Abstract
Wood falls on the ocean floor form chemosynthetic ecosystems that remain poorly studied compared with features such as hydrothermal vents or whale falls. In particular, the microbes forming the base of this unique ecosystem are not well characterized and the ecology of communities is not known. Here we use wood as a model to study microorganisms that establish and maintain a chemosynthetic ecosystem. We conducted both aquaria and in situ deep-sea experiments to test how different environmental constraints structure the assembly of bacterial, archaeal and fungal communities. We also measured changes in wood lipid concentrations and monitored sulfide production as a way to detect potential microbial activity. We show that wood falls are dynamic ecosystems with high spatial and temporal community turnover, and that the patterns of microbial colonization change depending on the scale of observation. The most illustrative example was the difference observed between pine and oak wood community dynamics. In pine, communities changed spatially, with strong differences in community composition between wood microhabitats, whereas in oak, communities changed more significantly with time of incubation. Changes in community assembly were reflected by changes in phylogenetic diversity that could be interpreted as shifts between assemblies ruled by species sorting to assemblies structured by competitive exclusion. These ecological interactions followed the dynamics of the potential microbial metabolisms accompanying wood degradation in the sea. Our work showed that wood is a good model for creating and manipulating chemosynthetic ecosystems in the laboratory, and attracting not only typical chemosynthetic microbes but also emblematic macrofaunal species.
Introduction
Deep-sea chemosynthetic ecosystems build upon the use of reduced chemicals by microorganisms and develop on and around benthic features such as hydrothermal vents, cold seeps, whale falls or woods falls (Jorgensen and Boetius, 2007). Woods transported from land to sea sink once waterlogged and create unique ecosystems on the ocean floor. The refractory nature of wood, which becomes quickly anaerobic in seawater (Yucel et al., 2013), leads to a slow degradation of its organic matter and the production of sulfide (Laurent et al., 2013) typical for chemosynthetic ecosystems. Wood falls ecosystems have been the focus of a number of research projects as they are thought to have a key role in the dispersal and evolution of deep-sea chemosynthetic organisms (Distel et al., 2000; Kiel et al., 2009), and because they are home for an emblematic fauna harboring microbial symbionts (Dubilier et al., 2008). The free-living microorganisms that digest wood or produce sulfide remain, however, poorly studied.
Little is currently known about microbial communities that colonize the wood itself. Wood fall bacterial communities that were early characterized according to their physical action on the wood matrix (Mouzouras et al., 1988; Jurgens et al, 2003), and first described through the isolation of cultivable representatives (Cundell and Mitchell, 1977; Austin et al., 1979), were precisely identified only recently by molecular tools. A large cloning and sequencing effort gave the first insights into the diversity of natural wood fall bacterial and archaeal assemblages (Fagervold et al., 2012). It demonstrated the presence of sequences associated to possible fermenters and sulfide-oxidizing bacteria, as well as the co-occurrence of free-living sulfate-reducing bacteria. Archaea have also been detected on wood falls, especially on samples with substantial signs of decay (Palacios et al., 2009). They are potentially involved in the production of methane (Fagervold et al., 2012), but the observation of the association of a giant Thaumarchaeota with sulfur-oxidizing bacteria (Muller et al., 2010) suggests the presence of diverse archaeal metabolisms on wood falls. The first molecular descriptions of wood microbes gave clues to potential metabolisms associated with the degradation of wood in the marine environment. Cellulose could be hydrolyzed into simple sugars by bacteria or fungi. Marine fungi have the potential to degrade cellulose and lignin (Raghukumar et al., 1994; Pointing et al., 1998), but their presence on marine wood falls has rarely been documented (Raghukumar, 2012; Rämä et al., 2014). Simple sugars will in turn be fermented to small carbon molecules that could then be used by sulfate reducers and methanogens, which represent the last step of the anaerobic degradation of organic matter. Finally, the sulfide produced by sulfate-reducing bacteria can be re-oxidized aerobically or with nitrate as electron acceptor, thus setting the basis for the wood fall chemosynthetic ecosystems.
The first insight into the ecology of wood falls bacteria communities was published recently through experimental deployments of woods in the sea (Palacios et al., 2009; Bienhold et al., 2013; Fagervold et al., 2013). The results showed how local environmental features could influence bacterial community assembly, but contrasting results have been reported. In one case, similar communities were present in woods situated at geographically distant sites and at different depths of immersion (Palacios et al., 2009). While in other cases, the location of the wood inside a submarine canyon (Fagervold et al., 2013; Bessette et al., 2014), in the vicinity of carbonate crust (Bienhold et al., 2013), or its colonization by wood-boring Xylophaga bivalves (Fagervold et al., 2014) strongly structured the microbial communities. The type of wood may also be important in structuring sunken wood bacterial communities (Palacios et al., 2009; Fagervold et al., 2013). Data on community ecology remain, however, rare, as they require experimental approaches that are particularly challenging in the deep sea. Evidences of possible differences in community composition between the different parts of the wood are, for instance, lacking, and there are no direct evidences of temporal succession in the microbial communities. Previous studies relied on samples collected solely after several month of immersion (Palacios et al., 2009; Bienhold et al., 2013; Fagervold et al., 2013), which made it difficult to study the temporal dynamics of microbial wood colonization and community assembly.
Microbial community assembly can theoretically be dictated by neutral processes, such as genetic drift or immigration of species with similar traits and fitness, but assembly can also be ruled by species sorting, which is the selection of species by environmental conditions (Langenheder and Székely, 2011). We hypothesize that in wood fall ecosystems, the local environmental conditions are key factors for structuring communities with time, as the degradation of organic matter by bacterial consortia, and the production of reduced chemicals will continuously transform the habitat.
The major aim of this study was to determine how environmental conditions dictate community assembly during the colonization and establishment of a deep-sea chemosynthetic ecosystem. We use wood as a model ecosystem that can easily be manipulated and monitored in aquaria, and compared the results to in situ experiments from deep-sea deployments. In order to encompass diverse environmental constraints on community assembly, we focus on both a spatial scale, represented by wood microhabitats (wood bark, inside the wood and surface of the wood), and temporal scale that is the immersion time and the effect of wood species (pine vs oak). We also compared woods from a natural environment (submarine canyon) with woods from an artificial environment (aquarium). We used pyrosequencing for a precise description of the bacterial, archaeal and fungal community structure and diversity, and measured changes in wood lipid content and sulfide concentrations to assess the potential activity of microorganisms.
Material and methods
Experimental design
Aquaria experiment
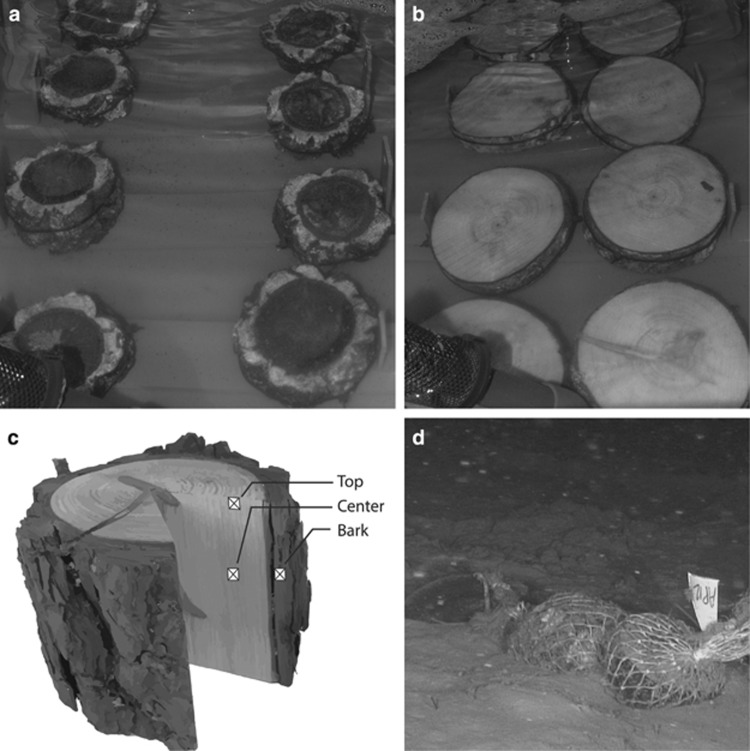
Wood pieces used for the experiments originated from pine and oak trees planted in 1980s at the Banyuls sur Mer botanical garden. Eight pieces of oak (Quercus suber) and eight pieces of pine (Pinus pinea) woods (10 × 16 cm; Figures 1a and b) cut from a tree branch were incubated in tanks filled with natural seawater at 13±1 °C in the dark, simulating the temperature and light conditions of the deep Mediterranean Sea (Zavatarielli and Mellor, 1995). These tanks were filled continuously with oxygenated water pumped directly 30 m away from the coast at 4 m depth.
Figure 1.
Experimental design with eight pieces of oak (a) and eight pieces of pine (b) wood incubated in the dark in aquaria filled with natural seawater thermostated at 13 °C. (c) Position of the different microhabitat wood samples: the bark sample is at the interface between bark and seawater, the top sample is at the interface between sapwood and seawater and the center sample has not been in direct contact with seawater. (d) Pine and oak woods deployed in situ at 520 m depth in the Lacaze-Duthiers canyon in the NW Mediterranean Sea.
Pieces of wood were collected five times during a 307-day incubation period after 30, 78, 121, 240 and 307 days. Wood was sampled with a saw washed with ethanol and rinsed with sterile water. Samples were collected from the following three locations on each wood piece: center, surface and bark (Figure 1c). These three types of samples were chosen to represent different potential microbial microhabitats. The wood center was not in direct contact with seawater, the surface constituted an interface between seawater and wood, and finally the bark, also at the interface of seawater, which had a different chemical composition than wood. Samples were frozen immediately after collection in liquid nitrogen and stored at −80 °C.
In situ experiment
Twelve pieces of pine and oak wood cut from the same tree as the one used for the aquaria experiment were immersed in the Lacaze–Duthiers canyon in the Gulf of Lions (NW Mediterranean Sea), 25 km away from shore (42° 32° 26°° N, 03° 25° 9°° E) at 520 m depth (Figure 1d) in November 2010. Wood immersion and recovery were conducted with the ROV ‘Super Achille' deployed from the research vessel Minibex. Three pieces of pine and three pieces of oak were recovered after 210 days and again after 365 days of immersion. The wood pieces were strongly degraded by wood-boring bivalves, thus we could not sample the three different wood microhabitats. Therefore, only the center was sampled following the same protocol as for the aquarium experiment and was stored immediately at −80 °C.
DNA extraction and pyrosequencing
Wood samples were powdered with a Tissuelyzer (RETSCH Mixer Mill, Retsch Inc., MM301; Palacios et al., 2009). Stainless steel grinding jars (25 ml, Retsch Inc., Newtown, PA, USA; MM 400), containing one stainless steel ball, and the sample were kept in liquid nitrogen for 5 min, then powdered for 1:30 min at a frequency of 25 Hz. This step was repeated three times. The powder was recovered and placed inside 2 ml Eppendorf tubes. One tube was used for DNA extraction and the other was reserved for biochemical analysis. DNA extraction was performed with the Power plant MoBio kit (MOBIO Laboratories, Carlsbad, CA, USA) following an adapted version of the MoBio protocol (Fagervold et al., 2013). The extraction was followed by a cleanup step by isopropanol precipitation before resuspending DNA in TE buffer.
A portion of the 16 S rRNA gene was amplified using the Bacteria-specific primers 28 F (5′-TTTGATCNTGGCTCAG-3′) and 519 R (5′-GTNTTACNGCGGCKGCTG-3′), and the Archaea primers 340 f (5′-CCTACGGGGYGCASCAG-3′ Ovreas et al., 1997) and 806r (5′-GGACTACNNGGGTATCTAAT-3′ Yu et al., 2005). A portion of the fungal internal transcribed spacer (ITS) was amplified with primers ITSF1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′ Gardes and Bruns, 1993) and ITS4R (5′-TCCTCCGCTTATTGATATGC-3′ White et al., 1990). We sequenced 42 samples for Bacteria (30 from the aquarium and 12 from the canyon). For Archaea, all samples were first screened by PCR and only 12 samples gave positive results and were sequenced (nine from the aquarium and three from the canyon). The same 12 samples were sequenced for Fungi (Supplementary Table 1). Pyrosequencing was conducted by a commercial laboratory (Research and Testing Laboratory, Lubbock, TX, USA) on a Roche 454 FLX (Brandford, CT, USA) using commercially prepared Titanium reagents. Sequences have been deposited to the GenBank Sequence Read archive under number SRR1564096 for bacterial sequences and SRR1958899 for archaeal and fungal sequences.
Sequence data analysis
Sequences were processed following the standard operating procedure in Mothur (Schloss et al., 2011). Briefly, sequences were denoised using Mothur's implementation of PyroNoise (Quince et al., 2011) with default parameters, and chimeras were removed using uchime (Edgar et al., 2011). Sequences were grouped into operational taxonomic units (OTUs) at a 97% similarity cutoff, and taxonomy was assigned using the Silva database (v. 108; Pruesse et al., 2007) for Bacteria and Archaea and the UNITE database (v6; Kõljalg et al., 2013) for Fungi, based on a representative sequence of each OTU, chosen as the most abundant sequence of an OTU. Finally, data sets were resampled down to an equal number of 1663 sequences per sample for Bacteria, 2000 sequences for Archaea and 750 sequences for Fungi. Three archaeal samples were excluded from the analysis because they had too few sequences (Supplementary Table 1).
Diversity metrics and statistics
We calculated rarefaction curves and the Shannon diversity index (H°) in Qiime (Caporaso et al., 2010b). We also estimated phylogenetic diversity by computing the Net Relatedness Index (NRI) (Webb, 2000) using the picante package in R (Kembel et al., 2010). NRI calculations are based on the mean pairwise distance that measures branch length between each OTU in a phylogenetic tree. The tree was build using representative sequence of each OTU aligned with Pynast (Caporaso et al., 2010a). The phylogenetic tree was built using the FastTree algorithm (Price et al., 2010). NRI significance was tested against a null model in which OTUs are randomly drawn from the sequence pool.
An unweighted pair group method with arithmetic mean cluster analysis was conducted based on the Bray–Curtis dissimilarity index. The strength of the cluster's nodes was assessed by jackknife analysis and the significant difference between larger clusters was tested by running a SIMPROF analysis (Clarke et al., 2008; Yoshioka, 2008). To test the effect of microhabitat and time on the bacterial communities, we used a permutational multivariate analysis of variance (ADONIS) using a Bray–Curtis distance matrices (Anderson, 2001). The significance of the test was verified with a Monte Carlo permutation (9999 times).
Analysis of variance, the Wilcoxon and the Mann–Whitney statistical test were computed to test whether lipid concentrations varied with time. Differences were considered significant when P<0.05. All statistics were computed with the R software (V 2.15.13, Vienna, Austria).
Association network
Associations between bacterial OTUs were characterized by computing the maximal information coefficient between each pair of abundant OTUs (Reshef et al., 2011). Maximal information coefficient provides a score that represents the strength of a relationship between data pairs. The matrix of maximal information coefficient values >0.4, which reduces the false discovery rate (P=5 × 10−5), and corresponding to positive linear correlations, was used in Cytoscape 3.1.0 to visualize the network of associations (Smoot et al., 2011) using the spring-embedded algorithm (Kamada and Kawai, 1989). Subnetworks of OTUs were defined with the hierarchical clustering algorithm HC-PIN (Wang et al., 2011) and the modularity of each subnetwork was calculated. Modularity is the sum of the differences between the number of edge within the subnetwork and the square number of edges that linked another subnetwork (Clauset et al., 2004).
Lipid concentrations
We quantified lipid concentrations to compare pine and oak wood characteristics. Lipids used as reserves characterize pine as a fat tree compared with oak that uses starch, a glucose polymer, for storage (Fengel and Wegener, 1984). Powdered woods were freeze dried before analysis and total lipids were determined for each sample on triplicates using the sulfophosphovanillin assay (Barnes and Blackstock, 1973). Briefly, lipids were extracted from 25 mg of dried material with 1.5 ml of chloroform/methanol (2v:1v) for 20 min. An aliquot of the lipid extract was evaporated to dryness and hydrolyzed with concentrated sulfuric acid. The phosphoric acid–vanillin reagent was added to 100 μl of hydrolysate and the optical density at 520 nm was read on a PerkinElmer Lambda 25 spectrophotometer (Waltham, MA, USA). Concentrations were estimated using cholesterol as a standard (Barnes and Blackstock, 1973). Variability within triplicates was on average 6.5% (ranging from 0.6 to 16%).
Sulfide measurements
Discrete measurements of sulfide were performed on wood pieces using a voltammetric three-electrode cell with Au/Hg electrode as working electrode (Luther et al., 2001). The electrode was placed in contact with the wood surface during these measurements. Each measurement consisted of five scans following a 10-s cleaning step, and five such measurements were performed on a chosen point on the experimental substrate (Yucel et al., 2013).
Results
Wood lipid and sulfide concentrations
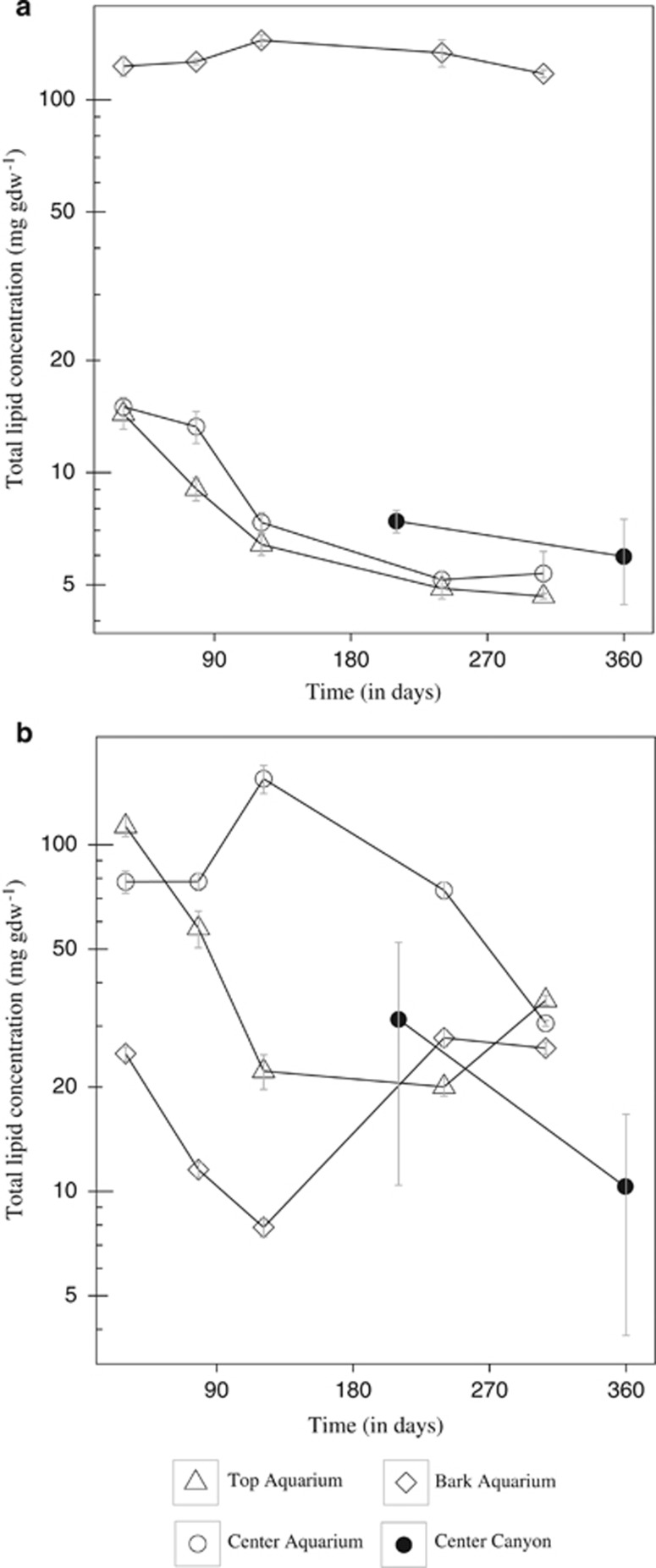
In the aquaria experiment, three pine and oak wood microhabitats (the bark, the center and the top of the wood) were monitored to test whether the different microhabitats changed with time. Oak bark lipid concentrations, which were 10 times higher than in the top and center of the wood (Figure 2a), did not vary significantly along the course of the experiment. In contrast, center and top lipid concentrations decreased significantly (top: analysis of variance, F=406.83, P<0.01, n=15; center: analysis of variance, F=90.031, P<0.01, n=15), especially during the first 121 days of incubation, and were reduced by more than half after 307 days. Pine samples (Figure 2b) contained significantly more lipids than oak in all three microhabitats (Wilcoxon, W=1291, P<0.01, n=90). On the first sampling point, lipid concentrations were higher in the top and center than in the bark (Figure 2b). As in the oak, lipid concentrations mostly decreased with time in the wood center, but no significant trends were detected in the other microhabitats.
Figure 2.
Total lipid concentrations and standard errors in oak (a) and pine (b) woods over time in different microhabitats (bark, top and center) in aquarium experiments and in situ canyon deployments. The y axis is in log scale.
In the canyon experiment, we monitored the center of pine and oak wood after 210 and 365 days of immersion. We also observed a decrease of lipid concentrations with time in both pine (Mann–Whitney test, W= 63, P=0.050, n=18) and oak samples (analysis of variance, F=4.15, P=0.058, n=18).
In the aquaria experiment, discrete measurements of sulfide concentrations were done on the wood during the first four months of the experiment when a white biofilm developed on the wood surface (Supplementary Figure 1). Sulfide was never detected on the bare wood surface itself but significant amounts were measured on the wood biofilms (Supplementary Table 2). Sulfide was always detected on the oak biofilm with values ranging from 2 to 46 μM after 78 days. On pine biofilms, sulfide was detected frequently after 21 days of incubation but less often later in the experiment (Supplementary Table 2). After 122 days, the biofilm disappeared and no more sulfide measurements were done.
Community and phylogenetic diversity
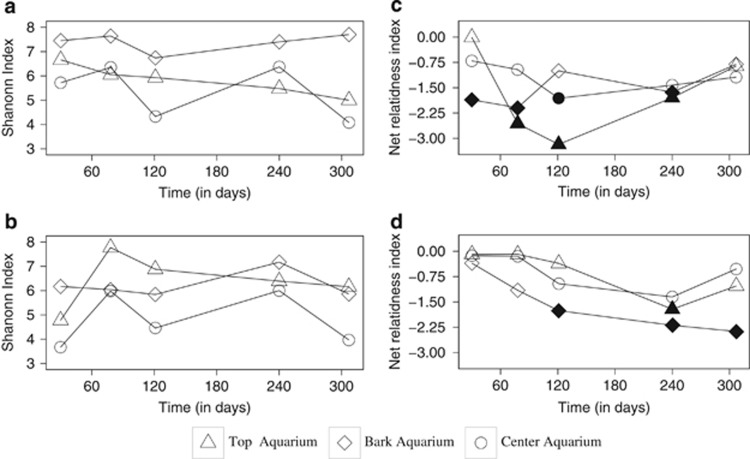
Pyrosequencing produced a total of 410 923 raw bacterial sequences from 42 samples gathered from the aquaria and in situ experiments. A total of 164 386 sequences remained after denoising and 154 604 after chimera checking (Supplementary Table 1). Sequences clustered at a similarity threshold of 97% grouped into 7326 OTUs. Bacteria rarefaction curves for pine and oak samples (Supplementary Figure 2) showed that our sequencing effort did not cover the full community diversity. In pine samples from the aquarium, the bark was the most diverse microhabitat with a Shannon index that varied between 7 and 8 (Figure 3a). The lowest value occurred after 121 days. In the top microhabitats, diversity decreased with time from 6.5 to <5. The center microhabitat showed large variations in diversity: days 30, 121 and 307 had a lower diversity than days 78 and 240. Oak center microhabitat had the same patterns of diversity as pine. However, the oak bark was less diverse, and the oak top microhabitat diversity increased strongly between days 30 and 78 to a value of 8, exceeding what was observed in pine (Figure 3b). Shannon diversity indexes were always higher in the canyon than in the aquarium (Supplementary Table 1).
Figure 3.
Shannon index (a, b) and NRI (c, d) for bacterial communities in the bark, (⋄) top (▵) and center (○) microhabitats in pine (a, c) and oak (b, d) woods. Filled symbols indicate significant NRI values.
Phylogenetic diversity (Figures 3c and d) was calculated with the NRI on aquaria samples. Positive NRI values indicate a clustered phylogeny, whereas a negative index indicates overdispersion with OTUs less related to each other. The patterns of phylogenetic diversity were different than the patterns observed for community diversity. In pine, the three microhabitats showed different patterns. The largest variation was observed in the top with lowest values after 121 days (Figure 3c). In oak, the NRI decreased with incubation time in the three microhabitats but showed significant values for the bark only (Figure 3d). In top and middle, the NRI decreased during the first 240 days before increasing again after 307 days.
For Archaea, a total of 11 609 sequences remained for the diversity analysis and grouped into 50 OTUs. The diversity was lower than for Bacteria with a Shannon index ranging from 1.43 to 1.73 (Supplementary Table 1).
For fungal communities, 12 735 amplified sequences remained in total. Fungal diversity was also lower than Bacteria with a Shannon index ranging from 0.62 to 2.92 (Supplementary Table 1).
Similarity between communities
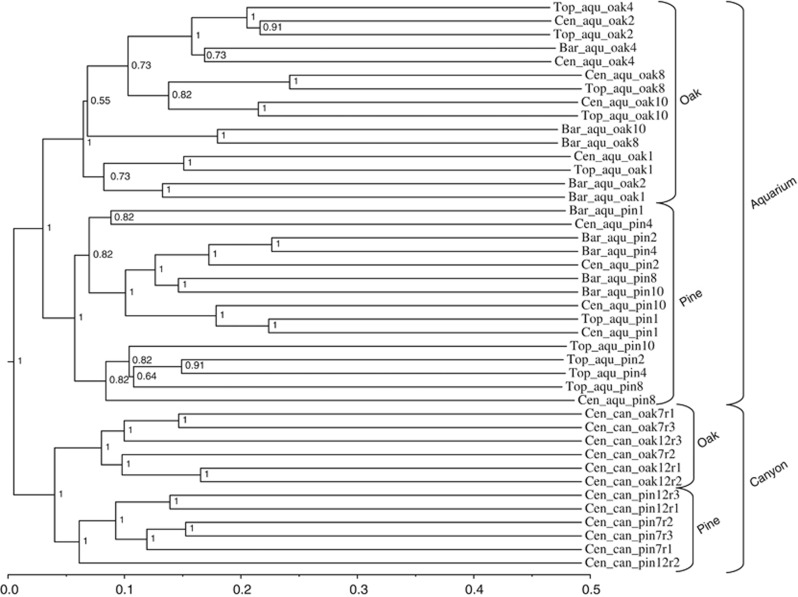
For Bacteria, the cluster analysis based on Bray–Curtis similarity split the communities in four significant groups (SIMPROF, P>0.01): two from the canyon and two from the aquarium experiment (Figure 4). Within the canyon and the aquarium, the two subgroups clustered according to the type of wood (pine vs oak).
Figure 4.
Cluster analysis based on the Bray–Curtis index showing similarity between bacterial community assemblies in top (Top), center (Cen) and bark (Bar) microhabitats in pine and oak woods from the aquarium experiment (aqu) and in situ canyon deployments (can). Numbers indicate the time of incubation in month, and r1, r2 and r3 indicate replicate samples in the canyon.
Within the aquarium, oak samples were significantly separated in the following three main groups: one containing samples incubated for 30 days, one with samples incubated for 78 and 121 days, and one with samples incubated for 240 and 307 days. For the pine samples, two main clusters were distinguished. All bark microhabitat samples were grouped in one of the cluster, and all but one top samples were grouped in the other cluster. In comparison with bark and top, center samples were more spread through the clusters (Figure 4). The nonparametric multivariate ADONIS test confirmed the significant effect of time on oak bacterial communities (F=2.8, P=0.0046, R2=18%) and the significant effect of microhabitat on pine bacterial communities (F= 2.0, P=0.0106, R2=25%).
For the canyon, samples could not be separated into different microhabitats owing to the advanced degradation state of the wood. For both pine and oak, immersion time appeared to separate the communities as most replicated samples taken after 210 days grouped away from 365 days samples (Figure 4). It should be noted that the entire canyon cluster was composed of long branches indicative of large differences between community compositions.
For Archaea, we detected sequences in oak only. Samples were grouped in two clusters that separated canyon from aquarium samples (Supplementary Figure 3). In the aquarium, the bark microhabitat samples grouped together away from the top microhabitats. After 240 days, Archaea were detected in the bark only.
For Fungi, the aquarium samples after the first month grouped away from communities sampled after 78 and 240 days (Supplementary Figure 4), and samples from 78 days center and top grouped separately from samples from the bark after 78 and 240 days.
Community composition
Bacteria communities
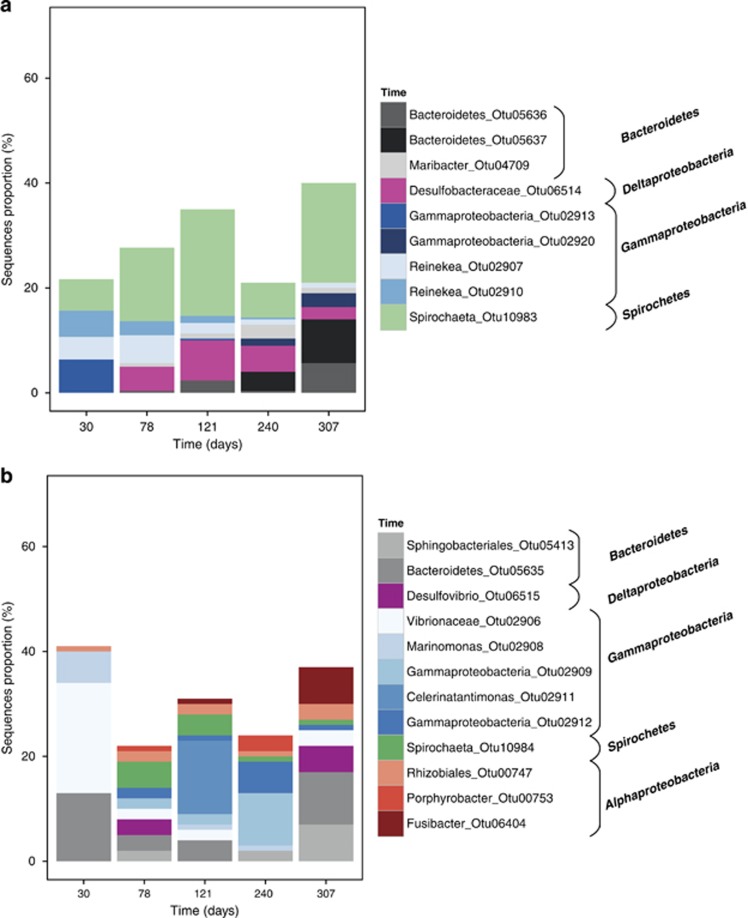
Among the abundant OTUs found in aquarium oak samples incubated for 30 days, a large number of bacterial sequences were affiliated to Reinekea marinisedimentorum (98% similarity). Reinekea marinisedimentorum produce acids from glucose under aerobic and anaerobic conditions, and are able to reduce nitrate (Romanenko et al., 2004; Figure 5). A deep-branching Gammaproteobacteria was also abundant as well as Spirocheta sequences only distantly related to cultivated strains. The relative number of Spirochaeta sequences increased after 78 and 240 days in all microhabitats (Supplementary Figures 5 and 6). These sequences were represented by one single OTU distantly related to any cultivated strain (Otu10983). Desulfobacteraceae sequences appeared after 78 days, as well as few sequences affiliated to the Maribacter genus. After 240 and 307 days, an uncultured Gammaproteobacteria OTU was detected that increased in abundance concomitant with a decrease of Desulfobacteraceae sequences after 307 days. The proportion of Bacteroidetes sequences increased from 240 to 307 days.
Figure 5.
Taxonomic affiliation and proportion of the five most abundant OTUs in each sample for oak (a) and pine (b) woods from the aquarium. The three wood microhabitats are grouped together. Details are shown in Supplementary Figure 5.
Pine samples from the aquarium (Supplementary Figure 7) did not present any clear temporal trend of OTU dynamics when pooling the three microhabitats together (Figure 5). A total of four OTUs were present in all pine samples (Figure 5): an unknown Rhizobiales (Otu747), an unknown Bacteroidetes (Otu5635), a Vibrio, which was closed to Vibrio aestuarinus with 95% similarity (Otu2906), and an OTU similar (99%) to Desulfovibrio piezophilus (Otu6515), a sulfate reducer originally isolated from a deep-sea wood fall in the Mediterranean Sea (Khelaifia et al., 2011). Among other pine OTUs, the few that had a close match to database sequences were Alteromonas stellipolaris (99% similarity; Otu2918), which is a chemoheterotrophic aerobic bacterium that uses cellulose degradation downstream products as a carbon source (Van Trappen et al., 2004). Marinomonas mediterranea (99% similarity; Otu2908) was found especially in the bark (Supplementary Figure 5), which is aerobic and motile sugar oxidizer (Solano and Sanchez-Amat, 1999). Interestingly, OTUs found in the center microhabitat were often distantly related to known strains (ca. 90%). Pine and oak wood shared two abundant OTUs, one belonging to Spirochaetes (Otu10984) and the other belonging to Bacteroidetes (Otu5635).
In the canyon samples (Supplementary Figure 8), the largest difference in community composition was observed between the type of wood and then between incubation times (Figure 3). Regarding the type of wood, Flavobacterales, Methylophiliales and Alteromonadales were typical for pine samples, whereas the Rhodobacterales and Spirochaetales class characterized the oak.
Archaea and fungi communities
Most of the archaeal sequences were only distantly related to known sequences. Among the ones that we could identify more precisely, otu35 (Supplementary Figure 9) was 99% similar to Methanococcoides alaskensis, a methanogen that uses trimethylamine as catabolic substrate with methane as end-product (Singh et al., 2005). Its sequence was more abundant after 78 days in the top microhabitats in the aquaria experiment (Supplementary Figure 9). The other abundant known sequence in the top was a marine group I Thaumarchaeota (otu1) belonging to the Nitrospumilus genus that is commonly found in seawater (Könneke et al., 2005). After 240 days in the aquarium, Archaea were present only in the bark microhabitat with a dominance of Thaumarchaeota similar to sequences previously found in marine sponges and sediments.
Canyon Archaea were different from the ones detected in the aquarium (Supplementary Figure 9). Sequences belonged to the lake dagow sediment and rice cluster V cluster of Euryarchaeota (Barberán et al., 2011) and were distantly related (⩽95%) to sequences found in marine sediments.
The most abundant fungal sequences (otu001; Supplementary Figure 10) detected after 30 days of incubation in the aquaria was identified as a Phialemonium (100% similarity), a genus that has been previously found in marine surface waters (Oliveira et al., 2013). After 78 days in the aquaria, the bark community was characterized by two Ascomyta (otu2 and otu9; Supplementary Figure 10). They had low similarity (⩽95%) to sequences from the database. The otu2 was identified as a Leotiomycetes, a class containing marine fungi found in cold seeps (Nagahama et al., 2011) and hydrothermal vents (Burgaud et al., 2009). Other OTUs (otu3, 7 and 11) belonging to the Ascomyta phylum were also present in the center and top, but only otu11 could be precisely assigned (97% similarity) to sequences from spring water (Oliveira et al., 2013). In the Canyon after 210 days, the most abundant fungal OTU was an Ascomyta that was similar to sequences found in the sea and mangrove sediments with 100% similarity, and another OTU that was 99% similar to a sequence from methane hydrate-bearing deep-sea marine sediments (Lai et al., 2007).
Association network
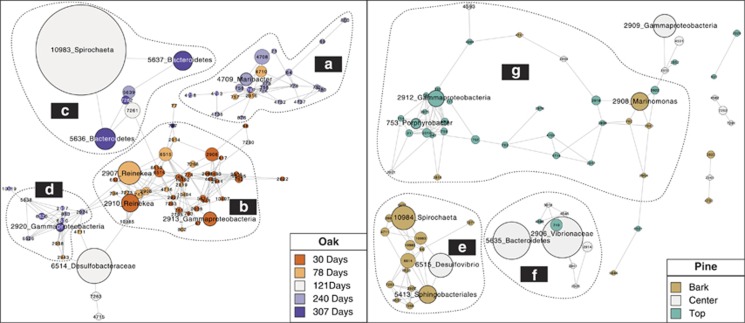
A network was constructed to identify OTUs that were closely associated to each other in the aquaria experiments. In oak, four subnetworks could be distinguished by the hierarchical clustering algorithm HC-PIN (Wang et al., 2011), which mathematically dissociates groups of OTU into subnetworks. The subnetwork (Figure 6a) with the highest modularity (∞, that is, densest connections between nodes) was composed of 23 OTUs, and 71% of these OTUs were more abundant after 240 days of incubation (Figure 6a). Most of them belonged to Alphaproteobacteria (48%) and Flavobacteria (38%). Forty other OTUs formed a second subnetwork with a modularity of 12 (Figure 6b). This group was composed of a majority of OTUs that were abundant after 30 (63%) or 78 days (30%) in the aquarium. This subnetwork was more diverse than the previous one and was dominated by Gammaproteobacteria (28%) and Alphaproteobacteria (23%). The third subnetwork (Figure 6c) revealed by the algorithm was smaller with only seven nodes and a modularity of 8. Three OTUs (5636, 5637 and 10 983) reach their maximum abundance between 121 and 307 days. The last subnetwork was made of 10 nodes composed of 50% of Gammaproteobacteria and 30% of Deltaproteobacteria, among which D. piezophilus (6520; Figure 6d). In summary, we identified one large subnetwork made of OTUs that reached their highest sequence abundance per sample at the beginning of the experiment (b), then a subnetwork with maximum OTU sequence abundance after 240 days of experiment (a) and, finally, two subnetworks that were not clearly associated to incubation time. It should be noted that many abundant OTUs did not appear in any subnetwork because they did not show specific associations to other OTUs, probably because they were ubiquitous across the different samples.
Figure 6.
Network showing associations between OTUs identified by maximal information coefficient values. Nodes represent abundant OTUs. Numbers indicate OTU names. Taxonomic assignment is given for the largest OTUs. Node size is proportional to the number of sequences in each OTU. The left panel presents the oak sample network from the aquarium and the right panel represents the pine samples from the aquarium. Dashed lines delineate subclusters defined by the HC-PIN algorithm. Colors indicate OTUs typical for the different incubation times or microhabitats. a–g indicates subnetworks discriminated by the HC - PIN analysis.
Looking at the same projection for pine, we observed a first subnetwork (modularity=∞) made of 17 nodes, among which 94% reached their highest sequence number in the bark microhabitat (Figure 6e). These OTUs were mostly composed of Deltaproteobacteria and one of them (6515) was 99% similar to D. piezophilus as mentioned earlier. There were also three OTUs related to Spirochaetales (10 983, 10984 and 10985), which are known to ferment simple sugars such as glucose in acetate, CO2 and H2, and Oceanospiralles (2908). Those two orders contained OTUs that were similar to those found in the oak (Spirochaeta littoralis and M. mediteranea). Subnetwork 2 (modularity=∞) contained nine OTUs and seven of them reached maximum sequence abundance in the center microhabitat (Figure 6f). Five OTUs belonged to the Gammaproteobacteria and one OTU belonged to the Vibrionales order (2906). The third and last subnetwork was the largest with 31 OTUs and a modularity of 32.5. Close to 71% of the OTUs from this cluster reached a maximum sequence number in the top microhabitat. A total of 13 OTUs belonged to Gammaproteobacteria and 9 belonged to Alphaproteobacteria. A noteworthy OTU (4593) in this subnetwork was related to an Arcobacter able to oxidize sulfide.
In summary, the networks showed that most of the abundant OTU in oak varied together based on incubation time, while in pine, microhabitat was the strongest structuring factor for microbial communities. However, some interactions between microhabitats were detected. Microbes from the bark had few associations to OTUs from the center microhabitat, and most of the abundant center microhabitat OTUs had some connections to OTUs abundant in the top. The top microhabitat, as seen in the subnetwork g, was connected to OTUs abundant in the bark and center.
Discussion
Our deployment of wood substrates in marine waters demonstrated that the patterns of microbial colonization of a same object, wood, changed markedly depending on the environmental context. The location, which encompasses factors such as the biological, chemical and physical properties of water, and the vicinity of sediments were the strongest structuring factors as illustrated by the difference observed between woods incubated in deep-sea water in the canyon and wood incubated in coastal water in the aquarium. Moreover, our results also showed that within a given environment, the factors controlling microbial communities differed. First, the type of wood was the strongest controlling factor as illustrated by the difference between pine and oak wood communities in both canyon and aquarium, and as shown earlier (Palacios et al., 2009; Fagervold et al., 2014). Then, communities changed between wood microhabitats, within the same wood, as seen in pine in the aquarium. The temporal scale appeared, however, to have a stronger impact on community assembly in oak wood, which had communities changing with time of incubation rather than between microhabitats. It shows that under similar environmental conditions, in this case, wood incubated over time in the same aquarium, the pattern of microbial colonization is influenced by different constraints (time vs microhabitat) depending on the substrate (pine vs oak).
The comparison of phylogenetic diversity between communities in the different microhabitats of the wood also highlighted a scale-dependent control of community structure. In oak, communities' phylogenetic diversity changed temporally. Communities were always more phylogenetically clustered after 30 days of incubation, indicating that they were more related to each other in the beginning of the experiment. This pattern suggests that microbial community assembly was shaped by species sorting (that is, regulated by the abiotic environment; Webb et al., 2002). In this case, only one specific fraction of the environmental bacterial pool is able to colonize and persist in the wood matrix when colonization starts. This abiotic selection process is called habitat filtering (Webb et al., 2002) and was shown earlier to structure wood fall communities on the sea floor (Fagervold et al., 2012). Interestingly, the level of habitat filtering decreased with time. It reached a minimum after 240 days, which was indicative of an overdispersed phylogeny. Dispersion suggests that new microhabitats became available selecting for more distantly related bacterial species that would be more ecologically different. This scenario is interpreted as an increased importance of competitive exclusion for community structuring. Only species that do not compete with each other, by occupying distinct microhabitats, are able to develop in the environment. We can hypothesize that at this stage the full cascade of organic matter degradation is taking place from aerobic processes on the wood surface to anaerobic metabolisms in the wood center. After 240 days, values became more variable, suggesting that the wood environment underwent perturbations. The colonization by wood-boring species of the family Terenididae, which we observed in the wood after 240 days of incubation, transformed the wood matrix creating new microhabitats for microbial colonization (Bessette et al., 2014, Fagervold et al., 2014). In pine wood, the different microhabitats showed different dynamics of phylogenetic diversity, illustrating that the local spatial scale had an important role in structuring the phylogenetic structure of communities. The top microhabitats had the strongest variation of phylogenetic diversity with time, whereas the middle microhabitat changed less. Overall our data showed that the importance of habitat filtering changed with both time and microhabitats, illustrating the complexity of the ecological interactions shaping the wood fall chemosynthetic ecosystem.
The presence of different communities, in different microhabitats and/or at different times, formed the basis of microecosystems with distinct potential microbial metabolisms. Even though caution should be taken when inferring potential metabolisms from 16 S rRNA-based studies, the close similarity of some of our sequences to cultivated strains indicated the potential for sugar degradation, fermentation, sulfate reduction, methanogenesis and sulfide oxidation in the wood. In contact with seawater, aerobic bacteria such as Marinomonas or other heterotrophs may be degrading the sugars originating from the cellulose. Cellulose breakdown is likely of bacterial origin as illustrated by the presence of cellulose degrading Clostridiales (Ljungdahl and Eriksson, 1985; Shiratori et al., 2006), but fungi, the major wood degrader in terrestrial environment (Ljungdahl and Eriksson, 1985; Raghukumar, 2008), could also participate in the process. We were able to directly detect fungi and to assign them to fungal groups without using long incubations (Azevedo et al., 2010; Pang et al., 2011) or culture methods (Rämä et al., 2014). Whether these were able to degrade cellulose remains to be shown, as none of our ITS sequences had perfect match to cultivated strains. Nevertheless, our data give a rare indication of fungal sunken wood colonization in the sea and add to recent reports of high fungal diversity on sea floor logs (Rämä et al., 2014). Deeper in the wood, where oxygen disappears, anaerobic fermenters belonging to Spirochaeta, Bacterioidales and Vibrionales probably take advantage of the sugar released from the cellulose. After 78 days, a sulfidic stage was observed with the presence of Desulfobacterales and Desulfovibrionales in the center. Desulfovibrionales are sulfate-reducing bacteria that probably take advantage of fermentation end-products. The production of sulfide inside the wood has been documented earlier (Yucel et al., 2013), and our discrete measurements of sulfide concentrations on the wood surface indicated that sulfate reduction also took place in our experiments. Methanogenesis is the other main end process of the anaerobic degradation of organic matter (Canfield, 1993). The presence of 16 S rRNA gene sequences from methanogens in the top of the wood after 78 days suggests that methane could also be produced when wood is degraded in marine water. The potential for methanogenesis was not detected later in the incubation as only archaeal sequences belonging to possible ammonia oxidizing Thaumarcheaota, common on the surface of marine sediments, were found on the bark at the end of the experiment. The metabolism of the canyon wood Archaea belonging to the lake dagow sediment and rice cluster V clusters of the Euryarchaeota phylum (Barberán et al., 2011) is not known.
The difference between canyon and aquarium bacterial communities could be linked to several factors such as the source of the microbial colonizers, the influence of the sample location on the wood matrix itself and different wood degradation kinetics between canyon and aquarium. Our results confirm previous reports showing that woods incubated in different environments had different bacterial communities (Bienhold et al., 2013; Fagervold et al., 2013) They contrast, however, with a fingerprinting-based study that showed high similarity between woods incubated at different geographical locations or depths (Palacios et al., 2009).
Interestingly, despite the large difference observed between canyon and aquarium, some sequences, such as the one identified as belonging to D. piezophilus, were common to both the natural and the artificial environments. The fact that D. piezophilus, originally found in deep-sea habitats, is able to colonize poorly connected environments, highlights that even rare bacterial species have an extraordinary dispersal capability in the sea. The presence of abundant sequences of D. piezophilus in our aquarium woods also indicates that our experiments in cold water in the dark were able to attract such emblematic species. It also demonstrates that D. piezophilus, thought to be a typical piezophile (Khelaifia et al., 2011; Pradel et al., 2013), may also thrive under atmospheric pressure conditions.
The clear differences in term of community composition between oak and pine in both the aquarium and the canyon could be due to different wood chemical composition. Our finding confirms earlier results indicating that different types of wood may harbor different microbial communities (Palacios et al., 2009; Fagervold et al., 2014). Pine wood contained higher concentrations of lipids than oak wood. These lipids used as reserves characterize pine as a fat tree compared with oak that uses starch, a glucose polymer, for storage (Fengel and Wegener, 1984). The kind of substrate available for bacterial community thus differed between wood species. We hypothesize that the significant initial decrease in the oak wood lipid content was due to bacterial or fungal activity. We could not, however, identify by 16 S rRNA gene analysis the specific bacterial group harboring lipase, the enzyme allowing lipid hydrolysis, as it is common across bacterial lineages (Arpigny and Jaeger, 1999).
Conclusion
We provide a direct demonstration that wood falls are dynamic microecosystems with high spatial and temporal community turnover. Community assembly was dictated by different constraints that depended on the scale of observation. The most illustrative example was the difference between pine communities, apparently structured spatially according to the wood microhabitat, and oak communities, which changed more significantly with time. Changes in community assembly were reflected by changes in phylogenetic diversity that could be interpreted as shifts between assemblies ruled by species sorting to assemblies structured by competitive exclusion. These ecological interactions mirrored the dynamics of the potential microbial metabolisms accompanying wood degradation in the sea. Finally, we showed that wood is a good model for creating and manipulating potential chemosynthetic ecosystem in the laboratory, and attracting not only typical chemosynthetic microbes but also emblematic macrofaunal species.
Acknowledgments
The present work was supported by the Agence Nationale de la Recherche (ANR) project MICADO (ANR-11JSV7-003-01), the UPMC (University Pierre and Marie Currie) and Fondation TOTAL as part of the chair ‘Extreme marine environments, Biodiversity, and Global change', and by the CNRS GDRE DIWOOD. We thank the crew of the RV ‘Minibex' (COMEX) for their assistance during fieldwork. We are grateful to Julie Rius and Benjamin Falgas from the botanical garden ‘Biodiversarium' in Banyuls sur Mer for their kind and precious help in providing pine and oak wood.
The authors declare no conflict of interest
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Anderson MJ. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46. [Google Scholar]
- Arpigny J, Jaeger K. (1999). Bacterial lipolytic enzymes: classification and properties. Biochem J 343: 177–183. [PMC free article] [PubMed] [Google Scholar]
- Austin B, Allen D, Zachary A, Belas M, Colwell R. (1979). Ecology and taxonomy of bacteria attaching to wood surfaces in a tropical harbor. Can J Microbiol 25: 447–461. [DOI] [PubMed] [Google Scholar]
- Azevedo E, Rebelo R, Caeiro MF, Barata M. (2010). Diversity and richness of marine fungi on two Portuguese marinas. Nova Hedwigia 90: 3–4. [Google Scholar]
- Barberán A, Fernàndez-Guerra A, Auguet JC, Galand PE, Casamayor EO. (2011). Phylogenetic ecology of widespread uncultured clades of the Kingdom Euryarchaeota. Mol Ecol 20: 1988–1996. [DOI] [PubMed] [Google Scholar]
- Barnes H, Blackstock J. (1973). Estimation of lipids in marine animals and tissues: detailed investigation of the sulphophosphovanilun method for ‘total'lipids. J Exp Mar Biol Ecol 12: 103–118. [Google Scholar]
- Bessette S, Fagervold SK, Romano C, Martin D, Le Bris N, Galand PE. (2014). Diversity of bacterial communities on sunken woods in the Mediterranean Sea. J Mar Sci Technol 22: 60–66. [Google Scholar]
- Bienhold C, Ristova PP, Wenzhöfer F, Dittmar T, Boetius A. (2013). How deep-sea wood falls sustain chemosynthetic life. Plos One 8: e53590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgaud G, Le Calvez T, Arzur D, Vandenkoornhuyse P, Barbier G. (2009). Diversity of culturable marine filamentous fungi from deep-sea hydrothermal vents. Environ Microbiol 11: 1588–1600. [DOI] [PubMed] [Google Scholar]
- Canfield DE. (1993). Organic matter oxidation in marine sediments. In Wollast R, Mackenzie FT, Chou L (eds) Interactions of C, N, P and S Biogeochemical Cycles and Global Change. Springer: New York, NY, USA, pp 333–363. [Google Scholar]
- Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. (2010. a). PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010. b). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke KR, Somerfield PJ, Gorley RN. (2008). Testing of null hypotheses in exploratory community analyses: similarity profiles and biota-environment linkage. J Exp Mar Biol Ecol 366: 56–69. [Google Scholar]
- Clauset A, Newman ME, Moore C. (2004). Finding community structure in very large networks. Phys Rev E 70: 066111. [DOI] [PubMed] [Google Scholar]
- Cundell AM, Mitchell R. (1977). Microbial succession on a wooden surface exposed to the sea. Int Biodeterior 13: 67–73. [Google Scholar]
- Distel DL, Baco AR, Chuang E, Morrill W, Cavanaugh C, Smith CR. (2000). Do mussels take wooden steps to deep-sea vents? Nature 403: 725–726. [DOI] [PubMed] [Google Scholar]
- Dubilier N, Bergin C, Lott C. (2008). Symbiotic diversity in marine animals: the art of harnessing chemosynthesis. Nat Rev Microbiol 6: 725–740. [DOI] [PubMed] [Google Scholar]
- Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagervold S, Bessette S, Romano C, Martin D, Plyuscheva M, Le Bris et al. (2013). Microbial communities associated with the degradation of oak wood in the Blanes submarine canyon and its adjacent open slope (NW Mediterranean). Prog Oceanogr 118: 137–143. [Google Scholar]
- Fagervold SK, Galand PE, Zbinden M, Gaill F, Lebaron P, Palacios C. (2012). Sunken woods on the ocean floor provide diverse specialized habitats for microorganisms. FEMS Microbiol Ecol 82: 616–628. [DOI] [PubMed] [Google Scholar]
- Fagervold SK, Romano C, Kalenitchenko D, Borowski C, Nunes-Jorge A, Martin D et al. (2014). Microbial communities in sunken wood are structured by wood-boring bivalves and location in a submarine canyon. Plos One 9: e96248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fengel D, Wegener G. Wood: Chemistry, Ultrastructure, Reactions. De Gruyter: Berlin, Germany,. (1984). [Google Scholar]
- Gardes M, Bruns TD. (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 2: 113–118. [DOI] [PubMed] [Google Scholar]
- Jorgensen BB, Boetius A. (2007). Feast and famine—microbial life in the deep-sea bed. Nat Rev Microbiol 5: 770–781. [DOI] [PubMed] [Google Scholar]
- Jurgens J, Blanchette RA, Carlson D. (2003) Evaluating the Wooden Remnants of the Tektas burnu Shipwreck. The Metropolitan Museum of Art: NY, USA. [Google Scholar]
- Kamada T, Kawai S. (1989). An algorithm for drawing general undirected graphs. Inform Process Lett 31: 7–15. [Google Scholar]
- Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD et al. (2010). Picante: R tools for integrating phylogenies and ecology. Bioinformatics 26: 1463–1464. [DOI] [PubMed] [Google Scholar]
- Khelaifia S, Fardeau M-L, Pradel N, Aussignargues C, Garel M, Tamburini C et al. (2011). Desulfovibrio piezophilus sp. nov., a piezophilic, sulfate-reducing bacterium isolated from wood falls in the Mediterranean Sea. Int J Syst Evol Microbiol 61: 2706–2711. [DOI] [PubMed] [Google Scholar]
- Kiel S, Amano K, Hikida Y, Jenkins RG. (2009). Wood-fall associations from Late Cretaceous deep-water sediments of Hokkaido, Japan. Lethaia 42: 74–82. [Google Scholar]
- Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AF, Bahram M et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol 22: 5271–5277. [DOI] [PubMed] [Google Scholar]
- Könneke M, Bernhard AE, José R, Walker CB, Waterbury JB, Stahl DA. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546. [DOI] [PubMed] [Google Scholar]
- Lai X, Cao L, Tan H, Fang S, Huang Y, Zhou S. (2007). Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME J 1: 756–762. [DOI] [PubMed] [Google Scholar]
- Langenheder S, Székely AJ. (2011). Species sorting and neutral processes are both important during the initial assembly of bacterial communities. ISME J 5: 1086–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent MC, Le Bris N, Gaill F, Gros O. (2013). Dynamics of wood fall colonization in relation to sulfide concentration in a mangrove swamp. Mar Environ Res 87-88: 85–95. [DOI] [PubMed] [Google Scholar]
- Ljungdahl LG, Eriksson K-E. Ecology of Microbial Cellulose Degradation. Advances in Microbial Ecology. Springer: New York, NY,. (1985). pp 237–299. [Google Scholar]
- Luther GW III, Glazer BT, Hohmann L, Popp JI, Taillefert M, Rozan TF et al. (2001). Sulfur speciation monitored in situ with solid state gold amalgam voltammetric microelectrodes: polysulfides as a special case in sediments, microbial mats and hydrothermal vent waters. J Environ Monitor 3: 61–66. [DOI] [PubMed] [Google Scholar]
- Mouzouras R, Jones EG, Venkatasamy R, Holt DMicrobial Decay of Lignocellulose in the Marine Environment. Thomson MF, Sarojini R, Nagabhushanam R. Marine Biodeterioration. Oxford and IBH Publishing: New Delhi, India,. (1988). pp 329–335. [Google Scholar]
- Muller F, Brissac T, Le Bris N, Felbeck H, Gros O. (2010). First description of giant Archaea (Thaumarchaeota) associated with putative bacterial ectosymbionts in a sulfidic marine habitat. Environ Microbiol 12: 2371–2383. [DOI] [PubMed] [Google Scholar]
- Nagahama T, Takahashi E, Nagano Y, Abdel-Wahab MA, Miyazaki M. (2011). Molecular evidence that deep-branching fungi are major fungal components in deep-sea methane cold-seep sediments. Environ Microbiol 13: 2359–2370. [DOI] [PubMed] [Google Scholar]
- Oliveira B, Barreto Crespo M, San Romao M, Benoliel M, Samson R, Pereira V. (2013). New insights concerning the occurrence of fungi in water sources and their potential pathogenicity. Water Res 47: 6338–6347. [DOI] [PubMed] [Google Scholar]
- Ovreas L, Forney L, Daae FL, Torsvik V. (1997). Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16 S rRNA. Appl Environ Microbiol 63: 3367–3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios C, Zbinden M, Pailleret M, Gaill F, Lebaron P. (2009). Highly similar prokaryotic communities of sunken wood at shallow and deep-sea sites across the oceans. Microb Ecol 58: 737–752. [DOI] [PubMed] [Google Scholar]
- Pang K-L, Chow RK, Chan C-W, Vrijmoed LL. (2011). Diversity and physiology of marine lignicolous fungi in Arctic waters: a preliminary account. Polar Res 30: 5859. [Google Scholar]
- Pointing S, Vrijmoed L, Jones E. (1998). A qualitative assessment of lignocellulose degrading enzyme activity in marine fungi. Bot Mar 41: 293–298. [Google Scholar]
- Pradel N, Ji B, Gimenez G, Talla E, Lenoble P, Garel M et al. (2013). The first genomic and proteomic characterization of a deep-sea sulfate reducer: insights into the piezophilic lifestyle of Desulfovibrio piezophilus. Plos One 8: e55130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. Plos One 5: e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J et al. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35: 7188–7196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ, Turnbaugh PJ. (2011). Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghukumar C, Raghukumar S, Chinnaraj A, Chandramohan D, D'souza T, Reddy C. (1994). Laccase and other lignocellulose modifying enzymes of marine fungi isolated from the coast of India. Bot Mar 37: 515–524. [Google Scholar]
- Raghukumar C. (2008). Marine fungal biotechnology: an ecological perspective. Fungal Divers 31: 19–35. [Google Scholar]
- Raghukumar C. Biology of Marine Fungi, vol. 3. Springer: Berlin, Germany,. (2012). [Google Scholar]
- Rämä T, Nordén J, Davey ML, Mathiassen GH, Spatafora JW, Kauserud H. (2014). Fungi ahoy! Diversity on marine wooden substrata in the high North. Fungal Ecol 8: 46–58. [Google Scholar]
- Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ et al. (2011). Detecting novel associations in large data sets. Science 334: 1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanenko LA, Schumann P, Rohde M, Mikhailov VV, Stackebrandt E. (2004). Reinekea marinisedimentorum gen. nov., sp. nov., a novel gammaproteobacterium from marine coastal sediments. Int J Syst Evol Microbiol 54: 669–673. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Gevers D, Westcott SL. (2011). Reducing the effects of PCR amplification and sequencing artifacts on 16 S rRNA-based studies. Plos One 6: e27310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratori H, Ikeno H, Ayame S, Kataoka N, Miya A, Hosono K et al. (2006). Isolation and characterization of a new Clostridium sp. that performs effective cellulosic waste digestion in a thermophilic methanogenic bioreactor. Appl Environ Microbiol 72: 3702–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N, Kendall MM, Liu Y, Boone DR. (2005). Isolation and characterization of methylotrophic methanogens from anoxic marine sediments in Skan Bay, Alaska: description of Methanococcoides alaskense sp. nov., and emended description of Methanosarcina baltica. Int J Syst Evol Microbiol 55: 2531–2538. [DOI] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. (2011). Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics 27: 431–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solano F, Sanchez-Amat A. (1999). Note: Studies on the phylogenetic relationships of melanogenic marine bacteria: proposal of Marinomonas mediterranea sp. nov. Int J Syst Bacteriol 49: 1241–1246. [DOI] [PubMed] [Google Scholar]
- Van Trappen S, Tan T-L, Yang J, Mergaert J, Swings J. (2004). Alteromonas stellipolaris sp. nov., a novel, budding, prosthecate bacterium from Antarctic seas, and emended description of the genus Alteromonas. Int J Syst Evol Microbiol 54: 1157–1163. [DOI] [PubMed] [Google Scholar]
- Wang J, Li M, Chen J, Pan Y. (2011). A fast hierarchical clustering algorithm for functional modules discovery in protein interaction networks. IEEE-ACM T Comput Biol Bioinform 8: 607–620. [DOI] [PubMed] [Google Scholar]
- Webb CO. (2000). Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am Nat 156: 145–155. [DOI] [PubMed] [Google Scholar]
- Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. (2002). Phylogenies and Community Ecology. Annu Rev Ecol Syst 33: 475–505. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. PCR Protocols: a Guide to Methods and Applications. Academic Press: New York, USA,. (1990). pp 315–322. [Google Scholar]
- Yoshioka PM. (2008). Misidentification of the Bray-Curtis similarity index. Mar Ecol Prog Ser 368: 309–310. [Google Scholar]
- Yu Y, Lee C, Kim J, Hwang S. (2005). Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng 89: 670–679. [DOI] [PubMed] [Google Scholar]
- Yucel M, Galand PE, Fagervold SK, Contreira-Pereira L, Le Bris N. (2013). Sulfide production and consumption in degrading wood in the marine environment. Chemosphere 90: 403–409. [DOI] [PubMed] [Google Scholar]
- Zavatarielli M, Mellor GL. (1995). A numerical study of the Mediterranean Sea circulation. J Phys Oceanogr 25: 1384–1414. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.