Abstract
The genetic composition of marine microbial communities varies at the microscale between particle-associated (PA; >1.6 μm) and free-living (FL; 0.2–1.6 μm) niches. It remains unclear, however, how metabolic activities differ between PA and FL fractions. We combined rate measurements with metatranscriptomics to quantify PA and FL microbial activity in the oxygen minimum zone (OMZ) of the Eastern Tropical North Pacific, focusing on dissimilatory processes of the nitrogen (N) cycle. Bacterial gene counts were 8- to 15-fold higher in the FL compared with the PA fraction. However, rates of all measured N cycle processes, excluding ammonia oxidation, declined significantly following particle (>1.6 μm) removal. Without particles, rates of nitrate reduction to nitrite (1.5–9.4nMNd−1) fell to zero and N2 production by denitrification (0.5–1.7nMNd−1) and anammox (0.3–1.9nMNd−1) declined by 53–85%. The proportional representation of major microbial taxa and N cycle gene transcripts in metatranscriptomes followed fraction-specific trends. Transcripts encoding nitrate reductase were uniform among PA and FL fractions, whereas anammox-associated transcripts were proportionately enriched up to 15-fold in the FL fraction. In contrast, transcripts encoding enzymes for N2O and N2 production by denitrification were enriched up to 28-fold in PA samples. These patterns suggest that the majority of N cycle activity, excluding N2O and N2 production by denitrification, is confined to a FL majority that is critically dependent on access to particles, likely as a source of organic carbon and inorganic N. Variable particle distributions may drive heterogeneity in N cycle activity and gene expression in OMZs.
Introduction
The genetic diversity and activity of pelagic marine microorganisms vary in complex patterns in response to environmental conditions and interspecies interactions. Such variation is best understood at the scale of meters, for example, over depth gradients of light or over regional (kilometer) gradients in nutrient availability. Microbial communities are also structured over much smaller spatial scales, notably the micron-scale distances separating free-living (FL) cells from those on the surfaces or within the diffusive boundary layer of suspended or sinking organic particles (Stocker, 2012). Extensive research has identified consistent taxonomic compositional differences between FL and particle-associated (PA) bacterial and archaeal communities (DeLong et al., 1993; Hollibaugh et al., 2000; Ganesh et al., 2014), as well as differences in total microbial abundance, production and enzyme activity (Simon et al., 2002; Grossart et al., 2003, 2007). Surprisingly, most studies analyzing microbial genomic or metagenomic composition or rates of specific metabolic processes have either focused on microbes within a single biomass size fraction (typically the 0.2–3.0-μm range) or on bulk water samples. Such studies risk excluding important information about contributions from PA cells, larger non-PA cells or metabolic substrates in PA fractions. Furthermore, only a single study (Satinsky et al., 2014) has examined community gene expression (metatranscriptomes) in PA versus FL communities. When coupled to measurements of metabolic rates, size-fractionated metatranscriptomics can help identify the drivers, magnitudes and spatial scales of biogeochemistry in the oceans.
The extent to which microbial metabolisms are partitioned between PA and FL communities remains uncharacterized for many ocean regions of importance to global biogeochemical cycles. These include the marine oxygen minimum zones (OMZs) where microbial communities control key steps in nitrogen (N), carbon and sulfur transformations. OMZs form in poorly ventilated regions where microbial respiration fueled by high productivity in the overlying waters creates a layer of oxygen depletion. The largest OMZs occur in areas of persistent nutrient upwelling, as in the Eastern Tropical North Pacific (ETNP) off Mexico (Karstensen et al., 2008). Encompassing ~12 × 106 km2 of shelf and off-shelf waters south of Baja California, the ETNP OMZ is the largest of the major permanent OMZs (41% of total OMZ area; Paulmier and Ruiz, 2009), with dissolved O2 concentrations falling below the detection limit (<0.1 μm) at mid-water depths (~150–750 m; Cline and Richards, 1974; Tiano et al., 2014).
Oxygen drawdown in OMZs significantly restructures the pelagic ecosystem, resulting in microbe-dominated communities expressing diverse microaerophilic or anaerobic metabolisms. These include the reductive processes of denitrification and anaerobic ammonia oxidation with nitrite (anammox), whose combined activity in OMZs contribute up to half of oceanic N loss as N2 or N2O gas (Codispoti et al., 2001; Gruber, 2004). Research in the eastern Pacific OMZs, notably in the anoxic OMZ off Chile and Peru, indicates that anammox and denitrification are linked to varying degrees with a diverse set of other microbial N transformations, including the production of nitrite and nitrate via nitrification along the oxycline and dissimilatory nitrate reduction to ammonium within the OMZ (Lam et al., 2009; Kalvelage et al., 2013). OMZ N cycle processes also mediate key steps in other elemental cycles, for example, the fixation of carbon by anammox and nitrifying autotrophs, the remineralization of organic carbon and nutrients by denitrifying heterotrophs, and the oxidation of reduced sulfur compounds by chemoautotrophic denitrifiers (Ulloa et al., 2012).
The distributions of these networked metabolic interactions at multiple spatial scales are becoming understandable for some OMZs. Surveys combining marker gene counts and rate measurements have suggested mesoscale patterns in N cycling along coastal to offshore gradients in the Eastern Tropical South Pacific (ETSP) OMZ off Chile and Peru (Kalvelage et al., 2013), as well as clear depth-specific variation in N cycle genes, transcripts, and rates (Thamdrup et al., 2006; Dalsgaard et al., 2012; Stewart et al., 2012). Recently, analysis of microbial metagenomes in the ETSP OMZ also identified a non-uniform distribution of N cycle genes among filter size fractions (Ganesh et al., 2014). Genes for aerobic ammonia oxidation at OMZ boundaries and for anammox at the OMZ core were consistently overrepresented in the FL fraction (0.2–1.6 μm). In contrast, genes mediating different steps of denitrification showed variable distributions between size fractions. Genes encoding NarG, part of the enzyme catalyzing nitrate reduction to nitrite, were abundant and evenly distributed between PA (>1.6 μm) and FL fractions within the OMZ, whereas genes mediating the two terminal steps of denitrification, the reduction of nitric oxide to nitrous oxide (norB) and nitrous oxide to dinitrogen (nosZ), exhibited a fourfold increase in relative abundance in PA compared with FL metagenomes. These data highlight a potential separation of linked metabolic processes at varying spatial scales, potentially over micron gradients separating PA and FL communities.
The proportional contribution of PA N metabolism to bulk process rates in OMZs remains unclear, notably for the larger ETNP OMZ. Despite recent studies describing the diversity and activity of key functional taxonomic groups in the ETNP (Podlaska et al., 2012; Rush et al., 2012; Beman et al., 2012, 2013), this region remains under-explored from a microbiological and molecular perspective. Assessing the actual biochemical significance of microspatial separation of microbial processes in OMZs requires quantification of metabolic rates and gene expression among size fractions from diverse OMZ regions. We combine community transcription profiling and rate measurements to quantify size fraction partitioning of key steps of the marine N cycle. These data, coupled with taxonomic analysis of genes and transcripts and estimates of bacteria abundance among size fractions, indicate a significant contribution of particles and PA bacteria to OMZ N cycling.
Materials and methods
Molecular analysis
Sample collection
Samples were collected from the ETNP OMZ during the OMZ Microbial Biogeochemistry Expedition cruise (R/V New Horizon, 13–28 June 2013). Seawater was sampled from six depths spanning the upper oxycline (30 m), lower oxycline (85 m) oxic–nitrite interface (91 m), secondary chlorophyll maximum (100 m), secondary nitrite maximum (125 m) and OMZ core (300 m) at Station 6 (18° 54.0° N, 104° 54.0° W) on 19 June. Collections were made using Niskin bottles on a rosette containing a Conductivity Temperature Depth profiler (Sea-Bird SBE 911plus, Sea-Bird Electronics Inc., Bellevue, WA, USA) equipped with a Seapoint fluorometer (Seapoint Sensors Inc., Exeter, NH, USA) and SBE43-dissolved oxygen sensor (Sea-Bird Electronics Inc.). High-resolution Switchable Trace amount OXygen sensors (STOX) were also mounted to the rosette to quantify oxygen at nanomolar concentrations (Revsbech et al., 2009; Supplementary Methods). Samples were also collected from 20, 40, 90, 100, 125 and 300 m during a replicate cast on the same day and used to count bacterial 16S rRNA genes.
Size-fractionated biomass was collected for RNA analysis by sequential inline filtration of seawater (~10–15 l) through a nylon disk filter (47 mm, 30 μm pore size, Millipore, Bellerica, MA, USA), a glass fiber disc filter (GF/A, 47 mm, 1.6 μm pore size, Whatman, GE Healthcare Bio-sciences, Pittsburgh, PA, USA) and a primary collection filter (Sterivex, 0.22 μm pore size, Millipore) via peristaltic pump. Replicate filters for DNA were collected from equivalent water volumes following RNA collection. Water samples (45 ml each) for 16S rRNA gene counts were filtered through 0.2 μm disc filters (25 mm, cellulose nitrate, Whatman). Disc filters were transferred to cryovials containing RNA stabilizing buffer (25 mM sodium citrate, 10 mM EDTA and 70 g ammonium sulfate per 100 ml solution, pH 5.2) for RNA samples or lysis buffer (50 mM Tris-HCl, 40 mM EDTA and 0.73 M sucrose) for DNA samples. Sterivex filters were filled with either RNA stabilizing buffer or lysis buffer, capped and flash-frozen (RNA samples). Filters were stored at −80 °C. Less than 20 min elapsed between sample collection (water on deck) and fixation in buffer.
DNA and RNA extraction
DNA was extracted from disc and Sterivex filters using a phenol/chloroform protocol as in Ganesh et al. (2014). Cells were lysed by adding lysozyme (2 mg in 40 μl of lysis buffer per filter) directly to the disc filter containing cryovial or to the Sterivex cartridge, sealing the caps/ends and incubating for 45 min at 37 °C. Proteinase K (1 mg in 100 μl lysis buffer with 100 μl 20% SDS) was added, and cryovials and cartridges were resealed and incubated for 2 h at 55 °C. The lysate was removed and DNA was extracted once with phenol:chloroform:isoamyl alcohol (25:24:1) and once with chloroform:isoamyl alcohol (24:1), and then concentrated by spin dialysis using Ultra-4 (100 kDa, Amicon, Millipore) centrifugal filters.
RNA was extracted from disc and Sterivex filters using a modification of the mirVana miRNA Isolation kit (Ambion, Life Technologies, Carlsbad, CA, USA) as in Stewart et al. (2012). Filters were thawed on ice and RNA stabilizing buffer was removed by pipette from cryovials or expelled via syringe from Sterivex cartridges and discarded. Cells were lysed by adding lysis buffer and miRNA homogenate additive (Ambion) directly to the cryovial or cartridge. Following vortexing and incubation on ice, lysates were transferred to RNase-free tubes and processed via acid–phenol/chloroform extraction according to the kit protocol. The TURBO DNA-free kit (Ambion) was used to remove DNA and the extract was purified using the RNeasy MinElute Cleanup Kit (Qiagen, Hilden, Germany).
16S rRNA gene counts
Quantitative PCR was used to count bacterial 16S rRNA genes in bulk seawater (biomass >0.2 μm) and in 0.2–1.6 and 1.6–30 μm size fractions. Extracts from the >30 μm fraction did not contain measureable DNA (via Qubit) and failed to yield signals during quantitative PCR with diluted and undiluted extracts. We were not able to sum counts from size-fractionated samples to obtain counts per volume because exact volumes of filtered seawater were not recorded. We instead used samples of fixed volume from a replicate cast (‘Sample collection' section above) to quantify total gene copies per ml of seawater. Because equal water volumes passed through sequential filters during size fractionation, counts from size-fractionated samples were used to quantify the ratio of copies in the 0.2–1.6-μm fraction to those in the 1.6–30-μm fraction (FL/PA ratio); counts from the >30 μm fraction are assumed to be negligible.
Quantitative PCR used TaqMan-based reagents and universal bacterial 16S primers 1055 f and 1392r, as in Ritalahti et al. (2006) and Hatt et al. (2013). Tenfold serial dilutions of DNA from a plasmid carrying a single copy of the 16S rRNA gene (from Dehalococcoides mccartyi; Ritalahti et al., 2006) were included on each quantitative PCR plate and used to generate standard curves. Assays were run on a 7500 Fast PCR System and a StepOnePlus Real-Time PCR System (Applied Biosystems, Life Technologies). All samples were run in triplicate (20 μl each) and included 1 × TaqMan Universal PCR Master Mix (Life Technologies), 300 nm of primers, 300 nm of TaqMan MGB probe (Life Technologies) and 2 μl of template DNA. Thermal cycling involved incubation at 50 °C for 2 min to activate uracil-N-glycosylase, followed by 95 °C for 10 min to inactivate uracil-N-glycosylase, denature template DNA and activate the AmpliTaq Gold polymerase, followed by 40 cycles of denaturation at 95 °C (15 s) and annealing at 60 °C (1 min).
16S rRNA gene amplicon sequencing
High-throughput sequencing of dual-indexed PCR amplicons encompassing the V4 region of the 16S rRNA gene was used to assess bacterial community composition in all filter fractions. Despite low DNA yields (above), 16S gene fragments were amplifiable from the >30-μm fraction. The diversity of amplicons generated using Archaea-specific primers was not evaluated. Amplicons were synthesized using Platinum PCR SuperMix (Life Technologies) with primers F515 and R806 (Caporaso et al., 2011). Both forward and reverse primers were barcoded and appended with Illumina-specific adapters according to Kozich et al. (2013). Thermal cycling involved: denaturation at 94 °C (3 min), followed by 30 cycles of denaturation at 94 °C (45 s), primer annealing at 55 °C (45 s) and primer extension at 72 °C (90 s), followed by extension at 72 °C for 10 min. Amplicons were analyzed by gel electrophoresis to verify size (~400 bp) and purified using the QIAQuick PCR Clean-Up Kit (Qiagen). Amplicons from different samples were pooled at equimolar concentrations and sequenced on an Illumina MiSeq (Illumina, San Diego, CA, USA) using a 500 cycle kit with 5% PhiX as a control.
cDNA synthesis and metatranscriptome sequencing
Shotgun Illumina sequencing of community cDNA was used to characterize gene expression in 0.2–1.6 μm and 1.6–30 μm biomass fractions. Most samples of the >30-μm fraction did not yield sufficient RNA for sequencing; this fraction was excluded from meta-omic analysis. Community RNA was prepared for sequencing using the ScriptSeq v2 RNA-Seq Library preparation kit (Epicenter, Madison, WI, USA). Briefly, cDNA was synthesized from fragmented total RNA (rRNA was not removed) using reverse transcriptase and amplified and barcoded using ScriptSeq Index PCR Primers (Epicenter) to generate single-indexed cDNA libraries. cDNA libraries were pooled and sequenced on an Illumina MiSeq using a 500 cycle kit (Supplementary Table S1). Metagenomes from coupled DNA samples were analyzed to allow standardization of taxon transcript levels relative to abundance in the DNA pool. These data sets were generated in a separate study (in review, NCBI accession SRP044185) using the Nextera XT DNA Sample Prep kit and paired-end MiSeq sequencing as above for cDNA samples.
Sequence analysis—16S rRNA gene amplicons
Amplicons were analyzed using QIIME (Caporaso et al., 2010) following standard protocols. Barcoded sequences were de-multiplexed and filtered to remove low-quality reads (Phred score <25). Paired-end sequences were merged using custom scripts incorporating the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html) and USEARCH algorithm (Edgar, 2010), with criteria of minimum 10% overlap and 95% nucleotide identity within the overlapping region. Merged sequences were clustered into operational taxonomic units at 97% sequence similarity using open-reference picking with the UCLUST algorithm (Edgar, 2010), with taxonomy assigned to representative operational taxonomic units from each cluster using the Greengenes database (DeSantis et al., 2006). Operational taxonomic unit counts were rarefied (10 iterations) and alpha diversity was quantified at a uniform sequence depth (n=6506) using the phylogenetic diversity metric of Faith (1992). To compare community composition between samples, sequences were aligned using the PyNAST aligner (Caporaso et al., 2010) and beta diversity was calculated using the weighted Unifrac metric (Lozupone and Knight, 2005). Sample relatedness based on Unifrac was visualized with a two-dimensional principal coordinate analysis.
Sequence analysis—metatranscriptomes and metagenomes
Analysis of protein-coding genes and transcripts followed that of Stewart et al. (2012) and Ganesh et al. (2014). Reads were filtered by quality and merged as described above. rRNA transcripts were identified using riboPicker (Schmieder et al., 2012) and removed. Merged non-rRNA sequences were queried via BLASTX against the NCBI-nr database (November 2013). BLASTX matches to bacterial and archaeal genes (>bit score 50) were retained to evaluate marker genes of dissimilatory N metabolism: ammonia monooxygenase (amoC), nitrite oxidoreductase (nxrB), hydrazine oxidoreductase (hzo), nitrate reductase (narG), nitrite reductase (nirK+nirS), nitric oxide reductase (norB) and nitrous oxide reductase (nosZ). N gene transcript abundances were normalized based on gene length and expressed as a proportion of the abundance of transcripts matching the gene encoding RNA polymerase subunit B (rpoB), as has been done in studies of diverse bacteria (for example, Schumann et al., 2010; Ceja-Navarro et al., 2014; Dalsgaard et al., 2014; Eldholm et al., 2014). Although rpoB expression can vary (Vandecasteele et al., 2001), rpoB appears to be one of the more stably expressed housekeeping genes (Sue et al., 2004; Sihto et al., 2014). Furthermore, it has been shown that rpoB can be a proxy of bulk mRNA transcription level for bacteria (Milohanic et al., 2003; Sue et al., 2004)—rpoB-normalized values therefore reflect transcription of a target gene relative to a housekeeping gene under a given condition/sample.
The taxonomic composition of protein-coding genes and transcripts was determined using MEtaGenome ANalyzer 5 (Huson et al., 2011) based on the annotations of BLASTX-identified genes, according to the NCBI taxonomy. The proportional contribution of a taxon (Phyla and Class levels) to the RNA pool was calculated as an RNA:DNA ratio, calculated as in Frias-Lopez et al. (2008) and Stewart et al. (2012) to account for variation in taxon abundance in the DNA pool: (protein-coding RNA reads per taxon/total protein-coding RNA reads)/(protein-coding DNA reads per taxon/total protein-coding DNA reads). For subsets of transcripts matching specific functional taxa (anammox genera, Nitrospina), genes differentially expressed between FL and PA fractions were identified using baySeq (Hardcastle and Kelly, 2010) as in Ganesh et al. (2014). Full details regarding sequence analysis are in the Supplementary Methods.
Amplicon and metatranscriptome sequences are available through NCBI under BioProject ID PRJNA263621
Biochemical analysis
Water for rate measurements was taken from Niskin bottles immediately after arrival on deck and transferred to 2 l glass bottles. Bottles were overflowed (three volume equivalents) and sealed without bubbles using deoxygenated butyl rubber stoppers (De Brabandere et al., 2014). Bottles were stored in the dark at in situ temperature until experimentation (<6 h). Each bottle was purged with a gas mixture of helium and carbon dioxide (800 p.p.m. carbon dioxide) for ~20 min. Under a slight overpressure, water was dispensed into 12 ml exetainers (Labco, Lampeter, Ceredigion, UK) either directly or through helium-flushed inline filter holders, and immediately capped with deoxygenated lids.
Rates were measured for two biomass fractions: bulk water containing all particles (no filtration) and water without particles >1.6 μm. Particles were excluded via filtration as described above. Exetainers were subsequently amended with 15N-labeled substrate by injection through the septum. Three 15N amendments were carried out for all samples: 15NH4+ addition (to 5 μm concentration), 15NO2− addition (5 μm) and 15NO3− addition (15 μm). Headspaces of 2 ml were introduced into each exetainer and flushed twice with the helium–carbon dioxide mixture as in De Brabandere et al. (2014). For each experiment, triplicate exetainers were preserved with 100 μl of 50% (w/v) ZnCl2 at the start of the incubation and again at the end point (27 h). Oxygen concentration was <80 nm in all treatments. The act of filtering did not affect exetainer oxygen content (Supplementary Methods).
The production of 14N15N and 15N15N was determined on a gas chromatography isotope ratio mass spectrometer as in Dalsgaard et al. (2012). Rates of N2 production by anammox and denitrification were calculated using the equations in Thamdrup and Dalsgaard (2002). Nitrite oxidation was determined from the production of 15NO3− from incubations with 15NO2−. After removal of unused 15NO2− using sulfamic acid, 15NO3− was converted to 15NO2− with cadmium and then to N2 with sulfamic acid (Füssel et al., 2012; McIlvin and Altabet 2005). Ammonia oxidation and nitrate reduction rates were determined as production of 15NO2− from incubations with 15NH4+ or 15NO3−. 15NO2− produced during incubations was converted to N2 with sulfamic acid. N2 was then analyzed on the gas chromatography isotope ratio mass spectrometer. Rates for all processes were calculated from the slope of the linear regression of 14N15N or 15N15N with time. T-tests were applied to determine whether rates were significantly different from zero (P<0.05). Non-significant rates are presented as not detected.
Nitrate, nitrite and ammonium concentrations were determined using standard protocols, as described in the Supplementary Methods. The N deficit estimates the amount of fixed N loss and was calculated as: N deficit=(DIN)–(16 × [PO43−] × 0.86), where DIN=[NO3−]+[NO2−]+[NH4+]. The value of 0.86 accounts for the release of PO43− from organic N remineralized during denitrification (Codispoti et al., 2001).
Results and discussion
Particle removal significantly altered N cycle rates and gene expression, suggesting physiological variation at the microscale between water column and PA niches. These patterns are discussed below relative to oxygen and N concentrations, bacterial abundance and community taxonomic composition.
Oxygen and nitrogen
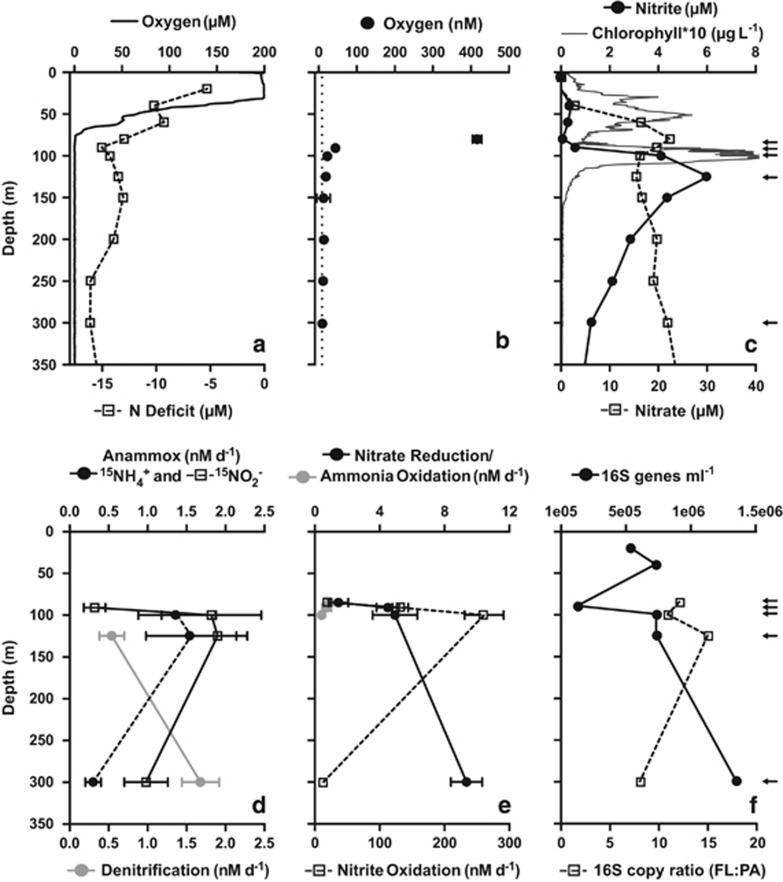
Oxygen concentrations recorded by the SBE43 sensor were highest (~200 μm) in surface waters and declined steeply in the oxycline between 30 and 85 m (Figure 1a). From 90 to 800 m, concentrations measured by STOX sensors were <50 nm, with the majority close to or below the detection limit (Figure 1b) representing the OMZ core. Oxygen concentrations at the six depths sampled for size fractionation work varied markedly, from 200 μm at 30 m to 229 nm at 85 m, representing the upper and lower limits of the oxycline, respectively, 43 nm at the oxic–nitrite interface (91 m), 21 nm at the secondary chlorophyll maximum (100 m) and 14 nm at the secondary nitrite maximum (125 m), before reaching the detection limit (9 nm) at 150 m and staying undetectable to 300 m in the OMZ core (Figure 1b). Oxygen profiles coincided with patterns in nitrate, nitrite and the N deficit. As commonly observed in anoxic OMZs (Ulloa et al., 2012), a prominent secondary nitrite maximum (up to 6.1 μm) was observed as oxygen declined, coinciding with local nitrate and N deficit minima (Figures 1a and c). Ammonium concentrations remained below 155 nm throughout the profile. Chlorophyll peaked at 0.6 and 0.8 μg l−1 at 50 and 100 m, respectively, indicating the primary and secondary chlorophyll maxima characteristic of OMZs (Figure 1c; Ulloa et al., 2012).
Figure 1.
Vertical profiles of hydrochemical parameters, bulk N cycling process rates and bacterial 16S rRNA gene counts at Station 6, off Manzanillo, Mexico on 19 June 2013. The N deficit (a, open squares) is shown relative to oxygen concentrations, measured during the upcast profile with both a SBE43 sensor (a; solid line) and STOX sensors (b), and to profiles of nitrite, nitrate and chlorophyll concentration (c). The dashed line in b indicates the detection limit of the STOX sensors (~9 nm). N cycle process rates (d, e) are bulk rates (no size fractionation) in nM N per day. (f) Quantitative PCR-based counts of 16S rRNA gene copies per ml of bulk seawater (all biomass >0.2 μm) and the ratio of gene counts in the FL (0.2–1.6 μm) biomass fraction to counts in the PA (1.6–30 μm) fraction. Arrows (c, f) indicate the depths sampled for rate measurements. Error bars represent the standard error.
16S rRNA gene abundance
Counts of bacterial 16S rRNA genes varied by an order of magnitude with depth (Figure 1f). Bulk seawater counts declined from ~6 × 105 per ml in the primary chlorophyll maximum and upper oxycline to 1.3 × 105 at the oxic–nitrite interface (90 m), before increasing to a maximum of 1.4 × 106 at the OMZ core. This pattern agrees with trends from other OMZ sites, where particulate backscattering and bacterial load show local maxima in the photic zone and again in the OMZ (Spinrad et al., 1989; Naqvi et al., 1993; Whitmire et al., 2009). At all depths, gene numbers were substantially higher in the 0.2–1.6 μm (FL) fraction compared with the 1.6–30 μm (PA) fraction, with FL/PA ratios from 8 to 15 (Figure 1f). Elevated microbial biomass in FL compared with PA fractions has been observed in diverse ocean regions (Cho and Azam 1988; Karl et al., 1988; Turley and Stutt 2000; Ghiglione et al., 2009). Here, FL/PA was lowest at 300 m, suggesting an increased contribution of PA or large bacteria within the OMZ core, possibly related to increases in particle load or size.
Community composition—16S rRNA gene amplicons
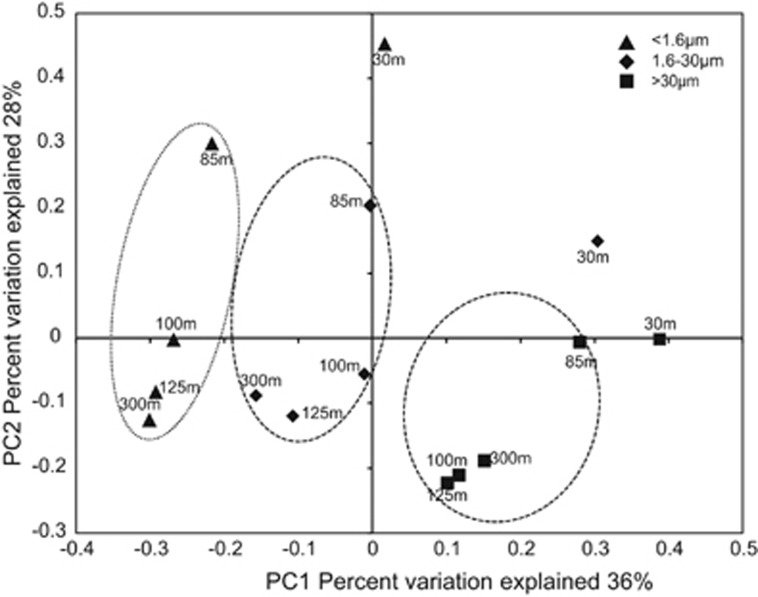
Community 16S rRNA gene diversity varied substantially among biomass fractions and depths. Unifrac-based clustering indicated that size class was a stronger predictor of community relatedness compared with depth, notably for samples from the OMZ layer (100, 125 and 300 m) (Figure 2). Phylogenetic diversity differed widely among fractions, with values in the two PA fractions up to 50% greater than those for the corresponding FL fraction (Supplementary Figure S1). Elevated diversity in PA communities has been reported for the ETSP OMZ (Ganesh et al., 2014) and may be driven by increased niche richness due to substrate heterogeneity on particles, or to the exposure of particles during sinking to diverse communities in different biogeochemical zones. Here, phylogenetic diversity in all size fractions peaked at the secondary chlorophyll maximum, similar to trends reported for non-fractionated samples (biomass >0.2 μm) from ETNP sites north of the study area, where taxon richness peaked just below the oxycline, before declining with depth (Beman and Carolan, 2013). That study identified deoxygenation as a driver of bulk community diversity, whereas our results suggest that diversity estimates vary depending on the filter fraction examined.
Figure 2.
Principle coordinate analysis of community taxonomic relatedness based on 16S rRNA gene amplicons, as quantified by the weighted Unifrac metric. Samples from different filter size fractions are circled (excluding outliers from the 30-m depth).
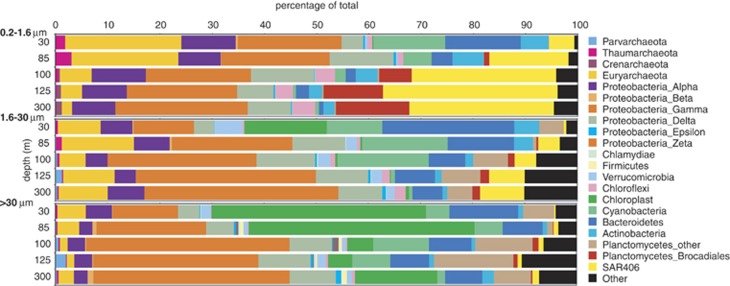
Several microbial clades with roles in N cycling were proportionally enriched in FL relative to PA fractions (Figure 3; Supplementary Figure S2). Sequences matching ammonia-oxidizing Thaumarchaeota and nitrite-oxidizing Nitrospina (classified as Deltaproteobacteria in Supplementary Figure S3; Teske et al., 1994) peaked in relative abundance (2.2–2.9% of total) in the FL fraction from the lower oxycline before declining with depth. Abundances of both groups decreased by 50–60% in the 1.6–30-μm PA fraction relative to the FL fraction and were negligible in the >30-μm PA fraction. Sequences matching the anammox Planctomycete Candidatus Scalindua (Order Brocadiales) were also confined primarily to the FL fraction, peaking at 14.1% of total FL amplicons at 300 m, compared with 0.3–1.5% in PA fractions. Anammox bacteria were previously shown to be enriched in the smallest size fractions in studies of the Chilean OMZ (Ganesh et al., 2014), the Black Sea (Fuchsman et al., 2012), and an OMZ off Costa Rica (Kong et al., 2013), but have also been detected in direct contact with particles in oxygen-depleted waters over the Namibian shelf (Woebken et al., 2007).
Figure 3.
ETNP OMZ bacterial community diversity revealed by 16S rRNA gene Illumina sequencing. The abundances of major bacterial divisions are shown as a percentage of total identifiable 16S rRNA gene sequences. ‘Other' includes 34 divisions, including unassigned sequences.
The composition of the ETNP bacterial community with potential roles in coupled N and sulfur cycles differed from that of other OMZs and varied among size fractions. The sulfur-oxidizing and denitrifying Gammaproteobacterial SUP05 clade, a ubiquitous inhabitant of low-oxygen waters (Wright et al., 2012), accounted for <0.2% of amplicons in ETNP samples. ETNP samples were instead enriched in members of the uncultured SAR406 and Deltaproteobacterial SAR324 lineages (Figure 3), as well as Gammaproteobacteria of the Thiohalorhabdales, Chromatiales and Thiotrichales (Supplementary Figure S3 and Supplementary Information), all of which may contain sulfur-oxidizing members. The SAR406 cluster was particularly abundant in FL samples, contributing 22% (average) of FL sequences, compared with 2.8% in PA fractions (Figure 3). SAR406 may participate in sulfur cycling via dissimilatory polysulfide reduction to sulfide or sulfide oxidation (Wright et al., 2014), although pathways for denitrification and carbon fixation have not been unambiguously identified. The SAR324 lineage, which includes members with genes for sulfur chemolithoautotrophy and nitrite reduction (Swan et al., 2011; Sheik et al., 2014), was also enriched in FL samples, representing 5–8% of sequences at OMZ depths, compared with <0.5% in PA fractions (Supplementary Figure S3). FL enrichment of SAR406 and SAR324 is consistent with an autotrophic lifestyle, as autotrophs would not require organic particles for carbon acquisition. Whereas sulfide-oxidizing autotrophs in other OMZs are capable of using oxidized N compounds as terminal oxidants (Walsh et al., 2009; Canfield et al., 2010), the extent to which ETNP clades participate in sulfur-driven denitrification remains unclear.
Many microbial groups common to low-oxygen waters, including those discussed above, were either at low abundance or absent from the >30-μm PA fraction. This fraction was instead enriched in taxa known to associate with the surfaces or guts of plankton or polymer aggregates (for example, fecal pellets), notably Gammaproteobacteria related to Vibrio, Pseudomonas and Alteromonas (Supplementary Figure S3; Rao et al., 2005; Hunt et al., 2008; Ivars-Martinez et al., 2008). Microorganisms on particles >30 μm are likely a minor component of the bulk OMZ microbial community, and may be relatively transient members, passing through low-oxygen depths attached to sinking aggregates or zooplankton.
RNA:DNA ratios
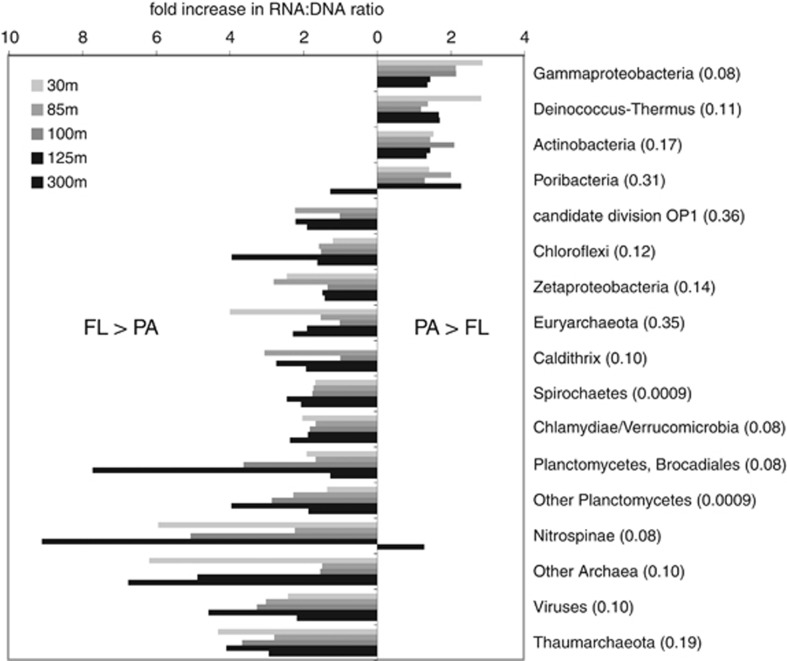
The taxonomic identities of protein-coding genes and transcripts suggest differences in the proportional contributions of specific taxa to bulk transcript pools in FL versus PA (1.6–30 μm) communities. RNA/DNA ratios for several groups with roles in N cycling, including anammox bacteria (Brocadiales), ammonia-oxidizing Thaumarchaeota and nitrite-oxidizing Nitrospina bacteria, were consistently elevated in FL relative to PA communities (Figure 4). These groups were also proportionately enriched in 16S gene pools from FL samples. In contrast, groups such as the Actinobacteria and Gammaproteobacteria exhibited greater RNA/DNA ratios in the PA fraction. Similar patterns were observed for ratios based on 16S rRNA gene amplicons and transcripts from metatranscriptomes (rRNA:rDNA; Supplementary Figure S4). Although RNA:DNA ratios, based either on target genes or community gene pools, have been used as proxies for metabolic activity (Campbell and Yu, 2011; Hunt et al., 2013; Satinsky et al., 2014), linking ratios to activity is tenuous. Notably, as RNA/DNA ratios are based on proportional abundances, increases in taxon representation in the transcriptome can reflect either increases in the activity of the target taxon or decreases in the activity of other organisms. Furthermore, using ratios to infer activity is challenging for broad divisions such as the Gammaproteobacteria that contain taxonomically and functionally diverse members, as the relationship between cell RNA content and activity is unlikely to be consistent across members or activity gradients (Blazewicz et al., 2013). Here, the composition of Gammaproteobacteria at the Order level varied widely (Supplementary Figure S3), highlighting the possibility that taxon-specific differences confounds interpretation of Phylum-level ratios. Studies at finer levels of taxonomic resolution utilizing transcriptome mapping to reference genomes coupled with measurements of absolute transcript numbers are necessary to confirm shifts in taxon-specific activity between FL and PA niches. The latter could reflect differences in substrate conditions or the proportions of live versus dead cells between fractions.
Figure 4.
Fold increase in taxon RNA/DNA ratios between PA (1.6–30 μm) and FL (0.2–1.6 μm) communities. Plot shows the fold increase in ratios for major taxonomic groups identified by the NCBI annotations of protein-coding genes and transcripts in coupled metatranscriptome and metagenome data sets. Values right of zero indicate higher ratios in PA communities (PA/FL>1). Values left of zero indicate higher ratios in FL communities (FL/PA>1). The plot shows only groups with relative abundance in metagenome data sets greater than 0.2% of total sequences, and only groups with an average (across all five depths) fold increase >1.5. Numbers in parentheses are false discovery rate q-values for t-test comparisons of PL and FL ratios, calculated as in Storey and Tibshirani (2003).
Bulk biochemical rates
N transformation rates in bulk water (no filter fractionation) varied among processes and over depth gradients. Rates of nitrate reduction to nitrite increased with depth and diminishing in situ oxygen concentration, with measured rates from 1.5 to 9.4 nm N d−1 (Figure 1e). The reduction of nitrite to N2 via denitrification was slower and measurable only below 125 m, with a maximum of 1.7 nm N d−1 at 300 m (Figure 1d). Anammox was measurable between 91 and 300 m at rates from 0.3 to 1.9 nm N d−1, with the highest rates in the secondary nitrite maximum. Anammox rates after 15NO2− addition were consistently higher than those after 15NH4+ addition, similar to trends in De Brabandere et al. (2014) working in the ETSP OMZ. The largest discrepancy in 15NO2− versus 15NH4+-based anammox rates occurred at 300 m, the depth where maximum denitrification rates were measured (Figure 1d), suggesting that nitrite shunting by denitrifiers could explain the offset in anammox rates, as proposed by De Brabandere et al. (2014). In this interpretation, incubations with 15NH4+ reflect true anammox rates, while incubations with 15NO2− overestimate the process. Our measured N loss rates are similar to those reported by Babbin et al. (2014) for stations close to ours (0.8–4.7 and 0.9–3.3 nm N d−1 for anammox and denitrification, respectively), and are generally consistent with those recorded in other OMZs (Lam and Kuypers, 2011). Dissimilatory nitrate reduction to ammonium was not detected in any of the samples. Ammonia oxidation was detected only at 100 m or shallower, with maximum rates at 85 m (0.92 nm N d−1), the shallowest and most oxygenated of the depths where rates were measured (Figure 1e). These rates are at the lower end of those previously observed in the Gulf of California and ETNP (0–348 nm d−1; Beman et al., 2012) and in the ETSP OMZ (0.2–89 nm d−1; Kalvelage et al., 2013). Nitrite oxidation was detected at all depths except 125 m, at rates (13–261 nm d−1; Figure 1e) up to an order of magnitude higher than those for other processes, with rates peaking in the secondary chlorophyll maximum, suggesting a decoupling between ammonia and nitrite oxidation. These values are consistent with nitrite oxidation rates measured in the Gulf of California and ETNP (0–213 nm d−1; Beman et al., 2013), the ETSP (5–928 nm d−1; Kalvelage et al., 2013) and the Benguela upwelling (14–372 nm d−1; Füssel et al., 2012), further confirming the activity of nitrite oxidizers at nanomolar oxygen levels (Füssel et al., 2012).
Size-fractionated biochemical rates and gene expression
Nitrate reduction and denitrification
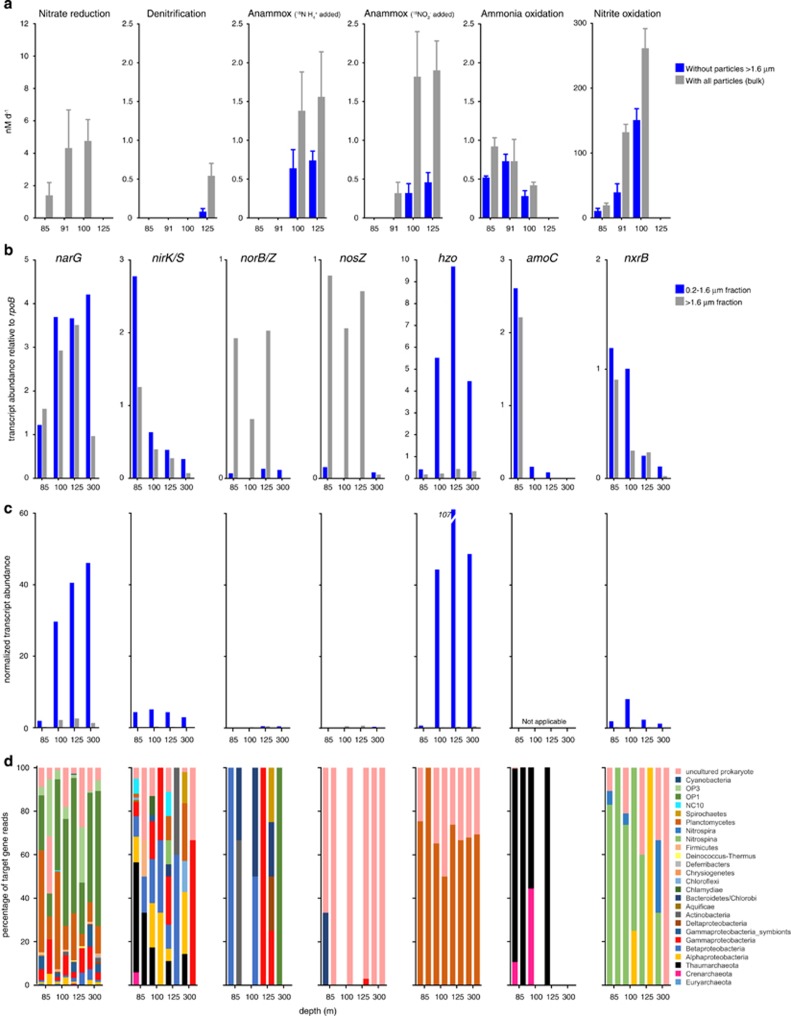
Rates of nitrate reduction to nitrite were undetectable after removal of particles (>1.6 μm) by filtration (Figure 5a). Filtration therefore removed either a significant proportion of total nitrate-reducing cells or a critical substrate source fueling FL nitrate reducers. Although the proportional abundances of transcripts encoding nitrate reductase (narG) were uniform among filter fractions (Figure 5b), suggesting roughly equivalent contributions of nitrate reduction to fraction-specific metabolism in PA and FL niches, total transcript counts were still considerably higher in the FL fraction (Figure 5c). The taxonomic identities of narG transcripts spanned a wide phylogenetic range (Figure 5d), with sequences related to anammox bacteria and candidate divisions OP3 and OP1 particularly highly represented in both fractions, which is consistent with results from ETSP OMZ metagenomes (Ganesh et al., 2014). The distribution of narG suggests that declines in nitrate reduction after filtration were likely driven primarily by the removal of particulate organic matter (OM) for use by diverse FL nitrate reducers and to a minor extent by the removal of particle-attached nitrate reducers.
Figure 5.
Measured rates and marker gene transcript levels for major dissimilatory N cycle processes. Process rates (a) are shown for incubations with particles (no filtration) and without particles measuring 1.6 μm or larger. Zero values indicate non-significant rates and error bars represent the standard errors. Note variation in y axis scales. (b) Marker gene transcript abundances in FL (0.2–1.6 μm) and PA (1.6–30 μm) filter fractions. Abundances are calculated as read count per gene per kilobase of gene length, and shown as a proportion of the abundance of transcripts matching the universal, single-copy gene rpoB. A value of 1 indicates abundance equal to that of rpoB. Note variation in y axis scales. (c) Multiplies the values in b by the counts of bacterial 16S genes in Figure 1f and the FL/PA ratio (for FL counts only), with the values then divided by 1 000 000 for presentation. The resulting values do not reflect absolute counts, but provide an approximation of how variation in bacterial abundance (inferred by proxy from 16S counts) is predicted to affect comparisons of absolute transcript counts across samples, assuming that absolute transcript counts scale with bacterial load. Scaled amoC values are not provided in c, as FL:PA ratios were determined using bacterial-specific primers, and amoC transcripts were dominated exclusively by archaeal sequences; FL/PA ratios do not reflect archaeal contributions. Patterns in c should be interpreted cautiously, as they could change if RNA content per unit biomass varies substantially among samples. (d) The taxonomic affiliations of marker gene transcripts based on NCBI annotations of genes identified as top matches (> bit score 50) via BLASTX.
Rates of N2 production by denitrification also declined significantly (by 85%) following particle removal. Transcripts encoding enzymes for nitrite reduction (nirK/S) and N2O and N2 gas production (nosZ and norB, respectively) were at low abundance compared with those of nitrate reduction and anammox (Figure 5c), and showed contrasting size fraction distributions. Transcripts for NirK/S were at relatively uniform proportional abundances in PA and FL data sets. These transcripts reflected a diverse assemblage, but were notably enriched at the lower oxycline and oxic–nitrite interface by Thaumarchaeota nirK sequences (Figure 5d). NO-forming Nir activity in ammonia-oxidizing Thaumarchaeota is not well understood (Lund et al., 2012), but may have roles in both energy generation and nitrite detoxification, with the latter of potential relevance in nitrite-replete OMZs. In contrast, nosZ and norB transcripts were enriched up to 28-fold in PA fractions, particularly at upper OMZ depths (Figure 5b), and affiliated with a community taxonomically distinct from that catalyzing upstream steps of denitrification (Figure 5d), as also reported in Dalsgaard et al. (2014). These results support metagenome data from the Chilean OMZ (Ganesh et al., 2014) and from an estuarine site (Smith et al., 2013) showing nor/nos enrichment in PA fractions, suggesting a conserved PA niche. After scaling based on 16S gene counts (a proxy for cell counts), absolute counts of nosZ and norB transcripts were estimated to be comparable between size fractions (Figure 5c), suggesting that filtration removes a significant proportion of the N2O- and N2-forming denitrifier community.
Anammox
Anammox rates declined 53–100% without particles. These declines may indicate that particle removal eliminates an important substrate source for FL anammox bacteria. Particles could facilitate anammox by providing (1) ammonium remineralized by heterotrophic PA bacteria consuming particulate OM (Shanks and Trent, 1979), (2) ammonium produced by anammox bacteria via oxidation of particulate OM with nitrate or nitrite (Kartal et al., 2007) or (3) nitrite produced by other (non-anammox) nitrate-reducing heterotrophs consuming particulate OM. Explanations (1) and (3) are consistent with the observed reduction in rates of nitrate reduction to nitrite, the most active respiratory process detected, in the absence of particles. We therefore hypothesize that particle-dependent nitrate or nitrite reduction by both FL and PA communities likely provides a critical supply of ammonium or nitrite for anammox, although it remains unclear whether anammox bacteria are ammonium- or nitrite-limited in OMZs (Lam and Kuypers, 2011; De Brabandere et al., 2014).
Two alternate hypotheses could explain the decline in anammox after filtration. First, filtration removed a significant fraction of total anammox-capable cells. This hypothesis is dismissed given the substantial enrichments of total 16S rRNA gene copies and Brocadiales 16S copies in the FL fraction. Second, anammox-capable cells in PA fractions, although a minor component of the total population, contributed disproportionately to bulk anammox rates, potentially overexpressing anammox enzymes compared with FL cells. The active transcription of anammox genes in the FL fraction argues against this hypothesis. Transcripts encoding hydrazine oxidoreductase, catalyzing hydrazine oxidation to N2 during anammox, were up to 15-fold higher in proportional abundance in FL samples (Figure 5b), consistent with the 14-fold enrichment of Brocadiales 16S genes in the FL fraction.
To further examine the potential that anammox bacteria alter their metabolism between FL and PA niches, we tested for differential gene expression between PA and FL data sets, focusing only on genes from known anammox genera (Supplementary Methods). After normalizing for variation in total transcripts matching anammox genera, PA and FL transcript profiles were well correlated (R2=0.81) and no genes (of 1392 total) were differentially expressed between fractions (P>0.05). However, testing for DE required grouping data sets from different depths as replicates, raising the possibility that depth-specific variation confounded the detection of DE between fractions. Despite this apparent uniformity in expression, marker genes for the anammox process itself (hzo and hzs encoding hydrazine synthase) were consistently, although not statistically, enriched (2–3 fold) in the FL fraction, suggesting an upregulation of anammox in the FL niche (Supplementary Figure S5). In contrast, transcripts encoding nitrate reductase NarGH were among those most abundant and enriched in PA profiles. Nar likely acts in reverse in anammox bacteria, converting nitrite to nitrate and helping replenish electrons lost during CO2 fixation via the acetyl-CoA pathway (Strous et al., 2006; Jetten et al., 2009). Here transcripts encoding key acetyl-CoA pathway genes (formate dehydrogenase and carbon monoxide dehydrogenase) were ~10-fold less abundant than Nar transcripts, and did not differ appreciably between PA and FL fractions. Alternatively, Nar in anammox bacteria may function as a traditional nitrate reductase during anaerobic growth on organic compounds (Güven et al., 2005), and it is plausible that oxidation of organics is enhanced in anammox bacteria on particles. The observed fold change differences in hzo, hzs and nar raise the possibility of microniche-specific gene expression by anammox-capable bacteria, but do not support the hypothesis that the anammox process itself is upregulated in PA compared with FL communities. Thus, the effect of filtration on anammox rates is best explained by dependence of FL anammox bacteria on the PA fraction for substrate.
Nitrification
Compared with denitrification and anammox, ammonia and nitrite oxidation were less affected by particle removal. Filtration decreased ammonia oxidation rates by ~25% in the lower oxycline and upper OMZ. At the oxic–nitrite interface, rates did not change following filtration. Nitrite oxidation decreased by ~50% without particles, although rates after filtration were still several orders of magnitude higher than those of other N processes. Transcripts encoding ammonia monooxygenase (amoC) were proportionately enriched in the FL fraction (excluding at 85 m) and affiliated predominantly with sequences annotated as Thaumarchaeota or as ‘uncultured crenarchaeote' (the latter were entered into NCBI before formal recognition of the ‘Thaumarchaeota' designation and likely should be re-annotated as Thaumarchaeota; Figure 5d). Transcripts encoding nitrite oxidoreductase (nxrB) did not show a strong size fraction-specific enrichment, consistent with nxrB gene distributions in metagenomes from the Chilean OMZ (Ganesh et al., 2014). NxrB transcripts affiliated primarily with sequences from the genome of Nitrospina gracilis (Lücker et al., 2013), supporting reports of high Nitrospina activity in the ETNP region (Beman et al., 2013). Analysis of only N. gracilis-affiliated transcripts did not reveal any genes that were differentially expressed between FL and PA fractions (P>0.05), although low N. gracilis read counts in PA data sets (5% those in FL data sets) likely confounded statistical detection. Indeed, nxr genes (all subunits combined) represented similar proportions of total N. gracilis counts in both fractions (2.5% and 3.2% in FL and PA, respectively), suggesting that expression of nitrite oxidation genes was not substantially altered between PA and FL niches.
Conclusions
Filter fractionation significantly altered N metabolism rates, confirming a critical role for particles and PA microorganisms in OMZ N cycling. A strong dependence on particles may help explain heterogeneity of denitrification and anammox rates among OMZ sites and time points (Dalsgaard et al., 2012; De Brabandere et al., 2014; Kalvelage et al., 2013), notably if particle load varies over depth and distance from shore or between times of high versus low export from the photic zone. Our data also suggest differences in how OMZ microbes interact with particles. A significant fraction of some functional groups, notably N2O- and N2-producing denitrifiers, are recovered in PA fractions, suggesting direct adherence to particles, or potentially larger cell sizes. A tight coupling to particle surfaces could facilitate access to organic carbon for respiration, as well as oxidized N compounds produced by upstream steps of the denitrification pathway. Indeed, enhanced hydrolytic activity of particulate organic substrates has been demonstrated in diverse marine waters (Hoppe et al., 1993; Taylor et al., 2009; Yamada et al., 2012). Denitrifiers are generally known as facultative anaerobes (Zumft, 1997) and populations of denitrifiers on OMZ particles may develop during sinking through the oxycline, switching to denitrification upon transition to the OMZ core.
In contrast, groups such as obligately anaerobic anammox bacteria are recovered almost exclusively within the FL fraction, but appear no less dependent on particles. Anammox cells presumably rely on particles indirectly as sources of nitrite and ammonium, produced by nitrate or nitrite reducers consuming particulate OM. Although anammox cells have been recovered in direct contact with particles in other systems (Woebken et al., 2007), enrichment of anammox genes and transcripts in OMZ FL fractions suggests a relatively loose physical association with particles, although it is possible that some particle-adhered cells are dislodged into the FL fraction during filtration. It is plausible that the proportion of the anammox community adhered to particles varies depending on local substrate conditions and oxygen content. Direct adherence may enable access to anoxic microniches and be favored at upper OMZ depths where water column oxygen content is closer to the threshold for anammox inhibition (~900 nm; Dalsgaard et al., 2014).
The potential for OMZ microbes to alter their physiology between FL and PA niches remains unresolved. Anammox Planctomycetes and nitrite-oxidizing Nitrospina, two groups recovered in both FL and PA fractions and occupying relatively well-defined functional roles, did not exhibit significantly different expression profiles between fractions. This suggests that the physical act of filtration had a relatively minor effect on cell state. Nonetheless, certain genes exhibited large fold changes in relative abundance between fractions, raising the possibility that additional analyzes with higher replication per depth may reveal size fraction-specific transcriptional differences in certain taxa. Clarifying such patterns, potentially by sequencing deeply across biological replicates coupled with transcriptome mapping to reference genomes, may help estimate per-cell differences in biochemical processing at the microscale. However, the use of biomass fractionation to characterize the microspatial architecture of microbial activity remains challenging. As shown here, particle removal significantly disrupts bulk N cycle processes mediated by a FL majority, likely by severing important chemical and biological linkages between FL and PA niches.
Acknowledgments
We thank the crew of the R/V New Horizon for help in sample collection, Neha Sarode and Josh Parris for help in sequencing, Anni Glud for technical assistance and for preparing the STOX, and Peter Girguis, Kathy Barbeau and Forest Rohwer for providing essential equipment. This work was supported by the National Science Foundation (1151698 to FJS), the Sloan Foundation (RC944 to FJS), the Danish National Research Foundation DNRF53, the Danish Council of Independent Research and the European Research Council ‘Oxygen' grant (267233 supporting LAB, ML and BT).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Babbin AR, Keil RG, Devol AH, Ward BB. (2014). Organic matter stoichiometry, flux, and oxygen control nitrogen loss in the ocean. Science 344: 406–408. [DOI] [PubMed] [Google Scholar]
- Beman JM, Popp BN, Alford SE. (2012). Quantification of ammonia oxidation rates and ammonia-oxidizing archaea and bacteria at high resolution in the Gulf of California and eastern tropical North Pacific Ocean. Limnol Oceanogr 57: 711–726. [Google Scholar]
- Beman JM, Carolan MT. (2013). Deoxygenation alters bacterial diversity and community composition in the ocean's largest oxygen minimum zone. Nat Commun 4: 2705. [DOI] [PubMed] [Google Scholar]
- Beman JM, Leilei Shih J, Popp BN. (2013). Nitrite oxidation in the upper water column and oxygen minimum zone of the eastern tropical North Pacific Ocean. ISME J 7: 2192–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. (2013). Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J 7: 2061–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Yu L, Heidelberg JF, Kirchman DL. (2011). Activity of abundant and rare bacteria in a coastal ocean. Proc Natl Acad Sci USA 108: 12776–12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield DE, Stewart FJ, Thamdrup B, De Brabandere L, Dalsgaard T, Delong EF et al. (2010). A cryptic sulfur cycle in oxygen-minimum-zone waters off the Chilean coast. Science 330: 1375–1378. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ et al. (2011). Global patterns of 16 S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA 15: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceja-Navarro JA, Nguyen NH, Karaoz U, Gross SR, Herman DJ, Andersen GL et al. (2014). Compartmentalized microbial composition, oxygen gradients and nitrogen fixation in the gut of Odontotaenius disjunctus. ISME J 8: 6–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho BC, Azam F. (1988). Major role of bacteria in biogeochemical fluxes in the ocean's interior. Nature 332: 441–443. [Google Scholar]
- Cline JD, Richards FA. (1974). Oxygen deficient conditions and nitrate reduction in the eastern tropical North Pacific ocean. Limnol Oceanogr 17: 885–900. [Google Scholar]
- Codispoti LA, Brandes JA, Christensen JP, Devol AH, Naqvi SWA, Paerl HW et al. (2001). The oceanic fixed nitrogen and nitrous oxide budgets: moving targets as we enter the anthropocene? Sci Mar 65: 85–105. [Google Scholar]
- Dalsgaard T, Thamdrup B, Farias L, Revsbech NP. (2012). Anammox and denitrification in the oxygen minimum zone of the eastern South Pacific. Limnol Oceanogr 57: 1331–1346. [Google Scholar]
- Dalsgaard T, Stewart FJ, Thamdrup B, De Brabandere L, Revsbech NP, Ulloa O et al. (2014). Oxygen at nanomolar levels reversibly suppresses process rates and gene expression of anammox and denitrification in the oxygen minimum zone off northern Chile. mBio 5: e01966–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Brabandere L, Canfield DE, Dalsgaard T, Friederich GE, Revsbech NP, Ulloa O et al. (2014). Vertical partitioning of nitrogen-loss processes across the oxic–anoxic interface of an oceanic oxygen minimum zone. Environ Microbiol 16: 3041–3054. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Franks DG, Alldredge AL. (1993). Phylogenetic diversity of aggregate-attached vs free-living marine bacterial assemblages. Limnol Oceanogr 38: 924–934. [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16 S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Eldholm V, Norheim G, von der Lippe B, Kinander W, Dahle UR, Caugant DA et al. (2014). Evolution of extensively drug-resistant Mycobacterium tuberculosis from a susceptible ancestor in a single patient. Genome Biol 15: 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DP. (1992). Conservation evaluation and phylogenetic diversity. Biol Conserv 61: 1–10. [Google Scholar]
- Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW et al. (2008). Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci USA 105: 3805–3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchsman CA, Staley JT, Oakley BB, Kirkpatrick JB, Murray JW. (2012). Free-living and aggregate-associated Planctomycetes in the Black Sea. FEMS Microbiol Ecol 80: 402–416. [DOI] [PubMed] [Google Scholar]
- Füssel J, Lam P, Lavik G, Jensen MM, Holtappels M, Gunter M et al. (2012). Nitrite oxidation in the Namibian oxygen minimum zone. ISME J 6: 1200–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh S, Parris DJ, DeLong EF, Stewart FJ. (2014). Metagenomic analysis of size-fractionated picoplankton in a marine oxygen minimum zone. ISME J 8: 187–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiglione JF, Conan P, Pujo-Pay M. (2009). Diversity of total and active free-living vs particle-attached bacteria in the euphotic zone of the NW Mediterranean Sea. FEMS Microbiol Lett 299: 9–21. [DOI] [PubMed] [Google Scholar]
- Grossart HP, Hietanen S, Ploug H. (2003). Microbial dynamics on diatom aggregates in Øresund, Denmark. Mar Ecol Prog Ser 249: 69–78. [Google Scholar]
- Grossart HP, Tang KW, Kiørboe T, Ploug H. (2007). Comparison of cell-specific activity between free-living and attached bacteria using isolates and natural assemblages. FEMS Microbiol Lett 266: 194–200. [DOI] [PubMed] [Google Scholar]
- Gruber NThe dynamics of the marine nitrogen cycle and its influence on atmospheric CO2 variations Follows M, Oguz T The Ocean Carbon Cycle and Climate, NATO ASI Series. Kluwer: Dordrecht, Germany,. (2004). 97–148. [Google Scholar]
- Güven D, Dapena A, Kartal B, Schmid MC, Maas B, van de Pas-Schoonen K et al. (2005). Propionate oxidation by and methanol inhibition of anaerobic ammonium-oxidizing bacteria. Appl Environ Microbiol 71: 1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle TJ, Kelly KA. (2010). baySeq: Empirical Bayesian methods for identifying differential expression in sequence count data. BMC Bioinform 11: 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatt JK, Ritalahti KM, Ogles DM, Lebrón CA, Löffler FE. (2013). Design and application of an internal amplification control to improve Dehalococcoides mccartyi 16 S rRNA gene enumeration by qPCR. Environ Sci Technol 47: 11131–11138. [DOI] [PubMed] [Google Scholar]
- Hollibaugh T, Wong PS, Murrell MC. (2000). Similarity of particle- associated and free-living bacterial communities in northern San Francisco Bay, California. Aquat Microb Ecol 21: 102–114. [Google Scholar]
- Hoppe HG, Ducklow H, Karrasch B. (1993). Evidence for dependency of bacterial growth on enzymatic hydrolysis of particulate organic matter in the mesopelagic ocean. Mar Ecol Prog Ser 93: 277–283. [Google Scholar]
- Hunt DE, David LA, Gevers D, Preheim SP, Alm EJ, Polz MF et al. (2008). Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science 320: 1081–1085. [DOI] [PubMed] [Google Scholar]
- Hunt DE, Lin Y, Church MJ, Karl DM, Tringe SG, Izzo LK et al. (2013). Relationship between abundance and specific activity of bacterioplankton in open ocean surface waters. Appl Environ Microbiol 79: 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Mitra S, Ruscheweyh HJ, Weber N, Schuster SC. (2011). Integrative analysis of environmental sequences using MEGAN4. Genome Res 21: 1552–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivars-Martinez E, Martin-Cuadrado AB, D'Auria G, Mira A, Ferriera S, Johnson J et al. (2008). Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. ISME J 2: 1194–1212. [DOI] [PubMed] [Google Scholar]
- Jetten MS, Niftrik LV, Strous M, Kartal B, Keltjens JT, Op den Camp HJ et al. (2009). Biochemistry and molecular biology of anammox bacteria. Crit Rev Biochem Mol Biol 44: 65–84. [DOI] [PubMed] [Google Scholar]
- Kalvelage T, Lavik G, Lam P, Contreras S, Arteaga L, Loscher CR et al. (2013). Nitrogen cycling driven by organic matter export in the South Pacific oxygen minimum zone. Nat Geosci 6: 228–234. [Google Scholar]
- Karl DM, Knauer GA, Martin JH. (1988). Downward flux of particulate organic matter in the ocean: a particle decomposition paradox. Nature 332: 438–441. [Google Scholar]
- Karstensen J, Stramma L, Visbeck M. (2008). Oxygen minimum zones in the eastern tropical Atlantic and Pacific oceans. Prog Oceanogr 77: 331–350. [Google Scholar]
- Kartal B, Kuypers MMM, Lavik G, Schalk J, Op den Camp HJM, Jetten MSM et al. (2007). Anammox bacteria disguised as denitrifiers: nitrate reduction to dinitrogen gas via nitrite and ammonium. Environ Microbiol 9: 635–642. [DOI] [PubMed] [Google Scholar]
- Kong L, Jing H, Kataoka T, Buchwald C, Liu H. (2013). Diversity and spatial distribution of hydrazine oxidoreductase (hzo) gene in the oxygen minimum zone off Costa Rica. PLoS One 8: e78275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol 79: 5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P, Lavik G, Jensen MM, van de Vossenberg J, Schmid M, Woebken D et al. (2009). Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc Natl Acad Sci USA 106: 4752–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam P, Kuypers MMM. (2011). Microbial nitrogen cycling processes in oxygen minimum zones. Annu Rev Mar Sci 3: 317–345. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. (2005). UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microb 71: 8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lücker S, Nowka B, Rattei T, Spieck E, Daims H. (2013). The genome of Nitrospina gracilis illuminates the metabolism and evolution of the major marine nitrite oxidizer. Front Microbiol 4: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund MB, Smith JM, Francis CA. (2012). Diversity, abundance and expression of nitrite reductase (nirK-like genes in marine thaumarchaea. ISME J 6: 1966–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlvin MR, Altabet MA. (2005). Chemical conversion of nitrate and nitrite to nitrous oxide for nitrogen and oxygen isotopic analysis in freshwater and seawater. Anal Chem 77: 5589–5595. [DOI] [PubMed] [Google Scholar]
- Milohanic E, Glaser P, Coppée JY, Frangeul L, Vega Y, Vázquez-Boland JA et al. (2003). Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol Microbiol 47: 1613–1625. [DOI] [PubMed] [Google Scholar]
- Naqvi SWA, Kumar MD, Narvekar PV, De Sousa SN, George MD, Silva CD et al. (1993). An intermediate nepheloid layer associated with high microbial rates and denitrification in the northwest Indian Ocean. J Geophys Res 98: 16469–16479. [Google Scholar]
- Paulmier A, Ruiz Pino. (2009). Oxygen minimum zones (OMZs) in the modern ocean. Prog Oceanogr 80: 113–128. [Google Scholar]
- Podlaska A, Wakeham SG, Fanning KA, Taylor GTMicrobial community structure and productivity in the oxygen minimum zone of the eastern tropical North Pacific. Deep Sea Res Part I,. (2012) 66: 77–89. [Google Scholar]
- Rao D, Webb JS, Kjelleberg S. (2005). Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl Environ Microbiol 71: 1729–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revsbech NP, Larsen LH, Gundersen J, Dalsgaard T, Ulloa O, Thamdrup B et al. (2009). Determination of ultra-low oxygen concentrations in oxygen minimum zones by the STOX sensor. Limnol Oceanogr Methods 7: 371–381. [Google Scholar]
- Ritalahti KM, Amos BK, Sung Y, Wu Q, Koenigsberg SS, Löffler FE et al. (2006). Quantitative PCR targeting 16 S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl Environ Microbiol 72: 2765–2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush D, Wakeham SG, Hopmans EC, Schouten S, Sinninghe Damsté JS. (2012). Biomarker evidence for anammox in the oxygen minimum zone of the Eastern Tropical North Pacific. Org Geochem 53: 80–87. [Google Scholar]
- Satinsky BM, Crump BC, Smith CB, Sharma S, Zielinski BL, Doherty M et al. (2014). Microspatial gene expression patterns in the Amazon River Plume. Proc Natl Acad Sci USA 111: 11085–11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Lim YW, Edwards R. (2012). Identification and removal of ribosomal RNA sequences from metatranscriptomes. Bioinformatics 28: 433–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumann U, Edwards MD, Rasmussen T, Bartlett W, van West P, Booth IR et al. (2010). YbdG in Escherichia coli is a threshold-setting mechanosensitive channel with MscM activity. Proc Natl Acad Sci USA 107: 12664–12669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanks AL, Trent JD. (1979). Marine snow: microscale nutrient patches. Limnol Oceanogr 24: 850–854. [Google Scholar]
- Sheik CS, Jain S, Dick GJ. (2014). Metabolic flexibility of enigmatic SAR324 revealed through metagenomics and metatranscriptomics. Environ Microbiol 6: 304–317. [DOI] [PubMed] [Google Scholar]
- Sihto HM, Tasara T, Stephan R, Johler S. (2014). Validation of reference genes for normalization of qPCR mRNA expression levels in Staphylococcus aureus exposed to osmotic and lactic acid stress conditions encountered during food production and preservation. FEMS Microbiol Lett 356: 134–140. [DOI] [PubMed] [Google Scholar]
- Simon M, Grossart HP, Schweitzer B, Ploug H. (2002). Microbial Ecology of organic aggregates in aquatic ecosystems. Aquat Microb Ecol 28: 175–211. [Google Scholar]
- Smith MW, Allen LZ, Allen AE, Herfort L, Simon HM. (2013). Contrasting genomic properties of free-living and particle-attached microbial assemblages within a coastal ecosystem. Front Microbiol 4: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinrad RW, Glover H, Ward BB, Codispoti LA, Kullenberg G. (1989). Suspended particle and bacterial maxima in Peruvian coastal waters during a cold water anomaly. Deep-Sea Res 36: 715–733. [Google Scholar]
- Stewart FJ, Ulloa O, DeLong EF. (2012). Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ Microbiol 14: 23–40. [DOI] [PubMed] [Google Scholar]
- Stocker R. (2012). Marine microbes see a sea of gradients. Science 338: 628–633. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. (2003). Statistical significance for genomewide studies. Proc Natl Acad Sci USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW et al. (2006). Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440: 790–794. [DOI] [PubMed] [Google Scholar]
- Sue D, Fink D, Wiedmann M, Boor KJ. (2004). Sigma(B)-dependent gene induction and expression in Listeria monocytogenes during osmotic and acid stress conditions simulating the intestinal environment. Microbiology 150: 3843–3855. [DOI] [PubMed] [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D et al. (2011). Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the dark ocean. Science 333: 1296–1300. [DOI] [PubMed] [Google Scholar]
- Taylor GT, Thunell R, Varela R, Benitez-Nelson C, Scranton MI. (2009). Hydrolytic ectoenzyme activity associated with suspended and sinking organic particles within the anoxic Cariaco Basin. Deep Sea Res I 56: 1266–1283. [Google Scholar]
- Teske A, Alm E, Regan JM, Toze S, Rittmann BE, Stahl DA et al. (1994). Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J Bacteriol 176: 6623–6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamdrup B, Dalsgaard T. (2002). Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl Environ Microbiol 68: 1312–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thamdrup B, Dalsgaard T, Jensen MM, Ulloa O, Farias L, Escribano R et al. (2006). Anaerobic ammonium oxidation in the oxygen-deficient waters off northern Chile. Limnol Oceanogr 51: 2145–2156. [Google Scholar]
- Tiano L, Garcia Robledo E, Dalsgaard T, Devol A, Ward B, Uloa O et al. (2014). Oxygen distribution and aerobic respiration in the north and southeastern tropical Pacific oxygen minimum zones. Deep-Sea Res I 194: 173–183. [Google Scholar]
- Turley CM, Stutt ED. (2000). Depth-related cell-specific bacterial leucine incorporation rates on particles and its biogeochemical significance in the Northwest Mediterranean. Limnol Oceanogr 45: 419–425. [Google Scholar]
- Ulloa O, Canfield DE, DeLong EF, Letelier RM, Stewart FJ. (2012). Perspective: microbial oceanography of anoxic oxygen minimum zones. Proc Natl Acad Sci USA 109: 15996–16003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandecasteele SJ, Peetermans WE, Merckx R, Van Eldere J. (2001). Quantification of expression of Staphylococcus epidermidis housekeeping genes with Taqman quantitative PCR during in vitro growth and under different conditions. J Bacteriol 183: 7094–7101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh DA, Zaikova E, Howes CG, Song YC, Wright JJ, Tringe SG et al. (2009). Metagenome of a versatile chemolithoautotroph from expanding oceanic dead zones. Science 326: 578–582. [DOI] [PubMed] [Google Scholar]
- Whitmire AL, Letelier RM, Villagran V, Ulloa O. (2009). Autonomous observations of in vivo fluorescence and particle backscattering in an oceanic oxygen minimum zone. Optics Express 17: 21992–22004. [DOI] [PubMed] [Google Scholar]
- Woebken D, Fuchs BM, Kuypers MM, Amann R. (2007). Potential interactions of particle-associated anammox bacteria with bacterial and archaeal partners in the Namibian upwelling system. Appl Environ Microbiol 73: 4648–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JJ, Konwar KM, Hallam SJ. (2012). Microbial ecology of expanding oxygen minimum zones. Nat Rev Microbiol 10: 381–394. [DOI] [PubMed] [Google Scholar]
- Wright JJ, Mewis K, Hanson NW, Konwar KM, Maas KR, Hallam SJ et al. (2014). Genomic properties of Marine Group A bacteria indicate a role in the marine sulfur cycle. ISME J 8: 455–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada N, Fukuda H, Ogawa H, Saito H, Suzumura M. (2012). Heterotrophic bacterial production and extracellular enzymatic activity in sinking particulate matter in the western North Pacific Ocean. Front Microbiol 3: 379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft WG. (1997). Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61: 533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.