4 μm) morphotypes (for example, vibrios and spirilla) (doi: 10.5061/dryad.2gm06).
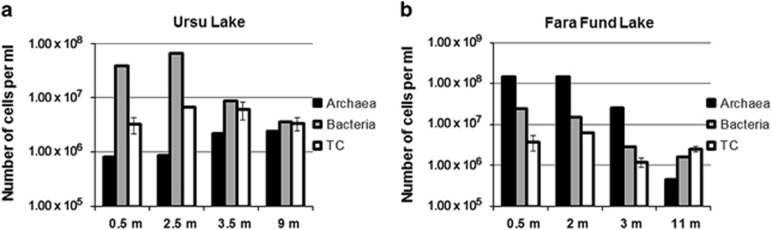
For both lakes, the maximum prokaryotic abundance, estimated by qPCR, was found within the euphotic zone and on average Fara Fund Lake (mean cell numbers per ml=4.56 × 107±28.2 × 104) harbored higher cell density than Ursu Lake (mean cell numbers per ml=1.54 × 107±7.4 × 104). In Ursu Lake the bacterial cell abundance ranged from 3.6 to 66.5 × 106 cells per ml, and the archaeal ones from 8.1 to 24.1 × 105 cells per ml (Figure 3). In this lake the bacterial density increased with depth between 0.5 and 2.5 m, but subsequently decreased steadily to the lowermost sampling point, whereas the archaeal numbers increased with depth throughout the water column. In Fara Fund Lake, the bacterial and archaeal densities ranged from 1.6 to 24.6 × 106 cells per ml, and from 4.4 × 105 to 1.4 × 108 cells per ml respectively (Figure 3). In Fara Fund Lake the bacterial numbers per ml gradually decreased from the surface to 11 m deep whereas the archaeal abundance slightly increased from 0.5 (archaeal cell numbers per ml=1.46 × 108±7 × 105) to 2 m depth (archaeal cell numbers per ml=1.49 × 108±6.2 × 105) to continuously decrease throughout the water column.
Figure 3.
Prokaryotic cell numbers per ml as estimated by quantitative PCR (black and gray columns) and DAPI counts (white columns) for the Ursu (a) and Fara Fund (b) lakes. The sampling depths from each lake are represented on the abscissa. The columns represent means, and the error bars represent their s.d. values. TC, total cell counts.
The results of standard Mantel tests performed on domain-specific qPCR data (Supplementary Table S2) revealed that archaeal abundances within Ursu Lake were significantly positively correlated with both salinity (R=0.949, P<0.05) and sulfate concentrations (R=0.672, P<0.05), whereas bacterial abundances were positively correlated with nitrite concentrations (R=0.84, P<0.05). The same tests indicated that the archaeal abundances in Fara Fund Lake were positively correlated with total carotenoid concentrations (R=0.993, P<0.05), whereas bacterial abundances were positively correlated with organic nitrogen concentrations (R=0.952, P<0.05) (Supplementary Table S2).
Prokaryotic α-diversity patterns
To compare the α-diversity indices across lakes and depths, we normalized the sequence number of each sample to 5411 for Archaea and 5953 for Bacteria (the fewest among the data set) through random resampling. Two Ursu Lake samples (0.5 and 2.5 m) were excluded from the Archaea data set, as they contained 267 and 340 reads respectively (Supplementary Table S1). Sequences with ≥97% similarity were grouped into OTUs and used for the diversity estimations. In addition, the normalized data set was shown to cover ∼98.3-–99.9% of the species richness as indicated by Good's estimator (Supplementary Table S3).
The α-diversity levels in Ursu Lake (for both Archaea and Bacteria) typically increased with depth as proven by each of the measurement indexes used: divergence-based phylogenetic diversity (Faith's phylogenetic diversity), OTU-based observed species or the quantitative Shannon's index (Table 3). Furthermore, these indices showed that α-diversity was higher for Bacteria than for Archaea. In Fara Fund the archaeal α-diversity increased from surface to the bottom of the lake for all the three indices used (phylogenetic diversity PD, observed species and Shannon), whereas the bacterial one decreased monotonously along the water column till 3 m, after which it rose at 11 m deep. The α-diversity metrics indicated that, in general, the Bacteria were more diverse than Archaea (Table 3).
Table 3. Archaeal and bacterial estimates of α-diversity metrics for the samples collected from Ursu and Fara Fund lakes.
| Lake | Depth (m) | α-Diversity indices |
||
|---|---|---|---|---|
| PD | OS | S | ||
| Archaea | ||||
| Ursu | 3.5 | 8.512 | 73 | 2.715 |
| Ursu | 9 | 8.821 | 93.14 | 3.346 |
| Fara Fund | 0.5 | 2.388 | 27.13 | 0.21 |
| Fara Fund | 2 | 3.637 | 46.23 | 0.302 |
| Fara Fund | 3 | 4.209 | 56.63 | 2.992 |
| Fara Fund | 11 | 10.418 | 124.71 | 4.062 |
| Bacteria | ||||
| Ursu | 0.5 | 12.9 | 112.42 | 4.644 |
| Ursu | 2.5 | 20.903 | 188.12 | 4.994 |
| Ursu | 3.5 | 27.31 | 250.02 | 4.996 |
| Ursu | 9 | 29.426 | 272.32 | 5.456 |
| Fara Fund | 0.5 | 16.468 | 124.26 | 3.741 |
| Fara Fund | 2 | 15.018 | 101.93 | 3.284 |
| Fara Fund | 3 | 9.782 | 55.23 | 2.299 |
| Fara Fund | 11 | 23.77 | 193.76 | 3.536 |
Abbreviations: OS, observed species; PD, phylogenetic diversity; S, Shannon.
Archaeal α-diversity of the two hypersaline lakes, measured as phylogenetic diversity (Table 3), was positively correlated with ammonium nitrogen (r=0.921, P<0.05), Cl− (r=0.844, P<0.05), Na+ (r=0.848, P<0.05), depth (r=0.884, P<0.05), total nitrogen (r=0.862, P<0.05) and HCO3− (r=0.848, P<0.05) and negatively with pH (r=−0.874, P<0.05) and organic nitrogen (r=−0.828, P<0.05) (Supplementary Table S4). In contrast, the bacterial phylogenetic diversity values were positively correlated with HCO3− (r=0.74, P<0.05) and total nitrogen (r=0.712, P<0.04) and negatively with dissolved oxygen (r=−0.792, P<0.05) and the redox potential (r=−0.773, P<0.05) (Supplementary Table S4).
Prokaryotic diversity and community structure in Ursu and Fara Fund lakes
We assigned taxonomy for the tags using the RDP classifier with the GreenGenes database, via QIIME software package. OTU (≥97% similarity clusters) rarefaction curves reached a clear asymptote (Supplementary Figure S1), and the Good's coverage estimator indicated that we sampled between 99.7% and 99.9% of the species richness in each sample (Supplementary Table S3). From the total number of sequences (Supplementary Table S1), ~3% could not be classified at domain rank and ~5.7% could not be attributed to any particular phylum.
Ursu Lake
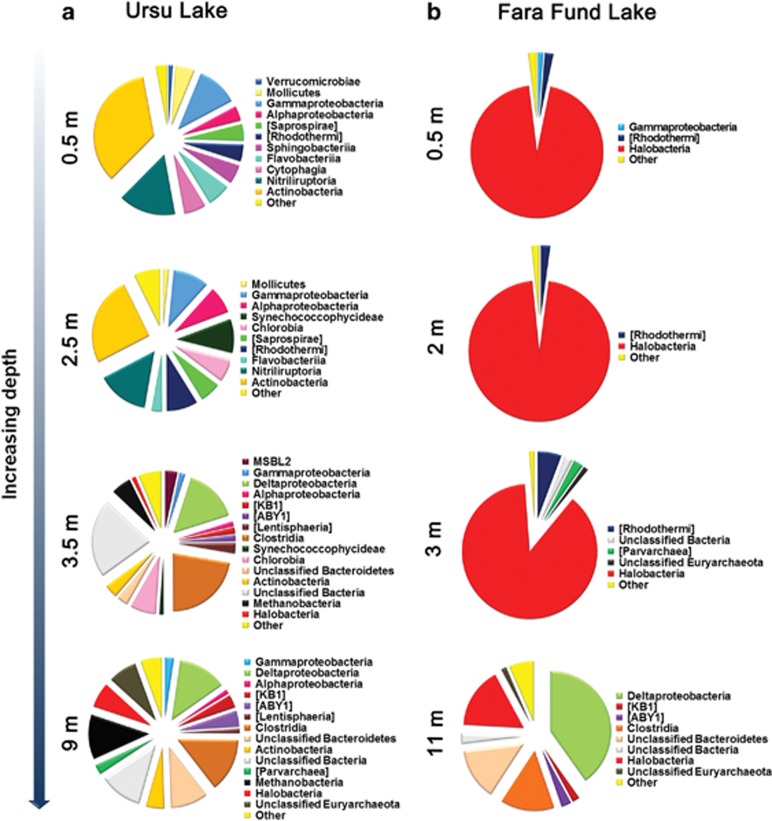
In Ursu Lake, the OTUs were found to belong to both prokaryotic domains, and their abundance increased continuously with depth, ranging from 180 at 0.5 m deep to 575 at the bottom of the lake (Supplementary Table S1). The taxonomy results showed that Ursu Lake harbored a wider spectrum of prokaryotic lineages than Fara Fund Lake, and that they were affiliated with 2 Archaea (that is, Euryarchaeota and Parvarchaeota), and 20 Bacteria phyla (that is, Actinobacteria, Bacteroidetes, Chlamydiae, Chlorobi, Cyanobacteria, Deferribacteres, Firmicutes, GN02, Gemmatimonadetes, Lentisphaerae, OD1, OP1, Planctomycetes, Proteobacteria, SR1, Spirochaetes, Tenericutes, Verrucomicrobia, WWE1 and Thermi) (Supplementary Table S5).
The fraction of archaeal reads present in the 0.5 and 2.5 m samples was low (between ~0.4% and 0.5% of total reads). Therefore, the prokaryotic assemblages at these depths will be considered as comprising exclusively of Bacteria. The majority of archaeal OTUs were classified within Euryarchaeota that covered ~8% of the sequences from 3.5 m and 26% from the ones in the 9 m sample. The same tendency was observed in Parvarchaeota that increased in abundance from the chemocline (~0.6%) to the monimolimnion (~2%) (Supplementary Table S5). The bacterial taxonomy at phylum level showed that the abundance of Proteobacteria remained relatively constant throughout the water column, ranging between ~16% and 19%. Even though their vertical distribution was relatively uniform, the samples from mixolimnion and the upper part of the chemocline preponderantly comprised Gammaproteobacteria and Alphaproteobacteria, in contrast to the lower part of the chemocline and monimolimnion where Deltaproteobacteria prevailed (Figure 4). Actinobacteria abundance decreased from 0.5 m (~50%) to 3.5 m (~4%) and slightly increased in the 9 m sample (~5%). A similar pattern was also observed for Bacteroidetes, whose abundance decreased from the mixolimnion (~26%) through chemocline (~19% and 5%, respectively) and increased afterwards in the monimolimnion (~11%). Furthermore, a significant part of the sequences from 3.5 m (~22%) and 9 m (~14%) were affiliated with Firmicutes (Supplementary Table S5).
Figure 4.
Class-level taxonomic profiles of the prokaryotic communities using 16S rRNA gene sequences.
Fara Fund Lake
Overall, the numbers of OTUs in Fara Fund Lake fluctuated between samples from 127 to 474 (Supplementary Table S1) and were found to appertain to both Archaea and Bacteria domains. The majority of the sequences in the data set were found to be affiliated to the phyla Euryarchaeota, Parvarchaeota, Proteobacteria, Bacteroidetes, Firmicutes, OD1 and OP1 (Supplementary Table S5). In addition to the prevailing phyla, OTUs belonging to Lentisphaerae, Deferribacteres, OP3 and Spirochaetes were present within the lake's water column.
The Archaea domain was found to dominate the mixolimnion and chemocline samples (that is, 0.5, 2 and 3 m) where it accounted for >90% of the sequences, in contrast with the monimolimnion, where it represented ~20% of the prokaryotic community reads (Supplementary Table S5). The Euryarchaeota phyla covered the vast majority of reads in the 0.5 m (~95%), 2 m (~96%) and 3 m (~89%) samples, and accounted for ~18% of the sequences in the monimolimnion. In contrast, the number of Parvarchaeota reads peaked at 3 m depth (~2%), while being poorly represented in the rest of the lake (0.02–0.7%). The percentage of Proteobacteria sequences decreased with depth from 1.9% in the 0.5 m sample to 0.57% in the 3 m sample, and increased afterwards to ~42% in the 11 m sample. Within the Proteobacteria, the most abundant class in the mixolimnion and chemocline was Gammaproteobacteria, substituted in monimolimnion by Deltaproteobacteria that covered ~40% of the total prokaryotic reads (Figure 4). A similar pattern was observed in the abundance of Bacteroidetes that increased from ~3% in the mixolimnion to ~15% at 11 m deep. Furthermore, a significant part of the sequences from the monimolimnion were affiliated with Firmicutes (~14.4%), OD1 (~2.7%) and OP1 (2%) (Supplementary Table S5).
Discussion
Water chemistry
The water chemistry of the lakes was found to be a reflection of halite diapirism and of the sedimentary input of clastic materials that shaped the Transylvanian Basin stratigraphy (Har et al., 2010). As a consequence, the raised levels of alkali and alkaline cations (for example, sodium, potassium, magnesium and calcium) found within the monimolimnia of both lakes could be attributed to halide dissolution (that is, halite) and to the weathering of tectosilicate (that is, anorthite and albite), inosilicate (that is, ferro-actinolite) and phyllosilicate (that is, vanadian and barian muscovite, clinochlore) under acidic pH conditions (doi: 10.5061/dryad.2gm06).
Both Ursu and Fara Fund lakes presented a perennially strong stratification of the water column with contrasting chemical compositions between the mixolimnion and monimolimnion. The higher particulate iron concentrations found in the lower parts of the water column were most probably caused by the sedimentation of ferric oxyhydroxides within the oxic strata. Even though the suboxic conditions may favor their reductive dissolution, with the generation of dissolved Fe2+, the presence of HS− in the monimolimnion might induce FeS formation, thereby restricting its role as terminal electron acceptor for organic matter oxidation (Cappellen et al., 1998; Hurtgen et al., 1999). Ammonium reached the highest concentrations in the deep anoxic layers of the lakes, indicating a higher rate of mineralization than assimilation, coupled with a higher stability in the acidic pH range. The depth profiles of ammonium concentration were comparable for both Ursu and Fara Fund lakes, and similar to the ones described in other stratified lakes (Auguet et al., 2012; La Cono et al., 2013; Yau et al., 2013). The vertical profiles of nutrients (sulfate, sulfide, nitrite, nitrate and phosphate) were found to be a result of sedimentation, biogeochemical cycling or conservative mixing (Pasche et al., 2009), and were also comparable with those found in other studied lakes (Lepère et al., 2010; La Cono et al., 2013; Marteinsson et al., 2013).
Dissolved CH4 concentrations in Ursu and Fara Fund lakes (Table 2) were found to be within the typical range observed in the monimolimnion of several types of lakes (for example, Schubert et al., 2010), including the hypersaline Big Soda Lake (Iversen et al., 1987). The monimolimnion of a lake typically shows CH4 with a stable C isotope composition (δ13CCH4) that is higher (13C-enriched) compared with that in the deeper anoxic CH4-producing layers (generally <−65‰), and lower (13C-depleted) compared with that in the shallower water (>−50‰ see, for example, Iversen et al., 1987), because of moderate CH4 oxidation. The monimolimnion δ13CCH4 values of Ursu and Fara Fund (from −59.7 to −63.1‰ Table 2) are consistent with this general trend. The slight increase of δ13CCH4 values at lower depths (a difference of +3.4‰ from 11 to 9 m in the Ursu Lake, and +0.9‰ from 13 to 11 m in the Fara Fund Lake) combined with a practically constant CH4 concentration could suggest that the eventual CH4 oxidation by methanotrophs is minimal within the monimolimnion. The values confirm a microbial origin, likely related to a fermentation pathway, as typically found in lakes (see, for example, Whiticar et al., 1986). The absence of higher n-alkane hydrocarbon gases (C2–C6) suggests the lack of significant geological seepage of thermogenic gas from the lake sediments (typically 13C-enriched, with δ13CCH4 >−50‰). But seepage of microbial geological (fossil) methane, which is C2–C6 free and isotopically indistinguishable from modern biologic CH4, cannot be excluded. Such a microbial seepage, releasing CH4 with δ13C <−60‰ is in fact quite common in the Transylvanian Basin (Etiope et al., 2009; Spulber et al., 2010; Ionescu, 2015). In this respect, radiocarbon analyses of CH4 would be necessary to reveal a possible geological component of CH4 in the lakes. Given the high salinity of the water, it is likely that methane in Ursu and Fara Fund lakes is dominantly formed via a specific fermentation pathway such as methylotrophic methanogenesis (Kelley et al., 2012) the only metabolic pathway shown to occur at salinities comparable with those found in the lakes' monimolimnion (Andrei et al., 2012).
Vertical patterns in prokaryotic abundances
Although among the prokaryotes living in extreme hypersaline waters the most abundant are usually considered to be Archaea (Andrei et al., 2012), recently it has become indisputable that representatives of the domain Bacteria play key metabolic roles in high-salt ecosystems (Oren, 2012).
As expected, Archaea dominated the mixolimnion and chemocline samples from Fara Fund, harboring similar numbers as that reported within other meromictic salt lakes (Baricz et al., 2014). Even though the bacterial abundance was comparable to that typically described for high-salt environments (Demergasso et al., 2008; Baricz et al., 2014), the fact that they constituted ~78% of the monimolimnion community was unanticipated, as previous studies reported the dominance of Archaea at salinities >300 g l−1 (Ghai et al., 2011; Baricz et al., 2014). We assume that the increased abundance of Bacteria in the deeper water strata could be achieved by outcompeting Archaea in suboxic hypersaline conditions, as the majority of halophilic archaeal species are aerobic (Andrei et al., 2012). In Ursu Lake, a different vertical distribution pattern of prokaryotic abundance was observed, as Bacteria clearly dominated the water column communities. The relative abundance of Archaea in the 0.5 (~2%) and 2 m (1.2%) depth assemblages was minor, and could be attributed to the lower salinity levels found at these depths (that is, ~70 g l−1). Similar findings were reported in other saline lakes (Yau et al., 2013; Baricz et al., 2014) or shallow ponds (Ghai et al., 2011), and are supported by the fact that most haloarchaeal species lyse at salt concentrations of <100 g l−1 (Andrei et al., 2012). As the salinity increased over 240 g l−1 (at 3.5 m depth) the proportion of the Archaea in the communities was higher, ranging between ~20% (at 3.5 m depth) and ~40% (at 9 m depth), but nonetheless lower than the bacterial one. We argue that although the salinity premise was supported, the suboxic environment favored thriving of the Bacteria. Overall, the dissimilar prokaryotic abundance patterns found in the two lakes could be attributed to their different physicochemical milieus, as well as the different metabolic requirements of Archaea and Bacteria domains (as shown by correlation analyses; Supplementary Table S4).
Prokaryotic diversity
A general ecological principle states that as the factors in the environment become more extreme, the species that might be harbored will be less diverse (Frontier, 1985). Nevertheless, even though the reproducibility of this pattern is uncertain for microbial communities, several studies found evidence for a declining tendency in prokaryotic diversity along increasing salinity gradients (Benlloch et al., 2002; Hollister et al., 2010). Strikingly, both archaeal and bacterial diversity increased along with the salinity values from surface to the bottom of the Ursu Lake. In Fara Fund Lake, the increase of archaeal diversity paralleled the salinity trend, whereas the bacterial one decreased from surface through chemocline and subsequently rose in the deeper water strata. The decrease of Bacteria diversity from surface to 3 m deep recorded in Fara Fund Lake overlapped with an increased abundance of Salinibacter sequences (~5%), indicating that it outcompeted the other taxa. Thus, the observed shift could be attributed to changes in the bacterial ecological niches through the euphotic zone.
The lake's monimolimnion may be considered a more ‘extreme' environment by comparison with the mixolimnion, for the reason that it has higher concentrations of salts (>300 g l−1), sulfide (26–114 mg l−1) and ammonium (>30 mg l−1) that have accumulated as a result of extended meromixis. In spite of the metabolic constraints possibly associated with these factors, the highest prokaryotic diversity was found to be present here. The elevated phylogenetic diversity may be attributed to the downward metabolite fluxes that favor niche diversity through the maintenance of various nutrient sources. Furthermore, visual and microscopy observations showed increased levels of suspended particulate and floc matter in the lakes' monimolimnia that may contribute to increased phylogenetic divergence through habitat diversification (Tang et al., 2009). Another hypothesis may be that low oxygen environments have the capacity to maintain a higher variety of energetic pathways than oxygen-rich environments, leading to retention of higher ecological diversity coupled with lowered interspecific competition (Humayoun et al., 2003). This postulation is corroborated by our results that indicate strong negative correlations between Bacteria diversity and dissolved oxygen concentrations (Supplementary Table S4). Furthermore, the prokaryotic taxonomic composition showed that oxygen gradients have an obvious effect of separating aerobic from anaerobic taxa, allowing oxygen-sensitive nitrogen and sulfur (or methane) cycles to occur in the monimolimnion.
Community physiology inferences in Ursu and Fara Fund lakes
The taxonomy results showed a clear vertical separation throughout the water column of both Archaea and Bacteria taxa (Figure 4). Furthermore, these findings are corroborated with other studies that detected disparate communities in the mixolimnion versus monimolimnion of other meromictic lakes (Barberán and Casamayor, 2011; Lauro et al., 2011; Comeau et al., 2012). In addition, the overall organization of the prokaryotic communities was found to be typical for stratified lakes with regard to depth distribution of photosynthetic and redox conditions (Lauro et al., 2011; Comeau et al., 2012; Edberg et al., 2012; Lentini et al., 2012; Yau et al., 2013).
Although 16S metabarcoding analyses do not provide direct evidence on the functional status of detected phylotypes, useful information might be deduced from the abundance and distribution of 16S gene reads belonging to major metabolic groups (Supplementary Figure S2 and Supplementary Table S6). Furthermore, suppositions on the putative metabolic activity could also be inferred from the stratification of environmental parameters and nutrient profiles.
Oxygenic photoautotrophic cyanobacteria (mainly Synechococcus spp.) together with significant populations of unicellular algae (Máthé et al., 2014) and anoxygenic photosynthesizers (Supplementary Table S6) might be responsible for the primary productivity in the upper layer (down to 2.5 m deep) of Ursu Lake. We consider that the accumulated organic carbon might be subsequently metabolized by both heterotrophic aerobic (Actinobacteria, Bacteroidetes, Gammaproteobacteria and Planctomycetes) and anaerobic Bacteria (Halanaerobiales and Clostridiales) as well as Archaea (Halobacteriales). In comparison with Ursu Lake, the photic zone of Fara Fund Lake hosts smaller communities of phototrophic microorganisms as shown by the low chlorophyll a content reported in this study (see Table 1) and by previous works (Keresztes et al., 2012). Thanks to our microscopy and molecular analyses (doi: 10.5061/dryad.2gm06), we consider that oxygen levels in the euphotic zone of Fara Fund Lake were mostly generated by the photosynthetic activity of Dunaliella algae (see also Somogyi et al., 2014). Microbial communities capable of organic carbon degradation were found to be distributed along the water column of the lake, spreading from the oxic (Halobacteriales and Nanohaloarchaeota) to the suboxic layers (Halanaerobacter, Halanaerobium and Halorhabdus) (Supplementary Table S6).
Taxonomic analyses revealed that the coexistence of phototrophy with anaerobic sulfur oxidation might be facilitated by the presence of purple sulfur bacteria and green sulfur bacteria in Ursu Lake and by purple sulfur bacteria of Ectothiorhodospiraceae family (that is, mainly Halorhodospira spp.) in Fara Fund Lake (Supplementary Table S6). In Ursu Lake, 16S reads belonging to purple sulfur bacteria of the Class Chromatiales (Chromatiaceae and Ectothiorhodospiraceae) were found in the epilimnion and upper chemocline, whereas those affiliated to green sulfur bacteria of the Class Chlorobia (Prosthecochloris) were found to be prevalent within the O2/HS− (redox) transition zone (as also shown by Máthé et al., 2014). The occurrence of aerobic sulfur-oxidizing bacteria in the moderately saline mixolimnion of Ursu Lake might be an indicator of an active aerobic or microaerophilic sulfur-oxidation metabolism performed by members of Thiotrichales (for example, Thiomicrospira) and Chromatiales (for example, Halothiobacillus). In both lakes, the abundance of 16S sequences indicated that two Deltaproteobacteria families (Desulfohalobiaceae and Desulfobacteraceae) dominated by far the monimolimnetic sulfate-reducing communities (Supplementary Table S6).
We discovered that several bacterial taxa, if active, could be involved in various steps of nitrogen cycling (Supplementary Figure S2) such as nitrogen assimilation (carried out by members of the Rhodospirillaceae family) and heterotrophic denitrification (performed by representatives of Halomonadaceae and Rhodospirillaceae). In addition, in Fara Fund Lake we found sequences related to extremely halophilic denitrifying lithoautotrophic sulfur-oxidizing bacteria Thiohalorhabdus sp. (Sorokin et al., 2008). Potential for ammonification could be inferred in Ursu Lake where we found 16S reads assigned to Desulfurispirillum alkaliphilum (family Chrysiogenaceae), an anaerobic dissimilatory nitrate-reducing and halotolerant bacterium that utilizes elemental sulfur as electron acceptor (Sorokin et al., 2007).
As for methanotrophy, the only taxa capable of methane metabolization (Methylococcales) were detected at moderate salinity solely in Ursu Lake (Supplementary Table S6).
Furthermore, the Fe/Mn metabolism seemed to be faintly supported by 16S reads belonging to Deferribacteraceae (genus Deferribacter) and Desulfuromonadaceae (primarily Desulfuromonas) that were detected below the chemocline of Ursu Lake (Supplementary Table S6). However, most of these anaerobic bacteria were identified in marine and estuarine sediments and are capable of using Fe3+, Mn4+, nitrate or sulfur as electron acceptors, playing a role in the organic matter diagenesis (Roden and Lovley, 1993). In spite of the fact that 16S rRNA gene sequences belonging to Deferribacteres were identified in other hypersaline habitats (Ferrer et al., 2012), no evidence that these iron reducers could function under high-salt concentration was shown to date.
Although both lakes were found to harbor prokaryotes capable of biogeochemical cycling (carbon, sulfur and nitrogen), the inferred physiology of Fara Fund Lake's microbiota was shown to be less diverse than the one found in Ursu Lake. This finding may be attributed to the site-specific conditions and larger environmental gradients (for example, salinity and oxidoreduction potential) found in Ursu Lake that supposedly favored higher niche variety and metabolic diversity. By matching taxa to known taxon-specific biogeochemical functions, we were able to identify a close correspondence between the functional specialists and environmental gradients. Although a coherent picture could be sketched for several steps of C, S and N cycling in Ursu and Fara Fund lakes (for example, carbon fixation and organic carbon degradation, sulfur oxidation and sulfate reduction and nitrogen assimilation and denitrification), other aspects of ecosystem element cycling were beyond the purpose of this study and remained to be addressed by more specific approaches (for example, in situ measurements of the production/consumption rate for nutrients, strain isolation, targeting of functional marker genes and shotgun metagenomics).
Methanogenesis
Biogenic methane production in hypersaline conditions is thermodynamically challenging because of bioenergetic constraints (Oren, 2011). We found that sequences matching taxa attributed to methanogenesis were present in the suboxic strata of the lakes, and represented between ~0.6 (in Fara Fund Lake) and ~12% (in Ursu Lake) of the prokaryotic communities. Most of the reads were classified to the Methanobacteriales or the euryarchaeal candidate division MSBL1. As the majority of halophilic methanogenic Archaea are classified within Methanosarcinales (Andrei et al., 2012), with Methanobacteriales usually being hydrogenotrophic (Bonin and Boone, 2006), we searched for related DNA sequences from other environments (Supplementary Table S7). The most abundant OTUs belonging to Methanobacteriales or MSBL1 were highly similar (95–99%) to unclassified sequences recovered from other hypersaline meromictic lakes, solar salterns sediments or deep sea-hypersaline anoxic basins, indicating the existence of a yet-undescribed methanogenic group present in extreme hypersaline habitats. Although this assumption is supported by a recent study on Medee brines (Yakimov et al., 2013), in which MSBL1 candidate division was shown to be responsible for the methanogenic fermentation of trimethylamine, further experiments using cultivation and 14C-assimilation are needed to unambiguously describe the methanogenesis in these high-salt ecosystems.
Ultrasmall, uncultivated archaeal lineages thriving in the chemocline of Fara Fund Lake
The Archaeal communities from the oxic zone of Fara Fund were found to be typical for extreme hypersaline aquatic environments, in terms of community composition, as they were dominated by Halobacteriales phylotypes (Ghai et al., 2011). Aside from a clear separation from the monimolimnion assemblages, the communities also showed a vertical variation. As the samples from 0.5 and 2 m depths were mainly dominated by Halobacteriaceae reads, related to an environmentally derived DNA sequence from a solar saltern (Baati et al., 2010), the 3 m depth sample contained a high abundance of reads belonging to the MSP41 group (~47.7%). Moreover, a V4 SSU rDNA phylogenetic tree suggested that the 3 m community consists of OTUs related to candidate genera Nanosalinarum, Nanosalina and Haloredivivus (Supplementary Figure S3) (Ghai et al., 2011; Narasingarao et al., 2012). In addition to the MSP41 group, which was recently proposed as phylum Nanohaloarchaeota, the community also contained OTUs related to the Parvarchaeota (2.3%) phylum (Rinke et al., 2013). Even though we know that the prokaryotes from the mentioned putative phyla have an ultrasmall cellular size and aerobic heterotrophic lifestyles (Baker et al., 2010; Narasingarao et al., 2012), their role and metabolic contribution to the microbial assemblages is uncertain. Furthermore, even though the nanohaloarchaea appear to be abundant in hypersaline environments and to have a worldwide distribution (Zhaxybayeva et al., 2013), data on the presence of Parvarchaeota in hypersaline lakes are missing. Thus, as far as we know, this is the first report of an archaeal community dominated by the MSP41 group, as well as the first one recording the co-occurrence of Nanohaloarchaeota and Parvarchaeota.
Conclusions
To our knowledge, the current metabarcoding study represents the first attempt to use deep amplicon sequencing to assess the extent of prokaryotic diversity occurring in hypersaline meromictic lakes. The employed polyphasic methodology provided a powerful snapshot of the vertical distribution of archaeal and bacterial communities, and could serve further as a framework for functional studies.
Even though both lakes are meromictic and extremely saline, their distinctive subtle limnological variances were found to be reflected in their prokaryotic communities' structure and composition. Thus, Ursu Lake was found to be dominated by Bacteria and to have a greater prokaryotic diversity than Fara Fund Lake that harbored an increased cell density and was dominated mostly by Archaea (within the oxic strata). Regardless of the energetic constraints imposed by the high ionic strength, both lakes were found to be colonized by diverse microbial communities whose physiological inferences generally pointed toward organic carbon degradation and sulfur reduction pathways.
In spite of the high salinity, the Bacteria were found to be more diverse than Archaea, indicating that our current view on microbial diversity in hypersaline ecosystems needs to be broadened. We also found that the prokaryotic communities harbored a wide variety of taxa, and contained a large proportion of OTUs with no close relatives, showing that these poorly investigated environments are potential sources of novel microorganisms. By matching taxa to known taxon-specific biogeochemical functions, we were able to sketch parts of biogeochemical cycles, but further functional studies are needed to describe nutrient cycles near the thermodynamic limits of life.
Acknowledgments
This work was supported by grants of the Romanian National Authority for Scientific Research, CNCS–UEFIS-CDI, project numbers PN-II-ID-PCE-2011-3-0546 and PN-II-ID-PCE-2011-3-0765. A-ŞA was supported by a POSDRU/159/1.5/S/132400 research scholarship; CC was supported by Grants PN 09-360201 and POSDRU/159/1.5/S/133391; AI was supported by a POSDRU/159/1.5/S/133391 doctoral scholarship; MSR and MP were supported by Oak Ridge National Laboratory (ORNL). ORNL is managed by UT-Battelle, LLC, for the US Department of Energy. We thank Zamin Yang and Dawn Klingeman for the help provided with Illumina amplicon preparation and sequencing. We are grateful to Tudor Tămaş for the mineralogical analysis, Zsolt G Keresztes for supporting unpublished data and to Dimitry Y Sorokin for his critical review of the manuscript. We are grateful to Daniela Buta (Ocna Sibiului) and Nagy Fülop János (Sovata) for the permission to enter the study areas.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alexe M. (2010) Studiul Lacurilor Sărate Din Depresiunea Transilvaniei ed Presa Universitară Clujeană: Cluj-Napoca, Romania, (in Romanian).
- Andrei A-Ş, Banciu HL, Oren A. (2012). Living with salt: metabolic and phylogenetic diversity of archaea inhabiting saline ecosystems. FEMS Microbiol Lett 330: 1–9. [DOI] [PubMed] [Google Scholar]
- Auguet JC, Triado-Margarit X, Nomokonova N, Camarero L, Casamayor EO. (2012). Vertical segregation and phylogenetic characterization of ammonia-oxidizing Archaea in a deep oligotrophic lake. ISME J 6: 1786–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baati H, Guermazi S, Gharsallah N, Sghir A, Ammar E. (2010). Novel prokaryotic diversity in sediments of Tunisian multipond solar saltern. Res Microbiol 161: 573–582. [DOI] [PubMed] [Google Scholar]
- Baker BJ, Comolli LR, Dick GJ, Hauser LJ, Hyatt D, Dill BD et al. (2010). Enigmatic, ultrasmall, uncultivated archaea. Proc Natl Acad Sci USA 107: 8806–8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberán A, Casamayor EO. (2011). Euxinic freshwater hypolimnia promote bacterial endemicity in continental areas. Microb Ecol 61: 465–472. [DOI] [PubMed] [Google Scholar]
- Baricz A, Coman C, Andrei A-Ş, Muntean V, Keresztes ZG, Păusan M et al. (2014). Spatial and temporal distribution of archaeal diversity in meromictic, hypersaline Ocnei Lake (Transylvanian Basin, Romania). Extremophiles 18: 399–413. [DOI] [PubMed] [Google Scholar]
- Benlloch S, López-López A, Casamayor EO, Øvreås L, Goddard V, Daae FL et al. (2002). Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environ Microbiol 4: 349–360. [DOI] [PubMed] [Google Scholar]
- Biderre-Petit C, Jézéquel D, Dugat-Bony E, Lopes F, Kuever J, Borrel G et al. (2011). Identification of microbial communities involved in the methane cycle of a freshwater meromictic lake. FEMS Microbiol Ecol 77: 533–545. [DOI] [PubMed] [Google Scholar]
- Boehrer B, Schultze M. (2009). Density stratification and stability. In: Likens GE (ed) Encyclopedia of Inland Waters. Elsevier: Oxford, pp 583–593. [Google Scholar]
- Bonin AS, Boone DR. (2006). The order Methanobacteriales. In: Dworkin M, Falkow S, Rosenberg E, Schleifer K-H, Stackebrandt E (eds) The Prokaryotes, Volume 3: Archaea. Bacteria: Firmicutes, Actinomycetes. Springer-VerlagNew York, NY, USA, pp 231–243. [Google Scholar]
- Bowman JP, McCammon SA, Rea SM, McMeekin TA. (2000). The microbial composition of three limnologically disparate hypersaline Antarctic lakes. FEMS Microbiol Lett 183: 81–88. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010. a). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. (2010. b). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellen PV, Viollier E, Roychoudhury A. (1998). Biogeochemical cycles of manganese and iron at the oxic-anoxic transition of a stratified marine basin (Orca Basin, Gulf of Mexico). Environ Sci Technol 32: 2931–2939. [Google Scholar]
- Clesceri LS, Greenberg AE, Eaton AD. (1999) Standard Methods for the Examination of Water and Wastewater, 20th edn. APHA, AWWA, WEF: Washington, USA. [Google Scholar]
- Comeau AM, Harding T, Galand PE, Vincent WF, Lovejoy C. (2012). Vertical distribution of microbial communities in a perennially stratified Arctic lake with saline, anoxic bottom waters. Sci Rep 2: 604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristea A, Andrei A-Ş, Baricz A, Muntean V, Banciu HL. (2014). Rapid assessment of carbon substrate utilization in the epilimnion of meromictic Ursu Lake (Sovata, Romania) by the BIOLOG EcoplateTM approach. Studia UBB Biologia 59: 41–53. [Google Scholar]
- Demergasso C, Escudero L, Casamayor EO, Chong G, Balagué V, Pedrós-Alió C. (2008). Novelty and spatio-temporal heterogeneity in the bacterial diversity of hypersaline Lake Tebenquiche (Salar de Atacama). Extremophiles 12: 491–504. [DOI] [PubMed] [Google Scholar]
- Edberg F, Andersson AF, Holmström SJ. (2012). Bacterial community composition in the water column of a lake formed by a former uranium open pit mine. Microb Ecol 64: 870–880. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996–998. [DOI] [PubMed] [Google Scholar]
- Einen J, Thorseth IH, Ovreås L. (2008). Enumeration of Archaea and Bacteria in seafloor basalt using real-time quantitative PCR and fluorescence microscopy. FEMS Microbiol Lett 282: 182–187. [DOI] [PubMed] [Google Scholar]
- Etiope G, Feyzullayev A, Baciu CL. (2009). Terrestrial methane seeps and mud volcanoes: a global perspective of gas origin. Mar Petrol Geol 26: 333–344. [Google Scholar]
- Ferrer M, Werner J, Chernikova TN, Bargiela R, Fernández L, La Cono V et al. (2012). Unveiling microbial life in the new deep-sea hypersaline Lake Thetis. Part II: a metagenomic study. Environ Microbiol 14: 268–281. [DOI] [PubMed] [Google Scholar]
- Frontier S. (1985). Diversity and structure in aquatic ecosystems. Oceanogr Mar Biol 23: 253–312. [Google Scholar]
- Ghai R, Pašić L, Fernández AB, Martin-Cuadrado A-B, Mizuno CM, McMahon KD et al. (2011). New abundant microbial groups in aquatic hypersaline environments. Sci Rep 1: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habicht KS, Miller M, Cox RP, Frigaard N-U, Tonolla M, Peduzzi S et al. (2011). Comparative proteomics and activity of a green sulfur bacterium through the water column of Lake Cadagno, Switzerland. Environ Microbiol 13: 203–215. [DOI] [PubMed] [Google Scholar]
- Hamilton TL, Bovee RJ, Thiel V, Sattin SR, Mohr W, Schaperdoth I et al. (2014). Coupled reductive and oxidative sulfur cycling in the phototrophic plate of a meromictic lake. Geobiology 12: 451–468. [DOI] [PubMed] [Google Scholar]
- Hammer UT. (1986) Saline Lake Ecosystems of the World vol. 59. Dr W Junk Publishers: Dordrecht, The Netherlands, p 15. [Google Scholar]
- Har N, Rusz O, Codrea V, Barbu O. (2010). New data on the mineralogy of the salt deposit from Sovata (Mureş County-Romania). Carpath J Earth Env 5: 127–135. [Google Scholar]
- Hollister EB, Engledow AS, Hammett AJM, Provin TL, Wilkinson HH, Gentry TJ. (2010). Shifts in microbial community structure along an ecological gradient of hypersaline soils and sediments. ISME J 4: 829–838. [DOI] [PubMed] [Google Scholar]
- Humayoun SB, Bano N, Hollibaugh JT. (2003). Depth distribution of microbial diversity in Mono Lake, a meromictic soda lake in California. Appl Environ Microbiol 69: 1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtgen MT, Lyons TW, Ingall ED, Cruse AM. (1999). Anomalous enrichments of iron monosulfide in euxinic marine sediments and the role of H2S in iron sulfide transformations: examples from Effingham Inlet, Orca Basin, and the Black Sea. Am J Sci 299: 556–588. [Google Scholar]
- Huson DH, Scornavacca C. (2012). Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol 61: 1061–1067. [DOI] [PubMed] [Google Scholar]
- Ionescu A. (2015). Geogenic methane in petroliferous and geothermal areas in Romania: origin and emission to the atmosphere. PhD thesis, Faculty of Environmental Science and Engineering. Babes-Bolyai University: Cluj-Napoca. [Google Scholar]
- Iversen N, Oremland RS, Klug MJ. (1987). Big Soda Lake (Nevada).3. Pelagic methanogenesis and anaerobic methane oxidation. Limnol Oceanogr 32: 804–814. [Google Scholar]
- Kelley CA, Poole JA, Tazaz AM, Chanton JP, Bebout BM. (2012). Substrate limitation for methanogenesis in hypersaline environments. Astrobiology 12: 89–97. [DOI] [PubMed] [Google Scholar]
- Keresztes ZG, Felföldi T, Somogyi B, Székely G, Dragoş N, Márialiget K et al. (2012). First record of picophytoplankton diversity in Central European hypersaline lakes. Extremophiles 16: 759–769. [DOI] [PubMed] [Google Scholar]
- La Cono V, La Spada G, Arcadi E, Placenti F, Smedile F, Ruggeri G et al. (2013). Partaking of Archaea to biogeochemical cycling in oxygen-deficient zones of meromictic saline Lake Faro (Messina, Italy). Environ Microbiol 15: 1717–1733. [DOI] [PubMed] [Google Scholar]
- Lane DJ. (1991). 16S/23S rRNA sequencing. In Stackebrandt E, Goodfellow M (eds) Nucleic Acid Techniques in Bacterial Systematics. John Wiley & Sons Ltd: United Kingdom, pp 115–175. [Google Scholar]
- Lauro FM, DeMaere MZ, Yau S, Brown MV, Ng C, Wilkins D et al. (2011). An integrative study of a meromictic lake ecosystem in Antarctica. ISME J 5: 879–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentini V, Gugliandolo C, Maugeri TL. (2012). Vertical distribution of Archaea and Bacteria in a meromictic lake as determined by fluorescent in situ hybridization. Curr Microbiol 64: 66–74. [DOI] [PubMed] [Google Scholar]
- Lepère C, Masquelier S, Mangot J-F, Debroas D, Domaizon I. (2010). Vertical structure of small eukaryotes in three lakes that differ by their trophic status: a quantitative approach. ISME J 4: 1509–1519. [DOI] [PubMed] [Google Scholar]
- Lopes F, Viollier E, Thiam A, Michard G, Abril G, Groleau A et al. (2011). Biogeochemical modeling of anaerobic vs. aerobic methane oxidation in a meromictic crater lake (Lake Pavin, France). Appl Geochem 26: 1919–1932. [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL. (2013). Practical innovations for high-throughput amplicon sequencing. Nat Methods 10: 999–1002. [DOI] [PubMed] [Google Scholar]
- Marteinsson VT, Rúnarsson Á, Stefánsson A, Thorsteinsson T, Jóhannesson T et al. (2013). Microbial communities in the subglacial waters of the Vatnajökull ice cap, Iceland. ISME J 7: 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12. [Google Scholar]
- Máthé I, Borsodi AK, Tóth EM, Felföldi T, Jurecska L, Krett G et al. (2014). Vertical physico-chemical gradients with distinct microbial communities in the hypersaline and heliothermal Lake Ursu (Sovata, Romania). Extremophiles 18: 501–514. [DOI] [PubMed] [Google Scholar]
- McDonald D, Clemente JC, Kuczynski J, Rideout JR, Stombaugh J, Wendel D et al. (2012. a). The Biological Observation Matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A et al. (2012. b). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G, Waal EC, Uitterlinden AG. (1993). Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol 59: 695–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasingarao P, Podell S, Ugalde JA, Brochier-Armanet C, Emerson JB, Brocks JJ et al. (2012). De novo metagenomic assembly reveals abundant novel major lineage of Archaea in hypersaline microbial communities. ISME J 6: 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. (2011). Thermodynamic limits to microbial life at high salt concentrations. Environ Microbiol 13: 1908–1923. [DOI] [PubMed] [Google Scholar]
- Oren A. (2012). Approaches toward the study of halophilic microorganisms in their natural environments: who are they and what are they doing? In: Vreeland RH (ed) Advances in Understanding the Biology of Halophilic Microorganisms. Springer: New York, pp 1–33. [Google Scholar]
- Pasche N, Dinkel C, Müller B, Schmid M, Wüest A, Wehrli B. (2009). Physical and biogeochemical limits to internal nutrient loading of meromictic Lake Kivu. Limnol Oceanogr 54: 1863–1873. [Google Scholar]
- Pouliot J, Galand PE, Lovejoy C, Vincent WF. (2009). Vertical structure of archaeal communities and the distribution of ammonia monooxygenase A gene variants in two meromictic High Arctic lakes. Environ Microbiol 11: 687–699. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. (2010). FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Peplies J, Glöckner FO. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28: 1823–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: (Database issue) D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437. [DOI] [PubMed] [Google Scholar]
- Roden EE, Lovley DR. (1993). Dissimilatory Fe(III) reduction by the marine microorganism Desulfuromonas acetoxidans. Appl Environ Microbiol 59: 734–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert CJ, Lucas FS, Durisch-Kaiser E, Stierli R, Diem T, Scheidegger O et al. (2010). Oxidation and emission of methane in a monomictic lake (Rotsee, Switzerland). Aquat Sci 72: 455–466. [Google Scholar]
- Somogyi B, Vörös L, Pálffy K, Székely G, Bartha C, Keresztes ZG. (2014). Picophytoplankton predominance in hypersaline lakes (Transylvanian Basin, Romania). Extremophiles 18: 1075–1084. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Foti M, Tindall BJ, Muyzer G. (2007). Desulfurispirillum alkaliphilum gen. nov. sp. nov., a novel obligately anaerobic sulfur-and dissimilatory nitrate-reducing bacterium from a full-scale sulfide-removing bioreactor. Extremophiles 11: 363–370. [DOI] [PubMed] [Google Scholar]
- Sorokin DY, Tourova TP, Galinski EA, Muyzer G, Kuenen JG. (2008). Thiohalorhabdus denitrificans gen. nov., sp. nov., an extremely halophilic, sulfur-oxidizing, deep-lineage gammaproteobacterium from hypersaline habitats. Int J Syst Evol Microbiol 58: 2890–2897. [DOI] [PubMed] [Google Scholar]
- Spulber L, Etiope G, Baciu C, Malos C, Vlad SN. (2010). Methane emission from natural gas seeps and mud volcanoes in Transylvania (Romania). Geofluids 10: 463–475. [Google Scholar]
- Tang XM, Gao G, Qin B, Zhu L, Chao J, Yang G. (2009). Characterization of bacterial communities associated with organic aggregates in a large, shallow, eutrophic freshwater lake (Lake Taihu, China). Microb Ecol 58: 307–322. [DOI] [PubMed] [Google Scholar]
- Trüper HG, Schlegel HG. (1964). Sulfur metabolism in Thiorhodaceae. 1. Quantitative measurements on growing cells of Chromatium okenii. Antonie van Leeuwenhoek 30: 225–238. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. (2007). Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JJ, Koren O, Hugenholtz P, DeSantis TZ, Walters WA, Caporaso JG et al. (2011). Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J 6: 94–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetzel RG, Likens GE. (1991) Limnological Analyses. Springer-Verlag: New York, p 39. [Google Scholar]
- Wetzel RG, Likens GE. (1995) Limnological Analysis, 2nd edn. Springer-Verlag: New York, NY, USA. [Google Scholar]
- Whiticar MJ, Faber E, Schoell M. (1986). Biogenic methane formation in marine and freshwater environments: CO2 reduction vs acetate fermentation – isotope evidence. Geochim Cosmochim Acta 50: 693–709. [Google Scholar]
- Yakimov MM, La Cono V, Slepak VZ, La Spada G, Arcadi E, Messina E et al. (2013). Microbial life in the Lake Medee, the largest deep-sea salt-saturated formation. Sci Rep 3: 3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau S, Lauro FM, Williams TJ, Demaere MZ, Brown MV, Rich J et al. (2013). Metagenomic insights into strategies of carbon conservation and unusual sulfur biogeochemistry in a hypersaline Antarctic lake. ISME J 7: 1944–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhaxybayeva O, Stepanauskas R, Mohan NR, Papke RT. (2013). Cell sorting analysis of geographically separated hypersaline environments. Extremophiles 17: 265–275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.