Abstract
Proteorhodopsins (PR) are retinal-binding membrane proteins that function as light-driven proton pumps to generate energy for metabolism and growth. Recently PR-like genes have been identified in some marine eukaryotic protists, including diatoms, dinoflagellates, haptophytes and cryptophytes. These rhodopsins are homologous to green-light-absorbing, ATP-generating PRs present within bacteria. Here we show that in the oceanic diatom Pseudo-nitzschia granii, PR-like gene and protein expressions increase appreciably under iron limitation. In a survey of available transcriptomes, PR-like genes in diatoms are generally found in isolates from marine habitats where seasonal to chronic growth limitation by the micronutrient iron is prevalent, yet similar biogeographical patterns are not apparent in other phytoplankton taxa. We propose that rhodopsin-based phototrophy could account for a proportion of energy synthesis in marine eukaryotic photoautotrophs, especially when photosynthesis is compromised by low iron availability. This alternative ATP-generating pathway could have significant effects on plankton community structure and global ocean carbon cycling.
Microbial rhodopsins are a large family of proteins capable of carrying out solar energy conversion or photosensory transduction in diverse prokaryotic and eukaryotic microorganisms. Although proton-pumping rhodopsins were first identified in halophilic archaea (Oesterhelt and Stoeckenius, 1971), it was their discovery in marine heterotrophic bacteria that gained attention by challenging the assumption of chlorophyll a as the single most important light-capturing pigment in ocean surface waters (Beja et al., 2000). All rhodopsins contain retinal as their chromophore, which is bound to a lysine residue inside seven transmembrane helices. The retinal linkage to a protonated Schiff base is important to the function of all types of rhodopsins. Upon illumination, the bound retinal molecule undergoes light-induced isomerization that results in either the translocation of a proton (or other ion such as chloride) across the membrane or in signaling to transducer proteins (Spudich et al., 2000).
The response of eukaryotic phytoplankton to iron enrichment in iron-limited waters of the NE Pacific Ocean was recently investigated using comparative metatranscriptome analyses (Marchetti et al., 2012). Among the genes differentially expressed in the iron-enriched community relative to the ambient, iron-limited community were several eukaryotic phytoplankton-associated PR-like genes that were highly expressed. Most notably, PR-like genes in diatoms, an important member of the ambient phytoplankton community and dominant after the addition of iron, were highly expressed under iron limitation. Diatoms account for as much as half of total marine primary productivity, yet in vast regions of our oceans, their growth is limited by iron. In iron-limited diatoms, 50–100% of the cellular iron requirement is within components of the photosynthetic electron carriers (Raven et al., 1999; Strzepek and Harrison, 2004). Therefore, the use of PR, which requires substantially less iron than photosynthesis, for ATP production would be advantageous under such conditions and is consistent with these findings as well as those hypothesized by others (Raven, 2009).
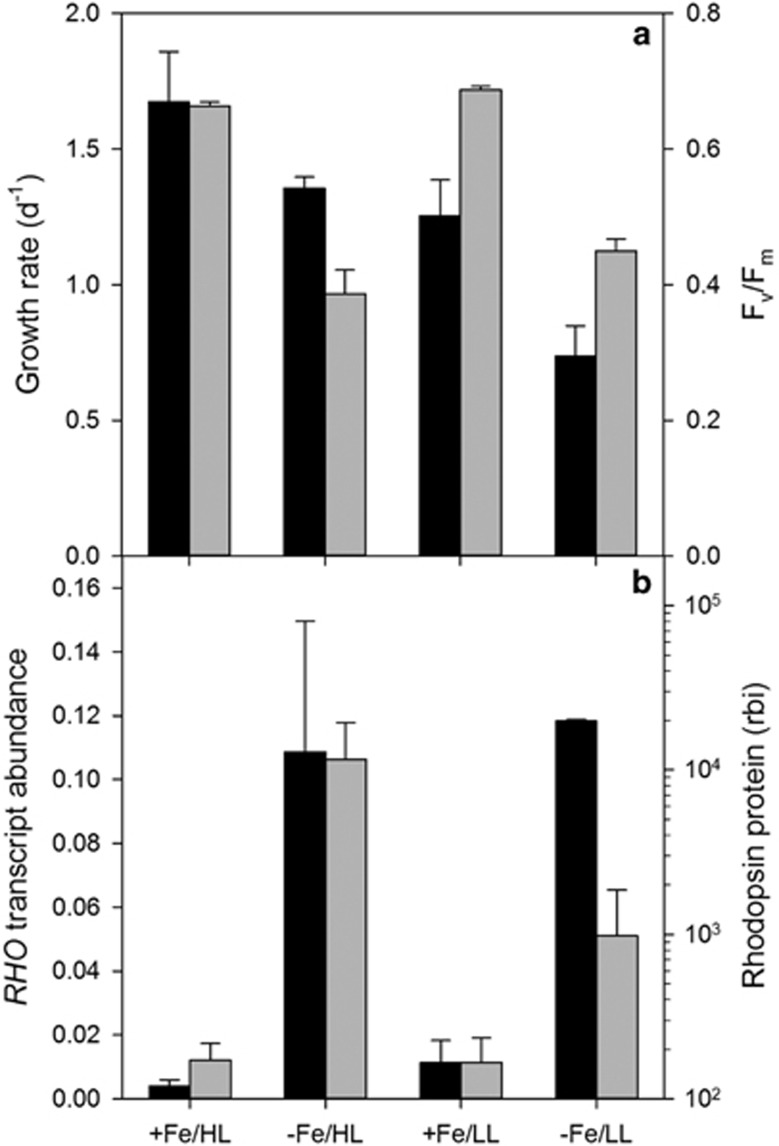
Subsequently, the sequencing of the transcriptome of the oceanic diatom Pseudo-nitzschia granii, a member of a diatom genus that commonly dominates after iron additions to high nutrient, low chlorophyll (HNLC) regions (de Baar et al., 2005), also revealed the presence of a PR-like gene. The P. granii rhodopsin gene (RHO) encodes for the expected seven transmembrane α-helices along with a retinal-binding pocket and a diagnostic residue required for proton transport, making it highly likely that the RHO is proton-pumping rather than sensory (Marchetti et al., 2012). We investigated whether iron and light limitation influenced the expression patterns of P. granii RHO and the subsequent protein abundance. Iron and/or light-limited P. granii both displayed a reduction in specific growth rate, whereas photosynthetic efficiency (Fv/Fm) was primarily affected by iron status (Figure 1a). The expression of P. granii RHO was significantly influenced by iron availability (P<0.001) but was not significantly affected by light availability either under iron-replete or iron-limited conditions (two-way analysis of variance). P. granii grown under iron-limited conditions exhibited an approximate order of magnitude increase in RHO transcript abundance relative to iron-replete conditions (Figure 1b). Similar expression patterns were found at the protein level, confirming that the changes in gene expression have functional consequences (Figure 1b). These results suggest the increase in P. granii rhodopsin is specific to iron-limited growth conditions and is not a general response to growth rate reduction.
Figure 1.
Growth characteristics and rhodopsin gene expression and protein abundance in P. granii. (a) P. granii growth rates (black bars) and Fv/Fm (gray bars) under Fe-replete, high light (+Fe/HL), Fe-limited, high light (−Fe/HL), Fe-replete, low light (+Fe/LL) and Fe-limited, low light (+Fe/LL) conditions. (b) P. granii RHO transcript abundance (relative to actin; black bars) and rhodopsin protein abundance (gray bars) under the growth conditions described in a. Protein abundances are provided in relative band intensities. Error bars are the standard deviations of the means (n=3 for transcript abundance and n=2 for protein abundance). See Supplementary Information for methods.
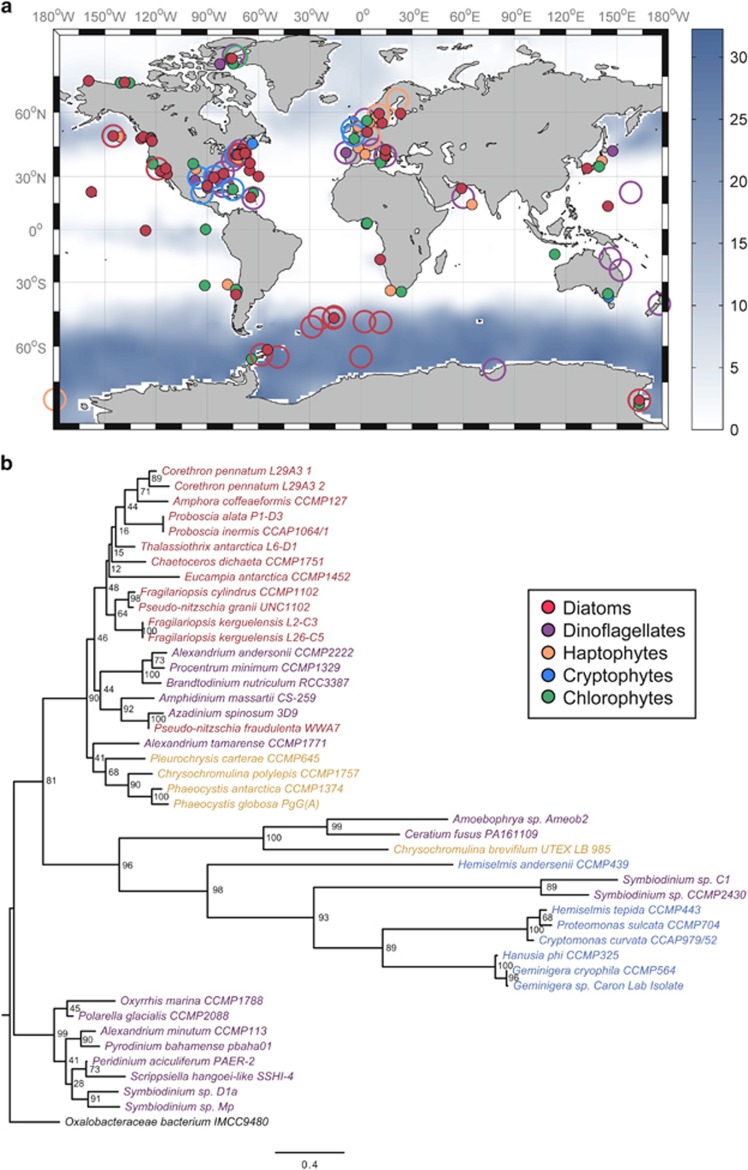
To gain a biogeographical perspective on eukaryotic PR distributions, we searched sequenced genomes and transcriptomes of marine protists isolated throughout the world's oceans, taking advantage of the greatly expanded species coverage offered by the Marine Microbial Eukaryote Transcriptome Sequencing Project (Keeling et al., 2014). There appears to be a geographical influence on distributions of PR-containing diatoms, as the majority of these isolates are from marine environments where seasonal to widespread chronic iron limitation is prevalent, such as in the Southern Ocean, the North Pacific Ocean and California coastal upwelling regime (Figure 2a). PR genes were found in other members of the eukaryotic phytoplankton including dinoflagellates (Lin et al., 2010; Slamovits et al., 2011), cryptophytes and haptophytes, but their distribution within these groups did not appear related to iron availability, although for some taxa a scarcity of isolates from regions of low iron availability limits our ability to make firm conclusions.
Figure 2.
Biogeography and phylogenetic analysis of rhodopsin-containing marine protists. (a) The locations where strains have been isolated in which a rhodopsin gene homolog is present (large open circles) or absent (small closed circles) within their transcriptomes or whole-genome sequences. Isolates are grouped according to phylum, with the exception of including a diatom grouping. Background represents climatological annual surface nitrate concentration (μmoll−1) where high surface nitrate concentrations often reflect iron limitation. Nutrient data are from the World Ocean Atlas (Garcia et al., 2009). (b) Maximum likelihood tree of assembled rhodopsin genes from marine protists. Species name colors correspond to groups as listed in a. Node labels show percent consensus support for tree arrangement and branch length represents the amount of divergence between nodes (substitutions/site). Proteorhodopsin from the proteobacterium Oxalobacteraceae bacterium IMCC9880 was used as the outgroup. See Supplementary Information for methods.
Rhodopsin-like genes in marine protists are most structurally similar to xanthorhodopsins from bacteria that contain a light-harvesting carotenoid antenna. When compared with prokaryotes, all eukaryotic PR genes group together, thus making them appear to be monophyletic (Figure 2b and Figure 5 in Marchetti et al., 2012). Their similarity to PRs within bacteria along with the dispersed presence amongst protist species suggests an initial acquisition from bacteria through horizontal gene transfer followed by subsequent gene losses. Acquisition of this gene may have been a critical evolutionary step towards survival in iron-limited regions for the diverse diatom taxa that have adapted to thrive in these waters (for example, Pseudo-nitzschia, Fragilariopsis, Corethron, Proboscia and so on). The observed trend of PR-containing diatoms being primarily confined to HNLC regions of the oceans (Figure 2a) is highly supportive of the idea that they may have a competitive advantage in low iron environments due to the acquisition of PR.
Taken together, our findings suggest a potential role for diatom PRs under iron limitation could be to produce ATP for continued survival until iron becomes available for growth. If a source of reductant other than water is readily available (Fuhrman et al., 2008), synthesis of ATP by PR might also provide an alternative means of carbon fixation independent of O2-evolving photosynthesis. A direct and significant influence of PR on carbon cycling independent of O2-evolving mechanisms will be contingent on finding an alternative and suitable inorganic electron donor at the ocean surface with sufficient abundance or turn over to influence carbon fixation. However, PR could also influence carbon cycling indirectly by maintaining seed populations of PR-containing diatoms under chronic iron stress. Thus rhodopsin-based phototrophy in diatoms and other eukaryotic phytoplankton could have far-reaching implications for our estimates of carbon fixation derived through O2 super-saturation measurements or indirectly through preferentially selecting rhodopsin-containing phytoplankton. Future studies should further elucidate the distributions of PR within marine eukaryotic phytoplankton and the extent to which rhodopsin-based phototrophy is used relative to O2-evolving photosynthesis, which if shown to be significant, could ultimately necessitate a rethinking of our understanding of the major metabolic processes affecting the ocean carbon cycle.
Acknowledgments
MMETSP was funded in part by the Gordon and Betty Moore Foundation through grant no. 2637 to the National Center for Genome Resources. Culture work was funded in part by NSF EAGER grant OCE0946260 to EV Armbrust and AM. Protein work was supported by NSF grant 1041034 to BMH. We thank I Oleinikov and C Moreno for assistance with diatom culturing.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Beja O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP et al. (2000). Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289: 1902–1906. [DOI] [PubMed] [Google Scholar]
- de Baar HJW, Boyd PW, Coale KH, Landry MR, Tsuda A, Assmy P et al. (2005). Synthesis of iron fertilization experiments: from the iron age in the age of enlightenment. J Geophys Res-Oceans 110: C09S16. [Google Scholar]
- Fuhrman JA, Schwalbach MS, Stingl U. (2008). Proteorhodopsins: an array of physiological roles? Nat Rev Microbiol 6: 488–494. [DOI] [PubMed] [Google Scholar]
- Garcia HE, Locarmini RA, Boyer TP, Antonov JI, Zweng MM, Baranova OK et al. (2009). World Ocean Atlas, Volume 4: nutrients (phosphate, nitrate, silicate). In: Levitus S (ed.), NOAA Atlas NESDIS 71. USA Government Printing Office: Washington, DC, USA, p 398. [Google Scholar]
- Keeling PJ, Burki F, Wilcox HM, Allam B, Allen EE, Amaral-Zettler LA et al. (2014). The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): illuminating the functional diversity of eukaryotic ife in the oceans through transcriptome sequencing. PLoS Biol 12: e1001889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Zhang H, Zhuang Y, Tran B, Gill J. (2010). Spliced leader-based metatranscriptomic analyses lead to recognition of hidden genomic features in dinoflagellates. Proc Natl Acad Sci USA 107: 20033–20038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti A, Schruth DM, Durkin CA, Parker MS, Kodner RB, Berthiaume CT et al. (2012). Comparative metatranscriptomics identifies molecular bases for the physiological responses of phytoplankton to varying iron availability. Proc Natl Acad Sci USA 109: E317–E325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oesterhelt D, Stoeckenius W. (1971). Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nat New Biol 233: 149–152. [DOI] [PubMed] [Google Scholar]
- Raven JA. (2009). Functional evolution of photochemical energy transformations in oxygen-producing organisms. Funct Plant Biol 36: 505–515. [DOI] [PubMed] [Google Scholar]
- Raven JA, Evans MCW, Korb RE. (1999). The role of trace metals in photosynthetic electron transport in O2-evolving organisms. Photosynth Res 60: 111–149. [Google Scholar]
- Slamovits CH, Okamoto N, Burri L, James ER, Keeling PJ. (2011). A bacterial proteorhodopsin proton pump in marine eukaryotes. Nat Commun 2: 183. [DOI] [PubMed] [Google Scholar]
- Spudich JL, Yang CS, Jung KH, Spudich EN. (2000). Retinylidene proteins: Structures and functions from archaea to humans. Annu Rev Cell Dev Biol 16: 365–392. [DOI] [PubMed] [Google Scholar]
- Strzepek R, Harrison P. (2004). Photosynthetic architecture differs in coastal and oceanic diatoms. Nature 431: 689–692. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.