Abstract
To test whether protist grazing selectively affects the composition of aquatic bacterial communities, we combined high-throughput sequencing to determine bacterial community composition with analyses of grazing rates, protist and bacterial abundances and bacterial cell sizes and physiological states in a mesocosm experiment in which nutrients were added to stimulate a phytoplankton bloom. A large variability was observed in the abundances of bacteria (from 0.7 to 2.4 × 106 cells per ml), heterotrophic nanoflagellates (from 0.063 to 2.7 × 104 cells per ml) and ciliates (from 100 to 3000 cells per l) during the experiment (∼3-, 45- and 30-fold, respectively), as well as in bulk grazing rates (from 1 to 13 × 106 bacteria per ml per day) and bacterial production (from 3 to 379 μg per C l per day) (1 and 2 orders of magnitude, respectively). However, these strong changes in predation pressure did not induce comparable responses in bacterial community composition, indicating that bacterial community structure was resilient to changes in protist predation pressure. Overall, our results indicate that peaks in protist predation (at least those associated with phytoplankton blooms) do not necessarily trigger substantial changes in the composition of coastal marine bacterioplankton communities.
Introduction
Marine bacteria are a cornerstone component of the microbial loop, acting as a trophic link—transforming dissolved organic carbon into particulate matter—to higher trophic levels (Azam et al., 1983). Owing to the central role played by bacteria in modulating organic carbon fluxes, it is essential to know what factors control their abundance and production in the sea. Two main factors are recognized for regulating standing stocks of aquatic bacteria: resource supply, such as organic carbon or inorganic nutrients (often named ‘bottom-up' control), and predation by bacterivores and mortality because of viruses (‘top-down' control). In most environments, these mechanisms are constantly at play, and integrating over extended time, growth is balanced by losses. Still, pronounced imbalances in growth versus loss rates over relatively more limited spatiotemporal scales can lead to pronounced dynamics in bacterial abundance. However, as both types of mechanisms can become important under different conditions (Thingstad, 2000), there is still not an accepted general theory of the regulation of bacterial stocks and production in marine systems (Jürgens and Massana, 2008). Differences in susceptibility to bacterivory among bacteria have been suggested as a key aspect that could influence the regulatory mechanisms of marine bacteria and the relation between bottom-up and top-down controls (Jürgens and Massana, 2008).
Prey cell size has an important role in determining bacterial susceptibility to bacterivory, as shown by several empirical studies reporting a preference for larger bacterial prey by cultured and natural assemblages of flagellates (Andersson et al., 1986; Gonzalez et al., 1990; Jürgens and Güde, 1994; Kinner et al., 1998) and ciliates (Fenchel, 1980; Epstein et al., 1992; Simek et al., 1995). The physiological state of bacterial cells has also been suggested as a key factor in grazing selectivity (Del Giorgio and Gasol, 2008), and preferential grazing of the more active cells within a community by protist grazers has been repeatedly observed (Del Giorgio et al., 1996; Pernthaler et al., 1997; Simek et al., 1997; Tadonléké et al., 2005; Sintes and Del Giorgio, 2014). This is probably related to the general positive relation between cell size and activity in marine bacteria (Gasol et al., 1995; Hahn and Höfle, 2001; Matz and Jürgens, 2001; Matz et al., 2002; Corno and Jürgens, 2006), suggesting that larger bacterioplankton cells are also usually the most active ones. Moreover, there could be a concentration-dependent component, where grazing might be different on abundant versus non-abundant populations (Boenigk et al., 2002). Besides size, differences in grazing behavior because of differences in prey type (algal versus bacterial cells) and prey characteristics have been reported (Fu et al., 2003). Also, the physiological state and the ability of bacteria to develop grazing resistance mechanisms (e.g., motility, toxin release, exopolymer formation) can affect prey selection by protists (Pernthaler, 2005).
Given that different marine bacterial lineages may possess distinct grazing-influencing properties (i.e., size, activity, cell surface properties or other grazing resistance mechanisms), selective protistan grazing should in theory affect particular bacterial populations. In some studies, changes in predation pressure on bacteria are reflected in changes in bacterial community composition (Pernthaler et al., 1997; Hahn and Höfle, 1999; Jürgens et al., 1999; Šimek et al., 1999; Langenheder and Jürgens, 2001; Šimek et al., 2002). In other studies, interactions of bacterial prey—protistan predators did not result in changes in bacterial community composition (Suzuki, 1999; Massana and Jurgens, 2003; Yokokawba and Nagata, 2005), indicating that selective grazing was largely independent of bacterial phylogeny. This is in contrast with clear evidence of selective grazing on the more active bacteria (Del Giorgio et al., 1996; Vaqué et al., 2001; Longnecker et al., 2010). These latter reports were based on the analyses at a broad phylogenetic level using fingerprinting techniques and FISH (fluorescence in situ hybridization with class/phylum level probes), which provides a limited view of the actual changes in bacterial community composition and diversity occurring at a higher taxonomic resolution. Advances in recent years in sequencing capabilities allow us to examine in more detail bacterial diversity shifts in response to protist grazing pressure.
We studied the responses in bacterial abundance, activity (at the bulk and single-cell levels), community composition and diversity to pronounced shifts in the protistan grazing regimen in a mesocosm experiment with Mediterranean Sea water to which inorganic (ammonia, phosphorus) and organic (glucose) nutrients were added. This experiment has already provided significant insights into the ecology of marine bacteria and their responses to nutrient enrichments and viruses (Allers et al., 2007; Sandaa et al., 2009; Tanaka et al., 2009). Using FISH with family-level probes, Allers et al. (2007) revealed a temporal succession where Alteromonadaceae responded to the experimental manipulations and profited from allochthonous glucose inputs, whereas Rhodobacteraceae benefited from the subsequent nutrient-induced phytoplankton bloom. The data suggested a potential grazing effect on Alteromonadaceae based on a quasi-simultaneous increase of nanoflagellate abundance. However, although much was learned from the overall experiment, the issue of possible effects of selective grazing had not been resolved. In this work, we build on the major findings of that experiment and specifically focus on the treatment (i.e., glucose+phosphate) that showed the strongest response in the abundance of protist grazers (heterotrophic nanoflagellates). Thus, we expected that it would be the treatment with the most pronounced effect of grazing pressure. Our objective was to decipher whether protistan grazing on marine bacterioplankton was selective and to identify potential selection criteria (e.g., bacterial size, physiological state, taxonomy). In particular, we wanted to address the unresolved question of whether increased protist predation triggers diversity changes in marine bacterioplankton communities. We therefore combined the use of high-throughput sequencing techniques (454 pyrosequencing of the 16S rRNA gene)—to quantify changes in bacterial community structure and diversity—with the estimation of grazing rates (with fluorescently labeled bacteria (FLB)), changes in bacterial, flagellate and ciliate stocks, and shifts in bacterial cell size and physiological state (using the CTC (5-cyano-2,3-diotolyl tetrazolium) and NADS (nucleic acid double-staining) methodologies).
Materials and methods
Experimental setup
Two 200 l mesocosms were filled with surface water from the Blanes Bay Microbial Observatory (Mediterranean Sea 41°40′N, 2°48′E) on October 2004. These mesocosms were amended daily with 2 μm NH4Cl to prevent N limitation and were initially supplied with 50 nm of phosphate (KH2PO4) and 13.25 μm of glucose and incubated for 8 days at in situ temperature in a 12:12 h light–dark cycle per regimen. All mesocosms were mixed two times a day by hand-held stirring, and samples for bacterial abundance and production, phytoplankton biomass (chlorophyll a), heterotrophic nanoflagellate (HNF) and ciliate abundance were taken once a day at the same time. Samples for bacterial community composition (454 tag pyrosequencing of the 16S rRNA gene) were collected every second day.
Chlorophyll a concentration
Chlorophyll a concentrations were determined fluorometrically (Parsons et al., 1984). Subsamples of 150 ml were filtered through glass fiber filters (GF/F; Whatman, Maidstone, UK) and subsequently extracted in 90% acetone overnight at 4 °C in the dark, and then analyzed with a Turner Designs fluorometer (Sunnyvale, CA, USA).
Prokaryotic abundance, nucleic acid-content, membrane-compromised bacteria (NADS) and actively respiring bacteria (CTC) determined by flow cytometry
Bacterial abundance was estimated with a Becton Dickinson FACScalibur flow cytometer (BD Bioscience, Franklin Lakes, NJ, USA) as described previously (Gasol and Giorgio, 2000). Samples of 1 ml were fixed with 1% buffered paraformaldehyde solution (pH 7.0) plus 0.05% glutaraldehyde, incubated for 10 min at room temperature and then stored in liquid nitrogen. For total bacterial cell counts, 200 μl of these subsamples were stained with a dimethyl sulfoxide-diluted SYBR Green I stock solution (10:1; Molecular Probes, Eugene, OR, USA) at a final concentration of 10x. Staining was carried out for 10 min in the dark. For flow cytometric analyses, 10 μl of a solution of yellow-green latex beads (size, 1 μm; final concentration, 106 ml−1; Polyscience, Washington, PA, USA) was added to each sample as an internal standard. Bacterial abundances were determined from the ratios of cells to beads. Bacteria were identified by their signatures in a plot of side scatter versus green fluorescence (FL1). High and low nucleic acid-content cells (HNAs and LNAs) were separated in the scatter plot of side scatter—FL1 (Gasol et al., 1999). Picocyanobacteria were discriminated in a plot of FL1 versus red fluorescence (FL3). Bacterial cell sizes were estimated from nucleic acid-derived fluorescence scaled to that of reference beads, and converted to cell size with an empirically derived calibration with image-analyzed DAPI (4′,6-diamidino-2′-phenylindole)-stained cells (Gasol and Giorgio, 2000).
Membrane-intact and -damaged prokaryotic cells (named ‘live' and ‘dead' for simplicity) were enumerated in non-fixed samples following the NADS protocol (Gregori et al., 2001; Falcioni et al., 2008). NADS+, green cells (assumed to be live, with intact membranes), and NADS−, red cells (assumed to be inactive, with compromised cell membranes), were identified by simultaneous double staining with a membrane-permeable (SYBR Green; Molecular Probes) and impermeable (propidium iodide) probe. Immediately after collection, samples (0.4 ml) were incubated in the dark with 4 μl of SYBR Green (10x final conc.) and 4 μl of propidium iodide (10 μg ml−1 final) for 15 min (Falcioni et al., 2008). NADS+ and NADS− cells were enumerated by flow cytometry and differentiated in a scatter plot of FL1 (green)–FL3 (red emission after blue-light excitation). Samples for prokaryotic abundance and NADS were run at a flow rate of 18–20 μl min−1.
Bacteria with an enhanced respiration rate were determined with CTC (Polysciences), which intercalates in the electron transport chain of respiring prokaryotes and gets reduced to a red-fluorescence formazan salt and is taken to indicate the prokaryotes with high respiring rates (Sherr et al., 1999; Gasol and Giorgio, 2000). Unfixed samples were incubated with 5 mm CTC (fresh stock solution of 50 mm) for 90 min, in the dark and at room temperature and run at high speed (50 μl min−1) in a FACSCalibur Becton Dickinson flow cytometer (Becton-Dickinson, Franklin Lakes, NJ, USA) with a blue (440 nm) laser. CTC-positive cells (hereafter CTC+) were then enumerated using the FL2 vs FL3 dot plot (see Gasol and Arístegui (2007) for details).
Flagellate and ciliate abundance
Samples (100 ml) for HNF quantification were preserved as described previously (Simek et al., 1995). Subsamples of 5–15 ml from these samples were stained with DAPI (final concentration, 1 μg ml−1) and passed through 0.8 μm Poretics polycarbonate filters (GE Osmonics, Minnetonka, MN, USA) by gentle vacuum filtration. Nanoflagellates were enumerated via epifluorescence microscopy using ultraviolet and counterchecking with blue-light excitation. Duplicate samples were analyzed within 24 h after preservation and at least 200 individual HNF cells were counted per sample.
Ciliate abundances were determined using the Utermöhl method. One liter of sample was fixed with acidic lugol (2% final concentration). After sedimentation of 100 ml aliquots for 24–48 h, ciliates were counted in an inverted microscope (Zeiss, Jena, Germany). Average ciliate size was determined after measuring all cells recorded per sample (between 100 and 3000 cells).
Bacterial production
Bacterial activity was estimated from the incorporation rate of [3H]leucine as described by Kirchman et al. (1985) and adapted by Smith and Azam (1992). Samples were incubated with 40 nm of [3H]leucine in microcentrifuge tubes in the dark for 1 h at ambient temperatures. Trichloroacetic acid-killed samples were used as controls. Bacterial heterotrophic production was calculated as Leu incorporation rate times the standard 3.1 kg C mol per leucine conversion factor.
Grazing experiments and grazing experiments
FLB were prepared from a Brevundimonas diminuta (syn. Pseudomonas diminuta) strain obtained from the Spanish Type Culture Collection (Burjassot, València, Spain). B. diminuta has already been used several times to prepare FLB (e.g., Vazquez-Dominguez et al., 1999; Vaqué et al., 2004; Unrein et al., 2014) because of their small size (0.065 μm3) well mimicking typical sizes of the indigenous bacterial populations. FLB were produced by scraping cells from agar plates, suspending them in carbonate–bicarbonate buffer (CO3Na2-HCO3Na, pH 9.5), and staining them by incubation with 100 pg ml−1 of DTAF (5-([4,6 dichlorotriazin-2yl] amino)-fluorescein) for 2 h in a water bath at 60 °C (Sherr et al., 1987). Stained cells were rinsed with filtered (<0.2 μm) carbonate–bicarbonate buffer, resuspended, centrifuged five times (10 min, 10 000 r.p.m.) and pelletized to prevent the transfer of leftover dye to the natural samples. The cell suspensions were kept frozen (−20 °C) until use. Before addition to the experimental containers, the FLB working solution was thawed and gently sonicated three rounds of 10 s with the microtip at 35% of power output to prevent cell clustering (Sonic dismembrator DYNATECH, ARTEK Systems Co., Formingdale, NY, USA; Model 300).
Total bacterivory by organisms <100 μm was estimated from the disappearance rate of FLB (Vazquez-Dominguez et al., 1999). Subsamples of the mesocosms were incubated in 2 -l Nalgene bottles in duplicate. The bottles were kept in the culture chamber for 24 h at the same conditions experienced by the mesocosms (20 °C, photoperiod 1130–1230 hours light–dark, 150 μmol m−2 s−1). Bacteria and FLB abundances were counted immediately after the addition of FLB (T0), and at 6 and 24 h. A control bottle was also run for each mesocosm to test for nonpredatory disappearance of FLB (e.g., fluorescence losses or FLB attachment on the bottle wall). Controls were established with 0.2 μm filtered sample water, amended with FLB and maintained at the same conditions as described above. Calculations of total consumed bacteria (g, grazing rate) were obtained following the mathematical model of Salat and Marrasé (1994) and the correction by Unrein et al. (2007), as g=−(1/t) Ln (Ft/F0), where t is the incubation time and Ft and F0 the abundances of FLB at the final and initial times (FLB per ml). Net bacterial growth rate (a) was calculated as a=(1/t) Ln (Nt/N0), where Nt and N0 are the natural bacterial abundances at the end and start of the experiment (bact. per ml). The decrease of FLB in control bottles was checked in every experiment and corrected, if necessary as explained by Unrein et al. (2007) by calculating: gC=−(1/t) Ln (cFt/cF0), where gC is the apparent grazing rate in the control bottles (per day); t the incubation time; cFt the abundance of FLB at final time and cF0 the abundance of FLB at initial time in the controls (FLB per ml). Then, corrected grazing is G=[(g−gC)/a] (Nt−N0), where G is the total grazing (bact. per ml). We divided by t to obtain the total grazing rate (bact. per ml per day). Measurements used for running this model were done at time zero and after 3 and 5 days of incubaction. Grazing rates were measured in the mesocosms at days 0, 3 and 6. They were measured in the considered mesocosms as well as in other treatments (explained in Allers et al., 2007), making a total of 17 measurements. We used these points to create a multiple regression model that explained 78% of the variance as: GR (bact. per ml per day)=−2.32 × 105+0.31 × Bt (cells per ml)+42.46 HNF (cells per ml), N=17, P<0.0001, where Bt is bacterial abundance and HNF is the abundance of heterotrophic nanoflagellates. This model predicted, for the two replicated mesocosms considered here, values at times 0, 3 and 6 that were within 20% of the observed value, and in addition could be used to estimate grazing pressure at the other times for which we had no measurements.
DNA sampling collection and extraction
Samples of microbial community DNA for subsequent 454 tag pyrosequencing analysis of 16S rRNA were collected at the beginning of the experiment (day 0) and on every second day until day 8. Approximately 700 ml of the samples were filtered through a 0.2 μm polycarbonate filters (diameter, 47 mm; Durapore; Millipore, Billerica, MA, USA) and stored frozen at −70 °C in sucrose buffer (0.75 m sucrose, 40 mm EDTA, 50 mm Tris, pH=8.3). A combined treatment with enzymes (lysozyme, proteinase K) and enzyme/phenol–chloroform was used to extract the DNA as described previously (Riemann et al., 2000). DNA was resuspended in TE buffer (10 mm Tris, 1 mm EDTA, pH 8.0) and quantified fluorometrically (PicoGreen; Molecular Probes).
PCR and sequencing preparation
Partial bacterial 16S rRNA genes were amplified for pyrosequencing using a primer cocktail containing the degenerate primers 530 F (5′-GTGCCAGCMGCNGCGGTA-3′) (Dowd et al., 2008) but with TA added at the 3′ end to increase specificity, and 1061 R (5′-CRRCACGAGCTGACGAC-3′) (Dowd et al., 2008) labeled with specific hexamers for each sample for amplification (Sjöstedt et al., 2012). The PCR products were excised from the agarose gel, purified with QIAquick Gel Extraction Kit (Qiagen) and concentrated with QIAquick PCR Purification Kit (Qiagen). Addition of adaptor and pyrosequencing on a Roche GS FLX TITANIUM (Roche Applied Science) were performed at LGC Genomics (Berlin, Germany) according to the manufacturer's instructions.
Sequence analysis
A total of 192 771 sequences (average length 395 bp) were obtained and analyzed following the approach described previously (Fierer et al., 2008; Hamady et al., 2008; Lauber et al., 2009) using the QIIME (Quantitative Insights Into Microbial Ecology) pipeline (http://qiime.org). Low-quality sequences and sequences shorter than 200 bp were removed. Denoising was carried out via the n3phele cloud (http://www.n3phele.com) working with the QIIME toolkit. Singletons were not included in further analyses. Using this conservative approach, the final number of sequences remaining was 117 571 (average length 506 bp; 14 696 sequences per sample). Similar sequences were binned into operational taxonomic units (OTUs) using UCLUST (Edgar, 2010) with a minimum pairwise identity of 97%. These sequences resulted in a final OTU table consisting of 256 OTUs (excluding singletons), with the number of OTUs per sample ranging from 67 to 210. Representative sequences for each OTU were aligned with PyNAST, the taxonomic identity of each phylotype determined using the Green-Genes database (DeSantis et al., 2006), and a tree built using FastTree (Price et al., 2009). Rarefaction, to a subsampling depth determined by the minimum number of sequences in the samples (i.e. 3780 sequences), was performed in QIIME on all samples to standardize the sequencing effort. Alpha-diversity was expressed as the Shannon index. Hierarchical cluster analysis was performed and visualized in R 3.1.2 (R Core Team, 2015) with Bray–Curtis distances calculated from relative abundances of OTUs (normalized sequence reads). Sequences have been deposited in GenBank under the accession numbers KP988571–KP989708.
Statistical analyses
The relations between biological variables were examined by means of correlation analysis computing Pearson's pairwise statistics. Data were log transformed to equalize variance, and normality was checked with a Shapiro–Wilks test before Pearson's correlations were calculated.
For testing the correlation between changes in grazing rate and %BT with shifts in bacterioplankton community composition, we performed permutational analysis of variance (PERMANOVA) on the same Bray–Curtis distances as above, using the package Vegan (Oksanen et al., 2011) in R 3.1.2 (R Core Team, 2015).
Results
Temporal dynamics of phytoplankton biomass and abundance of bacteria, HNF and ciliates
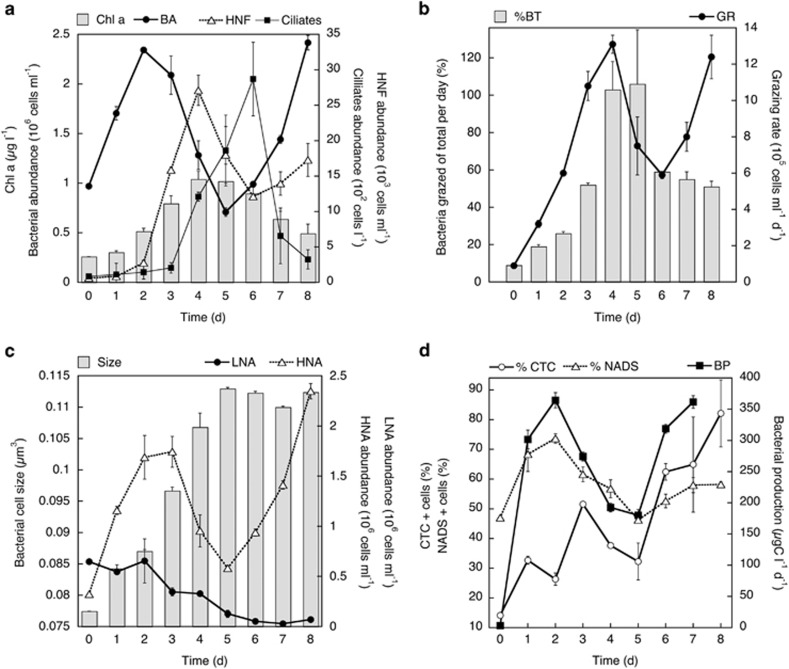
Chlorophyll a concentrations increased threefold (from 0.3 to 1 μg l−1) during the first 4–5 days of incubation, and then decreased by half towards the end of the experiment (Figure 1a). Bacterial abundance increased by 2.4-fold (from 1 × 106 to 2.4 × 106 cells per ml) after 2 days, and then decreased by >3-fold (from 2.4 × 106 to 0.7 × 106 cells per ml) at day 5 (Figure 1a). Subsequently, bacterial abundance increased at the end of the experiment, reaching similar abundances as in the first peak (i.e., ∼2.4 × 106 cells per ml at days 2 and 8). The abundance of HNF followed a similar pattern as that of bacterial cell numbers, but with a time lag of around 1–2 days (Figure 1a). HNF peaked after 4 days, increasing up to 45-fold (from 0.063 to 2.7 × 104 cells per ml), and then decreased during the next 2 days to less than half (from 2.7 × 104 to 1.2 × 104 cells per ml). At the end of the experiment, a second bloom of HNF occurred. The abundance of ciliates followed HNF cell numbers with a 1-day lag (Figure 1a). Ciliates peaked after 5–6 days of incubation increasing by 30-fold (from 100 to 3000 cells per l), and then decreased to low abundances towards the end of the experiment (Figure 1a).
Figure 1.
Average temporal variation in (a) chlorophyll a concentration, bacterial abundance, HNFs and ciliates, (b) grazing rates (GRs) and corresponding percentage of total bacteria grazed per day (%BT), (c) bacterial cell size, abundance of LNA and HNA cells and (d) single-cell and bulk activity estimates including the percentage of actively respiring (CTC-positive) and viable (NADS-positive) cells and bulk bacterial production during the mesocosm experiments with water from the Bay of Blanes. (a) The means of duplicate treatments and vertical bars show their range of values (the minimum and maximum values of the duplicates).
Temporal dynamics of grazing, production, size and proportion of active bacteria
The reduction of bacterial abundance coincided with a one order of magnitude increase in the grazing rates (from 1 × 106 cells per ml per day at time 0 to 13 × 106 cells per ml per day at day 4) and in the percentage of bacteria being grazed per day by HNF (from 9% at time 0 to 126% at day 4) (Figure 1b). Afterwards, the proportion of bacteria grazed decreased by half (to 59%) at day 6, allowing the second increase in bacterial abundance towards the end of the experiment.
The dynamics in abundance of HNAs mirrored that of the total bacterial abundance and production, whereas the LNAs decreased in abundance almost constantly until day 6 (by 13-fold, from 6.5 × 105 to 0.5 × 105 cells per ml) and remained low for the rest of the experiment (Figure 1c). The relative proportion of HNA cells doubled from 34% at the initial time to 68% at day 1, and went on increasing up to 84% by day 3. After a decreasing trend on days 4 and 5, the percentage of HNA cells increased again towards the end of the experiment to account for around 98% of the cells. The estimated average bacterial size increased by 1.5-fold (from 0.077 to 0.113 μm3) during the first 5 days, and then remained roughly constant for the rest of the experiment (Figure 1c). The percentage of HNA cells was positively correlated to the average size of all bacterial cells (Pearson's r=0.85, N=9, P=0.0035).
Bacterial production followed a similar pattern as bacterial abundance (but with even stronger changes in magnitude), increasing over two orders of magnitude (from 3 to 379 μg per C l per day) after 2 days, afterwards decreasing by around half until day 5 (to 183 μg per C l per day). Finally, production increased again towards the end of the experiment, reaching similar rates as during the two blooms observed at days 2 and 7, that is, ∼370 μg per C l per day (Figure 1d).
The percentage of highly respiring cells (CTC+) followed a similar development as the HNA cells, with the lowest values at the initial time (14%) and the highest (82%) at the end of the experiment (Figure 1d). The abundance of CTC+ and HNA cells were positively correlated throughout the experiment (N=9, Pearson's r=0.91, P=0.0006). The proportion of ‘live cells' (as determined by the NADS protocol) relative to the total number of bacterial cells firstly elevated, then decreased by ca. 20% from day 2 to day 5 (from ∼70% to 46%) and again increased to 58% towards the end of the experiment (Figure 1d). The percentage of NADS+ cells was inversely correlated to HNF abundance with a 1-day lag (N=8, Pearson's r=0.94, P=0.0005).
Dynamics of bacterial community composition and diversity indexes
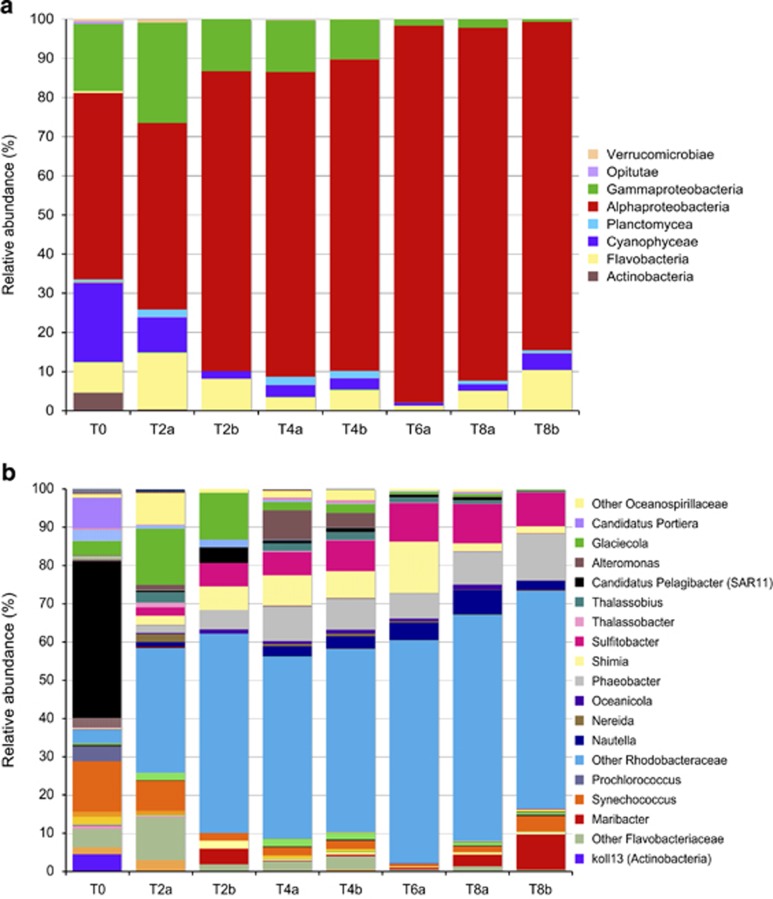
At the class level, Alphaproteobacteria (48%), Cyanobacteria (20%), Gammaproteobacteria (17%), Flavobacteria (8%) and Actinobacteria (4%) were the most abundant members in the initial water (Figure 2a). Interestingly, although there was not a strong shift in community composition at the class level during the study, a progressive change in the relative abundance of the major classes was found, where Alphaproteobacteria increased (from 48% at time 0 to 85–90% relative abundance at day 8) at the expense of Cyanobacteria and Gammaproteobacteria.
Figure 2.
Average temporal variation in the percentage of relative abundance of taxonomical (a) classes and (b) genera or deeper taxonomic level possible at the initial time (T0), at day 2 (T2), 4 (T4), 6 (T6) and 8 (T8) in duplicate mesocosms (1, 2). Some groups of sequences could not be characterized down to the genera level but were included in this figure as well because they were abundant (e.g., Other Oceanospirillaceae, Other Rhodobacteraceae and so on). Not all groups were included in the legend of the plots because of a lack of space. Full information on the relative abundance of all members is presented in Supplementary Table S1. Note that some sequences could not be resolved deeper than the class level, but the deepest resolved taxonomic level was included in this plot.
At a finer taxonomic resolution (like at the OTU level, defined as a level of 16 S rRNA gene sequence identity of 97%), more shifts than at the class level were observed (Figure 2b). Although the Gammaproteobacteria class decreased in relative abundance with time, not all Gammaproteobacteria subgroups decreased uniformly through the experiment. For instance, the Gammaproteobacteria genus Glaciecola increased in relative abundance from time 0 to day 2 (from ∼3% to 12–14%), before decreasing again to <3% from day 4 onwards. The Gammaproteobacteria genus Alteromonas increased in relative abundance, from 0.2% at time 0 to 4–7% at day 4, decreasing afterwards to values of <0.5%. However, more pronounced differences were found among Alphaproteobacteria subgroups. At the beginning of the experiment, the relative abundance of OTUs of the Alphaproteobacteria Rhodobacteraceae family increased by more than one order of magnitude (from 3% at time 0 to >50% at day 2). The Rhodobacteraceae family was widely diverse, represented by many different OTUs (belonging to members of the genera Nautella, Nereida, Oceanicola, Phaeobacter, Shimia, Sulfitobacter, Thalassobius and Other Rhodobacteraceae), which became abundant along the experiment (Figure 2b and Supplementary Table S1). Simultaneously, the relative abundance of Alphaproteobacteria SAR11 cluster (Cand. Pelagibacter) decreased from 41% at time 0 to <4% at day 2. Among the Flavobacteria, OTUs of the genus Maribacter became relatively more abundant at days 2 and 8, coinciding with the peaks of bacterial abundance and production.
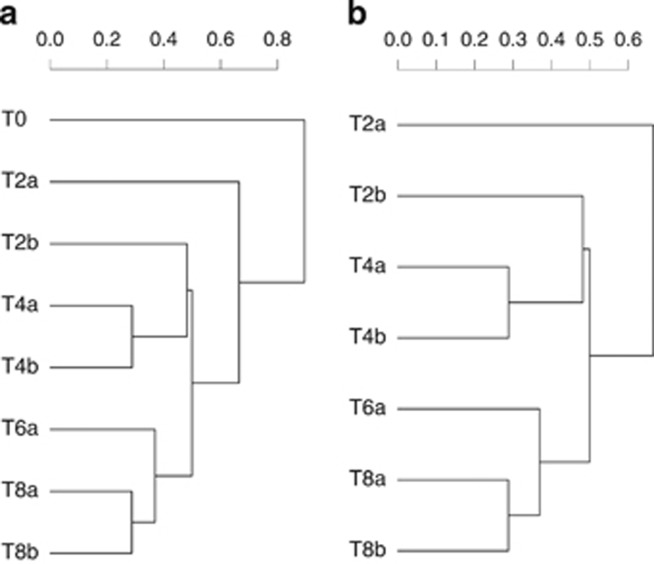
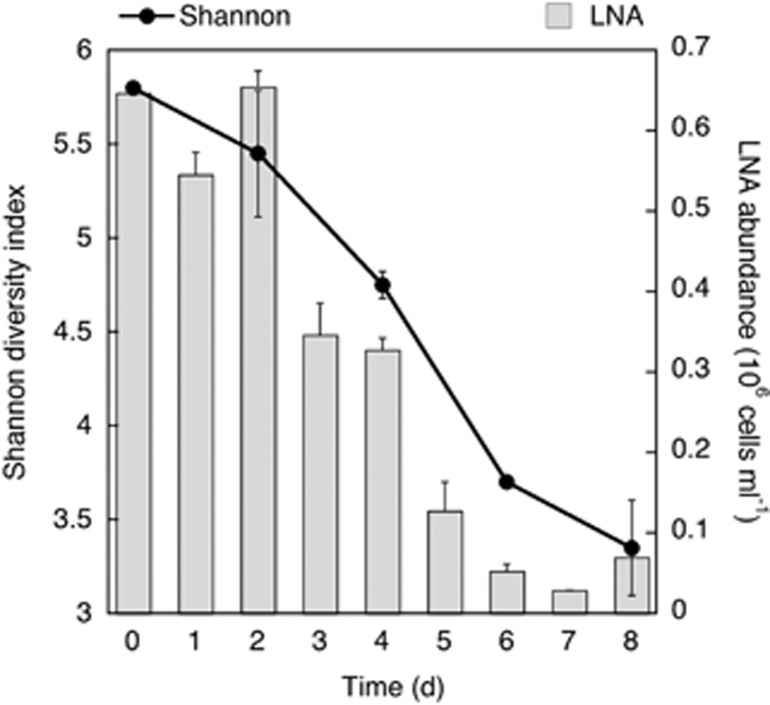
A gradual change in bacterial community composition was revealed by the hierarchical clustering analysis (Figure 3a), where only the initial time (T0) seemed to be skewed from the other days. To study in more detail the relation between bacterial community composition and grazing, we performed the same hierarchical clustering analysis but excluding T0 (Figure 3b). This analysis showed a gradual temporal change, unrelated to grazing rates (PERMANOVA, R2=0.08309, P=0.05824, n=7). Thus, despite the above-mentioned temporal differences in the relative abundance of specific bacterial groups, the overall community structure remained relatively stable from day 2 onwards. Moreover, analysis of the alpha-diversity metrics (Shannon index) also indicated a smooth transition in bacterial diversity during the experiment, with a progressive decrease in diversity with time (Figure 4), in striking contrast with the very large shifts in protozoan abundances, grazing rates, bacterial production and the measures of single-cell activity (CTC, NADS and %HNA).
Figure 3.
Bray–Crustis distances hierarchical clustering analysis of bacterioplankton communities during the mesocosm experiments (a) including the initial time (T0) and (b) excluding T0.
Figure 4.
Average temporal variation in the Shannon diversity index and the abundance of LNA cells during the experiments.
Discussion
The bloom of phytoplankton (reflecting the inorganic nutrient enrichment) together with the supply of dissolved organic carbon, positively stimulated bacterial abundance and activity that were followed by a sequential increase in the abundance of HNF and ciliates. After this initial bottom-up stimulation, a strong top-down control was manifested by the marked changes observed in the temporal dynamics of bacteria, HNF, ciliates and their bulk grazing rates. These results are in agreement with the role of protist bacterivory as a dominant bacterioplankton loss factor (Pedrós-Alió et al., 2000; Jürgens and Massana, 2008). Although FLB-based methods might not always be a perfect proxy for grazing pressure, particularly on motile cells (González et al., 1993), we found a strong direct correlation (N=9, Pearson's r=0.89, P=0.0011) between HNF abundance and FLB disappearance rate (grazing rates) (Figures 1a vs b), suggesting that the major factor determining the disappearance of bacteria in the experiment was the direct ingestion and digestion by HNF. These coinciding trends sharply contrast with, for example, the nonsignificant trends in FLB disappearance rates when compared with the abundance of ciliates (which in some cases are important bacterivores). Now, bearing in mind the strong grazing pressure on bacteria observed in this experiment, the next question is whether that grazing pressure was selective—and thereby shaped the phenotypic and genetic composition of the bacterioplankton community—or not.
Despite the strong shifts observed in cell numbers, activities and grazing rates (Figure 1), the bacterial community composition remained relatively stable from day 2 onwards throughout the experiment, not presenting significant shifts in community structure at the class level (Figure 2). It is worth mentioning that most of the studies carried out with FISH usually target this level (class–order). When analyzed at a higher level of taxonomic resolution (i.e., genus), there was more variability in the taxonomic composition between time points, but we still did not find a pronounced and well-defined change in community composition coinciding with the strong grazing event (Figure 3). A decrease in the proportion of the dominant bacteria (e.g., Rhodobacteraceae) at the days of intense grazing would be indicative of taxon-selective grazing, whereas a gradual change in community composition and diversity—disconnected from shifts in abundances, grazing and metabolic rates—would be expected if grazing was not phylotype-selective/specific. Indeed, we found a progressive temporal development in the relative abundance of some specific members (dominated by the Alphaproteobacteria class, in general, and the Rhodobacteraceae family, in particular) and a decrease in others (Gammaproteobacteria), which was unrelated to the strong fluctuations of grazing and production rates and cell numbers. This succession pattern between Alteromonadaceae (Gammaproteobacteria) and Rhodobacteraceae was also found using FISH (Allers et al., 2007) and DGGE (denaturing gradient gel electrophoresis; Sandaa et al., 2009).
In this study, we provide diversity data and information on population dynamics at the level of specific OTUs. The level of sequencing depth in this study allowed the detection of OTUs, making up around 0.03% of the bacterial community, or about 264 cells out of a million. The strongest change in the most abundant OTUs occurred in the first 2 days of the experiment, when HNFs were still at a very low abundance, but bacterial cells had already doubled, as a response to the beginning of the phytoplankton bloom. In that transition, only 20% of the most abundant OTUs were shared between time 0 and day 2 (Table 1). This change is consistent with the decrease in the number of OTUs belonging to the SAR11 clade and increase in Glaciecola or Rhodobacteraceae populations benefitting from improved nutrient availabilities. Afterwards, the number of shared abundant OTUs between consecutive time points became gradually higher (Table 1), suggesting a more stable community composition.
Table 1. Fifteen most abundant OTUs at any given time during the experiment ordered by rank according to the relative abundance of reads.
| Rank | T0 | T2 | T4 | T6 | T8 |
|---|---|---|---|---|---|
| 1 | Candidatus Pelagibacter | Glaciecola | Unc. Rhodobacteraceae | Unc. Rhodobacteraceae | Unc. Rhodobacteraceae |
| 2 | Synechococcus | Unc. Rhodobacteraceae | Unc. Rhodobacteraceae | Shimia | Sulfitobacter |
| 3 | Candidatus Pelagibacter | Unc. Oceanospirillaceae | Phaeobacter gallaeciensis | Sulfitobacter | Nautella |
| 4 | Candidatus Portiera | Unc. Rhodobacteraceae | Shimia | Phaeobacter gallaeciensis | Unc. Rhodobacteraceae |
| 5 | Glaciecola | Synechococcus | Sulfitobacter | Unc. Rhodobacteraceae | Unc. Rhodobacteraceae |
| 6 | Prochlorococcus marinus | Unc. Flavobacteriaceae | Alteromonas marina | Unc. Rhodobacteraceae | Maribacter |
| 7 | koll13 | Shimia | Unc. Rhodobacteraceae | Thalassobius gelatinovorus | Shimia |
| 8 | koll13 | Nereida | Unc. Pirellulaceae | Unc. Rhodobacteraceae | Unc. Rhodobacteraceae |
| 9 | Chroococcales | Sulfitobacter | Nautella | Unc. Oceanospirillaceae | Synechococcus |
| 10 | Rhodospirillaceae | Unc. Flavobacteria | Unc. Oceanospirillaceae | Synechococcus | Oceanicola |
| 11 | Candidatus Pelagibacter | Unc. Pirellulaceae | Unc. Flavobacteriaceae | Glaciecola | Phaeobacter gallaeciensis |
| 12 | Oceanospirillales | Phaeobacter gallaeciensis | Thalassobius | Unc. Rhodobacteraceae | Unc. Rhodobacteraceae |
| 13 | Candidatus Pelagibacter | Unc. Rhodobacteraceae | Unc. Rhodobacteraceae | Nautella | Glaciecola |
| 14 | Candidatus Pelagibacter | Thalassobacter | Thalassobius gelatinovorus | Unc. Rhodobacteraceae | Unc. Rhodobacteraceae |
| 15 | Unc. Flavobacteriaceae | Alteromonas marina | Unc. Rhodobacteraceae | Maribacter | Unc. Pirellulaceae |
|
Shared OTUs with previous time point |
|||||
| Glaciecola | Unc. Rhodobacteraceae | Unc. Rhodobacteraceae | Unc. Rhodobacteraceae | ||
| Synechococcus | Unc. Rhodobacteraceae | Shimia | Sulfitobacter | ||
| Unc. Flavobacteriaceae | Phaeobacter gallaeciensis | Sulfitobacter | Nautella | ||
| Shimia | Phaeobacter gallaeciensis | Unc. Rhodobacteraceae | |||
| Sulfitobacter | Unc. Rhodobacteraceae | Unc. Rhodobacteraceae | |||
| Alteromonas marina | Unc. Rhodobacteraceae | Maribacter | |||
| Unc. Rhodobacteraceae | Thalassobius gelatinovorus | Shimia | |||
| Unc. Pirellulaceae | Unc. Rhodobacteraceae | Unc. Rhodobacteraceae | |||
| Unc. Flavobacteriaceae | Unc. Oceanospirillaceae | Synechococcus | |||
| Unc. Rhodobacteraceae | Phaeobacter gallaeciensis | ||||
| Nautella | Unc. Rhodobacteraceae | ||||
| Glaciecola | |||||
| Unc. Rhodobacteraceae | |||||
| % of shared OTUs | 20 | 60 | 73 | 87 | |
The OTUs shared between consecutive time points are indicated.
The only indication of potential taxon-selective grazing could be the pronounced shift observed in the relative abundance of two different Alteromonadaceae OTUs (i.e., Glaciecola and Alteromonas) between days 2 and 4. However, the lack of further evidences supporting that possibility (e.g., PERMANOVA analysis), together with the more continuous temporal dynamics of all the other OTUs, points towards a more prominent dominant role of a non phylotype-selective grazing. Diversity indexes also showed evidence of a progressive change in diversity that did not coincide with the abrupt shifts observed in bacterial, HNF and ciliate numbers and grazing rates (no significant correlation were found between the Shannon index and any of those parameters). These results suggest that increased grazing pressure did not produce significant shifts in bacterial community composition and diversity, therefore indicating that HNF grazing on marine bacteria was not strongly selective from a point of view of particular bacterial taxa (i.e., protists consumed most bacterial phylotypes indiscriminately).
The lack of effects of grazing on bacterial community composition is in contrast to previously observed changes of bacterial community composition in response to changes in predation pressure in model chemostat systems (Pernthaler et al., 1997; Hahn and Höfle, 1999), food web manipulation experiments (Jürgens et al., 1999; Šimek et al., 1999; Langenheder and Jürgens, 2001) and seasonal plankton successions in lakes (Pernthaler et al., 2004). However, our results are in agreement with reports where the impact of changing predation regimes on bacterial community composition was studied in marine systems (Suzuki, 1999; Massana and Jurgens, 2003; Yokokawa and Nagata, 2005). The main difference between these previous studies and ours is that, in the present one, we focused on the response of bacterial communities to grazing during a phytoplankton bloom (not during steady state non-bloom conditions). Moreover, the use of high-throughput sequencing techniques allowed us to authenticate, with a higher taxonomical resolution compared with that in these previous works, the nonspecific grazing behavior of HNF on marine bacterial communities.
Although protist grazing did not seem to be selecting for specific bacterial taxa, we found evidence that grazing could be selective in terms of the size and activity level of the bacterial prey. The abundance of HNAs mirrored the dynamics of the total abundance of bacteria. In contrast, the abundance of LNAs decreased almost linearly with time (until day 6) independently of the strong shifts that were occurring in the total abundance of bacteria and in the HNA fraction. This linear decrease in the abundance of LNA with time, independent of the ups and downs of bacterial and protist abundances and grazing rates, suggests that LNA cells were not responding to the shifts observed in HNF grazing, and could indicate that HNA cells were preferentially grazed over LNAs. HNAs were larger compared with LNAs according to the positive relation obtained between the size of bacterial cells and the proportion of HNAs (N=9, Pearson's r=0.85, P=0.0035). Moreover, the strong positive relation (N=9, Pearson's r=0.91, P=0.0006) found between the abundance of HNA and that of actively respiring (CTC+) cells indicated that HNA bacteria (the larger cells) were also the metabolically most active ones, explaining why they would be preferentially grazed over the LNAs. These results are in agreement with previous reports suggesting a clear correlation between bacterial size and activity (Gasol et al., 1995; Bernard et al., 2000), and with the size-selective grazing strategy generally followed by bacterivorous flagellates on planktonic bacterial assemblages (Chrzanowski and Simek, 1990; Gonzalez et al., 1990), by which the actively growing portion of the bacterial assemblage is highly susceptible to grazing and thereby preferentially eliminated (Sherr et al., 1992; Gasol et al., 1995).
If HNA and LNA populations were taxonomically different, preferential grazing on the HNA cells should generate a taxon-selective grazing. However, contrasting results have been reported, with some studies in coastal sites showing that the HNA and LNA populations are different, whereas others found similar taxonomic composition (Servais et al., 2003; Longnecker et al., 2005). Moreover, it is possible that the HNA cells growing (and then being grazed) in our experiment belonged to a limited number of taxa responding to the phytoplankton bloom by rapid growth, thus overriding the effect of the heavy HNF bacterivory; this is a life strategy suggested for freshwater bacteria of the genus Limnohabitans (Šimek et al., 2014). Such predominant responses of a few bacterial populations with strong growth capacity have been previously observed in response to phytoplankton blooms (Alonso-Sáez et al., 2007; Gilbert et al., 2011; Teeling et al., 2012), and would be consistent with the strong increase in relative abundance of two main taxa at the onset of the experiment (Glaciecola and Rhodobacteraceae; Figure 2b). Under these premises, a strong predation on HNA cells would not necessarily provoke a strong decrease in diversity. The observed decrease in LNA cells independently of the grazing pressure peaks could thus be explained bearing in mind that we are studying the response of the bacterial community to grazing during a phytoplankton bloom. We speculate that two parallel processes are taking place during this event; bacterial numbers (mostly HNA cells) show pronounced dynamics owing to grazing impact, and superimposed on that we observe changes in relative abundance of LNA cells as well as specific bacterial populations (defined as OTUs in 16S rRNA gene analyses) that would happen even if grazing were absent as a response to the changing conditions (e.g., nutrient/substrate concentration, ecological interactions between different bacterial members, and so on) during the experiment.
The decrease in the Shannon diversity index along this phytoplankton bloom experiment is consistent with the common negative diversity–productivity relationship found in aquatic ecosystems (Smith, 2007). This suggests that the overall diversity reduction was because of the stimulation of bacterial growth conditions associated with the phytoplankton bloom development (bottom-up), and that protist grazing subsequently acted on top of this diversity decrease. Our analysis indicated that LNA cells could have a major role in sustaining the diversity of marine bacterial communities (Figure 4). Accordingly, the strong positive relation between the Shannon diversity index and the abundance of LNA cells (N=5, Pearson's r=0.96, P=0.0044) suggested that the diversity among HNA cells growing along the experiment was not enough to compensate for the loss in diversity resulting from the continuous reduction of LNA cells. This progressive reduction in diversity could be related to the increase found in one Alphaproteobacteria family (i.e., the Rhodobacteraceae, increasing from 3% relative abundance at time 0 to >85% at day 8), at the expense of the Gammaproteobacteria, Cyanobacteria and SAR11 clade members. Several authors have found that SAR11 clade bacteria are large contributors to the LNA cells fraction in ocean environments (Fuchs et al., 2005; Longnecker et al., 2005; Mary et al., 2006; Vila-Costa et al., 2012). This is in agreement with the parallel reduction found in the number of LNA cells and in the relative abundance of SAR11 during the experiment. The resulting prevalence of Rhodobacteraceae in a community subjected to strong grazing pressure could be related to a smaller effect of HNF on this group compared with the others. However, as grazing losses did not seem to be phylotype-specific in this experiment, the resulting dominance of Rhodobacteraceae was most likely because of their reported genotypic and metabolic diversity (Brinkhoff et al., 2008) and their ability to take advantage of the environmental conditions associated with algal blooms (Eilers et al., 2001; Pinhassi et al., 2004; Allers et al., 2007; Baltar et al., 2007; Buchan et al., 2014) and the phytoplankton-derived DOC produced (Zubkov et al., 2001; Vila et al., 2004; Alonso and Pernthaler, 2006; Sarmento and Gasol, 2012), more than due to a superior grazing-avoidance capability. Thus, whether avoidance or lessening of grazing losses is a crucial ecological trait for bacteria, and thus a critical factor for the observed dominance of marine bacterial clusters such as Rhodobacteraceae, remains to be shown for marine systems.
In summary, strong oscillations were observed in the abundance of bacteria, HNF and ciliates, coincident with changes in grazing pressure and in the proportion of active and viable/damaged cells—yet this was not followed by strong shifts in bacterial community composition. In contrast, a progressive increase of some community members (mainly from the Rhodobacteraceae family) occurred during the experiment, resulting in a gradual decrease in diversity. Although not selective from a phylotype-specific point of view, protists were inferred to be selectively preying on active and larger cells and HNA-containing bacteria. Although LNA cells were not preferentially grazed, we hypothesize that they have an important role in the conservation of the bacterial community diversity. In conclusion, we found that, under phytoplankton bloom conditions, increased protist predation triggered strong changes in bacterial abundance and activity (both at the bulk and single-cell level), but did not trigger clear diversity changes in a coastal Mediterranean bacterioplankton community.
Acknowledgments
We thank all the people in project BASICS involved in this experiment, in particular, V Balagué, R Simó and M Vila-Costa for helping set up the experiment, J Masmitjà for the ciliate counts, J Felipe for help with cytometry and our missed friend J Jezbera, who participated in the grazing measurements. We also thank Markus V Lindh for insightful discussions. The mesocosm experiment was supported by EU project BASICS (EVK3-CT-2002-00078). This research was supported by projects from the European Science Foundation (EuroEEFG project MOCA) and the Swedish Research Council to JP. KŠ and KH were supported by a CSIC—CAS (Spanish-Czech) bilateral mobility project awarded to JMG and KŠ. JMG is supported by project DOREMI (CTM2012-34294) and the GRC 2014SGR/1179 from Generalitat de Catalunya, and FB by a University of Otago Research Grant.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Allers E, Gómez-Consarnau L, Pinhassi J, Gasol JM, Šimek K, Pernthaler J. (2007). Response of Alteromonadaceae and Rhodobacteriaceae to glucose and phosphorus manipulation in marine mesocosms. Environ Microbiol 9: 2417–2429. [DOI] [PubMed] [Google Scholar]
- Alonso C, Pernthaler J. (2006). Roseobacter and SAR11 dominate microbial glucose uptake in coastal North Sea waters. Environ Microbiol 8: 2022–2030. [DOI] [PubMed] [Google Scholar]
- Alonso-Sáez L, Balague V, Sà EL, Sanchez O, González JM, Pinhassi J et al. (2007). Seasonality in bacterial diversity in north-west Mediterranean coastal waters: assessment through clone libraries, fingerprinting and FISH. FEMS Microbiol Ecol 60: 98–112. [DOI] [PubMed] [Google Scholar]
- Andersson A, Larsson U, Hagström Å. (1986). Size-selective grazing by a microflagellate on pelagic bacteria. Mar Ecol Prog Ser 33: 51–57. [Google Scholar]
- Azam F, Fenchel T, Field JG, Gray JS, Meyer-Reil LA, Thingstad F. (1983). The ecological role of water-column microbes in the sea. Mar Ecol Prog Ser 10: 257–263. [Google Scholar]
- Baltar F, Arístegui J, Gasol JM, Hernández-León S, Herndl GJ. (2007). Strong coast–ocean and surface–depthgradients in prokaryotic assemblage structure and activity in a coastal transition zone region. Aquat Microb Ecol 50: 63–74. [Google Scholar]
- Bernard L, Courties C, Servais P, Troussellier M, Petit M, Lebaron P. (2000). Relationships among bacterial cell size, productivity, and genetic diversity in aquatic environments using cell sorting and flow cytometry. Microb Ecol 40: 148–158. [DOI] [PubMed] [Google Scholar]
- Boenigk J, Matz C, Jürgens K, Arndt H. (2002). Food concentration-dependent regulation of food selectivity of interception-feeding bacterivorous nanoflagellates. Aquat Microb Ecol 27: 195–202. [Google Scholar]
- Brinkhoff T, Giebel H-A, Simon M. (2008). Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Archiv Microbiol 189: 531–539. [DOI] [PubMed] [Google Scholar]
- Buchan A, LeCleir GR, Gulvik CA, González JM. (2014). Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nat Rev Microbiol 12: 686–698. [DOI] [PubMed] [Google Scholar]
- Chrzanowski TH, Simek K. (1990). Prey-size selection by freshwater flagellated protozoa. Limnol Oceanogr 35: 1429–1436. [Google Scholar]
- Corno G, Jürgens K. (2006). Direct and indirect effects of protist predation on population size structure of a bacterial strain with high phenotypic plasticity. Appl Environ Microbiol 72: 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Giorgio PA, Gasol JM. (2008). Physiological structure and single-cell activity in marine bacterioplankton. In: Kirchman DL (ed). Microbial Ecology of the Ocean, 2nd edn. Wiley-Liss: : New York NY, USA, pp 243–298. [Google Scholar]
- Del Giorgio PA, Gasol JM, Vaqué D, Mura P, Agusti S, Duarte CM. (1996). Bacterioplankton community structure: protists control net production and the proportion of active bacteria in a coastal marine community. Limnol Oceanogr 41: 1169–1179. [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K et al. (2006). Greengenes, a chimera-checked 16 S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72: 5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd SE, Sun Y, Wolcott RD, Domingo A, Carroll JA. (2008). Bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP) for microbiome studies: bacterial diversity in the ileum of newly weaned Salmonella-infected pigs. Foodborne Pathogen Dis 5: 459–472. [DOI] [PubMed] [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Eilers H, Pernthaler J, Peplies J, Glöckner FO, Gerdts G, Amann R. (2001). Isolation of novel pelagic bacteria from the German Bight and their seasonal contributions to surface picoplankton. Appl Environ Microbiol 67: 5134–5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein SS, Burkovsky IV, Shiaris MP. (1992). Ciliate grazing on bacteria, flagellates, and microalgae in a temperate zone sandy tidal flat: ingestion rates and food niche partitioning. J Exp Mar Biol Ecol 165: 103–123. [Google Scholar]
- Falcioni T, Papa S, Gasol JM. (2008). Evaluating the flow-cytometric nucleic acid double-staining protocol in realistic rituations of planktonic bacterial death. Appl Environ Microbiol 74: 1767–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenchel T. (1980). Suspension feeding in ciliated protozoa: feeding rates and their ecological significance. Microb Ecol 6: 13–25. [DOI] [PubMed] [Google Scholar]
- Fierer N, Hamady M, Lauber CL, Knight R. (2008). The influence of sex, handedness, and washing on the diversity of hand surface bacteria. Proc Natl Acad Sci 105: 17994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, O'Kelly C, Sieracki M, Distel DL. (2003). Protistan grazing analysis by flow cytometry using prey labeled by in vivo expression of fluorescent proteins. Appl Environ Microbiol 69: 6848–6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs BM, Woebken D, Zubkov MV, Burkill P, Amann R. (2005). Molecular identification of picoplankton populations in contrasting waters of the Arabian Sea. Aquat Microb Ecol 39: 145–157. [Google Scholar]
- Gasol JM, Arístegui J. (2007). Cytometric evidence reconciling the toxicity and usefulness of CTC as a marker of bacterial activity. Aquat Microb Ecol 46: 71–83. [Google Scholar]
- Gasol JM, Giorgio PAD. (2000). Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci Mar 64: 197–224. [Google Scholar]
- Gasol JM, Giorgio PAD, Massana R, Duarte CM. (1995). Active versus inactive bacteria: size-dependence in a coastal marine plankton community. Mar Ecol Prog Ser 128: 91–97. [Google Scholar]
- Gasol JM, Zweifel UL, Peters F, Fuhrman JA, Hagström Å. (1999). Significance of size and nucleic acid content heterogeneity as measured by flow cytometry in natural planktonic bacteria. Appl Environ Microbiol 65: 4475–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B et al. (2011). Defining seasonal marine microbial community dynamics. ISME J 6: 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez JM, Sherr EB, Sherr BF. (1990). Size-selective grazing on bacteria by natural assemblages of estuarine flagellates and ciliates. Appl Environ Microbiol 56: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González JM, Sherr EB, Sherr BF. (1993). Differential feeding by marine flagellates on growing versus starving, and on motile versus nonmotile, bacterial prey. Mar Ecol Prog Ser 102: 257–267. [Google Scholar]
- Gregori G, Citterio S, Ghani A, Labra M, Sgorbati S, Brown S et al. (2001). Resolution of viable and membrane-compromised bacteria in freshwater and marine waters based on analytical flow cytometry and nucleic acid double staining. Appl Environ Microbiol 67: 4662–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Höfle MG. (1999). Flagellate predation on a bacterial model community: interplay of size-selective grazing, specific bacteiral cell size, and bacterial community composition. Appl Environ Microbiol 65: 4863–4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Höfle MG. (2001). Grazing of protozoa and its effect on populations of aquatic bacteria. FEMS Microbiol Ecol 35: 113–121. [DOI] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK, Gold NJ, Knight R. (2008). Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 5: 235–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens K, Güde H. (1994). The potential importance of grazing-resistant bacteria in planktonic systems. Mar Ecol Prog Ser 112: 169–188. [Google Scholar]
- Jürgens K, Massana R. (2008). Protistan grazing on marine bacterioplankton. In: Kirchman DL (ed). Microbial Ecology of the Oceans, 2nd edn. Wiley: : Hoboken, NJ, USA, pp 383–441. [Google Scholar]
- Jürgens K, Pernthaler J, Schalla S, Amann R. (1999). Morphological and compositional changes in a planktonic bacterial community in response to enhanced protozoan grazing. Appl Environ Microbiol 65: 1241–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinner N, Harvey R, Blakeslee K, Novarino G, Meeker L. (1998). Size-selective predation on groundwater bacteria by nanoflagellates in an organic-contaminated aquifer. Appl Environ Microbiol 64: 618–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman D, K'Ness E, Hodson R. (1985). Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl Environ Microbiol 49: 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenheder S, Jürgens K. (2001). Regulation of bacterial biomass and community structure by metazoan and protozoan predation. Limnol Oceanogr 46: 121–134. [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. (2009). Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol 75: 5111–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker K, Sherr BF, Sherr EB. (2005). Activity and phylogenetic diversity of bacterial cells with high and low nucleic acid content and electron transport system activity in an upwelling ecosystem. Appl Environ Microbiol 71: 7737–7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longnecker K, Wilson MJ, Sherr EB, Sherr BF. (2010). Effect of top-down control on cell-specific activity and diversity of active marine bacterioplankton. Aquat Microb Ecol 58: 153–165. [Google Scholar]
- Mary I, Heywood JL, Fuchs BM, Amann R, Tarran GA, Burkill PH et al. (2006). SAR11 dominance among metabolically active low nucleic acidd bacterioplankton in surface waters among an Atlantic meridional transect. Aquat Microb Ecol 45: 107–113. [Google Scholar]
- Massana R, Jurgens K. (2003). Composition and population dynamics of planktonic bacteria and bacterivorous flagellates in seawater chemostat cultures. Aquat Microb Ecol 32: 11–22. [Google Scholar]
- Matz C, Boenigk J, Arndt H, Jürgens K. (2002). Role of bacterial phenotypic traits in selective feeding of the heterotrophic nanoflagellate Spumella sp. Aquatic microbial ecology 27: 137–148. [Google Scholar]
- Matz C, Jürgens K. (2001). Effects of hydrophobic and electrostatic cell surface properties of bacteria on feeding rates of heterotrophic nanoflagellates. Appl Environ Microbiol 67: 814–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet F, Kindt R, Legendre P, O'Hara R, Simpson G et al. (2011), Vegan: Community Ecology Package. R package version 1.17-5. Available at: http://CRAN.R-project.org/package=vegan.R-project.org/package=vegan last accessed 12 June 2015.
- Parsons T, Maita Y, Lalli C. (1984) A Manual of Chemical and Biological Methods for Seawater Analysis. Pergamon Press: : Oxford, UK. [Google Scholar]
- Pedrós-Alió C, Calderón-Paz JI, Gasol JM. (2000). Comparative analysis shows that bacterivory, not viral lysis, controls the abundance of heterotrophic prokaryotic plankton. FEMS Microbiol Ecol 32: 157–165. [DOI] [PubMed] [Google Scholar]
- Pernthaler J, Posch T, Simek K, Vrba J, Amann R, Psenner R. (1997). Contrasting bacterial strategies to coexist with a flagellate predator in an experimental microbial assemblage. Appl Environ Microbiol 63: 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler J, Zöllner E, Warnecke F, Jürgens K. (2004). Bloom of filamentous bacteria in a mesotrophic lake: identity and potential controlling mechanism. Appl Environ Microbiol 70: 6272–6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernthaler J. (2005). Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol 3: 537–546. [DOI] [PubMed] [Google Scholar]
- Pinhassi J, Sala MM, Havskum H, Peters F, Guadayol O, Malits A et al. (2004). Changes in bacterioplankton composition under different phytoplankton regimens. Appl Environ Microbiol 70: 6753–6766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. (2009). FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26: 1641–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2015) R: A Language and Environment for Statistical Computing [Internet]. R Foundation for Statistical Computing: Vienna, Austria, 2013. Available at: http://www.r-project.org. (last accessed 12 June 2015. [Google Scholar]
- Riemann L, Steward GF, Azam F. (2000). Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl Environ Microbiol 66: 578–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salat J, Marrasé C. (1994). Exponential and linear estimations of gazing on bacteria: effects of changes in the proportion of marked cells. Mar Ecol Prog Ser 104: 205–209. [Google Scholar]
- Sandaa RA, Gómez-Consarnau L, Pinhassi J, Riemann L, Malits A, Weinbauer MG et al. (2009). Viral control of bacterial biodiversity—evidence from a nutrient-enriched marine mesocosm experiment. Environ Microbiol 11: 2585–2597. [DOI] [PubMed] [Google Scholar]
- Sarmento H, Gasol JM. (2012). Use of phytoplankton-derived dissolved organic carbon by different types of bacterioplankton. Environ Microbiol 14: 2348–2360. [DOI] [PubMed] [Google Scholar]
- Servais P, Casamayor EO, Courties C, Catala P, Parthuisot N, Lebaron P. (2003). Activity and diversity of bacterial cells with high and low nucleic acid content. Aquat Microb Ecol 33: 41–51. [Google Scholar]
- Sherr BF, Giorgio PD, Sherr EB. (1999). Estimating abundance and single-cell characteristics of respiring bacteria via the redox dye CTC. Aquat Microb Ecol 18: 117–131. [Google Scholar]
- Sherr BF, Sherr EB, Fallon RD. (1987). Use of monodispersed, fluorescently labeled bacteria to estimate in situ protozoan bacterivory. Appl Environ Microbiol 53: 958–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr BF, Sherr EB, McDaniel J. (1992). Effect of protistan grazing on the frequency of dividing cells in bacterioplankton assemblages. Appl Environ Microbiol 58: 2381–2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simek K, Bobkova J, Macek M, Nedoma J. (1995). Ciliate grazing on picoplankton in a eutrophic reservoir during the summer phytoplankton maximum: a study at the species and community level. Limnol Oceanogr 40: 1077–1090. [Google Scholar]
- Simek K, Hartman P, Nedoma J, Pernthaler J, Springmann D, Vrba J et al. (1997). Community structure, picoplankton grazing and zooplankton control of heterotrophic nanoflagellates in a eutrophic reservior during the summer phytoplankton maximum. Aquat Microb Ecol 12: 49–63. [Google Scholar]
- Šimek K, Kojecká P, Nedoma J, Hartman P, Vrba J, Dolan JR. (1999). Shifts in bacterial community composition associated with different microzooplankton size fractions in a eutrophic reservoir. Limnol Oceanogr 44: 1634–1644. [Google Scholar]
- Šimek K, Nedoma J, Pernthaler J, Posch T, Dolan JR. (2002). Altering the balance between bacterial production and protistan bacterivory triggers shifts in freshwater bacterial community composition. Antonie Van Leeuwenhoek 81: 453–463. [DOI] [PubMed] [Google Scholar]
- Šimek K, Nedoma J, Znachor P, Kasalický V, Jezbera J, Hornňák K et al. (2014). A finely tuned symphony of factors modulates the microbial food web of a freshwater reservoir in spring. Limnol Oceanogr 59: 1477–1492. [Google Scholar]
- Sintes E, Del Giorgio P. (2014). Feedbacks between protistan single-cell activity and bacterial physiological structure reinforce the predator/prey link in microbial foodwebs. Front Microbiol 5: 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjöstedt J, Koch-Schmidt P, Pontarp M, Canbäck B, Tunlid A, Lundberg P et al. (2012). Recruitment of members from the rare biosphere of marine bacterioplankton communities after an environmental disturbance. Appl Environ Microbiol 78: 1361–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith VH. (2007). Microbial diversity–productivity relationships in aquatic ecosystems. FEMS Microbiol Ecol 62: 181–186. [DOI] [PubMed] [Google Scholar]
- Smith DC, Azam F. (1992). A simple, economical method for measuring bacterial protein synthesis rates in seawater using 3H-leucine. Mar Microb Food Webs 6: 107–114. [Google Scholar]
- Suzuki MT. (1999). Effect of protistan bacterivory on coastal bacterioplankton diversity. Aquat Microb Ecol 20: 261–272. [Google Scholar]
- Tadonléké R, Planas D, Lucotte M. (2005). Microbial food webs in boreal humic lakes and reservoirs: ciliates as a major factor related to the dynamics of the most active bacteria. Microb Ecol 49: 325–341. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Thingstad TF, Gasol JM, Cardelús C, Jezbera J, Sala MM et al. (2009). Determining the availability of phosphate and glucose for bacteria in P-limited mesocosms of NW Mediterranean surface waters. Aquat Microb Ecol 56: 81–91. [Google Scholar]
- Teeling H, Fuchs BM, Becher D, Klockow C, Gardebrecht A, Bennke CM et al. (2012). Substrate-controlled succession of marine bacterioplankton populations induced by a phytoplankton bloom. Science 336: 608–611. [DOI] [PubMed] [Google Scholar]
- Thingstad TF. (2000). Control of bacterial growth in idealized food webs. In: Kirchman DL (ed). Microbial Ecology of the Oceans. Wiley-Liss: : New York, NY, USA, pp 229–260. [Google Scholar]
- Unrein F, Gasol JM, Not F, Forn I, Massana R. (2014). Mixotrophic haptophytes are key bacterial grazers in oligotrophic coastal waters. ISME J 8: 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unrein F, Massana R, Alonso-Sáez L, Gasol JM. (2007). Significant year-round effect of small mixotrophic flagellates on bacterioplankton in an oligotrophic coastal system. Limnol Oceanogr 52: 456–469. [Google Scholar]
- Vaqué D, Casamayor EO, Gasol JM. (2001). Dynamics of whole community bacterial production and grazing losses in seawater incubations as related to the changes in the proportions of bacteria with different DNA-content. Aquat Microb Ecol 25: 163–177. [Google Scholar]
- Vaqué D, Agustí S, Duarte CM. (2004). Response of bacterial grazing rates to experimental manipulation of an Antarctic coastal nanoflagellate community. Aquat Microb Ecol 36: 41–52. [Google Scholar]
- Vazquez-Dominguez E, Peters F, Gasol JM, Vaqué D. (1999). Measuring the grazing losses of picoplankton: methodological improvements in the use of fluorescently tracers combined with flow cytometry. Aquat Microb Ecol 20: 119–128. [Google Scholar]
- Vila M, Simó R, Kiene RP, Pinhassi J, González JM, Moran MA et al. (2004). Use of microautoradiography combined with fluorescence in situ hybridization to determine dimethylsulfoniopropionate incorporation by marine bacterioplankton taxa. Appl Environ Microbiol 70: 4648–4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila-Costa M, Gasol JM, Sharma S, Moran MA. (2012). Community analysis of high-and low-nucleic acid-containing bacteria in NW Mediterranean coastal waters using 16 S rDNA pyrosequencing. Environ Microbiol 14: 1390–1402. [DOI] [PubMed] [Google Scholar]
- Yokokawa T, Nagata T. (2005). Growth and grazing mortality rates of phylogenetic groups of bacterioplankton in coastal marine environments. Appl Environ Microbiol 71: 6799–6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubkov MV, Fuchs BM, Archer SD, Kiene RP, Amann R, Burkill PH. (2001). Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ Microbiol 3: 304–311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.