Abstract
Subsurface microbial communities comprise a significant fraction of the global prokaryotic biomass; however, the carbon metabolisms that support the deep biosphere have been relatively unexplored. In order to determine the predominant carbon metabolisms within a 3-km deep fracture fluid system accessed via the Tau Tona gold mine (Witwatersrand Basin, South Africa), metagenomic and thermodynamic analyses were combined. Within our system of study, the energy-conserving reductive acetyl-CoA (Wood-Ljungdahl) pathway was found to be the most abundant carbon fixation pathway identified in the metagenome. Carbon monoxide dehydrogenase genes that have the potential to participate in (1) both autotrophic and heterotrophic metabolisms through the reversible oxidization of CO and subsequent transfer of electrons for sulfate reduction, (2) direct utilization of H2 and (3) methanogenesis were identified. The most abundant members of the metagenome belonged to Euryarchaeota (22%) and Firmicutes (57%)—by far, the highest relative abundance of Euryarchaeota yet reported from deep fracture fluids in South Africa and one of only five Firmicutes-dominated deep fracture fluids identified in the region. Importantly, by combining the metagenomics data and thermodynamic modeling of this study with previously published isotopic and community composition data from the South African subsurface, we are able to demonstrate that Firmicutes-dominated communities are associated with a particular hydrogeologic environment, specifically the older, more saline and more reducing waters.
Introduction
For over two decades, scientists have been characterizing the geochemistry and biological composition of the subsurface biosphere to depths as great as 3.6 km (Borgonie et al., 2011). These investigations have revealed that in sub-seafloor sedimentary environments, the microbial abundance appears to be positively correlated with total organic carbon (TOC) concentration (0.4–20 mol kg−1) (Lipp et al., 2008). In continental groundwaters and petroleum reservoirs, the microbial abundance and activity are related to proximity to interfaces with organic-rich strata (Chapelle and Lovley, 1990; Krumholz et al., 1997; Bennett et al., 2013). Within continental flood basalts or granitic intrusions, however, autotrophic communities relying upon dissolved inorganic carbon (DIC) may have the dominant role (Stevens and McKinley, 1995; Pedersen, 1997). The low TOC (0.01–0.1 mol kg−1) (Silver et al., 2012), dissolved organic carbon (DOC) (~400 μM) and organic acid (~40 μM) concentrations of deep fracture water at the Witwatersrand Basin (Onstott et al., 2006) call into question whether the subsurface microbial communities residing in these fractures are supported by these limited DOC pools or by the larger pools of alternative carbon sources such as biogenic or abiogenic CH4 (~20 mM), abiogenic C2-4 hydrocarbons (~500 μM) or DIC (DIC averaging 800 μM) (Kieft et al., 2005; Onstott et al., 2006; Sherwood Lollar et al., 2006). Thus far, it has been shown that some of the subsurface microbial communities are sustained by radiolytically derived H2 and sulfate (Lin et al., 2006b; Chivian et al., 2008). Obvious signatures of biodegradation of the abiogenic hydrocarbon pool, such as elevated isobutane to normal butane ratios (Ehrenberg and Jakobsen, 2001), are not apparent (Onstott et al., 2006).
Of the many biodiversity studies of fracture water samples that were based on 16S ribosomal RNA (rRNA) genes, one horizontal borehole located at 2.8 km below the surface (kmbls) in the Mponeng gold mine (MP104) was found to be comprised almost entirely (>99%) of a sulfate reducing chemoautolithotrophic firmicute, ‘Candidatus Desulforudis audaxviator' (Chivian et al., 2008). The subsurface residence time of the bulk fracture water of MP104 was on the order of tens of Ma and the Δ2HH2-H2O geothermometry suggested that the fracture water originated from a depth of 4.2 kmbls (Lin et al., 2006b). Other fracture water sites, ranging in depth from 0.6 to 3.2 kmbls and distributed over 300 km across the Basin, were found to contain assemblages of bacteria primarily consisting of Proteobacteria, Firmicutes, and, occasionally, a minor portion of Euryarchaeota based on the analyses of 16S rRNA genes (Takai et al., 2001; Baker et al., 2003; Moser et al., 2003; Onstott et al., 2003; Kieft et al., 2005; Gihring et al., 2006; Lin et al., 2006a; Silver et al., 2010; Davidson et al., 2011; Magnabosco et al., 2014). The surprisingly low to no observed archaeal 16S rRNA genes, the low TOC and DOC concentrations, and the low porosity (0.5% matrix, 0.01% fracture) of the Witwatersrand Basin sites distinguishes them from the more prominent archaeal communities often reported to inhabit the shallower, higher TOC bearing, higher porosity (50–90%) sub-seafloor sediments (Biddle et al., 2006). Instead, the carbon cycling pathways used in the TOC- and DOC-poor Witwatersrand Basin fluids may be more similar to the SLiME communities of the deep Columbia River basalt aquifer (Stevens and McKinley, 1995), the planktonic microbial communities found within the fracture water of the Proterozoic granite in the Fennoscandian Shield (Pedersen, 1997) and autotrophic communities of the Tablelands serpentinite (Brazelton et al., 2012).
A recent reevaluation of protein turnover times in the deep subsurface derived from the D/L aspartic acid racemization ratio estimated the maximum protein turnover times for the Witwatersrand Basin microbial communities to be 1–2 years (Onstott et al., 2014). This finding indicates that the deep crustal biosphere is metabolically more active than previously thought, so much so that the low DOC concentrations in the Witwatersrand Basin fracture water would be depleted in ~103 years. Given the voluminous size of the habitable, yet carbon poor, regions of the deep continental crust and the previous estimates of subsurface microbial turnover times (1–2 × 103 years; Whitman et al., 1998), the deep continental subsurface microbial carbon metabolism requires reevaluation from an interdisciplinary perspective. Here, we report the first integration of metagenomic analyses with traditional geochemical estimates of metabolic activity in the Witwatersrand Basin and the insight gained into the carbon metabolism of microbial communities inhabiting the deep, organic carbon-poor terrestrial subsurface.
Materials and methods
Geological setting
A sub-horizontal, diamond drilled borehole (TT107) ~100 m in length located on level 107 (depth=3048 mbls) of AngloGold Ashanti's Tau Tona Au Mine was sampled. The borehole intersects a fluid-filled fracture associated with the contact between the Witwatersrand Supergroup quartzite and Jeans Dyke and was sampled for metagenomic and geochemical analyses.
Sampling
Before sampling, fracture water and gas was allowed to flow from the sampling borehole to displace any potential contaminants introduced during borehole drilling. A stainless steel manifold with control valves was connected to the borehole, and PTFE tubing was attached to the ball valves on the sides of the manifold. Temperature, pH, conductivity and reduction potential (Eh) were measured from the water with handheld probes (Hanna Instruments, Carrollton, TX, USA), and dissolved O2, H2O2, Fe(II), total Fe and sulfide were measured using field test kits (Chemetrics Inc., Calverton, VA, USA). In order to obtain sufficient biomass for sequencing, an autoclaved 0.2 μm pleated GE Memtrex NY (MNY) filter cartridge (cat. no. MNY921AAS; 25 cm in length) was placed in a stainless steel casing and attached to the steel manifold. The filter was left to accumulate biomass for 13 days at an initial flow rate of 4 l min−1. On the 13 day, the filter was retrieved from the mine, stored on blue ice and transported back to the field station at the University of Free State where it was stored at −80 °C. In total, 110 950 l of water had passed trough the massive filter, which based on cell count-derived concentrations of 280 cells ml−1 (Simkus et al., 2015), accounts for the accumulation of approximately 3.1 × 1010 cells. The filter was transported back to Princeton using a MVE ZC 20/3V vapor shipper and stored at −80 °C until further processing.
Free energy flux
Samples for cation and anion analysis were collected using the protocols described in Lau et al. (2014). Mineral solubility, dissolved species activity, partial pressures and free energy of microbial reactions were calculated using the geochemical modeling program, The Geochemist's Workbench version 8.0 (Bethke, 2008). In order to elucidate whether or not the microorganisms are maintaining autotrophic, mixotrophic or heterotrophic lifestyles, we analyzed the steady-state free energy flux (FEF; Onstott, 2004) of 47 energy-yielding redox reactions to test the hypothesis that the more favorable the reaction, the more likely it would be to proceed within the biological community, if the necessary protein-encoding genes (PEGs) for a given metabolic pathway were present in the metagenome.
The steady-state FEF of a planktonic microorganism suspended in an aqueous solution was calculated using Geochemist's Workbench V8 Professional (Bethke, 2008) and the following relationship,
where r is the average radius of a cell (cm) and assumed to be 7.5 × 10−5 cm, D is the effective diffusion constant (cm2 s−1) of the limiting reactant corrected for the in situ temperature (Cussler, 2009), C is the concentration of the limiting reactant (mol cm−3) and ΔG is the in situ Gibbs-free energy of the limiting reactant (kJ mol−1). For microbial redox reactions that involve sessile microorganisms and a hard mineral phase (for example, Fe(OH)3 and pyrolusite), the effective diffusion constant (Deff) of the electron donor was adjusted according to the following equation,
where φ is porosity of the hard mineral phase, τ is tortuosity and δ is constrictivity. For the purpose of this study, the porosity of the Witwatersrand Basin rock units is ~0.5% (Silver et al., 2012), δ=1, and τ=2.59 (Takahashi et al., 2009).
DNA extraction and sequencing
DNA was extracted from the MNY filter as described in Lau et al., (2014). The DNA sample was sent to the Marine Biological Laboratory (MBL, Woods Hole, MA, USA) for whole-genome library prep using NuGen Ultralow Ovation system that included 18 cycles of PCR amplification of all adaptor-viable molecules (http://nugen.com). The metagenomic library (insert size ~160 bp) was then sequenced using an Illumina HiSeq 1000 system (Marine Biological Laboratory) for 101-bp paired-end reads. In total, 31 721 135 paired-end reads were generated. Sequences were then uploaded to the MG-RAST metagenomic pipeline (http://metagenomics.anl.gov) where they were merged and annotated according to Meyer et al. (2008). A total of 30 621 568 sequences passed the quality control of MG-RAST with an average sequence length of 158±8 bp (MG-RAST metagenome ID: 4529964.3).
Metagenome assembly and annotation
The MG-RAST quality control-passed reads were directly assembled using IDBA-ud (Peng et al., 2011) and only contigs longer than 200 bp were analyzed. Average coverage for each contig was calculated as the IDBA header designated read count multiplied by the average sequence length and divided by the length of the contig. Assembled contigs of length >200 bp were used for gene prediction using Prodigal and the ‘-p meta' function (Hyatt et al., 2012). Predicted open reading frames (ORFs) were then clustered at 90% identity using CD-HIT-EST (Fu et al., 2012). Annotations of the consensus ORFs were determined by using BLASTp against the m5nr database, collecting the top 10 hits with a maximum e-value threshold of 10−5. A consensus protein annotation was then selected using a majority vote script, whereas taxonomy of the ORF was declared as the best hit to the selected annotation. The abundance of clustered ORFs was calculated by summing the average coverage of all contributing contigs to each ORF. A complete summary of assembly and annotation statistics is provided in Table 1. Notably, only annotations related to bacteria and archaea were included in this analysis.
Table 1. Assembly and annotation statistics.
| Raw reads | 31 721 135 paired-end reads |
| Reads post QC | 30 621 568 (avg. length 158±8 bp) |
| Contigs | 62 968 |
| N50 of contigs | 3258 bp |
| Longest contig | 174 953 bp |
| Predicted ORFs | 126 317 |
| ORFs after 90% clustering | 116 827 |
| Percent of ORFs annotated | 84.6% |
Abbreviations: ORF, open reading frame; QC, quality control.
Taxonomic diversity
The taxonomic diversity of the TT107 fracture water was determined from the relative abundances of single copy PEGs (SC-PEG) in the assembled data set as they have been shown to greatly reduce the biases, such as horizontal gene transfer or gene duplication, associated with assigning taxonomy from total protein diversity analyses (Ciccarelli et al., 2006; Von Mering et al., 2007). The set of SC-PEGs used in this study are reported in Supplementary Table S1.
Metagenomic comparisons
The unassembled TT107 metagenome was compared with 15 subsurface metagenomes publicly available on MG-RAST (Supplementary Table S2). Subsystem hierarchies for each metagenome were downloaded from MG-RAST with thresholds of 60% identity, alignment lengths >25 and maximum e-value of 10−5. Annotations were then filtered to only include PEGs classified under the RAST subsystem hierarchy ‘carbon monoxide induced hydrogenase'. The MG-RAST reported abundances of these genes were then normalized (mean 0, variance 1) within each metagenome. The relative abundance of Firmicutes in each metagenome was calculated from all MG-RAST annotations (predicted proteins and ribosomal RNA genes).
Results and discussion
Geochemical characteristics
The water temperature during the 2-week sampling period ranged from 52.1 °C to 50.4 °C, the pH varied from 8.6 to 8.7, the Eh from −46 to −133 mV and the salinity remained constant at 0.20 ppt (Table 2). The δ2H and δ18O of the water lies very close to that reported for the Transvaal dolomite aquifer and the Global Meteoric Water Line (GMWL) (Lau et al., 2014), but is displaced to more negative values compared with the annual mean of modern precipitation, suggesting a paleometeoric origin consistent with other mine waters investigated in this part of the Witwatersrand Basin. 14C dating of the DIC indicates that the subsurface residence time of the fracture water is 1–6 kyr (Lau et al., 2014). The electron donors identified in the fracture water included formate, acetate, H2, CO, HS−, NH4+, CH4, ethane, propane and butane (Table 2). Electron acceptors were limited and lower in concentration than the electron donors and consisted primarily of SO42−, NO3−, HCO3− and O2 (Table 2). The total dissolved nitrogen was entirely comprised of NH4+ and NO3−, whereas formate and acetate comprised 50% of the DOC (Table 2). Of the 47 redox reactions potentially constrained by these measurements, the FEF of 31 reactions were calculated (Supplementary Table S3). The 16 remaining FEFs were not calculated because of the fact that either the reactants were below detection or the ΔG was positive.
Table 2. Geochemistry Data.
| Sampling Date | 11 Aug 2011 | 24 Aug 2011 |
|---|---|---|
| T (°C) | 52.1 | 50.4 |
| pH | 8.6 | 8.7 |
| Eh (mV) | −46 | −133 |
| TDS (ppt) | 0.20 | 0.20 |
| Cells ml−1 | 280a | 200b |
| Gases (M) | Cations (M) | Carbon Sources (M) | Metals (M) | ||||
|---|---|---|---|---|---|---|---|
| He | 6.07 × 10−4 | Na+ | 2.5 × 10−3 | DOC | 1.83 × 10−5 | Total Fe | 1.4 × 10−7 |
| H2 | 1.71 × 10−5 | K+ | 3.5 × 10−5 | DIC | 5.70 × 10−4 | Fe2+ (Chemet) | <8.9 × 10−7 |
| O2 | 1.52 × 10−4 | Mg2+ | 4.1 × 10−5 | DIC-Gas Solubility | 3.56 × 10−3 | Fe3+ (Chemet) | 2.7 × 10−6 |
| O2 (Chemet) | 6.25 × 10−6 | Ca2+ | 2.7 × 10−4 | Formate | 8.00 × 10−6 | Mn | 1.3 × 10−7 |
| N2 | 5.98 × 10−3 | Sr2+ | 3.2 × 10−6 | Acetate | 3.39 × 10−7 | Cr | 3.9 × 10−8 |
| CO | 9.1 × 10−5 | Ba2+ | 5.7 × 10−7 | Lactate | <1.10 × 10−6 | Co | 3.4 × 10−8 |
| CO2 | 1.71 × 10−5 | Al3+ | 2.2 × 10−6 | Propanoate | <1.40 × 10−6 | Ni | 1.2 × 10−6 |
| CH4 | 8.75 × 10−3 | Si4+ | 4.8 × 10−4 | ||||
| C2H6 | 1.40 × 10−4 | ||||||
| C3H8 | 2.05 × 10−5 | ||||||
| i-C4H10 | 1.75 × 10−6 | ||||||
| n-C4H10 | 3.00 × 10−5 | ||||||
| i-C5H12 | 1.76 × 10−6 | ||||||
| n-C5H12 | <1.48 × 10−6 | ||||||
| Non-Methane Hydrocarbons | 4.44 × 10−4 | ||||||
|
Anions (M) |
Sulfur Species (M) |
Nitrogen Species (M) |
|||||
| F− | 1.0 × 10−4 | SO42− | 1.04 × 10−4 | Total N | 3.21 × 10−6 | ||
| Cl− | 2.8 × 10−3 | S2O32− | 3.09 × 10−6 | NO2− | <2.2 × 10−6 | ||
| Br− | 5.6 × 10−6 | HS− | 7.48 × 10−8 | NO3− | 9.52 × 10−7 | ||
| PO43− | 3.5 × 10−7 | NH4+ | 3.88 × 10−6 | ||||
Based upon cell counts using DAPI (Simkus et al., 2015).
Based on the bacterial biomass calculated from 46 599 l of filter H2O resulting in 41.2 μg of PLFA (0.9 pg/ml) (Simkus et al., 2015).
Energy conservation with the acetyl-CoA pathway
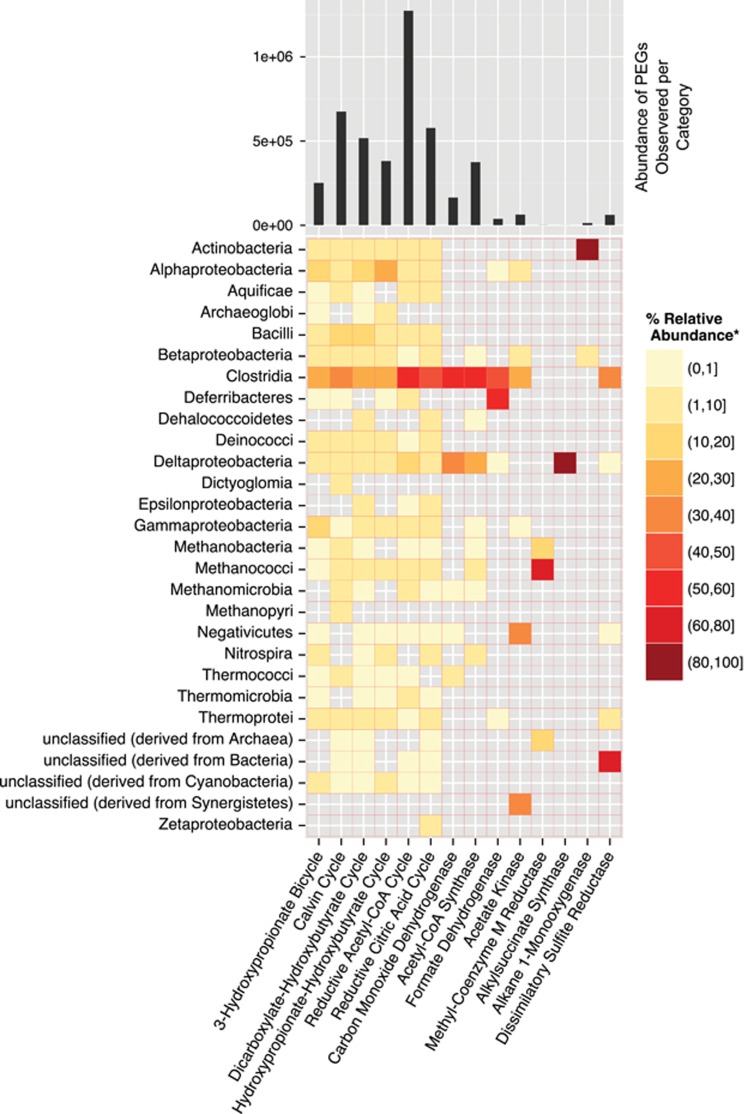
PEGs involved in six known carbon fixation pathways (Berg, 2011) were identified within the assembled metagenome (Figure 1) and the KEGG accessions of proteins used to identify the pathways are listed in Supplementary Table S4. Of these six carbon fixation pathways, the acetyl-CoA pathway was the most represented carbon fixation pathway in the metagenome (abundance=1 273 022) despite containing the lowest number of representative genes searched for in the metagenome (n=8; Supplementary Table S4).
Figure 1.
Carbon cycle-specific genes. The six carbon fixation pathways (3-hydtroxypropionate bicycle, Calvin cycle, dicarboxylate-hydroxybutyrate cycle, hydroxypropionate-hydroxybutyrate cycle, reductive acetyl-CoA cycle and reductive citric acid cycle) and other important C-cycling genes are listed on the x axis. The intensity of each block corresponds to the percent relative abundance (see legend) of each class (y axis) within a given individual gene/pathway (x axis). Gray squares indicate that no sequences were observed meeting the given criteria. At the top of each column is a bar chart that indicates the total abundance of each gene/pathway in the metagenome. The KEGG accessions for the genes used to annotate the six carbon fixation pathways are provided in Supplementary Table S4.
The dominance of the reductive acetyl-CoA pathway is, in part, because of the wide variety of organisms that have been reported to use this pathway. Such organisms include acetogens, methanogens, sulfate reducing bacteria (SRB), ammonia-oxidizing planctomycetes, an autotrophic archaeal SRB, Archaeoglobales (Berg, 2011), anaerobic facultative autotrophs (Schauder et al., 1988) and, reversibly, with heterotrophs using carbon monoxide dehydrogenase (CODH) and acetyl-CoA synthase to oxidize acetyl-CoA (Rabus et al., 2006). Callaghan et al. (2012) have also postulated that a reversed Wood-Ljungdahl pathway is carried out during the complete oxidation of alkanes under anaerobic conditions. The ΔG of many of the dominant anaerobic reactions hover close to the ~70 kJ (mol)−1 required for synthesis of ATP (Supplementary Table S3), suggesting that this fracture water system is energy limited. Therefore, in addition to the variety of metabolisms the acetyl-CoA pathway has been reported to participate, the dominance of the acetyl-CoA pathway is, likely, due to the fact that it is the preferred carbon fixation pathway of bacteria and archaea living close to the thermodynamic limit of life (Berg, 2011).
Of the PEGs identified in the acetyl-CoA pathway, 57.6% of the total calculated PEG abundance were related to the class Clostridia (Phylum: Firmicutes) and, despite having a low SC-PEG relative abundance (1%), Deltaproteobacteria-related acetyl-CoA pathway genes were enriched (18.8% of the eight indicative acetyl-CoA genes; Figure 1). At the individual PEG level, the majority of CODH and acetyl-CoA synthase PEGs, as well as formate dehydrogenase, an enzyme responsible for the oxidation of formate to CO2 in the acetyl-CoA pathway, are related to known SRB in the classes Clostridia and Deltaproteobacteria (Figure 1). Organisms within these two taxa, including ‘Ca. D. audaxviator', have been shown to exhibit both heterotrophic and autotrophic lifestyles, depending on the conditions (Londry et al., 2004; Rabus et al., 2006; Chivian et al., 2008). The flexibility of individual microorganisms to perform both heterotrophy and autotrophy has been suggested to be a critical survival adaptation for hydrogeologically isolated systems as the carbon substrate concentrations may change over time (Moser et al., 2005) and flexibility may also be an important feature of keystone species (for example, ‘Ca. D. audaxviator') inhabiting the energy-limited TT107 fracture water.
Absence of aerobic heterotrophy
Six of the 10 most favorable reactions identified by FEF analysis required O2 (Supplementary Table S3); however, in situ conditions were likely anoxic, despite the apparent detection of low amounts of dissolved O2. Oxygen was measured soon after the borehole was opened for sampling (soon after it was drilled) and the Eh decreased (−46 to −133 mV) during the 13-day sampling period (Table 2). Therefore, the initial measurement of O2 may reflect residual O2 introduced during drilling of the borehole and may not be representative of in situ conditions. Metagenomic evidence also fails to support the FEF-predicted aerobic activity. Particulate methane monooxygenase, soluable methane monooxygenase and alkane 1-monoxygenases PEGs were used as indicator PEGs for O2-driven redox reactions. No PEGs related to particulate methane monooxygenase and soluable methane monooxygenase were identified in the metagenome and only two alkane 1-monoxygenases PEGs, related to those of an actinobacterium and a betaproteobacterium, were identified (Figure 1). Furthermore, the acetyl-CoA pathway, which contains genes that function only under strict anoxia (Berg, 2011), were found to be abundant in the metagenome (Figure 1). Therefore, the genetic potential for aerobic heterotrophic reactions is low when compared with anaerobic reactions. Consequently, only the FEF of anaerobic reactions are reported in Table 3.
Table 3. Ten anaerobic reactions with the greatest free energy flux.
| ΔG (kJ mol –1)a | Free Energy Flux (kJ cell−1 s−1) | |
|---|---|---|
| 0.4Propane+SO42−→HS−+1.2HCO3−+0.2H++0.4H2O | −21 | −1.2 × 10−13 |
| CO+2H2O→H2+HCO3−+H+ | −25 | −1.1 × 10−13 |
| 0.3077n-Butane+SO42−→HS−+1.231HCO3−+0.2308H++0.3077H2O | −22 | −4.6 × 10−14 |
| H2+0.4H++0.4NO3−→0.2N2+1.2H2O | −191 | −3.4 × 10−14 |
| 3H2+CO→methane+H2O | −33 | −9.8 × 10−15 |
| 4H2+H++SO42−→>HS−+4H2O | −28 | −6.2 × 10−15 |
| n-Butane+4H2O→2acetic acid+5H2 | −13 | −4.4 × 10−15 b |
| 4H2+H++HCO3−→methane+3H2O | −12 | −2.7 × 10−15 |
| 4H2+2HCO3−+H+→acetate+4H2O | −10 | −2.2 × 10−15 |
| Acetate+1.6NO3−+0.6H+→2HCO3−+0.8H2O+0.8N2 | −181 | −2.1 × 10−15 |
kJ mol–1 of 2e−. A table of the ΔG and free energy flux of 31 reactions can be found in Supplementary Table S3.
The hydrolysis of butane represents an abiogenic reaction that is kinetically inhibited, but no known microorganisms has been isolated that performs this function.
Firmicutes-dominated community
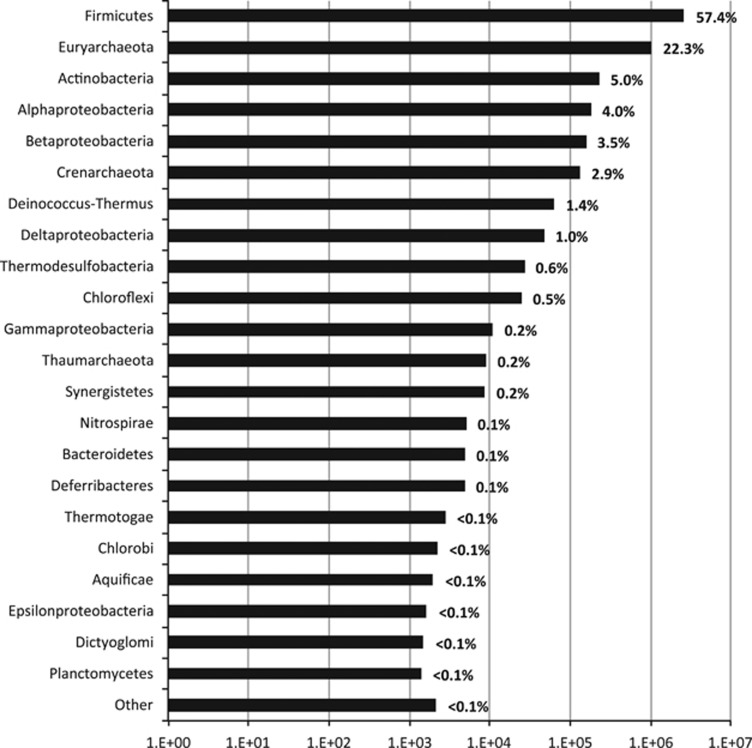
Based on the analysis of SC-PEGs, Gram-positive Firmicutes were the most frequently detected phylum (57.4%) followed by Euryarchaeota (22.3%) and smaller contributions of Proteobacteria (8.7%), Actinobacteria (5.0%), Crenarchaeota (2.9%) and Deinococcus-Thermus (1.4%) (Figure 2). Before this study, the only other fully analyzed metagenome from the Witwatersrand Basin was from a filter sample of MP104 located 3 km SW of TT107 (Chivian et al., 2008). Over 99% of MP104's reads were found to be related to a chemolithoautotrophic bacterium, ‘Ca. D. audaxviator', capable of performing sulfate reduction and utilizing the reverse acetyl-CoA (Wood-Ljungdahl) pathway for CO2 fixation. ‘Ca. D. audaxviator' also comprised 98–100% of the 96 bacterial clones library (90% of the total prokaryotic community) of the Dr938H3 borehole (Moser et al., 2005) located 1.7 km east of TT107. Although identified in the TT107 metagenome, ‘Ca. D. audaxviator' accounts for only 1.6% of SC-PEG genera, whereas the most abundant genus in TT107 was its closest known relative within the family Peptococcaceae, Desulfotomaculum (8.9% of SC-PEGs).
Figure 2.
Relative abundance of single copy protein encoding genes (SC-PEGs). The phylum (y axis) level abundances (x axis) of SC-PEGs are displayed. The percent relative abundance of each phylum is shown at the end of each bar. The following phyla are included in the ‘Other' designation: Korarchaeota, Acidobacteria, Chlamydiae, Fusobacteria, Fibrobacteres, Tenericutes and Cyanobacteria.
In contrast to the unprecedented low alpha diversity of MP104 reported by Chivian et al. (2008), 289 genera were observed in the assembled metagenome of TT107 and, in a study by Magnabosco et al. (2014) using V6 16S rDNA amplicons, 549 plus genera were identified in six different South African boreholes that sampled fracture fluids at depths of 1–3.1 km. It is important to note that of the six boreholes (including a borehole from the 109 level of Tau Tona) sampled by Magnabosco et al. (2014), all samples displayed a similar community composition that was heavily dominated by Proteobacteria and contained <10% relative abundance of Firmicutes. This is consistent with other clone-based bacterial 16S rRNA gene surveys of other South African fracture water sites (Gihring et al., 2006; Lin et al., 2006a; Silver et al., 2010) with the exception of two sites: Dr546bh1 (3.2 kmbls) (Moser et al., 2003) and Ev818H6 (2.24 kmbls) (Davidson et al., 2011). The 16S rRNA gene clone libraries for Dr546bh1 and Ev818H6 revealed Firmicutes-dominated communities but the numbers of clones were small, and thus, the full alpha diversity may not have been sampled. The dominance of Firmicutes-related bacteria in TT107, Dr938H3 and MP104 suggests that the subsurface microbial community compositions of the South African fracture waters range from Firmicutes-dominated to Proteobacteria-dominated communities.
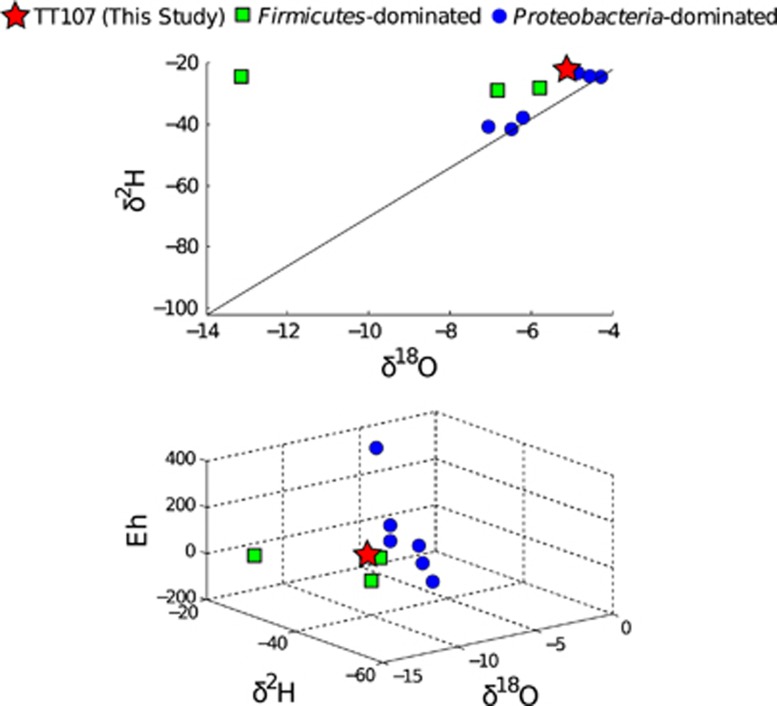
If we compare the δ2H versus δ18O of the fracture water for MP104, TT107, Dr938H3, Ev818H6, Dr546bh1 and the six sites sampled in the V6 amplicon study, we find that Firmicutes-dominated sites tend to be more removed from the GMWL than the six Proteobacteria-dominated V6 amplicon study sites, which sit on or near the GMWL (Figure 3). Location of fluids with respect to the GMWL is a key indicator of their origin and residence time in the subsurface. Fluids with relatively recent recharge from the surface (on a scale of tens of thousands of years) lie along the GMWL, reflecting their meteoric origin (Craig, 1961). In contrast, saline waters and brines that have been sequestered in the deep crystalline rocks of the Precambrian Shields worldwide have been shown to have δ18O and δ2H signatures well elevated over those of the GMWL (Lippmann-Pipke et al., 2011). The representation of the South African sites within the δ2H/δ18O plot suggests that the fracture fluids in this study represent mixtures of ancient hydrothermal fluids and paleometeoric water (Onstott et al., 2006; Lippmann-Pipke et al., 2011). Furthermore, it appears that fracture fluids that have seen a smaller amount of mixing with paleometeoric waters, in combination with lower Eh (Figure 3), select for Firmicutes from mixed populations of Proteobacteria and Firmicutes residing in the paleometeoric water infiltrating the crustal fractures.
Figure 3.
Community composition relative to geochemistry. Top: the δ18O (‰) versus δ2H (‰) of the six Proteobacteria-dominated sites included in Magnabosco et al. (2014) are shown in blue circles. Firmicutes-dominated sites (MP104, Dr938H3, Dr546bh1) are green squares and TT107 is shown as a red star. The black line indicates the GWML (Craig, 1961). Bottom: the δ18O (‰) versus δ2H (‰) versus Eh (mV) are shown using the same color scheme as (top). *The δ18O and δ2H for the Firmicutes-dominated Ev818H6 site were not measured but the geochemical similarity with other boreholes located on Ev818 suggests that the water is located far from the GWML (Davidson et al., 2011). The Eh of Ev818H6 was measured to be −128 mV (Davidson et al., 2011).
A similar pattern of community composition based on fluid origins has also been reported in the Fennoscandian Shield (Itävaara et al., 2011), a terrestrial serpentinite system in northern California (the Cedars) (Suzuki et al., 2013), the Tablelands Ophiolite (Brazelton et al., 2013) and other serpentinizing sites (Schrenk et al., 2013). In the Fennoscandian Shield, the microbial communities inhabiting 100-m increments of the Outokumpu deep borehole (up to1500 m depth) were reported. There, Proteobacteria dominated the oxygenated, shallow (0–900 m) samples, whereas Firmicutes dominated the deeper samples (900–1000 m, 1400–1500 m) of the borehole (Itävaara et al., 2011). In the case of terrestrial serpentinites (Brazelton et al., 2013; Schrenk et al., 2013; Suzuki et al., 2013), it has been reported that Firmicutes (and Chloroflexi in the case of Suzuki et al. (2013)) dominate serpentinite springs fed solely by deep groundwater, whereas the sites whose water was a mixture of deep and shallow groundwater contain communities dominated by Betaproteobacteria. In the shallow sites, it is expected that the Betaproteobacteria such as Hydrogenophaga make use of the abundant O2 and H2 associated with the paleometeoric waters, whereas the Firmicutes-dominated sites are characterized by organisms capable of dealing with the low abundance of oxidants and/or with the ability to switch between autotrophic and mixtrotrophic growth (Schrenk et al., 2013).
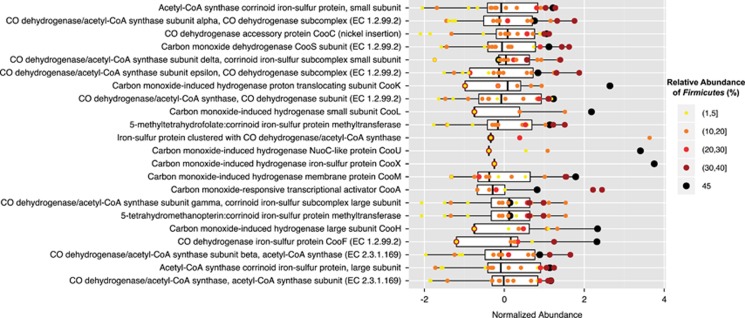
To explore whether or not the shift in the relative abundance of Firmicutes in subsurface microbial communities is associated with a change in metabolism, the abundance of PEGs related to the ‘carbon monoxide induced hydrogenase' category were analyzed in parallel with taxonomic data from 16 unassembled subsurface metagenomes (See Materials and methods: Metagenomic Comparisons section). Notably, the ‘carbon monoxide induced hydrogenase' category was selected because it is the category that is most directly related to the reductive acetyl-CoA pathway and, in turn, energy conservation (See Results and Discussion: Energy conservation with the acetyl-CoA pathway). From this analysis, we found that, in general, sites with a higher relative abundance of Firmicutes exhibited a higher abundance PEGs in this category (Figure 4). This trend further supports the hypothesis that organisms capable of using acetyl-CoA are selected for in energy-limited environments.
Figure 4.
The normalized abundances of PEGs within the ‘carbon monoxide induced hydrogenase' RAST category are shown. For each PEG (bars), the normalized abundance of that PEG in a given subsurface metagenomes is shown (dots). The size and color of the dot represents the percent relative abundance of Firmicutes identified in the metagenome. The TT107 metagenome (this study) is represented by a large, black circle (45% relative abundance of Firmicutes in the unassembled metagenome).
The recurring nature of the observation that distinct hydrogeological and geochemical niches select for Proteobacteria- versus Firmicutes-dominated communities in the South African subsurface, Fennoscandian Shield (Itävaara et al., 2011), and terrestrial serpentinites (Brazelton et al. 2013, Schrenk et al. 2013, Suzuki et al., 2013) suggests this may be a genuine feature of the subsurface biome and not an artifact of methodology. The observation raises the question of whether the geochemical and hydrogeological history of fracture fluids influences the nature of the in situ microbial community and how it develops. If such a correlation persists in future investigations, we may be able to predict a subsurface microbial community from geochemical measurements characterizing the subsurface environment. Such a tool would, in turn, improve our estimates of the distribution of microbes and our understanding of the controls on biodiversity in the terrestrial subsurface.
Biological carbon cycling at TT107
Methanogenesis
Aside from its distinctive bacterial community, TT107 also contains the highest relative abundance of Euryarchaeota (22%) yet reported in the South African subsurface (Gihring et al., 2006; Simkus et al., 2015). Methyl-coenzyme M reductase, the enzyme responsible for the final step in methanogenesis, was identified primarily within the classes Methanococci and Methanobacteria representing 71.5% and 16.7% of methyl-coenzyme M reductase PEGs, respectively (Figure 1). Although the methanogens in TT107 are relatively more numerous than other sites within the Witwatersrand Basin, the FEF for biological CH4 production (−9.8 × 10−15 kJ cell−1 s−1; Table 3) is less thermodynamically favorable than what has been reported for Dr938H3 (FEF as high as 1.0 × 10−13 kJ cell−1 s−1) (Moser et al., 2005) and Mponeng mine (FEF as high as 3.7 × 10−14 kJ cell−1 s−1) (Lin et al., 2006b). These differences reflect the higher H2 concentrations found in Dr938H3 and MP104 relative to that of TT107. Previously, Sherwood Lollar et al. (2006) has interpreted the low H2 concentration of South African fracture water as consistent with the depletion of H2 by active methanogenesis. This interpretation is further supported by the combination of the Δ14CCH4 value (−996±1‰), Δ14CDIC value (−496.6‰), the CH4 concentration (8.8 mM) and the DIC concentration (570 μM), which yields an estimate the in situ CH4 production rate by autotrophic methanogenesis of 8.7±2.3 nM CH4 yr−1 at TT107 (Simkus et al., 2015; Supplementary Table S5).
Anaerobic oxidation of hydrocarbons
This study determined that oxidation of short chain hydrocarbons via sulfate reduction is much more thermodynamically favorable than methanogenesis (Table 3). Notably, Clostridia and Deltaproteobacteria-related dissimilatory sulfite reductase PEGs for sulfate reduction were present in the metagenome (Figure 1) and could have an integral role to the coupling of sulfate reduction to propane (FEF=−1.2 × 10−13 kJ cell−1 s−1) and n-butane (FEF=−4.6 × 10−14 kJ cell−1 s−1) oxidation. Currently, little is known about the enzymes that catalyze anaerobic alkane oxidation and, to date, only two genomes, Desulfococcus oleovorans Hxd3 and Desulfatibacillum alkenivorans AK-01 (both Deltaproteobacteria), are available among anaerobic alkane oxidizers. Of these two organisms, only the mechanism of alkane activation for D. alkenivorans AK-01 is known (Callaghan et al., 2012). The activation of alkanes by D. alkenivorans AK-01 is catalyzed by an alkylsuccinate synthase that was also identified in the TT107 metagenome (Figure 1; Callaghan et al., 2008). As the complete oxidation of alkanes by D. alkenivorans AK-01 is believed to proceed by reversing the Wood-Ljungdahl (reductive acetyl-CoA) pathway, it is possible that anaerobic alkane oxidizers contribute to the elevated abundance of Deltaproteobacteria-related acetyl-CoA pathway genes described earlier.
Carboxydotrophy
CO oxidation coupled to the reduction of H2O to H2 was the second most powerful anaerobic reaction (Table 3; FEF=−1.1 × 10−13 kJ cell−1 s−1) and provides another role for the numerous CODH PEGs. A carboxydovore-related CODH with 94% nucleotide identity to Desulfotomaculum carboxydivorans CO-1-SRB was identified in the TT107 metagenome (Supplementary Table S6). D. carboxydivorans CO-1-SRB is a bacterium that was cultured in 100% pCO headspace and with the absence of any other electron donors, carbon substrates and sulfate (Parshina et al., 2005). Parshina et al. (2005) found that the D. carboxydivorans CO-1-SRB oxidized CO to CO2 while reducing H2O to H2 and, upon the addition of sulfate, switched to sulfate reduction. In TT107, Desulfotomaculum, is the most abundant annotated genus (relative abundance=8.9%) and, because of the fact that the oxidation of CO is the second most favorable anaerobic reaction in TT107 (Table 3), it is conceivable that, in addition to the fixation of CO2 via the acetyl-CoA pathway, the utilization of CO by carboxydivores is an important catabolic process in the TT107 fracture.
Acetogenesis
Acetogens are famous for their ability conserve energy (Müller, 2003) and Clostridia-related acetate kinase PEGs that may be representative of acetogenesis were identified (Figure 1). However, given that the ΔG for the acetogenic reaction is only −10 kJ (mol 2e−)−1 and the FEF is only 2.2 × 10−15 kJ cell−1 s−1 (two orders of magnitude lower than top two FEF values of Table 3), the acetate kinase PEGs, most likely, perform the function of phosphorylating acetate for biosynthesis rather than contributing biogenic acetate to the DOC pool.
Summary
The observation that the FEFs of hydrocarbon oxidation reactions coupled to sulfate reduction are far greater than those of methane- and acetate-generating processes suggests that the TT107 fracture water was likely dominated by mixotrophs or heterotrophs that utilize the oxidative acetyl-CoA pathway. The presence of alkylsuccinate synthase genes related to the anaerobic alkane oxidizer D. alkenivorans AK-01 and a carboxydovore-related CODH/acetyl-CoA synthase gene cluster provide a molecular explanation for how the highly favorable anaerobic hydrocarbon and CO oxidation reactions may proceed in TT107.
Further supporting these mixed modes of carbon metabolisms are the recently reported carbon isotope signatures of PLFA collected from TT107 (δ13CPLFA=−11 to −25.5‰ Δ14CPLFA=−633.2±11.3‰) (Simkus et al., 2015). When these values are compared with the carbon isotope values for DIC and CH4 (Simkus et al., 2015), we find that the Δ13CDIC-PLFA is 6.0‰ to 20.5‰, which is consistent with the preferential uptake of the lighter isotope of carbon from DIC by autotrophic microbial communities (Boschker and Middelburg, 2002). The Δ14C of TT107's PLFA (Δ14C PLFA=−633.2±11.3‰) (Simkus et al., 2015) is, however, lower than the Δ14C of the DIC (−496.6±2.1‰) (Lau et al., 2014) resulting in a Δ14CDIC-PLFA=143‰. This Δ14CDIC-PLFA indicates microbial consumption of carbon source(s) in addition to DIC, which are likely the abiogenic hydrocarbons. The use of the acetyl-CoA pathway as both an oxidative and reductive pathway is a likely explanation for the Δ13CDIC-PLFA and Δ14CDIC-PLFA patterns.
Conclusions
The combination of steady-state FEF calculations and metagenome analyses suggest that the TT107 fracture water is home to both organic and inorganic C utilizers. Firmicutes dominate the TT107 fracture water and distinguish it from Proteobacteria-dominated communities found in waters with younger residence times and higher Eh in the South African subsurface. The selection for Firmicutes has also been observed in other terrestrial subsurface sites and may be driven by the energy limitations that come with a lowered Eh of the fracture water. These results support the growing evidence that the deep subsurface biosphere is a more dynamic ecosystem than previously understood and that the bacterial and archaeal inhabitants have the capability to actively contribute and respond to the transformation of carbon within the deep subsurface.
Data availability
The FASTQ reads of the TT107 metagenome are available on MG-RAST (metagenome ID: 4529964.3) and can be accessed via http://metagenomics.anl.gov/linkin.cgi?metagenome=4529964.3.
Acknowledgments
We thank G Slater and D Simkus for insightful discussions on carbon isotope signatures. In addition, we would like to thank F Vermeulen, M Pienaar, A Munro, and the other staff from Tau Tona gold mine (AngloGold Ashanti Ltd) for providing access and assistance during sampling, T Tlalajoe and M Maleke for their assistance in sampling and field measurements, and the Marine Biological Laboratory for performing library prep and metagenomic sequencing. Subsurface research was funded by the National Geographic Society, National Science Foundation (NSF) and Natural Sciences and Engineering Research Council of Canada (NSERC) to BSL. NSF funding was provided to TLK (EAR-0948335), TCO (EAR-0948659), and CM (DGE-1148900). Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation. CM also received funding from the Center for Dark Energy Biosphere Investigations Graduate Fellowship and this article is C-DEBI Contribution 276.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Baker BJ, Moser DP, MacGregor BJ, Fishbain S, Wagner M, Fry NK et al. (2003). Related assemblages of sulphate-reducing bacteria associated with ultradeep gold mines of South Africa and deep basalt aquifers of Washington State. Environ Microbiol 5: 267–277. [DOI] [PubMed] [Google Scholar]
- Bennett B, Adams JJ, Gray ND, Sherry A, Oldenburg TBP, Huang H et al. (2013). The controls on the composition of biodegraded oils in the deep subsurface—part 3. The impact of microorganism distribution on petroleum geochemical gradients in biodegraded petroleum reservoirs. Org Geochem 56: 94–105. [Google Scholar]
- Berg IA. (2011). Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol 77: 1925–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke CM. (2008) Geochemical and Biogeochemical Reaction Modeling2nd edn. University of Illinois: : Urbana, IL. [Google Scholar]
- Biddle JF, Lipp JS, Lever MA, Lloyd KG, Sørensen KB, Anderson R et al. (2006). Heterotrophic archaea dominate sedimentary subsurface ecosystems off Peru. Proc Natl Acad Sci USA 103: 3846–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgonie G, García-Moyano A, Litthauer D, Bert W, Bester A, van Heerden E et al. (2011). Nematoda from the terrestrial deep subsurface of South Africa. Nature 474: 79–82. [DOI] [PubMed] [Google Scholar]
- Boschker HTS, Middelburg JJ. (2002). Stable isotopes and biomarkers in microbial ecology. FEMS Microbiol Ecol 40: 85–95. [DOI] [PubMed] [Google Scholar]
- Brazelton WJ, Morrill PL, Szponar N, Schrenk MO. (2013). Bacterial communities associated with subsurface geochemical processes in continental serpentinite springs. Appl Environ Microbiol 79: 3906–3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brazelton WJ, Nelson B, Schrenk MO. (2012). Metagenomic evidence for h(2) oxidation and h(2) production by serpentinite-hosted subsurface microbial communities. Front Microbiol 2: 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaghan AV, Morris BEL, Pereira IAC, McInerney MJ, Austin RN, Groves JT et al. (2012). The genome sequence of Desulfatibacillum alkenivorans AK-01: a blueprint for anaerobic alkane oxidation. Environ Microbiol 14: 101–113. [DOI] [PubMed] [Google Scholar]
- Callaghan AV, Wawrik B, Chadhain SMN, Young LY, Zylstra GJ. (2008). Anaerobic alkane-degrading strain AK-01 contains two alkylsuccinate synthase genes. Biochem Biophys Res Commun 366: 142–148. [DOI] [PubMed] [Google Scholar]
- Chapelle FH, Lovley DR. (1990). Rates of microbial metabolism in deep coastal plain aquifers. Appl Environ Microbiol 56: 1865–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivian D, Brodie EL, Alm EJ, Culley DE, Dehal PS, DeSantis TZ et al. (2008). Environmental genomics reveals a single-species ecosystem deep within Earth. Science 322: 275–278. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. (2006). Toward automatic reconstruction of a highly resolved tree of life. Science 311: 1283–1287. [DOI] [PubMed] [Google Scholar]
- Craig H. (1961). Isotopic variations in meteoric waters. Science (80-) 133: 1702–1703. [DOI] [PubMed] [Google Scholar]
- Cussler EL. (2009) Diffusion: Mass Transfer in Fluid Systems. Cambridge University Press: New York, NY, USA. [Google Scholar]
- Davidson MM, Silver BJ, Onstott TC, Moser DP, Gihring TM, Pratt LM et al. (2011). Capture of planktonic microbial diversity in fractures by long-term monitoring of flowing boreholes, Evander Basin, South Africa. Geomicrobiol J 28: 275–300. [Google Scholar]
- Ehrenberg SN, Jakobsen KG. (2001). Plagioclase dissolution related to biodegradation of oil in Brent Group sandstones (Middle Jurassic) of Gullfaks Field, northern North Sea. Sedimentology 48: 703–721. [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28: 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gihring TM, Moser DP, Lin L-H, Davidson M, Onstott TC, Morgan L et al. (2006). The distribution of microbial taxa in the subsurface water of the Kalahari Shield, South Africa. Geomicrobiol J 23: 415–430. [Google Scholar]
- Hyatt D, LoCascio PF, Hauser LJ, Uberbacher EC. (2012). Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 28: 2223–2230. [DOI] [PubMed] [Google Scholar]
- Itävaara M, Nyyssönen M, Kapanen A, Nousiainen A, Ahonen L, Kukkonen I. (2011). Characterization of bacterial diversity to a depth of 1500 m in the Outokumpu deep borehole, Fennoscandian Shield. FEMS Microbiol Ecol 77: 295–309. [DOI] [PubMed] [Google Scholar]
- Kieft TL, McCuddy SM, Onstott TC, Davidson M, Lin L-H, Mislowack B et al. (2005). Geochemically generated, energy-rich substrates and indigenous microorganisms in deep, ancient groundwater. Geomicrobiol J 22: 325–335. [Google Scholar]
- Krumholz LR, McKinley JP, Ulrich GA, Suflita JM. (1997). Confined subsurface microbial communities in Cretaceous rock. Nature 386: 64–66. [Google Scholar]
- Lau MCY, Cameron C, Magnabosco C, Brown CT, Schilkey F, Grim S et al. (2014). Phylogeny and phylogeography of functional genes shared among seven terrestrial subsurface metagenomes reveal N-cycling and microbial evolutionary relationships. Front Microbiol 5: 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L-H, Hall J, Onstott TC, Gihring T, Lollar BS, Boice E et al. (2006. a). Planktonic microbial communities associated with fracture-derived groundwater in a deep gold mine of South Africa. Geomicrobiol J 23: 475–497. [Google Scholar]
- Lin L-H, Wang P-L, Rumble D, Lippmann-Pipke J, Boice E, Pratt LM et al. (2006. b). Long-term sustainability of a high-energy, low-diversity crustal biome. Science 314: 479–482. [DOI] [PubMed] [Google Scholar]
- Lipp JS, Morono Y, Inagaki F, Hinrichs K-U. (2008). Significant contribution of archaea to extant biomass in marine subsurface sediments. Nature 454: 991–994. [DOI] [PubMed] [Google Scholar]
- Lippmann-Pipke J, Lollar BS, Niedermann S, Stroncik NA, Naumann R, van Heerden E et al. (2011). Neon identifies two billion year old fluid component in Kaapvaal Craton. Chem Geol 283: 287–296. [Google Scholar]
- Londry KL, Jahnke LL, Des Marais DJ. (2004). Stable carbon isotope ratios of lipid biomarkers of sulfate-reducing bacteria. Appl Environ Microbiol 70: 745–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnabosco C, Tekere M, Lau MCY, Linage B, Kuloyo O, Erasmus M et al. (2014). Comparisons of the composition and biogeographic distribution of the bacterial communities occupying South African thermal springs with those inhabiting deep subsurface fracture water. Front Microbiol 5: 679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer F, Paarmann D, D'Souza M, Olson R, Glass EM, Kubal M et al. (2008). The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9: 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser DP, Gihring TM, Brockman FJ, Fredrickson JK, Balkwill DL, Dollhopf ME et al. (2005). Desulfotomaculum and Methanobacterium spp. dominate a 4- to 5-kilometer-deep fault. Appl Environ Microbiol 71: 8773–8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser DP, Onstott TC, Fredrickson JK, Brockman FJ, Balkwill DL, Drake GR et al. (2003). Temporal shifts in the geochemistry and microbial community structure of an ultradeep mine borehole following isolation. Geomicrobiol J 20: 517–548. [Google Scholar]
- Müller V. (2003). Energy conservation in acetogenic bacteria. Appl Environ Microbiol 69: 6345–6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onstott TC. (2004). Impact of CO2 injections on deep subsurface microbial ecosystems and potential ramifications for the surface biosphere. CO2 Capture Storage Proj 2: 1207–1239. [Google Scholar]
- Onstott TC, Lin L-H, Davidson M, Mislowack B, Borcsik M, Hall J et al. (2006). The origin and age of biogeochemical trends in deep fracture water of the Witwatersrand Basin, South Africa. Geomicrobiol J 23: 369–414. [Google Scholar]
- Onstott TC, Magnabosco C, Aubrey AD, Burton AS, Dworkin JP, Elsila JE et al. (2014). Does aspartic acid racemization constrain the depth limit of the subsurface biosphere? Geobiology 12: 1–19. [DOI] [PubMed] [Google Scholar]
- Onstott TC, Moser DP, Pfiffner SM, Fredrickson JK, Brockman FJ, Phelps TJ et al. (2003). Indigenous and contaminant microbes in ultradeep mines. Environ Microbiol 5: 1168–1191. [DOI] [PubMed] [Google Scholar]
- Parshina SN, Sipma J, Nakashimada Y, Henstra AM, Smidt H, Lysenko AM et al. (2005). Desulfotomaculum carboxydivorans sp. nov., a novel sulfate-reducing bacterium capable of growth at 100% CO. Int J Syst Evol Microbiol 55: 2159–2165. [DOI] [PubMed] [Google Scholar]
- Pedersen K. (1997). Microbial life in deep granitic rock. FEMS Microbiol Rev 20: 399–414. [Google Scholar]
- Peng Y, Leung HCM, Yiu S-M, Chin FYL. (2011). Meta-IDBA: a de novo assembler for metagenomic data. Bioinformatics 27: i94–i101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabus R, Hansen TA, Widdel F. (2006). Dissimilatory sulfate- and sulfur-reducing prokaryotes. The Prokaryotes. Springer: New York, NY, USA, pp 659–768. [Google Scholar]
- Schauder R, Preuß A, Jetten M, Fuchs G. (1988). Oxidative and reductive acetyl CoA/carbon monoxide dehydrogenase pathway in Desulfobacterium autotrophicum. Arch Microbiol 151: 84–89. [Google Scholar]
- Schrenk MO, Brazelton WJ, Carolina N, Lang SQ. (2013). Serpentinization, carbon, and deep life. Rev Mineral 75: 575–606. [Google Scholar]
- Sherwood Lollar B, Lacrampe-Couloume G, Slater GF, Ward J, Moser DP, Gihring TM et al. (2006). Unravelling abiogenic and biogenic sources of methane in the Earth's deep subsurface. Chem Geol 226: 328–339. [Google Scholar]
- Silver BJ, Onstott TC, Rose G, Lin L-H, Ralston C, Sherwood-Lollar B et al. (2010). In situ cultivation of subsurface microorganisms in a deep mafic sill: implications for SLiMEs. Geomicrobiol J 27: 329–348. [Google Scholar]
- Silver BJ, Raymond R, Sigman DM, Prokopeko M, Lollar BS, Lacrampe-Couloume G et al. (2012). The origin of NO 3- and N 2 in deep subsurface fracture water of South Africa. Chem Geol 294: 51–62. [Google Scholar]
- Simkus DN, Slater GF, Sherwood Lollar B, Wilkie K, Kieft TL, Magnabosco C et al. (2015). Variations in microbial carbon sources and cycling in the deep continental subsurface. GEOCHIM COSMOCHIM AC.
- Stevens TO, McKinley JP. (1995). Lithoautotrophic microbial ecosystems in deep basalt aquifers. Science (80-) 270: 450–455. [Google Scholar]
- Suzuki S, Ishii S, Wu A, Cheung A, Tenney A, Wanger G et al. (2013). Microbial diversity in the Cedars, an ultrabasic, ultrareducing, and low salinity serpentinizing ecosystem. Proc Natl Acad Sci USA 110: 15336–15341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Seida Y, Yui M. (2009). 3D X-ray CT and diffusion measurements to assess tortuosity and constrictivity in a sedimentary rock. Diffus Org 11: 1–11. [Google Scholar]
- Takai K, Moser DP, DeFlaun M, Onstott TC, Fredrickson JK. (2001). Archaeal diversity in waters from deep South African gold mines. Appl Environ Microbiol 67: 5750–5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Mering C, Hugenholtz P, Raes J, Tringe SG, Doerks T, Jensen LJ et al. (2007). Quantitative phylogenetic assessment of microbial communities in diverse environments. Science (80-) 315: 1126–1130. [DOI] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. (1998). Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95: 6578–6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The FASTQ reads of the TT107 metagenome are available on MG-RAST (metagenome ID: 4529964.3) and can be accessed via http://metagenomics.anl.gov/linkin.cgi?metagenome=4529964.3.