Abstract
Members of the archaeal Miscellaneous Crenarchaeotic Group (MCG) are among the most successful microorganisms on the planet. During its evolutionary diversification, this very diverse group has managed to cross the saline–freshwater boundary, one of the most important evolutionary barriers structuring microbial communities. However, the current understanding on the ecological significance of MCG in freshwater habitats is scarce and the evolutionary relationships between freshwater and saline MCG remains poorly known. Here, we carried out molecular phylogenies using publicly available 16S rRNA gene sequences from various geographic locations to investigate the distribution of MCG in freshwater and saline sediments and to evaluate the implications of saline–freshwater transitions during the diversification events. Our approach provided a robust ecological framework in which MCG archaea appeared as a core generalist group in the sediment realm. However, the analysis of the complex intragroup phylogeny of the 21 subgroups currently forming the MCG lineage revealed that distinct evolutionary MCG subgroups have arisen in marine and freshwater sediments suggesting the occurrence of adaptive evolution specific to each habitat. The ancestral state reconstruction analysis indicated that this segregation was mainly due to the occurrence of a few saline–freshwater transition events during the MCG diversification. In addition, a network analysis showed that both saline and freshwater MCG recurrently co-occur with archaea of the class Thermoplasmata in sediment ecosystems, suggesting a potentially relevant trophic connection between the two clades.
Introduction
Over the course of the last decade, the rapid accumulation of genetic data has fundamentally changed our understanding of ecosystem functioning by unraveling new abundant microbial groups (mostly represented by uncultivated microbes) involved in important ecological processes such as aerobic ammonia oxidation in Thaumarchaeota (Konneke et al., 2005), anaerobic ammonia oxidation in Planctomycetales (Dalsgaard et al., 2003; Kuypers et al., 2003) and light-energy conversion using proteorhodopsin in SAR86 (Beja et al., 2000). However, most of these abundant microbial groups escape our capacity to understand their ecology and physiology. This is for instance the case of the Miscellaneous Crenarchaeotic Group (MCG), a sister clade of the archaeal phyla Thaumarchaeota and Aigarchaeota, which appears to be particularly abundant and widespread in marine sediments (Biddle et al., 2006, 2008; Durbin and Teske, 2012; Kubo et al., 2012; Lloyd et al., 2013; Lazar et al., 2014), where it accounts on average for 12% of total prokaryotic cells in CARD-FISH counts (Kubo et al., 2012) and for 30% of all clones in archaeal 16S rRNA gene libraries (Fry et al., 2008). Considering that half of the microbial cells in the oceans are found in sediments (Kallmeyer et al., 2012) and that oceans covers ~70% of the surface of the planet, members of the MCG may be one of the most successful lineages on the Earth. This success extends beyond the limits of the marine sediment habitat as MCG 16S rRNA gene sequences are also present in other natural habitats such as freshwater plankton and sediment, and hydrothermal vents (see Figure 3-group C2 in Auguet et al., 2010).
The widespread distribution of MCG suggests a great versatility in their physiological capabilities. That is in agrement with the discovery of a functional bacteriochlorophyll a synthase (bchG) gene in a MCG fosmid, conferring metabolic plasticity to adapt to various environments (Meng et al., 2009). However, the lack of cultured MCG representatives has hampered a better understanding of the metabolic potential of the group. First evidence for heterotrophy based on buried organic carbon was brought by Biddle et al. (2006), who observed assimilation of sedimentary organic compounds by MCG archaea using isotopic analysis. Assimilation of organic carbon by MCG archaea have recently been corroborated in incubation experiments of estuarine sediments by stable isotope probing (Webster et al., 2010; Seyler et al., 2014). Recent findings from metagenomic data (Meng et al., 2014) and single-cell genomics (Lloyd et al., 2013) provided indirect evidence that members of the MCG lineage may be involved in the degradation of aromatic compounds (for example, protocatechuate) and detrital proteins, respectively. This heterogeneity in the potential substrates used by MCG archaea reflects the extremely high genomic diversity of the lineage (Meng et al., 2014), hindering a full understanding on the ecological role and influence of MCG in biogeochemical cycles.
Indeed, members of the MCG are very divergent (up to 24%, based on 16S rRNA gene identity, Kubo et al., 2012) and the whole lineage has been proposed as a new archaeal phylum named Bathyarchaeota (Meng et al., 2014). Although a comprehensive phylogeny subdividing the MCG lineage into 17 monophyletic subgroups has recently been reported (Kubo et al., 2012), many questions remain unanswered concerning the distribution and functions of these subgroups in the different habitats. Of particular interest is the ability of MCG to overcome one of the most important evolutionary barriers structuring microbial communities, that is, salinity (Auguet et al., 2010; Lozupone and Knight, 2007) and to colonize both marine and freshwater habitats (Casamayor et al., 2001; Biddle et al., 2006; Lehours et al., 2007; Llirós et al., 2008, 2010; Auguet et al., 2010, 2012; Bhattarai et al., 2012; Borrel et al., 2012; Buckles et al., 2013; Fillol et al., 2015). In addition to the fact that the abundance of MCG in freshwater habitats is still not well estimated, no study has yet been conducted to investigate the evolutionary relationships between marine and freshwater MCG, and whether or not distinct MCG subgroups have evolved along the transition between marine and freshwater habitats.
In the present investigation, we used publicly available archaeal 16S rRNA gene sequences from globally distributed studies to (1) examine the ecological significance of MCG within archaeal communities in both freshwater and marine sediments and (2) investigate the hypothesis of a relationship between diversification and habitat during the evolution of the MCG lineage and the transition between marine and freshwater sediments. In addition, we used network analysis in an attempt to unveil potential syntrophic and/or mutualistic interactions in which MCGs would be involved and, by association, in the potential metabolisms they might harbor.
Materials and methods
Data set constructions
A multistep Biopython homemade script was used as wrapper to the Entrez Programming Utilities to search and retrieve archaeal 16S rRNA gene sequences from the GenBank NCBI-nr database (before January 2014). The Esearch utility was used to capture DNA sequences matching the following string ‘16S AND 600:2000[Sequence Length] AND archaea[Organism]AND rrna[Feature key] AND isolation_source[All fields] NOT genome OR chromosome OR plasmid'. In a following step, Efetch retrieved the entries found by Esearch and stored them (97668 sequences) in a GenBank formatted flat file to get further access to environmental information. Next, the script checked and removed sequences that were not ribosomal, too short (that is, <600 bp) and those that lacked the isolation source tag. To retain only sequences retrieved from marine or freshwater sediment habitats, sequences were filtered by isolation source using the string: ‘sediment | sediments'. We ended with 17 839 archaeal sequences collected from 358 studies. Variations in sampling efforts and methodologies among studies were homogenized by clustering archaeal sequences at a 97% and 90% identity threshold using MOTHUR (Schloss et al., 2009). Studies with less than 10 representative sequences were discarded from downstream analysis. The final data set contained 12 120 sequences distributed in 207 studies/sites: 153 from marine sediment, 13 from hypersaline sediment and 41 from freshwater sediments (see Supplementary Table 1). These sequences were clustered into 7098 operational taxonomic unit (OTUs) (97% cutoff) and 3427 OTUs (90% cutoff).
Phylogenetic analysis
Representative OTU sequences of each study were aligned in MOTHUR (Schloss et al., 2009) and imported into ARB software (Ludwig et al., 2004; http://www.arb-home.de) loaded with the Greengenes database (version gg_13_5, http://greengenes.secondgenome.com/). A base frequency filter was applied to exclude highly variable positions before adding sequences to the maximum parsimony backbone tree using the parsimony quick add marked tool implemented in ARB, thereby maintaining the overall tree topology provided by default. The MCG phylogenetic tree was constructed using 2781 16S rRNA gene sequences that were dereplicated at 90% identity and classified into 1120 OTUs. Twelve sequences were finally excluded from the tree because of poor alignment score.
Affiliations of MCG sequences to each MCG subgroups were carried out through reference sequences of the 17 MCG subgroups (Kubo et al., 2012) used as phylogenetic anchors. For the construction of the MCG tree, phylogenetic inference was carried out with RAxML version 7.7.1 (Stamatakis et al., 2008) that estimates large phylogenies by maximum likelihood. The best phylogenetic tree estimated by the GTRCAT model with 1000 bootstrap replicates was drawn with iTOL (Letunic and Bork, 2007).
Statistical analyses
Phylogenetic-based (according to the evolutionary distances captured by the ML tree) and taxon-based analyses (in which taxa were picked at a defined level and then treated as equally divergent) were run separately.
For the phylogenetic-based analyses, distance matrices were constructed using UniFrac, a beta diversity metric that quantifies community similarity based on the phylogenetic relatedness (Lozupone et al., 2006; Hamady et al., 2010). Principal coordinate analysis plots were used to represent the ordering relationships obtained from the UniFrac distance matrices. We used permutational Manova based on 1000 permutations (McArdle, 2001) with function Adonis of the vegan package in R (Oksanen et al., 2008) to assess the source of variation in the UniFrac matrix.
The phylogenetic diversity (PD) index was calculated as the sum of the branch length associated with the 16S rRNA gene sequences within each site (Faith, 1992). To correct for unequal number of sequences, we calculated the mean PD of 1000 randomized subsamples of each habitat (Barberan and Casamayor, 2010).
The phylogenetic structure was evaluated with the phylogenetic species variability index for each study (Helmus et al., 2007). Phylogenetic species variability estimates PD as the variance of a trait evolving under a neutral model. The value is 1 when all species are phylogenetically unrelated (that is, a star phylogeny) and approaches 0 as species become more related. To test statistically whether marine and freshwater archaeal communities were composed of species that were more or less related to each other than expected, we compared the mean observed phylogenetic species variability with distributions of mean null values (1000 iterations) using two different randomization procedures. Null model 1 maintains species occurrence, whereas null model 2 maintains habitat species richness (Helmus et al., 2007). All these analyses were run with the R package picante (Kembel et al., 2008).
In the taxon-based approach, a table of lineage relative abundance was constructed by considering all the clusters or divisions immediately subordinate to the main archaeal phyla and provided by default in the Greengenes tree. This table was further used to examine the species abundance distribution (SAD) patterns of each lineage and determine the ecological importance of the MCG lineage in archaeal sediment communities. The index of dispersion for each archaeal lineage was calculated as the ratio of the variance to the mean abundance multiplied by the occurrence. This index was used to model whether lineages follows a Poisson distribution (that is, stochastic distribution), falling between the 2.5% and 97.5% confidence interval of the χ2 distribution (Krebs, 1999).
A multivariate regression tree (MRT) was computed using the R package mvpart (De'Ath, 2002) to represent the relationship between the table of lineage relative abundances and the environmental matrix.
We used the indicator value (IndVal) index, which combines relative abundance and relative frequency of occurrence (Dufrene and Legendre, 1997) to identify archaeal lineages as analogous to the concept of ‘indicator species'.
Ancestral state reconstruction (ASR)
We performed an ASR to test the hypothesis of a relationship between diversification and salinity in MCG. For each MCG OTU (90% cutoff), character state for salinity was coded as follow: 1=freshwater, 2=saline and 3=hypersaline. ASR was performed using Mesquite 2.75 (Maddison and Maddison, 2011) with the Mk1 model (maximum likelihood) and the package ape in R. The transitions among each state were not assumed to occur at equal rates, as the transitions between both extremes of salinity require more cellular adaptations than saline–freshwater or saline–hypersaline transitions.
Co-occurrence network construction
Associations between MCG OTUs and other archaeal OTUs in the sediment habitat were inferred from an undirected co-occurrence network. Pairwise score between archaeal 90% OTUs represented by more than five sequences was computed using Spearman's rank correlations. Only co-occurrences corresponding to correlations with a coefficient (rho)>0.6 and a statistical significance (P-value)<0.01 were considered for further analysis. Non-random co-occurrence patterns were tested with the checkerboard score (C-score) under a null model preserving site frequencies (Stone and Roberts, 1990; Gotelli and McCabe, 2002). A C-score calculated for each pair of archaeal OTUs was compared with the C-score computed for 5000 randomly assembled null matrices. To avoid biases affecting raw C-score values (that is, OTU number, abundance…), the standardized effect size (SES) was calculated (Sridhar et al., 2012). Because the C-score is an inverse indicator of the frequency of co-occurrence, positive SES values indicate less co-occurrence than expected by chance (that is, predominance of segregation within communities) and vice versa for negative values (that is, predominance of facilitation). If co-occurrences were not different from what was expected by chance, values of SES should fall between −2 and 2.
The network was visualized with the gephi software (Bastian et al., 2009). Nodes represented archaeal OTUs at 90% identity and edges represented the significant correlations between them. Network characterization was performed using a set of overall network topological indices (that is, node degree distribution, average node connectivity, average path length, diameter, clustering coefficient and modularity) and network indices for individual nodes (that is, degree, closeness centrality and betweenness; Newman, 2003). All analyses were run using the R packages vegan (Oksanen et al., 2008) and igraph (Csardi and Nepusz, 2006).
Results and Discussion
The MCG: a characteristic lineage for sediment habitats
The MCG lineage has a cosmopolitan distribution in natural ecosystems (Inagaki et al., 2003; Fry et al., 2008; Auguet et al., 2010; Kubo et al., 2012; Lloyd et al., 2013) but has been mostly recovered from marine anoxic sediments habitat (Fry et al., 2008; Kubo et al., 2012; Lloyd et al., 2013). Nonetheless, large variations in their relative abundance (from 1% to 100%) were observed at global scale (Lloyd et al., 2013). In turn, current knowledge on the MCG distribution pattern in freshwater sediments is very limited. Consequently, a more rigorous ecological framework is needed before claiming for a major ecological significance of MCG archaea in general sedimentary habitats.
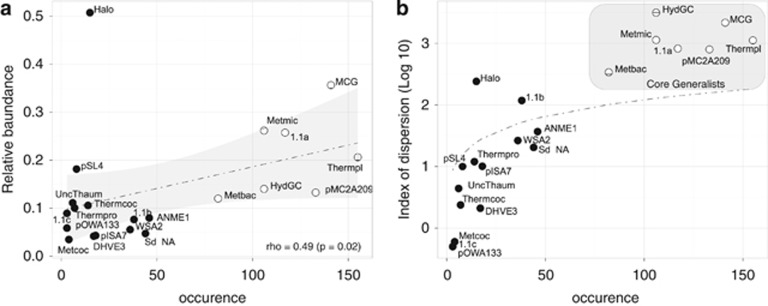
For this purpose, 7098 archaeal OTUs (97% cutoff) belonging to 21 archaeal lineages and from 207 sediments worldwide distributed were analyzed in an abundance vs occurrence plot (Figure 1a). We observed a significant positive relationship between mean relative abundance and occurrence (that is, number of sites in which archaeal groups were detected) showing that widespread archaeal lineages were more locally abundant than those archaeal classes with a more restricted distribution with the exception of Haloarchaea. One explanation for this trend, one of the most robust in macroecology (Gaston et al., 2000), is related with the alternative concept of ‘jack-of-all-trades is master of all' (Brown, 1984; Gaston et al., 1997; Verberk et al., 2010), in which cosmopolitan species that can tolerate a large spectrum of environmental conditions and use a broad range of resources become locally dominant. This concept seems to characterize very well members of the MCG lineage both in terms of resources (Biddle et al., 2008; Lloyd et al., 2013; Meng et al., 2014) and distribution. The MCG lineage was one of the most frequent archaeal lineage found in the sediment habitat as it occurs in 68% (141 sites) of the 207 sediments analyzed. It was also one of the most abundant lineages as it represented on average 36%±22% of the OTUs in each study where MCG were detected.
Figure 1.
Species abundance distribution (SAD) pattern of archaeal classes in the clone libraries analyzed. (a) Occurrence of archaeal lineages (number of studies in which a given lineage was found) plotted against its average abundance across these studies. A significant positive distribution–abundance relationship is observed. Core lineages (in white) were defined as those appearing in >75 studies and satellite lineages (in black) occurred in less than 50 studies. (b) Occurrence of each archaeal lineage plotted against its dispersion index. The line depicts the 2.5% confidence limit of the χ2 distribution: lineages falling bellow this line follow a Poisson distribution and are randomly dispersed in space. ANME, anaerobic methanotroph; DHVE3, Deep Hydrothermal Vent Euryarchaeota-3; Halo, Halobacteria; MCG, Miscellaneous Crenarchaeotic Group; Metbac, Methanobacteria; Metmic, Methanomicrobia; Metcoc, Methanococci; SAGMEG, South African Gold Mine Euryarchaeotic Group; Thermcoc, Thermococci; Thermprot, Thermoprotei; Thermpl, Thermoplasmata; UncThaum, Uncultured Thaumarchaeota; 1.1a, Thaumarchaeota 1.1a; 1.1.b, Thaumarchaeota 1.1b; 1.1.c, Thaumarchaeota 1.1c.
As previously observed when investigating SAD patterns (Magurran and Henderson, 2003; Verberk et al., 2010; van der Gast et al., 2011), a discontinuity in the occurrence vs relative abundance plot (Figure 1a) separated archaeal lineages into two groups: one group composed of 7 persistent/abundant core lineages where MCG were included (that is, lineages detected in more than 75 libraries) and another with 14 rare/less abundant satellite lineages (that is, lineages detected in less than 50 libraries). To statistically support this empirical SAD partition, the index of dispersion for each archaeal class was compared with a model assuming a stochastic distribution (Poisson model) falling between the 2.5% and 97.5% confidence limit of the χ2 distribution (Krebs, 1999). Plotting dispersion indices against occurrence confirmed that MCG represented a core lineage in the sediment habitat and was not randomly distributed through sediment samples. In turn, most satellite lineages fell below the 2.5% confidence limit line indicating random distribution (Figure 1b).
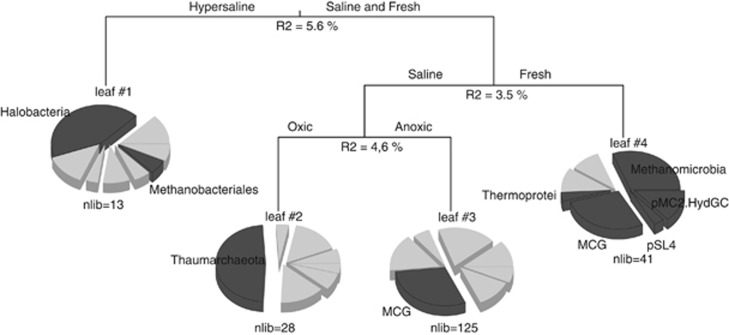
Thus, according to its broad distribution, MCG is a core generalist lineage in contrast to the specialist Haloarchaea restricted to a specific type of sediment. To illustrate this point and to link the abundance of the different lineages to environmental data, a MRT analysis was carried out (Figure 2). The analysis showed a four-leaf tree ordination explaining 13% of the phylogenetic lineage variance and primarily based on salinity for the two first nodes. Samples clustered in the leaves of the tree merely in function of their source habitat (that is, hypersaline, freshwater, surface and deep marine sediments). Pie charts in Figure 2 show in detail how the relative abundance of each phylogenetic group contributed to the separation and composition of the leaves. Changes in the relative abundance of the MCG lineage, which was particularly abundant in freshwater and anoxic marine sediments, accounted for most variation in the MRT tree (Figure 2). Calculation of the IndVal index on each leaf of the MRT tree showed that most core lineages previously identified are indicator lineages (P<0.01) for one single leaf (color label in Figure 2). The MCG was the only lineage to be indicator for two leaves, saline anoxic sediments and freshwater sediments, the most frequent and abundant type of sediments on the Earth.
Figure 2.
Multivariate Regression Tree (MRT) relating the abundance of each archaeal lineage to environmental variables. The model explained 13.7% of the total variance of the taxonomic composition of archaeal communities. Pies under each leaf represent the mean of normalized archaeal lineage abundance for each lineage significantly correlated with environmental parameters. Dark grey sections identify significant indicator lineages according to the IndVal index calculated on each leaf (P<0.01). nlib=number of clone libraries.
Topology of the MCG phylogenetic tree
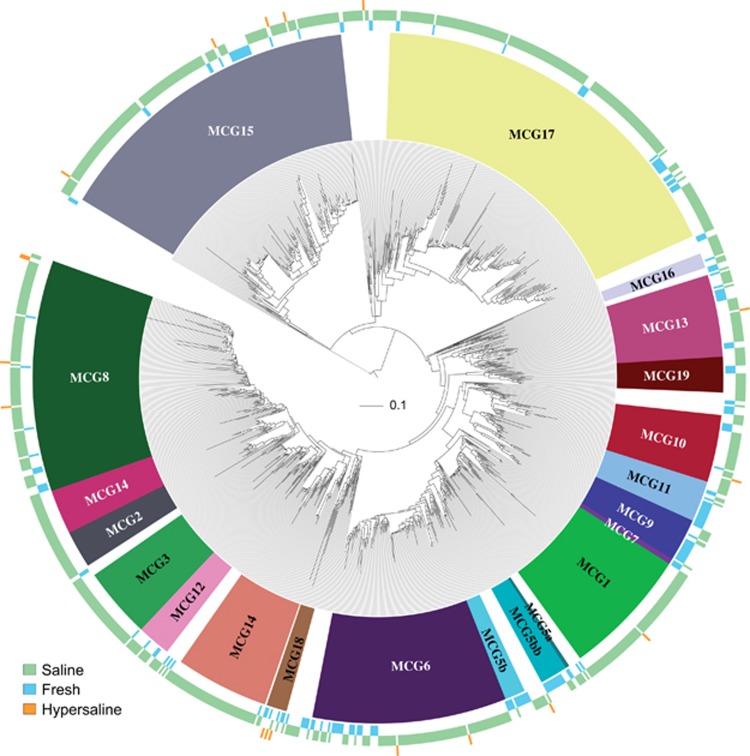
The phylogenetic tree of the entire MCG lineage contained 1120 OTUs (Figure 3). The lowest identity of the most divergent sequences was 75%, agreeing with published values (Kubo et al., 2012). This extremely low intragroup identity supports that MCG is one of the most phylogenetically diverse archaeal groups. The tree topology fairly agreed with that proposed by Kubo and co-workers, with most sequences clustering into the 18 subgroups already described (12% of sequences remained unclassified). However, differences were observed within subgroup5b, now split into 5b and 5bb, and the identification of two new clusters (MCG-18 and MCG-19), which were supported by high bootstrap values (96% and 89%, respectively). Accordingly, the MCG lineage comprised 21 clades that grouped sequences from uncultured representatives mainly from marine and freshwater sediments and less frequently from hypersaline environments (see color coding in Figure 3). On the basis of this monophyly and deep branching position in the overall archaeal tree several authors have recently proposed that MCG would be considered as a new archaeal phylum called Bathyarchaeota (Rinke et al., 2013, Meng et al., 2014).
Figure 3.
Maximum likelihood phylogenetic tree of the Miscellaneous Crenarchaeotic Group using available 16S rRNA gene sequences from GenBank NCBI-nr database dereplicated at 90%. Subgroup nomenclature from MCG-1 to MCG-17 follows those described Kubo et al. (2012). New subgroups identified in the current study are labeled as MCG-5bb, MCG-18 and MCG-19. Uncolored leaves identify sequences not assigned to any subgroup (that is, unclassified). Outer colored circles indicate sequence origin, as follows: freshwater (blue), saline (green) and hypersaline (orange). Tree was drawn using the web-based interactive tree of life.
Evolutionary relationships between freshwater and marine MCG lineages
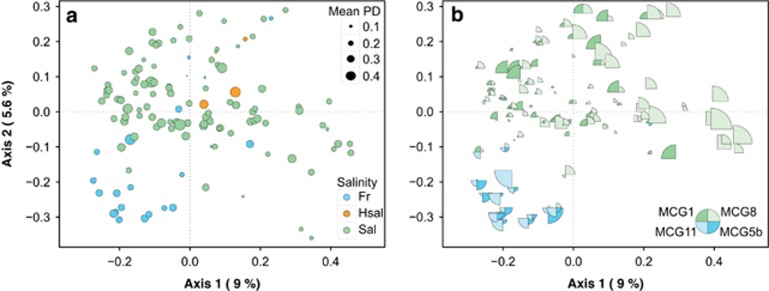
Our previous results suggested a key ecological role for the MCG lineage both in marine and freshwater sediments. Considering the high PD within the MCG lineage composed of 21 subgroups, one may wonder whether these subgroups have a widespread distribution or, in contrast, if distinct evolutionary subgroups occur in marine and freshwater sediments. To test these hypotheses, the MCG assemblages were sorted into an ordination plot according to the phylogenetic community similarity (Figure 4a). The mean observed phylogenetic species variability value (0.77) was significantly lower (that is, more phylogenetically clustered) than the null distribution for both model 1 (0.86, P<0.05) and model 2 (0.86, P<0.05), indicating a non-random sampling of phylotypes from the sequence pools. As non-random evolutionary processes were detected, we tested their association with environmental features (that is, salinity, oxic status and temperature). The observed segregation of freshwater and saline MCG assemblages in the ordination plot (Figure 4a) was statistically tested using permutational analysis of variance, which confirmed that salinity was the environmental variable explaining most of the variation in the phylogenetic MCG structure (R2=0.04, P<0.001). Secondarily, oxic status and temperature also had a significant effect (R2<0.02, P<0.01 for both variables). These results suggest that distinct evolutionary subgroups occurred in the marine and the freshwater sediment. Freshwater/marine segregation has been observed in diverse microorganisms (archaea, bacteria, micro-eukaryotes and viruses) confirming that the marine–freshwater frontier is an important colonization barrier (Logares et al., 2009). The strongest evidence for this pattern in archaea has been brought by a meta-analysis showing that the consistent phylogenetic separation of marine and freshwater archaea was based on characteristic lineages adapted to a specific habitat (Auguet et al., 2010).
Figure 4.
(a) Principal coordinate analysis (PCoA) plot obtained using a weighted UniFrac distance matrix calculated on 140 clone libraries containing >10 representative MCG sequences (97% cutoff). Libraries are colored according to salinity (see legend). Bubble size indicates the mean phylogenetic diversity (PD) index of 1000 randomized subsamples of each study. (b) Same as in a, but showing the relative abundance of MCG-IL (size of sectors) with the highest IndVal values in each study (MCG-1 and MCG-8 in green for marine sediments; MCG-5b and MCG-11 in blue for freshwater sediments).
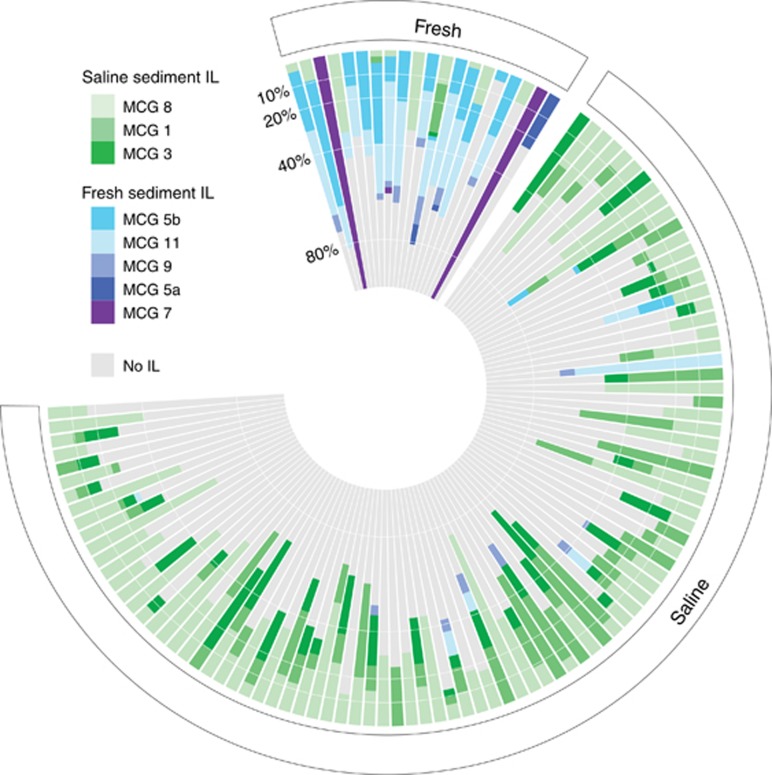
To identify which MCG subgroups were segregated by salinity, we calculated the IndVal index for each subgroup within the freshwater and saline sediments. Overall, 8 of 21 MCG subgroups (3 saline and 5 freshwater) showed significant IndVal values (P<0.01). Particularly, MCG subgroups 1 and 8 (for marine sediments) and 11 and 5b (for freshwater sediments) had the highest IndVal values and shaped the dissimilarity in MCG assemblages between these habitats (Figure 4b). Indeed, MCG subgroups 11 and 5b (Fillol et al., 2015), which dominated freshwater sediments, were almost never detected in saline sediments and vice versa for subgroups MCG-1 and MCG-8. Regardless of the habitat, these MCG indicator lineages (MCG-ILs) represented key members of MCG assemblages in term of relative abundance as they accounted, on average, for 44% of total MCG sequences in each study (Figure 5). Figure 5 illustrates the strong segregation of MCG-IL between marine and freshwater sediments. At a more local scale, Lazar et al. (2014) found a vertical segregation of MCG subgroups as a function of sulfide concentration and redox potential within marine sediments. These correlations between the distribution of MCG subgroups and global or local environmental conditions suggest the occurrence of adaptive evolution specific to each habitat.
Figure 5.
Polarplot showing the relative abundance of MCG-IL in freshwater and marine sediments.
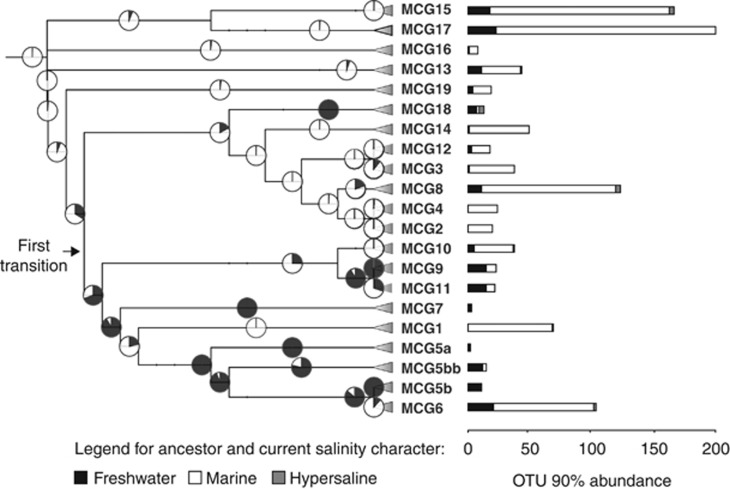
How shifts in salinity corresponding to the marine–freshwater boundary determine the distribution and evolution of prokaryotes is not fully understood, but differences in salt concentration can affect both energetic costs (related to osmoregulation) and metabolic pathways, and ultimately limit the environmental transitions (Oren, 2001). Here, we supported the influence of salinity on the diversification and specification of MCGs by the ASR using habitat salinity of each OTU (Figure 6). ASR analysis showed a significant correlation between patterns of diversification and salinity where MCG occurred. Assuming a saline representative as the most probable last common ancestor, this analysis supported the hypotheses of an evolutionary progression for MCGs from saline to freshwater sediments. Although ancient, colonization of freshwater sediment occurred more recently than for saline sediments in a first transition and subsequent diversification events that gave rise to all freshwater MCG-IL (Figure 6). Overall, and in agreement with other microbial lineages (Logares et al., 2009, 2010), a few environmental transition events (that is, switch from a freshwater ancestor to a saline descendant and vice versa) seemed to have occurred during the diversification of MCG. This result may explain why freshwater and saline sediments harbored evolutionary distinct communities of MCG.
Figure 6.
Ancestral state reconstruction (ASR) of salinity range for MCG. Pie charts on the nodes show the relative likelyhoods of the three states: freshwater (black), marine (white) and hypersaline (grey). Bar charts at the right indicate the current salinity state for MCG OTUs (90% cutoff).
Co-occurrence and potential MCG–Thermoplasmata synthrophy
To get a first insight in the potential syntrophic and/or mutualistic interactions in which MCGs may be involved and, by association, in the potential metabolisms they may harbor, we constructed a co-occurrence network based on strong and significant Spearman correlations. As illustrated by recent studies, microbial co-occurrence patterns can help to unveil ecologically meaningful interactions between species (Horner-Devine et al., 2007; Steele et al., 2011). Network approaches also showed that co-occurring species are often organized into groups, or modules, of functional significance (Chaffron et al., 2010; Barberan et al., 2012; Vick-Majors et al., 2014). After OTU clustering at 90% identity and filtering the data set for optimizing network sensitivity and specificity, network inference was calculated on 99 OTUs distributed across 205 studies. We first checked for the existence of non-random co-occurrence patterns in the network by using the SES of the C-score metric (Gotelli and McCabe, 2002; Horner-Devine et al., 2007; López et al., 2013). We observed a SES value of 2.45 (P<0.001, C-scorenorm=0.78) indicating non-random network structure and the existence of fewer co-occurrences than expected by chance (that is, more segregated taxa). Segregation of taxa seems to be an idiosyncratic feature for biological assemblages across domains of life (Webb, 2000; Kembel and Hubbell, 2006; Horner-Devine and Bohannan, 2006; Horner-Devine et al., 2007). Four major causal mechanisms (not mutually exclusive) have been proposed to explain segregation: competition, habitat filtering, historical effects and neutral processes (Diamond, 1975; Bell, 2005; Horner-Devine et al., 2007). The results obtained above and the low number of significant negative correlations in the network may indicate a dominance of habitat filtering and historical factors over competition in the explanation of the segregation of archaeal taxa observed here.
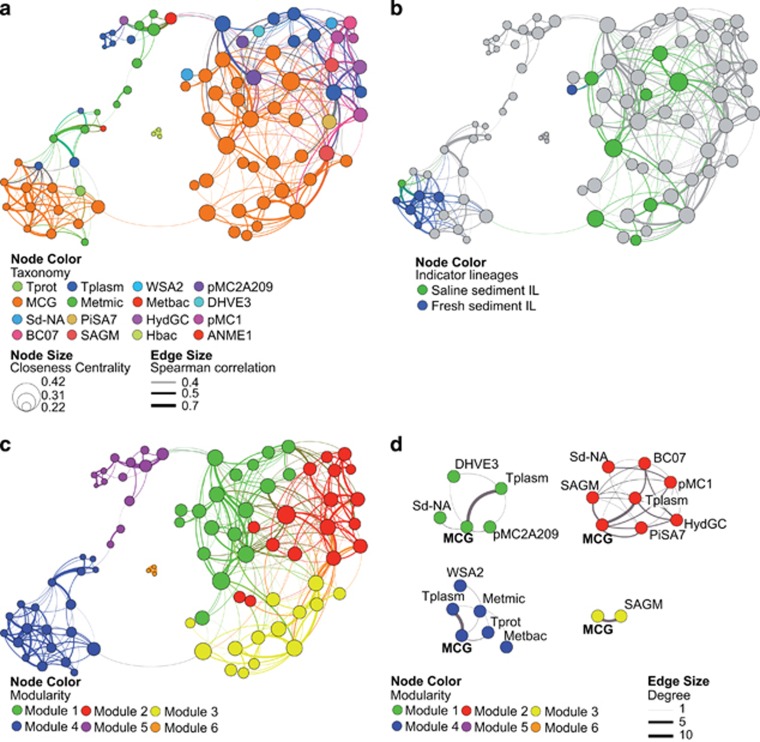
The sediment archaeal network (Figure 7a) consisted of 92 nodes and 373 edges and presented the typical topology for microbial network (Chaffron et al., 2010; Steele et al., 2011; Barberan et al., 2012; Deng et al., 2012): scale-free (that is, node connectivity distribution not different from a power law model, see Supplementary Figure 1), small-world (clustering coefficient of 0.56 and average path length of 3.66) and modular (modularity of 0.55). Within the resulting network, MCG was by far the most represented archaeal lineage accounting for 47% of the nodes and 53% of the edges (Figure 7a). Calculation of network indices for individual nodes further corroborated the pivotal role of the MCG lineage in the structuration of the network (Table 1). Indeed, looking at the ranking of nodes for the 10 highest values of each index, MCG nodes were the most represented and particularly for closeness centrality, which has been recently related to the concept of keystone species (Berry and Widder, 2014). In this concept initially developed for macro-organisms, keystone species are commonly understood as the ‘backbone' of the community on which the stability of the entire system depends (Paine, 1969). Recently, prokaryotes involved in major biogeochemical processes such as sulfates reducers (Pester et al., 2010) or primary degraders of refractory substrates (Ze et al., 2012) have been regarded as keystone species. Our results suggest that members of the MCG lineage might have a keystone role in archaeal sediment communities.
Figure 7.
(a) Co-occurrence OTU network based on correlation analysis. Each node denotes an archaeal OTU 90%. Node size is proportional to the closeness centrality (that is, the average shortest path of this node to any other node) and node color denotes taxonomic classification (see Figure 2 for abbreviations). Edge lines between nodes represent significant co-occurrences relationships. Edge size indicates the strength of Spearman correlation among nodes. (b) Same network as in a, but nodes are colored according to MCG-Indicator Lineages, as follows: green: indicator MCG lineages for saline sediments (MCG-1, MCG-3 and MCG-8); blue: indicator MCG lineages for freshwater sediments (MCG-5a, MCG-5b, MCG-7, MCG-9 and MCG-11). (c) Same network as in a, but nodes are colored by modules. (d) Sub-network modules clustering all OTUs of the same lineage colored by modularity. Edge size indicates the number of connections (degree). Only the sub-networks where MCG are present are displayed.
Table 1. Values of network indices and taxonomy for the first 10 nodes of each index.
| Lineage | Closeness centrality | Lineage | Betweeness centrality | Lineage | Degree |
|---|---|---|---|---|---|
| MCG | 0.42 | MCG | 0.32 | MCG | 25 |
| MCG | 0.41 | MCG | 0.31 | MCG | 25 |
| MCG | 0.39 | MCG | 0.20 | Thermpl | 21 |
| MCG | 0.39 | Thermpl | 0.19 | MCG | 19 |
| MCG | 0.38 | Metmic | 0.12 | SAGMEG-1 | 19 |
| Thermpl | 0.38 | MCG | 0.09 | SAGMEG-1 | 18 |
| Thermpl | 0.38 | HydGC | 0.08 | PiSA7 | 17 |
| MCG | 0.37 | MCG | 0.07 | MCG | 16 |
| pMMCGA209 | 0.36 | ANME1 | 0.07 | HydGC | 16 |
| SAGMEG-1 | 0.36 | MCG | 0.07 | pMMCGA209 | 15 |
Abbreviations: ANME1, anaerobic methanotroph1; MCG, Miscellaneous Crenarchaeotic Group; Metmic, Methanomicrobia; SAGMEG, South African Gold Mine Euryarchaeotic Group; Thermpl, Thermoplasmata.
Identifying MCG-IL within the network (Figure 7b) revealed its ecological structure and the presence of fresh and saline sub-networks. The structure of both sub-networks was very similar with a clear disconnection of typical methanogenic lineages (that is, essentially Methanomicrobia and Methanobacteria) from other lineages within each sub-network (Figure 7a). Notably, the lack of co-occurrence between MCG members and typical methanogens agrees with the fact that MCG are likely not methanotrophs (Kubo et al., 2012). Modularity analysis revealed a more complex structure (particularly for the saline network) with the presence of six modules (Figure 7c), that is, clusters of OTUs that are highly connected within the module but with very few connections outside the module. In agreement with previous works (Freilich et al., 2010; Chaffron et al., 2010; Faust and Raes, 2012), modules could be considered as ecological and/or functional niches as suggested by modules 4, 5 and 6, which represented freshwater, hypersaline and saline-methanogen sub-networks, respectively. MCG were present in four of the six modules and, interestingly, they systematically co-occurred more often with members of the Thermoplasmata as indicated by the high number of interconnections (that is, indicated by higher degree values; Figure 7d). As stated previously, many co-occurrences may not be the reflection of true biological interactions but the result of niche overlap (Barberan et al., 2012). However, the recurrence of MCG–Thermoplasmata associations in different modules representing different ecological niches was compelling and pointed toward a potential syntrophy. Additional support for this idea came from the potential metabolisms of these lineages related to the remineralization of carbon in sediments. The Thermoplasmata studied here matched the Marine Benthic Group D, the Terrestrial Miscellaneous Euryarchaeotal Group and the new order Methanomassiliicoccales (Iino et al., 2013). The latter archaeal group, the seventh order of methanogens, are methylotrophic methanogens obtaining energy and carbon from methanol and methylated amines (Paul et al., 2012; Dridi et al., 2012; Iino et al., 2013; Borrel et al., 2014). Although no information on the potential metabolism of the Terrestrial Miscellaneous Euryarchaeotal Group group is available, recent findings from metagenomic data (Meng et al., 2014; Castelle et al., 2015) and single-cell genomics (Lloyd et al., 2013) suggested that members of the MCG and Marine Benthic Group D lineages may be involved in the degradation of detrital proteins both in marine and continental sediments. The high PD of the MCG lineage invalidates any general assumption of potential metabolisms shared between all MCG subgroups (the specialization of certain subgroups towards freshwater and marine habitats is a good example of this). However, it is tempting to speculate on a system where MCG and Marine Benthic Group D act as primary degraders of detrital proteins producing more labile compounds for other microorganisms such as methylated substrates for methylotrophs.
To better understand if members of the MCG and Thermoplasmata lineages work in syntrophy or simply shared the same resources, further analyses involving culturing and metagenomic approaches targeting these poorly described branches of life are needed. Further research will surely provide clues to resolve not only the ecological role of two of the most important archaeal groups in sediment ecosystems but also the impact of their activity in the carbon cycle of marine sediments, which are considered the largest reservoir of carbon on the Earth.
Acknowledgments
This research was funded by projects ARCANOX (ref. CGL2009-13318-C02-02) and ARCOS (ref. CGL2012-33033) to CMB, and DARKNESS CGL2012-32747 to EOC from the Spanish Ministry of Economy and Competitiveness (MINECO). J-CA was funded by project METMIC from the Pyrenean Work Community and the Aquitaine region.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Auguet JC, Barberan A, Casamayor EO. (2010). Global ecological patterns in uncultured Archaea. ISME J 4: 182–190. [DOI] [PubMed] [Google Scholar]
- Auguet JC, Triado-Margarit X, Nomokonova N, Camarero L, Casamayor EO. (2012). Vertical segregation and phylogenetic characterization of ammonia-oxidizing Archaea in a deep oligotrophic lake. ISME J 6: 1786–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberan A, Bates ST, Casamayor EO, Fierer N. (2012). Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6: 343–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberan A, Casamayor EO. (2010). Global phylogenetic community structure and beta-diversity patterns of surface bacterioplankton metacommunities. Aquat Microb Ecol 59: 1–10. [Google Scholar]
- Bastian M, Heymann S, Jacomy M. (2009) International AAAI Conference on Weblogs and Social Media. San José: : California, USA. [Google Scholar]
- Beja O, Suzuki MT, Koonin EV, Aravind L, Hadd A, Nguyen LP et al. (2000). Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ Microbiol 2: 516–529. [DOI] [PubMed] [Google Scholar]
- Bell G. (2005). The co-distribution of species in relation to the neutral theory of community ecology. Ecology 86: 1757–1770. [Google Scholar]
- Berry D, Widder S. (2014). Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol 5: 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattarai S, Ross KA, Schmid M, Anselmetti FS, Burgmann H. (2012). Local conditions structure unique archaeal communities in the anoxic sediments of Meromictic Lake Kivu. Microb Ecol 64: 291–310. [DOI] [PubMed] [Google Scholar]
- Biddle JF, Fitz-Gibbon S, Schuster SC, Brenchley JE, House CH. (2008). Metagenomic signatures of the Peru Margin subseafloor biosphere show a genetically distinct environment. Proc Natl Acad Sci USA 105: 10583–10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sorensen KB, Anderson R et al. (2006). Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci USA 103: 3846–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrel G, Lehours AC, Crouzet O, Jezequel D, Rockne K, Kulczak A et al. (2012). Stratification of archaea in the deep sediments of a freshwater Meromictic Lake: vertical shift from methanogenic to uncultured archaeal lineages. PLoS ONE 7: e43346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrel G, Parisot N, Harris HMB, Peyretaillade E, Gaci N, Tottey W et al. (2014). Comparative genomics highlights the unique biology of Methanomassiliicoccales, a Thermoplasmatales-related seventh order of methanogenic archaea that encodes pyrrolysine. BMC Genomics 15: 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JH. (1984). On the relationship between abundance and distribution of species. Am Nat 124: 255–279. [Google Scholar]
- Buckles LK, Villanueva L, Weijers JWH, Verschuren D, Damste JSS. (2013). Linking isoprenoidal GDGT membrane lipid distributions with gene abundances of ammonia-oxidizing Thaumarchaeota and uncultured crenarchaeotal groups in the water column of a tropical lake (Lake Challa, East Africa). Environ Microbiol 15: 2445–2462. [DOI] [PubMed] [Google Scholar]
- Casamayor EO, Muyzer G, Pedros Alio C. (2001). Composition and temporal dynamics of planktonic archaeal assemblages from anaerobic sulfurous environments studied by 16S rDNA denaturing gradient gel electrophoresis and sequencing. Aquat Microb Ecol 25: 237–246. [Google Scholar]
- Castelle CJ, Wrighton KC, Thomas BC, Hug LA, Brown CT, Wilkins MJ et al. (2015). Genomic expansion of domain archaea highlights roles for organisms from new phyla in anaerobic carbon cycling. Curr Biol 25: 690–701. [DOI] [PubMed] [Google Scholar]
- Chaffron S, Rehrauer H, Pernthaler J, von Mering C. (2010). A global network of coexisting microbes from environmental and whole-genome sequence data. Genome Res 20: 947–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi G, Nepusz T. (2006). The igraph software package for complex network research. InterJournal Complex Systems, 1695.
- Dalsgaard T, Canfield DE, Petersen J, Thamdrup B, Acuna-Gonzalez J. (2003). N-2 production by the anammox reaction in the anoxic water column of Golfo Dulce, Costa Rica. Nature 422: 606–608. [DOI] [PubMed] [Google Scholar]
- De'Ath G. (2002). Multivariate regression trees: a new technique for modeling species-environment relationships. Ecology 83: 1105–1117. [Google Scholar]
- Deng Y, Jiang Y-H, Yang Y, He Z, Luo F, Zhou J. (2012). Molecular ecological network analyses. BMC Bioinformatics 13: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond JM. (1975) Assembly of species communities. In: Cody ML, Diamond JM (eds). Ecology and Evolution of Communities. Harvard University Press: : Cambridge, Massachusetts, USA. [Google Scholar]
- Dridi B, Fardeau ML, Ollivier B, Raoult D, Drancourt M. (2012). Methanomassiliicoccus luminyensis gen. nov., sp nov., a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol 62: 1902–1907. [DOI] [PubMed] [Google Scholar]
- Dufrene M, Legendre P. (1997). Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol Monogr 67: 345–366. [Google Scholar]
- Durbin AM, Teske A. (2012). Archaea in organic-lean and organic-rich marine subsurface sediments: an environmental gradient reflected in distinct phylogenetic lineages. Front Microbiol 3: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith DP. (1992). Conservation evaluation and phylogenetic diversity. Biol Conserv 61: 1–10. [Google Scholar]
- Faust K, Raes J. (2012). Microbial interactions: from networks to models. Nat Rev Microbiol 10: 538–550. [DOI] [PubMed] [Google Scholar]
- Fillol M, Sànchez-Melsió A, Gich F, Borrego CM. (2015). Diversity of Miscellaneous Crenarchaeotic Group archaea in freshwater karstic lakes and their segregation between planktonic and sediment habitats. FEMS Microbiol Ecol e-pub ahead of print 22 February 2015; doi:10.1093/femsec/fiv020. [DOI] [PubMed]
- Freilich S, Kreimer A, Meilijson I, Gophna U, Sharan R, Ruppin E. (2010). The large-scale organization of the bacterial network of ecological co-occurrence interactions. Nucleic Acids Res 38: 3857–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry JC, Parkes RJ, Cragg BA, Weightman AJ, Webster G. (2008). Prokaryotic biodiversity and activity in the deep subseafloor biosphere. FEMS Microbiol Ecol 66: 181–196. [DOI] [PubMed] [Google Scholar]
- Gaston KJ, Blackburn TM, Greenwood JJD, Gregory RD, Quinn RM, Lawton JH. (2000). Abundance-occupancy relationships. J Appl Ecol 37: 39–59. [Google Scholar]
- Gaston KJ, Blackburn TM, Lawton JH. (1997). Interspecific abundance-range size relationships: an appraisal of mechanisms. J Anim Ecol 66: 579–601. [Google Scholar]
- Gotelli NJ, McCabe DJ. (2002). Species co-occurrence: A meta-analysis of J. M. Diamond's assembly rules model. Ecology 83: 2091–2096. [Google Scholar]
- Hamady M, Lozupone C, Knight R. (2010). Fast UniFrac: facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J 4: 17–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmus MR, Bland TJ, Williams CK, Ives AR. (2007). Phylogenetic measures of biodiversity. Am Nat 169: E68–E83. [DOI] [PubMed] [Google Scholar]
- Horner-Devine MC, Bohannan BJM. (2006). Phylogenetic clustering and overdispersion in bacterial communities. Ecology 87: S100–S108. [DOI] [PubMed] [Google Scholar]
- Horner-Devine MC, Silver JM, Leibold MA, Bohannan BJM, Colwell RK, Fuhrman JA et al. (2007). A comparison of taxon co-occurrence patterns for macro- and microorganisms. Ecology 88: 1345–1353. [DOI] [PubMed] [Google Scholar]
- Iino T, Tamaki H, Tamazawa S, Ueno Y, Ohkuma M, Suzuki K et al. (2013). Candidatus Methanogranum caenicola: a Novel Methanogen from the Anaerobic Digested Sludge, and Proposal of Methanomassiliicoccaceae fam. nov and Methanomassiliicoccales ord. nov., for a Methanogenic Lineage of the Class Thermoplasmata. Microbes Environ 28: 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki F, Suzuki M, Takai K, Oida H, Sakamoto T, Aoki K et al. (2003). Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl Environ Microbiol 69: 7224–7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallmeyer J, Pockalny R, Adhikari RR, Smith DC, D'Hondt S. (2012). Global distribution of microbial abundance and biomass in subseafloor sediment. Proc Natl Acad Sci USA 109: 16213–16216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kembel S, Ackerly D, Blomberg S, Cowan P, Helmus M, Webb C. (2008), Picante: Tools for integrating phylogenies and ecology. Version 0.4-0 http://picante.r-forge.r-project.org/. [DOI] [PubMed]
- Kembel SW, Hubbell SP. (2006). The phylogenetic structure of a neotropical forest tree community. Ecology 87: S86–S99. [DOI] [PubMed] [Google Scholar]
- Konneke M, Bernhard AE, de la Torre JR, Walker CB, Waterbury JB, Stahl DA. (2005). Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437: 543–546. [DOI] [PubMed] [Google Scholar]
- Krebs CJ. (1999) Ecological Methodology 2nd edn. Addison-Wesley Educational Publishers, Inc: New York, NY, USA. [Google Scholar]
- Kubo K, Lloyd KG, F Biddle J, Amann R, Teske A, Knittel K. (2012). Archaea of the Miscellaneous Crenarchaeotal Group are abundant, diverse and widespread in marine sediments. ISME J 6: 1949–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuypers MMM, Sliekers AO, Lavik G, Schmid M, Jorgensen BB, Kuenen JG et al. (2003). Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature 422: 608–611. [DOI] [PubMed] [Google Scholar]
- Lazar CS, Biddle JF, Meador TB, Blair N, Hinrichs KU, Teske AP. (2014). Environmental controls on intragroup diversity of the uncultured benthic archaea of the Miscellaneous Crenarchaeotal Group lineage naturally enriched in anoxic sediments of the White Oak River Estuary (North Carolina, USA). Environ Microbiol 17: 2228–2238. [DOI] [PubMed] [Google Scholar]
- Lehours AC, Evans P, Bardot C, Joblin K, Gerard F. (2007). Phylogenetic diversity of archaea and bacteria in the anoxic zone of a meromictic lake (Lake Pavin, France). Appl Environ Microbiol 73: 2016–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. (2007). Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23: 127–128. [DOI] [PubMed] [Google Scholar]
- Llirós M, Casamayor EO, Borrego CM. (2008). High archaeal richness in the water column of a freshwater sulfurous karstic lake along an interannual study. FEMS Microbiol Ecol 66: 331–342. [DOI] [PubMed] [Google Scholar]
- Lliros M, Gich F, Plasencia A, Auguet JC, Darchambeau F, Casamayor EO et al. (2010). Vertical distribution of ammonia-oxidizing crenarchaeota and methanogens in the epipelagic waters of Lake Kivu (Rwanda-Democratic Republic of the Congo). Appl Environ Microbiol 76: 6853–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd KG, Schreiber L, Petersen DG, Kjeldsen KU, Lever MA, Steen AD et al. (2013). Predominant archaea in marine sediments degrade detrital proteins. Nature 496: 215–218. [DOI] [PubMed] [Google Scholar]
- Logares R, Brate J, Bertilsson S, Clasen JL, Shalchian-Tabrizi K, Rengefors K. (2009). Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol 17: 414–422. [DOI] [PubMed] [Google Scholar]
- Logares R, Brate J, Heinrich F, Shalchian-Tabrizi K, Bertilsson S. (2010). Infrequent Transitions between Saline and Fresh Waters in One of the Most Abundant Microbial Lineages (SAR11). Mol Biol Evol 27: 347–357. [DOI] [PubMed] [Google Scholar]
- López RP, Valdivia S, Rivera ML, Rios RS. (2013). Co-occurrence patterns along a regional aridity gradient of the subtropical andes do not support stress gradient hypotheses. PLoS ONE 8: e58518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Hamady M, Knight R. (2006). UniFrac - An online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics 7: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Knight R. (2007). Global patterns in bacterial diversity. Proc Natl Acad Sci USA 104: 11436–11440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar et al. (2004). ARB: a software environment for sequence data. Nucl Acids Res 32: 1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. (2011), Mesquite: a modular system for evolutionary analysis. Version 2.75: Available from: http://mesquiteproject.org.
- Magurran AE, Henderson PA. (2003). Explaining the excess of rare species in natural species abundance distributions. Nature 422: 714–716. [DOI] [PubMed] [Google Scholar]
- McArdle B. (2001). Fitting multivariate models to community data: A comment on distance-based redundancy analysis. Ecology 82: 290–297. [Google Scholar]
- Meng J, Wang FP, Wang F, Zheng YP, Peng XT, Zhou HY et al. (2009). An uncultivated crenarchaeota contains functional bacteriochlorophyll a synthase. ISME J 3: 106–116. [DOI] [PubMed] [Google Scholar]
- Meng J, Xu J, Qin D, He Y, Xiao X, Wang FP. (2014). Genetic and functional properties of uncultivated MCG archaea assessed by metagenome and gene expression analyses. ISME J 8: 650–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ. (2003). The structure and function of complex networks. Siam Rev 45: 167–256. [Google Scholar]
- Oksanen J, Kindt R, Legendre P, O'hara B, Simpson GL, Stevens MHH. (2008), Vegan: Community Ecology Package. version 1.11-4 http://vegan.r-forge.r-project.org.
- Oren A. (2001). The bioenergetic basis for the decrease in metabolic diversity at increasing salt concentrations: implications for the functioning of salt lake ecosystems. Hydrobiologia 466: 61–72. [Google Scholar]
- Paine RT. (1969). A note on trophic complexity and community stability. Am Nat 103: 91. [Google Scholar]
- Paul K, Nonoh JO, Mikulski L, Brune A. (2012). ‘Methanoplasmatales,' Thermoplasmatales-Related Archaea in Termite Guts and Other Environments, Are the Seventh Order of Methanogens. Appl Environ Microbiol 78: 8245–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pester M, Bittner N, Deevong P, Wagner M, Loy A. (2010). A 'rare biosphere' microorganism contributes to sulfate reduction in a peatland. ISME J 4: 1591–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature 499: 431–437. [DOI] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyler LM, McGuinness LM, Kerkhof LJ. (2014). Crenarchaeal heterotrophy in salt marsh sediments. ISME J 8: 1534–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar H, Srinivasan U, Askins RA, Canales-Delgadillo JC, Chen CC, Ewert DN et al. (2012). Positive Relationships between Association Strength and Phenotypic Similarity Characterize the Assembly of Mixed-Species Bird Flocks Worldwide. Am Nat 180: 777–790. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. (2008). A Rapid Bootstrap Algorithm for the RAxML Web-Servers. Syst Biol 75: 758–771. [DOI] [PubMed] [Google Scholar]
- Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY et al. (2011). Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J 5: 1414–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone L, Roberts A. (1990). The checkerboard score and species distributions. Oecologia 85: 74–79. [DOI] [PubMed] [Google Scholar]
- van der Gast CJ, Walker AW, Stressmann FA, Rogers GB, Scott P, Daniels TW et al. (2011). Partitioning core and satellite taxa from within cystic fibrosis lung bacterial communities. ISME J 5: 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verberk W, van der Velde G, Esselink H. (2010). Explaining abundance-occupancy relationships in specialists and generalists: a case study on aquatic macroinvertebrates in standing waters. J Anim Ecol 79: 589–601. [DOI] [PubMed] [Google Scholar]
- Vick-Majors TJ, Priscu JC, Amaral-Zettler LA. (2014). Modular community structure suggests metabolic plasticity during the transition to polar night in ice-covered Antarctic lakes. ISME J 8: 778–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb CO. (2000). Exploring the phylogenetic structure of ecological communities: An example for rain forest trees. Am Nat 156: 145–155. [DOI] [PubMed] [Google Scholar]
- Webster G, Rinna J, Roussel EG, Fry JC, Weightman AJ, Parkes RJ. (2010). Prokaryotic functional diversity in different biogeochemical depth zones in tidal sediments of the Severn Estuary, UK, revealed by stable-isotope probing. FEMS Microbiol Ecol 72: 179–197. [DOI] [PubMed] [Google Scholar]
- Ze XL, Duncan SH, Louis P, Flint HJ. (2012). Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 6: 1535–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.