Abstract
Marine microbial communities experience daily fluctuations in light and temperature that can have important ramifications for carbon and nutrient cycling. Elucidation of such short time scale community-wide dynamics is hindered by system complexity. Hypersaline aquatic environments have lower species richness than marine environments and can be well-defined spatially, hence they provide a model system for diel cycle analysis. We conducted a 3-day time series experiment in a well-defined pool in hypersaline Lake Tyrrell, Australia. Microbial communities were tracked by combining cultivation-independent lipidomic, metagenomic and microscopy methods. The ratio of total bacterial to archaeal core lipids in the planktonic community increased by up to 58% during daylight hours and decreased by up to 32% overnight. However, total organism abundances remained relatively consistent over 3 days. Metagenomic analysis of the planktonic community composition, resolved at the genome level, showed dominance by Haloquadratum species and six uncultured members of the Halobacteriaceae. The post 0.8 μm filtrate contained six different nanohaloarchaeal types, three of which have not been identified previously, and cryo-transmission electron microscopy imaging confirmed the presence of small cells. Notably, these nano-sized archaea showed a strong diel cycle, with a pronounced increase in relative abundance over the night periods. We detected no eukaryotic algae or other photosynthetic primary producers, suggesting that carbon resources may derive from patchily distributed microbial mats at the sediment-water interface or from surrounding land. Results show the operation of a strong community-level diel cycle, probably driven by interconnected temperature, light abundance, dissolved oxygen concentration and nutrient flux effects.
Introduction
Microorganisms exist in complex communities defined by networks of synergistic and competitive interactions. However, little is known about many of the microorganisms that inhabit natural systems or the factors that influence their abundance patterns. Organism abundance patterns may vary in response to viral or eukaryotic predation, temperature, dissolved oxygen concentration, solution chemistry and light. Notably, some of these environmental factors occur in a nonrandom, often temporally coupled manner (Baliga, 2008; Tagkopoulos et al., 2008) and many are linked to the day-to-night (diel) cycle. Such short-term fluctuations can have important ramifications for carbon and nutrient cycling (Ottesen et al., 2013). Further, information about how the relative abundances of uncultivated organisms vary over short time scales, particularly over the day-to-night cycle, could provide insight into their metabolic requirements and their relationship to other organisms (Ottesen et al., 2014).
Although most environments have high levels of diversity, habitats with low microbial diversity have proven to be excellent models to examine the ecology of native microbial consortia (Tyson et al., 2004; Allen and Banfield, 2005; Ram et al., 2005; Lo et al., 2007; Raes and Bork, 2008; Wilmes and Bond, 2009). One such model includes the aquatic microbial consortia that inhabit hypersaline lakes, salt ponds and solar salterns. Evaporative processes result in solutions close to or at salt saturation and selection for an extremely halophilic microbial community with low species richness. This type of model system has been historically well studied (Oren, 2008), and advances in sequencing technology are enabling a deeper understanding of these communities and their ecology (Ghai et al., 2011, Emerson et al., 2012; Narasingarao et al., 2012). Previously, total microbial community composition of these extreme hypersaline habitats was described as stable over months to years (Gasol et al., 2004; Rodriguez-Brito et al., 2010, Ghai et al., 2011). Yet, recent studies have begun to identify these communities as much more dynamic (Emerson et al., 2013a). Metagenomic analyses performed on samples collected over hours and days can provide insight into short-term population dynamics. Coupled with core lipid inventories, these tools are capable of providing community-wide assessments of changes in composition. As a quantitative tool, core lipid analysis can provide an important complement to metagenomic methods. Combining these methods enables a description of community composition, the identification of novel organisms and insight into abundance pattern dynamics.
Lake Tyrrell, Australia, is a thalassohaline system (one in which the water is compositionally similar to concentrated seawater, with NaCl being the major salt). The Lake Tyrrell ecosystem is ideal for research on hypersaline microbial communities as it is hydrologically, geochemically and biologically well characterized (Macumber, 1992; Williams, 2001; Narasingarao et al., 2012; Heidelberg et al., 2013; Podell et al., 2013; Emerson et al., 2013a, 2013b; Podell et al., 2014).
Here we analyzed the core lipid composition of water samples collected across 3 day-night cycles from Lake Tyrrell to monitor abundances of bacteria, archaea and eukaryotes, combined with metagenomic data to resolve the community compositions at species and strain levels. Cryo-transmission electron microscopy (TEM) based characterization was used to examine the presence of ultra-small cells, given prior detection of the presence of Nanohaloarchaea at this site (Narasingarao et al., 2012). In combination, results from these analyses indicate the operation of a diel cycle in planktonic bacterial and archaeal abundance levels, with proliferation of both Halobacteria and Nanohaloarchaea over the night period.
Materials and methods
Sampling site
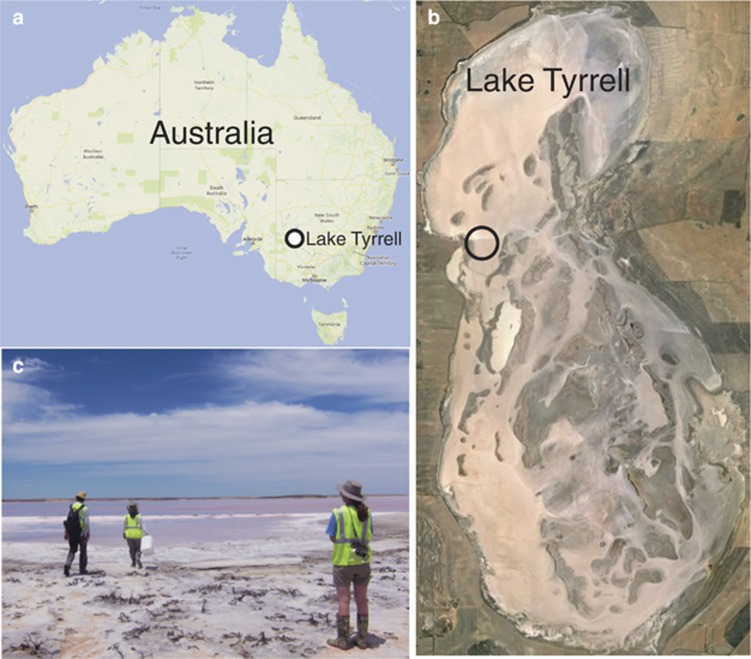
Lake Tyrrell, located in the Murray Basin in western Victoria, Australia has a surface area of ~160 km2. In summer, water evaporates, leaving a halite crust of up to 7-cm thick and residual brines with salt concentrations generally >330 g l−1. Halite re-solution and precipitation occurs throughout the year (Macumber, 1992). The sampling site was a shallow (<20- cm deep), wind-mixed pool separated from the main lake by a salt barrier (Figure 1).
Figure 1.
Location of the thallasohaline Lake Tyrell metagenomic sampling site. (a) Map showing the location of Lake Tyrrell, NW Victoria, Australia, indicated by a circle. (b) Sampling location in Lake Tyrrell, indicated by a circle. (c) Picture of the pool used for the time series study.
Sample collection
In January 2010 (Austral summer), water samples were taken for both lipid and metagenomic analysis from the shallow pool. For lipid analysis, samples were taken every 6–10 h over a period of 3 days, resulting in a total sampling time of 66 h (7–9 January 2010). In the field, water was transferred into sterilized 20 L plastic containers using a sterilized hand water pump. After transfer to the field laboratory (30 min), the water for lipid analysis was immediately filtered over 0.7 μm pore size glass fiber disc filters without binder (Millipore APFF14250, Bayswater, VIC, Australia, Æ d=142 mm, 90% porosity) using a peristaltic pump. The amount of filtered water varied between 2 and 3 L, depending on when filters clogged with biomass. The loaded filters were packed into combusted aluminum foil, transported on dry ice and stored in the laboratory at −30 °C or −80 °C before analysis in March 2010.
For metagenomic analysis, we collected five samples from the shallow pool during the 3-day time series experiment. Surface water samples were collected using a bilge pump. Planktonic cells were recovered by size exclusion filtration. Water samples were passed through a 20 μm Nytex prefilter and then sequentially filtered through polyethersulfone, 142-mm diameter membrane filters (Pall Corporation, Port Washington, NY, USA) of decreasing pore sizes (3 μm, 0.8 μm, 0.1 μm). Filters were placed in 50-ml centrifuge tubes with 10-ml DNA lysis buffer (100 μl TE buffer, 200 μl 1M EDTA, 200 μl 0.5 M EGTA and 10 ml DI water). Samples were stored on dry ice for a maximum of 6 days, followed by −80 °C freezer storage. Biogeochemical analysis was conducted as described in Heidelberg et al. (2013).
Ion analysis
Water samples were analyzed for inorganic cations and anions. For cation analysis, a Varian Vista AX CCD Simultaneous ICP-AES (Varian, Palo Alto, CA, USA) instrument was used. Anion analysis was carried out on a Dionex Ion Chromatograph Series 4500i (Dionex, Sunnyvale, CA, USA).
Lipid analysis
Filter quarters containing the biomass of approximately 500 ml of lake water were saponified in 10 ml 80:20 (v/v) methanol:aqueous potassium hydroxide solution (4.45 mol l−1) at 80 °C for 5 h. The neutral lipids were removed from the aqueous solution by liquid–liquid extraction with n-hexane in three steps. Acidic compounds, including fatty acids (FAs), were obtained after acidification with HCl to pH 2–3. The released FAs were transformed into trimethylsilyl ether derivatives using N,O-bis-trimethylsilyl-trifluoroacetamide. In a second method, lipids were transesterified by treatment with trimethylsulfoniumhydroxide in methanol modified from Butte (1983). Quantification of saponified whole-cell core lipid concentrations were measured by gas chromatography-flame ionization detector (Supplementary methods S1.1, Table 1, Supplementary Figure 2). More information, including information about lipid identification and quantification errors, can be found in the Supplementary Methods.
Table 1. Core lipid concentrations in μg l−1 released by alkaline hydrolysis.
| Identified compounds |
Time (h) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 12 | 18.3 | 24.2 | 36.8 | 46.8 | 56 | 65 | |
| Unassigned lipids | |||||||||
| C24:0 alcohol | 0.2 | 0.0 | 0.9 | 0.6 | 0.3 | 1.0 | 0.3 | 1.4 | 0.3 |
| Phytanol | 4.7 | 3.4 | 2.9 | 2.3 | 3.0 | 3.0 | 3.3 | 4.0 | 4.3 |
| Phytanoic acid | 1.6 | 1.5 | 2.3 | 2.5 | 2.2 | 2.4 | 2.7 | 2.4 | 2.8 |
| Bacterial FAs | |||||||||
| iso-C15:0 FA | 3.9 | 3.4 | 4.6 | 4.3 | 4.0 | 5.0 | 4.9 | 5.8 | 4.2 |
| anteiso-C15:0 FA | 1.8 | 2.2 | 2.0 | 2.0 | 2.1 | 2.2 | 2.4 | 3.0 | 2.6 |
| iso-C16:0 FA | 0.6 | 0.6 | 1.0 | 1.2 | 0.8 | 0.9 | 0.9 | 0.7 | 0.5 |
| cis-C16:1 FA | 1.0 | 0.8 | 1.1 | 1.3 | 1.1 | 1.4 | 1.4 | 1.4 | 1.1 |
| trans-C16:1 FA | 5.4 | 4.7 | 5.0 | 4.6 | 4.5 | 5.6 | 6.1 | 6.1 | 4.7 |
| C16:0 FA | 8.6 | 8.1 | 10.3 | 7.0 | 7.6 | 10.0 | 8.8 | 11.2 | 8.3 |
| iso-C17:0 FA | 2.2 | 1.9 | 1.9 | 1.8 | 1.8 | 2.0 | 2.3 | 2.1 | 1.7 |
| C17:0 2-OH FA | 0.2 | 0.0 | 0.3 | 0.6 | 0.0 | 0.6 | 0.4 | 0.5 | 0.4 |
| cis-C18:1 FA | 1.9 | 0.9 | 1.1 | 0.9 | 1.0 | 1.3 | 1.4 | 0.4 | 0.6 |
| trans-C18:1 FA | 3.2 | 2.7 | 3.0 | 2.8 | 2.8 | 3.6 | 3.9 | 3.8 | 2.8 |
| C18:0 FA | 1.6 | 4.4 | 6.6 | 2.9 | 1.6 | 5.6 | 4.0 | 8.6 | 2.2 |
| Sum FAs | 30.4 | 29.7 | 36.9 | 29.4 | 27.3 | 38.2 | 36.5 | 43.6 | 29.1 |
| Archaeols | |||||||||
| DPG (C20/C20) | 66.2 | 59.9 | 56.6 | 52.2 | 60.4 | 54.3 | 59.7 | 60.1 | 54.2 |
| PGS (C25/C20) | 13.7 | 12.8 | 9.4 | 7.7 | 10.9 | 9.2 | 11.2 | 11.6 | 13.1 |
| Sum archaeal lipids | 79.9 | 72.7 | 66.0 | 59.9 | 71.3 | 63.5 | 70.9 | 71.7 | 67.3 |
| Sum all lipids | 116.9 | 107.2 | 109.1 | 94.8 | 104.1 | 107.9 | 113.6 | 122.9 | 103.9 |
| (n-C18:0+n-C16:0 FAs)/(DPG+PSG) | 0.13 | 0.17 | 0.26 | 0.17 | 0.13 | 0.25 | 0.18 | 0.28 | 0.16 |
| n-C18:0/n-C16:0 | 0.19 | 0.54 | 0.64 | 0.41 | 0.21 | 0.56 | 0.45 | 0.77 | 0.27 |
| DPG/PSG | 4.8 | 4.7 | 6.1 | 6.8 | 5.6 | 5.9 | 5.3 | 5.2 | 4.1 |
Abbreviations: DPG, diphytanyl glycerol; FA, fatty acid; PGS, phytanylsesterterpanyl glycerol.
Statistical analysis of lipid data
Multivariate and univariate statistics were performed using the R programming environment (R_Core_Team, 2005; with the packages vegan and gplots). Absolute abundances of lipids (μg l−1) were used to calculated principal coordinate analysis based on Bray Curtis index, hierarchical clustering based on Euclidean dissimilarity, multi-response permutation procedure (implemented in the vegan package) based on Bray Curtis index and 1000 permutations and permutational multivariate analysis of variance (Adonis test implemented in vegan package) also based on Bray Curtis index and 1000 permutations. Metadata variables for multi-response permutation procedure and/or permutational multivariate analysis of variance were either categorical (day versus night) or continuous (temperature, day time starting 0600 hours).
Lipid abundances were individually tested for day versus night differences using an analysis of variance and correlated with either temperature or daytime using a Pearson correlation. Corrections for multiple testing were achieved by applying Benjamini–Hochberg correction (Benjamini and Hochberg, 1995) and are explicitly mentioned.
DNA extraction and sequencing
Community genomic DNA was extracted from 15 samples (five samples of each of three filter sizes) using a phenol:chloroform extraction protocol. Samples were sent to the J Craig Venter Institute (JCVI, Rockville, MD, USA) for library construction and sequencing (one sample per lane of paired-end, 100-bp insert size, Illumina, San Diego, CA, USA). Community genomic DNA extracted from one 0.7 μm glass fiber filters used for lipid analysis was also sequenced so that lipid information could be directly linked to community composition. The objective was to establish the relative abundances of the major organism groups that contributed to the lipid signal. Sequencing of DNA extracted from this filter fraction was performed at the Genomics Facility, Monash University Sunway Campus, Malaysia (Illumina MiSeq, 310-bp insert size).
Assembly and annotation of metagenomic samples
A total of 6.15 Gbp of sequence was obtained from the 3.0-μm filters and 1.389 Gbp from the 0.8-μm filters. Assembly and annotation focused on the 10 Gbp of high-quality sequence obtained from samples collected on 0.1-μm filters (Table 2). Each sample's sequence was individually assembled using IDBA-UD (Peng et al., 2012). Gene prediction was carried out using the meta-Prodigal option of Prodigal (Hyatt et al., 2010). Functional annotation was performed using BLASTp to compare predicted protein sequences with the KEGG and UniRef90 databases, giving priority to reciprocal best BLAST hits, and then protein motifs were annotated using InterproScan (Quevillon et al., 2005) using an e value ⩽1e-10 with ⩾70% coverage of the length of the protein.
Table 2. Description of 0.1-μm filter samples.
| Sample name | Filter size (μm) | Temperature at collection (°C) | Date | Time collected | Gbp |
|---|---|---|---|---|---|
| LT2010_0.1_AM_1 | 0.1 | 19.5 | 7 January 2010 | 7:45 | 1.36 |
| LT2010_0.1_PM_1 | 0.1 | 32 | 7 January 2010 | 20:00 | 2.73 |
| LT2010_0.1_AM_2 | 0.1 | 21 | 8 January 2010 | 8:00 | 3.82 |
| LT2010_0.1_PM_2 | 0.1 | 44.5 | 9 January 2010 | 16:15 | 2.17 |
A total of 6.2 Gbp DNA sequence was obtained from cells collected on the 0.7-μm filter. Sequences were assembled and analyzed using the same methods as described above for the 0.1-μm filter samples. The community composition of the 0.7 μm was profiled using contig coverage, GC content and phylogenetic signal.
Phylogenetic analysis based on a ribosomal protein tree
Ribosomal protein sequences were chosen for phylogenetic analysis from the set that are single copy and are unlikely to undergo lateral gene transfer (Sorek et al., 2007; Wu and Eisen, 2008). We used ribosomal proteins L2P, L3P, L4P, L5P, L6P, L14P, L15P, L18P, L22P, L24P, S3, S8, S10, S17 and S19, as these are encoded in a syntenic block on most microbial genomes (Hug et al., 2013). The use of the syntenic block allows multi-gene concatenated alignments to be made without the requirement for genome binning (Hug et al., 2013). All ribosomal protein blocks were identified in the assembled sequences based on BLASTp annotations. We required at least 8 out 15 of the ribosomal proteins for a given scaffold to be included in the analysis. If identical sequences were found (zero substitutions) in different samples, the scaffold with the greatest number of predicted proteins was chosen as the representative of that taxon. These gene data sets and a reference set of all published Euryarchaeotal genomes, as well as representatives across the archaeal tree of life, were aligned using MUSCLE (Edgar, 2004a, 2004b). The alignments were manually trimmed to remove single-taxon insertions and then concatenated to form a 15-gene alignment. A phylogenetic tree was constructed from the concatenated alignment using RAxML under the PROTGAMMALG substitution model with 100 bootstrap resamplings (Stamatakis, 2006).
A separate bacterial concatenated ribosomal protein tree was constructed for the Bacteroidetes phylum, as described for the archaea above. All published Bacteroidetes genomes, as well as all Bacteroidetes-affiliated scaffolds with a minimum of 8 out of 15 of the selected ribosomal proteins, were included in the tree.
Estimation of time series organism abundance patterns
The relative organism abundance in each sample, and estimates of the time series abundance patterns across four successful 0.1 μm metagenomic samples, were determined by using Bowtie and Bowtie2 (Langmead et al., 2009, Langmead and Salzberg, 2012) to map reads to the scaffolds encoding the ribosomal protein sequences (used to construct the concatenated ribosomal protein tree, see above). No mismatches were allowed, paired-end information was not used, and only the best hits were reported. Coverage was calculated by multiplying the total number of reads mapped by the read length (100 bp), and dividing by individual scaffold length. Coverage was normalized to account for different numbers of reads per sample. The normalized coverage was converted to a per-sample percentage and graphed.
Cryogenic TEM
Samples for cryo-TEM were prepared on site using a portable cryo-plunge device as described in Comolli et al. (2012). The program ImageJ (NIH, http://rsb.info.nih.gov/ij/) was used for analysis of the two-dimensional image projections.
Cell counts
Water samples were collected in the field in duplicate 20 ml 1.2 N HCl acid washed Nalgene bottles that had been rinsed three times with small amounts of lake water before being filled. Samples were preserved with 1% (final vol:vol) formalin for later enumeration. Samples (volumes of between 0.5 and 1 ml) were filtered onto a 0.2 μm Nuclepore black polycarbonate 25 mm diameter filter backed with a supporting 0.2 μm pore size cellulose nitrate filter and stained with an acridine orange solution (0.1% in 5% formalin, 0.22 μm filtered). Filters with cells were transferred to a microscope slide and enumerated using the acridine orange direct counting method (Hobbie et al., 1977). An Olympus BX60 epifluorescence microscope (Olympus America, Center Valley, PA, USA) with 100X objective and blue excitation generated using a 150-W xenon lamp (Opti-Quip, Highland Mills, NY, USA). Ten fields per replicate sample were counted.
Results
Over the sampling period (7–9 January 2010), water temperatures ranged between 19.5 °C and 45.4 °C and air temperatures between 19 °C and 48 °C. There was a warming trend over the 3 days of collection, with the daily water temperature varying by 18.4 °C on average. Coolest temperatures occurred in the early morning with the warmest measured in the late afternoon. Weather station data from Ouyen, Victoria (40 km NW of Lake Tyrrell) show that the daily temperatures during the sample collection period were among the highest recorded in the area over the last 57 years.
Solution chemistry reflected the ongoing evaporation over the course of the experiment, with concentrations of magnesium, strontium, potassium, lithium and boron ions increasing by ~20% (Supplementary Table S1, Supplementary Figure S1). The solution was circumneutral (pH~7.16). As previously described (Heidelberg et al., 2013; Podell et al., 2014), the major ions in the water were Na+, Cl−, Mg2+ and SO42−. Sodium and chloride were the only ions with stable concentrations over the sampling period, indicating precipitation of halite at constant saturation level. By comparison with the typical progression of precipitation of different elements in saline salterns (Herrmann et al., 1973), it is possible to infer that at the time of sampling precipitation of gypsum was ongoing and likely almost finished. Water samples were also analyzed for aluminum, cobalt, chromium, copper, molybdenum, vanadium and iron, and the anions fluoride, phosphate and nitrate, but these were generally below detection limits. Concentrations of these ions were not significantly associated with changes in the community lipid profile (Supplementary Table S2). The concentration of manganese (0.23–0.68 mg l−1) was enriched compared with Pacific Ocean water (0.06–0.17 μg kg−1) (Klinkhammer and Bender, 1980), and it was the only ion that showed a strong increase (~200%) over the 66-h period (Supplementary Table S1).
Core lipid analysis of water filtrates yielded diverse alcohols and FAs (Table 1, Supplementary Figure S3). The most abundant lipids were isoprenoidal building blocks of archaeal cytoplasm membranes, diphytanylglycerol (archaeol) and phytanylsesterterpanyl glycerol (Table 1), suggesting dominance of archaeal organisms. In contrast, sterols and other lipids diagnostic of eukaryotes were below detection limits in all samples. The second most abundant class of lipids were the FAs, which in the absence of eukaryotes, are assigned to be nearly quantitatively bacterial. These findings are consistent with metagenomic analyses of the 0.7-μm filter at time point 0 h, which indicate that the sample contained ~7% bacteria, 87% archaea (Euryarchaeota) and 6% Nanohaloarchaea (for detailed information, see Supplementary Table S4). At t=0 h, the most abundant FAs were n-C16:0 followed by trans-C16:1, iso-C15:0, trans-C18:1, cis-C18:1 and n-C18:0. Saturated FAs were generally more abundant than their unsaturated counterparts, and FAs with an even carbon number were more abundant than odd-numbered homologs.
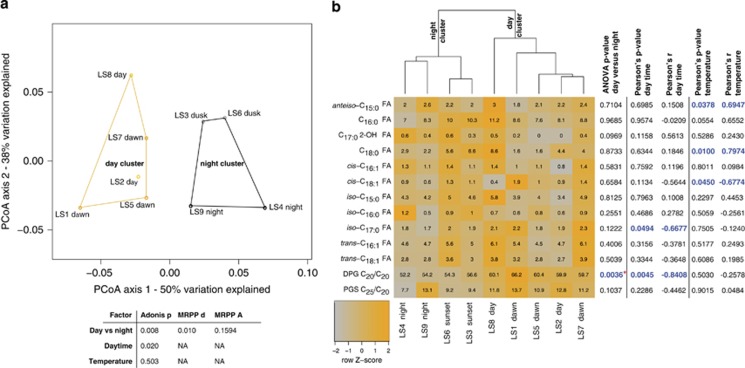
The detected core lipids provide quantitative information about archaeal and bacterial biomass. The absolute abundances of membrane lipids showed clear changes during day–night cycles over the 66-h period (Figure 2c). Although the absolute abundances of bacterial unsaturated and branched FAs showed little diel variation (Supplementary Figure S3), the combined abundance of saturated FAs n-C16:0 and n-C18:0 co-varied with water temperature, exhibiting a trend of increasing concentrations during daylight hours and decreasing concentrations after dark (Figure 2c). For example, on the first day, the sum of all bacterial FAs increased by 21% during the daylight period and decreased by 26% during the night (Table 1). Strong diurnal variations were also observed for the ratio of FA n-C18:0/n-C16:0 FAs, which fluctuated by a factor of 3 to 4, tracking with changes in water temperature (Table 1, Figure 2b). The overall microbial community profiled using lipid abundances showed a significant difference between day and night samples in Adonis testing, multi-response permutation procedure, principal coordinate analysis and hierarchical clustering (Figure 3). Multivariate statistics, as shown in the principal coordinate analysis plot, did not display a significant effect of temperature on the entire community profile (Adonis P-value; Figure 3a), yet abundances of specific bacterial lipids did show significant correlations with temperature (Figure 3b). Specifically, bacterial FA cis-C18:1 showed a significant decrease with temperature increase, C18:0 and anteiso-C15:0 both showed a statistically significant linear increase with temperature.
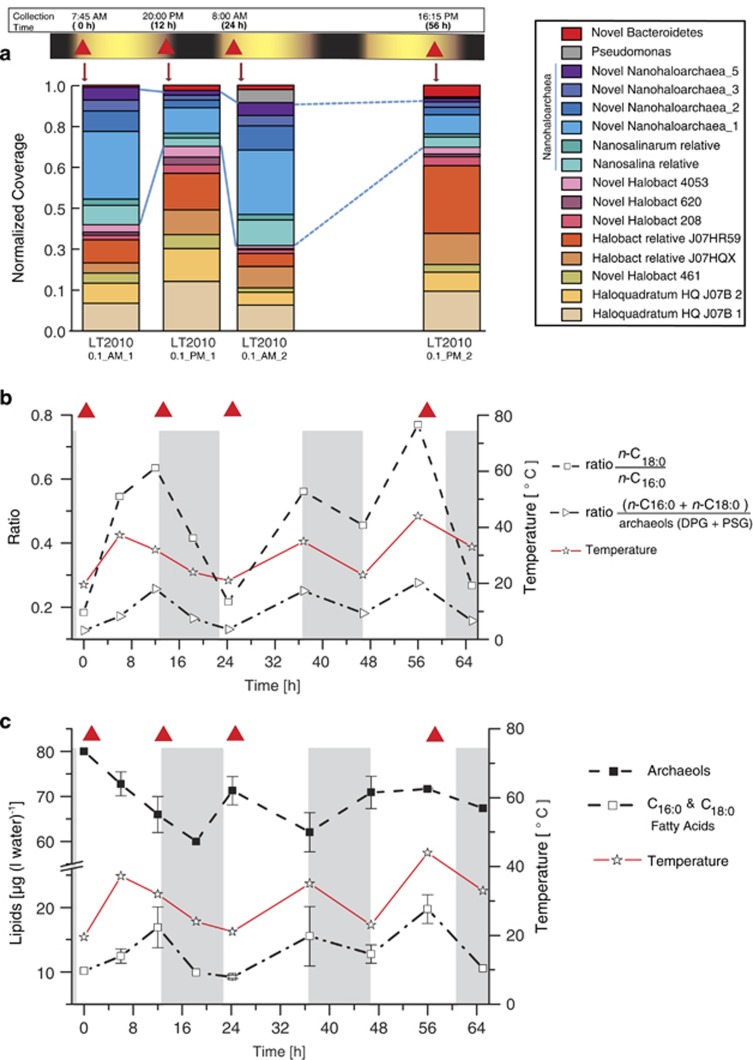
Figure 2.
Results for GC-FID analysis of trimethylsilyl (TMS) derivatives from hydrolyzed filter samples and organism relative abundance in 0.1-μm filter metagenomic data sets. (a) Diel cycle trends in organism relative abundance in the hypersaline Lake Tyrell. Organism abundance was estimated from depth of metagenomic sequence coverage for marker scaffolds encoding ribosomal protein genes. The relative abundances of the 16 operational taxonomic units are displayed as stacked bar charts for the four 0.1-μm filter sample data sets. The collection time of each sample is shown at the top and highlighted with red triangles. The legend defined the taxonomic assignment for each operational taxonomic unit. Coverage was calculated by multiplying the total number of reads mapped by read length and dividing by individual scaffold length. The coverage was normalized to account for different numbers of reads per sample and converted to a per-sample percentage. (b) The ratio of the absolute abundance of n-C18:0 over n-C16:0 FAs (open boxes) highlighting diurnal variation of average bacterial membrane lipid chain length, and ratio of the abundance of bacterial versus archaeal lipids (open triangles). In (b) and (c), water temperature on each sampling point is given in °C and gray areas show hours without daylight. Points without error bars indicate that no replicate filtrates were collected at these time points because of sampling conditions in the field. Red triangles indicate time of collection of 0.1-μm filter sample used for metagenomic analyses. (c) Absolute lipid concentrations from saponified 0.7-μm filter samples. Curves show the sum of C16:0 and C18:0 FAs [μg (l lake water)-1] of bacterial origin, and archaeols (diphytanylglycerol (DPG) and phytanylsesterterpanyl glycerol (PSG)) of archaeal origin.
Figure 3.
Results of multivariate and univariate statistical analyses of lipid abundances. (a) Principal coordinate analysis (PCoA) based on the Bray-Curtis index derived from absolute lipid abundances (LS, lipid sample). The ordination shows a separation of day and night samples along PCoA axis 1. Adonis P-values (Adonis p) and multi-response permutation procedure (MRPP) delta (MRPP d) are significant (<0.05) for these groupings and suggest a significant difference in community structure between day (day and dawn) and night (night and dusk) samples. Actual difference in the sample grouping is estimated by the chance corrected within-group agreement (MRPP A). The continuous variables daytime and temperature were also tested, whereas daytime was associated with a significant change in the community composition based on lipid abundances. (b) Heatmap combined with hierarchical clustering (based on Euclidean distance) and individual univariate analysis for each lipid signature. Samples show grouping into a day and night cluster and confirm findings from PCoA, Adonis and MRPP. Abundance of diphytanylglycerol (DPG) C20/C20 was significantly between day and night samples (*P-value was robust to Benjamini–Hochberg correction). Significant correlations with daytime or temperature were found for anteiso-C15:0 FA, C18:0 FA, cis-C18:1 FA, iso-C17:0 FA, and DPG C20/C20, all show in blue.
Archaeal core lipids diphytanylglycerol and phytanylsesterterpanyl glycerol showed an opposite trend to bacterial FAs, with the highest concentrations in the early morning and declining abundances during the day (Figure 2c). Abundances of the archaeal lipid diphytanylglycerol C20/C20 significantly increased when comparing night-to-day samples (analysis of variance P-value of 0.0036, robust to Benjamini–Hochberg correction) and a Pearson's correlation showed that the abundance of these archaeal lipids are negatively correlated with daytime (along with the abundance of the bacterial lipid iso-C17:0; Figure 3b). During the first day, archaeal core lipid concentrations decreased by 25%, reaching a minimum value at 0145 hours (18 h into the time series) followed by an increase of 19% toward dawn. The inverse diurnal behavior of the two microbial domains is particularly evident in the ratio of total bacterial to archaeal lipids, which increased by up to 58% during daylight hours and declined by up to 32% during the night (Table 1), whereas the ratio of bacterial n-C16+n-C18 over archaeal diphytanylglycerol+phytanylsesterterpanyl glycerol even doubled during the first day and halved in the subsequent dark hours (Figure 2b).
All organisms were sampled from the photic zone. Based on epifluorescence microscopic cell counts, we estimate an average planktonic cell concentration of 6 × 108 cells ml-1. We extracted between 1.84 and 51.87 μg of high-quality DNA from 0.1 , 0.8 and 3.0-μm filters collected at five time points for sequencing. As a result of a sequencing failure (reason unknown), only four 0.1 μm, one 0.8 μm and three 3.0 μm samples, out of 15 samples, generated sequencing information of high enough quality for analysis. This data set is augmented by sequence data from the 0.7-μm filter. Between 0.045 and 3.829 Gbp of DNA sequence information was generated from microbial populations from individual 0.1-μm filters, 1.39 Gbp from the 0.8-μm filter, 1.09–3.15 Gbp from the 3.0-μm filters and 5.72 Gbp from the 0.7-μm filter used for lipid analysis.
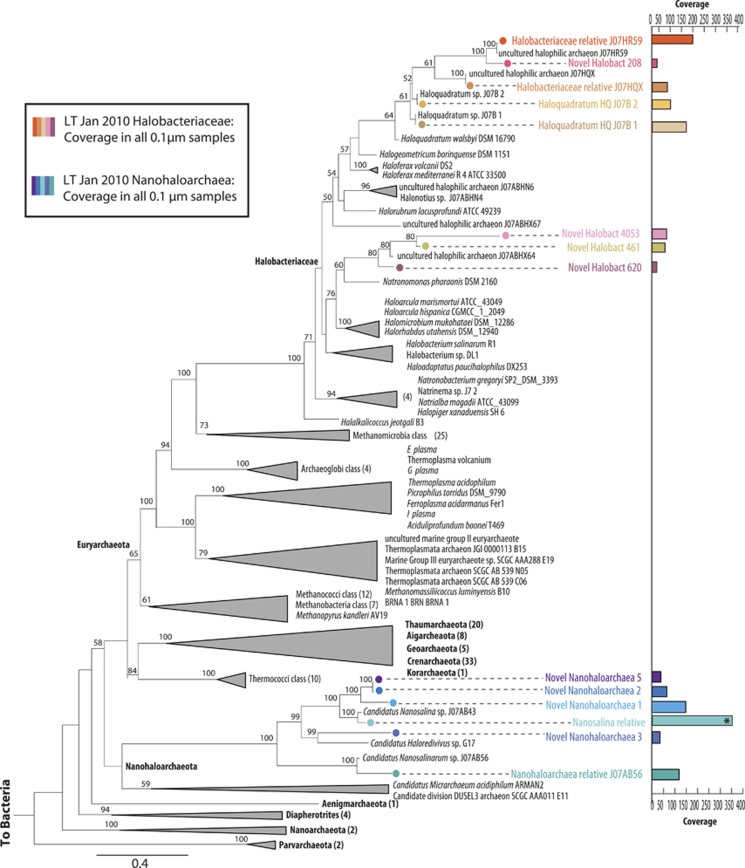
We used the expectation maximization iterative reconstruction of genes from the environment (EMIRGE) method (Miller et al., 2011) to reconstruct 80 near-complete 16 S ribosomal RNA gene sequences from all of the genomic data sets (Supplementary Methods S2, Supplementary Table S3, Supplementary Figure S4). When the 80 sequences were divided into groups based on taxonomic assignment using the RDP classifier (Wang et al., 2007), 12 common taxa were resolved. We used read mapping to track the abundances of these 12 groups over four time points, focusing on the four 0.1-μm filter metagenomic data sets. The results revealed short-term fluctuations in community composition. The relative abundance levels of two out of the three Nanohaloarchaea increased over the night period (teal and blue-green in Supplementary Figure 4). The pattern for the third Nanoharchaeal group was unclear. The coexisting Haloquadratum, Halorubrum, Halobacteriaceae and Salinibacter organisms showed an opposite trend, with relative abundances increasing during the day. However, ribosomal protein sequences provided higher-resolution taxonomic information about community membership than EMIRGE. Given the interest in detecting the presence of Nanohaloarchaea, we focused analyses on the 0.1-μm filter samples. We identified 16 distinct assembled ribosomal blocks from the 0.1-μm filter samples that were used as proxies for different organism types. Fifteen of the 16 organisms were detected in all 0.1-μm data sets. The concatenated ribosomal protein tree indicated that 14 of these organisms sampled on the 0.1-μm filter were archaea (Figure 4), all of which belong to previously undescribed species.
Figure 4.
The 15 ribosomal protein concatenated phylogeny places the 14 archaeal operational taxonomic units as novel organisms compared with previously sequenced genomes in 0.1-μm samples. RAxML phylogenetic tree with 100 bootstrap resamplings rooted at the split between archaea and bacteria. Numbers in parentheses indicate number of sequences included in the analysis that are not show. Fourteen distinct archaeal ribosomal blocks were identified in all of the 0.1-μm data sets, all of which are novel at least at the species level. Organism colors are consistent with Figure 2. Six are Nanohaloarchaea (basal archaeal group on the tree), all shown in purple and a gradient of blue. The organism most closely related to Candidatus Nanosalina sp. J07AB43 is the most abundant organism on the 0.1-μm filters (in teal on this tree and marked with an asterisk).
Six archaeal groups were classified as members of the candidate Phylum Nanohaloarchaeota (Rinke et al., 2013) based on clustering with previously reported Nanohaloarchaeotes; three of these are novel and represent a sister clade to the previously described Nanosalina lineage. One sequence type groups with Candidatus Nanosalina sp. J07AB43, one groups with Candidatus Nanosalinarum sp. J07AB56 (Narasingarao et al., 2012) and the third with Candidatus Halorevidivivus sp. SPG17 (Ghai et al., 2011). The population most closely related to Candidatus Nanosalina sp. J07AB43 was the most abundant organism type on the 0.1-μm filters (352 × average coverage in the metagenome; marked with an asterisk and in teal in Figure 4).
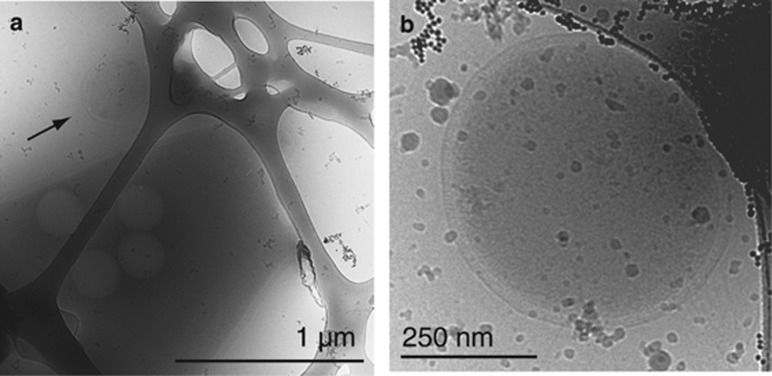
The Nanohaloarchaea were predicted to have small cell size based on enrichment on 0.1-μm filters, small genome size and microscopic identification based on group-specific fluorescence in situ hybridization (Narasingarao et al., 2012). Cryo-TEM imaging of planktonic cells recovered from lake water on the 0.1-μm filter directly verifies the presence of very small cells (<500 nm in diameter). Other than small cell size, features of these organisms, presumed to be the Nanohaloarchaea, include an inner membrane, outer membrane and periplasmic space dotted with small contrast elements (Figures 5a and b).
Figure 5.
Cryo-transmission electron microscopy (TEM) images of planktonic cells recovered from lake water on the 0.1-μm filters confirm the presence of very small cells, likely members of the Nanohaloarchaea. In all images, the electron microscopy grid is evident in the background. (a) Cryo-TEM of an organism presumed to be a nanohaloarchaeon next to a larger putative Haloquadratum-like organism. (b) An organism presumed to be a member of the Nanohaloarchaea because of its small size and small contrast elements in the inner membrane, outer membrane and perisplasmic space, as have been seen in the ARMAN nanoarchaea (Comolli and Banfield, 2014).
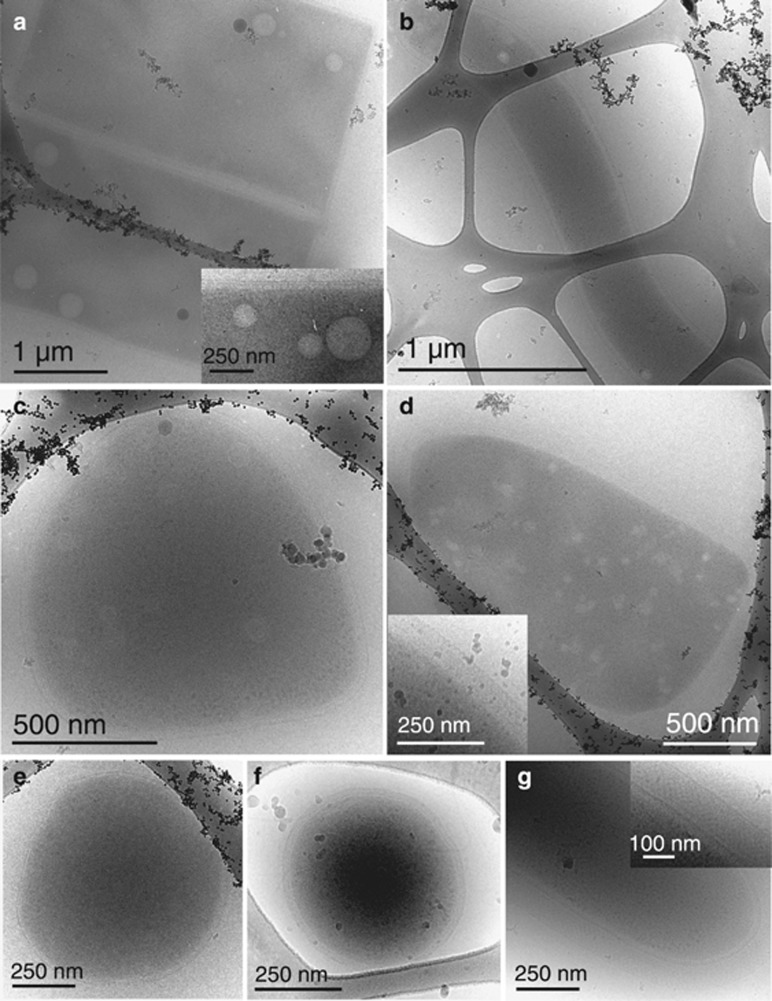
Also represented on the 0.1-μm filters were eight uncultured members of the Halobacteriaceae family, two of which are in the Haloquadratum genus (one closely related to Haloquadratum DSM 16790). The remaining Halobacteriaceae are related to Halorubrum, Halonotius and Natronomonas spp. and to genotypes reported previously from Lake Tyrrell by Podell et al. (2013, 2014) in a prior metagenomic study (Figure 4). Rectangular prism-shaped cells with large vesicles and a three-layer cell wall were evident in cryo-TEM images of planktonic cells recovered from lake water. Based on prior TEM studies (Comolli et al., 2012), we infer that these cells belong to the genus Haloquadratum (Figure 6a). Other prominent morphologies included rod-shaped and triangular cells (Figures 6b and d).
Figure 6.
Other prominent morphologies surveyed at Lake Tyrrell via cryo-transmission electron microscopy (TEM) imaging. (a) Rectangular prism-shaped cells with large vesicles and a three-layer cell wall, likely in the Haloquadratum genus, (b) long rod-like, (c) cocci with vesicles, (d) triangular with multiple vesicles and a three-layer outer membrane, (e) small cocci, (f) an organism presumed to be a nanohaloarchaeon and (g) rod-like organism with an exceptionally thick S-layer-like outer surface.
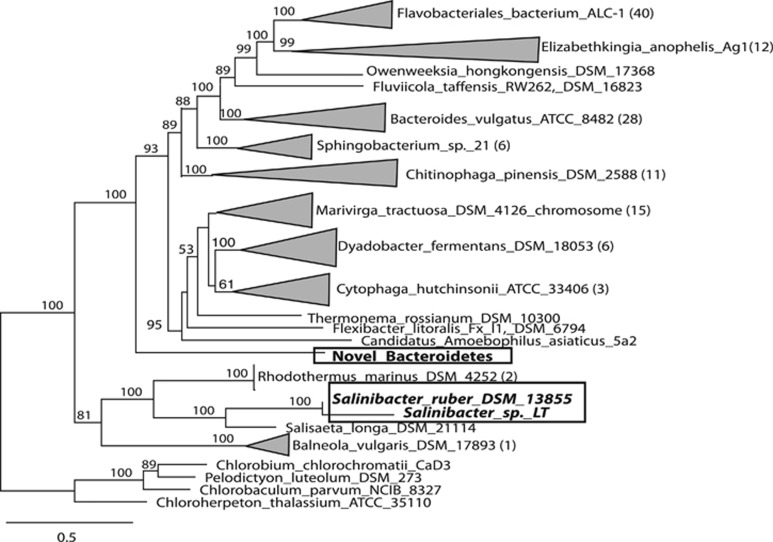
In the 0.1-μm filters only 2 of the 16 ribosomal protein blocks were associated with bacteria. One of these was associated with bacteria in the phylum Bacteroidetes. Interestingly, this organism is not in the same class as Salinibacter, a well-known halophile. Based on the length of the branch and the placement as a deep outgroup of several Bacteroidetes classes in the concatenated ribosomal protein tree for this phylum, this organism is likely a member of a genomically uncharacterized class in the Bacteroidetes (Figure 7). Sequences corresponding to Pseudomonadales (average coverage 14x) were present in only two samples and are possible contaminants.
Figure 7.
A 15 ribosomal protein concatenated phylogeny for the phylum Bacteroidetes. Rooted, RAxML phylogenetic tree with 100 bootstrap resamplings. Numbers in parentheses indicate number of sequences included in the analysis that are not show. This tree places the Lake Tyrell Bacteroidetes sp. as a novel member of a previously unsequenced class (bold). All sequenced Bacteroidetes genomes were included in the phylogeny. The boxes delineate the positions of the Lake Tyrell Bacteroidetes sp. and Salinibacter sp. (bold), the known halophiles within the Bacteroidetes, and highlight the evolutionary distance between these two groups.
Mapping of reads from individual 0.1-μm filter samples to the 16 scaffolds encoding the ribosome subunit protein sequences (representing 16 organism types), confirmed an increase in the relative abundance of all six Nanohaloarchaea in samples collected early in the morning and a decrease in relative abundance in the samples collected at the end of the day (Figure 2a). All of the coexisting Halobacteria (Euryarchaeota) and the Bacteroidetes showed the opposite trend, with relative abundances increasing during the day and decreasing after the night (Figure 2a). Yet, the lipid data establish relatively constant whole community cell abundance levels over the experiment. The combination of lipid and metagenomic data shows an overall preference in archaea for nighttime, relative to bacteria, which increase during the daytime. The metagenomic data from the 0.1-μm filter data clearly establish that Nanohaloarchaea exhibit a much stronger nighttime preference than do the other halophilic archaea (Figure 2a).
Discussion
The diel cycle (day and night) is one of the most conspicuous features of life at and near the Earth's surface. Previous studies have demonstrated that individual microorganisms are capable of maintaining an estimate of environmental time (Mori and Johnson, 2001), and that this ability is of critical adaptive value to the cell (Johnson, 2005). For many years, it was reasoned that since rapidly dividing microorganisms double in less than a day, a timing or predictive mechanism that extended longer than its doubling period would be of no use (Johnson et al., 2008). Disproving this notion, Huang et al. (1990) demonstrated that the phototrophic marine cyanobacterium Synechococcus elongatus displays a robust circadian rhythm. More recently, it was shown that photoheterotrophic Halobacterium synchronize their physiologies with a light–dark cycle (Whitehead et al., 2009). Evidence of this diel behavior has been found in individual organisms throughout the microbial world, pointing to its critical role in the ecology and evolution of microbial communities. Being able to anticipate the temporal interrelationships with changes in environmental factors and to prepare a response allows the organism to function optimally in a defined environmental space (Baliga, 2008). Complicating this, microorganisms exist in complex microbial communities defined by networks of synergistic and competitive interactions. As Tsai et al. (2012) demonstrated, while Synechococcus spp. does have an internal physiological diel cycle, community interactions also impact its day–night patterns. Similarly, Otteson et al. (2014) recently reported the existence of diel cycles in the expression of many gene transcripts of naturally occurring marine heterotrophic bacterioplankton groups. Adding to this work, this study points to the existence of diel cycles in microbial communities in extreme, hypersaline acquatic environments, making them potential model systems for diel cycle analysis. In addition, our work suggests the importance of community-wide approaches when exploring diel changes in microbial communities.
In Lake Tyrrell, both bottom-up (nutrients and/or physico-chemical parameters) and top-down (protistan grazing or viral predation) ecological processes may contribute to microbial community dynamics over short time periods. Although the diverse and dynamic viral assemblages of Lake Tyrrell have been well characterized (Emerson et al., 2012, 2013a, 2013b), metagenomic data do not indicate the occurrence of diel viral blooms over the time period studied here. Heterotrophic nanoflagellates can have significant impacts on microbial community turnover in hypersaline environments (Park et al., 2003). A few microbial eukaryotes have been described in Lake Tyrrell. The heterotrophic nanoflagellate, Colpodella edax was observed to actively graze on the green algae, Dunaliella spp. (Heidelberg et al., 2013). However, eukaryotic lipids were below detection limits in all samples in this study and eukaryotes were not detected in metagenomic data. Thus, we conclude neither grazing nor viral predation explain the observed diel patterns.
We detected striking shifts in the proportions of bacterial and archaeal lipids in day and night cycles. Statistical analyses confirmed a significant difference in lipid abundance-based community structure when comparing day-to-night samples. In addition, statistical analyses indicate a potential for oscillation, a gradual and repeating change in the microbial community across the diel cycle (Adonis P-value; Figure 3a). The observed cycles are likely driven by the interconnection of temperature, light abundance, dissolved oxygen concentration and nutrient flux effects. These shifts do not appear to be controlled by salt concentrations as the change in ion concentrations is a continuous process (over the longer term drying cycle), decoupled from daily temperature fluctuations (Supplementary Table S2).
Nutrient fluxes could drive changes in community composition. As we do not detect photosynthetic primary producers in the lake water (cyanobacterial abundance levels were <0.1%, based on the 0.7-μm filter) carbon compounds that support heterotrophic growth must be remotely derived. Potential sources include groundwater that has percolated through surrounding natural vegetation and farmland or the microbial mats that occur at many locations at the lake-sediment interface, in some cases beneath the salt crust. Although some carbon compounds are likely sourced from the surrounding land or groundwater, it is unlikely that the supply would show a strong diel fluctuation. Externally derived organic carbon to the pool is more likely to occur as diel pulses if derived from phototrophic organisms in mats. However, as both heterotrophic bacteria and archaea should respond in similar ways to such inputs, we suggest that carbon supply is not the primary driver of the observed microbial diel cycle. If carbon fluxes are linked to diel changes, it may be as a feedback, for example, because of release of lysate following in situ death of specific organisms, with cell death caused by changes in the physical environment.
Light variation may contribute to the diel signal, as several halophilic archaea are photoheterotrophic. However, the advantage of light energy does not offset the apparent disadvantages of daylight growth for archaea. Thus, it seems likely that the increasingly archaea-dominated ecosystem during the night and increase in bacterial abundance during the day is controlled by another factor.
Although lipid-based statistical analyses suggest that temperature is not the main factor driving the changes in the community relationships of the different samples collected, three individual bacterial FAs showed a significant correlation with the temperature measured. Hence, we suggest that one of the important drivers of the diel cycle for bacteria in the system studied here may also be the large daily variation in water temperature. Bacteria can adapt the physical properties of existing cytoplasmic membranes by introduction of double bonds in cis or trans configuration or introduction of cyclopropyl rings and methyl groups. Yet, cyclopropyl rings and systematic changes in methylation were not observed in the evaporating brine pool, even during temperature fluctuations of >20 °C (Supplementary Figure S3). However, the absolute abundance of cis-C18:1 was inversely correlated with temperature (Figure 3b) suggesting insertion of a double bond into membrane lipids in response to dropping temperatures. Moreover, bacteria can also stabilize their membranes at high temperatures through elongation of membrane lipids (Marr and Ingraham, 1962). This may explain the substantial increase in the ratio of n-C18:0/n-C16:0 FAs that tracks with the diurnal rise in water temperature (Table 1, Figure 2b). Surprisingly, little is known about membrane adaptation mechanisms of archaea to temperature. Insertion of more pentacyclic rings can increase thermal tolerance in archaea with membrane-spanning glycerol diphytane glycerol tetra ether lipids (Derosa et al., 1980), but these compounds were not detected (Supplementary Methods 1.3).
We computed the minimum cell doubling times required to account for the highest absolute increases of total bacterial and archaeal lipids during the diurnal cycles (assuming cells maintain a constant average size and lipid content) and found them to be ~19 h for bacteria and ~24 h for archaea. This is slightly faster than previously reported, yet compares well with known doubling times from laboratory culturing studies for halophilic archaea (Coker and DasSarma, 2007; Pedros-Alio et al., 2000).
Metagenomic tools allowed new exploration of Nanohaloarchaeal diversity. Nano-sized archaea were first described from hot springs environments (Huber et al., 2002), then in acid mine drainage (Baker and Banfield, 2003; Comolli et al., 2009) and subsequently in hypersaline environments (Narasingarao et al., 2012). More recently, single cell sequencing results suggested the existence of other clades of nano-sized archaea (Rinke et al., 2013). There is genomic and physical evidence that both the hyperthermophile Nanoarchaeum equitans and the acidophilic ARMAN lineage organisms depend on an associated organism for many metabolic needs (Huber et al., 2002; Comolli et al., 2009; Podar et al., 2013). Both have been co-cultivated with other organisms (Igniococcus in the case of Nanoarchaeum equitans; biofilm communities in the case of ARMAN).
In this study, Nanohaloarchaeal genomes (both published and presented here) were analyzed through the ggKbase interactive platform (http://ggkbase.berkeley.edu), and they do not appear to encode cytochrome C oxidase, suggesting that these organisms cannot use oxygen. In fact, the encoded metabolic potential is suggestive of a limited, fermentation-based metabolism. An anaerobic lifestyle was unexpected in the shallow (<20-cm deep), well-mixed lake environment. However, a diel pattern of non-planktonic mat photosynthesis could drive fluctuations in O2 concentrations, with a tendency to lower concentrations at night that could favor Nanohaloarchaea over other archaea. It is possible that Nanohaloarchaea depend on metabolites derived from coexisting organisms. This could involve uptake of dissolved compounds, possibly released by cell lysis, or a more direct interaction. The cell lysate-based metabolism of the archaeon Halobacterium salinarum has been linked to release of dissolved organic matter by nocturnal programmed cell death of phototrophic algae Dunaliella salina (Orellana et al., 2013). The absence of eukaryotes in our experiment suggests that another explanation is needed to explain the Nanohaloarchaeal dynamics. A symbiotic association between the Nanohaloarchaea (with their small genomes and small cell size) and halophilic archaea is possible, and is consistent with the night preference for both organisms. Death of archaeal, particularly Nanohaloarchaeal, cells during the day, possibly caused by temperature or ultraviolet stress (Ugalde et al., 2011), could releases nutrients that may promote the growth of bacteria.
Overall, a key finding of this study is the value in combining metagenomic and community lipid compositional information. Lipid data provided clear evidence of the existence of a diel cycle in relative abundances of bacteria and archaea and provided insight into adaptation mechanisms. However, lipid data cannot resolve the behaviors of specific types of organisms. In contrast, metagenomic coverage statistics identified from the 0.1-μm filter samples provided up to strain-level resolution of relative abundance levels for Halobacteria and Nanohaloarchaea, but the effects of total cell number change and different growth rates could not be deconvoluted. By combining the observations, it becomes clear that Nanohaloarchaea show a stronger nighttime preference than Halobacteria, although the behaviors may be coupled through dependence of Nanohaloarchaea on Halobacteria for basic nutritional requirements.
Acknowledgments
We thank Elizabeth Scott (USC) and Nadine Eisenkolb (USC) for fieldwork assistance. Funding for this work was provided by: NSF MCB Award #0626526 to JFB, EEA, and KBH. KA was supported by the ESPM Departmental Block grants, UC Berkeley Chancellors Fellowship, and the Andrew W Mellon Foundation fellowship. LRC was supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under contract no. DE-AC02-05CH11231. Cheetham Salt Works (Sea Lake, Australia) provided permission to collect samples.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allen EE, Banfield JF. (2005). Community genomics in microbial ecology and evolution. Nat Rev Microbiol 3: 489–498. [DOI] [PubMed] [Google Scholar]
- Baker BJ, Banfield JF. (2003). Microbial communities in acid mine drainage. FEMS Microbiol Ecol 44: 139–152. [DOI] [PubMed] [Google Scholar]
- Baliga NS. (2008). Systems biology - the scale of prediction. Science 320: 1297–1298. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. (1995). Controlling the false discovery rate — a practical and powerful approach to multiple testing. J Roy Stat Soc B Met 57: 289–300. [Google Scholar]
- Butte W. (1983). Rapid method for the determination of fatty-acid profiles from fats and oils using trimethylsulfonium hydroxiden for trans-esterification. J Chromatogr 261: 142–145. [Google Scholar]
- Coker JA, DasSarma S. (2007). Genetic and transcriptomic analysis of transcription factor genes in the model halophilic Archaeon: coordinate action of TbpD and TfbA. BMC Genet 8: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comolli LR, Baker BJ, Downing KH, Siegerist CE, Banfield JF. (2009). Three-dimensional analysis of the structure and ecology of a novel, ultra-small archaeon. ISME J 3: 159–167. [DOI] [PubMed] [Google Scholar]
- Comolli LR, Duarte R, Baum D, Luef B, Downing KH, Larson DM et al. (2012). A portable cryo-plunger for on-site intact cryogenic microscopy sample preparation in natural environments. Microsc Res Tech 75: 829–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comolli LR, Banfield JF. (2014). Inter-species interconnections in acid mine drainage microbial communities. Front Microbiol 5: 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derosa M, Esposito E, Gambacorta A, Nicolaus B, Bulock JD. (1980). Effects of temperature on ether lipid-composition of Caldariella-Acidophila. Phytochemistry 19: 827–831. [Google Scholar]
- Edgar RC. (2004. a). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform 5: 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004. b). MUSCLE: multiple sequence alignment with improved accuracy and speed. 2004 IEEE Computational Systems Bioinformatics Conference, Proceedings. Stanford, CA, pp 728–729. [Google Scholar]
- Emerson JB, Thomas BC, Andrade K, Allen EE, Heidelberg KB, Banfield JF. (2012). Dynamic viral populations in hypersaline systems as revealed by metagenomic assembly. Appl Environ Microb 78: 6309–6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JB, Andrade K, Thomas BC, Norman A, Allen EE, Heidelberg KB et al. (2013. a). Virus-host and CRISPR dynamics in archaea-dominated hypersaline Lake Tyrrell, Victoria, Australia. Archaea 2013: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerson JB, Thomas BC, Andrade K, Heidelberg KB, Banfield JF. (2013. b). New approaches indicate constant viral diversity despite shifts in assemblage structure in an Australian hypersaline lake. Appl Environ Microb 79: 6755–6764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasol JM, Casamayor EO, Joint I, Garde K, Gustavson K, Benlloch S et al. (2004). Control of heterotrophic prokaryotic abundance and growth rate in hypersaline planktonic environments. Aquat Microb Ecol 34: 193–206. [Google Scholar]
- Ghai R, Pasic L, Fernandez AB, Martin-Cuadrado AB, Mizuno CM, McMahon KD et al. (2011). New abundant microbial groups in aquatic hypersaline environments. Sci Rep-Uk 1: 135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidelberg KB, Nelson WC, Holm JB, Eisenkolb N, Andrade K, Emerson JB. (2013). Characterization of eukaryotic microbial diversity in hypersaline Lake Tyrrell, Australia. Front Microbiol 4: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann AG, Knake D, Schneide. J, Peters H. (1973). Geochemistry of modern seawater and brines from salt pans - main components and bromine distribution. Contrib Mineral Petr 40: 1–24. [Google Scholar]
- Hobbie JE, Daley RJ, Jasper S. (1977). Use of nuclepore filters for counting bacteria by fluorescence microscopy. Appl Environ Microb 33: 1225–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TC, Tu J, Chow TJ, Chen TH. (1990). Circadian-rhythm of the prokaryote Synechococcus Sp Rf-1. Plant Physiol 92: 531–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber H, Hohn MJ, Rachel R, Fuchs T, Wimmer VC, Stetter KO. (2002). A new phylum of archaea represented by a nanosized hyperthermophilic symbiont. Nature 417: 63–67. [DOI] [PubMed] [Google Scholar]
- Hug LA, Castelle CJ, Wrighton KC, Thomas BC, Sharon I, Frischkorn KR et al. (2013). Community genomic analyses constrain the distribution of metabolic traits across the Chloroflexi phylum and indicate roles in sediment carbon cycling. Microbiome 1: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. (2010). Prodigal: prokaryotic gene recognition and translation initiation site identification. Bmc Bioinformatics 11: 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH. (2005). Testing the adaptive value of circadian systems. Circadian Rhythms 393: 818–837. [DOI] [PubMed] [Google Scholar]
- Johnson CH, Mori T, Xu Y. (2008). A cyanobacterial circadian clockwork. Curr Biol 18: R816–R825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkhammer GP, Bender ML. (1980). The distribution of manganese in the Pacific-Ocean. Earth Planet Sc Lett 46: 361–384. [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. (2012). Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo I, Denef VJ, VerBerkmoes NC, Shah MB, Goltsman D, DiBartolo G et al. (2007). Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature 446: 537–541. [DOI] [PubMed] [Google Scholar]
- Macumber PG. (1992). Hydrological processes in the Tyrrell Basin, southeastern Australia. Chem Geol 96: 1–18. [Google Scholar]
- Marr AG, Ingraham JL. (1962). Effect of temperature on composition of fatty acids in Escherichia Coli. J Bacteriol 84: 1260–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CS, Baker BJ, Thomas BC, Singer SW, Banfield JF. (2011). EMIRGE: reconstruction of full-length ribosomal genes from microbial community short read sequencing data. Genome Biol 12: R44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Johnson CH. (2001). Independence of circadian timing from cell division in cyanobacteria. J Bacteriol 183: 2439–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasingarao P, Podell S, Ugalde JA, Brochier-Armanet C, Emerson JB, Brocks JJ et al. (2012). De novo metagenomic assembly reveals abundant novel major lineage of archaea in hypersaline microbial communities. ISME J 6: 81–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orellana MV, Pang WL, Durand PM, Whitehead K, Baliga NS. (2013). A role for programmed cell death in the microbial loop. Plos One 8: e6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A. (2008). Microbial life at high salt concentrations: phylogenetic and metabolic diversity. Saline Syst 4: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen EA, Young CR, Eppley JM, Ryan JP, Chavez FP, Scholin CA et al. (2013). Pattern and synchrony of gene expression among sympatric marine microbial populations. Proc Natl Acad Sci USA 110: E488–E497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottesen EA, Young CR, Gifford SM, Eppley JM, Marin R, Schuster SC et al. (2014). Multispecies diel transcriptional oscillations in open ocean heterotrophic bacterial assemblages. Science 345: 207–212. [DOI] [PubMed] [Google Scholar]
- Park JS, Kim HJ, Choi DH, Cho BC. (2003). Active flagellates grazing on prokaryotes in high salinity waters of a solar saltern. Aquat Microb Ecol 33: 173–179. [Google Scholar]
- Pedros-Alio C, Calderon-Paz JI, MacLean MH, Medina G, Marrase C, Gasol JM et al. (2000). The microbial food web along salinity gradients. FEMS Microbiol Ecol 32: 143–155. [DOI] [PubMed] [Google Scholar]
- Peng Y, Leung HC, Yiu SM, Chin FY. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics (Oxford, England) 28: 1420–1428. [DOI] [PubMed] [Google Scholar]
- Podar M, Makarova KS, Graham DE, Wolf YI, Koonin EV, Reysenbach AL. (2013). Insights into archaeal evolution and symbiosis from the genomes of a nanoarchaeon and its inferred crenarchaeal host from Obsidian Pool, Yellowstone National Park. Biol Direct 8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podell S, Ugalde JA, Narasingarao P, Banfield JF, Heidelberg KB, Allen EE. (2013). Assembly-driven community genomics of a hypersaline microbial ecosystem. Plos One 8: 1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podell S, Emerson JB, Jones CM, Ugalde JA, Welch S, Heidelberg KB et al. (2014). Seasonal fluctuations in ionic concentrations drive microbial succession in a hypersaline lake community. ISME J 8: 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R et al. (2005). InterProScan: protein domains identifier. Nucleic Acids Res 33: W116–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R_Core_Team. (2005). R: a language and environment for statistical computing In: Computing RFfS (ed).
- Raes J, Bork P. (2008). Systems microbiology — timeline — molecular eco-systems biology: towards an understanding of community function. Nat Rev Microbiol 6: 693–699. [DOI] [PubMed] [Google Scholar]
- Ram RJ, VerBerkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake RC et al. (2005). Community proteomics of a natural microbial biofilm. Science 308: 1915–1920. [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng JF et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Brito B, Li LL, Wegley L, Furlan M, Angly F, Breitbart M et al. (2010). Viral and microbial community dynamics in four aquatic environments. Isme Journal 4: 739–751. [DOI] [PubMed] [Google Scholar]
- Sorek R, Zhu YW, Creevey CJ, Francino MP, Bork P, Rubin EM. (2007). Genome-wide experimental determination of barriers to horizontal gene transfer. Science 318: 1449–1452. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics (Oxford, England) 22: 2688–2690. [DOI] [PubMed] [Google Scholar]
- Tagkopoulos I, Liu YC, Tavazoie S. (2008). Predictive behavior within microbial genetic networks. Science 320: 1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AY, Gong GC, Sanders RW, Chiang KP, Huang JK, Chan YF. (2012). Viral lysis and nanoflagellate grazing as factors controlling diel variations of Synechococcus spp. summer abundance in coastal waters of Taiwan. Aquat Microb Ecol 66: 159–167. [Google Scholar]
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM et al. (2004). Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428: 37–43. [DOI] [PubMed] [Google Scholar]
- Ugalde JA, Podell S, Narasingarao P, Allen EE. (2011). Xenorhodopsins, an enigmatic new class of microbial rhodopsins horizontally transferred between archaea and bacteria. Biol Direct 6: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microb 73: 5261–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead K, Pan M, Masumura K, Bonneau R, Baliga NS. (2009). Diurnally entrained anticipatory behavior in archaea. Plos One 4: 5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams WD. (2001). Anthropogenic salinisation of inland waters. Hydrobiologia 466: 329–337. [Google Scholar]
- Wilmes P, Bond PL. (2009). Microbial community proteomics: elucidating the catalysts and metabolic mechanisms that drive the Earth's biogeochemical cycles. Curr Opin Microbiol 12: 310–317. [DOI] [PubMed] [Google Scholar]
- Wu M, Eisen JA. (2008). A simple, fast, and accurate method of phylogenomic inference. Genome Biol 9: R151. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.